Abstract
Intestinal stem cells (ISCs) maintain regenerative capacity of the intestinal epithelium. Their function and activity are regulated by transcriptional changes, yet how such changes are coordinated at the genomic level remains unclear. The Cohesin complex regulates transcription globally by generating topologically-associated DNA domains (TADs) that link promotor regions with distant enhancers. We show here that the Cohesin complex prevents premature differentiation of Drosophila ISCs into enterocytes (ECs). Depletion of the Cohesin subunit Rad21 and the loading factor Nipped-B triggers an ISC to EC differentiation program that is independent of Notch signaling, but can be rescued by over-expression of the ISC-specific escargot (esg) transcription factor. Using damID and transcriptomic analysis, we find that Cohesin regulates Esg binding to promoters of differentiation genes, including a group of Notch target genes involved in ISC differentiation. We propose that Cohesin ensures efficient Esg-dependent gene repression to maintain stemness and intestinal homeostasis.
Research organism: D. melanogaster
Introduction
Intestinal stem cells (ISCs) contribute to epithelial homeostasis in the gastrointestinal tract due to their capacity to proliferate and renew old or damaged tissue (Casali and Batlle, 2009; Biteau et al., 2011; Signer and Morrison, 2013; Barker, 2014). ISCs self-renew and supply cells that differentiate into different lineages needed to perform all essential functions of the intestinal epithelium (Jiang and Edgar, 2011; Li and Jasper, 2016). ISC maintenance and proliferative activity thus influences intestinal homeostasis, as well as the general fitness and healthy life span (or healthspan) of the organism (Li and Jasper, 2016). While many intrinsic and extrinsic signals that govern ISC maintenance and function have been described (Casali and Batlle, 2009; Biteau et al., 2011; Signer and Morrison, 2013; Barker, 2014), little is known about how different elements of these regulatory networks interact to coordinate ISC proliferation and differentiation programs.
Drosophila melanogaster has proven to be a powerful system to dissect conserved signaling pathways, cellular networks, genetic, epigenetic and environmental factors influencing ISC function (Jasper, 2015; Li and Jasper, 2016). The intestinal epithelium in the fly midgut consists of ISCs, enteroblasts (EBs), enterocytes (ECs) and enteroendocrine (EEs) cells (Miguel-Aliaga et al., 2018). ISCs are the main mitotic cells in this tissue and can give rise to all cell types performing essential secretory, absorptive and immune roles (Ayyaz and Jasper, 2013; Li and Jasper, 2016; Liu and Jin, 2017). A number of highly conserved and well-studied signaling cascades control ISC proliferation and differentiation in response to outside stimuli (Li and Jasper, 2016; Miguel-Aliaga et al., 2018). Infection or stress, for example, triggers ISC proliferation through the activation of JAK/STAT, JNK, and EGFR signaling pathways (Biteau et al., 2008; Jiang and Edgar, 2009; Jiang et al., 2009; Buchon et al., 2010; Lin et al., 2010; Liu et al., 2010; Jiang et al., 2011). Notch activity limits ISC proliferation and is required for EB differentiation into ECs (Ohlstein and Spradling, 2007; Kapuria et al., 2012; Guo and Ohlstein, 2015). Intrinsic regulators of ISC maintenance include the master transcription factor Escargot (Esg), which positively regulates expression of stemness genes, but inhibits expression of differentiation factors, including of the transcription factor Nubbin/Pdm1 (Korzelius et al., 2014; Loza-Coll et al., 2014). It remains unclear how these and other extrinsic and intrinsic factors interact with the chromatin status of ISCs, and how chromatin regulation influences ISC activity and differentiation.
Chromatin is organized in a hierarchical fashion into highly dynamic domains (Filion et al., 2010; Sexton and Cavalli, 2015). Architectural proteins shape DNA and together with other epigenetic mechanisms control transcriptional responses in a cell type-dependent manner (Peric-Hupkes et al., 2010; Phillips-Cremins et al., 2013; Adam et al., 2015; Beagan et al., 2016; Huang et al., 2016; Neems et al., 2016; Schmitt et al., 2016). Cohesin, a ring-like structure formed by a complex of proteins (Smc1, Smc3, Rad21 and SA1/SA2 proteins) that interact with DNA, is a key element in genome organization (Peters et al., 2008; Feeney et al., 2010; Singh and Gerton, 2015). During mitosis, Cohesin encircles paired chromosomes and holds them together until late anaphase, when Rad21 is cleaved by the protease Separase to allow chromosome segregation towards centrosomes (Haarhuis et al., 2014; Morales and Losada, 2018). This ensures synchronous resolution and correct inheritance of chromatids and protects cells from aneuploidy (Onn et al., 2008; Haarhuis et al., 2013; Singh and Gerton, 2015). While only a minor amount (up to 13%) of Cohesin has been reported to be sufficient for chromosome cohesion (Heidinger-Pauli et al., 2010) most of Cohesin appears to be involved in chromatin organization in interphase cells via organization of topologically-associated domains (TADs) by means of loop extrusion (Dekker and Mirny, 2016; Rao et al., 2017; Wutz et al., 2017; Yuen and Gerton, 2018). Within TADs, Cohesin interacts with transcription factors and brings promotors into proximity with enhancers (Schaaf et al., 2013; Merkenschlager and Nora, 2016; Novo et al., 2018). Cohesin deficiency results in the elimination of TADs and alters transcription, supporting its crucial function in gene expression (Pauli et al., 2010; Dorsett and Ström, 2012; Zuin et al., 2014; Lupiáñez et al., 2015; Rao et al., 2017). Cohesin deficiency has further been shown to cause prominent changes in genomic organization, leading to transcription being dependent on local transcriptional elements and cryptic promotors rather than on distant enhancer elements (Schwarzer et al., 2017).
In the course of normal differentiation, as well as under stress conditions, stem cells reorganize their chromatin to adapt transcription to differentiation and activation programs, and to achieve new cellular functions (Cantone and Fisher, 2013; Rinaldi and Benitah, 2015; Adam and Fuchs, 2016). Such chromatin remodeling happens in embryonic and neural stem cells during differentiation into other cell types (Pękowska et al., 2018), precipitating dramatic transcriptional changes. Whether and how DNA conformation and higher order chromatin structures also regulate stemness and lineage determination in SCs of barrier epithelia, however, has not been explored. In this study, we investigate how Cohesin-dependent chromatin organization regulates ISCs using the fly intestine as a model. We define a transcriptional program mediated by Cohesin and find that Cohesin is essential for ISC maintenance by regulating the transcriptional output of Esg.
Results
Rad21 regulates ISC proliferation and differentiation
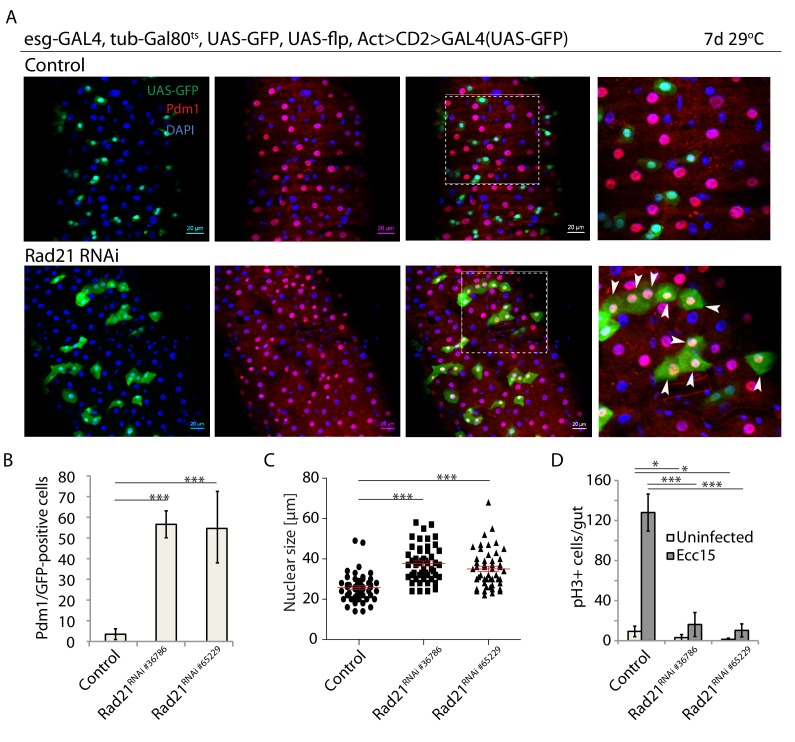
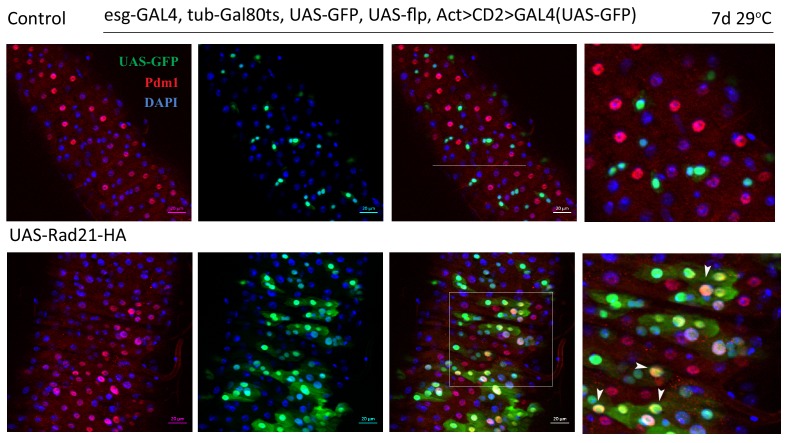
The Rad21 subunit of the Cohesin complex has been shown to act as a tumor suppressor and promoter of pro-inflammatory myeloid polarization in the hematopoietic system, while acting to preserve stemness and prevent differentiation of progenitor cells in the epidermis (Mullenders et al., 2015; Noutsou et al., 2017; Cuartero et al., 2018). These observations indicate that Cohesin may selectively regulate different types of somatic stem cells, and that further characterization of its role in stem cell behavior in vivo is warranted. Using the Drosophila ISC lineage as a model, we asked whether loss of Cohesin would influence stem cells in a barrier epithelium. We performed lineage tracing using the esg-FlpOut system (Jiang et al., 2009), which allows temperature-inducible Flp-mediated excision of a transcriptional STOP sequence in ISCs to achieve inherited expression of Gal4, and as a consequence expression of UAS-linked transgenes, in ISCs and their daughter cells. Using this system, knock down of Rad21 (using two independent shRNA constructs targeting Rad21) resulted in clones mostly containing individual cells with large nuclei and expressing Pdm1, indicating that Rad21 knock down triggered ISC differentiation into enterocytes (ECs) (Lee et al., 2009; Beebe et al., 2010; Mathur et al., 2010; Korzelius et al., 2014) (Figure 1A and B; Figure 1—figure supplement 1). Rad21-deficient cells became larger (Figure 1—figure supplement 2) and their nuclei were significantly bigger than nuclei of WT ISCs, supporting the notion of their terminal differentiation into ECs (Figure 1C). While Cohesin-depleted clones showed a significant increase in Pdm1 positive cells (Figure 1), they were not in fact smaller with respect to the numbers of cells per clone. Their size was about equal compared to wild-type clones, with some minor variability in the exact number of cells seen in different guts (Figure 1—figure supplement 1). Elimination of the Rad21 immunohistochemistry signal in ISCs after Rad21 knock down confirmed efficient protein downregulation (Figure 1—figure supplement 3).
Figure 1. Rad21 knock down leads to premature ISC differentiation.
(A) esg-FlipOut (F/O) midguts expressing UAS-GFP alone (control) or expressing rad21RNAi. Samples were stained for GFP and Pdm1. (B) Quantification of GFP-positive/Pdm1-positive cells from A. (n = 771, 543 and 389) ANOVA. (C) Analysis of nuclei size from A (n = 50). Mann-Whitney Test. (D) Quantification of the number of mitotic pH3-positive cells/midgut in the guts expressing UAS-EYFP alone (control) or expressing rad21RNAi with and without Ecc15 infection. (n = 6–8), ANOVA. *p<0.05, ***p<0.001. Differentiated cells are labeled with white arrowheads. Scale bars, 20 µm.
Figure 1—figure supplement 1. Esg-FlipOut (F/O) midguts expressing UAS-GFP alone (control) or expressing rad21RNAi.
Figure 1—figure supplement 2. Esgts midguts expressing UAS-EYFP alone (control) or expressing rad21RNAi.
Figure 1—figure supplement 3. A Esgts midguts expressing UAS-EYFP alone (control) or expressing rad21RNAi.
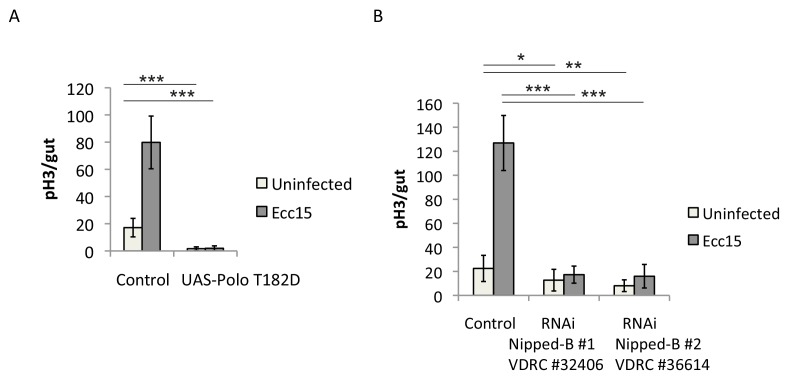
ISC differentiation is normally associated with abrogation of cell proliferation and reduction of mitotic markers. Consistent with ectopic ISC differentiation, loss of Rad21 inhibited ISC proliferation in response to infection by the mild enteropathogen Erwinia carotovora carotovora 15 (Ecc15), as assessed by the quantification of phospho-Histone H3+ cells in the gut (Figure 1D). This premature differentiation phenotype in Rad21 deficient Drosophila ISCs is consistent with loss of epidermal progenitor cells in mice due to aberrant differentiation after Rad21 knock down (Noutsou et al., 2017).
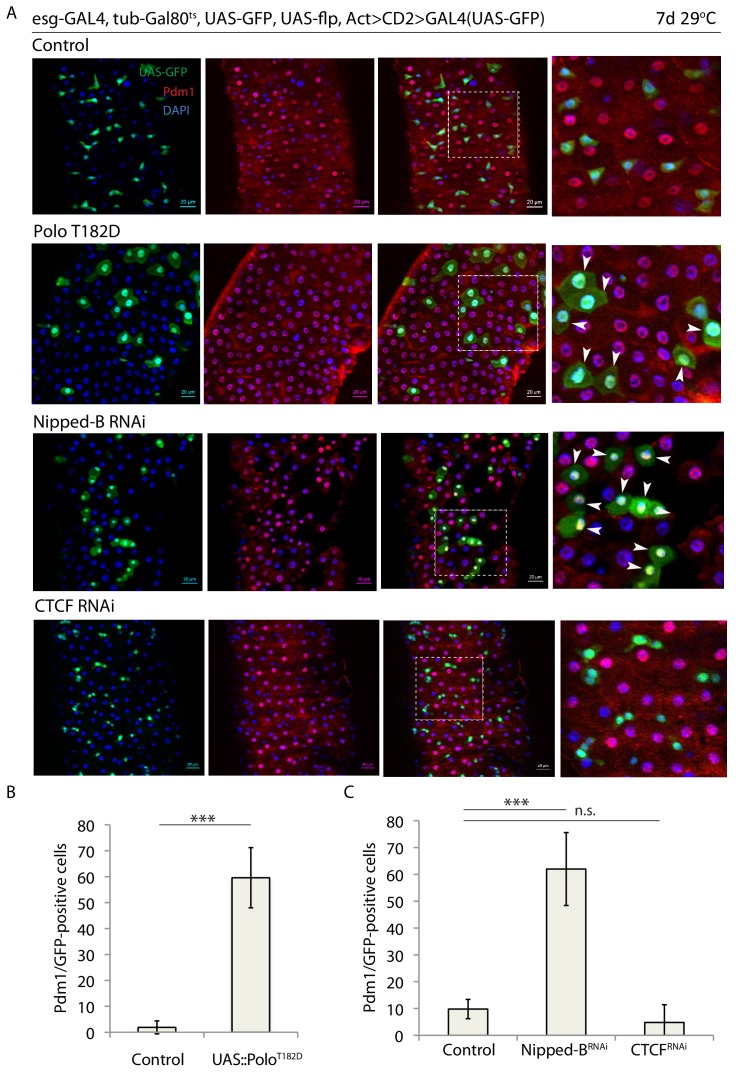
Rad21/Cohesin is loaded onto chromosomes by the kollerin complex, which includes Nipped-B, and on chromosome arms is removed during mitotic prophase by Polo kinase, likely via phosphorylation of its SA1/SA2 subunits, while the centromeric pool of Cohesin is removed during the metaphase to anaphase transition by Separase (Hauf et al., 2005; Dorsett, 2009). We asked whether perturbing Rad21 loading or promoting Cohesin release would be sufficient to phenocopy the effects of Rad21 knockdown. To perturb Cohesin loading or release, we knocked down Nipped-B or over-expressed the constitutively active Polo mutant poloT182D (the Drosophila homologue of the mammalian Plk1T210D allele that leads to Cohesin release from chromatin while also delaying mitosis) (Sumara et al., 2002; van de Weerdt et al., 2005) in ISCs, and analyzed differentiation and proliferation (Figure 2). Similar to reducing rad21 expression, these perturbations induced premature ISC differentiation, indicated by ectopic Pdm1 expression (Figure 2A–C) and inhibition of mitotic activity (Figure 2—figure supplement 1). Interestingly, depletion of the chromatin-shaping and transcriptional repressor and insulator CTCF (CCCTC-binding factor) (Sexton and Cavalli, 2015) did not trigger ISC differentiation (Figure 2A–C). These data suggest that loading of Rad21/Cohesin onto chromatin is crucial for ISC maintenance and to prevent premature differentiation of ISCs.
Figure 2. Binding of Rad21 to chromatin is crucial for ISC maintenance.
(A) esg-F/O midguts expressing UAS-GFP alone (control) or expressing UAS-PoloT182D, nippedBRNAi or CTCFRNAi. Samples were stained for GFP and Pdm1. (B and C) Quantification of GFP-positive/Pdm1-positive cells from C. (B n = 640 and 200; C n = 758, 408 and 622), ANOVA. ***p<0.001, n.s. not significant. Differentiated cells are labeled with white arrowheads. Scale bars, 20 µm.
Figure 2—figure supplement 1. Overexpression of PoloT182D or downregualtion of Nipped-B trigger ISC differentiation.
It is important to emphasize that ISCs divide at a very low rate in homeostatic conditions (Figures 1–4). The esg-FlpOut system we used allows determining the number of divisions that have occurred in our experiments. As shown in Figures 1A and 2A, in control flies, ISCs have for the most part, not divided (single labeled cells) or divided only once (doublets) in the course of the experiments. Rad21 knockdown, Polo activation, or NippedB knockdown resulted in cell differentiation regardless of whether ISCs had divided (clones with more than one labeled cell), or not (single labeled cell).
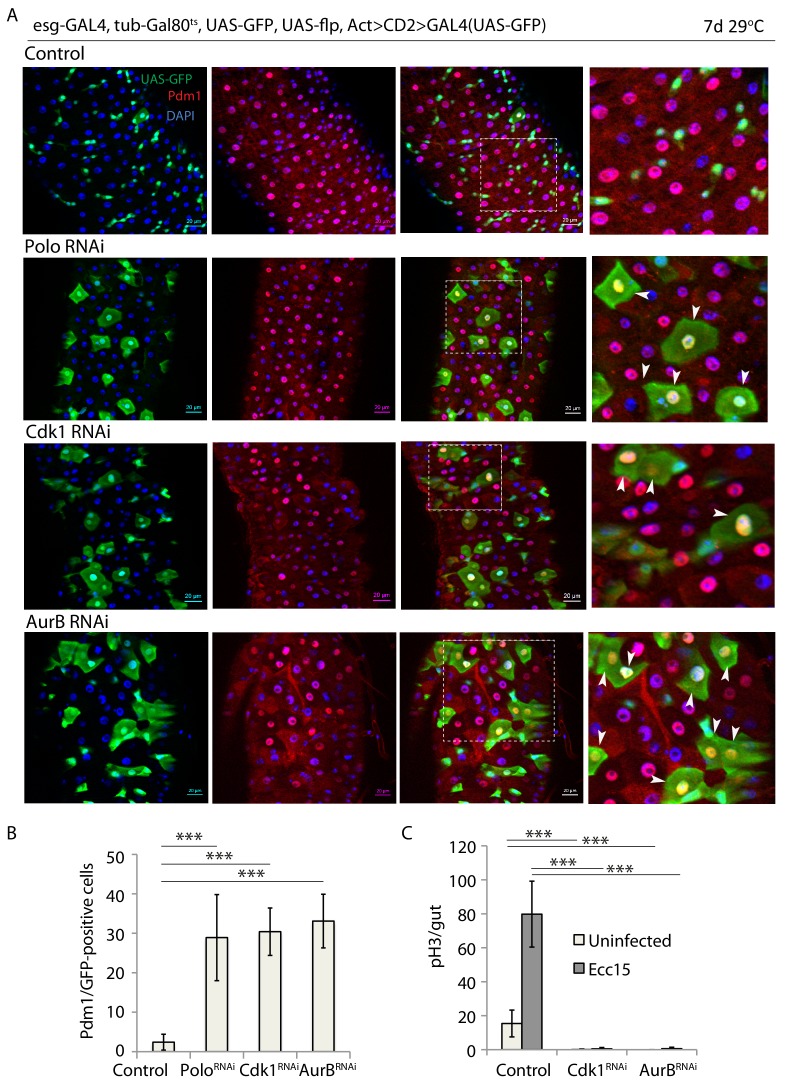
Figure 4. Knock down of cell cycle regulators leads to premature ISC differentiation.
(A) esg-F/O midguts expressing UAS-GFP alone (control) or expressing poloRNAi. cdk1RNAi and aurBRNAi. Samples were stained for GFP and Pdm1. (B) Quantification of GFP-positive/Pdm1-positive cells from A. (n = 609, 369, 500, 482). (C) Quantification of the number of mitotic pH3-positive cells/midgut in esgts guts expressing UAS-GFP alone (control) or cdk1RNAi and aurBRNAi with and without Ecc15 infection (n = 7–11), ANOVA. ***p<0.001. Differentiated cells are labeled with white arrowheads. Scale bars, 20 µm.
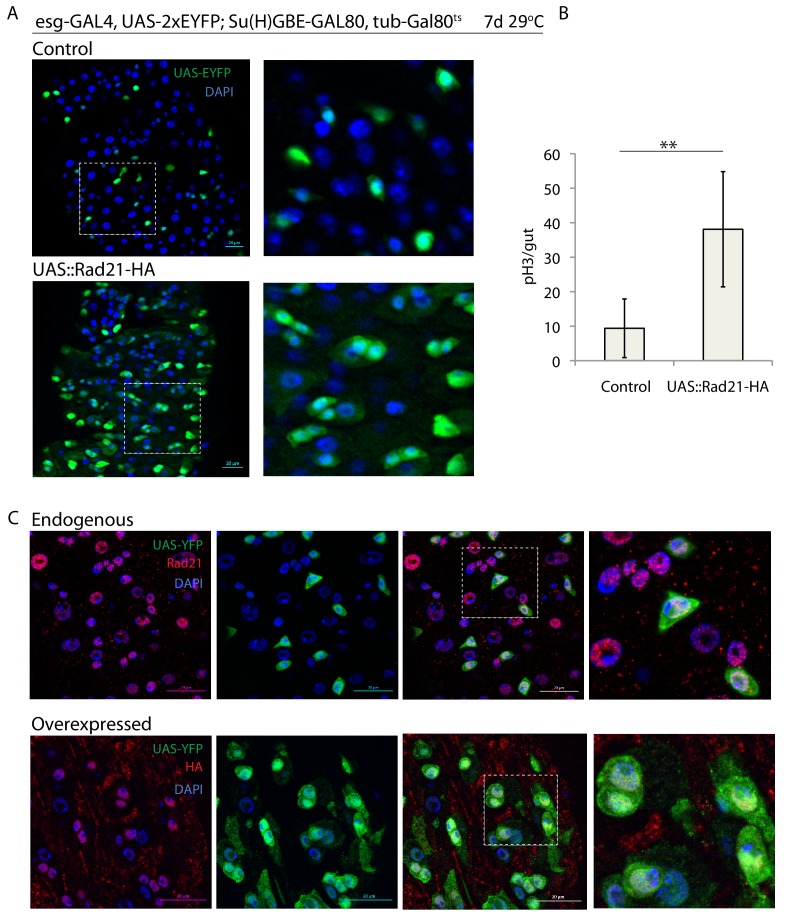
We further asked whether Rad21 over-expression would influence ISC behavior, or whether excessive Rad21 would only affect ISCs during mitosis, where its function in chromosome cohesion is important. Over-expression of Rad21-HA in ISCs using esg::Gal4 was sufficient to induce ISC proliferation in homeostatic conditions, suggesting that Rad21 can act in ISCs, at least in part, independently of chromatid cohesion (Figure 3A and B; over-expressed Rad21-HA localized to the nucleus, Figure 3C). Similarly, Rad21-HA overexpression in ISCs using esg-FlpOut system induced drastic cell overproliferation with formation of Pdm1-positive ECs (Figure 3—figure supplement 1). We therefore hypothesized that Rad21 influences ISC function via transcriptional regulation of proliferation and stemness genes in interphase, similar to its role in the mammalian epidermis (Noutsou et al., 2017).
Figure 3. Rad21 overexpression leads to increased ISC proliferation.
(A) esgts midguts expressing UAS-EYFP alone (control) or expressing UAS-Rad21-HA (B). Quantification of the number of mitotic pH3-positive cells/midgut in the guts from A (n = 8), (C). esgts midguts expressing UAS-EYFP alone (upper panels) or expressing UAS-Rad21-HA (lower panels). Samples were stained for GFP and either Rad21 (upper panels) or HA tag (lower panels). ANOVA. **p<0.01. Scale bars, 20 µm.
Figure 3—figure supplement 1. Esg-FlipOut (F/O) midguts expressing UAS-EYFP alone (control) or expressing UAS-Rad21-HA.
Mitotic failure impacts cohesin and ISC homeostasis
Rad21 and the Cohesin complex are critical for the accurate distribution of chromosomes during mitosis, and their perturbation has thus been implicated in aneuploidy (Haarhuis et al., 2014; Xu et al., 2014; Morales and Losada, 2018). Aneuploidy has recently been reported to cause ISC differentiation (Gogendeau et al., 2015) and also to promote dysplasia (Resende et al., 2018). These previous studies have reported conflicting results regarding the differentiation response of ISCs when aneuploidy is induced by knock down of spindle assembly checkpoint proteins: While depletion of bub3 was found to induce ISC differentiation and thus loss of ISCs (Gogendeau et al., 2015), depletion of BubR1, mad2, or mps1 all resulted in increased ISC proliferation and an accumulation of ISCs/EBs and EEs in the intestinal epithelium (Resende et al., 2018). Since depletion of all four factors results in aneuploidy, the differentiation response to Bub3 depletion seems to be a consequence of another function of Bub3, rather than the aneuploidy itself.
To better understand the effects of Rad21 perturbation in ISCs, and to differentiate possible aneuploidy-mediated phenotypes from effects caused by transcriptional changes in interphase, we directly compared the ISC phenotypes caused by aneuploidy with the effects of Rad21 knock down. We knocked down the essential mitosis regulators Cdk1, polo and aurora B (Godinho and Tavares, 2008) using esgF/O, and monitored ISC differentiation and proliferation. These perturbations caused premature ISC differentiation, indicated by ectopic Pdm1 and inhibited mitotic activity (Figure 4A–C) reminiscent of results in published reports in which the checkpoint kinase Bub3 was knocked down (Gogendeau et al., 2015). However, the frequency of Pdm1-positive cells after these perturbations (Figure 4B) was prominently lower compared to downregulation of Rad21 (Figure 1B). Furthermore, we assessed expression of the ISC marker Delta in Rad21 and aurB-depleted ISCs using a Dl-lacZ reporter. AurB RNAi led to a significant reduction in Delta-lacZ expression, but Rad21 knock down completely eliminated this expression, indicating a more complete loss of stemness following Rad21 depletion than in other aneuploidy-inducing conditions (Figure 5A and B). Since only a minor proportion of ISCs undergo mitosis under homeostatic conditions, these results further point to a cell-cycle independent role of Cohesin in ISCs. Finally, we compared Rad21 localization and expression between ISCs with deregulated mitosis (expressing poloT182D) and WT cells. Even though Rad21 was localized to the nucleus (Figure 5—figure supplement 1) its expression was significantly decreased in poloT182D–expressing ISCs compared to WT controls (Figure 5—figure supplement 1). Thus, cell cycle defects could lead to Rad21 downregulation further triggering ISC differentiation.
Figure 5. Downregulation of AurB and Rad21 differentially affect ISC maintenance.
(A) esg-F/O/Delta-lacZ midguts expressing UAS-GFP alone (control), aurBRNAi or expressing rad21RNAi. Samples were stained for GFP and b-gal. (B) Quantification of GFP-positive/Delta-lacZ-positive cells from A. (n = 1489, 1522, 733), ANOVA. **p<0.01, ***p<0.001. Scale bars, 20 µm.
Figure 5—figure supplement 1. Rad21 intenstiy is reduced in PoloT182D-expressing ISCs.
Transcriptional role of Rad21 in regulating ISCs
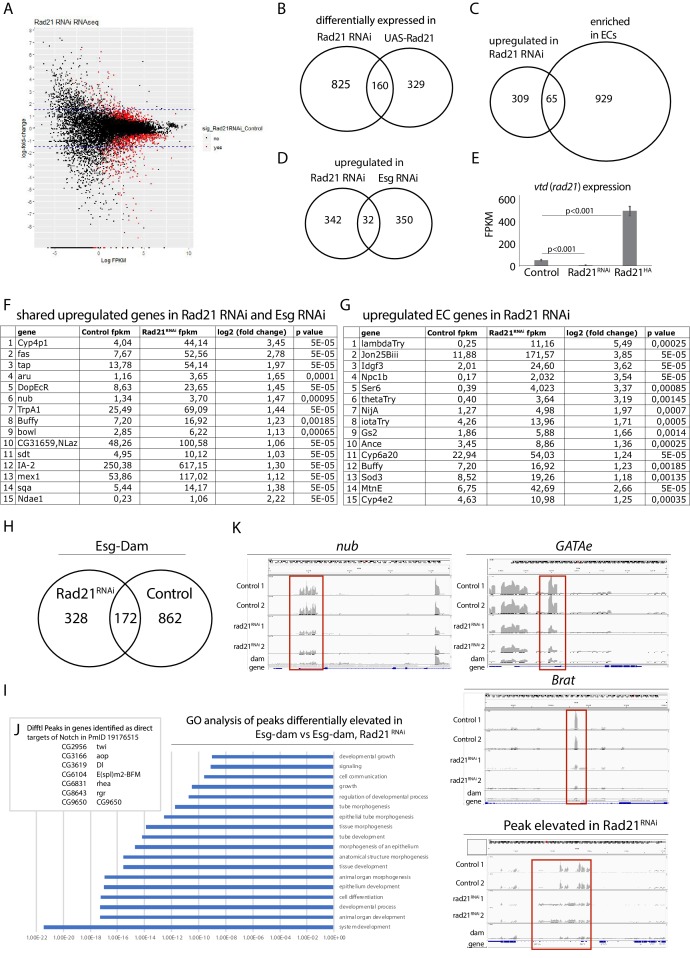
Rad21 is known to regulate transcription by shaping 3D structure of chromatin and bringing promotors and enhancers in close proximity to each other (Rudra and Skibbens, 2013). This is crucial for regulating transcription in interphase and for maintaining transcriptional memory after mitosis (Yan et al., 2013). These functions are dependent on recruitment of specific transcription factors and execution of particular transcriptional programs (Schaaf et al., 2013; Merkenschlager and Nora, 2016; Novo et al., 2018). We therefore hypothesized that Rad21 would regulate ISCs via recruitment of specific transcription factors to chromatin and promoting gene expression programs that maintain stemness and inhibit differentiation. To investigate how Rad21 regulates gene expression at a global level in ISCs, we conducted RNAseq analysis of FACS-sorted ISCs in which Rad21 was either downregulated (Rad21 RNAi) or overexpressed (UAS-Rad21-HA) for 3 and 7 days (Figure 6A–G). We used a previously described protocol for RNAseq analysis of FACS purified ISCs (Korzelius et al., 2014). Our data confirmed Rad21 knock down and overexpression, respectively, in ISCs (Figure 6E). We further observed that rad21 knockdown resulted in significant changes (q < 0.05 and Log2 equals or more then 1) in 985 genes against 489 genes in UAS-Rad21-HA samples (q < 0.05) with 160-gene overlap between conditions (Figure 6A and B, Figure 6—source data 1). Approximately 8,6% (32) of genes upregulated in Rad21 RNAi were also induced when the transcriptional repressor Esg was knocked down (Figure 6D and F), Figure 6—source data 1), including the transcription factor nubbin/pdm1, known to be repressed by Esg (Korzelius et al., 2014). EC-specific genes including Jon and trypsin family proteases, and ser6, were also enriched (approximately 17%, 65 genes) after Rad21 knock down (Dutta et al., 2015; Doupé et al., 2018) (Figure 6C and G, Figure 6—source data 1). Note that we did not observe upregulation of cell death markers after Rad21 knock down or over-expression (Figure 6, Figure 6—source data 1). These results were consistent with the differentiation phenotype of rad21-deficient ISCs (Figure 1), and led us to hypothesize that Rad21 may be required for Esg-mediated repression of differentiation genes. To test this hypothesis we performed a damID (Korzelius et al., 2014; Marshall et al., 2016) experiment to determine the genome-wide location of esg in wild-type and rad21 deficient ISCs. Using a previously described esg-dam construct, we observed 1034 significant peaks in esg-dam control samples, 862 of which were not observed in rad21 deficient ISCs, indicating that the vast majority of DNA interactions of esg in ISCs depend on functional rad21 (Figure 6H Figure 7 and Figure 6—source data 2). GO analysis of peaks appearing in Esg-Dam in contrast to Esg-Dam/Rad21 RNAi yielded a number of genes involved in ISC maintenance and differentiation pathways (Figure 6I and J) anticipated due to the function of esg, the lost interaction sites included locations close to differentiation and EC specific genes, such as nubbin/pdm1 (Figure 6K and Figure 6—source data 2).
Figure 6. RNAseq and DamID analysis of ISCs after downregulating or overexpressing Rad21.
(A) Transcriptomic analysis of ISCs expressing UAS-EYFP (Control) or Rad21RNAi or UAS-Rad21-HA. Scatter plot of gene expression regulation after Rad21 knock down. Red dots represent significant hits (false discovery rate q < 0.05). (B) Overlap between significantly differentially expressed genes found in Rad21 RNAi and UAS-Rad21-HA datasets, Fisher exact test (***p<0.001). (C) Overlap between genes upregulated in Rad21 RNAi dataset and EC-specific genes (Doupé et al., 2018), Fisher exact test (***p<0.001). (D) Overlap between genes upregulated in Rad21 RNAi and esg RNAi datasets, Fisher exact test (***p<0.001) (Korzelius et al., 2014). (E) Rad21 expression in Control, Rad21RNAi or UAS-Rad21-HA ISCs from (A). (F) Examples of EC genes upregulated after Rad21 knock down from C. (G) Examples of genes shared in Rad21 RNAi and esg RNAi datasets from D. (H) DamID analysis of ISCs expressing Esg-Dam (Control) and Esg-Dam/Rad21RNAi (Rad21 RNAi). Overlap between significant peaks found in Esg-Dam compared to Dam only and significant peaks in Esg-Dam/Rad21RNAi compared to Dam only. (I) GO analysis of peaks significantly elevated in Esg-Dam compared to Esg-Dam/Rad21 RNAi samples. (J) Differential peaks as direct targets of Notch from H. (K) Differential occupancy of nub, GATAe and Brat loci by Esg-Dam or Esg-Dam after Rad21 knock down. Dam only serves as control. Lower panel shows peaks elevated after Rad21 knock down.
Figure 6—figure supplement 1. qRT-PCR analysis of Pdm1 and Esg expression in ISCs after Rad21 knock down, ANOVA.

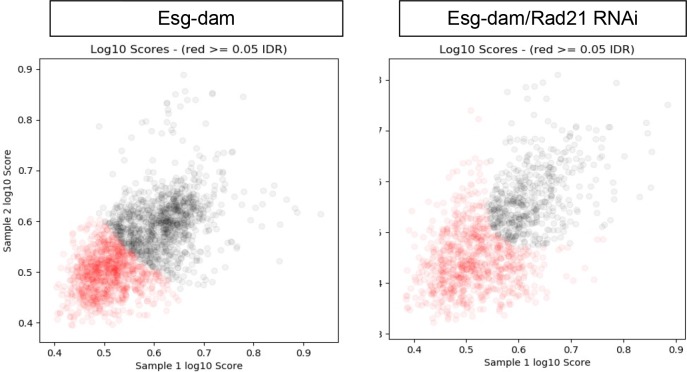
Figure 7. DamID analysis of ISCs expressing Esg-Dam (left) or Esg-Dam with Rad21RNAi (right).
Scatter plots showing reproducible peaks in repeat samples identified by the idr_tools pipeline. Black dots represent peaks with higher reproducibility (IDR <0.05).
Finally, to verify our transcriptome data we performed qRT-PCR on FACS-purified Rad21-depleted ISCs and compared those to controls. We assessed expression of Pdm1 and esg, and found that, consistent with immunofluorescence data, Pdm1 was strongly induced after Rad21 downregulation, while Esg mRNA levels did not change after Rad21 knock down (Figure 6—figure supplement 1).
Notch-independent stem cell differentiation in Rad21 loss-of-function conditions can be inhibited by esg over-expression
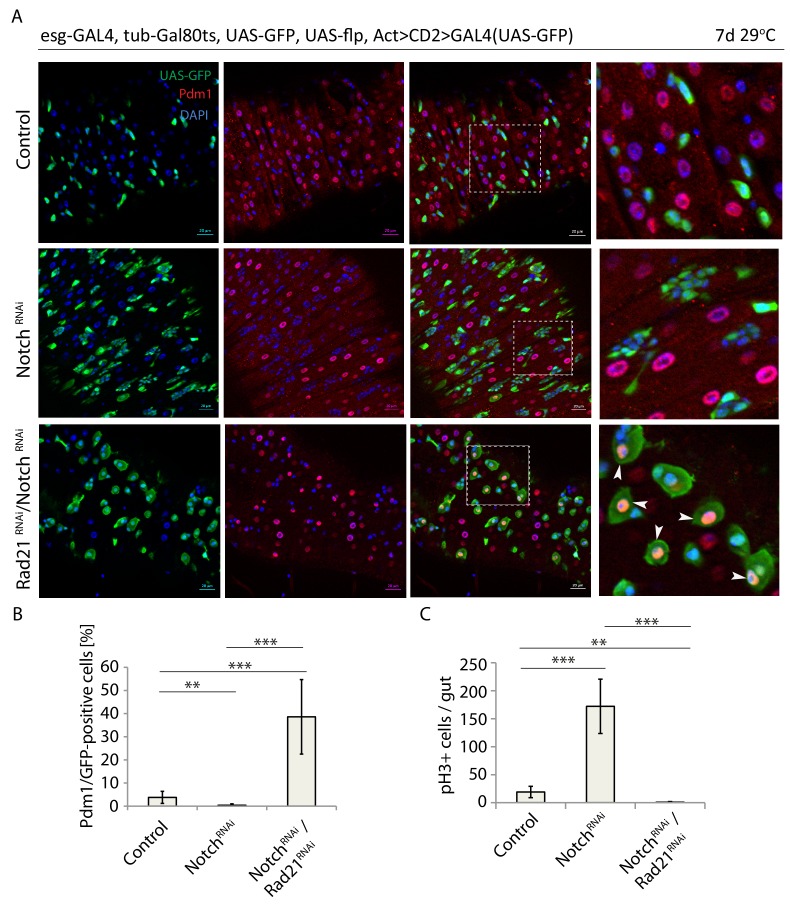
Altogether, our data indicated that Rad21 depletion results in induction of a transcriptional differentiation program. It remained unclear, however, how Rad21 interacts with canonical differentiation pathways in the ISC lineage. Since Notch signaling is the main pathway that initiates differentiation of ISCs into ECs (Ohlstein and Spradling, 2007; Kapuria et al., 2012; Guo and Ohlstein, 2015), we set to explore whether perturbing Notch signaling would interfere with ISC differentiation in Rad21 loss of function conditions. Loss of Notch in the ISC lineage perturbs differentiation, resulting in the formation of tumors consisting of ISCs and EEs. However, when Rad21 was knocked down in Notch-deficient ISCs, these cells still differentiated into Pdm1-positive cells (Figure 8A and B), and drastically inhibited ISC proliferation was observed (Figure 8C). ISC differentiation induced by loss of Rad21 is thus independent of Notch signaling.
Figure 8. Rad21 RNAi-induced ISC differentiation occurs independently of Notch signaling.
(A) esg-F/O midguts expressing UAS-GFP alone (control), notchRNAi or notchRNAi/rad21RNAi. Samples were stained for GFP and Pdm1. (B) Quantification of GFP-positive/Pdm1-positive cells from A. (n = 829, 2485, 376). (C) Quantification of the number of mitotic pH3-positive cells/midgut in esgts guts expressing UAS-GFP alone (control), notchRNAi or notchRNAi/rad21RNAi with and without Ecc15 infection (n = 6–7), ANOVA. **p<0.01, ***p<0.001. Differentiated cells are labeled with white arrowheads. Scale bars, 20 µm.
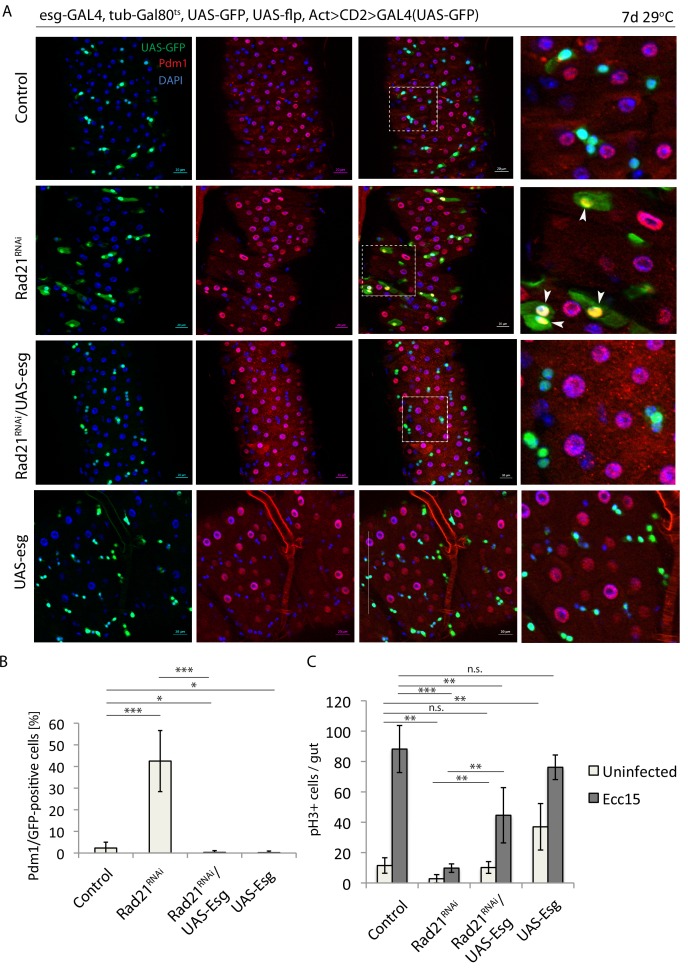
Esg is a well-understood regulator of ISC maintenance that represses the expression of differentiation genes (Korzelius et al., 2014; Loza-Coll et al., 2014). The inhibition of Delta expression and the induction of Pdm1, both known Esg targets, after Rad21 downregulation, as well as our damID experiment, indicated a possible role for Rad21 in the regulation of the Esg transcriptional program. To functionally evaluate this hypothesis, we assessed whether Esg overexpression would influence differentiation of ISCs in Rad21 loss of function conditions. Esg overexpression robustly inhibited ISC differentiation in these conditions, significantly reducing the number of Pdm1-positive cells and increasing the frequency of mitotic ISCs in homeostatic as well as regenerative conditions (after infection) (Figure 9). These results indicate that Rad21 plays a critical role in the Esg-mediated maintenance of the stem cell state in the ISC lineage (Figure 10). At the same time, however, these data indicate that the requirement for Rad21 can be overcome by elevating Esg expression levels, suggesting that Rad21 plays mostly an accessory role to ensure robust Esg-mediated gene regulation.
Figure 9. Escargot overexpression prevents Rad21 RNAi-induced ISC differentiation.
(A) esg-F/O midguts expressing UAS-GFP alone (control), expressing rad21RNAi, UAS-esgRNAi/rad21RNAi or UAS-esg. Samples were stained for GFP and Pdm1. (B) Quantification of GFP-positive/Pdm1-positive cells from A. (n = 491, 402, 391, 766), (C) Quantification of the number of mitotic pH3-positive cells/midgut in esgts guts expressing UAS-GFP alone (control), rad21RNAi, UAS-esg/rad21RNAi or UAS-esg with and without Ecc15 infection (n = 4–6), ANOVA. *p<0.05, **p<0.01, ***p<0.001, n.s. not significant. Differentiated cells are labeled with white arrowheads. Scale bars, 20 µm.
Figure 10. Model of how rad21 maintains ISC identity and proliferation via transcription factor esg.
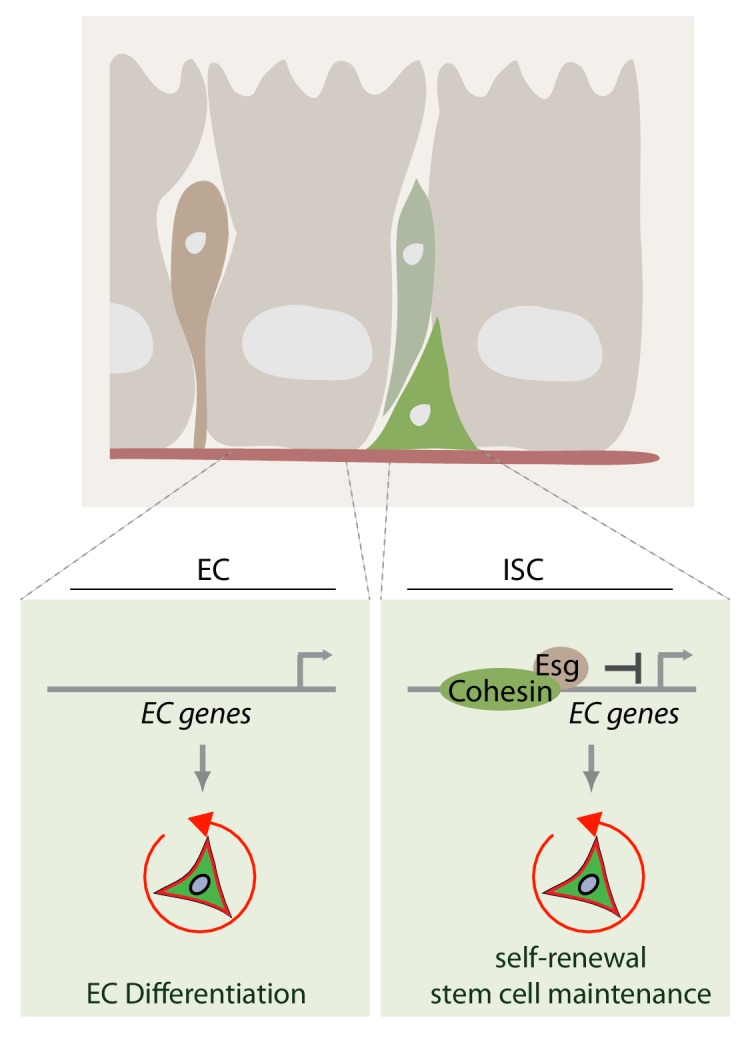
Rad21-containing Cohesin complex recruits esg to promotors of gene responsible for ISC maintenance and repression of differentiation (for example Pdm1). Rad21 loss triggers EC gene expression and ISC differentiation.
Discussion
Our data support a specific function for Cohesin-mediated chromatin regulation in the control of somatic stem cell biology. We find that both Rad21 gene function, as well as the activity of Nipped-B and Polo, previously described as regulators of Rad21 chromosome association, influence stem cell maintenance in the Drosophila ISC lineage. Our observations support the idea that the Cohesin complex interacts with interphase chromatin and supports selected regulators of gene expression.
It has been proposed that the 3-dimensional genome structure is more plastic in stem cells compared to differentiated progeny, thus facilitating re-shaping into the more stable chromatin organization that is critical for the establishment of a fully differentiated daughter cell with a specific transcriptional program (Filion et al., 2010; Sexton and Cavalli, 2015). According to this model, maintaining plasticity of genome organization is thus critical to maintain stemness and prevent premature differentiation. Our findings, together with other recent studies, indicate that Cohesin plays a central role in maintaining this plasticity. At the same time, these studies highlight the selectivity and cell-type specificity of Cohesin function in different stem cell lineages: Cohesin depletion increases differentiation of skin stem cells, but inhibits hematopoietic stem cell (HSC) differentiation (Mullenders et al., 2015; Galeev et al., 2016; Noutsou et al., 2017; Cuartero et al., 2018). In HSCs, Cohesin mediates recruitment of the transcription factor NFκB to chromatin and induces immune and pro-survival gene expression (Mullenders et al., 2015; Galeev et al., 2016; Cuartero et al., 2018). It has further been reported to control chromatin accessibility and activity of ERG, GATA2 and RUNX1 transcription factors in HSCs (Mazumdar et al., 2015). During hematopoiesis, the Polycomb protein ASLX1 interacts with Cohesin to maintain chromatid separation and transcription (Li et al., 2017). By mediating recruitment and transcriptional activity of the pluripotency factors Klf4, Esrrb and Sox2, Cohesin also regulates pluripotency of embryonic stem cells (Nitzsche et al., 2011).
In the mammalian intestine, elevated Rad21 expression has been implicated in loss of heterozygosity at the APC locus, as well as the activation of L1 retrotransposons (Xu et al., 2014). This effect suggests that Rad21 can also act as a tumor-promoting factor in the intestinal epithelium. This study did not observe defects in stem cell self-renewal efficiency in Rad21 heterozygotes, but did observe reduced expression of the ISC marker Lgr5 in heterozygous crypts. ISCs with homozygous Rad21 loss of function were not examined, however, and our results indicate that detailed lineage tracing of Rad21 mutant ISCs would be of interest in the mammalian ISC lineage.
Our work identifies Esg-mediated gene expression as the primary target of Cohesin in ISCs. Esg negatively regulates expression of differentiation factors such as Pdm1 and activates stemness genes including Delta and cell cycle proteins (Korzelius et al., 2014; Loza-Coll et al., 2014). Loss of Rad21 phenocopies the transcriptional consequences of Esg loss in ISCs, and it is striking that Esg overexpression is able to prevent ectopic ISC differentiation in the Rad21 loss of function background. This suggests that excess levels of Esg protein can exert the required repressive effect on differentiation genes even in the absence of Rad21-mediated genome re-organization. We propose that Cohesin supports Esg function by facilitating access of the transcription factor to promotor regions of differentiation genes, and that in the absence of cohesion, chromatin organization is relaxed, allowing ectopic expression of differentiation genes (Figure 10). By providing excess Esg protein, such promoters can then be repressed even in the absence of Rad21. Similar consequences of Cohesin perturbations have been reported in other cell types (Pauli et al., 2010; Dorsett and Ström, 2012; Zuin et al., 2014; Lupiáñez et al., 2015; Rao et al., 2017; Schwarzer et al., 2017).
Mechanistically, how may Cohesin regulate Esg-mediated gene expression? In other systems, Cohesin has been shown to regulate gene expression via interaction with the Mediator complex of Polymerase II (Kagey et al., 2010). Such a function of Cohesin in the transcriptional regulation complex of Esg is supported by the fact that depletion of Mediator subunits in ISCs can cause premature differentiation (Zeng et al., 2015), reminiscent of the Cohesin loss of function phenotype. Our damID data indicate that in the absence of rad21, esg loses its interaction with locations close to its target genes, but additional work is needed to dissect other mechanistic aspects of Cohesin-mediated transcriptional regulation in ISCs, such as transcription factor recruitment to chromatin, modifications and changes in chromatin conformation that lead to the transcriptional changes described here, and that ensure stem cell maintenance.
Aneuploidy has been recently linked to stem cell maintenance and proliferation (Gogendeau et al., 2015; Resende et al., 2018). Based on our findings and our observations, we propose that loss of Cohesin results in ISC differentiation independently of aneuploidy for a number of reasons: First, we find that both depleting Cohesin as well as promoting its release from DNA induces ISC differentiation. The various perturbations to that effect (two independent Rad21 shRNAs, Nipped B shRNA, T182D Polo mutant) elicited a significantly stronger differentiation phenotype compared to perturbations of other regulators of mitosis (AurB, Cdk1, Polo), indicating that the mitotic role of Cohesin could not completely account for the observed robust differentiation of ISCs into ECs (Figure 1, Figure 2 and Figure 3). Importantly, all of these perturbations resulted in a complete inhibition of proliferation, indicating a comparable level of cell cycle arrest (Figure 1D, Figure 2—figure supplement 1 and Figure 4C). Second, the fact that Nipped B knockdown also results in ISC differentiation strongly supports an aneuploidy-independent mechanism, as the binding of Cohesin to chromatin after completion of mitosis is mediated in interphase by the complex containing Nipped B. These data thus suggest that Cohesin binding to chromatin in interphase is critical for ISC maintenance (Figure 2A and C). Similar important roles of Cohesin in interphase cells have been recently described in mammals (Meisenberg et al., 2019). Third, ISCs are normally quiescent in unchallenged young fly midguts and enter mitosis at a very low rate (up to approximately 10–20%) (Figure 1D, Figure 2—figure supplement 1, Figure 3B and Figure 4C). Cohesin depletion in ISCs would thus only lead to aneuploidy in the small subset of ISCs that enter mitosis. In contrast, Cohesin depletion led to differentiation of around 60–70% of all ISCs (Figure 1B), and to an almost complete elimination of Delta-positive cells (to approximately 2%) (Figure 5B). Such a drastic effect does not reflect the frequency of cells entering mitosis. Finally, our RNAseq and DamID analyses suggest that Cohesin has profound impact on the transcriptome of ISCs, and significantly affects the transcriptional program regulated by Escargot (Figure 6). Accordingly, Cohesin depletion led to loss of Esg promotor binding, and significantly reduced Esg-regulated transcripts. Esg over-expression in Cohesin-depleted cells further significantly rescued the premature ISC differentiation (Figure 9) and restored homeostasis. It seems unlikely that these effects on Esg promoter loading and Esg-mediated gene regulation are merely an indirect consequence of aneuploidy. It is important to point out further that differentiation as a consequence of aneuploidy does not seem to be a robust phenotype: While depletion of bub3 was found to induce ISC differentiation and thus loss of ISCs, depletion of BubR1, mad2, or mps1 all result in increased ISC proliferation and an accumulation of ISCs/EBs and EEs in the intestinal epithelium (Gogendeau et al., 2015; Resende et al., 2018).
We anticipated that downregulation of the chromatin insulator CTCF might recapitulate the Rad21 RNAi phenotype but, on the contrary, we have not observed any detectable rise in ISC differentiation (Figure 2). Several recent studies on CTCF in Drosophila found little evidence of CTCF in chromosome domain/loop formation (Ramírez et al., 2018; Wang et al., 2018; Matthews and White, 2019). Other insulator protein complexes such as BEAF-32/CP190 or BEAF-32/Chromator have been implicated in TAD organization in Drosophila (Ramírez et al., 2018; Wang et al., 2018; Matthews and White, 2019). Therefore, it has been recently proposed that fly CTCF is not a major boundary definition protein but is rather a regulator of Hox or other gene expression (Gambetta and Furlong, 2018; Szabo et al., 2019).
Cohesin has been suggested to maintain transcriptional memory after mitosis (Yan et al., 2013). Contrary to original observations of complete Cohesin removal, some level of Cohesin has been detected in mitosis and suggested to mediate reloading of transcription factors onto DNA to resume gene expression in interphase (Yan et al., 2013) or maintain gene expression in mitosis (Teves et al., 2016). This would be especially relevant for stem cells highly dependent on persistent expression of stemness genes (Ferraro et al., 2016). Based on these reports and our data, it is possible that Rad21 in ISCs ensures reloading of Esg onto relevant promoters after mitosis to maintain stable expression of stemness genes and ensure self-renewal (Gogendeau et al., 2015) (Figure 10). Accordingly, we show that downregulation of mitotic factors and Rad21 release from chromatin leads to premature ISC differentiation. Furthermore, expression of constitutively active Polo, which is known to force cells into mitosis (Sumara et al., 2002; van de Weerdt et al., 2005), remove Cohesin from chromatin, and extend the duration of mitosis, triggers drastic ISC differentiation.
Recent evidence indicates that chromatin loops to a great extent could be reestablished shortly after Cohesin removal from chromatin (Rao et al., 2017). This could imply other mechanisms controlling chromatin organization and therefore regulating stem cell transcription and ‘bookmarking’ transcription sites in mitosis. Alternatively, this may suggest that Cohesin downregulation could lead to chromatin loop rearrangements even though partially preserving chromatin complexity but still switching transcriptional programs of stem cells towards differentiated progeny. It is also interesting that acute Cohesin depletion slightly inhibits but nevertheless preserves cell proliferation of developing neuroblasts (Mirkovic et al., 2019) indicating a complex and still understudied role of Cohesin in mitosis, which may include cell type-, stage- or organism-dependent differences in mitosis, involve other cellular processes and non-cell autonomous mechanisms. All together, our work supports the notion that characterizing chromatin dynamics in somatic stem cell lineages, and dissecting the multiple roles of Cohesin in stem cell maintenance, mitosis, and differentiation in a variety of stem cell lineages and organisms, is critical to understand somatic stem cell regulation and dysfunction.
Materials and methods
Fly stocks
The following stocks study were reported previously (Korzelius et al., 2014).
Esgts driver: y,w;esg‐GAL4/CyO;tub‐GAL80ts,UAS‐GFP/Tm6B, Esg‐Flip-Out (F/O) driver lines: w;esg‐Gal4,UAS‐GFP,tub‐Gal80ts/CyO;UAS‐flp,act >CD2>Gal4/Tm6B and w;esg-gal4, tub-Gal80ts, UAS-GFP/CyO,wg-lacZ;P{w[+mC]=UAS FLP.D}JD2/Tm6BEsg overexpression: y,w;UASt::esg/CyORad21 overexpression: w;UAS-Rad21-HA/Cyo (a gift from Stefan Heidmann, Bayreuth and Raquel Oliviera, Gulbenkian).Delta reporter: w;If/CyO,wglacZ; Dl05151-lacZ/Tm6B
DamID lines:Dam only: w;M{w[+mC]=hs .min(FRT.STOP1)dam}ZH-51C/CyO,wg-lacZEsg-Dam: w;esg-Dam(ZH-51C) M4M1/CyO, wg-lacZ;MKRS/Tm6BNotch knock down: UAS‐NotchRNAi.The following Drosophila lines were obtained from Bloomington Stock Collection (Indiana): Rad21 RNAi (#36786, #65229), Cdk1 RNAi (#28368), AurB RNAi (#58308), Polo T182D (#8434), Polo RNAi (#35146, #35770), Nipped-B (#32406, #36614), CTCF RNAi (#40850).Flies containing Gal80ts were raised at 18°C and used at the minimum of 3 days of age. Animals were then shifted to 29°C for 7 days unless indicated.
Infection with Erwinia carotovora
Bacteria were inoculated and grown overnight at 30 degrees, then collect and resuspended in 5% sucrose in water. Flies were shifted to 29°C for 7 days unless indicated to induce RNAi or transgene expression. Flies were then first starved for 4 hr in vials with water and subsequently flipped in new vials with Ecc15. Infections were conducted for 14–16 hr unless otherwise indicated. Esch experiment was conducted twice.
Antibodies
Chicken anti-GFP (A10262, Invitrogen), mouse monoclonal anti-GFP (sc-9996, B-2, Santa-Cruz) rabbit polyclonal anti-Pdm1 (kindly provided by Cai Yu, Singapore), rabbit polyclonal anti-Rad21/vtd (kindly provided by Dale Dorsett, Saint Louis), rabbit polyclonal anti-pH3 (sc-8656, Santa Cruz), mouse monoclonal anti- beta-galactosidase/LacZ (40-1a, DSHB), HA-tag mouse monoclonal antibody (HA-7, H3663, Sigma). Secondary antibodies: goat polyclonal anti-chicken, rabbit, mouse Alexa 488, Alexa 568-coupled (A11039, A11036, A11034, A11029, A11031, Invitrogen). DAPI was used to stain DNA.
Immunofluorescence
Protocol was adapted from the study described previously (Resnik-Docampo et al., 2017). Intestines from adult female flies were dissected into phosphate buffered saline (PBS) and fixed in in 4% paraformaldehyde (PFA) for 60 min at room temperature in nutating mixer. Guts were pemeabilized and washed by sequentially incubating in 0.5% Triton-x100 (Tx-100)/PBS, 0.5% Na-Deoxycholate (NaDoc), 0.3% Tx-100 for 10 min and blocked in 0.3% Tx-100/0.5 BSA/PBS for 30 min. Primary antibody in blocking solution was then added to guts and incubated overnight. Gut were further washed 4 times in 0.3% Tx-100 for 10 min. Then secondary antibody diluted in blocking solution was added, and guts were incubated for 2 hr. Afterward guts were washed 3 times in 0.3% Tx-100 for 10 min and incubated in DAPI for 5 min. Gut were subsequently washed once in 0.3% Tx-100 for 10 min and mounted in Vectashield (Vector Laboratories). Images were taken using Axiovert equipped with Apotome V or LSM710 confocal microscope and further analysed using Aviovision (Zeiss) and Image J software.
Quantification of pH3, Pdm1, DAPI, RAD21 and Delta signals
Images were taken using Apotome (Zeiss). Mitotic index in fly guts was scored by pH3-positive ISCs in esg[ts] crosses. Mounted guts were inspected under microscope and pH3-positive ISCs (labeled by UAS-GFP) were counted. 6–12 guts per sample were scored in each experiment; each experiment was performed 2–3 times (biological replicas). Pdm1 signals were quantified in posterior midguts from flies derived from EsgF/O crosses by Axiovision software (Zeiss). Pdm1 positive ISCs (labeled with UAS-GFP) were scored in controls and in RNAi midguts. Images from 8 to 10 guts within 2–3 experiments (biological replicas) were analysed and results were pulled and presented. Delta-positive ISCs were quantified as described for Pdm1. Size of DAPI-positive nuclei and Rad21 intensity were measured in posterior midguts using Image J. A line around each of 50 DAPI and Rad21-positive nuclei was drawn and either size or average pixel intensity were analysed using Image J.
RNA isolation
Intestines (50/sample) in four independent experiments were dissected in PBD and homogenized by incubating in elastase for 1 hr at 27 degrees with vigorous shaking and pipetting. Cell suspensions were then used for FACS sorting (FACS Aria) and gated based on UAS-GFP signal set by w1118 cells autofluorescence. 10.000–20.000 GFP-positive cells were then processed for RNA isolation using Arcturus PicoPure RNA Isolation Kit (KIT0202, KIT0204, Thermo) according to manufacturer’s instructions. RNA was subsequently used for cDNA preparation using QuantiTect-Reverse Transcription Kit (205311, Qiagen) or RNAseq.
qRT-PCR
PCR was performed 3 times using Thermo Scientific Maxima SYBR Green/ROX qPCR Master Mix (#K0222) according to manufacturer’s instructions. Pdm1 and Esg mRNA levels in rad21RNAi samples were normalized to housekeeping genes Gdh and Rpl32 and presented as ratio to controls (UAS-GFP). Primers: Gdh F: 5’-gctccgggaaaaggaaaa-3’, R: 5-tccgttaattccgatcttcg-3’; RpL32 F: 5’-atcgtgaagaagcgcaccaa-3’, R: 5’-tgtcgatacccttgggcttg-3’; Esg F: 5’-cgccagacaatcaatcgtaagc-3’, R: 5’-tgtgtacgcgaaaaagtagtgg-3’; Pdm1 (Nub) F: 5’-cgggataaatcgaaggaagc-3’, R: 5’-agtatttgatgtgtttgcgacttt-3’.
RNAseq
RNA-seq dual-indexed TrueSeq stranded mRNA kit was used to prepare RNA-seq libraries. Libraries sequenced on Illumina Hiseq machine using two different lanes. Read length is 51 bases and number of reads varied across samples from 8.7M reads to 64.1M reads. First we used Trimmomatic tool version 0.36 (Bolger et al., 2014) to trim and filter low quality reads. Then, we used Tophat2 version 2.1.1 (Kim et al., 2013) to align reads that passed quality control to the UCSC dam6 reference genome. The vast majority of the reads passed quality control and the overall read mapping rate is consistently between 96.7% and 97.4% across samples. To normalize between read counts and to find differentially expressed genes between Rad21RNAi, Rad21HA, and control we used Cufflink suite of tools version 2.2.1 (Trapnell et al., 2010; Roberts et al., 2011b; Roberts et al., 2011a; Trapnell et al., 2013).
Statistics
Data are presented as mean SD or SEM. Statistical analysis of two experimental groups was performed using one-way ANOVA test. For non-parametric distributed data, the Mann-Whitney-U-test or Fisher exact test was applied. Significance was considered at p<0.05. *p<0.05, **p<0.01, ***p<0.001.
DamID
Protocol was adapted from the study described previously (Marshall et al., 2016). Intestines (10–20/sample) in two independent experiments were dissected in PBD and used for extraction of genomic DNA using QIAamp DNA Micro Kit (56304, Qiagen). DNA was further digested by DpnI and cleaned via PCR purification. Digested DNA fragment were used for ligation of DamID adaptors and digestion by DpnII and PCR amplification. DNA was futher purified and sonicated to reduce fragment size. DNA quality was controlled by agarose gel and Agilent DNA Bioanalyser system. Samples were then used for library preparation and sequencing. NGS library was prepared using Illumina TruSeq nano 529 DNA kit. After library quality control sample were subsequently sequenced as 50 bp 530 single-end on an Illumina HiSeq2500.
DamID data analysis
Read alignment and peak calling: We used damid_pipeline script from Marshall OJ's damid_pipeline software version 1.4.4 (Marshall and Brand, 2015) to align fastq files to the Drosophila dm6 reference genome and to produce coverage files. Alignment rate was good and uniform across Dam control and Esg-Dam samples but was significantly lower for the Esg-Dam(rad21Δ) samples as shown in Table 1. Then, to identify DNA regions with higher mapping counts in Esg-Dam samples compared to a Dam-control sample we used find_peaks script from the same software. find_peaks was called with the option –fdr = 0.1, allowing peaks with relatively high level of false discovery rate. The pipeline was set to compare between samples according to Table 2. Reproducibility analysis: We used the idr tool version 2.02 to merge reproducible peaks in both replicates of Esg-Dam and two replicates of Esg-Dam(Rad21Δ). Gene annotation: To find genes in proximity to genomic regions identifies as reproducible peaks, we used the BioConductor annotatr R package version 1.8.0 (Cavalcante and Sartor, 2017). Differential binding analysis: To identify differential targets of esg between control and Rad21Δ backgrounds we first used mergePeaks tool from the HOMER software (Heinz et al., 2010) to merge peaks. We merged peaks that passed IDR analysis in Esg-Dam samples with peaks that passed IDR analysis in Esg-Dam(Rad21Δ) samples. mergePeaks resulted in a list of 1362 peaks. Then we used HOMER's getDifferentialPeaks tool to find 902 peaks that are enriched in Esg-Dam compared to Esg-Dam(Rad21Δ) and vice-versa, 328 peaks that are enriched in Esg-Dam(Rad21Δ) compared to Esg-Dam. Cutoff used for enrichment was fold-change of 1.2 or greater.
Table 1. Dam ID alignment.
Number of reads is uniform across samples, the sequence quality is good and uniform. Alignment rate is good and uniform across dam and Esg-Dam samples but significantly lower for the Esg-Dam/Rad21 samples.
| # reads | #reads containing no Ns | Overall alignment rate | |
|---|---|---|---|
| dam1 | 62,324,161 | 99.88 | 94.46% |
| dam2 | 61,289,068 | 99.88 | 95.17% |
| esg1 | 63,277,266 | 99.88 | 92.87% |
| esg2 | 64,279,981 | 99.88 | 92.41% |
| rad21esg1 | 66,375,925 | 99.88 | 56.21% |
| rad21esg2 | 63,063,034 | 99.88 | 79.86% |
Table 2. DamID peak calling.
| #low threshold peaks | #merged peaks | #peaks passing IDR cutoff of 0.05 | %peaks passing IDR cutoff of 0.05 | |
|---|---|---|---|---|
| esg-dam1 | 2521 | 1973 | 1034 | 52.4% |
| esg-dam2 | 2537 | |||
| rad21RNAi/esg-dam1 | 2387 | 1327 | 500 | 37.7% |
| rad21RNAi/esg-dam2 | 2175 |
Acknowledgements
We are indebted to Dale Dorsett, Stefan Heidmann, Raquel Oliviera and Cai Yu for reagents. We especially thank Jerome Korzelius for assistance in planning and executing experiments and for providing fly stocks. We also acknowledge support from the FLI core facilities Sequencing, FACS and Imaging. Work at the Buck Institute was funded by NIH R01 GM117412.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Aliaksandr Khaminets, Email: akhaminets@gmail.com.
Heinrich Jasper, Email: jasper.heinrich@gene.com.
Elaine Fuchs, Howard Hughes Medical Institute, The Rockefeller University, United States.
Utpal Banerjee, University of California, Los Angeles, United States.
Funding Information
This paper was supported by the following grant:
National Institutes of Health NIH R01 GM117412 to Heinrich Jasper, Tal Ronnen-Oron.
Additional information
Competing interests
No competing interests declared.
employee of Genentech, Inc, a member of the Roche group.
Author contributions
Conceptualization, Data curation, Software, Formal analysis, Supervision, Validation, Investigation, Visualization, Methodology, Project administration.
Resources, Data curation, Software, Formal analysis, Investigation, Visualization, Methodology.
Resources, Data curation, Investigation, Methodology.
Resources, Data curation, Investigation, Methodology.
Conceptualization, Data curation, Software, Supervision, Funding acquisition, Validation, Visualization, Methodology, Project administration.
Additional files
Data availability
Datasets generated during this study is included in Source data files.
The following previously published datasets were used:
David P Doupé, Owen J Marshall, Hannah Dayton, Andrea H Brand, Norbert Perrimon. 2018. Drosophila intestinal stem and progenitor cells are major sources and regulators of homeostatic niche signals. Gene Expression Omnibus. GSE101814
References
- Adam RC, Yang H, Rockowitz S, Larsen SB, Nikolova M, Oristian DS, Polak L, Kadaja M, Asare A, Zheng D, Fuchs E. Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature. 2015;521:366–370. doi: 10.1038/nature14289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam RC, Fuchs E. Stem cell fate selection. the yin and yang of chromatin dynamics. Trends in Genetics. 2016;32:89–100. doi: 10.1016/j.tig.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyaz A, Jasper H. Intestinal inflammation and stem cell homeostasis in aging Drosophila Melanogaster. Frontiers in Cellular and Infection Microbiology. 2013;3:98. doi: 10.3389/fcimb.2013.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nature Reviews Molecular Cell Biology. 2014;15:19–33. doi: 10.1038/nrm3721. [DOI] [PubMed] [Google Scholar]
- Beagan JA, Gilgenast TG, Kim J, Plona Z, Norton HK, Hu G, Hsu SC, Shields EJ, Lyu X, Apostolou E, Hochedlinger K, Corces VG, Dekker J, Phillips-Cremins JE. Local genome topology can exhibit an incompletely rewired 3D-Folding state during somatic cell reprogramming. Cell Stem Cell. 2016;18:611–624. doi: 10.1016/j.stem.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beebe K, Lee WC, Micchelli CA. JAK/STAT signaling coordinates stem cell proliferation and multilineage differentiation in the Drosophila intestinal stem cell lineage. Developmental Biology. 2010;338:28–37. doi: 10.1016/j.ydbio.2009.10.045. [DOI] [PubMed] [Google Scholar]
- Biteau B, Hochmuth CE, Jasper H. JNK activity in somatic stem cells causes loss of tissue homeostasis in the aging Drosophila gut. Cell Stem Cell. 2008;3:442–455. doi: 10.1016/j.stem.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B, Hochmuth CE, Jasper H. Maintaining tissue homeostasis: dynamic control of somatic stem cell activity. Cell Stem Cell. 2011;9:402–411. doi: 10.1016/j.stem.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon N, Broderick NA, Kuraishi T, Lemaitre B. Drosophila EGFR pathway coordinates stem cell proliferation and gut remodeling following infection. BMC Biology. 2010;8:152. doi: 10.1186/1741-7007-8-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantone I, Fisher AG. Epigenetic programming and reprogramming during development. Nature Structural & Molecular Biology. 2013;20:282–289. doi: 10.1038/nsmb.2489. [DOI] [PubMed] [Google Scholar]
- Casali A, Batlle E. Intestinal stem cells in mammals and Drosophila. Cell Stem Cell. 2009;4:124–127. doi: 10.1016/j.stem.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Cavalcante RG, Sartor MA. Annotatr: genomic regions in context. Bioinformatics. 2017;33:2381–2383. doi: 10.1093/bioinformatics/btx183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuartero S, Weiss FD, Dharmalingam G, Guo Y, Ing-Simmons E, Masella S, Robles-Rebollo I, Xiao X, Wang YF, Barozzi I, Djeghloul D, Amano MT, Niskanen H, Petretto E, Dowell RD, Tachibana K, Kaikkonen MU, Nasmyth KA, Lenhard B, Natoli G, Fisher AG, Merkenschlager M. Control of inducible gene expression links cohesin to hematopoietic progenitor self-renewal and differentiation. Nature Immunology. 2018;19:932–941. doi: 10.1038/s41590-018-0184-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Mirny L. The 3D genome as moderator of chromosomal communication. Cell. 2016;164:1110–1121. doi: 10.1016/j.cell.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D. Cohesin, gene expression and development: lessons from Drosophila. Chromosome Research. 2009;17:185–200. doi: 10.1007/s10577-009-9022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D, Ström L. The ancient and evolving roles of cohesin in gene expression and DNA repair. Current Biology. 2012;22:R240–R250. doi: 10.1016/j.cub.2012.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doupé DP, Marshall OJ, Dayton H, Brand AH, Perrimon N. Drosophila intestinal stem and progenitor cells are major sources and regulators of homeostatic niche signals. PNAS. 2018;115:12218–12223. doi: 10.1073/pnas.1719169115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D, Dobson AJ, Houtz PL, Gläßer C, Revah J, Korzelius J, Patel PH, Edgar BA, Buchon N. Regional Cell-Specific transcriptome mapping reveals regulatory complexity in the adult Drosophila midgut. Cell Reports. 2015;12:346–358. doi: 10.1016/j.celrep.2015.06.009. [DOI] [PubMed] [Google Scholar]
- Feeney KM, Wasson CW, Parish JL. Cohesin: a regulator of genome integrity and gene expression. Biochemical Journal. 2010;428:147–161. doi: 10.1042/BJ20100151. [DOI] [PubMed] [Google Scholar]
- Ferraro T, Esposito E, Mancini L, Ng S, Lucas T, Coppey M, Dostatni N, Walczak AM, Levine M, Lagha M. Transcriptional memory in the Drosophila embryo. Current Biology. 2016;26:212–218. doi: 10.1016/j.cub.2015.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, van Steensel B. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galeev R, Baudet A, Kumar P, Rundberg Nilsson A, Nilsson B, Soneji S, Törngren T, Borg Å, Kvist A, Larsson J. Genome-wide RNAi screen identifies cohesin genes as modifiers of renewal and differentiation in human HSCs. Cell Reports. 2016;14:2988–3000. doi: 10.1016/j.celrep.2016.02.082. [DOI] [PubMed] [Google Scholar]
- Gambetta MC, Furlong EEM. The insulator protein CTCF is required for correct Hox Gene Expression, but Not for Embryonic Development in Drosophila. Genetics. 2018;210:129–136. doi: 10.1534/genetics.118.301350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho S, Tavares AA. A role for Drosophila polo protein in chromosome resolution and segregation during mitosis. Cell Cycle. 2008;7:2529–2534. doi: 10.4161/cc.7.16.6439. [DOI] [PubMed] [Google Scholar]
- Gogendeau D, Siudeja K, Gambarotto D, Pennetier C, Bardin AJ, Basto R. Aneuploidy causes premature differentiation of neural and intestinal stem cells. Nature Communications. 2015;6:8894. doi: 10.1038/ncomms9894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Ohlstein B. Stem cell regulation. bidirectional notch signaling regulates Drosophila intestinal stem cell multipotency. Science. 2015;350:aab0988. doi: 10.1126/science.aab0988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarhuis JH, Elbatsh AM, van den Broek B, Camps D, Erkan H, Jalink K, Medema RH, Rowland BD. WAPL-mediated removal of cohesin protects against segregation errors and aneuploidy. Current Biology. 2013;23:2071–2077. doi: 10.1016/j.cub.2013.09.003. [DOI] [PubMed] [Google Scholar]
- Haarhuis JH, Elbatsh AM, Rowland BD. Cohesin and its regulation: on the logic of X-shaped chromosomes. Developmental Cell. 2014;31:7–18. doi: 10.1016/j.devcel.2014.09.010. [DOI] [PubMed] [Google Scholar]
- Hauf S, Roitinger E, Koch B, Dittrich CM, Mechtler K, Peters JM. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLOS Biology. 2005;3:e69. doi: 10.1371/journal.pbio.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidinger-Pauli JM, Mert O, Davenport C, Guacci V, Koshland D. Systematic reduction of cohesin differentially affects chromosome segregation, condensation, and DNA repair. Current Biology. 2010;20:957–963. doi: 10.1016/j.cub.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Liu X, Li D, Shao Z, Cao H, Zhang Y, Trompouki E, Bowman TV, Zon LI, Yuan GC, Orkin SH, Xu J. Dynamic control of enhancer repertoires drives lineage and Stage-Specific transcription during hematopoiesis. Developmental Cell. 2016;36:9–23. doi: 10.1016/j.devcel.2015.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasper H. Exploring the physiology and pathology of aging in the intestine of Drosophila melanogaster. Invertebr Reprod Dev. 2015;59:51–58. doi: 10.1080/07924259.2014.963713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Patel PH, Kohlmaier A, Grenley MO, McEwen DG, Edgar BA. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 2009;137:1343–1355. doi: 10.1016/j.cell.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Grenley MO, Bravo MJ, Blumhagen RZ, Edgar BA. EGFR/Ras/MAPK signaling mediates adult midgut epithelial homeostasis and regeneration in Drosophila. Cell Stem Cell. 2011;8:84–95. doi: 10.1016/j.stem.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Edgar BA. EGFR signaling regulates the proliferation of Drosophila adult midgut progenitors. Development. 2009;136:483–493. doi: 10.1242/dev.026955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Edgar BA. Intestinal stem cells in the adult Drosophila midgut. Experimental Cell Research. 2011;317:2780–2788. doi: 10.1016/j.yexcr.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapuria S, Karpac J, Biteau B, Hwangbo D, Jasper H. Notch-mediated suppression of TSC2 expression regulates cell differentiation in the Drosophila intestinal stem cell lineage. PLOS Genetics. 2012;8:e1003045. doi: 10.1371/journal.pgen.1003045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzelius J, Naumann SK, Loza-Coll MA, Chan JS, Dutta D, Oberheim J, Gläßer C, Southall TD, Brand AH, Jones DL, Edgar BA. Escargot maintains stemness and suppresses differentiation in Drosophila intestinal stem cells. The EMBO Journal. 2014;33:2967–2982. doi: 10.15252/embj.201489072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WC, Beebe K, Sudmeier L, Micchelli CA. Adenomatous polyposis coli regulates Drosophila intestinal stem cell proliferation. Development. 2009;136:2255–2264. doi: 10.1242/dev.035196. [DOI] [PubMed] [Google Scholar]
- Li Z, Zhang P, Yan A, Guo Z, Ban Y, Li J, Chen S, Yang H, He Y, Li J, Guo Y, Zhang W, Hajiramezanali E, An H, Fajardo D, Harbour JW, Ruan Y, Nimer SD, Yu P, Chen X, Xu M, Yang FC. ASXL1 interacts with the cohesin complex to maintain chromatid separation and gene expression for normal hematopoiesis. Science Advances. 2017;3:e1601602. doi: 10.1126/sciadv.1601602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Jasper H. Gastrointestinal stem cells in health and disease: from flies to humans. Disease Models & Mechanisms. 2016;9:487–499. doi: 10.1242/dmm.024232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G, Xu N, Xi R. Paracrine unpaired signaling through the JAK/STAT pathway controls self-renewal and lineage differentiation of Drosophila intestinal stem cells. Journal of Molecular Cell Biology. 2010;2:37–49. doi: 10.1093/jmcb/mjp028. [DOI] [PubMed] [Google Scholar]
- Liu W, Singh SR, Hou SX. JAK-STAT is restrained by notch to control cell proliferation of the Drosophila intestinal stem cells. Journal of Cellular Biochemistry. 2010;109:992–999. doi: 10.1002/jcb.22482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Jin LH. Tissue-resident stem cell activity: a view from the adult Drosophila gastrointestinal tract. Cell Communication and Signaling. 2017;15:33. doi: 10.1186/s12964-017-0184-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loza-Coll MA, Southall TD, Sandall SL, Brand AH, Jones DL. Regulation of Drosophila intestinal stem cell maintenance and differentiation by the transcription factor escargot. The EMBO Journal. 2014;33:2983–2996. doi: 10.15252/embj.201489050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, Santos-Simarro F, Gilbert-Dussardier B, Wittler L, Borschiwer M, Haas SA, Osterwalder M, Franke M, Timmermann B, Hecht J, Spielmann M, Visel A, Mundlos S. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–1025. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall OJ, Southall TD, Cheetham SW, Brand AH. Cell-type-specific profiling of protein-DNA interactions without cell isolation using targeted DamID with next-generation sequencing. Nature Protocols. 2016;11:1586–1598. doi: 10.1038/nprot.2016.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall OJ, Brand AH. damidseq_pipeline: an automated pipeline for processing DamID sequencing datasets. Bioinformatics. 2015;31:3371–3373. doi: 10.1093/bioinformatics/btv386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur D, Bost A, Driver I, Ohlstein B. A transient niche regulates the specification of Drosophila intestinal stem cells. Science. 2010;327:210–213. doi: 10.1126/science.1181958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews NE, White R. Chromatin architecture in the fly: living without CTCF/Cohesin loop extrusion?: alternating chromatin states provide a basis for domain architecture in Drosophila. BioEssays : News and Reviews in Molecular, Cellular and Developmental Biology. 2019;41:e1900048. doi: 10.1002/bies.201900048. [DOI] [PubMed] [Google Scholar]
- Mazumdar C, Shen Y, Xavy S, Zhao F, Reinisch A, Li R, Corces MR, Flynn RA, Buenrostro JD, Chan SM, Thomas D, Koenig JL, Hong WJ, Chang HY, Majeti R. Leukemia-Associated cohesin mutants dominantly enforce stem cell programs and impair human hematopoietic progenitor differentiation. Cell Stem Cell. 2015;17:675–688. doi: 10.1016/j.stem.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisenberg C, Pinder SI, Hopkins SR, Wooller SK, Benstead-Hume G, Pearl FMG, Jeggo PA, Downs JA. Repression of transcription at DNA breaks requires cohesin throughout interphase and prevents genome instability. Molecular Cell. 2019;73:212–223. doi: 10.1016/j.molcel.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkenschlager M, Nora EP. CTCF and cohesin in genome folding and transcriptional gene regulation. Annual Review of Genomics and Human Genetics. 2016;17:17–43. doi: 10.1146/annurev-genom-083115-022339. [DOI] [PubMed] [Google Scholar]
- Miguel-Aliaga I, Jasper H, Lemaitre B. Anatomy and physiology of the digestive tract of Drosophila melanogaster. Genetics. 2018;210:357–396. doi: 10.1534/genetics.118.300224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkovic M, Guilgur LG, Tavares A, Passagem-Santos D, Oliveira RA. Induced aneuploidy in neural stem cells triggers a delayed stress response and impairs adult life span in flies. PLOS Biology. 2019;17:e3000016. doi: 10.1371/journal.pbio.3000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales C, Losada A. Establishing and dissolving cohesion during the vertebrate cell cycle. Current Opinion in Cell Biology. 2018;52:51–57. doi: 10.1016/j.ceb.2018.01.010. [DOI] [PubMed] [Google Scholar]
- Mullenders J, Aranda-Orgilles B, Lhoumaud P, Keller M, Pae J, Wang K, Kayembe C, Rocha PP, Raviram R, Gong Y, Premsrirut PK, Tsirigos A, Bonneau R, Skok JA, Cimmino L, Hoehn D, Aifantis I. Cohesin loss alters adult hematopoietic stem cell homeostasis, leading to myeloproliferative neoplasms. The Journal of Experimental Medicine. 2015;212:1833–1850. doi: 10.1084/jem.20151323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neems DS, Garza-Gongora AG, Smith ED, Kosak ST. Topologically associated domains enriched for lineage-specific genes reveal expression-dependent nuclear topologies during myogenesis. PNAS. 2016;113:E1691–E1700. doi: 10.1073/pnas.1521826113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitzsche A, Paszkowski-Rogacz M, Matarese F, Janssen-Megens EM, Hubner NC, Schulz H, de Vries I, Ding L, Huebner N, Mann M, Stunnenberg HG, Buchholz F. RAD21 cooperates with pluripotency transcription factors in the maintenance of embryonic stem cell identity. PLOS ONE. 2011;6:e19470. doi: 10.1371/journal.pone.0019470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noutsou M, Li J, Ling J, Jones J, Wang Y, Chen Y, Sen GL. The cohesin complex is necessary for epidermal progenitor cell function through maintenance of Self-Renewal genes. Cell Reports. 2017;20:3005–3013. doi: 10.1016/j.celrep.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novo CL, Javierre BM, Cairns J, Segonds-Pichon A, Wingett SW, Freire-Pritchett P, Furlan-Magaril M, Schoenfelder S, Fraser P, Rugg-Gunn PJ. Long-Range enhancer interactions are prevalent in mouse embryonic stem cells and are reorganized upon pluripotent state transition. Cell Reports. 2018;22:2615–2627. doi: 10.1016/j.celrep.2018.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B, Spradling A. Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science. 2007;315:988–992. doi: 10.1126/science.1136606. [DOI] [PubMed] [Google Scholar]
- Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE. Sister chromatid cohesion: a simple concept with a complex reality. Annual Review of Cell and Developmental Biology. 2008;24:105–129. doi: 10.1146/annurev.cellbio.24.110707.175350. [DOI] [PubMed] [Google Scholar]
- Pauli A, van Bemmel JG, Oliveira RA, Itoh T, Shirahige K, van Steensel B, Nasmyth K. A direct role for cohesin in gene regulation and ecdysone response in Drosophila salivary glands. Current Biology. 2010;20:1787–1798. doi: 10.1016/j.cub.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pękowska A, Klaus B, Xiang W, Severino J, Daigle N, Klein FA, Oleś M, Casellas R, Ellenberg J, Steinmetz LM, Bertone P, Huber W. Gain of CTCF-Anchored chromatin loops marks the exit from naive pluripotency. Cell Systems. 2018;7:482–495. doi: 10.1016/j.cels.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Gräf S, Flicek P, Kerkhoven RM, van Lohuizen M, Reinders M, Wessels L, van Steensel B. Molecular maps of the reorganization of genome-nuclear Lamina interactions during differentiation. Molecular Cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Tedeschi A, Schmitz J. The cohesin complex and its roles in chromosome biology. Genes & Development. 2008;22:3089–3114. doi: 10.1101/gad.1724308. [DOI] [PubMed] [Google Scholar]
- Phillips-Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, Lajoie BR, Bell JS, Ong CT, Hookway TA, Guo C, Sun Y, Bland MJ, Wagstaff W, Dalton S, McDevitt TC, Sen R, Dekker J, Taylor J, Corces VG. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Bhardwaj V, Arrigoni L, Lam KC, Grüning BA, Villaveces J, Habermann B, Akhtar A, Manke T. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nature Communications. 2018;9:189. doi: 10.1038/s41467-017-02525-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huang SC, Glenn st hilaire B, Engreitz JM, Perez EM, Kieffer-Kwon KR, Sanborn AL, Johnstone SE, Bascom GD, Bochkov ID, Huang X, Shamim MS, Shin J, Turner D, Ye Z, Omer AD, Robinson JT, Schlick T, Bernstein BE, Casellas R, Lander ES, Aiden EL. Cohesin loss eliminates all loop domains. Cell. 2017;171:e24. doi: 10.1016/j.cell.2017.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resende LP, Monteiro A, Brás R, Lopes T, Sunkel CE. Aneuploidy in intestinal stem cells promotes gut dysplasia in Drosophila. The Journal of Cell Biology. 2018;217:3930–3946. doi: 10.1083/jcb.201804205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnik-Docampo M, Koehler CL, Clark RI, Schinaman JM, Sauer V, Wong DM, Lewis S, D'Alterio C, Walker DW, Jones DL. Tricellular junctions regulate intestinal stem cell behaviour to maintain homeostasis. Nature Cell Biology. 2017;19:52–59. doi: 10.1038/ncb3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi L, Benitah SA. Epigenetic regulation of adult stem cell function. FEBS Journal. 2015;282:1589–1604. doi: 10.1111/febs.12946. [DOI] [PubMed] [Google Scholar]
- Roberts A, Pimentel H, Trapnell C, Pachter L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics. 2011a;27:2325–2329. doi: 10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
- Roberts A, Trapnell C, Donaghey J, Rinn JL, Pachter L. Improving RNA-Seq expression estimates by correcting for fragment Bias. Genome Biology. 2011b;12:R22. doi: 10.1186/gb-2011-12-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudra S, Skibbens RV. Cohesin codes - interpreting chromatin architecture and the many facets of cohesin function. Journal of Cell Science. 2013;126:31–41. doi: 10.1242/jcs.116566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf CA, Kwak H, Koenig A, Misulovin Z, Gohara DW, Watson A, Zhou Y, Lis JT, Dorsett D. Genome-wide control of RNA polymerase II activity by cohesin. PLOS Genetics. 2013;9:e1003382. doi: 10.1371/journal.pgen.1003382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt AD, Hu M, Jung I, Xu Z, Qiu Y, Tan CL, Li Y, Lin S, Lin Y, Barr CL, Ren B. A compendium of chromatin contact maps reveals spatially active regions in the human genome. Cell Reports. 2016;17:2042–2059. doi: 10.1016/j.celrep.2016.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer W, Abdennur N, Goloborodko A, Pekowska A, Fudenberg G, Loe-Mie Y, Fonseca NA, Huber W, Haering CH, Mirny L, Spitz F. Two independent modes of chromatin organization revealed by cohesin removal. Nature. 2017;551:51–56. doi: 10.1038/nature24281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Cavalli G. The role of chromosome domains in shaping the functional genome. Cell. 2015;160:1049–1059. doi: 10.1016/j.cell.2015.02.040. [DOI] [PubMed] [Google Scholar]
- Signer RA, Morrison SJ. Mechanisms that regulate stem cell aging and life span. Cell Stem Cell. 2013;12:152–165. doi: 10.1016/j.stem.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh VP, Gerton JL. Cohesin and human disease: lessons from mouse models. Current Opinion in Cell Biology. 2015;37:9–17. doi: 10.1016/j.ceb.2015.08.003. [DOI] [PubMed] [Google Scholar]
- Sumara I, Vorlaufer E, Stukenberg PT, Kelm O, Redemann N, Nigg EA, Peters JM. The dissociation of cohesin from chromosomes in prophase is regulated by Polo-like kinase. Molecular Cell. 2002;9:515–525. doi: 10.1016/S1097-2765(02)00473-2. [DOI] [PubMed] [Google Scholar]
- Szabo Q, Bantignies F, Cavalli G. Principles of genome folding into topologically associating domains. Science Advances. 2019;5:eaaw1668. doi: 10.1126/sciadv.aaw1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teves SS, An L, Hansen AS, Xie L, Darzacq X, Tjian R. A dynamic mode of mitotic bookmarking by transcription factors. eLife. 2016;5:e22280. doi: 10.7554/eLife.22280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nature Biotechnology. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Weerdt BC, van Vugt MA, Lindon C, Kauw JJ, Rozendaal MJ, Klompmaker R, Wolthuis RM, Medema RH. Uncoupling anaphase-promoting complex/cyclosome activity from spindle assembly checkpoint control by deregulating polo-like kinase 1. Molecular and Cellular Biology. 2005;25:2031–2044. doi: 10.1128/MCB.25.5.2031-2044.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Sun Q, Czajkowsky DM, Shao Z. Sub-kb Hi-C in D. Melanogaster reveals conserved characteristics of TADs between insect and mammalian cells. Nature Communications. 2018;9:188. doi: 10.1038/s41467-017-02526-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz G, Várnai C, Nagasaka K, Cisneros DA, Stocsits RR, Tang W, Schoenfelder S, Jessberger G, Muhar M, Hossain MJ, Walther N, Koch B, Kueblbeck M, Ellenberg J, Zuber J, Fraser P, Peters JM. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. The EMBO Journal. 2017;36:3573–3599. doi: 10.15252/embj.201798004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Yan Y, Deb S, Rangasamy D, Germann M, Malaterre J, Eder NC, Ward RL, Hawkins NJ, Tothill RW, Chen L, Mortensen NJ, Fox SB, McKay MJ, Ramsay RG. Cohesin Rad21 mediates loss of heterozygosity and is upregulated via wnt promoting transcriptional dysregulation in gastrointestinal tumors. Cell Reports. 2014;9:1781–1797. doi: 10.1016/j.celrep.2014.10.059. [DOI] [PubMed] [Google Scholar]
- Yan J, Enge M, Whitington T, Dave K, Liu J, Sur I, Schmierer B, Jolma A, Kivioja T, Taipale M, Taipale J. Transcription factor binding in human cells occurs in dense clusters formed around cohesin anchor sites. Cell. 2013;154:801–813. doi: 10.1016/j.cell.2013.07.034. [DOI] [PubMed] [Google Scholar]
- Yuen KC, Gerton JL. Taking cohesin and condensin in context. PLOS Genetics. 2018;14:e1007118. doi: 10.1371/journal.pgen.1007118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Han L, Singh SR, Liu H, Neumüller RA, Yan D, Hu Y, Liu Y, Liu W, Lin X, Hou SX. Genome-wide RNAi screen identifies networks involved in intestinal stem cell regulation in Drosophila. Cell Reports. 2015;10:1226–1238. doi: 10.1016/j.celrep.2015.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuin J, Dixon JR, van der Reijden MI, Ye Z, Kolovos P, Brouwer RW, van de Corput MP, van de Werken HJ, Knoch TA, van IJcken WF, Grosveld FG, Ren B, Wendt KS. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. PNAS. 2014;111:996–1001. doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]