Summary
The ability to process and store information in living cells is essential for developing next-generation therapeutics and studying biology in situ. However, existing strategies have limited recording capacity and are challenging to scale. To overcome these limitations, we developed DOMINO, a robust and scalable platform for encoding logic and memory in bacterial and eukaryotic cells. Using an efficient single-nucleotide-resolution read-write head for DNA manipulation, DOMINO converts the living cells DNA into an addressable, readable and writable medium for computation and storage. DOMINO operators enable analog and digital molecular recording for long-term monitoring of signaling dynamics and cellular events. Furthermore, multiple operators can be layered and interconnected to encode order-independent, sequential and temporal logic, allowing recording and control over the combination, order, and timing of molecular events in cells. We envision that DOMINO will lay the foundation for building robust and sophisticated computation-and-memory gene circuits for numerous biotechnological and biomedical applications.
Keywords: Dynamic genome engineering, DNA writing, molecular recording, cellular computation, DNA memory, base editing, synthetic gene circuits, logic circuits, analog and digital recording and computation
In Brief
Farzadfard et al. developed a DNA writing platform that uses the genome to execute analog and digital information processing and storage in bacterial and eukaryotic cells. The platform enables recording and programming of the sequence and timing of molecular events in living cells.
Introduction
Platforms that enable robust and scalable molecular recording and computation in living cells have broad biotechnological and biomedical applications, ranging from the study of signaling dynamics and cellular lineages in development and cancer, to building living biosensors and adaptive therapeutics, to encoding logic and programming cellular phenotypes (Farzadfard and Lu, 2018). With the advent of in vivo DNA writing technologies, several memory architectures have been described that utilize genomic DNA as a medium for information processing and storage in living cells (Farzadfard and Lu, 2014; McKenna et al., 2016; Perli et al., 2016; Roquet et al., 2016; Sheth et al., 2017). These technologies capture and record various biological information in the form of mutational signatures in DNA. However, unlike their silicon-based counterparts, which have access to large capacities of addressable memory registers, in vivo genetic memory architectures utilize rudimentary “Read” and “Write” operations and remain limited in their encoding capacity and scalability. As a result, these memory architectures lose their recording capacity after recording one or a few molecular events and cannot be used to continuously monitor signaling dynamics or histories of events over long periods. Furthermore, these technologies lack an inherent “Read” operation to interrogate and monitor DNA memory states on the fly. Consequently, it is challenging to interconnect and scale-up these architectures to achieve complex DNA-based logic and memory operations in living cells. Additionally, because of the requirements for host-specific DNA repair and genome editing mechanisms, these systems have been applicable only to a subset of organisms (Farzadfard and Lu, 2018).
To overcome these bottlenecks, we describe a highly efficient and robust molecular recording and DNA memory platform that uses genomic DNA as an addressable, readable and writable information storage and computation medium in living cells, much like a hard drive. This platform, called DNA-based Ordered Memory and Iteration Network Operator (DOMINO), leverages precise DNA writing with CRISPR base editors (Komor et al., 2016; Nishida et al., 2016) to manipulate DNA dynamically and efficiently with single-nucleotide resolution in bacterial and mammalian cells. DOMINO enables robust and long-term molecular recording of the intensity and duration of signals of interest (i.e., analog information) into DNA. Multiple DOMINO operators can be layered to build logic operators that control the sequence and timing of events in living cells in a scalable fashion. Specifically, we show that the order and combinations of DNA writing events can be coordinated and tuned by external inputs, allowing order-independent (e.g., IF EVER A AND IF EVER B), sequential (e.g., A AND THEN B), and temporal (e.g., A AND AFTER TIME X THEN B) logic and memory operations to be executed. Additionally, DOMINO can be combined with CRISPR-based gene regulation strategies, such as CRISPR interference (CRISPRi) (Qi et al., 2013) and CRISPR activation (CRISPRa) (Farzadfard et al., 2013; Gilbert et al., 2013), to achieve modular and versatile memory and gene regulatory functions. By leveraging this feature, we built a non-destructive DNA-state reporter genetic circuit in which mutational memory states generated in response to an incoming gRNA are converted into distinct levels of a transcriptional output. Thus, the state of this circuit can be monitored by functional assays, thus obviating the need for cell destruction and DNA sequencing for readouts. As such, DOMINO extends the utility of molecular recording beyond DNA write-only applications (for which the output can be read only by disruptive sequencing methods) and enables long-term recording and monitoring of in vivo molecular events. These advances address many limitations of current in vivo recording and computing technologies and pave the way towards next-generation memory architectures for information processing and storage in living cells.
Results
Designing the DOMINO Memory Architecture Using Base Editors as an Efficient Read-Write Head for Genomic DNA
We previously developed a moderately-efficient, addressable, and precise DNA writer for genomic DNA called SCRIBE, and demonstrated that it can be used as a molecular recorder with a wide dynamic range to record signal intensity and duration (i.e., analog information) into long-lasting DNA records (Farzadfard and Lu, 2014). However, the absence of an inherent “Write” operation, the relatively low DNA writing efficiency, and the requirement for host-specific factors limited the application of this Write-only system.
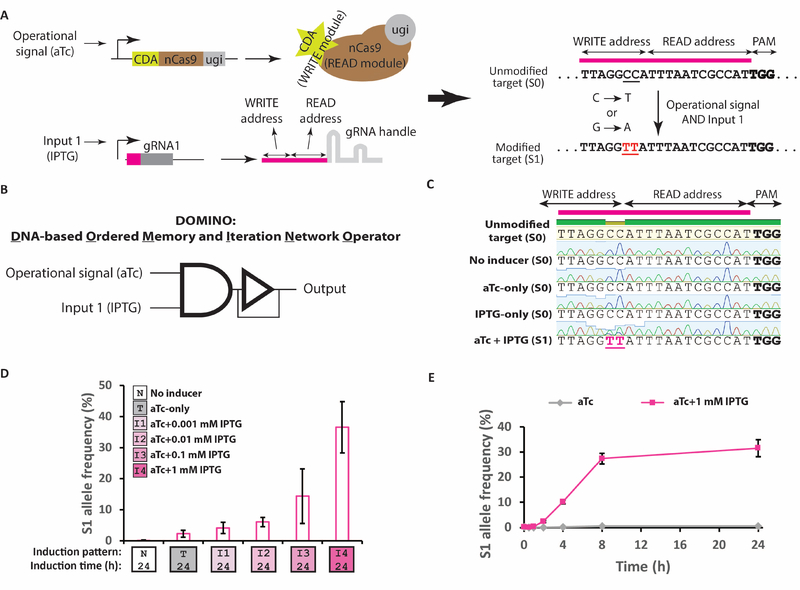
To address these limitations, we sought to leverage the base editing technology (Komor et al., 2016; Nishida et al., 2016; Tang and Liu, 2018) as a single-nucleotide resolution “read-write head” for genomic DNA to build dynamic and scalable memory architectures in living cells. The read-write head is composed of Cas9 nickase (nCas9, an addressable DNA “reader” module that is directed by gRNA to specific DNA targets and nicks them) fused to cytidine deaminase (CDA, a DNA “writer” module that edits the DNA) and uracil DNA glycosylase inhibitor (ugi, a peptide which improves the DNA writing efficiency by blocking cellular repair machinery). Once localized to the target based on the 12-bp gRNA seed sequence (“READ” address), the writer module can deaminate deoxycytidines (dC) in the vicinity of the 5’-end of the target (“WRITE” address), resulting in DNA lesions that are preferentially repaired to thymidines (dT) (Komor et al., 2016). Using CDA as the DNA writer module enables introduction of dC-to-dT mutations (or dG-to-dA mutations in the reverse complement strand) in the WRITE address, resulting in permanent records in DNA. In this memory architecture, an individual mutation or a group of mutations in a target site can be designated as a unique memory state for the corresponding memory register, and mutations introduced by DNA writing events can be considered as unidirectional transitions between DNA memory states (Figure 1A).
Figure 1 |. DOMINO design and molecular recording by this memory architecture in living cells.
A) Schematic representation of DOMINO memory architecture. B) A basic DOMINO operator can be symbolized as an AND gate because it requires the expression of both the DNA read-write head (i.e., CDA-nCas9-ugi controlled by the “operational signal”) and the gRNA (regulated by “Input 1”) with a downstream feedback delay operator (to illustrate the unidirectional and memory aspect of the operator). DOMINO operators can be layered to create a wide variety of memory and logic functions. C) Sanger chromatograms of the target locus in E. coli cells harboring the DOMINO operator shown in (B) after induction with different combinations of the inputs for 24 hours. Bold nucleotides on the target show the location of the NGG PAM sequence. Targeted nucleotides are underlined. D) The frequency of mutated allele (S1) in E. coli cells harboring the DOMINO operator shown in (B) induced with aTc and various dosages of the input (IPTG). E) The dynamics of S1 mutant accumulation in E. coli cells harboring the DOMINO operator shown in (B). Error bars indicate standard deviation for three biological replicates. See also Figure S1.
DNA writing events in this system can be controlled by internal or external inputs by placing the expression of both gRNA and CDA-nCas9-ugi under the control of inducible promoters. In this design, which forms the basis of DOMINO operators, the signal controlling the expression of CDA-nCas9-ugi, required for the overall circuit to function, can be considered as the “operational signal,” while the signals controlling the expression of individual gRNAs can be considered as independently controllable “inputs” (Figure 1B). To demonstrate the performance of an individual DOMINO operator, we placed CDA-nCas9-ugi and gRNA on separate cassettes under the control of anhydrotetracycline (aTc) and isopropyl β-D-1-thiogalactopyranoside (IPTG) inducible promoters, respectively. Inducing Escherichia coli (E. coli) cells harboring these two cassettes with both aTc and IPTG resulted in efficient dC-to-dT mutations at two dC residues within the gRNA WRITE window, demonstrating successful DNA writing by DOMINO (Figure 1C). No mutation was detected if the cultures were not induced or if they were induced by only one of the two inducers. These results demonstrate that DOMINO can be used as a precise DNA writer to efficiently and deterministically manipulate genomic DNA in response to signals of interest.
Molecular Recording by DOMINO
We first asked whether DOMINO operators can be used (in an analogous way to SCRIBE) to record the dynamics of transient transcriptional signals of interest into DNA. To assess this, we exposed E. coli cultures harboring the above-mentioned circuit to the operational signal (aTc) and various levels of the input (IPTG) and monitored the accumulation of mutant alleles in the population by Illumina High-Throughput Sequencing (HTS) over the course of 24 hours. The frequency of mutant allele (memory state S1) increased as the concentration of IPTG increased, demonstrating that the DOMINO operator recorded input levels in the form of the fraction of alleles in the S1 state (Figure 1D).
We then sought to study the dynamics of mutation accumulation. To this end, we initially induced the cultures with aTc for 4 hours, then diluted the cultures and added IPTG. A significant increase in the S1 allele frequency was detected as early as one hour after IPTG addition. Mutations accumulated linearly over the next 7 hours, demonstrating that the duration of exposure to the input could be recorded in DNA (Figure 1E). Mutant frequency, however, did not increase substantially after 8 hours as the cultures reached saturation, suggesting that mutations accumulated much faster in freshly diluted and actively growing cells. Consistent with the previous experiment, we did not observe significant accumulation of the mutant allele in cultures that were not exposed to the input (IPTG).
In these experiments, two mutable residues (CC) within the WRITE window of the gRNA were used, and the memory states were defined such that mutations in both residues were required to be considered as a state transition. Consistent with previous reports (Komor et al., 2016), we observed that residues in different positions along the WRITE window were edited with different dynamics (Figures S1A–B). These results suggest that the number of intermediate memory states, as well as the response dynamics can be tuned for each DOMINO operator by adjusting the number or position of mutable residues (dCs or dGs) within the WRITE window.
By replacing the IPTG-inducible promoter in the DOMINO operator with various inducible promoters, we further showed that various signals (having either biological or physiological relevance) could be recorded in DNA. These signals included sugars [arabinose (Ara)], heavy metals (Cu2+), and darkness, as well as several biomarkers of gastrointestinal inflammation: blood (heme), hydrogen peroxide, and nitric oxide (Figures S1C–H). These results demonstrate that DOMINO operators could be used as modular molecular recording devices to capture the dynamics of transcriptional signals of interest in DNA.
Layered Molecular Recording and Computation by DOMINO
Upon successful demonstration of molecular recording by DOMINO, we next sought to investigate whether multiple DOMINO operators could be used to concurrently record the dynamics of multiple signals and perform logic operations in living cells. Specifically, we theorized that due to the precise and well-defined nature of the mutational outcomes generated by DOMINO operators, information regarding multiple signals (e.g., logical features such as presence or absence, and analog features such as intensity or duration) could be recorded into nearby memory registers and the mutational state of these registers could be read by sequencing or on the fly with another DOMINO operator. These would allow DOMINO operators to be arrayed and interconnected in a highly scalable fashion such that the mutational outcome of one operator would serve as input for other operators. Such interconnected operators could be used to execute a series of order-independent and/or sequential unidirectional DNA writing events and build robust and complex forms of logic and memory operations to record and control the combination, order, and timing of molecular events in living cells.
1. Order-independent DOMINO Logic
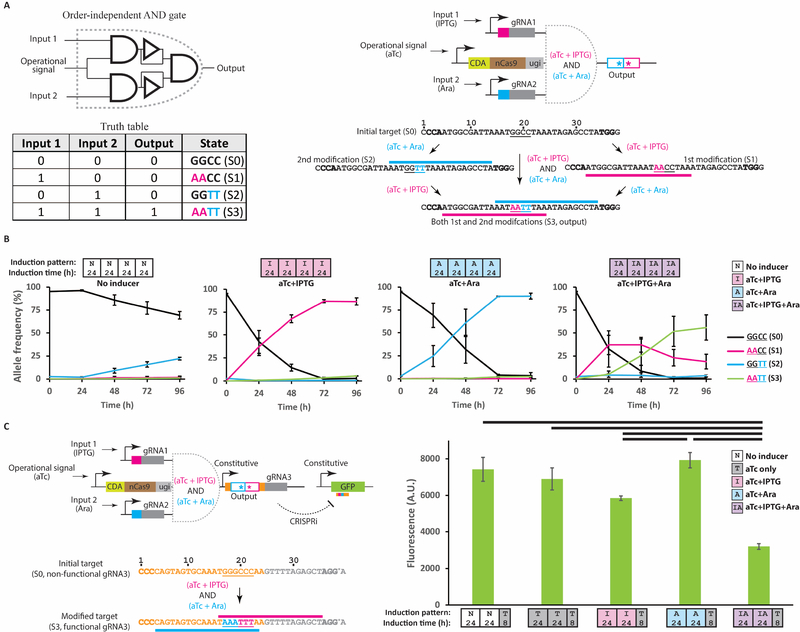
To demonstrate this concept, we set out to build a two-input order-independent AND logic gate, with which the A AND B logic is executed independent of the order of addition of the inputs, by layering two DOMINO operators as indicated in Figure 2A. In this design, two distinct gRNAs were used. One gRNA was placed under the control of an IPTG-inducible promoter and the other gRNA was placed under the control of an Ara-inducible promoter. In the presence of its corresponding inducer, each gRNA, once expressed, would direct the DNA read-write module (which itself is expressed in the presence of the operational signal, aTc) to its cognate target site, resulting in precise dC-to-dT mutations within its WRITE window.
Figure 2 |. Incorporating order-independent logic by DOMINO.
A) A schematic representation of the order-independent AND gate enabled by DOMINO where the output is ON only when both inputs have been present with any possible order. Induction of the circuit with either of the two inducers (IPTG or Ara) results in editing of the target and transition to an intermediate state (states S1 or S2, respectively). Induction of the circuit with both gRNAs results in generation of the doubly edited DNA sequence (state S3), which is designated as the ON state. B) Dynamics of allele frequencies obtained by HTS for the circuit shown in (A). E. coli cells were exposed to different combinations of the inducers for four days with serial dilution every 24 hours. Small boxes along the x-axis show the induction patterns and duration of induction used in each experiment. For example, the induction pattern of the last sample set ([IA][IA][IA][IA]) means that the samples were induced with aTc + IPTG + Ara for four days with dilutions every 24 hours. C) Coupling mutational outcome of DOMINO operators to downstream functional elements (e.g., output gRNA and GFP). Left: A schematic of the genetic circuit. In this example, AND logic is realized on the target DNA register (i.e., the output gRNA) while NAND logic is achieved on the output GFP reporter. A dA-to-dG mutation (shown by an asterisk) was introduced into the handle of the output gRNA in a position that was not essential for gRNA function (Briner et al., 2014). This modification was required to generate an NGG PAM motif for the binding of one of the input gRNAs. Right: Flow cytometry measurements for E. coli cells harboring the circuit shown on the left induced by various combinations of the inputs. Error bars indicate standard deviation for three biological replicates. See also Figure S2 and S3.
To assess the performance of the order-independent DOMINO AND gate, we induced E. coli cells harboring this circuit with different combinations of the inducers for several days with successive rounds of dilutions (to maintain the cells in an actively growing state) and analyzed allele frequencies at the target locus by HTS. In the presence of the operational signal (aTc) and either of the two inputs (IPTG or Ara), mutations accumulated in the target sites of the induced gRNA in a linear fashion within the population; the frequency of mutant alleles reached a plateau after 72 hours of induction (Figures 2B and S2A), corresponding to transitions from the unmodified state (state S0) to either of the two singly modified states (state S1 or S2). On the other hand, when cells were induced with both inputs (IPTG AND Ara), the target sites for both gRNAs were edited, resulting in the accumulation of doubly edited sites (state S3) in the target locus. Low levels of a singly mutated allele (state S2) accumulated in the absence of any induction, likely due to the leakiness of the Ara-inducible promoter (pBAD) (Meyer et al., 2019) (Figure S1C). Nevertheless, the doubly edited allele (state S3) accumulated only in the presence of both IPTG and Ara, indicating that robust AND logic can be achieved despite the leakiness of one of the input promoters.
In this experiment, we defined S0, S1, and S2 states as the OFF (“0”) states and S3 as the ON (“1”) state, which means that this system implements AND logic. However, these states are defined arbitrarily; the same circuit can be defined, for example, as a NAND gate if the unmodified state (S0) is designated as ON output and the modified states (S1, S2, and S3) are designated as OFF outputs. Alternatively, each of the four mutational states can be defined as a distinct output, in which case the circuit can be considered as a 2-input/4-output decoder.
The time required for transitioning between the unmodified and fully modified states can be considered as the “propagation delay” of the corresponding DOMINO operator. Each of the two DOMINO operators used in this experiment exhibited a propagation delay of ~3 days (3 dilution cycles). Notably, accumulation of the singly mutated alleles in the presence of the operational signal and individual inducer inputs followed a linear trend over the course of few days (Figure 2B). This feature means that DOMINO can be used to implement both analog and digital computing, because the continuous changes that occur within the propagation delay window can be used to implement analog computation, while fully converted states can be considered as transitions between digital states and so be used for digital computation.
While HTS offers a powerful way to quantify the outcome of DOMINO circuits, its relatively high cost motivated us to develop a strategy for using Sanger sequencing chromatograms to quantify position-specific mutant frequencies within a mixture of DNA species. This algorithm, named Sequalizer (for Sequence equalizer), normalizes Sanger chromatogram signals and calculates the difference between the normalized signals from a test sample and an unmodified reference to identify position-specific mutations. It then uses this calculated difference to estimate position-specific mutant frequencies at any given target position (see Supplementary Data S1). We validated the accuracy of this method by constructing a standard curve (based on known ratios of mutant and WT sequences) and then comparing the Sequalizer results with next-generation sequencing (Figure S2). The Sequalizer output, which is based on population-averaged Sanger sequencing results, provides an estimate of position-specific mutant frequencies in an entire population. Though Sequalizer does not always provide accurate absolute values of mutant frequencies, fold changes in estimated mutant frequencies are accurate (Figures S2B–D). Additionally, unlike HTS, Sequalizer output does not reveal the identities and frequencies of individual alleles in the population. Nevertheless, given the high specificity of the DNA writers and their predefined target sites, this approach can be used as a low-cost alternative to HTS to assess the performance of DOMINO and other precise genomeediting platforms.
We analyzed the samples obtained from the experiment shown in Figure 2B by Sanger sequencing and Sequalizer, in addition to HTS. As shown in Figure S2E, in samples induced with either of the two inputs, the frequencies of mutations in positions corresponding to the cognate target sites of the induced gRNA increased. On the other hand, in samples that were induced with both gRNAs, the mutation frequencies in the target sites of both gRNAs increased (state S3). These results demonstrate that Sequalizer results are consistent with those obtained by HTS and that Sequalizer could be used to accurately estimate changes in position-specific mutant frequencies obtained by HTS.
The output of DOMINO operators takes the form of DNA mutations that accumulate at a target site. These mutations can be directly read by sequencing or coupled with functional elements to control cellular phenotypes. For example, by flanking the input gRNA target site with a desired promoter and a gRNA handle, the output of a given DOMINO operator can be converted into downstream gRNA expression. The output gRNA can then be interconnected with other DOMINO operators to build more complex circuits, or alternatively, combined with CRISPR-based gene regulation strategies, such as CRISPRi and CRISPRa, to dynamically regulate cellular phenotypes. To demonstrate this concept, we engineered an AND gate by layering two DOMINO operators under the control of IPTG- and Ara-inducible promoters to edit a third gRNA as the output. In the presence of both inducers, the output gRNA was modified by both input gRNAs such that it could then bind to and repress a downstream reporter gene (GFP) (Figure 2C).
In addition to an AND gate, other logic operations can be readily implemented by DOMINO, by carefully positioning mutable residues on the targets or by designing the combinations and order of DNA writing events (Figures S3A–B). Furthermore, additional input gRNAs can be incorporated to achieve operators with more than two inputs, thus demonstrating the multi-input recording capacity and scalability of this approach (Figure S3C). Moreover, the mutational outcome of DOMINO operators, in addition to gRNAs, can be directed toward other regulatory and functional elements, such as promoters, ribosome-binding sites, start/stop codons, as well as active sites within proteins to tune the expression or activity of downstream components (Figure S3D).
2. Sequential DOMINO Logic
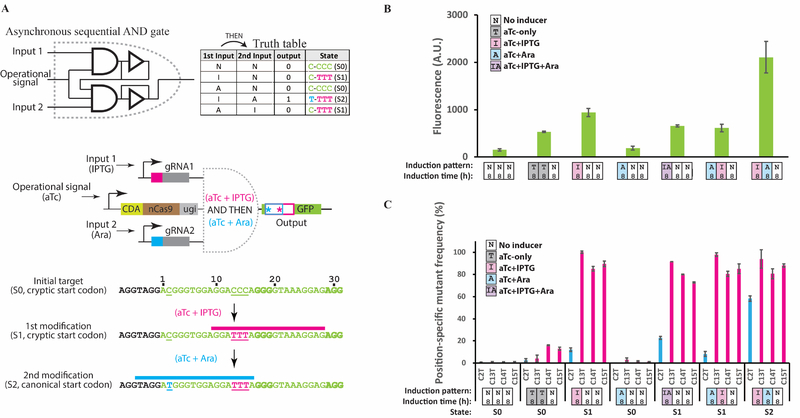
In addition to realizing order-independent logic, the order of DNA writing events executed by DOMINO operators can be carefully controlled to achieve sequential logic, which generates the desired outputs only when the correct order of inducers is added. For example, to achieve sequential logic, the gRNA output of one operator can be designed to serve as the input for a downstream operator (Figure S3B). This design can be used to functionally connect DOMINO operators that are not physically co-located. Alternatively, sequential logic can be achieved by overlapping mutable residues in the WRITE address of one operator with the READ address of a downstream operator (Figure 3). This design uses DNA mutations rather than cascades of gRNAs as a way to interconnect cis-encoded DOMINO operators, thus offering a highly compact and scalable strategy for encoding sequential logic.
Figure 3 |. Building sequential logic by DOMINO operators.
A) Sequential AND gate encoded with DOMINO operators. The output of a DOMINO operator was used as an input for another operator, which in turn mutates a non-canonical start codon (ACG) within the GFP ORF into a canonical (efficient) start codon (ATG), thus increasing GFP signal. The second gRNA (induced by Ara) can bind to and enact the start-codon mutation only after the first gRNA (induced by IPTG) has edited its target. B) GFP signal measured by flow cytometry for the circuit shown in (A). C) Position-specific mutation frequency obtained from Sequalizer analysis for the experiment shown in (A). Error bars indicate standard deviation for three biological replicates. See also Figure S3 and S4.
To demonstrate the latter strategy, we constructed an asynchronous sequential DOMINO AND gate in E coli, such that the sequential addition of the two inputs in the correct order (IPTG AND THEN Ara) would lead to the mutation of a cryptic start codon (ACG) into the canonical, more efficient start codon (ATG) in the GFP ORF, thus increasing the GFP signal (Figures 3A–B). We observed slight increases in GFP signal in cells that had been induced with the first inducer (i.e., IPTG) or those that had been co-induced with both inducers (Figure 3B). The former effect was likely caused by the leakiness of the Ara-inducible promoter (Figure S1C) while the latter was likely due to the simultaneous presence of both inducers in the media, which could produce, to some extent, sequential DNA mutations in the correct order. Nevertheless, the GFP signal was significantly higher when cells were exposed to the inducers in the correct order (IPTG AND THEN Ara). We further confirmed these results by analyzing Sanger sequencing chromatograms by Sequalizer. Consistent with the flow cytometry data, the highest level of mutation in the cryptic start codon (Figure 3C, blue bars) was achieved when the samples were induced with the correct order, indicating the execution of sequential AND logic. To further demonstrate that the order of different transcriptional events can be recorded as distinct mutational signatures in the DNA, we built two additional sequential logic circuits (Figure S4). These examples further demonstrate that sequential DOMINO logic circuits can be used to program and commit cells to defined states based on the order of inputs.
3. Temporal DOMINO Logic
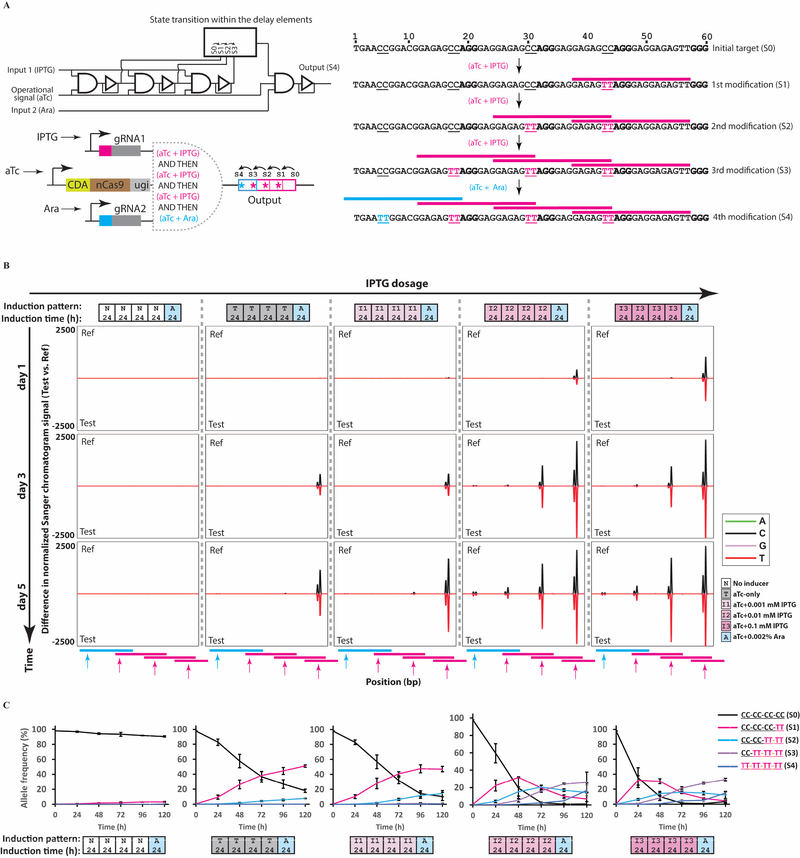
The above examples demonstrate that the sequence of DNA writing events mediated by DOMINO operators can be controlled by external cues. In addition to building sequential logic, such that the execution of events in a specified order leads to a desired output, the inherent propagation delay in DOMINO operators can be exploited to incorporate delays and temporal information into circuits, so that a desired output is produced only after a certain period of time has passed. Multiple DOMINO operators can be placed sequentially in an array to build longer delays. In a simple form, DOMINO delay operators can be built by constructing a series of overlapping repeats to act as target sites for a desired gRNA (Figure 4A). This repeat configuration allows the READ address of each gRNA operator site to overlap with the WRITE address of the previous gRNA. Initially, the gRNA can bind to the first (i.e., 3’-end) repeat but not to the upstream copies of the repeat that harbor dC residues (instead of dT) in the sequence corresponding to the gRNA READ address. Upon binding to the first repeat, the gRNA can recruit the read-write head to the first repeat, which can mutate the dC residues in the repeat immediately upstream of its binding site (i.e., the second repeat), thus converting that repeat to a new binding site for the same gRNA. This process is sequentially repeated to generate new binding sites for the same gRNA. Much like an array of physical domino pieces that fall one by one, each genome-editing event is initiated only after editing in the previous repeat has occurred, thus ensuring a sequential cascade of unidirectional DNA writing events over time. The output of the delay elements can be combined with additional logic operators and internal or external cues to create more complex forms of temporal logic.
Figure 4 |. Incorporating propagation delay and temporal logic into living cells.
A) A schematic of a temporal AND gate built by overlapping DOMINO operators, for which the timing between the two signals is important to execute the AND function (see text for details). B) E. coli cells harboring the circuit shown in (A) were exposed to different concentrations of the first inducer (IPTG) for 4 days with serial dilution after each day, followed by a one-day exposure to the second inducer (Ara). The propagation of the signal as manifested by sequential mutations in the repeat array was monitored by analyzing Sanger chromatograms with Sequalizer. C) Transitions between the memory states for samples shown in (B) assessed by HTS. Error bars indicate standard deviation for three biological replicates. See also Figure S5.
To demonstrate this concept, we placed three DOMINO delay elements into an array and linked the output of the array to a second DOMINO operator (Figure 4A). This design achieves temporal and sequential AND logic, because the first (i.e., IPTG-inducible) gRNA has to execute three consecutive DNA writing events before the Ara-inducible gRNA (corresponding to the last operator) can bind to and edit its target. We induced cells harboring this circuit with IPTG at various concentrations for 4 consecutive days followed by a final day of induction with Ara. Sequalizer and HTS analysis of these samples demonstrated a time- and IPTG-dosage-dependent accumulation of mutations in the target sites within repeats, corresponding to propagation of the signal through the repeat array (Figure 4B–C). By the end of the experiment, mutations in the target site of the second gRNA (shown by the blue arrow in Figure 4B) were detected only under conditions in which mutations had accumulated through the entire cascade, corresponding to the samples that had been induced with the high IPTG concentrations (i.e., 0.01 mM and 0.1 mM). The second gRNA, which is under the control of Ara, was only able to bind to and edit its target when the third copy of the repeat was edited by the first gRNA, thus demonstrating the desired time-dependent sequential logic. These results further demonstrate that, in addition to enacting delays in gene circuits, an array of DOMINO delay elements can be used as a multi-state memory register that undergoes step-wise transitions between discrete states in a time- and dosage-dependent fashion.
The timing and dynamics of transition between memory states (propagation delays) in DOMINO circuits can be further controlled by adjusting the writing efficiency (e.g., changing the position of mutable residues within the WRITE window (Figure S1A)) or tuned dynamically by external cues (i.e., stronger signal, Figure 4). Additionally, the number of memory states and the total delay can be programmed by changing the number of repeats (Figure S5). We envision that more complex versions of temporal logic, such as genetic counters and timers, can be constructed by integrating delay elements into multiple-input DOMINO operators.
Non-destructive DNA-State Reporters
Existing DNA-based molecular recording technologies rely on DNA sequencing as readout. As such, in order to retrieve the recorded information, recording has to be stopped. A unique feature of DOMINO compared to other memory platforms is that the DOMINO DNA read-write head can be further functionalized with additional effector domains (e.g., transcriptional activators and repressors) to achieve concurrent DNA writing and transcriptional regulation. This feature, combined with the precise and sequential DNA writing achieved by DOMINO, enables one to perform both genetic and epigenetic modulation and correlate DNA memory states with distinct functional outcomes that can be continuously monitored in living cells without disrupting the cells.
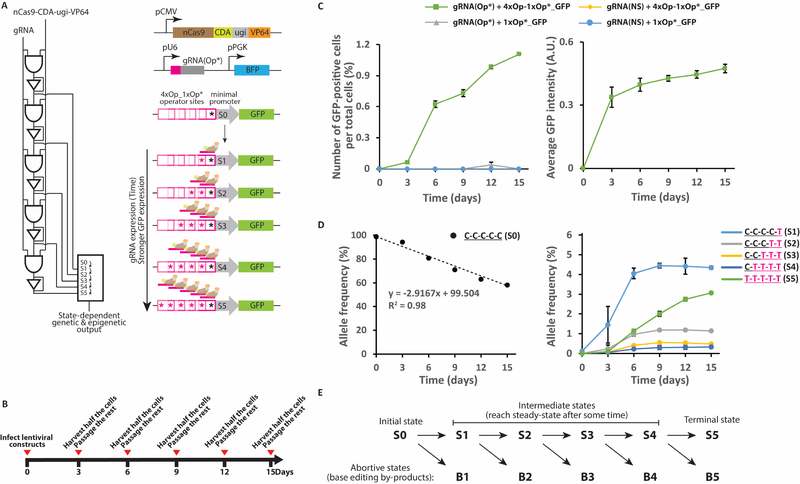
To demonstrate this concept, we constructed a non-destructive DNA-state reporter gene circuit in the human embryonic kidney (HEK) 293T cell line. We first made an array of four overlapping WT operator repeats (4xOp) and a downstream mutant repeat (1xOp*), which harbored a dC-to-dT mutation. We then placed this array upstream of a minimal Adenovirus Major Late Promoter (MLP) controlling the expression of GFP, in order to build the 4xOp_1xOp*_GFP reporter construct. As a control, we constructed the 1xOp*_GFP reporter, which previously had been shown to have negligible activity (Farzadfard et al., 2013). We also functionalized the DNA read-write head (nCas9-CDA-ugi) with a transcriptional activator domain (VP64) and cloned the nCas9-CDA-ugi-VP64 fusion construct along with either of the two reporter constructs into lentiviral vectors, which were subsequently introduced into HEK 293T cells. We then delivered a second lentiviral vector encoding either an Op*-specific gRNA [gRNA(Op*)] or a non-specific gRNA [gRNA(NS), as negative control] to these cells. Upon binding, gRNA(Op*) could mutate the critical dC residue in the adjacent upstream WT Op repeat, thus converting the Op repeat to a Op* sequence that could serve as a new binding site for the same gRNA; this strategy enables sequential rounds of mutation (i.e., Op to Op* conversion), gRNA binding, and transactivator recruitment events (Figure 5A).
Figure 5 |. Non-destructive DNA-state reporting circuit in human cells.
A) A schematic representation of the non-destructive DNA-state reporting circuit (see text for details). B) HEK 293T cells were transfected with the circuit shown in (A) via a two-step lentiviral delivery protocol and grown with serial passaging every three days as indicated. C) Left: The frequency of GFP-positive cells per total transfected cells Right: The average GFP signal intensities for GFP-positive cells harboring 4xOp_1xOP*_GFP and gRNA(Op*). D) Dynamics of the WT allele frequency (unmodified state S0, left) and modified states (S1-S5, right) in cultures harboring 4xOp_1xOp*_GFP and gRNA(Op*) assessed by HTS. E) A model describing the step-wise transition from S0 to S5 in this memory circuit. At later time points, the intermediate states (S1-S4) reach steady states while the initial (S0) and terminal (S5) are constantly consumed and produced, respectively. See also Figure S6.
We sequentially passaged cells harboring these circuits every three days for fifteen days (Figure 5B) and monitored both GFP expression (by microscopy, Figures 5C and S6A) and allele frequencies (by HTS, Figures 5D). The normalized frequency of GFP-positive cells and the GFP expression level of individual cells in cultures harboring the 4xOp_1xOp*_GFP reporter and gRNA(Op*) gradually increased over time, indicating the gradual activation of the reporter in these cells (Figure 5C). This data suggests that the number of bound transactivators, and thus, the number of activated repeats (i.e., Op*), which serve as operator sites for the chimeric read-write-transactivator protein, were increased in these cells. On the other hand, in cultures that were transfected with gRNA(NS), or those that contained the 1xOp*_GFP reporter, the GFP signal remained below the detection threshold.
These results were further confirmed by HTS analysis of the allele frequencies throughout the experiment. As shown in Figure 5D, the frequency of the WT allele (state S0) in cells containing the repeat array and gRNA(Op*) linearly decreased over time, indicating that the DNA writing circuit can be used as an analog recorder for the input gRNA. As expected, the final memory state (i.e., S5), which corresponds to the highest GFP expression state, increased steadily over time. Consistent with the GFP data, the first four memory states (S1 through S4) started to accumulate sequentially until they reached a plateau (i.e., the steady state) towards the end of the experiment (Figure 5E). No significant changes in allele frequencies were observed in cells that were transduced with a non-specific gRNA (Figure S6B). Together with the microscopy data, these results show that the analog properties of a signal, such as the duration of exposure to gRNA(Op*), can be faithfully and permanently recorded within the distribution of memory states of the DNA recorder. In this circuit, higher GFP-expression levels are correlated with the higher memory states. Thus, the GFP signal can be used to continuously monitor DNA memory states without requiring cellular destruction for sequencing. Furthermore, at the single cell level, this memory architecture constitutes a multi-bit digital recorder that associates a longer (or higher intensity of) incoming signal (i.e. gRNA expression) with transitions to higher memory states.
In this experiment, in addition to dC-to-dT mutations, we observed dC-to-dG and dC-to-dA mutations, albeit with lower frequencies than dC-to-dT mutations (Figure S6C). This observation is consistent with previous reports for mammalian cell lines (Komor et al., 2016; Nishida et al., 2016) and reflects the promiscuous repair outcome of deaminated dC (dU) lesions in these cells. Notably, in samples containing the 1xOp*_GFP reporter, frequency of the WT allele (state S0) decreased and frequency of the mutant alleles increased linearly over time (Figure S6C). Thus, even without having a repeat array, the accumulation of mutations in a specific target site can be used as an analog readout of an incoming signal.
In this experiment, we used VP64 as the transactivator domain. However, the activation level and dynamic range of the reporter output can be tuned by using stronger activator domains, such as VPR (Chavez et al., 2015). Alternatively, other effector domains, such as repressors (Farzadfard et al., 2013), DNA methyl transferases (Liu et al., 2016), acetyl transferases (Hilton et al., 2015), or other types of histone modification domains, could be used to implement more sophisticated forms of combined memory and gene regulation programs.
Discussion
By using a DNA read-write head that can manipulate genomic DNA with nucleotide resolution, DOMINO converts the genomic DNA into an addressable, readable, and writable medium for information processing and storage in living cells. Various orthogonal DOMINO operators can be built by simply changing the sequence of gRNAs, making the system highly scalable. Furthermore, since DOMINO enables manipulation of DNA with nucleotide resolution within a defined narrow window, compact multi-input operators can be readily created by targeting multiple gRNAs to nearby registers which can then be interrogated by a downstream operator. Unlike other precise DNA writing systems that require multiple proteins (e.g., recombinases) to encode memory, DOMINO uses small gRNAs and only one protein moiety, therefore minimizing metabolic burden on the cells. DOMINO enables molecular recording as well as highly compact and scalable logic and memory operators that, unlike previous strategies, can be used for both digital and analog computation in living cells. Thus, this scalable approach expands our capacity for molecular recording and computation in living cells and as a result, the ability to monitor and control cellular phenotypes.
Like other synthetic gene circuits, non-optimal performance of gene regulatory elements, such as leaky promoters, could negatively affect the performance of DOMINO. These limitations may be overcome with systematic optimizations, such as engineering reduced basal promoter activities via directed evolution strategies (Meyer et al., 2019), using engineered promoters with tighter activity (Arpino et al., 2013), or lowering the copy numbers of gene circuits (Lee et al., 2016).
In our experiments (Figure 1E), we detected the presence of a signal as fast as 1 hour (~2 generations) after the start of induction. The temporal dynamics of recording could be improved by using newer generations of base editors with higher editing efficiency (Koblan et al., 2018) or by making conditional DNA writing modules with faster response times (e.g., by incorporating signal-responsive RNA and protein motifs into the gRNA and the base editor, respectively). Combining the nucleotide-deaminase-based DNA writing modules with alternative DNA binding proteins as read modules (such as RNA polymerases) could also increase the temporal resolution and capacity of molecular recording.
Deterministic DOMINO operators and cascades rely on precise base-editing events for proper function. Our results show that when the CDA-nCas9-ugi head is used, the outcome of these operators in E. coli is almost exclusively in the form of dC-to-dT mutations (Figure S1). However, in human cells, other nucleotides (dG, and to a lesser extent, dA) are also generated, albeit at a lower rate than dT (Figure S6C). The promiscuous repair outcome of cytidine deaminases in mammalian cells results in by-products and abortive memory states that cannot undergo additional rounds of state transition in DOMINO cascades (Figure 5E), which in turn negatively affects the overall performance of the DOMINO circuits. Using DNA writer modules with higher DNA writing efficiency and purer mutational outcomes (such as adenosine base editors (Gaudelli et al., 2017)) could help to reduce the level of these by-products and improve the performance of deterministic and complex DOMINO circuits in mammalian cells. Furthermore, incorporating orthogonal DNA writing modules (e.g., cytidine and adenosine deaminases) into DOMINO should make reversible DNA writing possible, which has been challenging to achieve with previous DNA memory platforms. Reversible DNA writing would enable bidirectional cellular programs and pave the way for sophisticated biological state machines, cellular automata, and Turing machines that use the genomic DNA of living cells as a rewritable memory tape to perform advanced memory and computation operations.
Several CRISPR-Cas9-based strategies for recording biological events, such as signaling dynamics and cellular lineage histories, into DNA have been recently described (Kalhor et al., 2018; McKenna et al., 2016; Perli et al., 2016). These approaches rely on stochastic DNA memory states (i.e., indel mutations) that are generated by pseudorandom DNA writers mediated by Cas9-mediated double-strand DNA breaks and subsequent repair of these breaks by NHEJ. However, the recording capacity of these recorders is exhausted after recording a few molecular events due to the loss of gRNA target sites; these recorders are, therefore, not ideal for performing logic or long-term recording of signaling dynamics or event histories (Farzadfard and Lu, 2018). Moreover, since indel mutations (memory states) are stochastically generated by NHEJ, new mutations could destroy the previous mutations and thus overwrite the previous memory states, making it challenging to trace lineage histories. In addition, none of these strategies can be used in organisms lacking an efficient NHEJ repair pathway, such as prokaryotes. In contrast, mutational memory states generated by DOMINO are precise, unidirectional, position-specific, and minimally disruptive. These features ensure that previous mutations are preserved after each editing step and can be accurately traced.
In addition, the precise and predictable memory state transitions in DOMINO recorders enable memory states to be coupled to functional biological outcomes, such as changes in gene expression, thus obviating the need for sequencing as readouts for certain applications (Figure 5). Furthermore, DOMINO does not require double-strand DNA breaks or NHEJ; thus, it can function in both bacterial and mammalian cells in an autonomous and continuous fashion over many generations. We envision that the DNA records generated by DOMINO recording systems could be used to study signaling dynamics and event histories in their native contexts over many generations, as well as building gene circuits with artificial learning capacities. The promiscuous repair of dC lesions in mammalian cells could actually be beneficial for tracing cell lineages, as it can increase the number of potential memory states. Moreover, signal-responsive lineage maps with tunable resolution can be generated because the activity of DOMINO recorder can be modulated by internal or external signals of interest. Combining these recorders with single-cell sequencing, advanced barcoding schemes, and self-targeting guide RNAs (Perli et al., 2016) should pave the way toward more advanced recorders for long-time monitoring of signaling dynamics and cellular lineages (Farzadfard and Lu, 2018).
We envision that our long-term, compact, scalable, modular, and minimally disruptive genetic memory architectures will enable an unprecedented set of applications for building genetic programs and recording and controlling spatiotemporal molecular events in their native contexts. These applications could impact many different fields, including developmental biology, cancer, stem cell, and brain mapping. For example, DOMINO can be used to program the timing and progression of developmental stages within living animals, or to perform long-term lineage tracking experiments in mammals, which has not been feasible to date because of the lack of scalable and long-term methodologies. DOMINO recorders could be adapted to map neural activity by driving the activity of DNA writers with regulators that respond to neural activity. One could study the order and temporal nature of signaling events in their native contexts and robustly control cellular differentiation cascades ex vivo and in vivo. These recorders could be programmed to investigate tumor development and unveil the cellular and environmental cues involved in tumor heterogeneity. They could also be used to record arbitrary information into the DNA of living cells for DNA storage applications. Finally, living sensors could be designed to sense pathogens, toxins, or other signals within the body or in the environment and then later report on this information in detail.
STAR METHOD
CONTACT FOR REAGENT AND RESOURCE SHARING
Email contact for reagent and resource sharing: timlu@mit.edu
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Bacterial experiments were performed in E. coli MG1655 PRO strain (MG1655 strain that harbors PRO cassette (pZS4Int-lacI/tetR, Expressys) and expresses lacI and tetR at high levels) (Lutz and Bujard, 1997). Mammalian cell experiments were performed in HEK 293T cell line (ATCC CRL-11268).
METHOD DETAILS
Plasmid Construction
Standard molecular biology and cloning techniques, including ligation, Gibson assembly (Gibson, 2011) and Golden Gate assembly (Engler et al., 2008) were used to construct the plasmids. Chemically competent E. coli DH5α F’ lacIq (NEB) and E. cloni 10G (Lucigen) were used for cloning. Lists of plasmids, synthetic parts and sequencing primers used in this study are provided in Tables S1, S2, and S3, respectively. Plasmids and their corresponding maps will be available on Addgene.
Antibiotics and Inducers
For bacterial selection, antibiotics were used at the following concentrations: Carbenicillin (Carb, 50 μg/mL), Kanamycin (Kan, 20 μg/mL), and Chloramphenicol (Cam, 25–30 μg/mL). For the experiments shown in Figures 2C, S3B, and S4 different combinations of 200 ng/ml anhydrotetracycline (aTc), 0.1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) and 0.2% Arabinose (Ara) were used to induce the corresponding circuits. For the experiments shown in Figures S3D and S5, 250 ng/ml aTc and 0.005% Ara were used. For the experiment shown in Figure 3A, 150 ng/ml aTc, 0.1 mM IPTG, and 0.2% Ara were used. For all the other experiments, unless otherwise noted, 250 ng/ml aTc, 1 mM IPTG and 0.2% Ara were used. All concentrations are final concentrations.
Other inducers (CuSO4, Heme, H2O2, and NO) were used with final concentrations as indicated in Figures S1C–H. Diethylenetriamine/nitric oxide adduct was used as the source of NO. Defibrinated horse blood (Hemostat) was used as the source of Heme (Blood was lysed by first diluting 1:10 in simulated gastric fluid (SGF) (0.2% NaCl, 0.32% pepsin, 84 mM HCl, pH 1.2) before further dilution in culture media) (Mimee et al., 2018).
Bacterial Cell Experiments
Different plasmids expressing gRNAs and targets (listed in Table S1) were transformed into the reporter cells (MG1655 PRO) harboring aTc-inducible CDA-nCas9-ugi (for bacterial experiments, APOBEC1 CDA (Komor et al., 2016) was used as the writing module). Single transformant colonies were grown in appropriate antibiotics for 4–8 hours to obtain seed cultures. Seed cultures were diluted (1:100) in fresh media containing different combinations of the inducers and grown in 96-well plates (or tubes for the Dark/Light conditions shown in Figure S1E) with serial dilutions (if applicable) as indicated in induction patterns in corresponding figures. Samples for various measurements including HTS, Sequalizer, and flow cytometry were taken at indicated time points.
Mammalian Cell Culture
HEK 293T cells were grown in DMEM supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin at 37°C with 5% CO2.
Lentivirus Production
Lentiviral constructs were cloned in using the FUGW backbone (Addgene #25870) and packaged in HEK 293T cells by co-transfection with psPAX2 and pCMV-pVSV-G helper plasmids. In brief, 4.4×105 HEK 293T cells per well were seeded in a 6 well plate. After 24-hour incubation, media were replaced with 2mL fresh culture media containing FuGENE HD/DNA complexes. For FuGENE HD/DNA complex, 9 μL of FuGENE HD (Promega) was added to a mixture of 3 plasmids consisting of 0.1 μg of pCMV-VSV-G vector, 0.5 μg of lentiviral packaging psPAX2 vector, and 1 μg of lentiviral expression vector in 100 μL of Opti-MEM reduced serum medium (Thermo Fisher Scientific), followed by 20 minutes incubation at room temperature. Media of transfected cells were replaced with 2 mL of fresh culture media 18 hours post transfection. The supernatant containing newly produced viruses was collected at 48-hours post-transfection, and filtered through a 0.45 μm syringe filter (Pall Corporation, Ann Arbor, MI; Catalog #4614). Filtered lentiviruses were used to infect respective cell lines in the presence of polybrene (Sigma-Aldrich, 8 μg/mL). Successful lentiviral integration was confirmed by using lentiviral plasmid constructs constitutively expressing fluorescent proteins or antibiotic resistance genes to serve as infection markers.
Generation of Monoclonal Cell Lines
A lentiviral plasmid construct was made by placing the nCas9-CDA-ugi-VP64 fusion protein with nuclear localization signals linked to the Puromycin resistance gene with the P2A sequence under the control of constitutive CMV promoter (for mammalian experiments, PmCDA (Nishida et al., 2016) was used as the writing module). In addition, repeat arrays (4xOp_1xOp* or 1xOp*) were placed upstream of the minimal adenovirus major late promoter (MLP) promoter driving GFP and the resultant reporter constructs were cloned into the same lentiviral construct and packaged into viral particles. For generation of monoclonal cell lines, 4.4×105 HEK 293T cells per well were seeded in a 6 well plate. After 24-hour incubation, media were replaced with 4 mL of the viral supernatant to infect cells with lentiviruses in the presence of 8 μg/mL polybrene. The viral supernatant was replaced with 2 mL of fresh culture media 24 hours post transduction. Drug selection with Puromycin (Thermo Fisher Scientific, 7 μg/mL) was performed at 72 hours after transduction for 8 days. During drug selection, a confluent well or dish was expanded into a 10 cm dish or T-75 flask. The pooled population was then suspended at a concentration of 5 cells/mL in fresh cell culture media, and 100μL of cell suspension was transferred into each well of a 96-well plate. After 7-day undisturbed incubation, each well was assessed by a microscope, and monoclonal cells forming a colony were harvested and expanded.
Non-destructive DNA-State Reporter Experiment
On day 0, 440,000 clonal reporter cells per well were infected, in the presence of 8 μg/mL polybrene, with 4 mL of the viral supernatant with high titer lentiviral particles encoding the gRNAs driven by the U6 promoter in a 6-well plate in triplicate. Infection efficiency was more than 90% in every sample. The cells were harvested every 3 days until day 15 after the infection. Half of the harvested cells were seeded in a 6-well plate for further culture. One-fifth of the harvested cells was seeded in a glass bottom 6 well plate (MatTek Corporation, USA) in DMEM without phenol red supplemented with 10% FBS and 1% penicillin-streptomycin for microscopic analysis. The remaining cells were collected for next-generation sequencing.
Microscopy
Fluorescence microscopy images of cells in glass-bottom tissue culture plates were obtained at 12 hours after subculture by using the DeltaVision microscopy imaging system (Applied Precision) with a 20x objective lens.
Flow Cytometry
Bacterial cultures were diluted 1:10 and the GFP signal in diluted samples were measured using an LSR Fortessa II flow cytometer (Becton Dickinson, NJ) equipped with 488/FITC laser/filter set.
High-throughput Sequencing
For each bacterial sample, 5 μl of culture was resuspended in 15 μl of QuickExtract DNA Extraction Solution (Epicentre, WI) and lysed by a two-step protocol (15 minutes incubation at 65 °C followed by 2 minutes incubation at 98 °C). For each mammalian cell sample, cell pellet was resuspended in 40 μl of QuickExtract DNA Extraction Solution and lysed by a two-step protocol (30 minutes incubation at 65 °C followed by 16 minutes incubation at 98 °C). Target sites were PCR amplified using 2 μl of lysed bacterial cultures (or 2.5 μl of extracted mammalian cell lysate) as templates and the appropriate primers listed in Table S3. The obtained amplicons were used as templates in a second round of PCR to add Illumina barcodes and adaptors. The amplicons were then multiplexed and sequenced by Illumina platform.
Sanger Sequencing
For each sample, target sites were PCR amplified by target-specific primers and Sanger sequenced by Quintara Biosciences. The obtained chromatograms were analyzed by Sequalizer.
QUANTIFICATION AND STATISTICAL ANALYSIS
Flow Cytometry Data Analysis
All samples were uniformly gated (using the strategy indicated in Figure S2F) and the mean fluorescence and percent of GFP-positive cells were calculated by FACSDIVA and FlowJo (BD Biosciences). Experiments were performed in triplicates.
Microscopy Image Analysis
Microscopy images were analyzed by CellProfiler software and the number of GFP- and BFP-positive cells as well as GFP signal intensity in GFP-positive cells were measured using the ‘ColorToGray’, ‘IdentifyPrimaryObjects’ (for BFP), ‘IdentifyPrimaryObjects’ (for GFP), ‘MeasureObjectIntensity’, and ‘ExportToSpreadsheet’ modules. Image data were inspected for complete removal of false positive debris after the CellProfiler analysis. For each sample, the number of GFP-positive cells was normalized to the total number of gRNA-transduced cells by dividing the total number of GFP-positive cells to BFP-positive cells in 40 random fields of view. Experiments were performed in triplicates.
HTS Data Analysis
The obtained sequencing reads were demultiplexed and allele frequencies were calculated using a custom MATLAB script.
Sequalizer Analysis
The obtained Sanger chromatograms for each sample was analyzed by Sequalizer (Supplementary Data S1) as described in the supplementary materials, using chromatogram of the seed culture for the corresponding experiment as the reference.
DATA AND SOFTWARE AVAILABILITY
Software
Software from this study has been described under “QUANTIFICATION AND STATISTICAL ANALYSIS” section.
Data Resources
The accession number for the raw sequencing data reported in this paper is PRJNA509198.
Supplementary Material
Data S1 | Sequalizer script (written in MATLAB) used to analyses sanger sequencing chromatograms. Related to STAR Methods.
Highlights.
DOMINO operators enable analog recording and continuous monitoring of cellular events
Various forms of logic can be built by layering multiple DOMINO operators
DOMINO allows online monitoring of DNA memory states without need for sequencing
DOMINO can be coupled with gene regulation for advanced memory & computation operations
Acknowledgements
We thank Christina Harrison for helping with some of the early experiments in this project. This work was supported by the National Institutes of Health (P50 GM098792), the Office of Naval Research (N00014–13-1–0424), the National Science Foundation (MCB-1350625), the Defense Advanced Research Projects Agency, the MIT Center for Microbiome Informatics and Therapeutics, and NSF Expeditions in Computing Program Award 1522074. F.F. would like to thank the Schmidt Science Fellows Program, in partnership with the Rhodes Trust for their support.
Footnotes
Declaration of Interests
F.F. and T.K.L. have filed a patent application based on this work. TKL is a co-founder of Senti Biosciences, Synlogic, Engine Biosciences, Tango Therapeutics, Corvium, BiomX, and Eligo Biosciences. TKL also holds financial interests in nest.bio, Ampliphi, and IndieBio.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arpino JA, Hancock EJ, Anderson J, Barahona M, Stan GB, Papachristodoulou A, and Polizzi K (2013). Tuning the dials of Synthetic Biology. Microbiology 159, 1236–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briner AE, Donohoue PD, Gomaa AA, Selle K, Slorach EM, Nye CH, Haurwitz RE, Beisel CL, May AP, and Barrangou R (2014). Guide RNA functional modules direct Cas9 activity and orthogonality. Mol Cell 56, 333–339. [DOI] [PubMed] [Google Scholar]
- Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M,E,PRI, Lin S, Kiani S, Guzman CD, Wiegand DJ, et al. (2015). Highly efficient Cas9-mediated transcriptional programming. Nat Methods 12, 326–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C, Kandzia R, and Marillonnet S (2008). A one pot, one step, precision cloning method with high throughput capability. PLoS One 3, e3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzadfard F, and Lu TK (2014). Genomically encoded analog memory with precise in vivo DNA writing in living cell populations. Science 346, 1256272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzadfard F, and Lu TK (2018). Emerging applications for DNA writers and molecular recorders. Science 361, 870–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzadfard F, Perli SD, and Lu TK (2013). Tunable and multifunctional eukaryotic transcription factors based on CRISPR/Cas. ACS Synth Biol 2, 604–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, and Liu DR (2017). Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG (2011). Enzymatic assembly of overlapping DNA fragments. Methods Enzymol 498, 349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. (2013). CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton IB, D’Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, and Gersbach CA (2015). Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol 33, 510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhor R, Kalhor K, Mejia L, Leeper K, Graveline A, Mali P, and Church GM (2018). Developmental barcoding of whole mouse via homing CRISPR. Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koblan LW, Doman JL, Wilson C, Levy JM, Tay T, Newby GA, Maianti JP, Raguram A, and Liu DR (2018). Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nature biotechnology 36, 843–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor AC, Kim YB, Packer MS, Zuris JA, and Liu DR (2016). Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Gyorgy A, Cameron DE, Pyenson N, Choi KR, Way JC, Silver PA, Del Vecchio D, and Collins JJ (2016). Creating Single-Copy Genetic Circuits. Mol Cell 63, 329–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, Shu J, Dadon D, Young RA, and Jaenisch R (2016). Editing DNA Methylation in the Mammalian Genome. Cell 167, 233–247 e217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz R, and Bujard H (1997). Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res 25, 1203–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Findlay GM, Gagnon JA, Horwitz MS, Schier AF, and Shendure J (2016). Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science 353, aaf7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer AJ, Segall-Shapiro TH, Glassey E, Zhang J, and Voigt CA (2019). Escherichia coli “Marionette” strains with 12 highly optimized small-molecule sensors. Nature Chemical Biology 15, 196–204. [DOI] [PubMed] [Google Scholar]
- Mimee M, Nadeau P, Hayward A, Carim S, Flanagan S, Jerger L, Collins J, McDonnell S, Swartwout R, Citorik RJ, et al. (2018). An ingestible bacterial-electronic system to monitor gastrointestinal health. Science 360, 915–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K, Arazoe T, Yachie N, Banno S, Kakimoto M, Tabata M, Mochizuki M, Miyabe A, Araki M, Hara KY, et al. (2016). Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353. [DOI] [PubMed] [Google Scholar]
- Perli SD, Cui CH, and Lu TK (2016). Continuous genetic recording with self-targeting CRISPR-Cas in human cells. Science 353. [DOI] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, and Lim WA (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roquet N, Soleimany AP, Ferris AC, Aaronson S, and Lu TK (2016). Synthetic recombinase-based state machines in living cells. Science 353, aad8559. [DOI] [PubMed] [Google Scholar]
- Sheth RU, Yim SS, Wu FL, and Wang HH (2017). Multiplex recording of cellular events over time on CRISPR biological tape. Science 358, 1457–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, and Liu DR (2018). Rewritable multi-event analog recording in bacterial and mammalian cells. Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois C, Hong EJ, Pease S, Brown EJ, and Baltimore D (2002). Germline Transmission and Tissue-Specific Expression of Transgenes Delivered by Lentiviral Vectors. Science 295, 868–872. [DOI] [PubMed] [Google Scholar]
- Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, Sabatini DM, Chen ISY, Hahn WC, Sharp PA, et al. (2003). Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA (New York, NY) 9, 493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe ML (2005). SeqDoC: rapid SNP and mutation detection by direct comparison of DNA sequence chromatograms. BMC Bioinformatics 6, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1 | Sequalizer script (written in MATLAB) used to analyses sanger sequencing chromatograms. Related to STAR Methods.
Data Availability Statement
Software
Software from this study has been described under “QUANTIFICATION AND STATISTICAL ANALYSIS” section.
Data Resources
The accession number for the raw sequencing data reported in this paper is PRJNA509198.