Abstract
The fact that pharmacokinetic (PK) properties of drugs influence the interaction with protein targets is a principle known for decades. The same cannot be said for the opposite – namely that targets influence PK properties of drugs. Evidence confirming this possibility is first introduced here, as we show that certain protein families have a clear preference for drugs having specific PK properties. We investigate this by cross-referencing “druggable target” annotations for over 1,000 FDA approved drugs with their PK profile, as defined by the BDDCS criteria, then examining BDDCS preference for several major target protein families and therapeutic categories. Our findings may open a novel way to conduct drug discovery by focusing PK profiles at the very early stage of target selection.
Keywords: BDDCS, druggable genome, drug target, FDA solubility, extent of metabolism
Introduction
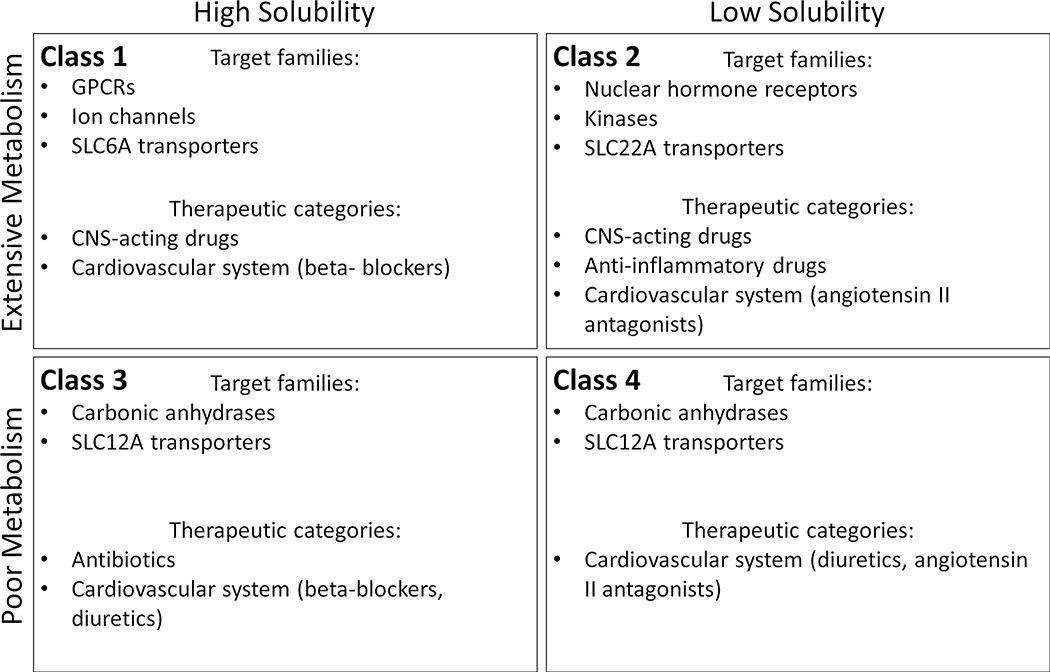
The advance of computational procedures and their integration with improving experimental technologies have resulted in more effective risk mitigation and improved patient safety. However, the process of developing new chemical entities (NCEs) continues to include sequential steps that span several refinement cycles over multiple years. When the lead NCEs fail to meet the anticipated target properties needed for progression to the next development step, the cycle undergoes another iteration or the project is closed. For example, lead identification and optimization remain sequential steps [1], and are often separated from target identification and validation [2]. This lack of integration between cycle steps, particularly at the level of data, information and knowledge transfer, impacts both the costs and time of drug discovery projects. One way to improve integration is to introduce an additional identification-optimization criterion by incorporating BDDCS (Biopharmaceutics Drug Disposition Classification System) criteria in early drug discovery by cross-referencing BDDCS with drug target information. The BDDCS [3] can be used to estimate pharmacokinetics (PK) properties [4] and to forecast drug disposition [5] issues for small molecule NCEs based on aqueous solubility (as defined by FDA criteria) and extent of metabolism [6] (see also Figure 1). Its usefulness, when combined with the “rule of 5” physico-chemical property criteria for drugs [7] was recently discussed [8]. Categorizing drugs into four BDDCS classes is an effective, easy to interpret tool for mapping PK properties and their relationships with other events such as transporters effects, drug-drug interaction (DDI) risk, etc. [9,10]. The BDDCS methodology and its relevance in early drug discovery are summarized in Box 1.
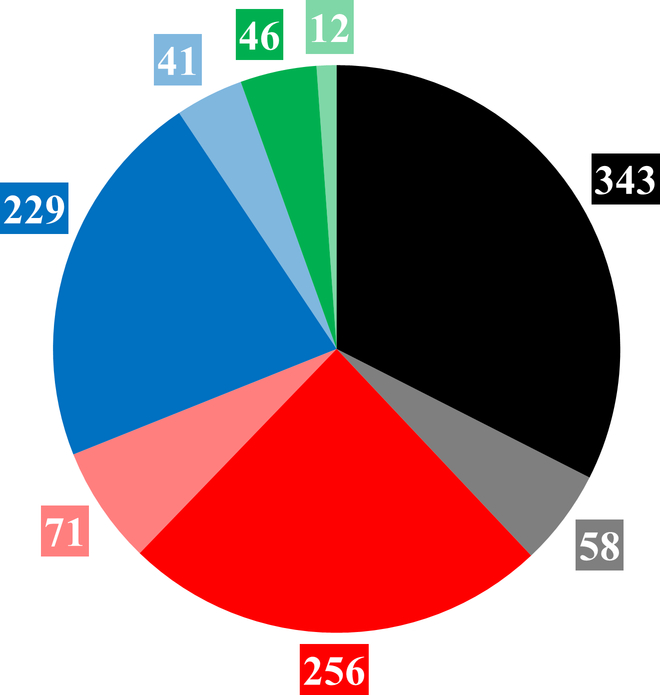
Figure 1.
BDDCS drugs count in the original Benet’s collection (plain color) and in the Hosey’s collection (transparent color). Class 1, 2, 3 and 4 are respectively depicted in black, red, blue and green. The 4 BDDCS classes are as follows: Class 1, high-solubility/high-metabolism; Class 2, low-solubility/high-metabolism; Class 3, high-solubility/low-metabolism; and Class 4, low-solubility/low-metabolism, respectively.
Box 1. The Biopharmaceutics Drug Disposition Classification System (BDDCS).
BDDCS is an adaption of an FDA classification system based on the extent of intestinal permeability and solubility of the highest approved dose strength over a pH range of 1–6.4 to facilitate waiver of in vivo bioequivalence studies for generic drugs and newly developed formulations of drugs previously approved by the regulatory agencies [6]. In contrast, BDDCS was developed to predict drug disposition of new molecular entities (NMEs) in humans, including the major route of elimination, the relative importance of enzymes and transporters in the elimination process and thus the ability to predict relevant drug-drug interactions (DDIs), the drug distribution into the brain and the potential for toxicity such as drug induced liver injury (DILI) [10]. BDDCS classification is based on the rate of membrane permeability, which distinguishes with remarkable accuracy NMEs that will be primarily eliminated by metabolism versus renal and biliary excretion of unchanged drug, and a simple 0.3 mg/ml solubility cut off to distinguish high and low solubility NMEs over the pH range 1–6.8, since early in drug development the highest approved dose strength will not be known [3]. In the BDDCS system, high permeability rate (extensively metabolized), highly soluble Class 1 NMEs can be expected to be extensively absorbed from the gastrointestinal tract, primarily eliminated by metabolism, transporter effects will be clinically insignificant, the NME will readily pass into the brain and DILI potential will be low and well predicted by preclinical animal studies. BDDCS Class 2, high membrane permeability rate, poorly soluble NMEs will also be highly absorbed and primarily eliminated by metabolism, but transporter effects can be rate limiting and cannot be ignored. DILI is primarily seen with BDDCS Class 2 NMEs and preclinical animal and in vitro studies may not be predictive. These NMEs, if substrates for efflux transporters, will not exhibit extensive central effects. BDDCS Class 3 and 4 NMEs will exhibit poor membrane permeability rate and will require transporters to achieve druggable pharmacokinetic characteristics. They will be primarily eliminated unchanged in the urine and bile and metabolic DDIs will not be a major source of concern.
Here, we report cross-referencing BDDCS categories for over 1,000 approved drugs to their mode-of-action (MoA) protein (“MoA_protein”) targets [11], as well as other targets that may be responsible for off-target effects. DrugCentral [12], the online drug compendium, was used to extract data for major protein families, particularly those that have been extensively explored and are considered “druggable” [13]. BDDCS classes, attributed to each drug, were assigned to the corresponding annotated MoA and off-target proteins, and thus for the associated protein families. This mapping reveals that certain protein families have higher preference for a specific BDDCS class. This information might be used to better understand the influence that drug targets may exert on defining the PK properties of NCEs. In the case of protein families that exhibit a clear BDDCS-class bias, these observations can reduce the sampling space to only chemicals that match specific PK properties, potentially streamlining the lead identification and optimization process. While the influence of PK properties on drug-target interactions, i.e., pharmacodynamics, has been amply studied, here we show that some protein drug target categories have themselves specific preference for drugs with certain PK profiles. Thus, pharmacodynamics influences pharmacokinetics as well. We further discuss BDDCS and target druggability integration in more detail for specific protein families in the context of ATC (Anatomic, Therapeutic and Chemical) classification codes and subcellular compartment location of proteins. To further verify differences in BDDCS class – MoA target category preference, we applied the Pearson’s chi-squared test to each data sample discussed in this work (data summarized in Figures 2 to 6G). For most of the examples, the p-values are below 0.05, suggesting that target preferences for a given BDDCS category are not aleatory (see Supplementary Table 1).
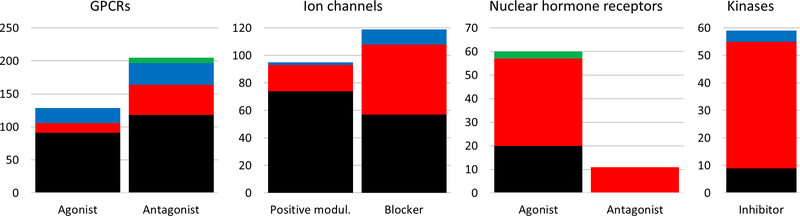
Figure 2.
BDDCS classes distributions for the major target families. On each plot, the vertical axis report the number of drug-target annotations and the horizontal axis displays the MoA. Class 1, 2, 3 and 4 are depicted in black, red, blue and green respectively.
BDDCS overview
For this study we combined a collection of over 900 drugs [3], with an additional 175 drugs [4]. BDDCS Class 0 drugs were not included in this study. The number of drugs belonging to the four BDDCS classes are summarized in Figure 1. During the 5-year gap that separates the two collections, there has been a 28% increase in BDDCS class 2 drugs, compared to 17% and 18% increase for class 1 and 3, respectively. Surprisingly, the overall under-represented class 4 drugs also increased by 26%, when compared to the 2011 collection. Additional temporal aspects (e.g., separating the collection into pre-1998 and post-1997 by approval date) are discussed below.
Druggable targets according to BDDCS
Analysis of major protein families
We examine the distribution of protein drug targets for those drugs that already have a BDDCS class annotation (“BDDCS_drug”), by focusing on six major target families: G-protein coupled receptors (GPCRs), ion channels, kinases, nuclear hormone receptors, enzymes and transporters, respectively. Note that enzymes and transporters relate here to the mechanism of action, not to the mechanism of drug disposition. We only considered those drug-protein pairs for which both MoA_protein and BDDCS_drug annotations were already present, i.e., known BDDCS_drug - MoA_protein pairs. MoA proteins are grouped by the most frequently annotated mechanism of action (e.g. agonist, antagonist, etc.) of the associated drugs, and displayed with one of four colors, depending on the BDDCS class of the corresponding drugs. The result of this analysis for GPCRs, ion channels, nuclear hormone receptors and kinases is summarized in Figure 2. The numbers on the vertical axis are drug-target annotations counts, not drug counts or protein counts. Supporting data are provided in the Supplementary Information file (Supplementary Table 2).
Uneven class distributions, which emerge when comparing GPCRs and ion channels against kinases and nuclear receptors, allow us to make some observations regarding drug lipophilicity and protein subcellular location. For GPCRs and ion channels, BDDCS class 1 drugs (high solubility, high metabolism) dominate, whereas nuclear hormone receptors and kinases seem to be targeted mostly by BDDCS class 2 (low solubility, high metabolism) drugs (see Box 1 for additional details on BDDCS). These data suggest that certain “druggable” target families have clear preference for a specific BDDCS class – and according to Pearson’s chi-squared test, these preferences are statistically significant (p = 2.2*10−16, Chi-squared = 147.62; see also Supplementary Table 1).
In our opinion, these differences can primarily be explained by the subcellular location of these proteins: GPCRs and ion channels are located primarily on the cell membrane, and appear to be better targeted by highly soluble, BDDCS Class 1 drugs. Kinases and nuclear receptors, on the other hand, have an intracellular location and are targeted by the less soluble, more lipophilic Class 2 drugs, i.e., drugs that are required to cross the cell membrane in order to exert their therapeutic action. This observation does not imply that low solubility is the primum mobile of drug-target interaction where kinases and nuclear receptors are concerned. Indeed, additional aspects need to be considered with respect to the specific hydrophilicity/hydrophobicity profile of the individual target’s binding site. Nevertheless, in accordance to these findings, it seems prudent to incorporate FDA solubility as one of the design factors when developing NCEs for these target families.
Analysis of protein sub-families
With respect to enzymes and transporters, BDDCS classes are more evenly distributed (data not shown), which is not surprising: Transporter active site(s) can be reached by both hydrophilic (from extracellular space or cytoplasm) and lipophilic (through the lipid bilayer) drugs [14]. Concerning enzymes, their mechanistic, substrate and cellular location diversity (compared to kinases) is far greater, and therefore BDDCS categories show less specificity. Since we did not find a clear BDDCS class preference for enzymes or transporters, we interrogated more specific protein sub-families, as summarized in Figure 3. Instances where the count of BDDCS_drug - MoA_Protein was under 5 were excluded for clarity. Supporting data are provided in the Supplementary Information file (Supplementary Tables 3 and 4).
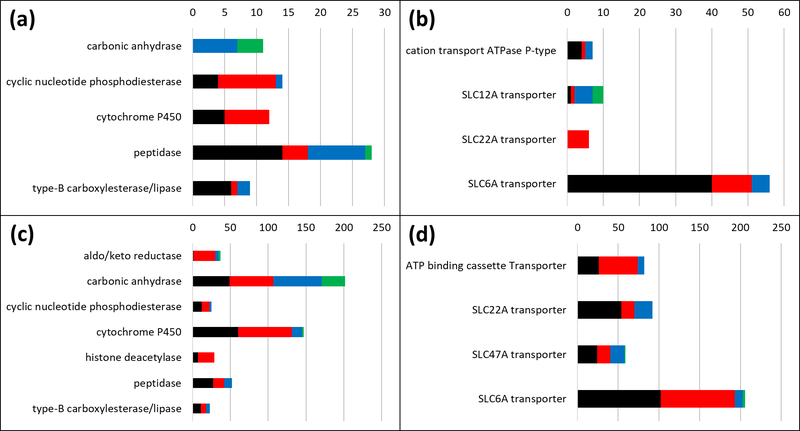
Figure 3.
BDDCS classes distributions for enzymes and transporters sub-families. (A) Drug-target records with annotated MoA for enzymes. (B) Drug-target records with annotated MoA for transporters. (C) Drug-target records without annotated MoA for enzymes. (D) Drug-target records without annotated MoA for transporters. On each plot, the horizontal axis reports the number of drugs and the vertical axis displays the sub-family name. Class 1, 2, 3 and 4 are depicted in black, red, blue and green respectively.
Only poorly metabolized (designated here as low metabolism) drugs are annotated as carbonic anhydrase (CAs) inhibitors (seven BDDCS class 3 and four BDDCS class 4 drugs – see also Figure 3A and Supplementary Tables 1 and 3). In terms of cellular compartment location, seven CAs are cytosolic and four are membrane-associated. With respect to our observations, class 3 and class 4 drugs target both cytosolic (CA1, CA2 and CA7) and membrane associated (CA4 and CA12) proteins. Low metabolism drugs (BDDCS class 3 and 4) are expected to have a low passive permeability (see Box 1). Hence, they are likely to access membrane-bound proteins, but not cytosolic proteins, by passive diffusion only. This may explain why membrane bound CAs are targeted by low metabolism drugs. How do Class 3 and 4 drugs reach cytosolic CAs? We suspect that, to overcome their low passive permeability, uptake transporters may be involved. However, the only evidence to support this hypothesis is a study where 2 CAs inhibitors, methazolamide and acetazolamide, Class 3 and Class 4 drugs respectively, are reported to have an affinity with two organic anion transporter proteins (OAT1 and OAT3) [15].
Most drugs targeting SLC6A (solute carrier 6A) transporters are BDDCS class 1 (high metabolism, high solubility; see Figure 3B, and Supplementary Tables 1 and 3). SLC6A transporter family proteins, which are, e.g., involved in transporting GABA and monoamine (dopamine, serotonin and norepinephrine) neurotransmitters, are expressed on both pre- and post-synaptic membranes in the central nervous system (CNS) and act as neurotransmitter re-uptake pumps. These are well-characterized drug targets for several neurological and psychiatric disorders, such as Parkinson’s, epilepsy and depression [16]. Preference for BDDCS class 1 drugs can perhaps be explained by the requirement that SLC6A drugs should be active at the synaptic interface, which is similar to the extracellular matrix, and perhaps by the lack of clinical effect of efflux transporters on these drugs. Efflux transporters represent a major xenobiotic elimination route for chemicals that pass the blood-brain barrier, but do not influence Class 1 drugs, most likely due to transporter saturation effects caused by the high degree of passive permeability and solubility of Class 1 drugs. According to our interpretation, drugs belonging to all other BDDCS categories, which do not exhibit the same permeability as class 1 drugs, could more effectively be removed from the brain and would therefore not be able to achieve a meaningful blockade on SLC6A transporters. The fact that BDDCS Class 1 drugs are optimal for reaching CNS targets has been noted before [17].
To compare BDDCS class relevance when examining clinical and in vitro data, we explored situations where a clear mechanism of action was not reported [11] for both enzymes (Figure 3C, and Supplementary Tables 1 and 4) and transporters (Figure 3D, and Supplementary Table 4). For carbonic anhydrases, “non-MoA” records show a relatively even distribution of BDDCS classes. The fact that only BDDCS classes 3 and 4 have a known MoA (Figure 3A) might be illustrative of the situation where measured in vitro activity may not be relevant in vivo. We can reach similar conclusions for SLC6A transporters, as the dominance of Class 1 drugs, which is relevant for MoA, shifts towards an even Class 1 / Class 2 distribution where in vitro data are concerned. Therefore, it seems prudent to conclude that situations where a specific BDDCS/MoA category is dominant can inform the likelihood of clinical success for NCE development.
In Figure 3B, we also show that SLC12A transporter inhibitors are preferentially targeted by BDDCS classes 3 and class 4, i.e., low metabolism drugs, similar to CAs. SLC12A transporters are expressed on kidneys cell membranes and are relevant in renal diseases [18]. According to definition, “low extent of metabolism” often implies that a high fraction of the drug is excreted unchanged in the urine; i.e., renal clearance plays a major role in their elimination. In this case, the drugs reach the organ where their intended drug targets are specifically expressed. In another example, SLC22A transporter inhibitors show preference for BDDCS Class 2 drugs. SLC22A are organic anion transporters expressed mainly in kidneys [19]. SLC22A12 (URAT1), which is expressed on the apical membrane of the proximal tubular cells, mediates the reabsorption of urate from primary urine into plasma. However, an abnormally high plasma level of urate, or hyperuricemia, causes gout. The Class 2 drug lesinurad is approved to treat gout by specifically blocking the URAT1 transporter. The SLC22A transporters preference for Class 2 low solubility drugs (Figure 3B) over Class 1 and 3 high solubility drugs (Figure 3D) may be explained by these targets location: High metabolism, low solubility drugs (Class 2) may have an optimal PK profile, in contrast to Class 1 drugs, which undergo hepatic clearance or Class 3 drugs, which require basolateral uptake to reach URAT1 or may involve drug-drug interactions.
Temporal Analysis
Using the date-of-first launch for each drug, we examined the influence of temporal trends on the distribution of BDDCS_drug - MoA_protein pairs for GPCRs, ion channels, kinases and nuclear hormone receptors (Supplementary Table 5 and Supplementary Figures 1 and 2, in Supplementary Information file). We separated the data into “drugs approved before 1998”, or “drugs approved after 1997”, respectively based on the year (1997) when the “rule of 5” landmark paper [7] was first published. All kinase inhibitors (except one) were approved after 1997. Most (53) BDDCS_drug – nuclear hormone receptor pairs are for drugs approved before 1998, compared to 23 after 1997. Therefore, we discuss GPCRs (234 pre-1998 and 90 post-1997) and ion channels (170 pre-1998 and 46 post-1997), respectively. While BDDCS Class 1 drugs remain the dominant category overall for both GPCRs and Ion Channels, there is a slight increase in BDDCS Class 2 drugs for GPCR antagonists launched after 1997 (Supplementary Figure 1), and a net increase in BDDCS Class 3 drugs for both GPCR agonists and ion channel positive modulators and blockers launched after 1997 (Supplementary Figures 1 and 2), respectively. BDDCS Class 1 drugs are less dominant in post-1997 drugs, both for GPCRs and Ion Channels. However, “high solubility” drugs (Class 1 and 3) remain dominant in the second time period (60 out of 90 BDDCS-GPCR drug pairs; and 33 out of 46 BDDCS-ion channel drug pairs, respectively).
Therapeutic categories according to BDDCS
To further explore the relationship between drug categories and BDDCS classes, we cross-referenced BDDCS-annotated drugs with ATC codes extracted from DrugCentral. The Anatomical Therapeutic Chemical (ATC) Classification System classifies each drug based on five different levels [20]. Briefly, the first ATC level is the anatomical component (organ or system of the human body) in which the drug acts; the second level indicates the therapeutic purpose; the third level describes pharmacological action; the fourth level is the chemical class of the drug in question; and the fifth level is the actual substance, respectively. A drug can have multiple ATC codes when it has multiple therapeutic uses. This can either refer to a different posology or to a different administration route. Examining the distribution of BDDCS classes from a pharmacological point of view, as summarized by ATC codes, can provide additional information regarding the BDDCS_drug - MoA_Protein relationships, particularly if specific pharmacological sub-categories appear to have a dominant BDDCS class. Such information could thus be used to guide the drug discovery process for cases involving that specific pharmacology, by focusing on a specific BDDCS class, or at least on a specific PK category, in the early stages of the process.
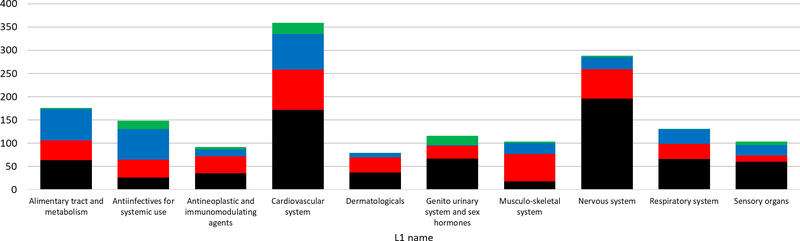
Drug counts, grouped by BDDCS, for each ATC Level 1 group are shown in Figure 4. We excluded groups having less than 50 drugs for simplicity. Figure 4 suggests that certain BDDCS classes have preference for specific anatomic components or organs; some of these are expected (e.g., Class 1 for nervous system (CNS) drugs), whereas others are not (e.g., Class 2 drugs for the musculo-skeletal system). This begs the question, is this preference conserved for lower ATC levels or not? To address this issue, we focused on those cases showing an uneven distribution among BDDCS classes.
Figure 4.
BDDCS drugs distributions on each ATC level 1 code. Class 1, 2, 3 and 4 are respectively depicted in black, red, blue and green.
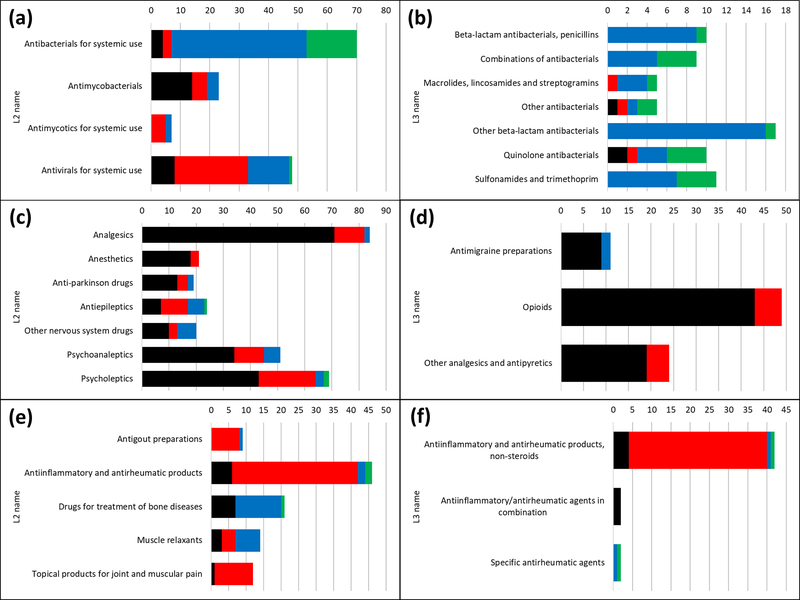
The majority of “antiinfectives for systemic use” drugs are low metabolism (Classes 3 and 4). By expanding ATC Level 1 into Level 2 (Figure 5A), we note that classes 3 and 4 pertain mostly to antibacterials (antibiotics), whereas most antivirals are BDDCS Class 2, and some are Class 3. As we expand into ATC Level 3 (Figure 5B), both class 3 and class 4 continue to be present in each subcategory. When developing new antibiotics, it seems prudent to give preference to molecules exhibiting BDDCS class 3 properties, since there are significantly fewer class 4 drugs, which suggests the approval process would be more problematic. Their low extent of metabolism could perhaps be explained by the fact many such antibacterials inhibit the formation of cross-links between bacterial cell wall peptides by blocking DD-transpeptidase, an enzyme that is accessible from the periplasm [21]. Regarding antivirals, BDDCS class assignment may be influenced by the life cycle stage of the targeted virus. Before viral entry, cell membrane proteins would be targeted (BDDCS Class 3), whereas cytoplasmic proteins or nucleic acids would be targeted at later stages (BDDCS Class 1 and 2).
Figure 5.
BDDCS distribution plots for antiinfectives for systemic use drugs: (A) ATC level 2 and (B) ATC level 3; nervous system drugs: (C) ATC level 2 and (D) ATC level 3; musculo-skeletal system drugs: (E) ATC level 2 and (F) ATC level 3. On each plot, the number of drugs is reported. Class 1, 2, 3 and 4 are respectively depicted in black, red, blue and green.
CNS-acting drugs are typically high metabolism (Class 1 or 2), and often high solubility as well (Class 1 only). As already stated, BDDCS Class 1 drugs have high brain permeability, which makes them ideal for CNS targets. Class 1 drugs preference is conserved over pharmacological subcategories as well (Figure 5C and 5D). Musculo-skeletal system drugs are equally interesting, since anti-inflammatory drugs are prevalently Class 2 (see Figure 5E and 5F). These drugs mainly target cyclooxygenases 1 (PTGS1) and 2 (PTGS2), which have intracellular location: on the Golgi apparatus, on the endoplasmic reticulum and in mitochondria, respectively.
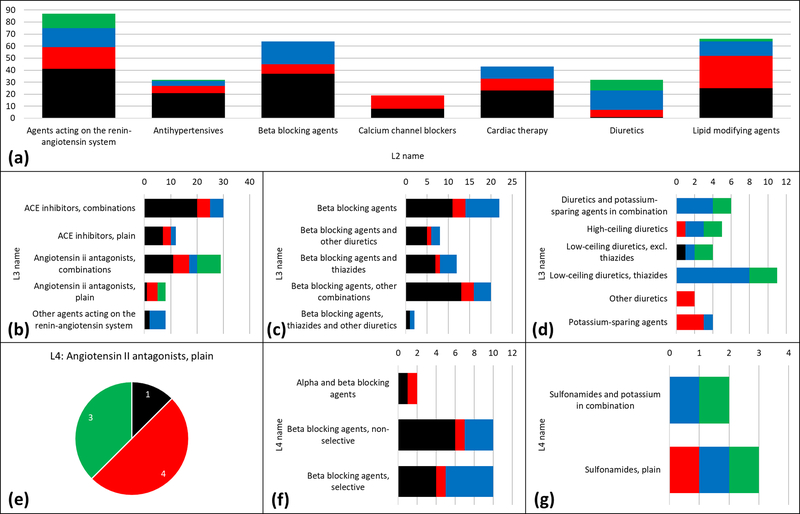
Contrary to the previous examples, drugs acting on the cardiovascular system have a more even BDDCS distribution. At the ATC Level 2, there are cases where high solubility (Class 1 and 3) prevails, e.g., for beta blocking agents, and where low metabolism prevails, e.g., for diuretics (Class 3 and 4). There is no clear separation for agents acting on the renin-angiotensin system (see Figure 6A). For beta-blockers and diuretics, ATC Level 3 (Figure 6C and 6D) and Level 4 (Figure 6F and 6G) do not provide additional information, as the prevailing classes have similar distribution. However, ATC Level 3 provides additional information for renin-angiotensin system inhibitors (Figure 6B). Yet, there are some categories for which even Level 4 shows the nearly equal BDDCS class distribution. Angiotensin receptor AT1 blockers are such an example (Figure 6E): Class 2 drugs are irbesartan, losartan potassium, tasosartan and telmisartan, whereas Class 4 drugs are candesartan cilexetil, eprosartan and valsartan; olmesartan medoximil is Class 1. This finding is in agreement with the different PK profiles for these drugs [22].
Figure 6.
(A) BDDCS distribution for cardiovascular system drugs by ATC level 2. (B) Level 3 distributions for agents acting on the renin-angiotensin system. (C) Level 3 distributions for beta blocking agents. (D) Level 3 distributions for diuretics (E) Pie chart of the unique ATC level 4 code of angiotensin II antagonists, plain. (F) Level 4 distributions for beta blocking agents (same name of level 3). (G) Level 3 distributions high-ceiling diuretics. On each plot, the number of drugs is reported. Class 1, 2, 3 and 4 are respectively depicted in black, red, blue and green.
Concluding remarks and future directions
In this article, we examined the relationship between BDDCS categories for over 1000 drugs and their annotated Mode-of-Action protein targets. The BDDCS distribution can show clear specificity for certain “druggable” target families or sub-families, as well as for specific pharmacological (ATC) drug categories. These results can often be rationalized in terms of protein tissue or compartment location. While this work is primarily descriptive, i.e., lacking a complete exploration across all proteins and pharmacological categories, there appears to be clear evidence that certain protein families are preferentially targeted by one, sometimes two BDDCS classes. The evidence is supported by Pearson’s chi-squared test (Supplementary Table 1). The preference for Class 1 drugs of GPCRs and ion channels, as opposed to the preference for Class 2 drugs of kinases and nuclear receptors, may be indicative of the way these target families “select” drugs with different solubility and hydrophobicity. The requirement for high solubility, detected by the temporal analysis, is clearly shown for GPCR- and ion channel-acting drugs. Furthermore, carbonic anhydrases and SLC12A transporters are preferentially targeted by low metabolism drugs (Class 3 and 4), whereas Class 1 are better suited to target SLC6A transporters and Class 2 drugs are better SLC22A transporters, respectively. The ATC drug classification system can also be used to show that, for certain therapeutic categories, a BDDCS Class is dominant.
We believe these concepts can lead to the development of an integrated “discovery” and “optimization” system that supports parallel property optimization that may significantly influence the drug design process. Indeed, these findings may help medicinal chemists by giving preference on lead series, or scaffolds, that meet specific PK profiles at the step of hit identification, and certainly lead optimization. In order to make available to the scientific community this type of analysis, we plan to create a web-app that medicinal chemists could use to study the BDDCS distribution for protein families or therapeutic drug categories of interest. The analysis described here could be extended in several ways. Future efforts could be aimed on exploring in more detail the location of protein targets and how BDDCS classes are distributed. Furthermore, the influence of the nature of the protein (drug target) binding sites on the BDDCS – target preferences described in this work could be examined by pharmacophore analyses of the known (e.g., X-ray crystallography) binding pockets for drugs having BDDCS/MoA target annotations.
Supplementary Material
Figure 7.
Biopharmaceutics Drug Disposition Classification System (BDDCS) class preference for certain protein families and therapeutic categories. Abbreviations: CNS, central nervous system; GPCR, G-protein-coupled receptor; SCL, solute carrier.
Acknowledgments
This work was supported by National Institutes of Health (NIH) Common Fund grants U24CA224370 (for IDG KMC), U24TR002278 (for IDG RDOC), and U01CA239108 and by National Cancer Institute (NCI) grant P30CA118100.
ABBREVIATIONS USED
- NCE
New Chemical Entity
- BDDCS
Biopharmaceutics Drug Disposition Classification System
- MoA
Mechanism of Action
- PK
pharmacokinetics
Biography
 Leslie Z. Benet, Professor and
former Chair (1978–1998) of Bioengineering and Therapeutic Sciences, UCSF,
received his AB, BS and MS from the University of Michigan, Ph.D. from UCSF. He is
the recipient of nine honorary doctorates and a member of the National Academy of
Medicine. His more recent honors include the 2015 ISSX North American Scientific
Achievement Award and the 2016 Remington Honor Medal of the American Pharmacists
Association. Dr. Benet has published over 590 scientific articles and book chapters,
holds 12 patents and served as editor of 7 books. He is listed by Clarivate
Analytics among the most highly cited pharmacologists worldwide with his
peer-reviewed publications cited more than 28,000 times. He first co-proposed the
BDDCS in 2005.
Leslie Z. Benet, Professor and
former Chair (1978–1998) of Bioengineering and Therapeutic Sciences, UCSF,
received his AB, BS and MS from the University of Michigan, Ph.D. from UCSF. He is
the recipient of nine honorary doctorates and a member of the National Academy of
Medicine. His more recent honors include the 2015 ISSX North American Scientific
Achievement Award and the 2016 Remington Honor Medal of the American Pharmacists
Association. Dr. Benet has published over 590 scientific articles and book chapters,
holds 12 patents and served as editor of 7 books. He is listed by Clarivate
Analytics among the most highly cited pharmacologists worldwide with his
peer-reviewed publications cited more than 28,000 times. He first co-proposed the
BDDCS in 2005.
 Dr. Bocci is a computational
chemist with 4+ years of experience as researcher in academia and 1-year of
experience in the pharmaceutical industry. He graduated from the Department of
Chemistry, University of Perugia, and completed his PhD in Cheminformatics under the
supervision of Prof. Gabriele Cruciani. Later he worked in the pharmacokinetic
division of the pharmaceutical company Laboratoires Servier. He conducts high-level
and multidisciplinary scientific research in drug discovery, cheminformatics,
bioinformatics, data analysis and pharmaceutical sciences. He has proven expertise
in the discovery of new active molecules, in ADME optimization and in transport
proteins modeling. Currently, he is a Post-doctoral Fellow at the University of New
Mexico (USA) in the group of Prof. Tudor I. Oprea.
Dr. Bocci is a computational
chemist with 4+ years of experience as researcher in academia and 1-year of
experience in the pharmaceutical industry. He graduated from the Department of
Chemistry, University of Perugia, and completed his PhD in Cheminformatics under the
supervision of Prof. Gabriele Cruciani. Later he worked in the pharmacokinetic
division of the pharmaceutical company Laboratoires Servier. He conducts high-level
and multidisciplinary scientific research in drug discovery, cheminformatics,
bioinformatics, data analysis and pharmaceutical sciences. He has proven expertise
in the discovery of new active molecules, in ADME optimization and in transport
proteins modeling. Currently, he is a Post-doctoral Fellow at the University of New
Mexico (USA) in the group of Prof. Tudor I. Oprea.
 Tudor Oprea, MD PhD is Professor
of Medicine and Pharmaceutical Sciences, and Chief, Translational Informatics
Division, at the Department of Internal Medicine, University of New Mexico School of
Medicine in Albuquerque, New Mexico (USA). He is also Guest Professor at the
Institute of Medicine, Gothenburg University in Sweden and at the Novo Nordisk
Foundation Center for Protein Research, University of Copenhagen, Denmark. To date,
Dr. Oprea has co-authored over 200 publications and book chapters, and 8 US patents.
Since 2014, Dr. Oprea is PI for the Illuminating the Druggable Genome Knowledge
Management Center, a NIH Common Fund initiative. Dr. Oprea’s work led to drug
repurposing clinical trials at UNM in two types of cancer, most notably R-ketorolac,
which shows promise in an open label clinical trial for ovarian cancer. His most
current research is in the development of validated machine learning and artificial
intelligence models for target and drug discovery, by combining numerical and
free-text information to model human health.
Tudor Oprea, MD PhD is Professor
of Medicine and Pharmaceutical Sciences, and Chief, Translational Informatics
Division, at the Department of Internal Medicine, University of New Mexico School of
Medicine in Albuquerque, New Mexico (USA). He is also Guest Professor at the
Institute of Medicine, Gothenburg University in Sweden and at the Novo Nordisk
Foundation Center for Protein Research, University of Copenhagen, Denmark. To date,
Dr. Oprea has co-authored over 200 publications and book chapters, and 8 US patents.
Since 2014, Dr. Oprea is PI for the Illuminating the Druggable Genome Knowledge
Management Center, a NIH Common Fund initiative. Dr. Oprea’s work led to drug
repurposing clinical trials at UNM in two types of cancer, most notably R-ketorolac,
which shows promise in an open label clinical trial for ovarian cancer. His most
current research is in the development of validated machine learning and artificial
intelligence models for target and drug discovery, by combining numerical and
free-text information to model human health.
References
- 1.Hughes JP et al. (2011) Principles of early drug discovery. British journal of pharmacology 162 (6), 1239–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schenone M et al. (2013) Target identification and mechanism of action in chemical biology and drug discovery. Nature chemical biology 9 (4), 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benet LZ et al. (2011) BDDCS applied to over 900 drugs. The AAPS journal 13 (4), 519–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hosey CM et al. (2016) BDDCS predictions, self-correcting aspects of BDDCS assignments, BDDCS assignment corrections, and classification for more than 175 additional drugs. The AAPS journal 18 (1), 251–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broccatelli F et al. (2012) BDDCS class prediction for new molecular entities. Molecular pharmaceutics 9 (3), 570–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen M-L et al. (2011) The BCS, BDDCS, and regulatory guidances. Pharmaceutical research 28 (7), 1774–1778 [DOI] [PubMed] [Google Scholar]
- 7.Lipinski CA (2004) Lead-and drug-like compounds: the rule-of-five revolution. Drug Discovery Today: Technologies 1 (4), 337–341 [DOI] [PubMed] [Google Scholar]
- 8.Benet LZ et al. (2016) BDDCS, the rule of 5 and drugability. Advanced drug delivery reviews 101, 89–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shugarts S and Benet LZ (2009) The role of transporters in the pharmacokinetics of orally administered drugs. Pharmaceutical research 26 (9), 2039–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benet LZ (2013) The role of BCS (biopharmaceutics classification system) and BDDCS (biopharmaceutics drug disposition classification system) in drug development. Journal of pharmaceutical sciences 102 (1), 34–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santos R et al. (2017) A comprehensive map of molecular drug targets. Nature reviews Drug discovery 16 (1), 19–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ursu O et al. (2018) DrugCentral 2018: an update. Nucleic acids research 47 (D1), D963–D970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oprea TI et al. (2018) Unexplored therapeutic opportunities in the human genome. Nature Reviews Drug Discovery 17 (5), 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bocci G et al. (2018) New insights in the in vitro characterisation and molecular modelling of the P-glycoprotein inhibitory promiscuity. European Journal of Pharmaceutical Sciences 121, 85–94 [DOI] [PubMed] [Google Scholar]
- 15.Hasannejad H et al. (2004) Interactions of human organic anion transporters with diuretics. Journal of Pharmacology and Experimental Therapeutics 308 (3), 1021–1029 [DOI] [PubMed] [Google Scholar]
- 16.Pramod AB et al. (2013) SLC6 transporters: structure, function, regulation, disease association and therapeutics. Molecular aspects of medicine 34 (2–3), 197–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Broccatelli F et al. (2012) Improving the prediction of the brain disposition for orally administered drugs using BDDCS. Advanced drug delivery reviews 64 (1), 95–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arroyo JP et al. (2013) The SLC12 family of electroneutral cation-coupled chloride cotransporters. Molecular Aspects of Medicine 34 (2), 288–298 [DOI] [PubMed] [Google Scholar]
- 19.Roth M et al. (2012) OATPs, OATs and OCTs: the organic anion and cation transporters of the SLCO and SLC22A gene superfamilies. British journal of pharmacology 165 (5), 1260–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The Anatomical Therapeutic Chemical (ATC) Classification System. https://www.whocc.no/atc/structure_and_principles/. [Google Scholar]
- 21.Tipper DJ and Strominger JL (1965) Mechanism of action of penicillins: a proposal based on their structural similarity to acyl-D-alanyl-D-alanine. Proceedings of the National Academy of Sciences of the United States of America 54 (4), 1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barreras A and Gurk-Turner C (2003) Angiotensin II receptor blockers In Baylor University Medical Center Proceedings (Vol. 16), pp. 123–126, Taylor & Francis; [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.