Abstract
Developed decades ago, traditional culture media were not intended to resemble the metabolic composition of human blood, and indeed poorly do so. Yet, despite what is now a clear recognition that environmental factors influence metabolism, such media remain standard to in vitro studies across virtually all areas of biological research. The recent development of physiologic media, like other efforts designed to address the modeling capacity of cell culture, holds immense potential to improve understanding and interpretation of diverse biological and pharmacological studies.
Introduction
‘It is to be hoped that an artificial medium will be found as satisfactory as the plasma, for the advantages are obvious if one can work with a known medium in the investigation of the many new problems, which suggest themselves.’ M.R. Lewis and W.H. Lewis (1911) [1]
Although a general network linking the thousands of reactions that comprise metabolic pathways has been mapped since the 1960s, there is a resurgence of interest in the metabolism of mammalian cells. Largely motivated by a growing recognition that this metabolic network is intertwined with basically every aspect of cell physiology, the study of metabolism now seemingly penetrates all areas of biology [2-10].
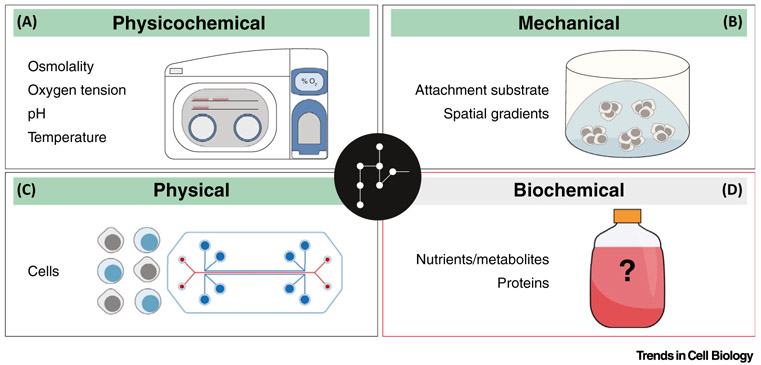
For over a century, the investigation of cells outside of their natural environment has been a fundamental objective of biological research. While fairly commonplace today, mammalian cell culture is a critical tool, whereby tissue-derived cells are placed in an artificial environment of controlled biochemical (nutrients, proteins), mechanical (cell attachment substrate if necessary), physical (cells themselves), and physicochemical (osmolality, oxygen tension, pH, temperature) conditions. Across diverse cell types that include human cancers, stem cells, and components of the immune system, cultured cells are used to examine various metabolic characteristics and points of intersection between metabolism and other processes [11-13]. Typical cell culture certainly offers an experimental system of unmatched scope, flexibility, and accessibility both to investigate cell physiology and to evaluate drug efficacy and toxicity. And implicit to the use of cultured cells for these aims is an expectation that in vitro phenotypes reasonably reflect in vivo cell behavior. As the influence of environmental factors on cell metabolism has become better recognized [14-19], ensuing consideration to themodeling capacity of culture conditions has indeed escalated over the past decade. For example, the use of airtight chambers that permit culture under hypoxic conditions reflective of those in solid tumors or – speculatively – under oxygen tensions reported elsewhere in the body (physicochemical) [20,21], the development of 3D culture systems intended to simulate extracellular matrix composition or spatial nutrient gradients within a tumor (mechanical) [22-25], and the design of co-culture or even organ-on-a-chip systems that aim to mimic possible interactions between different cell types or tissues (physical) [26-28]. However, although extracellular metabolites also influence metabolic networks, there has been relatively little investigation into modeling what are among the most manipulatable conditions (biochemical) of cell culture systems (Figure 1).
Figure 1. Summary of Controlled Cell Culture Conditions and Recent Efforts to Address the Modeling Capacity of in vitro Systems.
Cell culture provides an artificial environment of controlled biochemical, mechanical, physical, and physicochemical conditions that influence metabolism and other cellular processes. Airtight chambers can permit cell culture under more physiologically relevant oxygen tension levels (A), 3D organoids are intended to recapitulate tissue architecture (B), and various co-culture methods and organ-on-a-chip systems may better mimic possible interactions between different cell types or tissues/organs, respectively (C). However, there has been relatively less consideration for improving the modeling capacity of in vitro biochemical conditions, which are still primarily dictated by classic media that were developed generations ago (D).
History
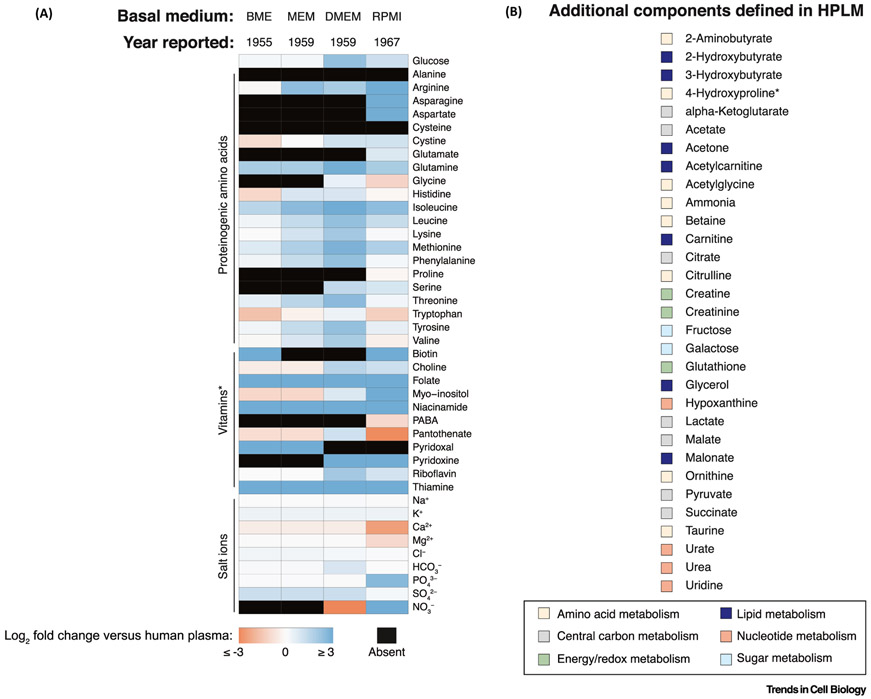
Efforts to develop chemically defined culture media were largely rooted in a desire for greater amounts of medium with less inherent complexity and variability than natural biological fluids and tissue extracts [29]. Through a series of systematic studies, each published in 1955, Harry Eagle first defined the nutrient requirements of two cell lines (HeLa, human carcinoma; L929, mouse fibroblast) [30-32]. While he then noted ‘The initial objective of these studies was the identification of the specific metabolites required for the growth of various cell types rather than the development of a chemically defined medium…’, Eagle nonetheless reported Basal Medium Eagle (BME) [33], which contained the 27 essential nutrients he identified through his initial studies, but at concentrations he determined as optimal for the growth of HeLa cells. Four years later, Eagle then described Minimal Essential Medium (MEM) [34], a BME derivative with increased amino acid availability intended both to promote the growth of several cell lines and to ‘…permit[s] the cultures to be kept for somewhat longer periods without refeeding’. In that same year, Dulbecco and Freeman communicated – via footnote – another BME derivative, now widely recognized as Dulbecco’s Modified Eagle Medium (DMEM) [35], which contained even greater boosts in amino acid (and vitamin) levels, and was used to support the culture of mouse embryonic cells. And less than a decade later, Moore and colleagues reported the composition of RPMI 1640 [36], a derivative of McCoy’s 5A Modified Medium [37]–itself based in part on BME – that was developed to promote the growth of human blood cells. It is remarkable to consider that most cell culture work [38] is still carried out using basal media (MEM, DMEM, RPMI 1640) that can be traced back to Eagle’s initial description of BME over 60 years ago. However, as these media were designed to support the proliferation of specific cell types rather than to model in vivo conditions, it should not be surprising that they poorly reflect the metabolite composition of human blood (Figure 2A). Moreover, as we appreciate that many cells in vivo are not in a proliferative state, primarily optimizing basal media to stimulate rapid cell growth is perhaps an inherently limited design objective.
Figure 2. Overview of Relative Basal Media Compositions.
(A) Heatmap depicting relative concentrations of defined basal media components compared to those in adult human plasma (log2-transformed fold changes). Components not present in a medium are marked as absent. Values used to generate the heatmap are defined elsewhere [42]. *RPMI further contains vitamin B12 (3.69 nM). (B) Additional HPLM components that are absent from the defined formulations of basal media described in panel (A). Designations used to group metabolites are not intended to be exclusive. *RPMI contains 4-Hydroxyproline at a concentration (153 μM) nearly eightfold that defined in HPLM. Abbreviations: BME, Basal Medium Eagle; DMEM, Dulbecco’s Modified Eagle Medium; HPLM, human plasma-like medium; MEM, Minimal Essential Medium; RPMI, Roswell Park Memorial Institute; denotes RPMI 1640.
Serum: The Black Box
As part of his BME description, Eagle noted that it ‘…did not permit growth unless a small amount of serum protein was added […] The function of the serum protein is not yet clear’. [33]. To establish a complete medium that supported cell growth, he therefore added to BME a dialyzed serum component and, later, similarly recommended that a 5–10% (unmodified or dialyzed) serum supplement be added to MEM as well [34]. And indeed, basal media are still usually supplemented with 10–20% serum from either fetal bovine or calf. Though initially less clear to Eagle, we now appreciate that serum contributes various growth factors, hormones, and trace elements needed to enable proliferation across a broad range of cell types. However, serum also adds an undefined and often unaccounted for cocktail of metabolites and lipids. And so, if motivated by a modeling-based objective, why not then eliminate this ‘black box’ component altogether? Although certain serum-free media have entirely defined recipes, they entail meticulous optimization and are often intended to support the growth of specific cell types, thereby all but eliminating the possibility of a broad use solution [39]. In addition, while the availability of basal media that further contain defined levels of certain proteins and trace elements offers an option meant to at least reduce serum requirements, such alternatives still require that serum addition be optimized for different cell types – as presumably often guided by relative cell growth rates. Moreover, as delivery of trace elements to complete media is otherwise mediated by protein carriers contributed from typical serum supplements, the defined addition of these essential nutrients or certain others (e.g. ascorbate) (method details in [40]) warrants careful management and/or proper chelation to avoid the induction of cell toxicity or oxidative stress (https://www.sigmaaldrich.com/life-science/cell-culture/learning-center/media-expert.html). Thus, current strategies intended to eliminate or greatly reduce serum concentrations in complete media can arguably compromise key advantages of cell culture and, in fact, may also elicit unintended biological consequences. In considering the other end of this “black box” spectrum, culturing cells in 100% (human) serum is certainly not a cost-effective option and, perhaps more importantly, would carry with it the inherent variability, complexity, and inflexibility that motivated initial efforts to develop chemically defined media over a century ago.
Toward Physiologic Media
Our current understanding of metabolism and other biological processes is largely derived from cells cultured in complete media, consisting of an undefined serum component and one of several defined basal media that poorly reflect the metabolic composition of human blood. Over the last decade, there have been described efforts to adjust the concentrations of defined media nutrients, typically one at a time in isolation, to better resemble physiologic levels. However, these modified media not only contained remaining nutrients at non-physiologic concentrations, but also lacked many metabolites now known to be present in human plasma. Though it is worth mention that one such example, serum-like modified Eagle’s Medium [41], was instead designed to adjust in parallel the defined levels of several nutrients and thus offered a greater relative step away from the traditional objectives of media development.
As first described just over 2.5 years ago, we ultimately applied a uniquely unbiased and bottom-up approach to systematically develop human plasma-like medium (HPLM) [42], which contains polar metabolites and salts at concentrations that represent average reported values for normal adult human plasma [43,44] (Figure 2B). Given our intent that HPLM be of broad utility to the scientific community, we supplement it with 10% dialyzed serum to add the various components required for cell growth across a broad range of cell types, while minimizing the contribution of polar metabolites at undefined levels. To differentiate from basal HPLM, we denote this complete medium as HPLM+dS, and I propose the use of a similar designation scheme when reporting complete media in general. It is worth noting that metabolite profiling RPMI 1640 supplemented instead with an equivalent concentration of unmodified serum (RPMI+S) revealed that fetal bovine serum also poorly mimics the polar metabolite composition of human plasma – a result that should be considered even when examining cells in complete media that contain reduced serum levels. And ultimately, other reported results established that, relative to both RPMI 1640 reference media, HPLM+dS extensively altered the metabolic landscape of cultured cells. As reported earlier this year, a second basal medium (Plasmax) [45] was independently developed with intent and design scheme analogous to ours, resulting in a formulation largely reflecting that of HPLM. Taken with additional relative metabolic differences described in cultured cells, this work is also certainly indicative of a growing appreciation for the utility of physiologic media going forward.
Limitations and Considerations
Since our initial description of HPLM, we have frequently faced two general questions (see Outstanding Questions). The first: what about lipids? There are a number of available lipophilic species with reported plasma concentrations above the threshold we set when first constructing HPLM. However, whereas dialysis permits a somewhat straightforward diluting out of polar metabolites from serum, charcoal stripping to remove serum lipids can deplete critical growth factors and hormones as well. Method development to address this issue is likely to continue. Further, given the low solubilities of most lipids, careful consideration and optimization is likely needed for potentially adding organic solvent to basal media or for verifying (unbound) media concentrations of free fatty acids that have been delivered via conjugation to a protein carrier. Given the sheer complexity of serum composition, it is also virtually impossible to assess (and thus to control) the entirety of consequences mediated by dialysis and de-lipidation methods. Therefore, serum treatment and concentration should at minimum be normalized for use in comparative cell culture studies.
Outstanding Questions.
Can we increase the biological complexity of physiologic media in a controlled and accessible manner?
Can the elapsed time between isolation and metabolic characterization of cells from the body be reduced to scales comparable to those typical for metabolite extraction from cultured cells?
How best do we extrapolate (or validate) cultured cell phenotypes in cases when biochemical conditions provided by (human) physiologic media cannot be recapitulated in mouse models?
Can physiologic media be further exploited to improve survival (and growth) rates of primary cells that have not been otherwise exposed to non-physiologic conventional media?
Given the heterogeneity of environmental conditions described for solid tumors, how best do we select, characterize, and benchmark those that may be most appropriate to model in potential non-plasma derivatives of physiologic media?
And the second question: how does culturing cells in HPLM compare to in vivo? I should first emphasize that no model can faithfully capture the full complexity of conditions encountered by cells in the human body–this truth effectively defines the term ‘model’. That said, it is reasonable to ask how the metabolic characteristics of cells in human circulation compare to those cultured in HPLM, and we indeed attempted to establish such a comparison. It is well appreciated that metabolite profiles are quite labile, and thus it is critical that extraction methods be performed as quickly as possible to avoid substantial metabolite loss due to residual transporter and enzyme activities. However, the time required to isolate even a crude mixture of mononuclear cells (MNCs) from whole blood (prior to immediate extraction), during which they are removed from the relevant in vivo metabolite environment, is substantially longer (roughly ten-fold) than that needed to perform analogous steps with cultured suspension cells. Therefore, while the plasma metabolite concentrations from our donor sample were expectedly comparable to those of HPLM, the levels of many metabolites were markedly reduced in MNCs extracted immediately following isolation from blood relative to those that were instead extracted following short-term culture in HPLM+dS (unpublished data). This suggests that assessing the relevant phenotypic comparison will require method development to address the current incompatibility between timescales for MNC isolation and the ensuing metabolite extraction desired. However, currently, given that recently described intraoperative methods have been elegantly used to determine the utilization patterns of certain nutrients within different human patient tumors [46,47], it could be interesting to potentially recapitulate such phenotypes using in vitro models that incorporate physiologic media and that, in turn, may be exploited to develop and test mechanistic hypotheses.
In addition, though there are several recognized caveats associated with using cells grown in mice to understand human cell behavior [48-50], mouse models undoubtedly provide many environmental conditions (e.g. mechanical, physical) that are more physiologically relevant than those offered via standard cell culture. This general disparity in environmental context between the two experimental systems often prompts a desire for ‘validation’ of in vitro findings in mouse models. However, while the plasma concentrations of glucose and many amino acids in mice are comparable to those in humans, the relative plasma levels of several other metabolites are quite different [42]. For instance, cell culture in HPLM+dS revealed an example of metabolic regulation mediated by a metabolite (uric acid) that, among basal media, was uniquely defined in HPLM and whose plasma concentrations differ by up to ten-fold between humans and mice. As a consequence of this unforeseen regulation, HPLM+dS could in fact influence the relative cytotoxicity of 5-fluorouracil, a classic chemotherapeutic that remains in wide use. This ensuing result effectively supported the growing awareness that environmental factors can impact drug responses [51] and, further, should perhaps motivate the future incorporation of physiologic media in (essential) upstream drug development efforts. Nonetheless, mice will remain key to most modeling pipelines of basic and translational biology research for the foreseeable future. And so, I think that perhaps the most pressing question then is how do we putatively extrapolate (and potentially corroborate) cell culture-derived phenotypes to mouse models if certain conditions in the former may now better mimic those in humans? There is unlikely a perfect solution to this question. Nevertheless, with the potential for physiologic media to become only more pervasive, we seemingly need to re-evaluate the often reflexive mentality to seek ‘validation’ in mice and instead, carefully consider how we best exploit – and when is most appropriate to integrate – what will continue to be evolving in vitro and in vivo model systems.
Lastly, it is worth noting that routine culture parameters (e.g., seeding density, passaging frequency) are often selected based on approximate observations or general recommendations (e.g., ATCC profiles) rather than on empirically determined growth curves or related metrics under conditions of interest. The composition of media in standard cell culture systems changes over time, as many metabolites are gradually depleted (or elevated) largely during the exponential phase of cell growth. The specific nature and time-dependence of these alterations differ between cell lines, and the availability of certain nutrients is undoubtedly a key factor that dictates the saturating cell density of a particular culture. Taken together, it is impractical to broadly assert how (if at all) the use of physiologic media will affect calibration of basic culture parameters, which instead, should be optimized for cells of interest via experimental (growth) characterization.
Concluding Remarks and Future Perspectives
We must recognize that the intent of physiologic media represents a sharp departure from applications-based objectives that have historically guided culture media development (see Outstanding Questions). The stimulation of cell growth is certainly chief among these and is one that will surely remain for more difficult to culture cell types (e.g. primary cells). Continued interest is also presumably expected for media optimized to meet other specific functions, including to: exploit cultured cells for production of biomolecules (e.g., virus, protein), establish new cell lines, and induce cell state transitions (e.g., differentiation). Beyond supporting basic cell survival and (in most cases) growth, the fundamental purpose of physiologic media is instead to better model in vivo conditions and should be independent of application outside of examining biological phenotypes in such contexts. For example, to revisit our understanding of metabolic regulation, preferences, and dependencies, as well as the influence of metabolite availability on other cellular processes. The use of physiologic media will also undoubtedly affect various omics-based and high-throughput screening data, which have been otherwise largely generated for cells cultured in traditional media. For instance, through the use of paired CRISPR-based screens performed in either HPLM+dS or RPMI+dS, we have begun to identify many genes whose loss-of-function differentially affects cell fitness in a medium-dependent manner (unpublished data). And finally, beyond serving their primary purpose, physiologic media –relative to classic formulations –may indeed offer potential advantages for addressing other applications noted above as well, such as prolonging the survival of cultured primary cells or increasing the success rate of cell line derivation from such samples.
There is also ample opportunity going forward to expand and manipulate the repertoire of physiologic media. Since shortly following our initial description of HPLM, we have actually already added to its reported composition four additional metabolites (acetylcarnitine, alpha-ketoglutarate, malate, and uridine), whose plasma concentrations were near our initial inclusion threshold. And of course, there is potential to similarly incorporate other components based on analogously relaxed design criteria or further quantitative characterization of human plasma. Among these are metabolites that have been described as immunosuppressive (e.g. adenosine, kynurenine) [18] or even as provided by the microbiome [52]. The inclusion of such compounds in defined media, like that of several HPLM components, has otherwise received little attention – primarily due to historically nutrientcentric and growth-promoting design goals. With continued technical advancements, the controlled incorporation of lipophilic species and proteins into physiologic media could become more easily accessible as well. In addition, while we initially designed HPLM to more closely reflect normal adult human plasma, it could be interesting to now create additional HPLM derivatives based on plasma conditions that are reflective of different diets (e.g., keto versus Western, targeted nutrient manipulations) [53-55] or (patho)physiologic states (e.g., human disease, aging). The integration of physiologic media and personalized medicine is a rather exciting future prospect as well, whereby the defined biochemical conditions of in vitro drug response models could be established through metabolite profiling of corresponding donor/patient plasma samples.
Physiologic media development can extend beyond mimicking conditions in human plasma as well. For example, the use of a medium designed to more closely resemble mouse plasma was reported earlier this year, serving as a useful means to compare phenotypes between cultured murine cells and those growing in the animal [56]. Similarly, an even more recently described metabolic characterization of interstitial fluid from distinct murine tumors revealed that such compositions can in part deviate from those of matched plasma [57], suggesting that media formulations guided by non-plasma in vivo conditions could too be possible for the culture of certain cell types. It will also be interesting to track how such techniques are potentially extrapolated and controlled for use with human samples and, further, how best to consider the anticipated heterogeneity of intratumoral environmental contexts.
And finally, while we have readily provided HPLM to many researchers, the eventual commercial distribution of physiologic media – made available without potential label as a ‘specialty’ reagent – should undoubtedly permit a far greater extent of accessibility and ensuing impact for the scientific community.
Without question, the pioneering efforts of Eagle and others ultimately enabled decades of cell culture and, in turn, countless advances in biological research and drug development. However, we should no longer instinctively pass to the next generation of scientists a bottle of basal medium in one hand and serum in the other, independent of experimental objective, but instead owing to precedence. While it is imperative that we continue to recognize, debate, and address key considerations of in vitro (and in vivo) model systems, we can also embrace the arrival and rise of physiologic media, which extend the modeling capacity of cell culture and can be exploited to uncover insights that would be otherwise difficult to identify through the use of existing model systems.
Highlights.
Environmental factors impact metabolic phenotypes and intertwined physiologic processes in mammalian cells.
Complete media that remain the workhorses of cell culture studies typically consist of a basal medium that poorly resembles conditions encountered in the human body and a largely undefined serum supplement.
The systematic and bottom-up design of new physiologic media represents an important advance toward improving the modeling capacity of cell culture methods.
The development and selection of culture media should now be guided by distinct applications or modeling-based objectives.
The immense promise for incorporation of physiologic media into basic and translational research must be balanced by careful considerations critical to the design, use, and continued evolution of experimental model systems.
Acknowledgments
I regret that due to space limitations, I was unable to cite many additional studies related to this topic. I would like to thank D.M. Sabatini, N.S. Chandel, and J. Rutter first for their encouragement and individual efforts to spread awareness for the importance of physiologic media; and also, for critical reading and discussion of the manuscript. I would also like to thank K. Huggler for assistance with illustrations. Described work in the laboratory of D.M. Sabatini was supported in part by fellowships to J.R.C from the American Cancer Society (PF-12-099-01-TBG) and the Koch Institute (Ludwig Postdoctoral Fellowship). J.R.C. is supported by the NIH/NCI (K22 CA225864).
References
- 1.Lewis M and Lewis WH (1911) The cultivation of tissues from chick embryos in solutions of NaCl, CaCl2, KC1 and NaHCO3. Anat. Rec 5, 277–293 [Google Scholar]
- 2.Campbell SL and Wellen KE (2018) Metabolic signaling to the nucleus in cancer. Mol. Cell 71, 398–408 [DOI] [PubMed] [Google Scholar]
- 3.López-Otí C et al. (2016) Metabolic control of longevity. Cell 166, 802–821 [DOI] [PubMed] [Google Scholar]
- 4.O’Neill LA et al. (2016) A guide to immunometabolism for immunologists. Nat. Rev. Immunol 16, nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pearce EL et al. (2013) Fueling immunity: insights into metabolism and lymphocyte function. Science 342, 1242454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rinschen MM et al. (2019) Identification of bioactive metabolites using activity metabolomics. Nat. Rev. Mol. Cell Biol 20, 353–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saxton RA and Sabatini DM (2017) mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valvezan AJ and Manning BD (2019) Molecular logic of mTORC1 signalling as a metabolic rheostat. Nat. Metab 1, 321–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vander Heiden MG and DeBerardinis RJ (2017) Understanding the intersections between metabolism and cancer biology. Cell 168, 657–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei P et al. (2018) The force is strong with this one: metabolism (over)powers stem cell fate. Trends Cell Biol. 28, 551–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buck MD et al. (2017) Metabolic instruction of immunity. Cell 169, 570–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeBerardinis RJ and Chandel NS (2016) Fundamentals of cancer metabolism. Sci. Adv 2, e1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Intlekofer AM and Finley LW (2019) Metabolic signatures of cancer cells and stem cells. Nat. Metab 1, 177–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cantor JR and Sabatini DM (2012) Cancer cell metabolism: one hallmark, many faces. Cancer Discov. 2, 881–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lyssiotis CA and Kimmelman AC (2017) Metabolic interactions in the tumor microenvironment. Trends Cell Biol. 27, 863–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muir A et al. (2018) Microenvironmental regulation of cancer cell metabolism: implications for experimental design and translational studies. Dis. Model. Mech 11, dmm035758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwörer S et al. (2019) Cancer metabolism drives a stromal regenerative response. Cell Metab. 29, 576–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singer K et al. (2018) Immunometabolism in cancer at a glance. Dis. Model. Mech 11, dmm034272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolpaw AJ and Dang CV (2017) Exploiting metabolic vulnerabilities of cancer with precision and accuracy. Trends Cell Biol. 28, 201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ast T and Mootha VK (2019) Oxygen and mammalian cell culture: are we repeating the experiment of Dr. Ox? Nat. Metab Published online August 5, 2019. 10.1038/s42255-019-0105-0. [DOI] [PubMed] [Google Scholar]
- 21.Garcia-Bermudez J et al. (2018) Aspartate is a limiting metabolite for cancer cell proliferation under hypoxia and in tumours. Nat. Cell Biol 20, 775–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carmona-Fontaine C et al. (2017) Metabolic origins of spatial organization in the tumor microenvironment. Proc. Natl. Acad. Sci. U. S. A. 114, 2934–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DelNero P et al. (2018) Cancer metabolism gets physical. Sci. Transl. Med 10, eaaq1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mihaylova MM et al. (2018) Fasting activates fatty acid oxidation to enhance intestinal stem cell function during homeostasis and aging. Cell Stem Cell 22, 769–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shamir ER and Ewald AJ (2014) Three-dimensional organotypic culture: experimental models of mammalian biology and disease. Nat. Rev. Mol. Cell Biol 15, nrm3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hassell BA et al. (2017) Human organ chip models recapitulate orthotopic lung cancer growth, therapeutic responses, and tumor dormancy in vitro. Cell Rep. 21,508–516 [DOI] [PubMed] [Google Scholar]
- 27.Mo X et al. (2019) HTiP: high-throughput immunomodulator phenotypic screening platform to reveal IAP antagonists as anti-cancer immune enhancers. Cell Chem. Biol 26, 331–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ronaldson-Bouchard K and Vunjak-Novakovic G (2018) Organs-on-a-chip: a fast track for engineered human tissues in drug development. Cell Stem Cell 22, 310–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morgan JF et al. (1950) Nutrition of animal cells in tissue culture. I. Initial studies on a synthetic medium. Proc. Soc. Exp. Biol. Med 73, 1–8 [DOI] [PubMed] [Google Scholar]
- 30.Eagle H (1955) The minimum vitamin requirements of the L and HeLa cells in tissue culture, the production of specific vitamin deficiencies, and their cure. J. Exp. Med 102, 595–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eagle H (1955) The specific amino acid requirements of a human carcinoma cell (strain HeLa) in tissue culture. J. Exp. Med 102, 37–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eagle H (1955) The specific amino acid requirements of a mammalian cell (strain L) in tissue culture. J. Biol. Chem 214, 839–852 [PubMed] [Google Scholar]
- 33.Eagle H (1955) Nutrition needs of mammalian cells in tissue culture. Science 122, 501–504 [DOI] [PubMed] [Google Scholar]
- 34.Eagle H (1959) Amino acid metabolism in mammalian cell cultures. Science 130, 432–437 [DOI] [PubMed] [Google Scholar]
- 35.Dulbecco R and Freeman G (1959) Plaque production by the polyoma virus. Virology 8, 396–397 [DOI] [PubMed] [Google Scholar]
- 36.Moore GE et al. (1967) Culture of normal human leukocytes. JAMA 199, 519–524 [PubMed] [Google Scholar]
- 37.McCoy TA et al. (1959) Amino acid requirements of the Novikoff hepatoma in vitro. Proc. Soc. Exp. Biol. Med 100, 115–118 [DOI] [PubMed] [Google Scholar]
- 38.Arora M (2013) Cell culture media: a review. Mater. Methods 3, 175 [Google Scholar]
- 39.Freshney RI (2010) Culture of Animal Cells: A Manual of Basic Technique and Specialized Applications, 6th edn (John Wiley and Sons; ), pp. 115–132 [Google Scholar]
- 40.Schoenfeld JD et al. (2017) O2 •– and H2O2-mediated disruption of Fe metabolism causes the differential susceptibility of NSCLC and GBM cancer cells to pharmacological ascorbate. Cancer Cell 31, 487–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tardito S et al. (2015) Glutamine synthetase activity fuels nucleotide biosynthesis and supports growth of glutamine-restricted glioblastoma. Nat. Cell Biol 17, 1556–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cantor JR et al. (2017) Physiologic medium rewires cellular metabolism and reveals uric acid as an endogenous inhibitor of UMP synthase. Cell 169, 258–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Psychogios N et al. (2011) The human serum metabolome. PLoS One 6, e16957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wishart DS et al. (2013) HMDB 3.0-the human metabolome database in 2013. Nucleic Acids Res. 41, D801–D807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Voorde J et al. (2019) Improving the metabolic fidelity of cancer models with a physiological cell culture medium. Sci. Adv 5, eaau7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Courtney KD et al. (2018) Isotope tracing of human clear cell renal cell carcinomas demonstrates suppressed glucose oxidation in vivo. Cell Metab. 28, 793–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hensley CT et al. (2016) Metabolic heterogeneity in human lung tumors. Cell 164, 681–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elsea SH and Lucas RE (2002) The mousetrap: what we can learn when the mouse model does not mimic the human disease. ILAR J. 43, 66–79 [DOI] [PubMed] [Google Scholar]
- 49.Mestas J and Hughes CC (2004) Of mice and not men: differences between mouse and human immunology. J. Immunol 172, 2731–2738 [DOI] [PubMed] [Google Scholar]
- 50.Willyard C (2018)The mice that grow human tumors. Nature 560, 156–157 [DOI] [PubMed] [Google Scholar]
- 51.Muir A and Vander Heiden MG (2018) The nutrient environment affects therapy. Science 360, 962–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zmora N et al. (2017) The role of the immune system in metabolic health and disease. Cell Metab. 25, 506–521 [DOI] [PubMed] [Google Scholar]
- 53.Gao X et al. (2019) Dietary methionine influences therapy in mouse cancer models and alters human metabolism. Nature 572, 397–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goncalves MD et al. (2018) Phosphatidylinositol 3-kinase, growth disorders, and cancer. New Engl. J. Med. 379, 2052–2062 [DOI] [PubMed] [Google Scholar]
- 55.Kanarek N et al. (2018) Histidine catabolism is a major determinant of methotrexate sensitivity. Nature 559, 632–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rodriguez AE et al. (2019) Serine metabolism supports macrophage IL-1 β production. Cell Metab. 29, 1003–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sullivan MR et al. (2019) Quantification of microenvironmental metabolites in murine cancers reveals determinants of tumor nutrient availability. eLife e44235, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]