Mechanical cues from matrix stiffness regulate endodermal fate of pluripotent stem cells via the long noncoding RNA 000458.
Abstract
During endoderm formation, cell identity and tissue morphogenesis are tightly controlled by cell-intrinsic and cell-extrinsic factors such as biochemical and physical inputs. While the effects of biochemical factors are well studied, the physical cues that regulate cell division and differentiation are poorly understood. RNA sequencing analysis demonstrated increases of endoderm-specific gene expression in hPSCs cultured on soft substrate (Young’s modulus, 3 ± 0.45 kPa) in comparison with hard substrate (Young’s modulus, 165 ± 6.39 kPa). Further analyses revealed that multiple long noncoding RNAs (lncRNAs) were up-regulated on soft substrate; among them, LINC00458 was identified as a stiffness-dependent lncRNA specifically required for hPSC differentiation toward an early endodermal lineage. Gain- and loss-of-function experiments confirmed that LINC00458 is functionally required for hPSC endodermal lineage specification induced by soft substrates. Our study provides evidence that mechanical cues regulate the expression of LINC00458 and induce differentiation of hPSC into hepatic lineage progenitors.
INTRODUCTION
The definitive endoderm (DE) gives rise to respiratory and gastrointestinal systems and their associated organs such as the liver, lung, pancreas, and thyroid (1). Genetic analysis revealed that Activin/Nodal signaling regulates mesoderm and endoderm specifications through increases in mesendoderm factors such as goosecoid (GSC) and eomesodermin (EOMES) (2–4). Moreover, the activation of Nodal signaling leads to the phosphorylation of receptor-regulated SMADs (SMAD2 and SMAD3), which translocate from the cytoplasm to the nucleus to regulate downstream targets responsible for endoderm specification such as SOX17 and FOXA2 (5). With the unique properties in self-renewal and differentiation, human pluripotent stem cells (hPSCs) are widely used for studying embryonic development (6). Various protocols have been developed to generate DE that can be further differentiated into downstream endodermal lineage cells, such as hepatocytes and pancreatic β cells, in vitro. However, efforts to produce those cells have yielded only modest successes. Accumulating evidence indicates that embryonic development is tightly regulated by conserved fate-decision modules, cell-cell contact, and reciprocal interactions with the microenvironment (7, 8). Interaction between extracellular matrix (ECM) and stem cells is mediated by membrane proteins such as cadherins and integrins, which play important roles in stem cell differentiation (9, 10). A recent study revealed that soft substrate stiffness has a positive effect on the induction of endoderm-related gene expression, implying that stem cell differentiation is sensitive to and regulated by physical inputs (11). These studies indicate that extrinsic mechanical cues can modulate stem cell specification. However, the mechanistic link between cell-extrinsic factors and lineage specification is poorly understood.
Long noncoding RNAs (lncRNAs) are transcripts longer than 200 nt, with limited coding potential (12). In contrast to microRNAs, which mainly function as posttranscriptional regulators in the cytoplasm, lncRNAs are found in both the cytoplasm and nucleus, suggesting that they may play a role in epigenetic modification and gene expression (13, 14). Long intergenic noncoding RNAs are key contributors to various biological processes and regulate stem cell lineage specification with examples as follows: lncRNA-RoR, an induced pluripotent stem cell (iPSC)–enriched lncRNA that is directly controlled by pluripotency factors (15); Bvht, an lncRNA required for rodent cardiovascular lineage commitment (16); RMST, which binds to SOX2 and activates neurogenic transcription factors (17); lnc-DC, which is highly expressed in human dendritic cells, regulating their differentiation (18); DEANR1, a key lncRNA in human endoderm differentiation (19); and linc-ADAL, an adipose tissue–enriched lncRNA that regulates adipocyte differentiation and modulates de novo lipogenesis (20). Consistent with these findings, a loss-of-function study using mouse embryonic stem cells revealed that lncRNAs regulate the pluripotent state and lineage differentiation (21). Thus, lncRNAs play critical roles in stem cell lineage specification and differentiation.
In this study, we examined the effects of controlled substrate stiffness, mimicking human tissue mechanics, on hPSC lineage specification. Through the integrative analysis of RNA sequencing (RNA-seq) data, we revealed that controlled matrix stiffness is able to induce endodermal lineage specification in hPSCs without the need of endoderm induction medium. LINC00458 is responsible for the endodermal specification by interacting with SMAD2/3 in the nucleus to modulate soft substrate–induced endodermal lineage commitment.
RESULTS
Lower substrate stiffness induces endodermal lineage commitment
To determine whether matrix stiffness regulates hPSC self-renewal and cell fate, we first fabricated substrates with four different levels of stiffness (Table 1) to mimic the range of human tissue rigidities (fig. S1, A and B), as well as the maximum stiffness of hydrogel. The hPSCs were then cultured on the substrates with mouse embryonic fibroblast–conditioned medium (MEF-CM) for 3 days (22), and changes in pluripotency markers, including POU5F1 and NANOG, were quantified. We observed significant down-regulation of pluripotency genes on soft substrates (3, 15, and 33 kPa) but not on hard substrates (165 and 363 kPa), compared to expression on tissue culture polystyrene surfaces (TCPSs) (fig. S1C). We chose 3 and 165 kPa for further examination, hereafter referred to as soft and hard substrates, respectively (Fig. 1A). We found that cells cultured on soft substrates (Fig. 1B and movies S1 to S3) exhibited a flat monolayer of petal and cobblestone-shaped cells. These kinds of cell morphological change is characteristic of hepatogenic differentiation of hPSCs as previously demonstrated (23), suggesting that the cells were undergoing endoderm differentiation. Moreover, with respect to previously defined cell polarity responses to ECM (24, 25), hPSCs had poorly aligned actin stress fibers with less prominent actin bundles in response to soft substrates (Fig. 1C). Collectively, these findings suggest that soft substrates decrease the polarization of cultured hPSCs and the expression of pluripotency genes.
Table 1. Young’s modulus (E) of polycrylamide hydrogels used in this study.
| Acrylamide (%) | Bis-acrylamide (%) | E ± SD (kPa) |
| 5 | 0.1 | 3 ± 0.45 |
| 7 | 0.2 | 15 ± 1.11 |
| 12 | 0.145 | 33 ± 1.83 |
| 12 | 6.0 | 165 ± 6.39 |
| 18.75 | 1.13 | 363 ± 42.42 |
Fig. 1. Soft substrate up-regulates endoderm-specific genes in hPSCs.
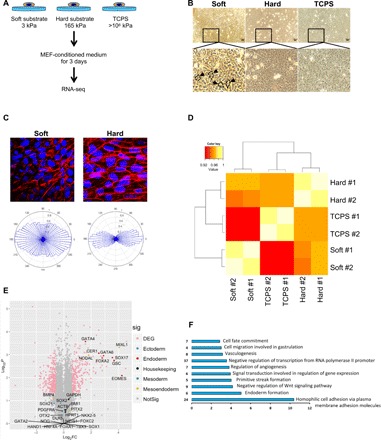
(A) Experimental setup for the examination of the effect of different substrate stiffnesses on hPSCs. (B) Morphology of hPSCs cultured on different substrate stiffnesses; arrows indicate typical petal and cobblestone-shaped cells. (C) hPSCs were stained for actin filaments on soft and hard substrates (top). Angular plots corresponding to soft and hard substrates are shown in the bottom compiled from 15 individual cells. (D) Hierarchical clustering analysis of mRNA expression in hPSCs cultured on soft and/or hard substrates and TCPS. (E) Volcano plot representative of the entire gene expression dataset (hPSCs cultured on soft versus hard substrates for 3 days). Gene groups are shown by different colors (light red, differentially expressed; blue, ectoderm; dark red, endoderm; black, housekeeping genes; green, mesoderm; yellow, mesoendoderm), and dots represent individual genes. Insignificant genes are in gray. DEG, differentially expressed genes. (F) Gene ontology (GO) terms highlighting the categories of up-regulated genes in the cells on soft substrates in comparison to those on hard substrates. FC, fold change.
To elucidate hPSC responses to substrate stiffness, we performed RNA-seq to analyze differential gene expression on different substrates. A total of 23,890 transcripts were successfully assembled based on reference gene annotations. An assessment of RNA from hPSCs on different substrates (soft, hard, and TCPS) showed that soft substrates differentially regulated RNA, compared to hard substrates or TCPS (Fig. 1D). A volcano plot revealed that compared to hard substrates, soft substrates induced endoderm-specific gene expression, such as EOMES (5), GSC (26), FOXA2 (27), GATA4 (28), GATA6 (28), and SOX17 (26, 28) (Fig. 1E). Gene ontology (GO) analysis of biological processes illustrated that soft substrates enriched endoderm formation, primitive streak formation, and cell migration involved in gastrulation, implying that the cells were undergoing endoderm differentiation (Fig. 1F). Moreover, the top two soft substrates mediated the down-regulation of ECM organization and cell adhesion relative genes (fig. S2 and Table 2). To further examine whether hPSCs on the soft substrate have a better potential to differentiate into functional hepatocytes, we examine the urea production on soft substrate and TCPS culture with our previously established three-step induction protocol (22). As shown in fig. S3, soft substrate significantly enhanced urea production compared to TCPS after differentiation induction for 12 days (fig. S3). Collectively, our data demonstrated that soft substrates induce hPSCs to exit the pluripotency stage and up-regulate endodermal genes. A combination of chemical and biophysical cues can further enhance endoderm specification.
Table 2. GO analysis of the top two differentially expressed genes.
| Category | Term | Count | P value | Genes | FDR |
| GOTERM_BP_ DIRECT |
GO:0030198~extracellular matrix organization |
33 | 1.43 × 10−16 | PDGFB, ADAMTSL4, CD47, COL9A1, NPHP3, COL9A3, LAMB2, COL7A1, ITGAV, SERPINE1, TGFBI, AGRN, LOX, THBS1, LOXL1, COL11A1, FGF2, SPP1, FN1, CYR61, COL4A2, HSPG2, CCDC80, ITGA3, SPARC, COL16A1, COL4A5, COL14A1, LAMA5, ITGA7, LAMC2, COL1A1, LAMC1 |
2.00 × 10−13 |
| GOTERM_BP_ DIRECT |
GO:0007155~cell adhesion | 046 | 4.52 × 10−14 | IGFBP7, SPOCK1, EDIL3, VCL, CDH6, APLP1, CDH8, ALCAM, CD47, SORBS3, LAMB2, SRPX, COL7A1, SORBS2, CTGF, ITGAV, TGFBI, COL12A1, THBS1, LOXL2, THBS3, SPP1, CYR61, ADAM9, FN1, HAPLN3, LPP, PODXL, MFGE8, ITGA3, PTPRU, MCAM, COL16A1, TINAGL1, CTNNA3, ITGA7, CNTN1, LAMC2, COL1A1, LAMC1, TGFB1l1, DST, FEZ1 |
7.96 × 10−11 |
In vitro, hPSCs can differentiate into anterior primitive streak (APS) and posterior primitive streak (PPS). The APS gives rise to DE and subsequently differentiates into endodermal tissues such as liver and pancreas, whereas the PPS forms the mesoderm (Fig. 2A). To determine whether soft substrates induce endoderm specification, we performed quantitative real-time polymerase chain reaction (qRT-PCR) analysis on the well-accepted stage-specific markers. Compared with hard substrate and TCPS, the APS marker genes EOMES and Brachyury (T) were highly up-regulated, and the PPS marker CDX2 was significantly down-regulated in hPSCs cultured in soft substrates (Fig. 2B). Consistent with these findings, the DE markers FOXA2 and SOX17 were also induced by the soft substrate (Fig. 2C). In contrast, the mesoderm markers FOXC2 and NKX2.5 were markedly lower in cells on soft gel (Fig. 2D). These results indicate that soft substrates induce hPSC endodermal lineage commitment in vitro. Furthermore, immunofluorescence staining demonstrated that, for cells on soft substrates, the percentage of SOX17+ cells was higher (30 ± 5.2%) at day 3, with a concomitant loss of expression of the key pluripotency marker NANOG. In contrast, on hard substrates and TCPS, the percentages of SOX17+ cells were significantly lower (2 ± 1.3% and undetectable), with high levels of NANOG expression (Fig. 2, E and F). Together, these results suggest that soft substrates, with a stiffness similar to that of human liver tissue, facilitate endodermal lineage specification without soluble factors.
Fig. 2. Soft substrate induces endodermal lineage commitment.
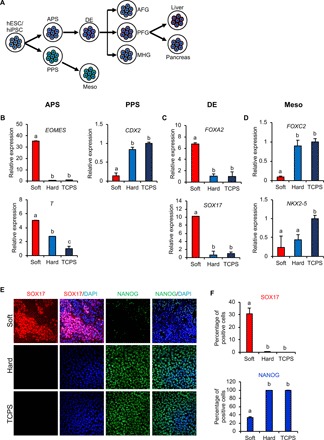
(A) A schematic drawing of the differentiation of hPSCs into liver and pancreatic cells. Meso, mesoderm; AFG, anterior foregut; PFG, posterior foregut; MHG, midgut/hindgut. Day 3 mRNA expression of hPSC genes involved in (B) APS/PPS, (C) DE, and (D) mesoderm differentiations. The results are presented as means ± SD of triplicates. One-way analysis of variance (ANOVA; n = 3 independent experiments). (E) Immunofluorescent staining and (F) quantification of SOX17 and NANOG in hPSCs grown on substrates with different stiffnesses. The results are presented as means ± SD of triplicates. One-way ANOVA (n = 3 independent experiments). Different letters indicate significant differences, and the same letters indicate no significant difference.
The lncRNA LINC01356/LINC00458 has a soft substrate–induced and cell type–specific expression signature
LncRNAs are key contributors to a variety of biological processes and regulate stem cell lineage specification. To explore their roles in substrate stiffness–mediated regulation of hPSC differentiation, we compared the long noncoding transcripts in our RNA-seq dataset between soft (Young’s modulus, 3 ± 0.45 kPa) and hard substrates (Young’s modulus, 165 ± 6.39 kPa) (Fig. 3A). Specific criteria (log2 fold change ≥ 1 or log2 fold change ≤ −1 and P < 0.05) were applied, and transcripts shorter than 200 nt were excluded. Among the top 20 differentially regulated lncRNAs, we confirmed that the DE-associated lncRNA1 (DEANR1, also known as linc00261), previously identified as important for endoderm differentiation (19), was up-regulated by soft substrates (Fig. 3B). These data support the idea that soft substrates not only induce endodermal gene expression but also trigger endoderm-related long noncoding transcript expression. Next, we proceeded to validate whether the top 20 differentially regulated lncRNAs from the deep sequencing results were correlated with substrate rigidity. It was confirmed that most of the differentially expressed lncRNAs showed a stiffness-associated expression signature in hPSCs (Fig. 3D and fig. S4), confirming that the expression of lncRNAs can be altered in response to substrate cues. LncRNAs have also been reported to be tissue specific (29). We compared the expression of lncRNAs in hPSCs with that in human adipose–derived stem cells and human monocytic leukemia cells (THP-1); both LINC01356 and LINC00458 were highly expressed in hPSCs (Fig. 3C), suggesting that expressions of these two lncRNAs are restricted to specific cell types during stem cell lineage commitment. To further investigate the dynamic changes in these two long intergenic noncoding RNAs, we determined the time courses of their expression over the 3-day culture. Both lncRNAs were induced by soft substrates as early as 4 hours and reached a maximum expression level at 16 hours (LINC00458) and 32 hours (LINC01356). To probe their potential functions, we determined their subcellular localizations in hPSCs by cellular fractionation. LINC00458 was predominantly localized to the nucleus, whereas LINC01356 was found in the cytoplasm (Fig. 3F). We therefore further focused on LINC00458 for functional and mechanistic studies because of its nuclear location, which may be involved directly in key cellular processes such as gene regulation.
Fig. 3. LINC01356 and LINC00458 are associated with soft matrix–induced and cell type–specific expression signatures.
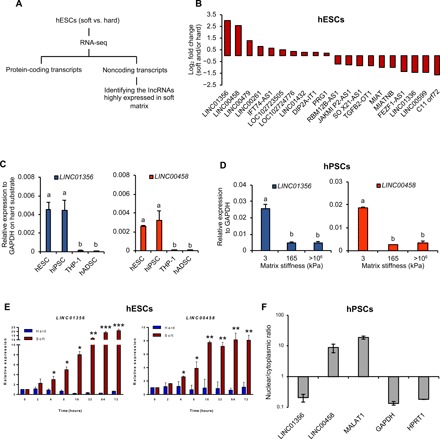
(A) Schematic plan to compare lncRNAs between soft and hard substrates. (B) Top 20 differentially regulated lncRNAs in hESCs cultured on soft or hard substrates. (C) LINC01356 and LINC00458 expressions in different human cells by qRT-PCR. hESC, human embryonic stem cell; hiPSC, human induced pluripotent stem cell; THP-1; monocytic leukemia cell; hADSC, human adipose–derived stem cell. The results are presented as means ± SD of triplicates. One-way ANOVA (n = 3 independent experiments). Different letters indicate significant differences, and the same letters indicate no significant difference. (D) Expression levels of LINC01356 and LINC00458 were determined by qRT-PCR after 3 days of culture on the indicated substrate stiffness. The results are presented as means ± SD of triplicates. One-way ANOVA (n = 3 independent experiments). Different letters indicate significant differences, and the same letters indicate no significant difference. (E) Induction of LINC01356 and LINC00458 expressions over 3 days. Cells were cultured on soft and hard substrates. RNA was isolated at the indicated time points and assessed by qRT-PCR. The results are presented as means ± SD of triplicates. ***P < 0.001, **P < 0.005, *P < 0.05, two-sided Student’s t test. (F) Total RNA was isolated from nuclear and cytoplasmic fractions and analyzed by qRT-PCR. MALAT1, metastasis-associated lung adenocarcinoma transcript 1.
Loss of LINC00458 impairs endoderm gene expression
To investigate the functional role of LINC00458 in lineage specification, we used locked nucleic acid (LNA) GapmeRs to knock down its expression in hPSCs cultured on soft substrates (Fig. 4A). LINC00458 knockdown significantly suppressed the mRNA and protein levels of the endoderm-specific genes FOXA2 and SOX17 (Fig. 4, B and C), as well as EOMES and GATA6 (Fig. 4B), suggesting an important role of LINC00458 in endoderm differentiation in response to soft substrates. In contrast, LINC00458 knockdown did not affect the pluripotency genes POU5F1, NANOG, and SOX2 when hPSCs were cultured on TSPC (fig. S5). Moreover, gain-of-function experiments indicated that overexpression of LINC00458 up-regulated endoderm gene expression (Fig. 4, D and E), as demonstrated by immunostaining. These results strongly support the notion that LINC00458 may play an important role in the soft substrate–induced hPSC to DE differentiation.
Fig. 4. LINC00458 is necessary for soft substrate–induced endoderm commitment.
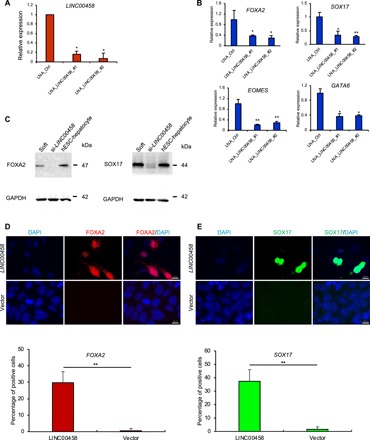
(A) hPSCs were cultured on TCPS and transfected with LNA GapmeRs targeting LINC00458 to determine knockdown efficiency. (B) hPSCs were cultured on soft substrate with LNA GapmeRs targeting LINC00458, and the expression of endodermal genes SOX17, FOXA2, EOMES, and GATA6 was analyzed by qRT-PCR. (C) Immunoblotting analysis of the FOXA2 and SOX17 expressions in hPSCs cultured on soft substrates with LNA GapmeRs targeting LINC00458. (D and E) Immunofluorescence detection of FOXA2 and SOX17 expressions in hPSCs cultured on hard substrates with LINC00458 transfection. The results are presented as means ± SD of triplicates. **P < 0.005, *P < 0.05, two-sided Student’s t test.
LINC00458 interacts with SMAD2/3, a core component of the endoderm-specific transcription factor
We next examined the underlying molecular mechanism through which LINC00458 regulates endoderm differentiation. Since lncRNAs can act in cis to regulate the expression of neighboring genes or in trans via diverse mechanisms (12, 30), we analyzed the role of LINC00458 in regulating its neighboring gene miR-1297. miR-1297 gene expression was not affected by LINC00458 knockdown, indicating that it does not act in a cis-regulatory manner (fig. S6). For the studies of trans-regulation, it has been demonstrated that hPSCs are driven toward endodermal differentiation through the activation of Activin/Nodal, which comprises the intracellular type I receptor that subsequently phosphorylates the important endoderm-specific transcriptional regulators SMAD2 and SMAD3 (2, 3). To test the hypothesis that LINC00458 interacts with SMAD2/3 to promote endoderm-specific gene expression, we first investigated the phosphorylation of SMAD2/3 in hPSCs cultured on soft substrates. As demonstrated by immunoblotting, SMAD2/3 [SMAD2 (Ser465/467)/SMAD3 (Ser423/425)] phosphorylation was increased in hPSCs on soft substrates as compared to hard substrates (Fig. 5A). Next, we tested the interaction between LINC00458 and SMAD2/3 during soft substrate–induced differentiation by RNA-binding protein immunoprecipitation using antibody against endogenous SMAD2/3 in cells on soft and hard substrates. In addition, we examined another member of the SMAD family, SMAD1, to test the interaction specificity. The results revealed that LINC00458 was enriched by ~80-fold in the SMAD2/3 complex when compared to that with an immunoglobulin G (IgG) control (Fig. 5B), whereas SMAD1 did not interact with LINC00458 (Fig. 5B). Furthermore, knocking down SMAD2/3 expression using siSMAD2 and siSMAD3 small interfering RNAs (siRNAs) (Fig. 5C) significantly reduced the interaction between LINC00458 and SMAD2/3 (Fig. 5D). Together, the results suggest that the soft substrate–induced LINC00458 regulates hPSC endoderm specification via a SMAD2/3-dependent pathway (Fig. 5E).
Fig. 5. LINC00458 interacts with SMAD3, a core component of the endoderm-specific transcription factor.
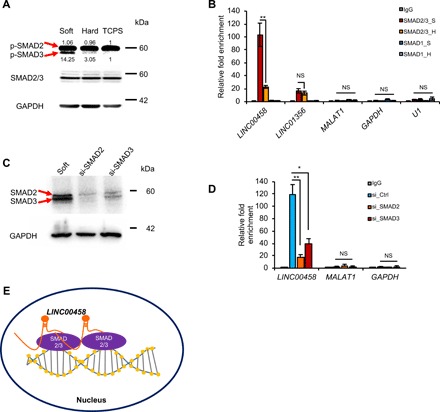
(A) Immunoblot of SMAD2/3 (SMAD2 (Ser465/467)/SMAD3 (Ser423/425) phosphorylation in hPSCs cultured on soft and/or hard substrates and TCPS. (B) qRT-PCR detection of LINC01356 and LINC00458 pulled down by SMAD2/3- or SMAD1-specific antibodies, as compared to that with immunoglobulin G (IgG), based on the RNA-binding protein immunoprecipitation (RIP) assay. SMAD2/3_S and SMAD2/3_H, soft and hard group samples, respectively, with anti-SMAD2/3 antibody; SMAD1_S and SMAD1_H, soft and hard group samples, respectively, with anti-SMAD1 antibody. (C) Immunoblot detection of the levels of SMAD2/3 in hPSCs on soft substrates transfected with SMAD2 or SMAD3 siRNAs. (D) qRT-PCR detection of LINC00458 and MALAT1 in RIP complex in hPSCs with SMAD2 or SMAD3 siRNAs. (E) A working model of soft substrate induces LINC00458 to interact with SMAD2/3, which promotes endoderm specification. The results are presented as means ± SD of triplicates. **P < 0.005, *P < 0.05, one-way ANOVA (n = 3 independent experiments). NS, not significant.
DISCUSSION
The hallmark of gastrulation during embryogenesis is polarization, which is highly regulated by the ECM. Using time-lapse microscopy, the ECM beneath the cells has been shown to serve as a motile substratum that places cells during early chick primitive streak formation and later during extension of the axis (31). In this study, we showed that tissue mechanics dictate the lineage specification and differentiation of pluripotent stem cells. Specifically, a substrate with a stiffness similar to the liver triggered the expression of endoderm-specific genes (EOMES, SOX17, and FOXA2), whereas hard substrates (similar to bone or TCPS) did not. A recent study suggested that stiffness-sensitive human lncRNAs can act as regulators of vascular smooth muscle cell function (32). Here, we identified lncRNA LINC00458 as a critical regulator of endodermal lineage commitment in response to ECM stiffness. We found that its expression was induced markedly in hPSCs cultured on soft substrates. A combination of stem cells, materials, and molecular signaling to regenerate tissues and organs could provide hope for patients suffering from incurable diseases. The generation of hepatocytes or pancreatic β cells from hPSCs was accomplished more than a decade ago, and it has been shown that the regenerated cells respond to their microenvironment not only in terms of chemical inputs but also in terms of physical cues. The lack of an appropriate physical environment might result in inefficient maturation of hepatocytes and β cells. Thus, it is critical to investigate the mechanisms through which physical cues such as substrate stiffness regulate stem cell lineage specification and differentiation. It is known that changes in tissue stiffness drive neuronal fate decisions (33) and that matrix rigidity modulates mouse hepatocyte function through hepatocyte nuclear factor 4 α (34).
LncRNAs can be detected in the cytoplasm, nucleus, or both. Thus, the knowledge of their subcellular localization can provide fundamental insights into their biological functions. Nuclear lncRNAs can have indirect regulatory effects on gene loci by acting as transcription-dependent activators, molecular guides or decoys, transcription cofactors, or scaffolds for chromatin-modifying complexes. Moreover, lncRNAs can fold into different complex structures and form ribonucleoprotein (RNP) complexes to regulate gene expression at transcriptional and posttranscriptional levels (35). For example, LINCRNA-p21 was found to form an lncRNA-RNP complex and interact with a nuclear factor to act as a global transcriptional repressor to facilitate p53-mediated apoptosis (36). We found that lncRNA LINC00458 is predominantly localized in the nucleus and interacts with SMAD2/3 in cells on soft substrates. We further demonstrated that LINC00458 exhibits a cell type–specific signature and is mainly expressed in the nucleus. Ablating LINC00458 in hPSCs diminished the expressions of endodermal markers (SOX17 and FOXA2), thus providing evidence that it plays a significant role for endodermal specification in response to ECM stiffness. These insights have expanded our knowledge of the cellular responses to mechanical cues that modulate endoderm specification in an lncRNA-dependent manner.
It has been shown that transforming growth factor–β (TGF-β) signaling is involved in the cellular processes including stem cell differentiation (37) and endothelial function (38) in response to mechanical factors. In addition, phosphorylation, localization, and transcriptional regulation of SMAD molecules are also tightly controlled by TGF-β signaling in response to mechanical cues. In this study, we observed that an enhancement in phosphorylation of the SMAD3 correlated with the LINC00458 when hPSCs were cultured on soft substrate. Enhanced amount of phosphorylated SMAD and LINC00458 expression increases the probability of interaction between each other. SMAD phosphorylation primarily takes place in the cytosol, and phosphorylated SMAD complex is then translocated and stabilized in the nucleus. We observed that LINC00458 was located primarily in the nucleus. Thus, future research efforts will be made to further clarify whether SMAD phosphorylation and LINC00458 up-regulation are directly controlled by TGF-β signaling, or lncRNAs per se serve as scaffold proteins or molecular sponges for modulating gene expression.
In this study, we observed an increased expression of LINC01356 in the cytosol. SMAD7 transcription has been reported to act as a negative regulator of SMAD signaling (39). Future studies are needed to address LINC01356 acts by activating SMAD2/3 or suppressing its inhibitor such as SMAD7. Understanding precisely how these lncRNAs are influenced by the substrate stiffness will undoubtedly guide researchers toward regenerative strategies for enhancing endodermal differentiation of hPSCs in vitro.
Current protocols to differentiate hPSCs toward hepatocytes commence with the initial facilitation of DE formation by Activin A/TGF-β. In a previous study, we showed that superior differentiation can be achieved with the addition of hepatocyte growth factor (HGF) at the early stage of DE induction rather than at the maturation stage. We demonstrated that HGF signaling can coordinate with Activin A and Wnt3a to promote DE formation. Furthermore, TGF-β1 showed a selective effect on fibroblast tractions on a stiff matrix (>13 kPa) but not on a soft matrix (<13 kPa) (40), suggesting that a soft matrix regulates mechano-transduction in parallel with TGF-β signaling. The levels of APS and DE markers in our study were increased in hPSCs in response to soft substrates without Activin A/TGF-β stimulation.
In summary, we discovered that the lncRNA LINC00458 mediates mechanically induced endodermal differentiation in hPSCs by directly interacting with the transcription factor SMAD2/3 in the nucleus to promote endodermal lineage–specific gene expression. LINC00458, as a specific regulator of endodermal lineage specification, may have potential relevance to clinical diseases involving endodermal lineages in development. Future efforts will be devoted to further study of the activation of the molecular signaling pathways regulated by lncRNAs in response to altered mechanical control of cell fate commitment and differentiation of hPSCs.
METHODS
Cell culture and LNA transfections
Before the study, human PSCs, including hiPSC (human foreskin fibroblast–derived iPS cell line, CFB46) and hESC (hES cell line, H9) (22), were cultured on feeder-free environments on plates coated with growth factor–reduced Matrigel (Thermo Fisher Scientific, Waltham, MA) in MEF-CM (22, 41) for over six passages to maintain pluripotency and to eliminate the possibility of contamination by MEFs. Cells were incubated at 37°C in 5% CO2 and passaged every 7 days. For culture with different matrix stiffness, cells were dissociated by ReLeSR treatment and plated onto surfaces with defined matrix stiffness in MEF-CM for 3 days or at indicated time points. To silence lncRNA expression, hPSCs were incubated 6 hours before transfection with reduced serum medium Opti-MEM I (Gibco, Waltham, MA) supplemented with 10 μM Y-27632 (Sigma-Aldrich, St. Louis, MO). The cells were then transfected with LNA GapmeRs (50 nM; Qiagen, MD) for 24 hours using Lipofectamine RNAiMax (Thermo Fisher Scientific, Waltham, MA). Fresh medium was then replaced, and the cells were transfected with LNA GapmeRs for another 24 hours. Throughout the study, hESCs were used for all the experiments, while hiPSCs were only used to further confirm the findings of expression (Fig. 3, C and D) and distribution (Fig. 3F) of target lncRNAs. For simplicity, Fig. 3 (D and F) shows only the data for hESCs.
Polyacrylamide hydrogel preparation
Compliant polyacrylamide hydrogel culture substrates were prepared as previously described (42) with a minor modification for the generation of reactive coverslips. Briefly, 18- or 25-mm no. 1 cover glass (Warner Instruments, Hamden, CT, USA) was etched with 0.1 N NaOH for 5 min, treated with (3-aminopropyl) trimethoxysilane (Sigma-Aldrich, St. Louis, MO, USA) for 30 min, and then washed with distilled water, followed by a 30-min exposure to 0.5% glutaraldehyde (Sigma-Aldrich). The reactive coverslips were washed several times with distilled water and air-dried before use. For hydrogel preparation, gel premix stock solutions were freshly prepared (42): 40% (w/v) acrylamide, 2% bis-acrylamide, 1% ammonium persulfate, and N,N,N′,N′-tetramethylenediamine (Sigma-Aldrich); details are presented in Table 1. For cross-linking ECM to the hydrogel, 200 μl of 2 mM N-sulfosuccinimidyl-6-[40-azido-20-nitrophenylamino] (Thermo Fisher Scientific, Waltham, MA, USA) in 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Sigma-Aldrich) buffer was dried onto all gel surfaces and exposed to an ultraviolet (UV) light cross-linker with 7500 J of energy. The gels were transferred to individual wells of six-well plates, hydrated in 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid buffer, and exposed to germicidal UV light for 20 min for sterilization. Then, 75 μl of Matrigel (Thermo Fisher Scientific, Waltham, MA, USA) was added onto parafilm, and the coverslips were inverted onto the Matrigel solution. After incubation overnight at 4°C, the Matrigel-coated coverslips were rinsed with sterile phosphate-buffered saline (PBS) before plating the cells.
RNA-seq experiments
RNA samples were extracted using TRIzol (Invitrogen, Carlsbad, CA) from hPSCs cultured on substrates of different stiffnesses. Following deoxyribonuclease I enzyme treatment, the RNA integrity number (RIN) was assessed using an Agilent 2200 TapeStation-RNA R6K Bioanalyzer (Agilent Technologies), and the high quality of RNA (RIN ≥ 8.8) was used. RNA-seq complementary DNA (cDNA) libraries were constructed following the standard Illumina protocol and validated using the Agilent 2200 TapeStation-D1000 assay (Agilent Technologies). cDNA libraries were quantitatively measured by qRT-PCR (Roche LightCycler 480 system) and a Qubit Fluorometer (Invitrogen, Carlsbad, CA, USA). The cDNA libraries were induced and sequenced on an Illumina NextSeq 500 platform in single-ended mode with 75 bp.
RNA-seq data analysis
Quality control on RNA-seq reads was conducted on the basis of the nucleotides’ Phred quality score using the FASTX-Toolkit version 0.0.13.2. Nucleotides with high-quality base calling (Phred quality score ≥ 20, which represents 99% accuracy of a base call) were included, and RNA reads longer than 35 nt were retained. Reads were then mapped to the University of California, Santa Cruz (UCSC) Human genome hg38 using TopHat2 (43) version 2.1.0 for transcriptome annotations. The transcripts were assembled, abundances were estimated, and normalization was performed with Cufflinks version 2.2.1 (44). Gene expression profiling was estimated using cuffdiff version 2.2.1. Genes were identified as being up- or down-regulated if the fold change was greater than 1.2 or less than 0.83, respectively, with P < 0.05. We selected gene lengths greater than 200 bp and gene annotations in UCSC described as noncoding RNA. To annotate differentially expressed genes, we performed GO and Kyoto Encyclopedia of Genes and Genomes pathway enrichment annotation using DAVID (database for annotations, visualization and integrated discovery) (45). GO enrichment analysis indicates the cellular process, biological regulation, and metabolic process most significantly enriched to select human target genes.
Western blotting
Whole-cell lysates were prepared using radioimmunoprecipitation assay buffer [50 mM tris-HCl (pH 7.5), 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, and 0.1% SDS, 5 mM EDTA] supplemented with protease and phosphatase inhibitors. Protein concentration was quantified by Bradford assays. Approximately 20 to 40 μg of protein lysate was separated by SDS–polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, and subsequently incubated with 5% bovine serum albumin in tris-buffered saline containing 0.1% Tween 20 for 45 min. Primary antibodies against SMAD2/3 (Cell Signaling, #5678), p-SMAD2/3 (Cell Signaling, #9510), and glyceraldehyde-phosphate dehydrogenase (GAPDH; Cell Signaling, #2118) were used to probe the membranes. Blots were then incubated with horseradish peroxidase–conjugated secondary antibodies and visualized with Pierce ECL Western blotting substrate (Thermo Fisher Scientific, Waltham, MA).
Immunofluorescence and time-lapse imaging
Cells were fixed for 10 min at 4°C in 4% paraformaldehyde in PBS, permeabilized with 0.1% Triton X-100 (Sigma-Aldrich, St. Louis, MO) for 10 min at room temperature, and blocked for 1 hour in PBS with 1% horse serum (Sigma-Aldrich, St. Louis, MO). The primary antibodies used were as follows: anti-NANOG (1:400; cat. no. 4903, Cell Signaling), anti-SOX17 (1:100; cat. no. 81778, Cell Signaling), anti-FOXA2 (1:100; cat. no. 685802, BioLegend), anti-SMAD2 (1:100; cat. no. 3122, Cell Signaling), anti-SMAD3 (1:100; cat. no. 9523, Cell Signaling), anti–p-SMAD2 (1:100; cat. no. 3108, Cell Signaling), and anti–p-SMAD3 (1:100; cat. no. 9520, Cell Signaling). The treated cells were subjected to three washes with PBS and further incubation with Alexa Fluor secondary antibodies (1:500; Jackson ImmunoResearch) for 1 hour at room temperature in the dark. The cells were then washed three times with PBS, with 4′,6-diamidino-2-phenylindole (Sigma-Aldrich, St. Louis, MO) added to the first wash to stain the nuclei. Images were acquired using a Confocal Zeiss LSM880. Intensity analysis was performed using ImageJ 1.51j8 (National Institutes of Health, MD, USA). For time-lapse imaging, cells were placed on an inverted microscope (Nikon, Ti-U inverted microscope system) and recorded every 2 hours for 72 hours in an environmental chamber maintained at 37°C with 5% CO2.
RNA isolation and qRT-PCR
The Direct-zol RNA MiniPrep (Zymo Research, Tustin, CA) and the Cytoplasmic and Nuclear RNA Purification Kit (Norgen Biotek, Thorold, ON) were used for total RNA/fraction RNA extraction, respectively. cDNA synthesis was performed with random primers according to the MMLV-RT (Moloney Murine Leukemia Virus-Reverse Transcriptase) (Promega, Madison, WI) manufacturer’s protocol. qRT-PCR analysis was performed with triplicate repeats using the CFX-Connect (Bio-Rad, Hercules, CA). Expression values of mRNA were calculated and normalized to those of the indicated housekeeping genes (GAPDH and HPRT1).
RNA-binding protein immunoprecipitation
RNA-binding protein immunoprecipitation was performed using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Burlington, MA) according to the manufacturer’s instructions. Specifically, 2 × 107 cells were washed in PBS and resuspended in RNA-binding protein immunoprecipitation lysis buffer (Millipore, Burlington, MA) containing protease and RNase inhibitors. Approximately 1 mg of protein was used for overnight immunoprecipitation with the primary antibody of interest or nonimmune species-matched IgG; 10% of the lysate was saved as preimmunoprecipitation input. After washes, the immune complexes and input were eluted, treated with proteinase K, and heated at 55°C for 30 min to digest the protein. RNA was purified with TRIzol reagent (Thermo Fisher Scientific, Waltham, MA) according to the supplier’s instructions. The RNA was resuspended in nuclease-free water, and the sample was used for reverse transcription using MMLV-RT (Promega, Madison, WI) according to the manufacturer’s protocol. The isolated RNA was quantified by qRT-PCR and normalized first to the preimmunoprecipitation input and then to the IgG control.
Statistical analysis
Data were presented as means ± SD. Comparison was made using one-way analysis of variance (ANOVA) among three or more than three groups. Different letters represented different levels of significance. Comparison between two groups was analyzed by two-tailed Student’s t test, and asterisks represented significance defined as *P < 0.05, **P < 0.01, or ***P < 0.001.
Supplementary Material
Acknowledgments
We thank A. E. T. Chiou and Y.-Q. Chen for technical advice and assistance. We also acknowledge the technical support provided by the Imaging Core Facility of Nanotechnology of the UST-NYMU. Funding: The authors acknowledge financial support from the Ministry of Science and Technology, Taiwan (MOST 105-2314-B-010-065-MY3, MOST 107-2314-B-010-015-MY3, MOST 107-2911-I-010-504, MOST 108-2911-I-010-502, MOST 108-2314-B-010-036-MY3, MOST 108-2321-B-010-006, MOST 108-2633-B-009-001, and MOST 108-2923-B-010-002-MY3). This study was also supported by the Aiming for the Top University Plan, a grant from the Ministry of Education, and NIH grants GM125379, HL121365, HL106579, and HL108735. This work was supported by “Development and Construction Plan” of the School of Medicine, National Yang-Ming University (107F-M01). Author contributions: Y.-F.C. designed and performed most of the experiments and wrote the draft manuscript. C.-H.C., M.Y.C., and H.-D.H. were responsible for the bioinformatics analysis. Y.-S.J.L. interpreted the data and revised the manuscript. S.C., O.K.L., and J.H.-C.H. designed the study and proofread the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/6/eaay0264/DC1
Fig. S1. Pluripotency markers are down-regulated in soft substrate.
Fig. S2. GO terms highlighting the characteristics of down-regulated genes in the soft substrates compared to that with hard substrates.
Fig. S3. Combination of soft substrate and chemical factors enhance the urea production of hPSC-derived hepatocytes.
Fig. S4. Most of the differentially regulated lncRNAs show a stiffness-associated expression signature in hPSCs.
Fig. S5. Knockdown of lncRNA LINC00458 does not affect pluripotency genes.
Fig. S6. The lncRNA LINC00458 does not act in a cis-regulatory manner.
Movie S1. Morphology of hPSCs upon soft substrate.
Movie S2. Morphology of hPSCs upon hard substrate.
Movie S3. Morphology of hPSCs upon TCPS.
REFERENCES AND NOTES
- 1.Zorn A. M., Wells J. M., Vertebrate endoderm development and organ formation. Annu. Rev. Cell Dev. Biol. 25, 221–251 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Attisano L., Wrana J. L., Smads as transcriptional co-modulators. Curr. Opin. Cell Biol. 12, 235–243 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Brennan J., Lu C. C., Norris D. P., Rodriguez T. A., Beddington R. S., Robertson E. J., Nodal signalling in the epiblast patterns the early mouse embryo. Nature 411, 965–969 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Gadue P., Huber T. L., Paddison P. J., Keller G. M., Wnt and TGF-beta signaling are required for the induction of an in vitro model of primitive streak formation using embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 103, 16806–16811 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teo A. K., Arnold S. J., Trotter M. W., Brown S., Ang L. T., Chng Z., Robertson E. J., Dunn N. R., Vallier L., Pluripotency factors regulate definitive endoderm specification through eomesodermin. Genes Dev. 25, 238–250 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Avior Y., Sagi I., Benvenisty N., Pluripotent stem cells in disease modelling and drug discovery. Nat. Rev. Mol. Cell Biol. 17, 170–182 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Gattazzo F., Urciuolo A., Bonaldo P., Extracellular matrix: A dynamic microenvironment for stem cell niche. Biochim. Biophys. Acta 1840, 2506–2519 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pacelli S., Basu S., Whitlow J., Chakravarti A., Acosta F., Varshney A., Modaresi S., Berkland C., Paul A., Strategies to develop endogenous stem cell-recruiting bioactive materials for tissue repair and regeneration. Adv. Drug Deliv. Rev. 120, 50–70 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Przybyla L., Lakins J. N., Weaver V. M., Tissue mechanics orchestrate Wnt-dependent human embryonic stem cell differentiation. Cell Stem Cell 19, 462–475 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shih Y. R., Tseng K. F., Lai H. Y., Lin C. H., Lee O. K., Matrix stiffness regulation of integrin-mediated mechanotransduction during osteogenic differentiation of human mesenchymal stem cells. J. Bone Miner. Res. 26, 730–738 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Jaramillo M., Singh S. S., Velankar S., Kumta P. N., Banerjee I., Inducing endoderm differentiation by modulating mechanical properties of soft substrates. J. Tissue Eng. Regen. Med. 9, 1–12 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rinn J. L., Chang H. Y., Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang K. C., Chang H. Y., Molecular mechanisms of long noncoding RNAs. Mol. Cell 43, 904–914 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quinn J. J., Chang H. Y., Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 17, 47–62 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Loewer S., Cabili M. N., Guttman M., Loh Y. H., Thomas K., Park I. H., Garber M., Curran M., Onder T., Agarwal S., Manos P. D., Datta S., Lander E. S., Schlaeger T. M., Daley G. Q., Rinn J. L., Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat. Genet. 42, 1113–1117 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klattenhoff C. A., Scheuermann J. C., Surface L. E., Bradley R. K., Fields P. A., Steinhauser M. L., Ding H., Butty V. L., Torrey L., Haas S., Abo R., Tabebordbar M., Lee R. T., Burge C. B., Boyer L. A., Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 152, 570–583 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ng S. Y., Bogu G. K., Soh B. S., Stanton L. W., The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol. Cell 51, 349–359 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Wang P., Xue Y., Han Y., Lin L., Wu C., Xu S., Jiang Z., Xu J., Liu Q., Cao X., The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 344, 310–313 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Jiang W., Liu Y., Liu R., Zhang K., Zhang Y., The lncRNA DEANR1 facilitates human endoderm differentiation by activating FOXA2 expression. Cell Rep. 11, 137–148 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X., Xue C., Lin J., Ferguson J. F., Weiner A., Liu W., Han Y., Hinkle C., Li W., Jiang H., Gosai S., Hachet M., Garcia B. A., Gregory B. D., Soccio R. E., Hogenesch J. B., Seale P., Li M., Reilly M. P., Interrogation of nonconserved human adipose lincRNAs identifies a regulatory role of linc-ADAL in adipocyte metabolism. Sci. Transl. Med. 10, eaar5987 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guttman M., Donaghey J., Carey B. W., Garber M., Grenier J. K., Munson G., Young G., Lucas A. B., Ach R., Bruhn L., Yang X., Amit I., Meissner A., Regev A., Rinn J. L., Root D. E., Lander E. S., lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477, 295–300 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y. F., Tseng C. Y., Wang H. W., Kuo H. C., Yang V. W., Lee O. K., Rapid generation of mature hepatocyte-like cells from human induced pluripotent stem cells by an efficient three-step protocol. Hepatology 55, 1193–1203 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siller R., Naumovska E., Mathapati S., Lycke M., Greenhough S., Sullivan G. J., Development of a rapid screen for the endodermal differentiation potential of human pluripotent stem cell lines. Sci. Rep. 6, 37178 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tojkander S., Gateva G., Lappalainen P., Actin stress fibers—Assembly, dynamics and biological roles. J. Cell Sci. 125, 1855–1864 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Prager-Khoutorsky M., Lichtenstein A., Krishnan R., Rajendran K., Mayo A., Kam Z., Geiger B., Bershadsky A. D., Fibroblast polarization is a matrix-rigidity-dependent process controlled by focal adhesion mechanosensing. Nat. Cell Biol. 13, 1457–1465 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Touboul T., Hannan N. R. F., Corbineau S., Martinez A., Martinet C., Branchereau S., Mainot S., Strick-Marchand H., Pedersen R., Di Santo J., Weber A., Vallier L., Generation of functional hepatocytes from human embryonic stem cells under chemically defined conditions that recapitulate liver development. Hepatology 51, 1754–1765 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Burtscher I., Lickert H., Foxa2 regulates polarity and epithelialization in the endoderm germ layer of the mouse embryo. Development 136, 1029–1038 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Schrode N., Saiz N., Di Talia S., Hadjantonakis A. K., GATA6 levels modulate primitive endoderm cell fate choice and timing in the mouse blastocyst. Dev. Cell 29, 454–467 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Necsulea A., Soumillon M., Warnefors M., Liechti A., Daish T., Zeller U., Baker J. C., Grützner F., Kaessmann H., The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 505, 635–640 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Ulitsky I., Bartel D. P., lincRNAs: Genomics, evolution, and mechanisms. Cell 154, 26–46 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voiculescu O., Bertocchini F., Wolpert L., Keller R. E., Stern C. D., The amniote primitive streak is defined by epithelial cell intercalation before gastrulation. Nature 449, 1049–1052 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Yu C. K., Xu T., Assoian R. K., Rader D. J., Mining the stiffness-sensitive transcriptome in human vascular smooth muscle cells identifies long noncoding RNA stiffness regulators. Arterioscler. Thromb. Vasc. Biol. 38, 164–173 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim S. N., Jeibmann A., Halama K., Witte H. T., Wälte M., Matzat T., Schillers H., Faber C., Senner V., Paulus W., Klämbt C., ECM stiffness regulates glial migration in Drosophila and mammalian glioma models. Development 141, 3233–3242 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Desai S. S., Tung J. C., Zhou V. X., Grenert J. P., Malato Y., Rezvani M., Español-Suñer R., Willenbring H., Weaver V. M., Chang T. T., Physiological ranges of matrix rigidity modulate primary mouse hepatocyte function in part through hepatocyte nuclear factor 4 alpha. Hepatology 64, 261–275 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun X., Haider Ali M. S. S., Moran M., The role of interactions of long non-coding RNAs and heterogeneous nuclear ribonucleoproteins in regulating cellular functions. Biochem. J. 474, 2925–2935 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huarte M., Guttman M., Feldser D., Garber M., Koziol M. J., Kenzelmann-Broz D., Khalil A. M., Zuk O., Amit I., Rabani M., Attardi L. D., Regev A., Lander E. S., Jacks T., Rinn J. L., A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142, 409–419 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Madl C. M., Mehta M., Duda G. N., Heilshorn S. C., Mooney D. J., Presentation of BMP-2 mimicking peptides in 3D hydrogels directs cell fate commitment in osteoblasts and mesenchymal stem cells. Biomacromolecules 15, 445–455 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou J., Li Y. S., Chien S., Shear stress-initiated signaling and its regulation of endothelial function. Arterioscler. Thromb. Vasc. Biol. 34, 2191–2198 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakao A., Afrakhte M., Morén A., Nakayama T., Christian J. L., Heuchel R., Itoh S., Kawabata M., Heldin N. E., Heldin C. H., ten Dijke P., Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling. Nature 389, 631–635 (1997). [DOI] [PubMed] [Google Scholar]
- 40.Marinkovic A., Mih J. D., Park J. A., Liu F., Tschumperlin D. J., Improved throughput traction microscopy reveals pivotal role for matrix stiffness in fibroblast contractility and TGF-β responsiveness. Am. J. Physiol. Lung Cell. Mol. Physiol. 303, L169–L180 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Braam S. R., Denning C., Matsa E., Young L. E., Passier R., Mummery C. L., Feeder-free culture of human embryonic stem cells in conditioned medium for efficient genetic modification. Nat. Protoc. 3, 1435–1443 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Hsieh W. T., Liu Y. S., Lee Y. H., Rimando M. G., Lin K. H., Lee O. K., Matrix dimensionality and stiffness cooperatively regulate osteogenesis of mesenchymal stromal cells. Acta Biomater. 32, 210–222 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S. L., TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trapnell C., Hendrickson D. G., Sauvageau M., Goff L., Rinn J. L., Pachter L., Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46–53 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang da W., Sherman B. T., Lempicki R. A., Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/6/eaay0264/DC1
Fig. S1. Pluripotency markers are down-regulated in soft substrate.
Fig. S2. GO terms highlighting the characteristics of down-regulated genes in the soft substrates compared to that with hard substrates.
Fig. S3. Combination of soft substrate and chemical factors enhance the urea production of hPSC-derived hepatocytes.
Fig. S4. Most of the differentially regulated lncRNAs show a stiffness-associated expression signature in hPSCs.
Fig. S5. Knockdown of lncRNA LINC00458 does not affect pluripotency genes.
Fig. S6. The lncRNA LINC00458 does not act in a cis-regulatory manner.
Movie S1. Morphology of hPSCs upon soft substrate.
Movie S2. Morphology of hPSCs upon hard substrate.
Movie S3. Morphology of hPSCs upon TCPS.


