Abstract
The amino acid serine plays diverse metabolic roles, yet bacteria actively degrade exogenously provided serine via deamination to pyruvate. Serine deamination is thought to be a detoxification mechanism due to the ability of serine to inhibit several biosynthetic reactions, but this pathway remains highly active even in nutrient‐replete conditions. While investigating the physiological roles of serine deamination in different growth conditions, we discovered that Escherichia coli cells lacking the sdaCB operon, which encodes the serine transporter SdaC and the serine deaminase SdaB, lyse upon glucose depletion in a medium containing no exogenous serine but all other amino acids and nucleobases. Unexpectedly, this lysis phenotype can be recapitulated by deleting sdaC alone and can be rescued by heterologous expression of SdaC. Lysis of ΔsdaC cells can be prevented by omitting glycine from the medium, inhibiting the glycine cleavage system, or by increasing alanine availability. Together, our results reveal that the serine transporter SdaC plays a critical role in maintaining amino acid homeostasis during shifts in nutrient availability in E. coli.
Keywords: amino acid transport, deamination, metabolism, serine
Our results reveal that the serine transporter SdaC plays a critical role in maintaining amino acid homeostasis during shifts in nutrient availability in Escherichia coli.

1. INTRODUCTION
The amino acid serine is a centrally important biomolecule, not only as a substrate for protein synthesis but also as a precursor of nucleotides, redox molecules, phospholipids, and other amino acids (Locasale, 2013; Stauffer, 2004). While the biochemical reactions involved in serine metabolism are well known, understanding of how cells regulate fluxes and intracellular concentrations of serine‐associated metabolites remains incomplete. Recent revelations that serine biosynthesis genes are amplified or up‐regulated in many cancers (Liao et al., 2016; Locasale et al., 2011; Mattaini, Sullivan, & Heiden, 2016; Possemato, 2011) and required for host colonization by bacterial pathogens (Gao et al., 2017; Yasuda et al., 2017) has led to renewed interest in elucidating the fundamental principles governing serine metabolism in all cells.
When exogenous serine is available, proliferating bacterial and mammalian cells deplete it rapidly, far faster than any other amino acid (Hosios et al., 2016; Selvarasu, 2009; Zhang, El‐Hajj, & Newman, 2010). This observation has been cited as evidence that replicating cells require large amounts of serine to generate biomass and functional metabolites such as one‐carbon units (Locasale, 2013; Newman & Maddocks, 2017). However, bacteria deaminate the majority of exogenous serine to produce pyruvate and ammonia (Netzer, Peters‐Wendisch, Eggeling, & Sahm, 2004; Ramotar & Newman, 1986) (Figure 1a), even though this pathway antagonizes growth (Ramotar & Newman, 1986) and most pyruvate‐derived carbon is ultimately excreted from the cell (Selvarasu et al., 2009). Serine deamination is clearly important because E. coli expresses at least one of its three serine deaminase enzymes at all times (Zhang & Newman, 2008), yet the metabolic benefits of this reaction remain unclear.
Figure 1.
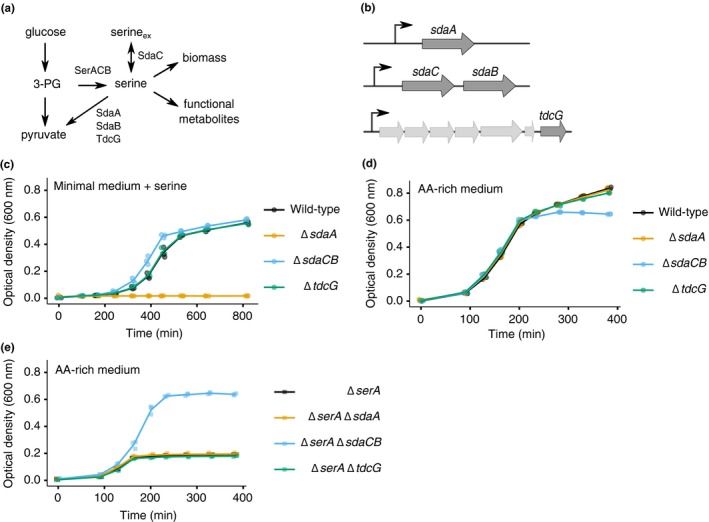
The role of serine deaminase operons in minimal versus rich media. (a) Schematic of metabolic reactions and relevant enzymes involved in serine biosynthesis and utilization. 3‐PG = 3‐phosphoglycerate. (b) Schematic of the three loci in the Escherichia coli genome that encode serine deaminase enzymes. (c) Growth curves of wild‐type and serine deaminase single deletion strains (sdaA::kan, ecMK101; sdaCB::cm, ecMK102; tdcG::cm, ecMK210) grown in minimal medium containing 1.5 mM serine. (d) Growth curves of wild‐type and serine deaminase single deletion strains described in (a) grown in AA‐rich medium containing 5 mM serine. (e) Growth curves of ΔserA strains with or without a serine deaminase deletion (serA::FRT, ecMK94; serA::FRT sdaA::kan, ecMK105; serA::FRT sdaCB::cm, ecMK106; serA::FRT tdcG::cm, ecMK211) grown in rich medium containing 5 mM serine. For all growth curves, three biological replicates are shown as points with their averages connected by lines. AA‐rich medium refers to a MOPS‐based defined medium containing all amino acids and nucleobases at defined concentrations (Neidhardt et al., 1974) (Materials and Methods). In the minimal medium, all amino acids and nucleobases were omitted from the AA‐rich medium except serine
Serine deamination in bacteria is considered a detoxification mechanism because free intracellular serine inhibits biosynthesis of isoleucine and aromatic amino acids (Hama, Sumita, Kakutani, Tsuda, & Tsuchiya, 1990; Saito, Ishida, & Yamada, 2003). Indeed, all three E. coli serine deaminases have Km values in the millimolar range (Burman, Harris, Hauton, Lawson, & Sawers, 2004; Cicchillo, 2004), consistent with a role in preventing serine excess (Zhang & Newman, 2008). Escherichia coli cells unable to deaminate serine become severely elongated in the presence of exogenous serine, suggesting that high serine levels inhibit cell division (Zhang et al., 2010; Zhang & Newman, 2008). This toxicity may stem from defects in cell wall production given that misincorporation of serine in place of alanine into peptidoglycan crosslinks impairs peptidoglycan biosynthesis (Parveen & Reddy, 2017). In all cases documented so far, toxicity occurs in the presence of exogenous serine; whether serine toxicity can occur in the absence of exogenous serine is not known.
Ribosome pausing is consistently observed at serine codons in bacterial ribosome profiling studies (Li, Oh, & Weissman, 2012; Martens, Taylor, & Hilser, 2015), indicating that intracellular serine concentrations or charging levels of seryl‐tRNAs may be low compared with other amino acids. Surprisingly, the proportion of charged to uncharged seryl‐tRNAs was observed to be much lower in rich media compared to minimal media (where exogenous serine is not available) (Avcilar‐Kucukgoze, 2016), suggesting that intracellular serine levels may be lower in nutrient‐rich conditions. Given that known serine toxicities should be less severe when all amino acids are available, the physiological benefits of maintaining low serine levels in rich media are unclear.
Here, we report that E. coli cells lacking the sdaCB operon, which encodes a serine transporter and a serine deaminase, lyse upon glucose depletion, thus revealing a role for sdaCB during adaptation to changes in nutrient availability. Unexpectedly, the serine transporter SdaC has a dominant role over the deaminase SdaB in mediating the observed phenotype. Lysis can be modulated by altering glycine or alanine levels in the growth medium, indicating the importance of maintaining a strict ratio of serine to other amino acids during metabolic shifts. Together, our results reveal that the serine transporter SdaC promotes amino acid homeostasis during adaptation to glucose limitation in E. coli.
2. MATERIALS AND METHODS
2.1. Growth media
Bacteria were grown in a MOPS‐based defined medium (Neidhardt, Bloch, & Smith, 1974) (adapted from the E. coli genome project [E. coli Genome Project]), containing 0.5% (28 mM) glucose as the carbon source unless otherwise noted. The 5X Supplement was replaced with a 5X solution containing 4 mM of each amino acid (0.8 mM final concentration) except serine, glycine and alanine, which were added separately. When needed, glycine and alanine were added to a final concentration of 0.8 mM and serine was added to a final concentration of 10 mM, unless otherwise noted. This medium is designated as AA‐rich medium throughout the text. The medium that produces lysis of ΔsdaC strains contains 0.025% (1.4 mM) glucose and no serine. For all growth curve experiments in AA‐rich medium, overnight cultures were grown in AA‐rich medium containing 0.5% glucose and 10 mM serine and diluted 1:100 into experimental media. For the minimal medium experiment (Figure 1c), nucleobases and amino acids were omitted. Overnight cultures were grown in minimal medium without serine and diluted 1:100 into minimal medium containing 1.5 mM serine. For cloning and strain construction, bacteria were grown in LB medium (BD/Difco) and the antibiotics carbenicillin (100 µg/ml), chloramphenicol (25 µg/ml), or kanamycin (25 µg/ml) were included when required.
2.2. Strain construction
All experiments were carried out with wild‐type Escherichia coli strains BW25113 (Datsenko & Wanner, 2000) or MG1655 (Blattner et al., 1997) and mutant derivatives constructed using the one‐step gene disruption method (Datsenko & Wanner, 2000; Datta, Costantino, & Court, 2006) (Table A1). Briefly, an antibiotic resistance cassette was amplified from plasmid pKD13 (kan) or pKD32 (cm) using primers containing gene‐specific 5′ extensions (Table A2). Each cassette was designed to remove the region from the start codon to the stop codon of the gene, or from the sdaC start codon to the sdaB stop codon in the case of the sdaCB deletion (see Table A2 for exact flanking sequences). Recombination was achieved by electroporating 500ng of cassette DNA into a strain harboring plasmid pSIM6. pSIM6‐harboring competent cells were prepared following growth to OD600 = 0.4–0.5 in LB and heat shock at 42°C for 15 min to induce expression of the λ‐Red recombinase machinery (Datta et al., 2006). When desired, the antibiotic resistance cassette was excised using pCP20 as described (Datsenko & Wanner, 2000), leaving an 84 bp scar sequence that contains stop codons in all six reading frames (denoted as ::FRT in the genotypes listed in Table A1) (Datsenko & Wanner, 2000). When making additional mutations in a strain already harboring a cassette or scar sequence, the antibiotic cassette was generated by colony PCR from a strain already containing the insertion using primers approximately 100 bp outside of the insertion. This strategy yields a cassette with longer homology arms and significantly reduced integration of the cassette into the locus of the first mutation. Successful strain construction was confirmed by colony PCR and Sanger sequencing.
2.3. Plasmid construction
pASEC1 (Addgene plasmid #53241), a very low‐copy expression vector (SC*101 ori, 3–4 copies per cell) used in our previous work (Subramaniam, Pan, & Cluzel, 2013), was used as a backbone for constructing the transcriptional reporters shown in Figure 4c. For each gene of interest, the promoter region was amplified from genomic DNA using the primers listed in Table A2 and inserted between the XhoI and EcoRI sites of pASEC1 via Gibson assembly. This strategy removes the pLtetO‐1 promoter and places the gene‐specific promoter directly upstream of the T7 ribosome binding site and yfp coding region. Successful construction was confirmed by colony PCR and Sanger sequencing.
Figure 4.
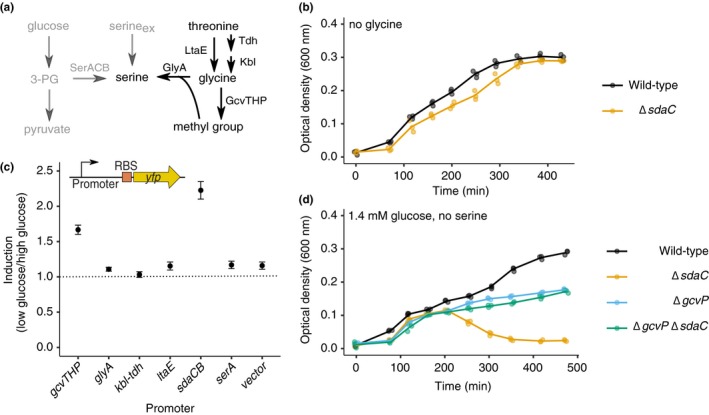
The glycine cleavage system promotes lysis upon glucose depletion. (a) Schematic of metabolic pathways involved in serine production. (b) Growth curves of wild‐type and ΔsdaC (sdaC::FRT, ecMK180) strains in AA‐rich medium with 1.4 mM glucose, no serine, and no glycine. (c) Relative YFP fluorescence (glucose‐depleted vs. glucose‐containing media) from wild‐type strains harboring transcriptional reporters for genes related to glycine and threonine catabolism (ecMK136, 137, 147, 172, 174, 176, 178). Raw fluorescence values were first normalized to culture OD, and relative fluorescence was calculated by dividing the YFP/OD signal (averaged over the three time points directly after glucose depletion occurs in the 1.4 mM glucose condition) from cells grown in media containing 1.4 mM versus 5.6 mM glucose. (d) Growth curves of wild‐type, ΔgcvP (gcvP::kan, ecMK129), ΔsdaC (sdaC::FRT, ecMK180), and ΔgcvP ΔsdaC (gcvP::cm sdaC::FRT, ecMK196) strains in AA‐rich medium with 1.4 mM glucose and no serine. For all growth curves, three biological replicates are shown as points with their averages connected by lines. For YFP induction, average and standard error over biological triplicates is shown
2.4. Growth and fluorescence measurements
Bacterial strains were streaked onto LB plates, and single colonies were used to inoculate overnight cultures of AA‐rich medium (with 10 mM serine) in triplicate in standard 96‐well plates (Costar 3595). Cultures were incubated overnight at 37°C with shaking (Titramax 100 shaker, 900 rpm), diluted 1:100 into experimental media, and returned to 37°C with shaking. Cell density (absorbance at 600 nm) and YFP fluorescence (excitation 504 nm and emission 540 nm) were monitored every 40–60 min using a Tecan plate reader (Infinite M1000 PRO). To minimize evaporation, wells on the edge of the plate were filled with water and plates were sealed with parafilm between readings.
2.5. Cell viability assays
At the indicated time points, 1 µl of each culture was serially diluted in 1× MOPS. 2 µl of each dilution was spotted onto an LB plate using a multichannel pipettor. Plates were photographed after incubation at 37°C for 18 hr.
2.6. Glucose assays
Cells were growth in 96‐well plates as described above. At the indicated time points, 900 µl of culture was passed through a 0.22 µm filter using a 1 ml syringe and stored at −20°C until processing. 100 µl of culture supernatant was mixed with 200 µl of glucose assay reagent (Sigma, GAHK20) in a 96‐well plate and incubated at room temperature for 15 min. Absorbance at 340nm was measured using a Tecan plate reader (Infinite M1000 PRO). Sample absorbance was normalized by subtracting the absorbance of 200 µl glucose assay reagent mixed with 100 µl water. Glucose quantities were calculated from a standard curve generated using the kit's glucose standard solution.
2.7. Microscopy
Cells harboring pASEC1 were grown in 96‐well plates as described above. At the indicated time points, 3 µl of culture was transferred to a poly‐L‐lysine‐coated microscope slide and covered with a coverslip. Cells were imaged on a DeltaVision Elite microscope at 60× magnification in the bright field and YFP channels. Cell number and cell area were measured using ImageJ software. YFP channel images (which only capture live cells) were converted to 16‐bit grayscale, and the threshold was adjusted to 61–255 to remove background. Cell area was calculated using the Analyze Particles function. Six fields per strain were imaged and quantified at each time point.
2.8. Live–dead staining and imaging
At the indicated time point, 5 µl of culture was mixed with a 1:100 dilution of a 1:1 mix of SYTO9 and propidium iodide from the BacLight Live/Dead bacterial viability kit (L7012, Thermo Fisher). The cells were stained by incubating at room temperature for 15 min and then placed on ice until imaging. 3 µl of the stained cultures were spotted between a coverglass and coverslip. Images were collected on a Leica DMi8 epifluorescence microscope with a 100× magnification oil‐immersion objective in the phase‐contrast, GFP and RFP channels. The images were opened using ImageJ software, and the contrast and brightness were adjusted using the Auto mode. Cells were manually identified using the phase‐contrast image and were then classified as live or dead from the corresponding fluorescence images. Five fields per strain were imaged and quantified at each time point.
2.9. Quantitative PCR
The 3 ml cultures were grown to mid‐exponential phase in AA‐rich medium (with 5 mM serine) and harvested by centrifugation at 3,000 g for 5 min. Cell pellets were resuspended in 250 µl 0.3 M sodium acetate and 10 mM EDTA (pH 4.5) and mixed with 250 µl acid phenol‐chloroform (pH 4.5) and 250 µl glass beads (G1877, Sigma). Samples were vortexed for 3 min at maximum speed and clarified by centrifugation at 12,000 g for 10 min at 4°C. The aqueous layer was collected, and RNA was purified using the Direct‐zol RNA purification kit (Zymo Research).
The 300 ng purified RNA was treated with DNase I (NEB) for 10 min at 37°C according to the manufacturer's instructions. Reverse transcription (RT) was performed using Maxima RT enzyme (EP0741, Thermo Fisher) and random hexamer primers using a 20 µl reaction volume. After incubation at 25°C for 10 min, 50°C for 30 min and 85°C for 5 min, samples were diluted 10‐fold and 2 µl of diluted sample was used as template for a 10 µl qPCR reaction in the next step. qPCR was performed using Maxima SYBR Green/ROX qPCR Master Mix (FERK0221, Thermo Fisher) according to the manufacturer's instructions. qPCR was performed in duplicate for each RT reaction using primers listed in Table A2. Negative RT controls were included to confirm the absence of DNA contamination. For each gene, CT values were converted to mRNA quantity by generating a standard curve from genomic DNA. gapA mRNA was used as internal reference to normalize all other mRNA levels.
3. RESULTS
3.1. Cells lacking sdaCB exhibit a growth defect during entry into stationary phase
The E. coli genome encodes three separate serine deaminases (Figure 1b), of which at least one is always expressed; SdaA predominates in nutrient‐poor conditions, SdaB predominates in nutrient‐rich conditions, and TdcG predominates during anaerobiosis (Shao & Newman, 1993; Zhang & Newman, 2008). SdaB is coexpressed with the predicted serine transporter SdaC (Shao, Lin, & Newman, 1994), but the functional interplay between these two proteins is unknown. To investigate the role of serine deamination in different growth conditions, we constructed strains of E. coli (BW25113) harboring deletions of sdaA, sdaCB or tdcG (Figure 1b, Table A1). [Note: for all strains mentioned in the text and figures, Δ indicates replacement of the gene's coding sequence by a kanamycin resistance cassette (kan), chloramphenicol resistance cassette (cm), or FRT scar sequence as described in Materials and Methods and as indicated in Table A1 and figure legends.] E. coli grown in minimal media containing serine require sdaA for survival (Su, Lang, & Newman, 1989; Su & Newman, 1991) because free serine inhibits the biosynthesis of some other amino acids (Hama et al., 1990; Saito et al., 2003). We confirmed that our ΔsdaA strain is unable to grow in MOPS minimal medium containing serine (Figure 1c). Under the same conditions, deletion of the sdaCB or tdcG genes did not negatively impact growth (Figure 1c), consistent with reports that these genes are not highly expressed in minimal media (Su & Newman, 1991).
To investigate the role of serine deamination in nutrient‐rich conditions, we monitored the growth of wild‐type and the three serine deaminase single mutant strains in a MOPS‐buffered defined medium supplemented with all amino acids and nucleobases (Neidhardt et al., 1974) (henceforth referred to as AA‐rich medium). All four strains exhibited identical growth kinetics until they began transitioning to stationary phase, at which point the optical density (OD) of the ΔsdaCB strain stopped increasing, resulting in a slight defect in overall growth yield (Figure 1d). This result suggested to us that sdaCB is not required for exponential growth in the presence of serine in AA‐rich medium but may become beneficial when specific nutrients are depleted. The growth yield defect of ΔsdaCB cells was smaller in a medium containing glycerol, rather than glucose, as the carbon source (Figure A1, panel a), suggesting that the effect may be glucose‐specific.
To estimate the effect of SdaC and SdaB activity on growth in AA‐rich medium, we performed the same experiment described above in a ΔserA background, in which cells are unable to synthesize serine de novo and growth yield is thus proportional to the availability of exogenous serine for biomass production. Indeed, growth of the ΔserA strain stopped abruptly after three hours (Figure 1e) and could be rescued by readdition of 5 mM serine (Figure A1, panel b), indicating that exogenous serine becomes depleted by mid exponential phase in our AA‐rich medium. If any of the serine deaminases are highly active in our experimental growth condition, then we expect the corresponding serine biosynthesis‐deaminase double mutant strains (Table A1) to have higher growth yield than the ΔserA single mutant because serine is not being converted to pyruvate and subsequently excreted from the cell as acetate. While the growth yield of the ΔserA ΔsdaA and ΔserA ΔtdcG strains was the same as the ΔserA strain, the ΔserA ΔsdaCB strain had a dramatically higher growth yield (Figure 1e), suggesting that SdaC/B activity competes with reactions that convert serine into biomass and consumes a significant amount of exogenous serine. Together, these results indicate that the sdaCB operon is highly active during growth in AA‐rich medium despite antagonizing biomass production and may benefit cells as they transition to nutrient deprivation.
3.2. Cells lacking sdaCB lyse upon glucose depletion
Because we observed a mild growth defect for the ΔsdaCB strain during entry into stationary phase in glucose‐containing AA‐rich medium, we hypothesized that sdaCB might aid cells in making the metabolic changes necessary to adapt when glucose is exhausted. To test this idea, we compared the growth of wild‐type and ΔsdaCB cells in modified AA‐rich medium in which availability of glucose was limited. In medium containing 1.4 mM glucose (20‐fold lower concentration than normal) and lacking serine, wild‐type cells grew normally to a final OD600 that was approximately 2.5‐fold lower than in AA‐rich medium (Figure 2a). In the same low‐glucose medium, the ΔsdaCB strain grew similarly to the wild‐type until approximately 150 min, at which point its optical density began decreasing, eventually dropping to background levels (Figure 2a). This phenotype was observed only when both glucose and serine were limiting; addition of either 5.6 mM glucose or 3 mM serine restored growth to wild‐type levels (Figure A2, panel a). Lysis of ΔsdaCB cells was also observed in a different E. coli genetic background (MG1655, Table A1), indicating that the phenotype is not strain‐specific (Figure A2, panel b).
Figure 2.
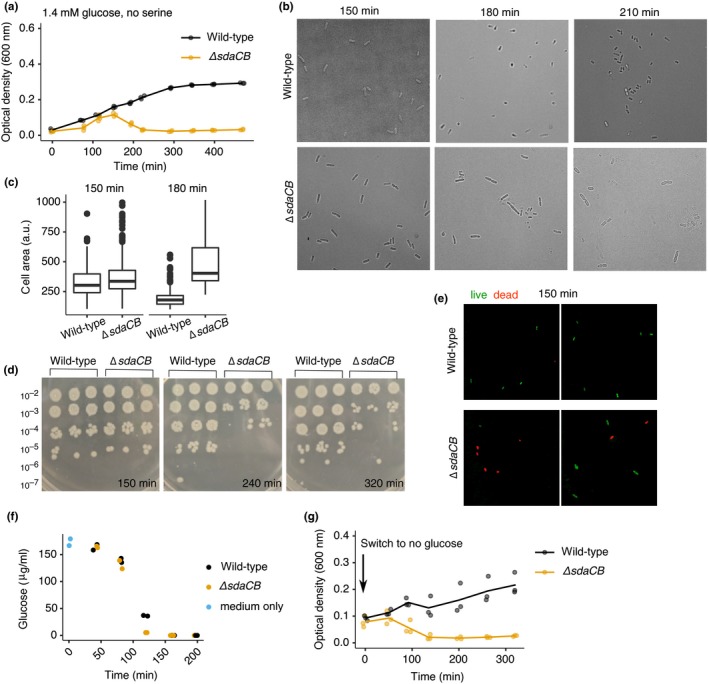
sdaCB prevents lysis upon glucose depletion. (a) Growth curves of wild‐type and ΔsdaCB (sdaCB::cm, ecMK102) strains in AA‐rich medium containing 0.025% (1.4 mM) glucose and no serine. (b) Bright‐field microscopy images of wild‐type and ΔsdaCB cells (sdaCB::cm, ecMK102; 60× magnification) grown as in (a) and transferred to poly‐L‐lysine‐coated microscope slides at the indicated time points. (c) Individual cell areas measured as described in Materials and Methods for wild‐type (n = 206, 150 min; n = 286, 180 min) and ΔsdaCB (sdaCB::cm, ecMK102; n = 316, 150 min; n = 50, 180 min) strains grown as in (a). (d) Colony‐forming unit (CFU) assays performed in biological triplicate at the indicated time points for cells grown in AA‐rich medium containing 0.025% (1.4 mM) glucose and no serine. 2 µl of each serial dilution was spotted onto an LB plate. (e) Live–dead staining of wild‐type and ΔsdaCB cells (sdaCB::cm, ecMK102; 100× magnification) grown as in (a) at the 150 min time point. Cells were stained and imaged by fluorescence microscopy as described in Materials and Methods. For each strain, two representative fields of view in the green (live) and red (dead) channels are shown out of the five fields of view that were quantified. 3% of wild‐type cells (one out of 39) and 43% of ΔsdaCB cells (17 out of 40) stained red and were classified as dead. (f) Glucose levels in sterile medium (1.4 mM glucose, no serine, blue points) and in culture supernatants at the indicated time points. Biological duplicates representative of three independent experiments are shown. (g) Growth curves of wild‐type and ΔsdaCB strains following removal of glucose. Overnight cultures were diluted 1:100 into AA‐rich medium with 2.8 mM glucose and no serine for 105 min. The cultures were then spun down briefly to remove the media, and cells were resuspended in AA‐rich medium with no glucose or serine prior to measurement of optical density over time. For all growth curves, three biological replicates are shown as points with their averages connected by lines
A downward trend in optical density is often indicative of cell lysis. Using microscopy, we observed fewer intact cells and accumulation of cellular debris for ΔsdaCB cells, but not wild‐type cells, after 150 min (Figure 2b). Interestingly, lysis was not preceded by significant morphological changes (Figure 2b), as has been observed in other studies of serine deaminase mutants (Zhang et al., 2010; Zhang & Newman, 2008). We did observe a decrease in cell size for the wild‐type strain (Figure 2c), which is expected for entry into stationary phase (Kolter, Siegele, & Tormo, 1993). By contrast, ΔsdaCB cells did not become smaller over time (Figure 2c), suggesting either that they failed to adapt to changing nutrient availability or that those ΔsdaCB cells that did shrink also lysed. To confirm that the drop in OD coincides with a loss of viability, we plated serial dilutions of wild‐type and ΔsdaCB cultures on LB plates, and observed that after 150–240 min, the ΔsdaCB cultures yielded approximately 100‐fold fewer colony‐forming units (CFU) than the wild‐type strain (Figure 2d). In addition, live/dead staining of bacteria at 150 min revealed that 43% of ΔsdaCB cells were dead at this time point (n = 40 cells), whereas only 3% of wild‐type cells were dead (n = 39 cells) (Figure 2e).
The lysis phenotype did not occur when glucose was replaced by the alternative carbon sources glycerol or pyruvate (Figure A2, panel c), suggesting a specific role for glucose depletion in causing lysis of ΔsdaCB cells. To determine whether the timing of lysis coincides with glucose depletion, we measured glucose levels in culture supernatants over time. We found that glucose levels dropped below detectable concentrations by 160 min, which corresponds with the time at which ΔsdaCB cells begin lysing (Figure 2f, Figure A2, panel d). To further test whether glucose depletion is the trigger for lysis of the ΔsdaCB strain, we grew cells in AA‐rich medium containing 2.8 mM glucose and no serine for 100 min and then switched them to the same medium containing no glucose. As predicted, the medium switch caused complete lysis of the ΔsdaCB strain, but not the wild‐type strain (Figure 2g). This result supports the conclusion that ΔsdaCB cells lyse specifically upon glucose depletion in the absence of exogenous serine.
3.3. sdaC is sufficient to prevent lysis
To gain insight into the mechanism by which loss of sdaCB causes lysis, we constructed sdaC and sdaB single deletion strains (Table A1). To minimize polar effects of removing sdaC on expression of sdaB, we excised the antibiotic cassette from the sdaC deletion strain using pCP20 (Datsenko & Wanner, 2000). This approach leaves an 84 bp scar sequence in place of the sdaC coding region that contains stop codons in all six reading frames (Datsenko & Wanner, 2000). We next measured the individual effects of sdaC and sdaB deletions on the lysis phenotype in medium with low glucose and no serine. Unexpectedly, the serine deaminase mutant, ΔsdaB, did not lyse upon glucose depletion, whereas the serine transporter mutant, ΔsdaC, did lyse (Figure 3a). qRT‐PCR analysis revealed that deletion of sdaC reduces sdaB expression (Figure A3, panel a) despite removal of the antibiotic cassette. To confirm that lysis of ΔsdaC cells was caused by loss of sdaC alone, rather than polar effects on expression of sdaB, we tested whether low‐copy plasmids (pSC*101 ori, 3–4 copies/cell, Table A1) (Lutz & Bujard, 1997) expressing sdaC or sdaB alone could prevent lysis. A plasmid expressing sdaC under the control of its native promoter completely rescued lysis of the ΔsdaCB strain (Figure 3b), whereas ΔsdaCB cells still lysed when harboring a plasmid expressing sdaB under the control of the sdaCB promoter (Figure 3b). Together, these results indicate that sdaC expression is sufficient to prevent lysis, regardless of whether sdaB is present (Figure 3b).
Figure 3.
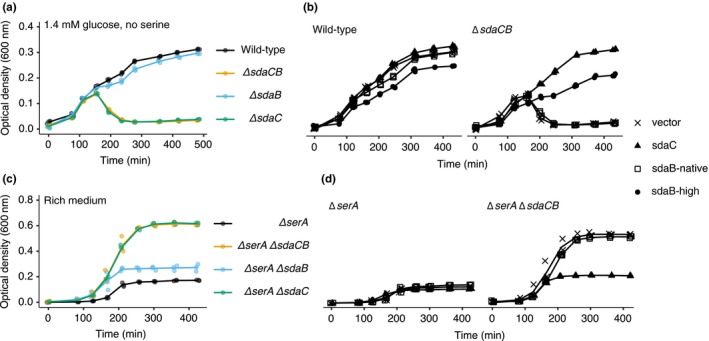
sdaC is sufficient to prevent lysis and antagonizes biomass production. (a) Growth curves of wild‐type, ΔsdaC (sdaC::FRT, ecMK180), ΔsdaB (sdaB::kan, ecMK163), and ΔsdaCB (sdaCB::cm, ecMK102) strains in AA‐rich medium with 1.4 mM glucose and no serine. (b) Growth curves of wild‐type and ΔsdaCB strains harboring a low‐copy plasmid expressing yfp (vector; ecMK136,141) or derivatives expressing sdaC under the control of its native promoter and leader sequence (sdaC; ecMK184,185), sdaB under the control of its native promoter (sdaB‐native; ecMK194,195) or sdaB under the control of the strong promoter pLtetO‐1 (sdaB‐high; ecMK187,188) (Table A1). Cells were grown in the same medium as in (a). (c) Growth curves of ΔserA strains with or without deletion of sdaB and/or sdaC (ecMK94, 106, 164, 181) in AA‐rich medium containing 5 mM serine. (d) Growth curves of ΔserA and ΔserA ΔsdaCB strains harboring a low‐copy plasmid expressing yfp (vector; ecMK140,142) or derivatives expressing sdaC under the control of its native promoter and leader sequence (sdaC; ecMK197, 198) or sdaB under the control of its native promoter (sdaB‐native; ecMK200, 201). Cells were grown in the same medium as in (c). For all growth curves, three biological replicates are shown as points with their averages connected by lines
Interestingly, a plasmid overexpressing sdaB under the control of the highly active pLtetO‐1 promoter (Table A1) (Lutz & Bujard, 1997) did prevent lysis of the ΔsdaCB strain (Figure 3b), indicating that overexpression of sdaB can substitute for loss of sdaC. To rule out the possibility that insufficiency of the ΔsdaB mutation to cause lysis is due to compensatory induction of another serine deaminase, we constructed a ΔsdaB ΔsdaA ΔtdcG triple deletion strain (Table A1); this strain also did not lyse upon glucose depletion (Figure A3, panel b). Finally, to rule out the possibility that a genetic determinant other than the sdaC coding region is responsible for preventing lysis, we constructed a derivative of the sdaC‐expressing plasmid containing a point mutation that introduces a premature stop codon after the 82nd amino acid of sdaC. This plasmid was unable to prevent lysis of ΔsdaCB cells (Figure A3, panel c).
To assess the effect of sdaC expression on biomass production, we returned to the ΔserA strain background that requires exogenously provided serine for normal growth yield. We observed that deletion of sdaC alone dramatically increased growth yield in the ΔserA background in AA‐rich medium, whereas deletion of sdaB alone only slightly increased yield (Figure 3c). To confirm that sdaC expression is sufficient to suppress growth, we performed the same experiment in strains harboring plasmids that express sdaC or sdaB from the sdaCB promoter, or the parent vector that constitutively expresses yfp (Table A1). The sdaC‐expressing plasmid reduced the growth yield of the ΔserA ΔsdaCB strain, whereas the vector and sdaB‐expressing plasmid had no effect (Figure 3d). These results indicate that sdaC expression significantly reduces growth yield in a strain unable to synthesize serine de novo–independently of sdaB, suggesting that SdaC diverts serine away from biomass production.
3.4. The glycine cleavage system promotes lysis upon glucose depletion
De novo serine biosynthesis begins with the glycolytic intermediate 3‐phosphoglycerate, which is converted to serine via three steps (Mattaini et al., 2016; Pizer, 1963; Stauffer, 2004) (Figure 1a). Because lysis of the ΔsdaC strain occurs when cells cannot obtain serine from the growth medium and have run out of its biosynthetic precursor (glucose), we reasoned that lysis may coincide with induction of alternative pathways for serine production (Figure 4a). The primary means of generating serine besides de novo synthesis is to catabolize other amino acids, such as glycine or threonine (Stauffer, 2004); thus, it could be that amino acid catabolism contributes to the lysis phenotype. In accordance with this hypothesis, the ΔsdaC strain did not lyse when glycine was omitted from the lysis‐promoting growth medium (1.4 mM glucose, no serine; Figure 4b).
In order to determine which biochemical pathway(s) involved in serine production might become more active upon glucose depletion, we constructed transcriptional reporters for operons known to be involved in generating serine from other amino acids: the glycine cleavage system (gcvTHP), serine hydroxymethyltransferase (glyA), threonine dehydrogenase (kbl‐tdh), and threonine aldolase (ltaE) (Figure 4a). As controls, we also constructed transcriptional reporters for the serine deaminase operon sdaCB and the serine biosynthesis gene serA. Promoters of interest were cloned upstream of the yfp translation initiation signals (RBS) and coding region in a low‐copy plasmid (Figure 4c, Table A1). We measured YFP levels over time in wild‐type strains harboring each reporter in two conditions: the lysis‐promoting medium (1.4 mM glucose, no serine) or the same medium containing additional glucose (5.6 mM). We compared YFP production from a given reporter over the time range at which ΔsdaC cells lyse in the low‐glucose condition (Figure 4c); cells grown in the higher glucose condition have not yet depleted the glucose supply by this time. The only amino acid catabolic operon that showed transcriptional induction upon glucose depletion was gcvTHP (Figure 4c), which encodes enzymes that extract a methyl group from glycine. This methyl group can then be combined with an additional glycine molecule by GlyA to produce serine (Stauffer, 2004). Of note, we also observed induction of sdaCB itself, consistent with the known regulation of this operon by catabolite repression (Hollands, Busby, & Lloyd, 2007) (Figure 4c).
Based on the role of glycine in promoting lysis (Figure 2a vs. Figure 4b) and the modest transcriptional activation of the glycine cleavage operon upon glucose depletion (Figure 4c), we hypothesized that glycine cleavage by GcvTHP contributes to lysis of the ΔsdaC strain. To test this idea, we deleted the gcvP gene, encoding glycine decarboxylase, in both wild‐type and ΔsdaC backgrounds (Table A1). In accordance with our hypothesis, gcvP deletion prevented lysis of the ΔsdaC strain (Figure 4d). Notably, ΔgcvP strains exhibited a growth defect relative to wild‐type following glucose depletion (Figure 4d), strengthening the notion that glycine cleavage is a major means of satisfying metabolic requirements when serine and glucose are absent.
3.5. The lysis phenotype is associated with a high serine to alanine ratio
We next sought to understand why production of serine through glycine cleavage might promote cell lysis upon glucose depletion. In bacteria, lysis is often indicative of a defect in cell wall biogenesis. A major component of the bacterial cell wall, peptidoglycan, is composed of glycan chains crosslinked by short peptides, and in E. coli and many other bacteria these peptide crosslinks have a high alanine content (Vollmer & Bertsche, 2008). It was recently shown that serine can be misincorporated into peptidoglycan crosslinks in place of alanine (Parveen & Reddy, 2017) when intracellular serine levels are high, likely due to weak substrate specificity of the enzyme that attaches L‐alanine to UDP‐N‐acetylmuramic acid in an early step of peptidoglycan biosynthesis (Liger, Blanot, & Heijenoort, 1991). Misincorporation of serine reduced the rate of peptidoglycan production and sensitized cells to β‐lactam antibiotics (Parveen & Reddy, 2017), indicating that it is deleterious. As a result, we hypothesized that lysis of the ΔsdaC strain could be due to a higher than normal serine to alanine ratio. Indeed, when we added 1.8 mM alanine to the lysis‐promoting growth medium (instead of the standard 0.8 mM in our AA‐rich medium), ΔsdaC cells no longer lysed (Figure 5a). Strikingly, omission of alanine from the lysis‐promoting medium led to lysis of the wild‐type strain (Figure 5b), further implicating a high serine to alanine ratio in triggering lysis upon glucose depletion. We also tested whether an orthogonal approach to altering alanine availability promotes lysis. Deletion of one of E. coli's three alanine‐synthesizing transaminases, alaA (Table A1), resulted in lysis of cells in which sdaCB was intact (Figure 5c). Finally, addition of equimolar pyruvate to the lysis‐promoting medium was insufficient to prevent lysis of ΔsdaC cells, reinforcing the notion that lysis is not caused by a lack of serine deamination products (Figure 5d).
Figure 5.
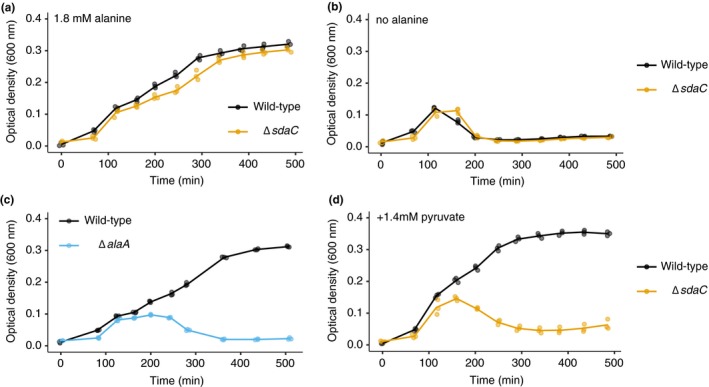
The lysis phenotype is associated with a high serine to alanine ratio. (a) Growth curves of wild‐type and ΔsdaC (sdaC::FRT, ecMK180) strains in AA‐rich medium with 1.4 mM glucose, no serine, and 1.8 mM alanine. (b) Growth curves of wild‐type and ΔsdaC (sdaC::FRT, ecMK180) strains in AA‐rich medium with 1.4 mM glucose, no serine, and no alanine. (c) Growth curves of wild‐type and ΔalaA (alaA::kan, ecMK165) strains in AA‐rich medium with 1.4 mM glucose, no serine, and 0.8 mM alanine. (d) Growth curves of wild‐type and ΔsdaC (sdaC::FRT, ecMK180) strains in AA‐rich medium with 1.4 mM glucose, no serine, 0.8 mM alanine, and 1.4 mM pyruvate. For all growth curves, three biological replicates are shown as points with their averages connected by lines
4. DISCUSSION
In this work, we discovered that glucose depletion in the absence of exogenous serine causes lysis of E. coli cells lacking the sdaCB operon. The lysis phenotype cannot be attributed to a requirement for serine deamination or its products, because (a) the lysis phenotype is independent of sdaB (Figure 3a,b), (b) a mutant lacking all three serine deaminases does not lyse (Figure A3, panel b), and (c) exogenous pyruvate is unable to prevent lysis of ΔsdaC cells (Figure 5d). Instead, we found SdaC to be the dominant activity in preventing lysis, thus revealing an unexpected role for serine transport during adaptation to changes in nutrient availability.
Lysis occurs upon glucose depletion in the absence of exogenously provided serine (Figure 2a) and is promoted by glycine and the glycine cleavage system (Figure 4). One possible explanation for these results is that exhaustion of primary serine sources (exogenous serine and its biosynthetic precursor, glucose) triggers catabolism of glycine (and possibly other amino acids) to satisfy cellular serine requirements and that this metabolic switch is surprisingly deleterious in the absence of sdaC. This scenario would suggest that serine produced via the GcvTHP/GlyA pathway has a greater potential to become toxic compared with serine synthesized de novo, possibly due to less stringent control of reaction rates leading to sudden accumulation of free serine. Indeed, we observed that ΔserA cells grown in glucose‐replete media do not appear to activate amino acid catabolic pathways upon exhaustion of exogenous serine, even though this should allow them to continue growing (Figure 1e). Although this situation, which is presumably due to glucose‐mediated repression (Wonderling & Stauffer, 1999), makes sense from an evolutionary perspective because wild‐type cells can produce serine from glucose (and amino acid precursors for producing serine are themselves useful as substrates for protein synthesis), our observation also implies that cells strongly prefer de novo synthesis to amino acid catabolism for endogenous production of serine.
In the above scenario, it is not immediately clear why SdaC can mitigate serine toxicity independent of SdaB. SdaC has been shown to import serine under physiological conditions (Hama, Shimamoto, Tsuda, & Tsuchiya, 1988; Shao et al., 1994), yet the medium that triggers lysis of ΔsdaC cells is devoid of exogenous serine. As a result, it might be that serine export is the function of SdaC that becomes essential upon glucose depletion. This notion is supported by our observations that (a) strong overexpression of the serine deaminase SdaB, which effectively removes serine from the cell, can compensate for loss of SdaC (Figure 3b) and (b) sdaC expression reduces the growth yield of a serine auxotroph (Figure 3d). SdaC is a proton/serine symporter that uses the energy gained from moving a proton down the electrochemical gradient (since cells are negative inside) to bring serine into the cell, often against its concentration gradient (Hama et al., 1988; Shao et al., 1994). However, because glucose depletion can reduce intracellular pH (Allison, Brynildsen, & Collins, 2011; Orij, Postmus, Ter Beek, Brul, & Smits, 2009; Peng et al., 2015) and secondary transporters often operate near thermodynamic equilibrium (Konings, Poolman, & Veen, 1994), it might be possible for glucose depletion to make the proton gradient favorable for serine export by SdaC, similar to the manner in which LacY reverses the direction of galactoside transport when the proton gradient is reduced (Guan & Kaback, 2009). As with LacY, this reversal could either be active (coupled to movement of H+) or via passive exchange (no movement of H+) (Guan & Kaback, 2009).
The other known serine transporters in E. coli are unlikely to export serine in the lysis‐promoting condition; the sodium‐serine symporter SstT would require a reversal of the sodium gradient and the anaerobic proton‐threonine/serine transporter TdcC is not expressed (Ogawa, Kim, Mizushima, & Tsuchiya, 1998). An alternative possibility is that SdaC is required to scavenge serine released from dead cells and/or exported by another protein, but our results are not consistent with serine starvation being the cause of lysis. In particular, omission of glycine or addition of alanine are not expected to increase intracellular serine availability. SdaC could also possess an additional serine detoxification function independent of its serine transport activity, but we consider this possibility unlikely due to the high homology of SdaC to well‐defined amino acid transporters such as TdcC (Shao et al., 1994).
Previous studies of E. coli lacking all three serine deaminases found that a triple mutant strain forms large, misshapen cells in glucose‐replete minimal media containing exogenous serine (Zhang et al., 2010; Zhang & Newman, 2008). These serine‐induced morphology defects occurred in the presence of sdaC, whereas we observe lysis of ΔsdaC cells but not ΔsdaB ΔsdaA ΔtdcG cells in our experimental condition (Figure 3b). Moreover, we did not observe any morphological changes in the sdaCB strain upon glucose depletion; the only noticeable difference between mutant and wild‐type strains was cell size (Figure 2c). Due to these significant differences in media composition and genetic determinants, it is unclear whether our lysis phenotype and the morphology defects observed by Zhang et al are related. In the case documented by Zhang et al, exogenous serine was responsible for the toxicity, whereas in our case endogenously produced serine may be the culprit. Interestingly, our observation that modulating alanine availability influences the lysis phenotype (Figure 5) is consistent with Zhang et al's report that serine toxicity could be prevented by alanine supplementation or overexpression of the alanine ligase, MurC (Zhang et al., 2010). Their hypothesis that free serine inhibits cell wall biogenesis by competing with alanine for MurC (Zhang et al., 2010) was recently corroborated by HPLC analysis directly demonstrating incorporation of serine into peptidoglycan crosslinks at the position normally occupied by alanine when intracellular serine levels are high (Parveen & Reddy, 2017). Serine incorporation was associated with a reduced rate of peptidoglycan production and sensitivity to β‐lactam antibiotics (Parveen & Reddy, 2017), indicating that the presence of serine in peptidoglycan crosslinks is indeed deleterious. Thus, it is possible that in our experimental condition glucose depletion and subsequent production of serine from glycine might cause a sudden imbalance between free serine and alanine levels that leads to toxic incorporation of serine into the cell wall.
An additional possibility consistent with our results is that altered amino acid homeostasis caused by changing glycine and alanine concentrations or mutating sdaC affects intracellular signaling and/or gene expression in a manner that avoids lysis. We did observe that the sdaCB mutant consumed glucose faster than the wild‐type strain (Figure 2f), so perhaps the cells have insufficient time to adapt to glucose depletion by synthesizing necessary biosynthetic enzymes. In this case, lysis might be caused by a shortage of enzymes and/or precursors needed for cell wall biogenesis. For example, expression of the major alanine racemase DadX, which catalyzes the production of D‐alanine from L‐alanine, is subject to catabolite repression in the presence of glucose (Wild, Hennig, Lobocka, Walczak, & Kłopotowski, 1985).
CONFLICT OF INTEREST
None declared.
AUTHOR CONTRIBUTIONS
MAK: involved in conceptualization, investigation, formal analysis, writing—original draft preparation; ARS: involved in conceptualization, investigation, writing—review and editing, supervision.
ETHICAL APPROVAL
None required.
ACKNOWLEDGMENTS
This work was supported by grant R35 GM119835 of the NIGMS (NIH) to ARS. MAK was supported by the Fred Hutch Chromosome Metabolism And Cancer Training Grant, NIH‐2T32CA9657‐26A1. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We thank Heungwon Park and Julio Vasquez for assistance with microscopy.
APPENDIX 1.
Figure A1.
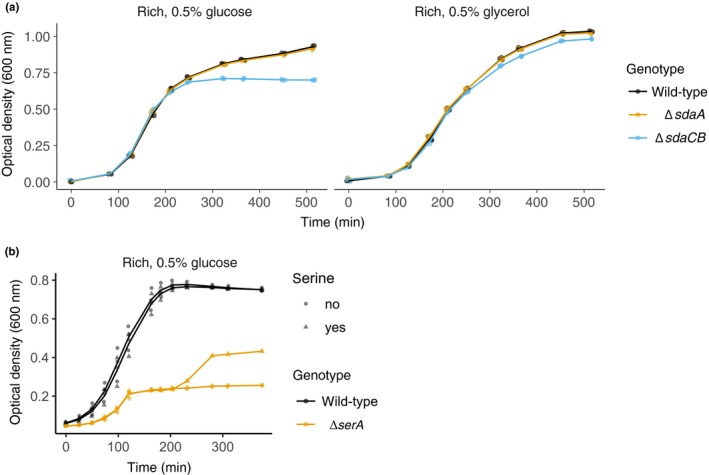
ΔsdaCB growth defect is specific to glucose‐containing media. (a) Growth curves of wild‐type and serine deaminase single deletion strains (sdaA::kan, ecMK101; sdaCB::cm, ecMK102) grown in AA‐rich medium containing 5 mM serine and 0.5% (56 mM) glycerol or 0.5% (28 mM) glucose as the carbon source. (b) Growth curves of wild‐type and (serA::FRT, ecMK94) strains in AA‐rich medium (0.5% glucose) containing 5 mM serine. After 200 min of growth, serine was added to a final concentration of 5 mM to three wells for each strain (triangles). For all growth curves, three biological replicates are shown as points with their averages connected by lines
Figure A2.
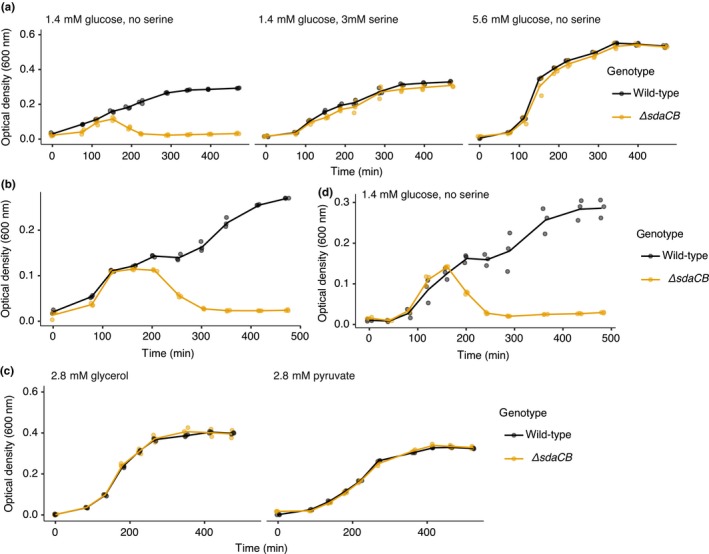
The lysis phenotype is specific to glucose depletion and also occurs in the MG1655 genetic background. (a) Growth curves of wild‐type and ΔsdaCB (sdaCB::cm, ecMK102) strains in AA‐rich medium containing 0.025% (1.4 mM) glucose and no serine (left), 1.4 mM glucose and 3 mM serine (center), or 5.6 mM glucose and no serine (right). Left panel is the same as in Figure 2a and is shown for comparison. (b) Growth curves of MG1655 and MG1655 ΔsdaCB (sdaCB::cm, ecMK209) strains in AA‐rich medium containing 0.025% (1.4 mM) glucose and no serine. (c) Growth curves of wild‐type and ΔsdaCB (sdaCB::cm, ecMK102) strains in AA‐rich medium with no glucose or serine, but containing 0.025% (2.8 mM) glycerol or pyruvate. (d) Growth curves corresponding to the glucose measurements in Figure 2e; wild‐type and ΔsdaCB (sdaCB::cm, ecMK102) strains in AA‐rich medium with no glucose and 1.4 mM serine. For all growth curves, three biological replicates are shown as points with their averages connected by lines
Figure A3.
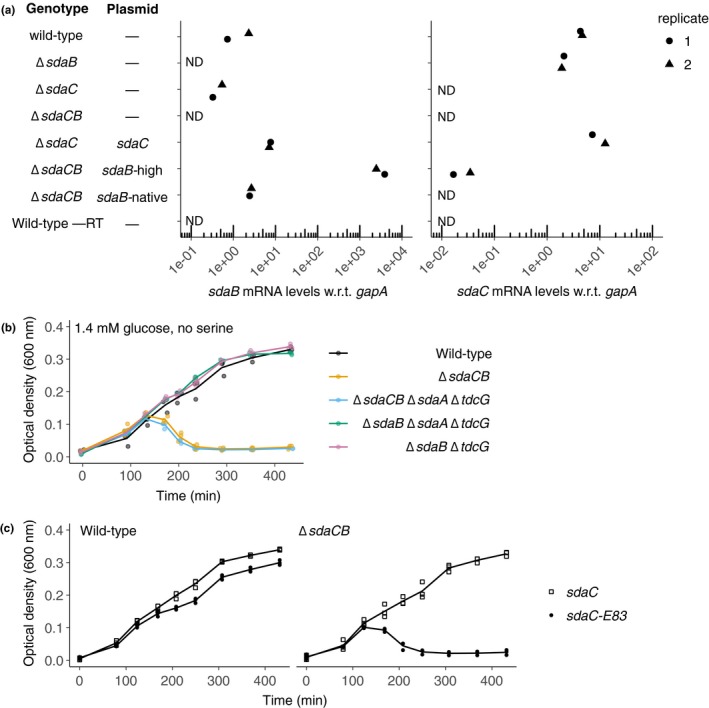
Expression levels of sdaC and sdaB in various mutant strains and lysis phenotypes of strains harboring different combinations of serine deaminase operon deletions and/or plasmids. (a) mRNA levels, as measured using qRT‐PCR, in indicated strains (from top to bottom: BW25113, ecMK163, 180, 102, 186, 188, 195) grown to midexponential phase in AA‐rich medium containing 5 mM serine. mRNA levels for sdaC and sdaB were normalized to the control gene gapA. ND = not detected, –RT = no reverse transcriptase. (b) Growth curves of strains harboring mutations in 0–3 of the serine deaminase loci (from top to bottom: BW25113, ecMK102, 228, 237, 236) in AA‐rich medium containing 0.025% (1.4 mM) glucose and no serine. (c) Growth curves of wild‐type and ΔsdaCB strains harboring plasmids expressing full‐length or truncated SdaC (premature stop codon at E83) in AA‐rich medium containing 0.025% (1.4 mM) glucose and no serine (ecMK136, 141, 221, 222). For growth curves, three biological replicates are shown as points with their averages connected by lines
Table A1.
Strains and plasmids used in this study
| Strain/Plasmid | Genotype | Source |
|---|---|---|
| Wild‐type Escherichia coli | BW25113 (F‐, DE(araD‐araB)567, lacZ4787(del)::rrnB‐3, LAM‐, rph‐1, DE(rhaD‐rhaB)568, hsdR514) | Datsenko and Wanner (2000) |
| ecMK94 | BW25113 serA::FRT | This work |
| ecMK101 | BW25113 sdaA::kan | This work |
| ecMK102 | BW25113 sdaCB::cm | This work |
| ecMK210 | BW25113 tdcG::cm | This work |
| ecMK105 | BW25113 serA::FRT sdaA::kan | This work |
| ecMK106 | BW25113 serA::FRT sdaCB::cm | This work |
| ecMK211 | BW25113 serA::FRT tdcG::cm | This work |
| ecMK136 | BW25113/pASEC1 | This work |
| ecMK137 | BW25113/pMKEC24 | This work |
| ecMK147 | BW25113/pMKEC32 | This work |
| ecMK172 | BW25113/pMKEC35 | This work |
| ecMK174 | BW25113/pMKEC36 | This work |
| ecMK176 | BW25113/pMKEC37 | This work |
| ecMK178 | BW25113/pMKEC30 | This work |
| ecMK163 | BW25113 sdaB::kan | This work |
| ecMK164 | BW25113 serA::FRT sdaB::kan | This work |
| ecMK180 | BW25113 sdaC::FRT | This work |
| ecMK181 | BW25113 serA::FRT sdaC::FRT | This work |
| ecMK129 | BW25113 gcvP::kan | This work |
| ecMK196 | BW25113 sdaC::FRT gcvP::cm | This work |
| ecMK184 | BW25113/pMKEC38 | This work |
| ecMK187 | BW25113/pMKEC39 | This work |
| ecMK194 | BW25113/pMKEC41 | This work |
| ecMK221 | BW25113/pMKEC43 | This work |
| ecMK140 | BW25113 serA::FRT/pASEC1 | This work |
| ecMK197 | BW25113 serA::FRT/pMKEC38 | This work |
| ecMK200 | BW25113 serA::FRT/pMKEC41 | This work |
| ecMK142 | BW25113 serA::FRT sdaCB::cm/pASEC1 | This work |
| ecMK198 | BW25113 serA::FRT sdaCB::cm/pMKEC38 | This work |
| ecMK201 | BW25113 serA::FRT sdaCB::cm/pMKEC41 | This work |
| ecMK141 | BW25113 sdaCB::cm/pASEC1 | This work |
| ecMK185 | BW25113 sdaCB::cm/pMKEC38 | This work |
| ecMK188 | BW25113 sdaCB::cm/pMKEC39 | This work |
| ecMK195 | BW25113 sdaCB::cm/pMKEC41 | This work |
| ecMK222 | BW25113 sdaCB::cm/pMKEC43 | This work |
| ecMK186 | BW25113 sdaC::FRT/pMKEC38 | This work |
| ecMK165 | BW25113 alaA::kan | This work |
| ecMK236 | BW25113 sdaB::FRT tdcG::cm | This work |
| ecMK237 | BW25113 sdaB::FRT tdcG::cm sdaA::kan | This work |
| ecMK228 | BW25113 sdaCB::FRT tdcG::cm sdaA::kan | This work |
| MG1655 | F‐, lambda‐, rph‐1 | Blattner et al. (1997) |
| ecMK209 | MG1655 sdaCB::cm | This work |
| pASEC1 | SC101* bla PLtetO1 T7 RBS yfp0 | Subramaniam et al. (2013) |
| pMKEC24 | SC101* bla PserA T7 RBS yfp0 | This work |
| pMKEC30 | SC101* bla PglyA T7 RBS yfp0 | This work |
| pMKEC32 | SC101* bla PsdaCB T7 RBS yfp0 | This work |
| pMKEC35 | SC101* bla Pkbl T7 RBS yfp0 | This work |
| pMKEC36 | SC101* bla PltaE T7 RBS yfp0 | This work |
| pMKEC37 | SC101* bla PgcvTHP T7 RBS yfp0 | This work |
| pMKEC38 | SC101* bla PsdaCB sdaC | This work |
| pMKEC39 | SC101* bla PLtetO1 T7 RBS sdaB | This work |
| pMKEC41 | SC101* bla PsdaCB sdaB | This work |
| pMKEC43 | SC101* bla PsdaCB sdaC E83* | This work |
| pKD13 | oriR6K bla FRT kan FRT | Datsenko and Wanner (2000) |
| pKD32 | oriR6K bla FRT cat FRT | Datsenko and Wanner (2000) |
| pSIM6 | SC101* repAts bla exo bet gam cI857ts | Datta et al. (2006) |
| pCP20 | SC101* repAts cI857ts bla cat FLP | Datsenko and Wanner (2000) |
Table A2.
Primers used in this study
| Primer | Purpose | Sequence(5′–3′) |
|---|---|---|
| oMK175 | tdcG::cm deletion cassette generation (long homology) | CAAGGCGATATGCGGTCTAC |
| oMK174 | tdcG::cm deletion cassette generation (long homology) | GATGACCGTGTTTATCACCG |
| oMK173 | sdaA::kan deletion cassette generation (long homology) | GCGTGATTGGCGAATTGTAC |
| oMK172 | sdaA::kan deletion cassette generation (long homology) | GGCATAATTCGTGAGCTGGC |
| oMK169 | Making E83* mutation in pMKEC38 | GCCCGGGTTTTTACCAGACAGTACG |
| oMK168 | Making E83* mutation in pMKEC38 |
CGTACTGTCTGGTAAAAACCCGGGCTAAGAC ATCACCGAGGTTGTAGAAG |
| oMK163 | sdaC‐qRT‐rev | TGCCATATCGCCGTACTCTT |
| oMK162 | sdaC‐qRT‐fwd | CACTGGAAACGCTGTCTCTG |
| oMK161 | sdaB‐qRT‐rev | CGGGTCACGTCTTTAAGCAG |
| oMK160 | sdaB‐qRT‐fwd | ATTGGCCCTTCCAGTTCTCA |
| oAS‐P6‐3I | gapA‐qRT‐for | GCTGAAGGCGAAATGAAAGG |
| oAS‐P6‐3J | gapA‐qRT‐rev | GTACCAGGATACCAGTTTCACG |
| oMK159 | Cloning pMKEC41 |
GGGATAATCTATCTTCCAAGTAGATTGTCCTT TCGCGCCGCTTTCGG |
| oMK158 | Cloning pMKEC41 | AATCTACTTGGAAGATAGATTAT |
| oMK157 | Cloning pMKEC39 |
ATTTGATGCCTCTAGACTCAGCTAATTAAGCT TTTAATCGCAGGCAACGATCTT |
| oMK156 | Cloning pMKEC38 |
TTGTTTAACTTTAAGAAGGAGATGGTACCATG ATTAGCGTATTCGATATTTTC |
| oMK155 | Cloning pMKEC38 |
ATTTGATGCCTCTAGACTCAGCTAATTAAGCT TTTAGCTGAACAGAGAGTAGAAG |
| oMK153 | Cloning pMKEC38 |
GGCACCGGAGGCTTTCGTCTTCACCTCGAGG GTCACTTACTCATCAACTC |
| oMK152 | Verification of chromosomal sdaC deletion | GGTCCAACGGTATGAGAACT |
| oMK151 | sdaC::kan deletion cassette generation | CGGAGGAAGCGCCGCCCGAAAGCGGCGCGA AAGGACATTCCGGGGATCCGTCGACC |
| oMK148 | Cloning pMKEC36 |
CCTTCTTAAAGTTAAACAAAATTATTGAATTCG GCATGTCCTTATTATGAC |
| oMK147 | Cloning pMKEC36 |
GGCACCGGAGGCTTTCGTCTTCACCTCGAGG ACGCGGTGATGAAGTGATC |
| oMK146 | Cloning pMKEC35 |
CCTTCTTAAAGTTAAACAAAATTATTGAATTCT GCGATTCTCCAGACAGGGC |
| oMK145 | Cloning pMKEC35 |
GGCACCGGAGGCTTTCGTCTTCACCTCGAGG TTTCCCTCCATTAGTGGGT |
| oMK144 | Cloning pMKEC37 |
CCTTCTTAAAGTTAAACAAAATTATTGAATTCC TTGTCCTCATTGAATAAGCGG |
| oMK143 | Cloning pMKEC37 |
GGCACCGGAGGCTTTCGTCTTCACCTCGAGA AATTTCTCCTCTGTTGTTTATTTG |
| oMK134 | Verification of chromosomal alaA deletion | GTTGCGCATGAGCGGCCAG |
| oMK133 | Verification of chromosomal alaA deletion | GCATGTAAAGCATATACACC |
| oMK132 | alaA::kan deletion cassette generation | CGCTGCCGGATGCAATCTCCGGCAGTGAAAT TAAGAATTCCGGGGATCCGTCGACC |
| oMK131 | alaA::kan deletion cassette generation | CGGCAGGGAGTGGCGATAACAGCAAAAAAG GTCAAGATTCGTGTAGGCTGGAGCTGCTTC |
| oMK130 | Verification of chromosomal sdaB deletion | GCGATGATCCTGTTCCTGATG |
| oMK126 | Verification of chromosomal serA deletion | GAAAGGCGGATGCAAATCC |
| oMK125 | Verification of chromosomal serA deletion | GACCTGCCCGTTGATTTTC |
| oMK121 | Verification of chromosomal gcvP deletion | CTGTCTGAGCACTTTTGTAC |
| oMK120 | Verification of chromosomal gcvP deletion | GCATACGAAGCATTGTTAG |
| oMK119 | gcvP::cm and gcvP::kan deletion cassette generation |
GTCATCTGACTAAAAAGGCGCCGAAGCGCCT TTAGAAAATTCCGGGGATCCGTCGACC |
| oMK118 | gcvP::cm and gcvP::kan deletion cassette generation |
GGTTCACAATTCACTGCACGTTTCAGGAACCA TCGCTCGTGTAGGCTGGAGCTGCTTC |
| oMK113 | Verification of chromosomal tdcG deletion | CGCAGGAGAATATTCATTC |
| oMK112 | Verification of chromosomal tdcG deletion | GAAGCCATCGCAGTACGTAG |
| oMK111 | tdcG::cm deletion cassette generation | GCACATTTGTGCACCCAAGGATGAAAGCTGA CAGCAATGATTCCGGGGATCCGTCGACC |
| oMK110 | tdcG::cm deletion cassette generation | CCGCTCCACTTCACTGAACGGCAATCCGAGG GTGTGGATGTGTAGGCTGGAGCTGCTTC |
| oMK109 | Verification of chromosomal sdaCB deletion | GATCAGGCGATAAGGGTAAC |
| oMK108 | Verification of chromosomal sdaCB deletion | GATTAAGCCATCGCCGATAGAC |
| oMK107 | sdaCB::cm deletion cassette generation | CGGGAAGAGGCCTCGCAAAACGAGGCCTTT GGAGAGCGAATTCCGGGGATCCGTCGACC |
| oMK106 | sdaCB::cm deletion cassette generation | GCTAAAAGCTGAATTATTTGCATTCCTCCAGG AGAAATAGGTGTAGGCTGGAGCTGCTTC |
| oMK105 | Verification of chromosomal sdaA deletion | GTAAGCGTATGGCGACAAAC |
| oMK104 | Verification of chromosomal sdaA deletion | GTGAATAGTTAAGCCAGTCG |
| oMK103 | sdaA::kan deletion cassette generation | GGGTATAAATTCGCCCATCCGTTGCAGATGG GCGAGTAAGAAGTAATTCCGGGGATCCGTCG ACC |
| oMK102 | sdaA::kan deletion cassette generation | CGTTACTGGAAGTCCAGTCACCTTGTCAGGA GTATTATCGTGTAGGCTGGAGCTGCTTC |
| oMK97 | Verification of kanR‐containing cassette integration | GCGCATCGCCTTCTATCGCC |
| oMK86 | Cloning pMKEC32 |
CCTTCTTAAAGTTAAACAAAATTATTGAATTCC TATTTCTCCTGGAGGAATGC |
| oMK84 | Cloning pMKEC30 |
CCTTCTTAAAGTTAAACAAAATTATTGAATTCC CGCATCTCCTGACTCAGC |
| oMK69 | Verification of cmR‐containing cassette integration | GATCTTCCGTCACAGGTAGG |
| oMK68 | Verification of cmR‐containing cassette integration | TTATACGCAAGGCGACAAGG |
| oMK66 | Verification of kanR‐containing cassette integration | GGAACACGGCGGCATCAGAG |
| oMK49 | Cloning pMKEC24 |
GGCACCGGAGGCTTTCGTCTTCACCTCGAGC GGTGTCAAAACCATTGTGAAATG |
| oMK48 | Cloning pMKEC24 |
CCTTCTTAAAGTTAAACAAAATTATTGAATTCT TACCCAATCCTGTCTTTTGAAATG |
| oMK47 | Cloning into pZS11 backbone | GAATTCAATAATTTTGTTTAACTTTAAGAAGG |
| oMK39 | Sequence verification of transcriptional reporters | GATCGTACGTTCTTGGAC |
| oMK38 | Sequence verification of transcriptional reporters | CAGGAAGGCAAAATGCCGC |
| oMK37 | Cloning pMKEC32 |
GGCACCGGAGGCTTTCGTCTTCACCTCGAGT CTCCGCTCCCCCGGTGACG |
| oMK33 | Cloning pMKEC30 |
GGCACCGGAGGCTTTCGTCTTCACCTCGAGA TGGTCTTCCTTTTTTTGCATC |
| oMK27 | Cloning into pZS11 backbone | CTCGAGGTGAAGACGAAAGCCTCCGGTGCC |
| oMF6 | Sequence verification of pZS11 plasmids | CAGTCTTTCGACTGAGCCTTTCGTTTTAT |
| oMF1 | Cloning into pZS11 backbone | GGTACCATCTCCTTCTTAAAGTTAAACAA |
Kriner MA, Subramaniam AR. The serine transporter SdaC prevents cell lysis upon glucose depletion in Escherichia coli . MicrobiologyOpen. 2020;9:e960 10.1002/mbo3.960
DATA AVAILABILITY STATEMENT
Raw data and scripts for performing all analyses and generating figures in this manuscript are publicly available at https://github.com/rasilab/mkriner_2018.
REFERENCES
- Allison, K. R. , Brynildsen, M. P. , & Collins, J. J. (2011). Metabolite‐enabled eradication of bacterial persisters by aminoglycosides. Nature, 473, 216–220. 10.1038/nature10069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avcilar‐Kucukgoze, I. , Bartholomäus, A. , Cordero Varela, J. A. , Kaml, R. F. , Neubauer, P. , Budisa, N. , & Ignatova, Z. (2016). Discharging tRNAs: A tug of war between translation and detoxification in Escherichia coli . Nucleic Acids Research, 44(17), 8324–8334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner, F. R. , Plunkett, G. 3rd , Bloch, C. A. , Perna, N. T. , Burland, V. , Riley, M. , … Shao, Y. (1997). The complete genome sequence of Escherichia coli K‐12. Science, 277, 1453–1462. [DOI] [PubMed] [Google Scholar]
- Burman, J. D. , Harris, R. L. , Hauton, K. A. , Lawson, D. M. , & Sawers, R. G. (2004). The iron‐sulfur cluster in the L‐serine dehydratase TdcG from Escherichia coli is required for enzyme activity. FEBS Letters, 576, 442–444. [DOI] [PubMed] [Google Scholar]
- Cicchillo, R. M. , Baker, M. A. , Schnitzer, E. J. , Newman, E. B. , & Krebs, C. (2004). Escherichia coli L‐serine deaminase requires a [4Fe‐4S] cluster in catalysis. The Journal of Biological Chemistry, 279, 32418–32425. [DOI] [PubMed] [Google Scholar]
- Datsenko, K. A. , & Wanner, B. L. (2000). One‐step inactivation of chromosomal genes in Escherichia coli K‐12 using PCR products. Proceedings of the National Academy of Sciences of the United States of America, 97, 6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta, S. , Costantino, N. , & Court, D. L. (2006). A set of recombineering plasmids for gram‐negative bacteria. Gene, 379, 109–115. 10.1016/j.gene.2006.04.018 [DOI] [PubMed] [Google Scholar]
- E. coli Genome Project . EZ rich defined medium. Retrieved from https://www.genome.wisc.edu/resources/protocols/ezmedium [Google Scholar]
- Gao, B. , Vorwerk, H. , Huber, C. , Lara‐Tejero, M. , Mohr, J. , Goodman, A. L. , … Hofreuter, D. (2017). Metabolic and fitness determinants for in vitro growth and intestinal colonization of the bacterial pathogen Campylobacter jejuni . PLOS Biology, 15, e2001390 10.1371/journal.pbio.2001390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, L. , & Kaback, H. R. (2009). Properties of a LacY ef ux mutant. Biochemistry, 48, 9250–9255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama, H. , Shimamoto, T. , Tsuda, M. , & Tsuchiya, T. (1988). Characterization of a novel L‐serine transport system in Escherichia coli . Journal of Bacteriology, 170, 2236–2239. 10.1128/jb.170.5.2236-2239.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama, H. , Sumita, Y. , Kakutani, Y. , Tsuda, M. , & Tsuchiya, T. (1990). Target of serine inhibition in Escherichia coli . Biochemical and Biophysical Research Communications, 168, 1211–1216. 10.1016/0006-291X(90)91157-N [DOI] [PubMed] [Google Scholar]
- Hollands, K. , Busby, S. J. W. , & Lloyd, G. S. (2007). New targets for the cyclic AMP receptor protein in the Escherichia coli K‐12 genome. FEMS Microbiology Letters, 274, 89–94. [DOI] [PubMed] [Google Scholar]
- Hosios, A. M. , Hecht, V. C. , Danai, L. V. , Johnson, M. O. , Rathmell, J. C. , Steinhauser, M. L. , … Vander Heiden, M. G. (2016). Amino acids rather than glucose account for the majority of cell mass in proliferating mammalian cells. Developmental Cell, 36, 540–549. 10.1016/j.devcel.2016.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolter, R. , Siegele, D. A. , & Tormo, A. (1993). The stationary phase of the bacterial life cycle. Annual Review of Microbiology, 47, 855–874. 10.1146/annurev.mi.47.100193.004231 [DOI] [PubMed] [Google Scholar]
- Konings, W. N. , Poolman, B. , & van Veen, H. W. (1994). Solute transport and energy transduction in bacteria. Antonie Van Leeuwenhoek, 65, 369–380. 10.1007/BF00872220 [DOI] [PubMed] [Google Scholar]
- Li, G.‐W. , Oh, E. , & Weissman, J. S. (2012). The anti‐Shine‐Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature, 484, 538–541. 10.1038/nature10965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, K.‐M. , Chao, T.‐B. , Tian, Y.‐F. , Lin, C.‐Y. , Lee, S.‐W. , Chuang, H.‐Y. , … Li, C.‐F. (2016). Overexpression of the PSAT1 gene in nasopharyngeal carcinoma is an indicator of poor prognosis. Journal of Cancer, 7, 1088–1094. 10.7150/jca.15258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liger, D. , Blanot, D. , & van Heijenoort, J. (1991). Effect of various alanine analogues on the L‐alanine‐adding enzyme from Escherichia coli . FEMS Microbiology Letters, 64, 111–115. [DOI] [PubMed] [Google Scholar]
- Locasale J. W. (2013). Serine, glycine and the one‐carbon cycle: Cancer metabolism in full circle. Nature Reviews. Cancer, 13, 572–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale, J. W. , Grassian, A. R. , Melman, T. , Lyssiotis, C. A. , Mattaini, K. R. , Bass, A. J. , … Vander Heiden, M. G. (2011). Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nature Genetics, 43, 869–874. 10.1038/ng.890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz, R. , & Bujard, H. (1997). Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1‐I2 regulatory elements. Nucleic Acids Research, 25, 1203–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens, A. T. , Taylor, J. , & Hilser, V. J. (2015). Ribosome A and P sites revealed by length analysis of ribosome profiling data. Nucleic Acids Research, 43, 3680–3687. 10.1093/nar/gkv200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaini, K. R. , Sullivan, M. R. , & Heiden, M. G. V. (2016). The importance of serine metabolism in cancer. The Journal of Cell Biology, 214, 249–257. 10.1083/jcb.201604085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt, F. C. , Bloch, P. L. , & Smith, D. F. (1974). Culture medium for enterobacteria. Journal of Bacteriology, 119, 736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netzer, R. , Peters‐Wendisch, P. , Eggeling, L. , & Sahm, H. (2004). Cometabolism of a nongrowth substrate: L‐serine utilization by Corynebacterium glutamicum . Applied and Environmental Microbiology, 70, 7148–7155. 10.1128/AEM.70.12.7148-7155.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, A. C. , & Maddocks, O. D. K. (2017). Serine and functional metabolites in cancer. Trends in Cell Biology, 27(9), 645–657. 10.1016/j.tcb.2017.05.001 [DOI] [PubMed] [Google Scholar]
- Ogawa, W. , Kim, Y.‐M. , Mizushima, T. , & Tsuchiya, T. (1998). Cloning and expression of the gene for the Na+‐coupled serine transporter from Escherichia coli and characteristics of the transporter. Journal of Bacteriology, 180, 6749–6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orij, R. , Postmus, J. , Ter Beek, A. , Brul, S. , & Smits, G. J. (2009). In vivo measurement of cytosolic and mitochondrial pH using a pH‐sensitive GFP derivative in Saccharomyces cerevisiae reveals a relation between intracellular pH and growth. Microbiology, 155, 268–278. 10.1099/mic.0.022038-0 [DOI] [PubMed] [Google Scholar]
- Parveen, S. , & Reddy, M. (2017). Identification of Y H (PgeF) as a factor contributing to the maintenance of bacterial peptidoglycan composition. Molecular Microbiology, 105, 705–720. [DOI] [PubMed] [Google Scholar]
- Peng, B. O. , Su, Y.‐B. , Li, H. , Han, Y. I. , Guo, C. , Tian, Y.‐M. , & Peng, X.‐X. (2015). Exogenous alanine and/or glucose plus kanamycin kills antibiotic‐resistant bacteria. Cell Metabolism, 21, 249–262. 10.1016/j.cmet.2015.01.008 [DOI] [PubMed] [Google Scholar]
- Pizer, L. I. (1963). The pathway and control of serine biosynthesis in Escherichia coli . The Journal of Biological Chemistry, 238, 3934–3944. [PubMed] [Google Scholar]
- Possemato, R. , Marks, K. M. , Shaul, Y. D. , Pacold, M. E. , Kim, D. , Birsoy, K. , … Sabatini, D. M. (2011). Functional genomics reveals serine synthesis is essential in PHGDH‐amplified breast cancer. Nature, 476, 346–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramotar, D. , & Newman, E. B. (1986). An estimate of the extent of deamination of L‐serine in auxotrophs of Escherichia coli K‐12. Canadian Journal of Microbiology, 32, 842–846. [DOI] [PubMed] [Google Scholar]
- Saito, K. , Ishida, S. , & Yamada, K. (2003). Inhibition of growth by L‐serine in Escherichia coli . The Bulletin of Mukogawa Women's University. Natural Science, 51, 51–53. [Google Scholar]
- Selvarasu, S. , Siak‐Wei Ow, D. , Lee, S. Y. , Lee, M. M. , Kah‐Weng Oh, S. , Karimi, I. A. , & Lee, D. Y. (2009). Characterizing Escherichia coli DH5α growth and metabolism in a complex medium using genome‐scale flux analysis. Biotechnology and Bioengineering, 102, 923–934. [DOI] [PubMed] [Google Scholar]
- Shao, Z. , Lin, R. T. , & Newman, E. B. (1994). Sequencing and characterization of the sdaC gene and identification of the sdaCB operon in Escherichia coli K12. European Journal of Biochemistry, 222, 901–907. [DOI] [PubMed] [Google Scholar]
- Shao, Z. , & Newman, E. B. (1993). Sequencing and characterization of the sdaB gene from Escherichia coli K‐12. European Journal of Biochemistry, 212, 777–784. 10.1111/j.1432-1033.1993.tb17718.x [DOI] [PubMed] [Google Scholar]
- Stauffer, G. V. (2004). Regulation of serine, glycine, and one‐carbon biosynthesis. EcoSal Plus, 1 10.1128/ecosalplus.3.6.1.2 [DOI] [PubMed] [Google Scholar]
- Su, H. S. , Lang, B. F. , & Newman, E. B. (1989). L‐serine degradation in Escherichia coli K‐12: Cloning and sequencing of the sdaA gene. Journal of Bacteriology, 171, 5095–5102. 10.1128/jb.171.9.5095-5102.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, H. , & Newman, E. B. (1991). A novel L‐serine deaminase activity in Escherichia coli K‐12. Journal of Bacteriology, 173, 2473–2480. 10.1128/jb.173.8.2473-2480.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam, A. R. , Pan, T. , & Cluzel, P. (2013). Environmental perturbations lift the degeneracy of the genetic code to regulate protein levels in bacteria. Proceedings of the National Academy of Sciences of the United States of America, 110, 2419–2424. 10.1073/pnas.1211077110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer, W. , & Bertsche, U. (2008). Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli . Biochimica Et Biophysica Acta (BBA) ‐ Biomembranes, 1778, 1714–1734. 10.1016/j.bbamem.2007.06.007 [DOI] [PubMed] [Google Scholar]
- Wild, J. , Hennig, J. , Lobocka, M. , Walczak, W. , & Kłopotowski, T. (1985). Identification of the dadX gene coding for the predominant isozyme of alanine racemase in Escherichia coli K12. Molecular & General Genetics: MGG, 198, 315–322. [DOI] [PubMed] [Google Scholar]
- Wonderling, L. D. , & Stauffer, G. V. (1999). The cyclic AMP receptor protein is dependent on GcvA for regulation of the gcv operon. Journal of Bacteriology, 181, 1912–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda, M. , Nagata, S. , Yamane, S. , Kunikata, C. , Kida, Y. , Kuwano, K. , … Okuda, J. (2017). Pseudomonas aeruginosa serA gene is required for bacterial translocation through Caco‐2 cell monolayers. PLoS ONE, 12 10.1371/journal.pone.0169367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , El‐Hajj, Z. W. , & Newman, E. (2010). Deficiency in l‐serine deaminase interferes with one‐carbon metabolism and cell wall synthesis in Escherichia coli K‐12. Journal of Bacteriology, 192, 5515–5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , & Newman, E. (2008). Deficiency in l‐serine deaminase results in abnormal growth and cell division of Escherichia coli K‐12. Molecular Microbiology, 69, 870–881. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw data and scripts for performing all analyses and generating figures in this manuscript are publicly available at https://github.com/rasilab/mkriner_2018.


