Abstract
Interactions between corals and their associated microbial communities (Symbiodiniaceae and prokaryotes) are key to understanding corals' potential for and rate of acclimatory and adaptive responses. However, the establishment of microalgal and bacterial communities is poorly understood during coral ontogeny in the wild. We examined the establishment and co‐occurrence between multiple microbial communities using 16S rRNA (bacterial) and ITS2 rDNA (Symbiodiniaceae) gene amplicon sequencing in juveniles of the common coral, Acropora tenuis, across the first year of development. Symbiodiniaceae communities in juveniles were dominated by Durusdinium trenchii and glynnii (D1 and D1a), with lower abundances of Cladocopium (C1, C1d, C50, and Cspc). Bacterial communities were more diverse and dominated by taxa within Proteobacteria, Cyanobacteria, and Planctomycetes. Both communities were characterized by significant changes in relative abundance and diversity of taxa throughout the year. D1, D1a, and C1 were significantly correlated with multiple bacterial taxa, including Alpha‐, Deltra‐, and Gammaproteobacteria, Planctomycetacia, Oxyphotobacteria, Phycisphaerae, and Rhizobiales. Specifically, D1a tended to associate with Oxyphotobacteria and D1 with Alphaproteobacteria, although these associations may represent correlational and not causal relationships. Bioenergetic modeling combined with physiological measurements of coral juveniles (surface area and Symbiodiniaceae cell densities) identified key periods of carbon limitation and nitrogen assimilation, potentially coinciding with shifts in microbial community composition. These results demonstrate that Symbiodiniaceae and bacterial communities are dynamic throughout the first year of ontology and may vary in tandem, with important fitness effects on host juveniles.
Keywords: 16S rRNA gene, co‐occurrence, coral, coral‐associated bacteria, ITS2 rDNA gene, juvenile, Symbiodiniaceae
We characterized the establishment and maintenance of bacterial and Symbiodiniaceae communities in the coral Acropora tenuis across the first year of juvenile ontogeny, identifying potentially key co‐occurrence patterns using network modeling. Bioenergetic modeling combined with physiological measurements of coral juveniles (surface area and Symbiodiniaceae cell densities) identified periods of carbon limitation and nitrogen assimilation, potentially coinciding with shifts in microbial community composition. These dynamic communities vary in tandem with important fitness effects on host juvenile corals.

1. INTRODUCTION
Scleractinian corals are key habitat engineers of one of the most ecologically diverse, economically valuable, but climatically vulnerable ecosystems on the planet (Bellwood, Hughes, Folke, & Nystrom, 2004). The sensitivity of reef‐building corals to contemporary rates of environmental change in an era of ever‐increasing anthropogenic influence on coastal ecosystems has led to alarming declines in coral reefs globally (De'ath & Fabricius, 2010; Hoegh‐Guldberg & Bruno, 2010; Hughes et al., 2003; Thompson & Dolman, 2010). A key contributor to coral health and survival under rapid environmental change is the corals' microbial community, particularly Symbiodiniaceae (Baird, Bhagooli, Ralph, & Takahashi, 2009) and potentially bacterial (Bourne, Morrow, & Webster, 2016) communities that respond more rapidly to change than their coral hosts (Torda et al., 2017). Although coral‐Symbiodiniaceae symbioses and coral‐bacterial associations have been extensively studied, few have focussed on the potential tripartite linkages among all three of these components of the coral holobiont. Documenting early stages in the development of coral‐microbe symbioses is an important step in identifying potential linkages in these complex symbioses. Such knowledge will lay the foundations for understanding how the corals' microbiome contributes to whole organism fitness and potential resilience when faced with the cumulative effects of ocean warming and other anthropogenic stressors.
Metazoans host large numbers of microbial associates that influence their host's phenotype in associations that range from mutualistic to parasitic (Salvucci, 2016). These associations are integral to host health and performance, although they may also influence each partner's evolutionary trajectories and adaptive potential (Hoang, Morran, & Gerardo, 2016; Torda et al., 2017). Increases in seawater temperatures of as little as 1–2°C above summer means disrupt the symbioses between corals and their microbes, resulting in mass coral bleaching (Hughes et al., 2018, 2017) and coral disease outbreaks (Brodnicke et al., 2019). Correlations between bacterial and Symbiodiniaceae community structures in some adult corals are likely due to coupled nutrient cycling (Bourne et al., 2013), for example, changes in carbon and nitrogen ratios in the environment (Cooper et al., 2011), which may influence symbiont community population dynamics within corals (Morris, Voolstra, Quigley, Bourne, & Bay, 2019). Thus, even fine‐scale changes in the diversity or abundance of symbionts can impact the coral host's physiology (Cunning, Silverstein, & Baker, 2015) due to variability in nutrient sharing and thermal tolerances across various partners (Karim, Nakaema, & Hidaka, 2015; Swain, Chandler, Backman, & Marcelino, 2016). In some cases, the co‐occurrence of some symbionts appears to be unrelated to host phylogenetic history or biogeographical area (Bernasconi, Stat, Koenders, & Huggett, 2018). In other cases, host/bacterial communities demonstrate phylosymbiotic patterns (e.g., members from Endozoicomonas) (O'Brien, Webster, Miller, & Bourne, 2019; Pollock et al., 2018). Overall, the intimacy of these associations suggests that the holobiont's biology can only be understood when the interactions of all members are considered (Bordenstein & Theis, 2015; Bosch & McFall‐Ngai, 2011; Gordon, Knowlton, Relman, Rohwer, & Youle, 2013).
The acquisition and development of bacterial and Symbiodiniaceae communities in juvenile varies throughout early stages of coral ontogeny (Abrego, van Oppen, & Willis, 2009a, 2009b; Fieth, Gauthier, Bayes, Green, & Degnan, 2016; Lema, Bourne, & Willis, 2014; Quigley, Bay, & Willis, 2017; Quigley, Willis, & Bay, 2017) likely regulated by both host and environmental factors (Poland & Coffroth, 2017; Quigley, Bay, et al., 2017; Quigley, Willis, et al., 2017; Souza et al., 2016). In the few species examined to date, the composition of bacterial communities is largely influenced by acquisition from the environmental pool and to a lesser extent by vertical, parental transmission (Apprill, Marlow, Martindale, & Rappé, 2009; Epstein, Torda, Munday, & van Oppen, 2019; Leite et al., 2017; Lema et al., 2014; Sharp, Distel, & Paul, 2012; Zhou et al., 2017). Once acquired, community shifts are common throughout ontogeny across a range of invertebrate groups (Douglas & Werren, 2016). For example, in corals, both Symbiodiniaceae (Abrego, van Oppen, & Willis, 2009b) and bacterial community shifts (Lema et al., 2014) have been demonstrated to be triggered by changes in immunocompetency or environmental conditions (Littman, Willis, & Bourne, 2009). Immune system development (Hoang et al., 2016) and host factors (e.g., quorum quenching; Grandclément, Tannières, Moréra, Dessaux, & Faure, 2016; Kuniya et al., 2015; Souza et al., 2016) also regulate endosymbiotic community flux in addition to environmental factors such as temperature, pCO2, or salinity (Glasl, Smith, Bourne, & Webster, 2019). The regulation of the microbiome's organization by the host during development (Davy, Allemand, & Weis, 2012) or inter‐symbiont competition (also partly host regulated) (Cunning et al., 2015) may also influence these shifts. Although a model for “holobiont” functioning and feedback loops has been proposed (Rohwer, Seguritan, Azam, & Knowlton, 2002), it has not been utilized for coral juvenile development. Despite the importance of these microbial communities and their capacity to enhance host acclimation and adaptation potential, there is limited understanding of how bacterial and Symbiodiniaceae communities potentially influence each other and co‐develop in association with host ontogeny in the wild.
In this study, we determined co‐occurrence patterns and population dynamics of Symbiodiniaceae and bacterial communities during the first year of life in juveniles of the hard coral species, Acropora tenuis, at an inshore location on the Great Barrier Reef. These data provide key baseline information of changes in microbial communities associated with juvenile coral ontogeny and shed light on key stages of symbiosis establishment and maintenance, with implications for host functioning and potential for adaptation.
2. MATERIALS AND METHODS
2.1. Spawning and larval rearing
Fourteen colonies of A. tenuis were collected from Geoffrey Bay, Magnetic Island (S 19°09.326′, E 146°51.861′) on the full moon in November 2014 (Great Barrier Reef Marine Park Authority Permit Number: G13/36318.1) and transported to the National Sea Simulator Facility (SeaSim) at the Australian Institute of Marine Sciences (AIMS). Corals were maintained in outside aquaria with constant flow through 0.1‐µm filtered seawater (FSW) and checked nightly for signs of imminent spawning. Gametes were collected on the fourth night after the full moon from 14 colonies. Eggs and sperm from each colony were mixed in equal proportions and left to fertilize for two hours, with periodic checks of fertilization. After >85% of the eggs had cleaved, fertilized eggs were gently collected and moved into 0.1‐µm FSW for a total of two washes to remove residual sperm. Once past the early gastrula stage, embryos were moved into three 500‐L culture tanks with a continuous, gentle flow of 26.5°C FSW, and low aeration. Aeration was then increased once embryos had matured into larvae.
2.2. Settlement
Approximately 3 months prior to spawning, 660 terracotta tiles were placed in large aquaria at the SeaSim for conditioning, that is, development of crustose coralline algae (CCA) communities and bacterial biofilms. Two days prior to larval settlement, the tiles were autoclaved at 2,000 kPa for 20 min at 180°C (Sabac T63) for sterilization. To help induce settlement and metamorphosis, CCA chips were removed from live rock, autoclaved, as described above, and kept at 4°C until ready for use. Once larvae were competent to settle (7 days postspawning), tiles were strung vertically on new flexible PVC aeration tubing in batches of 20, with 2.5‐cm PVC spacers used to separate tiles. Nine to ten strings of tiles were then hung vertically in each of the three 500‐L culture tanks. One milliliter of CCA slurry, composed of ~1 × 1 cm pieces of autoclaved CCA in FSW, was pipetted onto each tile to attract larvae and induce settlement. Larvae were left to settle for 11 days, during which time water flow (0.1‐µm FSW) was kept constant with low aeration to ensure optimal water conditions.
2.3. Field deployment
Settlement on each tile was assessed visually, and tiles containing juveniles were strung onto stainless steel rods in groups of 15 separated by PVC spacers. Nineteen days postspawning, 240 tiles, each with 40–200 juveniles, were transported to Magnetic Island in FSW. Nine rods (135 tiles) and 7 rods (105 tiles) were deployed at two sites at Magnetic Island (Geoffrey Bay and Nelly Bay), respectively, at a depth of approximately 6 m and secured with zip ties between pairs of vertical pickets hammered into the substratum. Fluorescent microscopy was used to confirm the absence of Symbiodiniaceae cells prior to field deployment. Juveniles were sampled from tiles on the 19th of each month from 8:30 to 10:30 a.m. The age of juveniles at each sampling time point was as follows: 40 days (T1), 71 (T2,), 111 (T3), 129 (T4), 167 (T5), 194 (T6), 316 (T7), and 380 (T8). Individual juveniles were placed into 100% ethanol or FSW. No samples were collected during the months of June, July, and August due to repeated sightings of a saltwater crocodile at the field site.
2.4. Surface area determination
Between 6 and 17 juveniles were photographed at each time point using a Leica microscope with a Leica MC170 HD lens (Switzerland) and the Leica Application Suite (v.4.4.0, Switzerland) for surface area determination. This method was adequate while the juveniles were flat. However, juveniles sampled in April began to present more complex branching patterns that could not be captured by the Leica Application Suite or were partially obscured while taking the photograph. 3D modeling, as used to estimate surface area of adult corals (Jones, Berkelmans, Berkelmans, van Oppen, Mieog, & Sinclair, 2008; Jones, Cantin, Berkelmans, Sinclair, & Negri, 2008), was then employed to capture the geometries of these more complex juvenile corals. Juveniles were measured using 3D scanning with the NextEngine scanner (NextEngine) and processed using NextEngine ScanStuidoPro (v. 2.0.2). Each juvenile was scanned twice using the 360° horizontal scan with 14 divisions and two bracket scans at 160,000 points per inch (HD resolution). Resulting.STL files were then imported into Geomagic Verify Viewer software (v. 2.0) to calculate surface area after trimming, polishing to fill holes, and then fusing. As described further below, surface area measurements were matched with Symbiodiniaceae cell counts and DNA extractions for each juvenile.
2.5. Symbiodiniaceae cell counts
Live juveniles were transported back to the SeaSim on tiles for size determination through either microscopy or 3D scanning. After size determination, juveniles were removed from tiles, and then spun down at 16,000 rcf for 90 s in seawater without fixative, after which the seawater was replaced with 5% formic acid and samples left for two hours for digestion. Eppendorf tubes containing juvenile tissue were then centrifuged again on the same settings, and the formic acid replaced with filtered seawater, well mixed, and then aliquoted into a hemocytometer for cell counts. Three hemocytometer plates were counted for each juvenile, corresponding to six‐well counts. The number of Symbiodiniaceae cells for each individual juvenile was then standardized to the surface area that had been measured for that exact juvenile (cells per mm2).
2.6. Symbiodiniaceae and bacterial community genotyping
Ten individual juveniles per time point were sequenced using Illumina technology targeting the Symbiodiniaceae ITS2 locus (n = 80). Within each time point, DNA was extracted from whole, single juveniles following SDS extraction (Wilson et al., 2002), including a bead‐beating step that was performed before water‐bath incubation using 1‐mm silica spheres (MPBio) on a FastPrep‐24 (MPBio) for three cycles of 30 s at 4.0 m/s, with the same DNA extract, sourced from the same individual used for cell counts, ITS2, and 16S rRNA gene sequencing. Library preparation and paired‐end Miseq sequencing (Illumina) were performed at the University of Texas at Austin's Genomics Sequencing and Analysis Facility (USA) using the following primers: ITS2alg‐F (5′‐TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGTGAATTGCAGAACTCCGTG) and ITS2alg‐R (3′‐TTCGTATATTCATTCGCCTCCGACAGAGAATATGTGTAGAGGCTCGGGTGCTCTG‐5′) (Pochon, Pawlowski, Zaninetti, & Rowan, 2001).
Bacterial sequencing was performed on the same DNA extracts and sequenced with the 16S_V1‐V3 (27f‐519r) primers (Bioplatforms Australia Marine Microbes primers): 27f (5′‐AGAGTTTGATCMTGGCTCAG) and 519R (5′ GWATTACCGCGGCKGCTG) at the Ramaciotti Centre for Genomics (Australia). Raw reads were analyzed using the DADA2 pipeline (Callahan et al., 2016). Full details and scripts can be found in Quigley, Willis, and Kenkel (2019). Cleaned and mapped sequence data were variance normalized, using the “DESeq2” package in R (Love, Huber, & Anders, 2014). Differential relative abundance analysis was performed with the same package using the contrast function to extract specific comparisons from the negative binomial generalized linear model. Nonmetric multidimensional scaling (NMDS) was performed on variance normalized ASVs (amplicon sequence variants) relative abundances using the packages “Phyloseq,” “vegan,” and “ellipse” with a Bray–Curtis dissimilarity matrix (McMurdie & Holmes, 2013; Murdoch, Chow, & Celayeta, 2007; Oksanen et al., 2013). NMDS analysis does not assume linear relationships between underlying variables and ASVs, resulting in distances between samples that are indicative of their similarity, with samples positioned closer in space being more similar (Ramette, 2007). The use of a Bray–Curtis matrix allows for the incorporation of the presence/absence of ASVs as well as their relative abundance. Permutational multivariate analysis of variance was used to determine if Symbiodiniaceae communities differed significantly between time points using the “adonis” function in “vegan.” Explanation of variance was calculated using the “stressplot” function in “vegan.” To collapse taxonomic complexity, mean percentage values were calculated at the level of bacterial Order and Symbiodiniaceae Class. Pearson's product‐moment correlation coefficients and respective p values were calculated per time point to test for co‐occurrence between each pairwise combination of bacterial and Symbiodiniaceae ASVs using the “Hmisc” package with the “rcorr” function (Harrell, 2007). Correlations were filtered to retain only those that were significant (p < .05).
2.7. Juvenile—Symbiodiniaceae bioenergetics modeling
Estimating juvenile‐Symbiodiniaceae bioenergetics was calculated using the “coRal” package (Cunning, Muller, Gates, & Nisbet, 2017). This model incorporates carbon and nitrogen budgets empirically measured across a range of coral taxa and Symbiodiniaceae associations. Assumptions include preferential sharing of nutrients and direct access to carbon by Symbiodiniaceae via host and nitrogen fluxes to the host via the environment. Time for model run was set from 0 to 380 days (T0: 9/11/2014; T8: 24/11/2015) with light varying from 0.3 to 42 µmol photons m2 d−1 to estimate changing light environments as juveniles grow from shaded crevices (Doropoulos et al., 2016) to more exposed, high‐light positions over time (Stimson, 1997). The final ratio of symbiont cells to host surface area for day 380 was used as a starting value (3,526.5 cells per mm2). Dissolved inorganic nitrogen (DIN) and heterotrophy and initial parameters were set to default values (Cunning et al., 2017).
2.8. Networks
Networks were constructed using the packages “igraph” and “phyloseq” (Csardi & Nepusz, 2006; McMurdie & Holmes, 2013). Igraph constructs networks by calculating Euclidean distances between points using nodes (ASVs in this case) and edges (line connections) derived from the relative abundance datasets. Relative abundances were converted to Bray–Curtis distances, with a maximum distance set at max.dist = 0.7–0.9, where separation was undirected.
3. RESULTS
3.1. Physiology
Juveniles exhibited exponential growth over the 380 days of sampling (Figure 1a), growing in size from 1.2 mm2 ± 0.07 (n = 6) in the first month to 635 mm2 ± 243.9 (n = 9) by the final time point. Symbiodiniaceae density was initially low, and variable in the first month (789 ± 624 cells per mm2) increased rapidly to ~2.2 × 104 cells per mm2 over the next 3 months, but then decreased to densities just below 104 cells per mm2 and appeared to stabilize at lower densities for the last two time points until juveniles were 380‐days old (Figure 1a).
Figure 1.
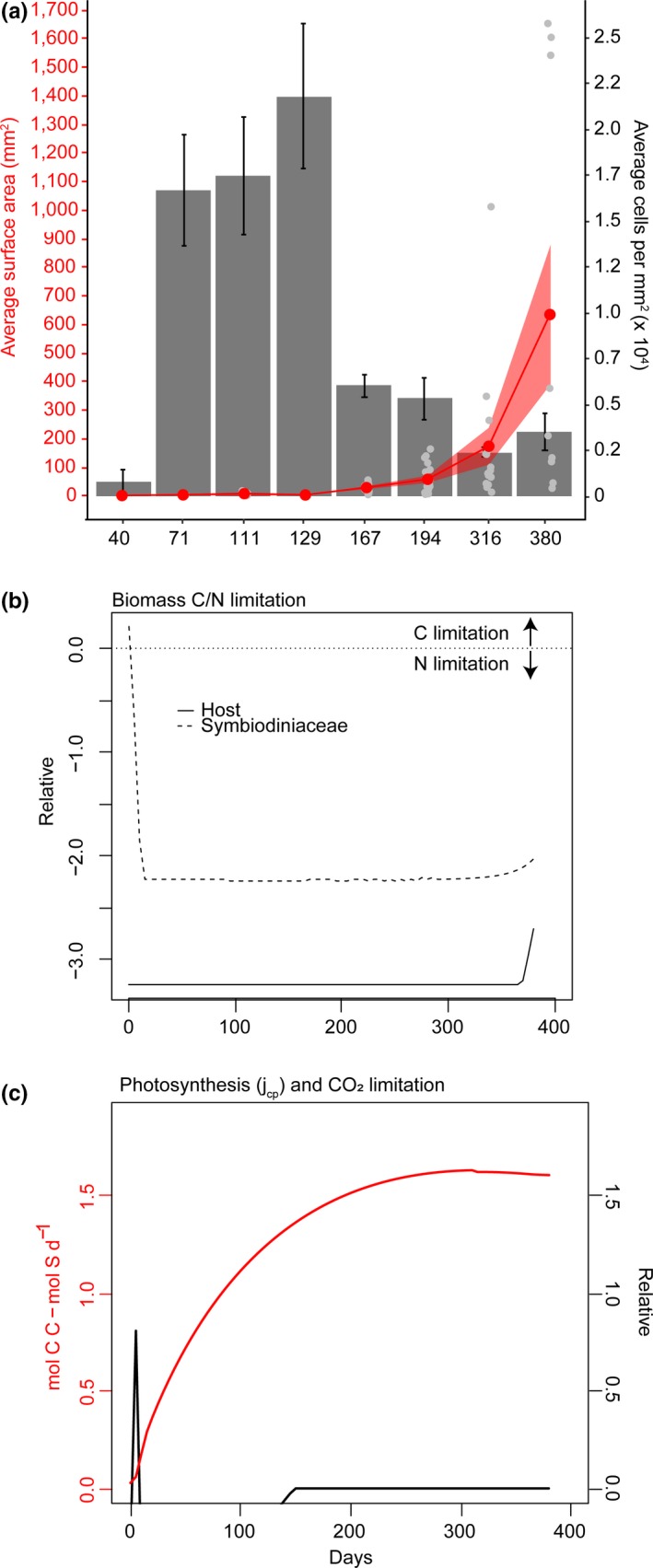
Physiological (a) and bioenergetic model outputs (b, c) for Acropora tenuis juveniles outplanted in the field for 1 year. (a) Average surface area of juveniles (red line; mm2, red point means ± SE) and Symbiodiniaceae cell densities (average cells in ×104, grey barplots ± SE). (b) Relative ratio of carbon to nitrogen limitation in the host and Symbiodiniaceae populations modelled over 0–400 days of growth in the field. Solid and dashed lines represent host and Symbiodiniaceae values, respectively. Relative ratios are limitation coefficients that are unbounded by zero, where negative values are indicative of formation rates that are lower than maximum production rates in either the host or symbiont (negative: nitrogen limitation over carbon; positive: carbon limitation over nitrogen; zero: neither is limiting). (c) Estimated CO2 limitation (black line) (relative units as this is a ratio of substrate input fluxes) and photosynthesis (red line; mol C C ‐ mol S d−1) for juveniles modelled over 400 days. Relative values represent limitation coefficients, with zero signifying that neither carbon nor nitrogen are limited (Cunning et al., 2017)
Bioenergetic modeling revealed that juveniles experienced an initial, brief period of carbon limitation during Symbiodiniaceae establishment (which is common at low symbiont densities typically seen during bleaching), but then intracellular nitrogen rapidly became limited as algal densities increased (Figure 1b). Initial carbon limitation was driven by CO2 limitation (relative units of the ratio of substrate input fluxes), which dissipated after ~130 days, potentially when symbiont densities equilibrated at around 104 cells per mm2. Photosynthesis increased rapidly with cell proliferation and then reached an asymptote at a “healthy” level of ~1.5 mol C C ‐ mol S d−1 (Figure 1c), after about 200 – 250 days of proliferation, coinciding with the point when growth became exponential in coral juveniles.
3.2. Symbiodiniaceae population shifts across ontogeny
Multiple Symbiodiniaceae from Cladocopium and Durusdinium associated with tissues of A. tenuis were highly abundant in the first year of juvenile development (Figure 2). At day 40 (T1), Symbiodiniaceae communities were variable among juveniles (light orange hull, Figure 2 NMDS inset 1), but converged across individuals throughout the remaining months, becoming the most conserved at the final time point (yellow hull, Figure 2). In general, Symbiodiniaceae communities associated with juveniles were dominated by D1 and D1a, across all time points sampled, with these taxa making up 25% and 17% of all Symbiodiniaceae ASVs. Other taxa from Durusdinium were also detected, but at lower relative abundances, including D5 and D9, followed by D6 and D2.2 (4.8–0.3% of ASVs). Taxa belonging to C1 (4.5%) were also prevalent over the sampling period, with C3, C15, C1d, C50, Cspc, and taxa from clades F and I detected in low but consistent abundances associated with A. tenuis juveniles. The Symbiodiniaceae community structure varied significantly over time (permutational multivariate analysis of variance (PERMANOVA): Df7, F = 1.73, R 2 = 0.14, p = .01), with time explaining 14% of the variance in community structure. These changes were driven by the varying dominant abundances of D1 and D1a as well as changes to the diversity in background Symbiodiniaceae through time. For example, juveniles were initially dominated at day 40 (T1) by D1 (31.5%) and lower abundances of D1a (8.5%). This was followed by a significant increase and co‐dominance of D1a with D1 until the last time points in which D1 re‐established dominance within the coral juveniles.
Figure 2.
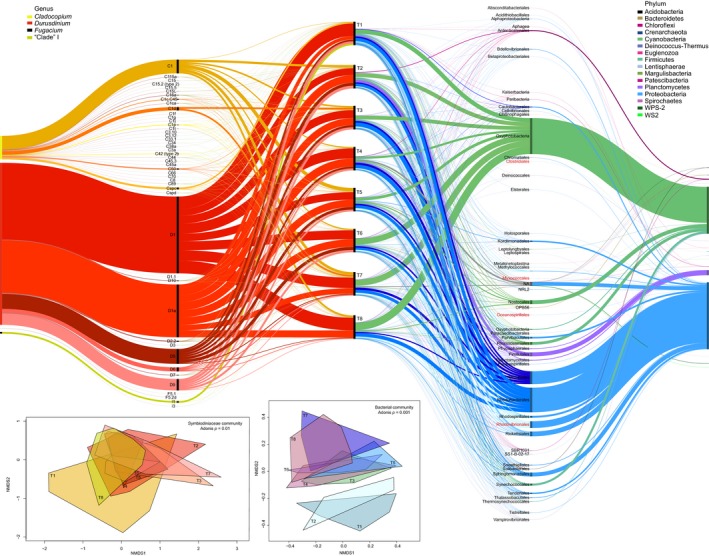
Alluvial plot of variance‐normalized relative abundances of Symbiodiniaceae (warm colors) and bacterial ASVs (cool colors) across one year of growth in Acropora tenuis juveniles outplanted to Magnetic Island. Timepoints (T1–T8) are represented in the middle section with black bars (time axis), where taxa are split to show the relative abundances of each at each of the time points. Abundances for Symbiodiniaceae taxa feed to the time axis from left and bacterial taxa feed in from the right, increasing in taxonomic resolution. Insets: Nonmetric multidimensional scaling (NMDS) of variance‐normalized abundances of Symbiodiniaceae (warm colors) and bacterial ASVs (cool colors) constructed using Bray‐Curtis distances. Only taxa greater than 0.01% are shown (hence rare taxa are excluded), with cut‐offs defined (Galand, Casamayor, Kirchman, & Lovejoy, 2009; Logares et al., 2014; Pedrós‐Alió, 2012). Bacterial taxa in red are those that have been identified as key mutualistic and parasitic mutualists in corals (Neave, Rachmawati, et al., 2016; Pollock et al., 2018)
3.3. Bacterial population shifts across ontogeny
Bacterial communities varied significantly among juveniles over time (PERMANOVA: Df7, F = 1.23, R 2 = 0.16, p = .001), with time explaining 16% of the variance in community structure. In comparison to Symbiodiniaceae communities, bacterial communities at the first and second time points were distinct relative to all other time points (Figure 2 NMDS inset 2, light blue and teal hulls), converged in diversity and abundance in T3 through T7, before diverging again at the final time point (T8) (dark pink hull).
A diverse bacterial community was associated with the early life stages of A. tenuis juveniles, with sequences affiliated across multiple taxa from the phyla Proteobacteria, Cyanobacteria, and Planctomycetes present at high relative abundances, although other key phyla were also detected, including Firmicutes and Bacteroidetes at lower abundances (Figure 2). Within the Proteobacteria, the dominant sequences were affiliated with both Alphaproteobacteria (43.5%, e.g., Rhodobacteria, Rhizobiales, Rickettsiales, Rhodovibrionales, Rhodospirillales, Caulobacterales, and Kordiimonadales) and Gammaproteobacteria (0.5%, e.g., Alteromonadales and Oceanospirillales). Specifically, 22, 39, and three ASVs were detected for Oceanospirillales (potential Endozoicomonas), Roseobacter, and Burkholderia. Sequences belonging to Litoribrevibacter, Balneatrix, Oleiphilus, and Kiloniellales (NCBI accession: KU688966.1) within Oceanospirillales (and related to the Endozoicomonas genus) were also commonly retrieved from the A. tenuis juveniles through the study period. ASVs from Oceanospirillales were associated with juveniles across sampling time points from T3 to T8, increasing in relative abundance from 0.03% to 0.2% at the final time point. Alphabacterial sequences belonging to the Roseobacter were detected in all time points at background abundances (0.2%–0.9%), while Burkholderia affiliated sequences were retrieved from two juveniles only at T6 and T7 (0.03%–0.08% relative abundance, respectively). Oxyphotobacteria (Cyanobacteria) was one of the most dominant overall (30%) ASVs associated with A. tenuis juveniles, becoming increasingly abundant throughout the year (T1: 3.4 to T8: 6.1%). Conversely, other taxa decreased in relative abundance throughout the year, including Pirellulales—Planctomycetes (T1: 0.04 to T8: 0.01%), Caulobacterales (T1: 0.3 to T8: 0.02%), and Rhodospirillales affiliated sequences (T1: 0.5 to T8: 0.2%).
3.4. Co‐occurrence
The first month of development for A. tenuis juveniles was typified by slow growth and initially low and variable Symbiodiniaceae cell densities. At this time point, Durusdinium taxa were more frequently correlated with specific bacterial taxa (e.g., Proteobacteria) compared to other Symbiodiniaceae genera (Figure 3). For example, D1 was highly positively correlated with Rhodobacterales (Alphaproteobacteria), but D1 was negatively correlated with D9. Co‐occurrence frequently occurred between D1, D1a, D9, D5, and sequences within the classes Alphaproteobacteria, Oxyphotobacteria, Gammaproteobacteria, Planctomycetacia, and Phycisphaerae (Figure 4a, co‐occurring nodes 1–3). Leptolyngbyales (Cyanobacteria) were also highly positively correlated with multiple Durusdinium sampled in T1 and T2. The most connected taxa were Sphingomonadales (degree = 635 connections), indicating many first‐degree connections to other taxa. Other important hubs included Ruegeria (Proteobacteria—Alphaproteobacteria) and Synechococcus (Oxyphotobacteria—Cyanobacteria). Similar to the correlation analysis, very few Cladocopium co‐occurred with bacterial taxa at this earliest time point.
Figure 3.
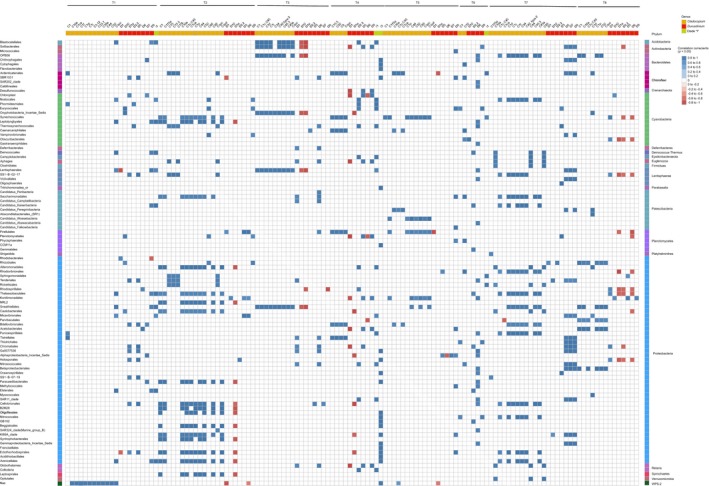
Heatmap of significant Pearson's product‐moment correlation coefficients (p < .05) between Symbiodiniaceae and bacterial taxa
Figure 4.
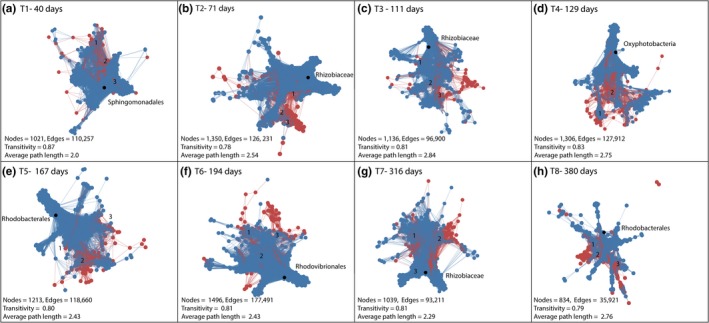
Network analysis of Symbiodinaceae and bacterial co‐occurance across 1 year of growth for Acropora tenuis juveniles
Juveniles remained small over the subsequent months, while Symbiodiniaceae cell densities increased rapidly. During these time points, connectedness between taxa was greater at T2 (71 days) and T3 (111 days) compared to T1, with the Rhizobiaceae (Proteobacteria) being the most connected taxa at both time points (Degree = 497 and 415, respectively). The diversity of correlations also increased after T1, with both Cladocopium and Durusdinium associating with many bacterial taxa at the later time points T2 and T3 (Figure 3). For example, Alphaproteobacteria, Planctomycetacia, and Oxyphotobacteria typically associated with Cladocopium (C1, C115a, C1c.C45, C1ca, C44, Cspd, and C50) (Figure 4b node 1) or D2.2, D5, and D1a (node 2) or D9 and D1 (node 3). While many more bacterial and Symbiodiniaceae taxa co‐occurred in T2 and T3 samples (Figure 4c), they were still centered around the same taxa detected in T1 samples (i.e., D1, D1a, Alphaproteobacteria, Planctomycetacia, Gammaproteobacteria, and Oxyphotobacteria) though with some additional taxa (C15.5). Co‐occurrence appeared to be more diverse for Cladocopium compared to Durusdinium (Figure 4c node 1). For example, multiple Cladocopium (e.g., C1cC45, C1j, C1, and C45.3) associated with many bacterial taxa (Alphaproteobacteria, Deltaproteobacteria, Gammaproteobacteria, Planctomycetacia, Oxyphotobacteria, and Phycisphaerae), whereas Durusdinium (D1, D1a, D2.2, D5, D9, and D10) associated with fewer bacteria (Alphaproteobacteria, Gammaproteobacteria, Phycisphaerae, and Oxyphotobacteria) (nodes 2 and 3).
Peak Symbiodiniaceae cell densities were reached by T4 (129 days) and corresponded to the beginning of the exponential growth phase in A. tenuis juveniles. During this time, the co‐occurrence of Cladocopium with bacterial taxa increased relative to earlier time points (e.g., C1, C1ca, C1p, C89, and Alphaproteobacteria, Gammaproteobacteria, Planctomycetacia) (Figure 4d node 1). Associations between D1, D1a, and Alphaproteobacteria remained abundant (Figure 4d node 2). Oxyphotobacteria (Cyanobacteria) exhibited the highest connectedness (Degree = 575).
A decrease and stabilization of Symbiodiniaceae densities associated with juveniles corresponded with accelerated growth rate at T5 (167 days) and T6 (194 days). During this time, associations between D1, D1a, multiple Cladocopium and Alphaproteobacteria, and Oxyphotobacteria were abundant (Figure 4e nodes 1 and 2, 4F node 1, node 3). Gammaproteobacteria and Oxyphotobacteria associated with a greater diversity of Durusdinium, including D1a, D3, D5, D6, and D9 (Figure 4f nodes 2 and 3). Rhodobacterales and Rhodovibrionales (Proteobacteria—Alphaproteobacteria) were the most connected nodes in T5 and T6 samples, respectively (Degree = 463, 568). Multiple Proteobacteria were also significantly positively correlated with Symbiodiniaceae I1 and I3 (e.g., Thalassobaculales, Sneathiellales, Micavibrionales, and Gammaproteobacteria, Figure 3).
Symbiodiniaceae cell densities decreased further as growth rates increased in T7 (316 days) and T8 (380 days), in which networks exhibited higher connectedness between bacterial and Symbiodiniaceae ASV taxa compared to other time points (Figure 4g across nodes 1–2, 4H node 1–2). These dense clusters of connectedness between ASVs consisted of a higher diversity of Cladocopium and Durusdinium (e.g., C1, C1p, C45, C1c, C115a, D1, D1a, D3, D2.2, D10 among others) associated with the same bacterial taxa represented at previous time points. However, some node clusters remained tightly associated with either Durusdinium (Figure 4g node 2, 4H node 1) or Cladocopium (4G node 3, 4H node 2). Additional bacterial diversity (Phycisphaerae and Deltaproteobacteria) was also associated with these clusters. Rhizobiaceae was the most connected taxon in T7 samples (Degree = 523), although connectedness decreased by almost half by the final sampling time point (Rhodobacterales, Degree = 268). Finally, the total number of nodes and edges was substantially lower at this final time point (nodes 834, edges = 35, 921, Figure 4a‐H), corresponding to reduced multidimensional space in the Symbiodiniaceae community (Figure 2 insets).
4. DISCUSSION
Changes in the relative abundances of constituent Symbiodiniaceae taxa allow corals to increase their thermal tolerance (Baker, 2014), where shifts to more heat‐tolerant communities significantly increase survival probability under various climate change scenarios (Logan, Dunne, Eakin, & Donner, 2014). However, the contribution of corals' bacterial partners and their influence on the Symbiodiniaceae community remains unclear. Elucidating symbiont establishment, maintenance, and community composition dynamics are critical as microbial partners may promote physiological resilience of the host; even as climate change is expected to cause the breakdown or disruption of symbioses (Kikuchi et al., 2016). For example, host fitness can improve if the faster‐growing microbial (e.g., bacterial, fungal, eukaryotic) community becomes more mutualistic (Pillai, Gouhier, & Vollmer, 2016); where net benefits to the host can be achieved through changes in background, cryptic microbes (Pillai, Gouhier, & Vollmer, 2014).
Understanding how host‐associated microbial communities develop will elucidate potential acclimatory and adaptive routes for the holobiont (Theis et al., 2016). Here, we show that bacterial and Symbiodiniaceae communities associated with the coral A. tenuis vary concurrently during the first year of ontogeny. Correlation and network analysis revealed key co‐occurrence groups within and across both Symbiodiniaceae and bacterial taxa during development. Acroporid juveniles at this site are known to be dominated by Durusdinium (Abrego et al., 2009b) and Proteobacteria (Alphaproteobacteria) (Lema et al., 2014). More specifically, we found A. tenuis juveniles were dominated by D1 and D1a over the year of sampling, with other taxa from Durusdinium and Cladocopium, including C. goreaui, detected at lower and persistent abundances. Proteobacteria (e.g., Alphaproteobacteria inclusive of Rhodobacteria, Rhizobiales, Rickettsiales, Rhodovibrionales, and Rhodospirillales; Gammaproteobacteria inclusive of Oceanospirillales), Cyanobacteria, and Planctomycetes were also highly abundant and prevalent across sampled juveniles.
4.1. The functional role of Durusdinium during juvenile A. tenuis development
Development in the first month for A. tenuis juveniles was typified by low Symbiodiniaceae cell densities, the dominance of Durusdinium compared to Cladocopium taxa, slow growth, and initial carbon limitation. Combined, these results are consistent with reports of slower growth in juveniles dominated by Durusdinium due to reduced nutrient sharing (Cantin, van Oppen, Willis, Mieog, & Negri, 2009). Later time points were typified by large increases in Symbiodiniaceae cell densities, which may explain the increased diversity and connectedness between Durusdinium and numerous bacterial taxa. The increased relative abundances of Durusdinium and Cladocopium may be linked to growth trade‐offs, in which juveniles benefit from increased photosynthate incorporation to achieve fast “escape size” growth during the transition from horizontal to vertical accretion (Abrego, Willis, & van Oppen, 2012; Cantin et al., 2009). The final time points (i.e., T7 and T8) suggest a consolidation of community diversity, as evidenced by increased network connectedness and decreased diversity, which potentially signals increased nutrient sharing (i.e., the dissipation of carbon limitation and stabilization of photosynthesis in our models) and consolidation of key symbiosis partnerships, which leads to exponential growth in juveniles.
Multiple Durusdinium types were dominant in A. tenuis juveniles, especially at the earliest time point. This initial dominance may be linked to the stress‐tolerance associated with this genus (Abrego, van Oppen, & Willis, 2009a, 2009b; reviewed in Quigley, Baker, Coffroth, Willis, & van Oppen, 2018). Durusdinium are commonly detected at higher abundances in marginal environments (Baums, Johnson, Devlin‐Durante, & Miller, 2010) and opportunistically increase in abundance during bleaching events (Guest et al., 2016; LaJeunesse, Smith, Finney, & Oxenford, 2009), with free‐living taxa detected at higher abundances in warmer‐inshore reefs (Quigley, Bay, et al., 2017; Quigley, Willis, et al., 2017). Adult corals that are Durusdinium‐dominated generally survive better under thermal stress events (Baker, Starger, McClanahan, & Glynn, 2004; Jones, Berkelmans, et al., 2008; Jones, Cantin, et al., 2008). Therefore, juveniles that establish symbiosis with Durusdinium may develop into more “resilient” adults by retaining the option to shuffle their symbiont community to one that performs better under more stressful environmental conditions (Jones, Berkelmans, et al., 2008; Jones, Cantin, et al., 2008). It may therefore be preferable for juveniles to establish symbiosis with this taxon under lower light, turbid environments (Finney et al., 2010; Reynolds, Bruns, Fitt, & Schmidt, 2008) as Durusdinium perform poorly in response to highlight and temperature in juveniles (Abrego, Ulstrup, Willis, & van Oppen, 2008). Alternatively, Durusdinium may be able to out‐compete other Symbiodiniaceae, though further work is needed to establish these functional competitive networks during early coral development.
4.2. Bioenergetics and physiology identify key periods of growth, carbon limitation, and nitrogen assimilation
Juveniles grew exponentially over the course of the year, on par with previously reported juvenile and adult growth rates (Bak, 1976; Cunning et al., 2017; Mieog, Olsen, et al., 2009; Mieog, van Oppen, Berkelmans, Stam, & Olsen, 2009). We also found that algal cell densities in juveniles were consistent with previous estimates (~1 × 104 cells/mm2, Abrego et al., 2012) and expectedly lower compared to adults (~1.4 × 106 cells/mm2, Mieog, Olsen, et al., 2009; Mieog, van Oppen, et al., 2009). Interestingly, Symbiodiniaceae densities increased rapidly between 40 and 71 days to ~2.2 × 104 cells per mm2 and then decreased to ~1 × 104. Bioenergetics indicate that photosynthesis reached “healthy” levels after this time point, suggesting that carrying‐capacity for juveniles may have been reached for this time of year (April), when water temperatures become cooler. This time point (T2) also corresponded with a large shift in the bacterial community structure and an increase in the number of significant positive correlations between Symbiodiniaceae and bacterial taxa. A previous study also found shifts within 2‐week‐old compared to 3‐month‐old juveniles, especially in taxa belonging to Roseobacter and Sphingomonadales sp. (Lema et al., 2014). Therefore, changes in the Symbiodiniaceae community and their abundances may drive changes in nutrient dynamics with flow‐on effects to bacterial community structure or vice versa, all of which have important implications for regulating host physiology and stress responses (Cunning & Baker, 2014).
4.3. Symbiodiniaceae and bacterial co‐occurrence networks associated with A. tenuis juvenile ontogeny
Overall, we found specific associations between Symbiodiniaceae Cladocopium goreaui, Durusdinium trenchii, and Durusdinium glynnii (hereafter referred to as C1, D1, and D1a for brevity; LaJeunesse et al., 2018), across multiple bacterial taxa, including Alphaproteobacteria, Deltaproteobacteria, Gammaproteobacteria, Planctomycetacia, Oxyphotobacteria, Phycisphaerae, and Rhizobiales. In particular, D1a tended to associate with Oxyphotobacteria and D1 with Alphaproteobacteria. Cladocopium were consistently highly correlated with Synechococcales and Thermosynechococcales (Cyanobacteria), Thalassobaculales, Sneathiellales, and Proteobacteria. Coral‐bacterial associations are known to be diverse and dynamic (Bourne et al., 2016) with previous studies also demonstrating some taxa such as Gammaproteobacteria associated with marine taxa that also establish partnerships with Symbiodiniaceae (Bourne et al., 2013). Recently, more specific associations have been reported across multiple coral species, including Rhodospirillaceae and C3/C1; Endozoicomonas (Gammaproteobacteria; Oceanospirillales) and C1, C1c, C43, C3u, C3, C43a, D1, and D1a; Pseudomonas/Alteromonadaceae/Flavobacteriaceae and C1, C3, D1, and D1a; Cyanobacteria/Pleurocapsa and C1, C3, A3, and B1; and finally, Vibrio spp. and C1, D1, and D1a (Bernasconi et al., 2018). Many of the bacterial taxa detected in this current study are considered part of a “healthy” community, including high abundances of Cyanobacteria and Proteobacteria (including Firmicutes and Bacteroidetes) (Littman, Willis, & Bourne, 2011).
We found evidence for the presence of three potentially key taxonomic groups in corals, Endozoicomonas‐related sequences (Oceanospirillales), Roseobacter, and Burkholderia. Endozoicomonas affiliated sequences have been found in adult corals and are in high abundance in healthy adults at this specific site on the GBR (Bourne, Iida, Uthicke, & Smith‐Keune, 2008; Neave, Apprill, Ferrier‐Pagès, & Voolstra, 2016), and large‐scale cophylogenetic analyses have identified Endozoicomonas as a key taxon in host evolution (Pollock et al., 2018). Tissue‐specific sequencing and fluorescence in situ hybridization have also found Endozoicomonas within the coral tissue, indicative of a true coral symbiont (Bourne et al., 2013; Neave et al., 2016; Wada et al., 2019). Generally, Alphaproteobacteria have been shown to be more dominant in the early life stages of corals, giving way to Gammaproteobacteria (including Endozoicomonas) later in life (Lema et al., 2014). This is consistent with patterns seen here, where multiple Alphaproteobacteria were retrieved at high relative abundance (Rhodobacteria, Rhizobiales, Rickettsiales, Rhodovibrionales, and Rhodospirillales), suggesting this is a key bacterial class for acquisition during coral ontogeny. Furthermore, we did not recover Endozoicomonas‐related sequences until T3, with the highest relative abundance in the final time point (T8). Roseobacter and Burkholderia—Paraburkholderia may also be important for juvenile health by way of nitrogen fixation (Damjanovic, Blackall, Webster, & van Oppen, 2017; Epstein et al., 2019; Leite et al., 2017), although more targeted sequencing is needed to determine if these represent true coral symbionts (e.g., Ainsworth et al., 2015).
Host‐microbiome interactions may also play key roles in influencing host evolution in marine ecosystems. Kiloniellales (mutualist), Myxococcales, and Clostridiaceae (parasitic) have all been implicated in host co‐evolution (Pollock et al., 2018). These taxa were also recovered in A. tenuis juveniles, including 300 ASVs identified as Kiloniellaceae (Proteobacteria—Alphaproteobacteria—Rhodovibrionales), and three within both Clostridiales (Family: Clostridiaceae) and Myxococcales. Other taxa detected here (Sphingomonadales, Rhizobiaceae, Rhodobacterales, Rhodovibrionales, and Rhodospirilaceae) have previously been identified as key taxa associated with C1, C3, D1, and D1a in adult corals globally (Bernasconi et al., 2018) and implicated in promoting host productivity and regulating symbiont community evenness in other aquatic and terrestrial ecosystems (Banerjee, Schlaeppi, & Heijden, 2018). Diazotrophic bacteria (Rhizobiales) were found to be key “hub” taxa associated with juvenile A. tenuis. Diazotrophs are key coral symbionts that potentially fix nitrogen for the host (Lema et al., 2014) and may contribute significantly to host nitrogen cycling (McDevitt‐Irwin, Baum, Garren, & Vega Thurber, 2017).
Changes in Symbiodiniaceae taxa may also lead to corresponding shifts in bacterial communities. For example, changes in nutrients produced or excreted by symbionts may induce changes in other symbiotic communities. There is some evidence of the influence of Symbiodiniaceae on bacterial communities, where increases in Symbiodinium (originally termed “clade A”) and decreases in Durusdinium (“clade D”) were correlated with increases in Vibrio spp. (Rouzé, Lecellier, Saulnier, & Berteaux‐Lecellier, 2016; but see Littman et al., 2009). However, co‐occurrence of Symbiodiniaceae and bacteria may not be present in all coral species (Bonthond et al., 2018). We also found that Durusdinium—bacterial co‐occurrence networks exhibited less bacterial diversity compared to networks with Cladocopium. Acropora tenuis juveniles infected with Durusdinium also harbored a reduced diversity of bacterial taxa compared to those infected with C1 (seven taxonomic groups vs. ten) (Littman et al., 2009). Interestingly, the flexibility of Symbiodiniaceae taxa to associate with different bacterial groups has been observed in adults as well, in which C1 associated with the greatest bacterial diversity, followed by D1a, C3, and D1 (Bonthond et al., 2018).
Concurrently sequencing the bacterial and Symbiodiniaceae communities from single juveniles allows predictions for how respective partners may influence each other and potential roles in host development (Bourne et al., 2013). However, it must be acknowledged that the associations detected here may represent secondary, indirect effects. For example, bacterial community shifts may be driven by mucus production derived from photosynthates or community patterns may reflect early colonists to the substrates around the juvenile corals (e.g., Leptolyngbyales—Cyanobacteria) and therefore not symbionts. Indeed, some of the taxa retrieved here (both bacterial and Symbiodiniaceae) may represent transient members of the community, contamination (Propionibacterium, Acinetobacter), or pathogens (Vibrio spp.) (Hammer, Sanders, & Fierer, 2019). The physical location of these symbionts is also important in understanding their role in the host coral, where Symbiodiniaceae reside within gastrodermal host cells (Davy et al., 2012), and the bacterial community is found throughout the outer mucosal layer, within tissues, and partitioned in the skeleton (Ainsworth et al., 2015; Wada et al., 2019). Given that the results presented here represent community patterns derived from whole juvenile sequencing, we are not able to partition where different bacterial taxa originate and can therefore not exclude the possibility that some members may be transient versus obligate symbionts (e.g., potentially those found exclusively in the host tissue). Targeted sequencing of Symbiodiniaceae cells themselves would elucidate co‐occurrence networks more clearly, in which bacterial taxa associated within or outside symbiont cells would suggest a direct symbiosis between the two (e.g., Ralstonia, Actinobacteria, and Proteobacteria; Ainsworth et al., 2015). Understanding the direct versus indirect interactions between Symbiodiniaceae and bacteria is important to elucidating the function behind these associations and remains to be resolved.
In summary, these data provide key insights into the establishment, maintenance, and potential interactions between bacterial and Symbiodiniaceae communities associated with coral juveniles in the wild. Co‐occurrence and network analysis identified novel associations between bacterial and Symbiodiniaceae taxa associated with the first year of A. tenuis development, including D1, D1a, C1, Alphaproteobacteria, Gammaproteobacteria, Oxyphotobacteria, and Rhizobiales. Together, these results elucidate stages in Symbiodiniaceae and bacterial community development that are potentially linked to early carbon and nitrogen dynamics associated with symbiosis establishment. Efforts should also be placed on assigning function to symbiont community structure (e.g., communities associated with heat tolerance or nutrient transport/cycling) and predicting how communities shift under projected climate change scenarios. Characterizing these communities is essential given the substantial acclimatory and adaptive potential Symbiodiniaceae and possibly bacterial taxa provide to their hosts.
CONFLICT OF INTEREST
None declared.
AUTHOR CONTRIBUTIONS
KQ and BW: conceived the experiment; KQ, CAR, and GT: performed the fieldwork; DB and BW: contributed reagents and materials; KQ: analyzed the data and wrote the first draft. All authors edited and reviewed the manuscript.
ETHICAL APPROVAL
This study was carried out with permission and in accordance with the recommendations from the Great Barrier Reef Marine Park Authority (Great Barrier Reef Marine Park Authority Permit Number: G13/36318.1 to BW).
ACKNOWLEDGMENTS
We thank Neal Cantin and Eric Matson for the use of their 3D scanning equipment.
Quigley KM, Alvarez Roa C, Torda G, Bourne DG, Willis BL. Co‐dynamics of Symbiodiniaceae and bacterial populations during the first year of symbiosis with Acropora tenuis juveniles. MicrobiologyOpen. 2020;9:e959 10.1002/mbo3.959
DATA AVAILABILITY STATEMENT
Sequencing data will be made available from Github (https://github.com/LaserKate).
REFERENCES
- Abrego, D. , Ulstrup, K. E. , Willis, B. L. , & van Oppen, M. J. H. (2008). Species–specific interactions between algal endosymbionts and coral hosts define their bleaching response to heat and light stress. Proceedings of the Royal Society B: Biological Sciences, 275(1648), 2273–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrego, D. , van Oppen, M. J. H. , & Willis, B. L. (2009a). Highly infectious symbiont dominates initial uptake in coral juveniles. Molecular Ecology, 18(16), 3518–3531. 10.1111/j.1365-294X.2009.04275.x [DOI] [PubMed] [Google Scholar]
- Abrego, D. , van Oppen, M. J. H. , & Willis, B. L. (2009b). Onset of algal endosymbiont specificity varies among closely related species of Acropora corals during early ontogeny. Molecular Ecology, 18(16), 3532–3543. [DOI] [PubMed] [Google Scholar]
- Abrego, D. , Willis, B. L. , & van Oppen, M. J. H. (2012). Impact of light and temperature on the uptake of algal symbionts by coral juveniles. PLoS ONE, 7(11), e50311 10.1371/journal.pone.0050311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apprill, A. , Marlow, H. Q. , Martindale, M. Q. , & Rappé, M. S. (2009). The onset of microbial associations in the coral Pocillopora meandrina . The ISME Journal, 3(6), 685–699. 10.1038/ismej.2009.3 [DOI] [PubMed] [Google Scholar]
- Baird, A. H. , Bhagooli, R. , Ralph, P. J. , & Takahashi, S. (2009). Coral bleaching: The role of the host. Trends in Ecology & Evolution, 24(1), 16–20. 10.1016/j.tree.2008.09.005 [DOI] [PubMed] [Google Scholar]
- Bak, R. P. M. (1976). The growth of coral colonies and the importance of crustose coralline algae and burrowing sponges in relation with carbonate accumulation. Netherlands Journal of Sea Research, 10(3), 285–337. 10.1016/0077-7579(76)90009-0 [DOI] [Google Scholar]
- Baker, A. C. (2014). Climate change: Many ways to beat the heat for reef corals. Current Biology, 24(24), R1166–R1168. 10.1016/j.cub.2014.11.014 [DOI] [PubMed] [Google Scholar]
- Baker, A. C. , Starger, C. J. , McClanahan, T. R. , & Glynn, P. W. (2004). Coral reefs: Corals' adaptive response to climate change. Nature, 430(7001), 741 10.1038/430741a [DOI] [PubMed] [Google Scholar]
- Banerjee, S. , Schlaeppi, K. , & Heijden, M. G. A. (2018). Keystone taxa as drivers of microbiome structure and functioning. Nature Reviews Microbiology, 1 10.1038/s41579-018-0024-1 [DOI] [PubMed] [Google Scholar]
- Baums, I. B. , Johnson, M. E. , Devlin‐Durante, M. K. , & Miller, M. W. (2010). Host population genetic structure and zooxanthellae diversity of two reef‐building coral species along the Florida Reef Tract and wider Caribbean. Coral Reefs, 29(4), 835–842. 10.1007/s00338-010-0645-y [DOI] [Google Scholar]
- Bellwood, D. R. , Hughes, T. P. , Folke, C. , & Nystrom, M. (2004). Confronting the coral reef crisis. Nature, 429(6994), 827–833. [DOI] [PubMed] [Google Scholar]
- Bernasconi, R. , Stat, M. , Koenders, A. , & Huggett, M. J. (2018). Global networks of Symbiodinium‐bacteria within the coral holobiont. Microbial Ecology, 10.1007/s00248-018-1255-4 [DOI] [PubMed] [Google Scholar]
- Bonthond, G. , Merselis, D. G. , Dougan, K. E. , Graff, T. , Todd, W. , Fourqurean, J. W. , & Rodriguez‐Lanetty, M. (2018). Inter‐domain microbial diversity within the coral holobiont Siderastrea siderea from two depth habitats. PeerJ, 6, e4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordenstein, S. R. , & Theis, K. R. (2015). Host biology in light of the microbiome: Ten principles of holobionts and hologenomes. PLOS Biology, 13(8), e1002226 10.1371/journal.pbio.1002226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch, T. C. G. , & McFall‐Ngai, M. J. (2011). Metaorganisms as the new frontier. Zoology, 114(4), 185–190. 10.1016/j.zool.2011.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne, D. G. , Dennis, P. G. , Uthicke, S. , Soo, R. M. , Tyson, G. W. , & Webster, N. (2013). Coral reef invertebrate microbiomes correlate with the presence of photosymbionts. The ISME Journal, 7(7), 1452–1458. 10.1038/ismej.2012.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne, D. , Iida, Y. , Uthicke, S. , & Smith‐Keune, C. (2008). Changes in coral‐associated microbial communities during a bleaching event. The ISME Journal, 2(4), 350 10.1038/ismej.2007.112 [DOI] [PubMed] [Google Scholar]
- Bourne, D. G. , Morrow, K. M. , & Webster, N. S. (2016). Insights into the coral microbiome: Underpinning the health and resilience of reef ecosystems. Annual Review of Microbiology, 70, 317–340. 10.1146/annurev-micro-102215-095440 [DOI] [PubMed] [Google Scholar]
- Brodnicke, O. B. , Bourne, D. G. , Heron, S. F. , Pears, R. J. , Stella, J. S. , Smith, H. A. , & Willis, B. L. (2019). Unravelling the links between heat stress, bleaching and disease: fate of tabular corals following a combined disease and bleaching event. Coral Reefs, 1–13. [Google Scholar]
- Callahan, B. J. , McMurdie, P. J. , Rosen, M. J. , Han, A. W. , Johnson, A. J. A. , & Holmes, S. P. (2016). DADA2: High‐resolution sample inference from Illumina amplicon data. Nature Methods, 13(7), 581–583. 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantin, N. , van Oppen, M. , Willis, B. , Mieog, J. , & Negri, A. (2009). Juvenile corals can acquire more carbon from high‐performance algal symbionts. Coral Reefs, 28(2), 405–414. 10.1007/s00338-009-0478-8 [DOI] [Google Scholar]
- Cooper, T. F. , Berkelmans, R. , Ulstrup, K. E. , Weeks, S. , Radford, B. , Jones, A. M. , … van Oppen, M. J. H. (2011). Environmental factors controlling the distribution of Symbiodinium harboured by the coral Acropora millepora on the Great Barrier Reef. PLoS ONE, 6(10), e25536 10.1371/journal.pone.0025536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csardi, G. , & Nepusz, T. (2006). The igraph software package for complex network research. InterJournal, Complex Systems, 1695(5), 1–9. [Google Scholar]
- Cunning, R. , & Baker, A. C. (2014). Not just who, but how many: The importance of partner abundance in reef coral symbioses. Frontiers in Microbiology, 5, 400 10.3389/fmicb.2014.00400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunning, R. , Muller, E. B. , Gates, R. D. , & Nisbet, R. M. (2017). A dynamic bioenergetic model for coral‐Symbiodinium symbioses and coral bleaching as an alternate stable state. Journal of Theoretical Biology, 431, 49–62. 10.1016/j.jtbi.2017.08.003 [DOI] [PubMed] [Google Scholar]
- Cunning, R. , Silverstein, R. N. , & Baker, A. C. (2015). Investigating the causes and consequences of symbiont shuffling in a multi‐partner reef coral symbiosis under environmental change. Proceedings of the Royal Society B: Biological Sciences, 282(1809), 20141725 10.1098/rspb.2014.1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D Ainsworth, T. , Krause, L. , Bridge, T. , Torda, G. , Raina, J.‐B. , Zakrzewski, M. , … Leggat, W. (2015). The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. The ISME Journal, 9(10), 2261–2274. 10.1038/ismej.2015.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damjanovic, K. , Blackall, L. L. , Webster, N. S. , & van Oppen, M. J. H. (2017). The contribution of microbial biotechnology to mitigating coral reef degradation. Microbial Biotechnology, 10(5), 1236–1243. 10.1111/1751-7915.12769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davy, S. K. , Allemand, D. , & Weis, V. M. (2012). Cell biology of cnidarian‐dinoflagellate symbiosis. Microbiology and Molecular Biology Reviews, 76(2), 229–261. 10.1128/MMBR.05014-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De'ath, G. , & Fabricius, K. (2010). Water quality as a regional driver of coral biodiversity and macroalgae on the Great Barrier Reef. Ecological Applications, 20(3), 840–850. 10.1890/08-2023.1 [DOI] [PubMed] [Google Scholar]
- Doropoulos, C. , Roff, G. , Bozec, Y. , Zupan, M. , Werminghausen, J. , & Mumby, P. J. (2016). Characterizing the ecological trade‐offs throughout the early ontogeny of coral recruitment. Ecological Monographs, 86(1), 20–44. [Google Scholar]
- Douglas, A. E. , & Werren, J. H. (2016). Holes in the hologenome: Why host‐microbe symbioses are not holobionts. MBio, 7(2), e02099‐15 10.1128/mBio.02099-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein, H. E. , Torda, G. , Munday, P. L. , & van Oppen, M. J. H. (2019). Parental and early life stage environments drive establishment of bacterial and dinoflagellate communities in a common coral. The ISME Journal, 13(6), 1635–1638. 10.1038/s41396-019-0358-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fieth, R. A. , Gauthier, M.‐E.‐A. , Bayes, J. , Green, K. M. , & Degnan, S. M. (2016). Ontogenetic Changes in the bacterial symbiont community of the tropical demosponge Amphimedon queenslandica: Metamorphosis is a new beginning. Frontiers in Marine Science, 3, 228 10.3389/fmars.2016.00228 [DOI] [Google Scholar]
- Finney, J. , Pettay, D. , Sampayo, E. , Warner, M. , Oxenford, H. , & LaJeunesse, T. (2010). The relative significance of host–habitat, depth, and geography on the ecology, endemism, and speciation of coral endosymbionts in the genus Symbiodinium . Microbial Ecology, 60(1), 250–263. 10.1007/s00248-010-9681-y [DOI] [PubMed] [Google Scholar]
- Glasl, B. , Smith, C. E. , Bourne, D. G. , & Webster, N. S. (2019). Disentangling the effect of host‐genotype and environment on the microbiome of the coral Acropora tenuis . PeerJ, 7, e6377 10.7717/peerj.6377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, J. , Knowlton, N. , Relman, D. A. , Rohwer, F. , & Youle, M. (2013). Superorganisms and holobionts. Microbe, 8(4), 152–153. 10.1128/microbe.8.152.1 [DOI] [Google Scholar]
- Grandclément, C. , Tannières, M. , Moréra, S. , Dessaux, Y. , & Faure, D. (2016). Quorum quenching: Role in nature and applied developments. FEMS Microbiology Reviews, 40, 86–116. 10.1093/femsre/fuv038 [DOI] [PubMed] [Google Scholar]
- Guest, J. R. , Low, J. , Tun, K. , Wilson, B. , Ng, C. , Raingeard, D. , … Steinberg, P. D. (2016). Coral community response to bleaching on a highly disturbed reef. Scientific Reports, 6, 20717 10.1038/srep20717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer, T. J. , Sanders, J. G. , & Fierer, N. (2019). Not all animals need a microbiome. FEMS Microbiology Letters, 366, fnz117 10.1093/femsle/fnz117 [DOI] [PubMed] [Google Scholar]
- Harrell, F. E. (2007). R package Harrell Miscellaneous (Hmisc) (3.2‐1). 3.2‐1. Retrieved from http://biostat.mc.vanderbilt.edu/s/Hmisc [Google Scholar]
- Hoang, K. L. , Morran, L. T. , & Gerardo, N. M. (2016). Experimental evolution as an underutilized tool for studying beneficial animal–microbe interactions. Frontiers in Microbiology, 7, 1444 10.3389/fmicb.2016.01444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoegh‐Guldberg, O. , & Bruno, J. F. (2010). The impact of climate change on the world's marine ecosystems. Science, 328, 1523–1528. 10.1126/science.1189930 [DOI] [PubMed] [Google Scholar]
- Hughes, T. P. , Anderson, K. D. , Connolly, S. R. , Heron, S. F. , Kerry, J. T. , Lough, J. M. , … Wilson, S. K. (2018). Spatial and temporal patterns of mass bleaching of corals in the Anthropocene. Science, 359(6371), 80–83. 10.1126/science.aan8048 [DOI] [PubMed] [Google Scholar]
- Hughes, T. P. , Baird, A. H. , Bellwood, D. R. , Card, M. , Connolly, S. R. , Folke, C. , … Kleypas, J. (2003). Climate change, human impacts, and the resilience of coral reefs. Science, 301(5635), 929–933. [DOI] [PubMed] [Google Scholar]
- Hughes, T. P. , Kerry, J. T. , Álvarez‐Noriega, M. , Álvarez‐Romero, J. G. , Anderson, K. D. , Baird, A. H. , … Berkelmans, R. (2017). Global warming and recurrent mass bleaching of corals. Nature, 543(7645), 373–377. [DOI] [PubMed] [Google Scholar]
- Jones, A. M. , Berkelmans, R. , van Oppen, M. J. H. , Mieog, J. C. , & Sinclair, W. (2008). A community change in the algal endosymbionts of a scleractinian coral following a natural bleaching event: Field evidence of acclimatization. Proceedings of the Royal Society B: Biological Sciences, 275(1641), 1359–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, A. M. , Cantin, N. E. , Berkelmans, R. , Sinclair, B. , & Negri, A. P. (2008). A 3D modeling method to calculate the surface areas of coral branches. Coral Reefs, 27(3), 521–526. 10.1007/s00338-008-0354-y [DOI] [Google Scholar]
- Karim, W. , Nakaema, S. , & Hidaka, M. (2015). Temperature effects on the growth rates and photosynthetic activities of Symbiodinium cells. Journal of Marine Science and Engineering, 3(2), 368–381. 10.3390/jmse3020368 [DOI] [Google Scholar]
- Kikuchi, Y. , Tada, A. , Musolin, D. L. , Hari, N. , Hosokawa, T. , Fujisaki, K. , & Fukatsu, T. (2016). Collapse of insect gut symbiosis under simulated climate change. MBio, 7(5), e01578‐16 10.1128/mBio.01578-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuniya, N. , Jimbo, M. , Tanimoto, F. , Yamashita, H. , Koike, K. , Harii, S. , … Watabe, S. (2015). Possible involvement of Tachylectin‐2‐like lectin from Acropora tenuis in the process of Symbiodinium acquisition. Fisheries Science, 81(3), 473–483. 10.1007/s12562-015-0862-y [DOI] [Google Scholar]
- LaJeunesse, T. C. , Parkinson, J. E. , Gabrielson, P. W. , Jeong, H. J. , Reimer, J. D. , Voolstra, C. R. , & Santos, S. R. (2018). Systematic revision of Symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Current Biology, 28(16), 2570–2580. 10.1016/j.cub.2018.07.008 [DOI] [PubMed] [Google Scholar]
- LaJeunesse, T. C. , Smith, R. T. , Finney, J. , & Oxenford, H. (2009). Outbreak and persistence of opportunistic symbiotic dinoflagellates during the 2005 Caribbean mass coral “bleaching” event. Proceedings of the Royal Society B: Biological Sciences, 276(1676), 4139–4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite, D. C. A. , Leão, P. , Garrido, A. G. , Lins, U. , Santos, H. F. , Pires, D. O. , … Peixoto, R. S. (2017). Broadcast spawning coral Mussismilia hispida can vertically transfer its associated bacterial core. Frontiers in Microbiology, 8, 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lema, K. A. , Bourne, D. G. , & Willis, B. L. (2014). Onset and establishment of diazotrophs and other bacterial associates in the early life history stages of the coral Acropora millepora . Molecular Ecology, 23(19), 4682–4695. [DOI] [PubMed] [Google Scholar]
- Littman, R. A. , Willis, B. L. , & Bourne, D. G. (2009). Bacterial communities of juvenile corals infected with different Symbiodinium (dinoflagellate) clades. Marine Ecology Progress Series, 389, 45–59. 10.3354/meps08180 [DOI] [Google Scholar]
- Littman, R. , Willis, B. L. , & Bourne, D. G. (2011). Metagenomic analysis of the coral holobiont during a natural bleaching event on the Great Barrier Reef. Environmental Microbiology Reports, 3(6), 651–660. 10.1111/j.1758-2229.2010.00234.x [DOI] [PubMed] [Google Scholar]
- Logan, C. A. , Dunne, J. P. , Eakin, C. M. , & Donner, S. D. (2014). Incorporating adaptive responses into future projections of coral bleaching. Global Change Biology, 20(1), 125–139. 10.1111/gcb.12390 [DOI] [PubMed] [Google Scholar]
- Love, M. I. , Huber, W. , & Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biology, 15(12), 1–21. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDevitt‐Irwin, J. M. , Baum, J. K. , Garren, M. , & Vega Thurber, R. L. (2017). Responses of coral‐associated bacterial communities to local and global stressors. Frontiers in Marine Science, 4, 262 10.3389/fmars.2017.00262 [DOI] [Google Scholar]
- McMurdie, P. J. , & Holmes, S. (2013). Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE, 8(4), e61217 10.1371/journal.pone.0061217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieog, J. C. , Olsen, J. L. , Berkelmans, R. , Bleuler‐Martinez, S. A. , Willis, B. L. , & van Oppen, M. J. H. (2009). The roles and interactions of symbiont, host and environment in defining coral fitness. PLoS ONE, 4(7), e6364 10.1371/journal.pone.0006364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieog, J. , van Oppen, M. , Berkelmans, R. , Stam, W. , & Olsen, J. (2009). Quantification of algal endosymbionts (Symbiodinium) in coral tissue using real‐time PCR. Molecular Ecology Resources, 9(1), 74–82. 10.1111/j.1755-0998.2008.02222.x [DOI] [PubMed] [Google Scholar]
- Morris, L. A. , Voolstra, C. R. , Quigley, K. M. , Bourne, D. G. , & Bay, L. K. (2019). Nutrient availability and metabolism affect the stability of coral–Symbiodiniaceae symbioses. Trends in Microbiology, 27(8), 678–689. 10.1016/j.tim.2019.03.004 [DOI] [PubMed] [Google Scholar]
- Murdoch, D. , Chow, E. D. , & Celayeta, J. M. F. (2007). Ellipse: Functions for drawing ellipses and ellipse‐like confidence regions. R Package Version 0.3‐5. [Google Scholar]
- Neave, M. J. , Apprill, A. , Ferrier‐Pagès, C. , & Voolstra, C. R. (2016). Diversity and function of prevalent symbiotic marine bacteria in the genus Endozoicomonas . Applied Microbiology and Biotechnology, 100(19), 8315–8324. 10.1007/s00253-016-7777-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien, P. A. , Webster, N. S. , Miller, D. J. , & Bourne, D. G. (2019). Host‐microbe coevolution: Applying evidence from model systems to complex marine invertebrate holobionts. MBio, 10(1), e02241–e2318. 10.1128/mBio.02241-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen, J. , Blanchet, F. G. , Kindt, R. , Legendre, P. , Minchin, P. R. , O'Hara, R. B. , … Wagner, H. (2013). Package ‘vegan’. Community Ecology Package, Version, 2(9). [Google Scholar]
- Pillai, P. , Gouhier, T. C. , & Vollmer, S. V. (2014). The cryptic role of biodiversity in the emergence of host–microbial mutualisms. Ecology Letters, 17(11), 1437–1446. 10.1111/ele.12349 [DOI] [PubMed] [Google Scholar]
- Pillai, P. , Gouhier, T. C. , & Vollmer, S. V. (2016). Ecological rescue of host‐microbial systems under environmental change. Theoretical Ecology, 10, 1–13. [Google Scholar]
- Pochon, X. , Pawlowski, J. , Zaninetti, L. , & Rowan, R. (2001). High genetic diversity and relative specificity among Symbiodinium‐like endosymbiotic dinoflagellates in soritid foraminiferans. Marine Biology, 139(6), 1069–1078. 10.1007/s002270100674 [DOI] [Google Scholar]
- Poland, D. M. , & Coffroth, M. A. (2017). Trans‐generational specificity within a cnidarian–algal symbiosis. Coral Reefs, 36, 1–11. 10.1007/s00338-016-1514-0 [DOI] [Google Scholar]
- Pollock, F. J. , McMinds, R. , Smith, S. , Bourne, D. G. , Willis, B. L. , Medina, M. , … Zaneveld, J. R. (2018). Coral‐associated bacteria demonstrate phylosymbiosis and cophylogeny. Nature Communications, 9(1), 4921 10.1038/s41467-018-07275-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley, K. M. , Baker, A. C. , Coffroth, M. A. , Willis, B. L. , & van Oppen, M. J. H. (2018). Bleaching resistance and the role of algal endosymbionts In: van Oppen M. J. H., Lough J. M. (Eds). Coral bleaching (pp. 111–151). Switzerland:Springer International Publishing. [Google Scholar]
- Quigley, K. M. , Bay, L. K. , & Willis, B. L. (2017). Temperature and water quality‐related patterns in sediment‐associated Symbiodinium communities impact symbiont uptake and fitness of juveniles in the genus Acropora. Frontiers in Marine Science, 4, 401. [Google Scholar]
- Quigley, K. M. , Willis, B. , & Bay, L. (2017). Heritability of the Symbiodinium community in vertically‐and horizontally‐transmitting broadcast spawning corals. Scientific Reports, 7, 8219 10.1038/s41598-017-08179-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley, K. M. , Willis, B. L. , & Kenkel, C. D. (2019). Transgenerational inheritance of shuffled symbiont communities in the coral Montipora digitata . Scientific Reports, 9(1), 13328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramette, A. (2007). Multivariate analyses in microbial ecology. FEMS Microbiology Ecology, 62(2), 142–160. 10.1111/j.1574-6941.2007.00375.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds, J. M. , Bruns, B. U. , Fitt, W. K. , & Schmidt, G. W. (2008). Enhanced photoprotection pathways in symbiotic dinoflagellates of shallow‐water corals and other cnidarians. Proceedings of the National Academy of Sciences of the United States of America, 105(36), 13674–13678. 10.1073/pnas.0805187105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohwer, F. , Seguritan, V. , Azam, F. , & Knowlton, N. (2002). Diversity and distribution of coral‐associated bacteria. Marine Ecology Progress Series, 243, 1–10. 10.3354/meps243001 [DOI] [Google Scholar]
- Rouzé, H. , Lecellier, G. , Saulnier, D. , & Berteaux‐Lecellier, V. (2016). Symbiodinium clades A and D differentially predispose Acropora cytherea to disease and Vibrio spp. colonization. Ecology and. Evolution, 6(2), 560–572. 10.1002/ece3.1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvucci, E. (2016). Microbiome, holobiont and the net of life. Critical Reviews in Microbiology, 42(3), 485–494. [DOI] [PubMed] [Google Scholar]
- Sharp, K. H. , Distel, D. , & Paul, V. J. (2012). Diversity and dynamics of bacterial communities in early life stages of the Caribbean coral Porites astreoides . The ISME Journal, 6(4), 790–801. 10.1038/ismej.2011.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza, D. T. , Genuário, D. B. , Silva, F. S. P. , Pansa, C. C. , Kavamura, V. N. , Moraes, F. C. , … Melo, I. S. (2016). Analysis of bacterial composition in marine sponges reveals the influence of host phylogeny and environment. FEMS Microbiology Ecology, 93, fiw204. [DOI] [PubMed] [Google Scholar]
- Stimson, J. (1997). The annual cycle of density of zooxanthellae in the tissues of field and laboratory‐held Pocillopora damicornis (Linnaeus). Journal of Experimental Marine Biology and Ecology, 214(1–2), 35–48. 10.1016/S0022-0981(96)02753-0 [DOI] [Google Scholar]
- Swain, T. D. , Chandler, J. , Backman, V. , & Marcelino, L. (2016). Consensus thermotolerance ranking for 110 Symbiodinium phylotypes: An exemplar utilization of a novel iterative partial rank aggregation tool with broad application potential. Functional Ecology, 31, 172–183. [Google Scholar]
- Theis, K. R. , Dheilly, N. M. , Klassen, J. L. , Brucker, R. M. , Baines, J. F. , Bosch, T. C. G. , … Bordenstein, S. R. (2016). Getting the hologenome concept right: An eco‐evolutionary framework for hosts and their microbiomes. mSystems, 1(2), e00028‐16 10.1128/mSystems.00028-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, A. A. , & Dolman, A. M. (2010). Coral bleaching: One disturbance too many for near‐shore reefs of the Great Barrier Reef. Coral Reefs, 29(3), 637–648. 10.1007/s00338-009-0562-0 [DOI] [Google Scholar]
- Torda, G. , Donelson, J. M. , Aranda, M. , Barshis, D. J. , Bay, L. , Berumen, M. L. , … Munday, P. L. (2017). Rapid adaptive responses to climate change in corals. Nature Climate Change, 7(9), 627–636. 10.1038/nclimate3374 [DOI] [Google Scholar]
- Wada, N. , Ishimochi, M. , Matsui, T. , Pollock, F. J. , Tang, S.‐L. , Ainsworth, T. D. , Bourne, D. G. (2019). Characterization of coral‐associated microbial aggregates (CAMAs) within tissues of the coral Acropora hyacinthus . BioRxiv, 576488 10.1101/576488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, K. , Li, Y. , Whan, V. , Lehnert, S. , Byrne, K. , Moore, S. , … Benzie, J. (2002). Genetic mapping of the black tiger shrimp Penaeus monodon with amplified fragment length polymorphism. Aquaculture, 204(3), 297–309. 10.1016/S0044-8486(01)00842-0 [DOI] [Google Scholar]
- Zhou, G. , Cai, L. , Yuan, T. , Tian, R. , Tong, H. , Zhang, W. , … Huang, H. (2017). Microbiome dynamics in early life stages of the scleractinian coral Acropora gemmifera in response to elevated pCO2. Environmental Microbiology, 19(8), 3342–3352. 10.1111/1462-2920.13840 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequencing data will be made available from Github (https://github.com/LaserKate).


