Abstract
Despite published evidence that IQS (2‐(2‐hydroxylphenyl)‐thiazole‐4‐carbaldehyde) is in fact aeruginaldehyde, a by‐product of the siderophore pyochelin biosynthesis or degradation and that the ambABCDE genes are not responsible for IQS synthesis, several authors, including in top review journals, perpetuate the wrong information. I hope that this short comment will clarify the situation once and for all.
Keywords: aeruginaldehyde, IQS, Pseudomonas aeruginosa, pyochelin, quorum sensing
Despite published evidence that IQS (2‐(2‐hydroxylphenyl)‐thiazole‐4‐carbaldehyde) is in fact aeruginaldehyde, a by‐product of the siderophore pyochelin biosynthesis or degradation and that the ambABCDE genes are not responsible for IQS synthesis, several authors, including in top review journals, perpetuate the wrong information. I hope that this short comment will clarify the situation once and for all.
1. COMMENTARY
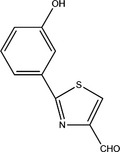
Pseudomonas aeruginosa is an important opportunistic pathogen causing a range of infections in immunocompromised individuals and in those with cystic fibrosis where it can establish itself in the lungs (Lyczak, Cannon, & Pier, 2000). The production of the majority of P. aeruginosa virulence factors is dependent on three linked quorum sensing systems, Las, Rhl, and PQS (Williams & Camara, 2009). Las and Rhl systems rely on N‐acyl‐homoserine lactones signal molecules whereby the Las system controls the Rhl system (Williams & Camara, 2009). A third system, involving the Pseudomonas quinolone signal (PQS) molecule, is also hierarchically dependent of Las (Williams & Camara, 2009). In Lee et al., 2013 published an article suggesting that there is a fourth quorum sensing signal molecule, termed IQS (2‐(2‐hydroxylphenyl)‐thiazole‐4‐carbaldehyde) (Lee et al., 2013), reviewed in 2015 (Lee & Zhang, 2015). They found that the production of pyocyanin, a phenazine whose production depends on the Rhl and PQS systems (Williams & Camara, 2009) is still observed in a las mutant under low phosphate condition (Lee et al., 2013). By screening a transposon mutant library, they found that the Las‐independent pyocyanin production was abolished in an ambB mutant (Lee et al., 2013). They further claimed that the ambABCDE genes are responsible for the biosynthesis of IQS (Lee et al., 2013). This last claim is wrong as explained in this commentary.
1.1. The ambABCDE genes are not involved in IQS synthesis
The ambABCDE genes code for proteins involved in the production of the antimetabolite L‐2‐amino‐4methoxy‐trans‐3‐butenoic acid (AMB) (Rojas Murcia et al., 2015) and could therefore not possibly be responsible for IQS production. AmbB and AmbE are nonribosomal peptide synthetases whose primary product is the l‐Ala‐l‐Glu‐l‐Ala tripeptide. AmbC and AmbD further modify the L‐Glu into AMB. AMB is released after removal of the two flanking L‐Ala residues and is excreted via AmbA, a LysE amino acid transporter (Rojas Murcia et al., 2015). AMB is produced by P. aeruginosa and causes a germination arrest in plants (Chahtane et al., 2018).
1.2. IQS is aeruginaldehyde, a by‐product of the siderophore pyochelin
A first indication that IQS could not be the product of AmbABCDE was the discovery that the IQS structure is identical to the structure of aeruginaldehyde, a compound produced by other Pseudomonas, including P. protegens and in Burkholderia thailandensis which do not have the amb genes cluster (Trottmann, Franke, Ishida, García‐Altares, & Hertweck, 2019; Ye et al., 2014). Aeruginaldehyde has been detected together with other compounds (aeruginoic acid, aeruginol, dihydroaeruginoic acid), which may be all products derived from the siderophore pyochelin biosynthetic pathway (Ye et al., 2014). In a very recent study, Trottmann et al. confirmed that aeruginaldehyde is derived from pyochelin since an incubation of the siderophore at 30°C resulted in the appearance of aeruginaldehyde (Trottmann et al., 2019). Ye et al. (2014) suggested that aeruginaldehyde could derive from dihydroaeruginoic acid (Dha) released from PchE during the assembly of the NRPS enzymes and the biosynthetic process (Ye et al., 2014). Interestingly, Maspoli, Wenner, Mislin, and Reimmann (2014) found in P. aeruginosa PA14 a genomic island containing genes for the biosynthesis of (enantio)pyochelin. Complementation of a pyochelin negative mutant with one NRPS gene led to the production of Dha (Maspoli et al., 2014). In a recent study, Sun et al. found that only the Rhl and PQS QS systems are involved in the regulation of phenazine production in a P. aeruginosa strain while mutants with deletions in amb or pch genes clusters were not affected in their production of phenazine compounds, excluding their possible participation in the QS circuitry (Sun, Zhou, Jin, Jiang, & He, 2016).
2. CONCLUSION
Despite the evidence presented here in this short commentary, still scientists publish articles, including reviews, mentioning IQS as the fourth quorum sensing molecule of P. aeruginosa being the product of AmbABCDE. The aim of this short comment is to clarify this point.
CONFLICT OF INTEREST
None declared.
AUTHOR CONTRIBUTIONS
PC: Conceptualization, Formal analysis, Writing—original draft, Writing—review and editing.
ETHICAL APPROVAL
None required.
ACKNOWLEDGMENTS
PC wishes to thank Dr. Christine Baysse and Prof. Sylvie Chevalier for critical reading of the manuscript. No funding declared.
Cornelis P. Putting an end to the Pseudomonas aeruginosa IQS controversy. MicrobiologyOpen. 2020;9:e962 10.1002/mbo3.962
DATA AVAILABILITY STATEMENT
Not applicable.
REFERENCES
- Chahtane, H. , Nogueira Füller, T. , Allard, P.‐M. , Marcourt, L. , Ferreira Queiroz, E. , Shanmugabalaji, V. , … Lopez‐Molina, L. (2018). The plant pathogen Pseudomonas aeruginosa triggers a DELLA‐dependent seed germination arrest in Arabidopsis . Elife, 7:e37082, 10.7554/eLife.37082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. , Wu, J. , Deng, Y. , Wang, J. , Wang, C. , Wang, J. , … Zhang, L.‐H. (2013). A cell‐cell communication signal integrates quorum sensing and stress response. Nature Chemical Biology, 9, 339–343. 10.1038/nchembio.1225 [DOI] [PubMed] [Google Scholar]
- Lee, J. , & Zhang, L. (2015). The hierarchy quorum sensing network in Pseudomonas aeruginosa . Protein Cell, 6, 26–41. 10.1007/s13238-014-0100-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyczak, J. B. , Cannon, C. L. , & Pier, G. B. (2000). Establishment of Pseudomonas aeruginosa infection: Lessons from a versatile opportunist. Microbes and Infection, 2, 1051–1060. [DOI] [PubMed] [Google Scholar]
- Maspoli, A. , Wenner, N. , Mislin, G. L. A. , & Reimmann, C. (2014). Functional analysis of pyochelin‐/enantiopyochelin‐related genes from a pathogenicity island of Pseudomonas aeruginosa strain PA14. BioMetals, 27, 559–573. 10.1007/s10534-014-9729-4 [DOI] [PubMed] [Google Scholar]
- Rojas Murcia, N. , Lee, X. , Waridel, P. , Maspoli, A. , Imker, H. J. , Chai, T. , … Reimmann, C. (2015). The Pseudomonas aeruginosa antimetabolite L ‐2‐amino‐4‐methoxy‐trans‐3‐butenoic acid (AMB) is made from glutamate and two alanine residues via a thiotemplate‐linked tripeptide precursor. Frontiers in Microbiology, 6, 170 10.3389/fmicb.2015.00170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, S. , Zhou, L. , Jin, K. , Jiang, H. , & He, Y.‐W. (2016). Quorum sensing systems differentially regulate the production of phenazine‐1‐carboxylic acid in the rhizobacterium Pseudomonas aeruginosa PA1201. Scientific Reports, 6, 30352 10.1038/srep30352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trottmann, F. , Franke, J. , Ishida, K. , García‐Altares, M. , & Hertweck, C. (2019). A pair of bacterial siderophores releases and traps an intercellular signal molecule: An unusual case of natural nitrone bioconjugation. Angewandte Chemie (International Ed. in English), 58, 200–204. 10.1002/anie.201811131 [DOI] [PubMed] [Google Scholar]
- Williams, P. , & Camara, M. (2009). Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: A tale of regulatory networks and multifunctional signal molecules. Current Opinion in Microbiology, 12, 182–191. 10.1016/j.mib.2009.01.005 [DOI] [PubMed] [Google Scholar]
- Ye, L. , Cornelis, P. , Guillemyn, K. , Ballet, S. , Christophersen, C. , & Hammerich, O. (2014). Structure revision of N‐mercapto‐4‐formylcarbostyril produced by Pseudomonas fluorescens G308 to 2‐(2‐hydroxyphenyl)thiazole‐4‐carbaldehyde [aeruginaldehyde]. Natural Products Communications, 9, 789–794. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


