In vitro differentiation of pluripotent stem cells provides a systematic platform to study development and disease. Recent advances in brain organoid technology have created new opportunities to investigate the formation and function of the human brain, under physiological and pathological conditions.
Keywords: Brain organoid, pluripotent stem cells, CRISPR, neurological disorders, evolution
Abstract
In vitro differentiation of pluripotent stem cells provides a systematic platform to study development and disease. Recent advances in brain organoid technology have created new opportunities to investigate the formation and function of the human brain, under physiological and pathological conditions. Brain organoids can be generated to model the cellular and structural development of the human brain, and allow the investigation of the intricate interactions between resident neural and glial cell types. Combined with new advances in gene editing, imaging, and genomic analysis, brain organoid technology can be applied to address questions pertinent to human brain development, disease, and evolution. However, the current iterations of brain organoids also have limitations in faithfully recapitulating the in vivo processes. In this perspective, we evaluate the recent progress in brain organoid technology, and discuss the experimental considerations for its utilization.
Dual Perspectives Companion Paper: Integrating CRISPR Engineering and hiPSC-Derived 2D Disease Modeling Systems, by Kristina Rehbach, Michael B. Fernando, and Kristen J. Brennand
Introduction
The human brain is one of the most complex organs in the human body. Much of our current knowledge about the molecular, cellular, and anatomical mechanisms of human brain development was gained by studying model organisms, which revealed that many aspects of brain development are evolutionarily conserved. However, the human brain is also substantially different from those of other species, including rodents and nonhuman primates, in size, shape, cellular composition, and ultimately, the capacity for higher cognitive function. Many of these differences are deeply rooted during fetal development and are already apparent at birth. At the structural level, the human brain, in particular the cerebral neocortex, is greatly expanded in size, driven by an increase in the number of neurons and glia as well as the overall cortical area (Bystron et al., 2008; Sun and Hevner, 2014). This expansion is accompanied by the emergence of extensive cortical folding.
At the cellular level, neural stem and progenitor cells undergo highly controlled proliferation and differentiation processes to generate the vast numbers of neurons and glia that make up the postnatal brain. It has been observed that human neural stem and progenitor cells have enhanced proliferative capacity, and are intrinsically more diverse than their counterparts in other species, such as the mouse and the rat (Rakic, 1974; Otani et al., 2016). Dysfunctions of these stem and progenitor cells have been linked to structural malformations of the brain (Walsh, 1999). Disruptions of their differentiated neuronal and glial progenies are associated with neurodevelopmental disorders ranging in severity (Hu et al., 2014). Many of these disorders have been proven difficult to study in animal models. Therefore, direct analysis of human neural cells and tissues is required to enhance our understanding of both conserved and nonconserved mechanisms governing normal and pathological human neurodevelopment.
The advent of human pluripotent stem cell (hPSC) technologies, including that of embryonic stem cells and induced pluripotent stem cells (iPSCs), opened a new avenue to study human neural development and disease in vitro. Directed differentiation of hPSCs into the neural lineage can now be efficiently performed in adherent 2D cultures (Zhang et al., 2001) (also see accompanying Dual Perspectives article by Rehbach et al.). It has long been observed that neural differentiation can occur efficiently in embryoid bodies, aggregates of PSCs cultured in suspension (Itskovitz-Eldor et al., 2000). Variants of the embryoid body-based differentiation methods were shown to yield complex neuroectodermal structures resembling the cerebral cortex and the optic cup, demonstrating the feasibility of recapitulating organogenesis in vitro using 3D differentiation and highlighting the ability of hPSCs and their progenies to self-organize (Eiraku et al., 2008; Nakano et al., 2012). Building on these efforts, and integrating successful methods developed to generate 3D intestinal epithelium structures from mouse intestinal stem cells (Sato and Clevers, 2013), Lancaster et al. (2013) developed protocols to generate cerebral organoids from hPSCs. Relying on the intrinsic propensity for hPSCs to differentiate toward the neural lineage, these 3D neural tissues harbor broad regional identity, such as the forebrain, midbrain, and hindbrain, and display complex structural organization reminiscent of the cytoarchitecture of the fetal human brain, including the formation of polarized cortical tissue and fluid-filled ventricles (Lancaster et al., 2013; Lancaster and Knoblich, 2014). Within these cerebral organoids, neuroepithelia are induced from PSCs, which give rise to neural stem and progenitor cells and undergo differentiation into diverse subtypes of neurons and glia. Single-cell RNA sequencing analysis revealed substantial similarities in gene expression profiles between cerebral organoid and fetal human cerebral cortex (Camp et al., 2015). It is worth noticing that non-neural lineages can also be derived in these self-patterned cerebral organoids due to unguided differentiation and insufficient inhibition of mesoderm and endoderm lineages (Camp et al., 2015; Quadrato et al., 2017; Ormel et al., 2018). Further incorporation of patterning cues allowed the generation of organoids with exclusive neural identity and organoids with specific brain-region or cell-type identities (Kadoshima et al., 2013; Mariani et al., 2015; Muguruma et al., 2015; Paşca et al., 2015; Sakaguchi et al., 2015; Qian et al., 2016).
The development of brain organoid technology expands the already diverse possibilities of using PSCs to study human neurobiology. Compared with adherent 2D neural differentiation, the process of brain organoid generation preserves the spatial architecture that forms as the primitive neural tissues self-organize, and allows the investigation of early human brain formation, a process difficult to observe in vivo and that diverges significantly between human and experimental organisms. Long-term culture of brain organoids encourages development of stable cell–cell interaction at physiological proximity and density, which is particularly relevant for the formation of meaningful connections and interactions between neuronal cells and among neurons and glia. In the short span of a few years, brain organoids have been used to model human brain development, neurological disorders, and evolution. The development of organoid technology also coincided with rapid advances in genome editing, single-cell genomics, tissue clearing, and imaging techniques, all of which are adaptable and well suited for application in brain organoids. Here, we explore the various use of brain organoid technologies and discuss experimental considerations for incorporating this tool into a discovery process.
Applications and limitations of brain organoid models
Studying human brain development
In vitro differentiation of hPSCs has revolutionized our ability to study early development and cell fate transition. In the absence of induction toward other lineages, PSCs adopt a default path into the neural fate (Muñoz-Sanjuán and Brivanlou, 2002). In 3D suspension culture, as in 2D adherent cultures, hPSCs can spontaneously differentiate into cell types characteristic of the primitive neuroepithelium (Eiraku et al., 2008). While self-assembly and self-patterning emerge in both types of cultures, continuous culturing of organoids in suspension without dissociation has the advantage of allowing uninterrupted self-organization into in vitro organogenesis (Fig. 1). This provides an opportunity to closely study this early developmental process, in particular the features that diverge between human and experimental organisms, such as the mouse.
Figure 1.
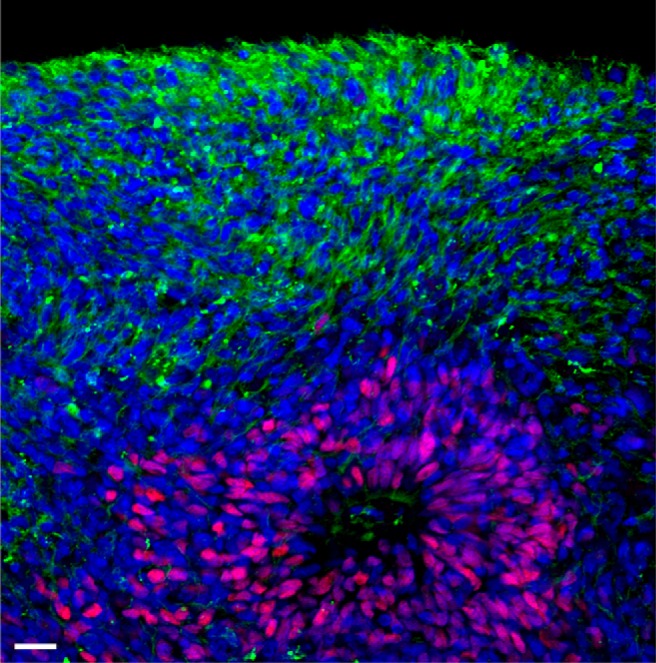
Cross section of a day 83 human forebrain organoid, showing MAP2+ neurons (green) and SOX2+ neural precursors (red), surrounding a ventricle-like structure. DAPI (blue). Scale bar, 20 μm.
It has been shown that, while brain organoids from both murine and human PSCs develop stratified structures resembling ventricular zone, subventricular zone, intermediate zone, and cortical plate, only human brain organoids develop outer subventricular zone-like structures (Eiraku et al., 2008; Kadoshima et al., 2013; Lancaster et al., 2013). This recapitulates the presence of the outer subventricular zone in vivo in the human, but not the mouse cortex. Within the germinal zones of human brain organoids, proliferative precursors resembling ventricular and outer radial glial cells are generated, in a temporal and spatial pattern consistent with in vivo human cortical development (Kadoshima et al., 2013; Lancaster et al., 2013). Of note, proliferation of precursors in the ventricular zone and subventricular zone persists for a prolonged period in the human brain organoids, compared with their mouse counterparts (Lancaster et al., 2013; Y. Li et al., 2017). As radial glial cells differentiate into intermediate progenitors, and later into neurons and glia, the cell types generated recapitulate the transcriptomic and epigenomic signatures of the human fetal brain (Camp et al., 2015; Luo et al., 2016; Quadrato et al., 2017; Amiri et al., 2018; Pollen et al., 2019). Differentiating cortical neurons display inside-out migratory patterns consistent with in vivo development, and establish rudimentary cortical layering according to their time of birth (Kadoshima et al., 2013; Lancaster et al., 2013).
To model long distance neuronal migration between distinct brain regions, brain organoids can be prepatterned to develop region-specific identity, after which different organoids can be cocultured to allow physical contact and fusion. Using this approach, directional migration of inhibitory neurons from ventral cortical organoids into dorsal cortical organoids has been observed (Bagley et al., 2017; Birey et al., 2017; Xiang et al., 2017; Sloan et al., 2018). Migration of human interneurons in brain organoids recapitulates the saltatory pattern seen during fetal human cortical development and is distinct from that of mouse interneurons (Birey et al., 2017).
As brain organoids mature with prolonged culture, functional neuronal connections are formed, allowing the observation of neuronal activity on the levels of cells and network. Spontaneous and evoked neural activity can be measured using calcium imaging (Lancaster et al., 2013; Paşca et al., 2015; Qian et al., 2016), current/voltage clamp (Paşca et al., 2015; Qian et al., 2016), and multielectrodes (Kawada et al., 2017; Quadrato et al., 2017; Giandomenico et al., 2019). Maturation of astrocytes (Paşca et al., 2015; Sloan et al., 2017) and oligodendrocytes (Madhavan et al., 2018; Marton et al., 2019) can also be seen, allowing the investigation of the development and function of these glial cell types.
Modeling human neurological disorders
An array of neural developmental and degenerative disorders has been successfully modeled using brain organoids. Because the course of brain organoid generation approximates the early steps of fetal brain formation, diseases that impact this developmental process were among the first to be successfully modeled. For example, several early studies modeled diseases that result in reduced neural structural formation, such as genetic forms of microcephaly (Lancaster et al., 2013; Jin et al., 2017; Omer Javed et al., 2018). After maternal exposure to Zika virus was linked to a range of fetal brain abnormalities, including microcephaly, lissencephaly, and cortical calcification, brain organoids were used to investigate the cause of the neuropathology and to identify potential therapeutic strategies (Garcez et al., 2016; Qian et al., 2016; Xu et al., 2016; Y. Li et al., 2017, 2019; Watanabe et al., 2017; Zhou et al., 2017). At the other end of the spectrum, enhanced and excessive growth of the fetal brain has been reported in human macrocephaly and polymicrogyria, including those caused by mutations in the AKT-mTOR signaling pathway (McBride et al., 2010; Lee et al., 2012; Poduri et al., 2012; Rivière et al., 2012). Genetically modified brain organoids with elevated AKT-mTOR signaling activities have been used to model these enhanced growth conditions (Y. Li et al., 2017). In addition, abnormal neural migration, which occurs in genetic forms of lissencephaly and periventricular heterotopia, caused by mutations in genes, such as DCHS1, FAT4, or PLEKHG6, has been modeled in brain organoids (Bershteyn et al., 2017; O'Neill et al., 2018; Klaus et al., 2019).
In addition to studying the mechanism underlying diseases involving well-defined brain malformation, organoids derived from hPSCs bearing causal mutations for neurodevelopmental and psychiatric diseases can be used to identify previously unrecognized abnormalities. For instance, altered neurogenesis and imbalance in neuronal and glial differentiation have been reported in organoid models of several neurodevelopmental disorders, including autism spectrum disorders (Mariani et al., 2015), Rett syndrome (Mellios et al., 2018), tuberous sclerosis (Blair et al., 2018), and DISC1-related schizophrenia (Ye et al., 2017; Srikanth et al., 2018).
Brain organoids have also proven advantageous in modeling neurodegenerative diseases and other neurological conditions. Because 3D culturing allows the formation of long-term cell–cell contacts at physiological density, it facilitates the retainment of pathological protein aggregates that are often the hallmark of neurodegenerative diseases. Increased accumulation of amyloid-β peptide and phosphorylated tau protein has been reported in organoid models of familial and sporadic forms of Alzheimer's disease (Raja et al., 2016; Lin et al., 2018), whereas increased levels of phosphorylated α-synuclein have been reported in an organoid model of Parkinson's disease with mutation in the LRRK2 gene (Kim et al., 2019). Accumulation of these protein aggregates has previously been difficult to detect and preserve using 2D adherent cultures. Organoids have also been used to model brain tumor initiation and progression. Introduction of known oncogenic mutations was shown to result in neoplastic cerebral organoids recapitulating features of glioblastoma and CNS primitive neuroectodermal tumors, enabling the study of the crucial first steps in the genetic transformation of normal neural stem cells into brain tumor initiating cells (Bian et al., 2018). In addition, transplantation of glioma stem cells from patient biopsy allowed the investigation of tumor invasion into host brain tissue, providing a novel human-based xenograft platform to assess disease mechanism and drug responses (Linkous et al., 2019).
Investigating the mechanisms of brain evolution
The development of PSC technology presents an unprecedented opportunity to perform interspecies comparative genomics studies. Our ability to derive neural cells and organoid tissues from these PSCs, combined with an ever-expanding toolkit for genome editing, allows functional interrogation of the underlying mechanisms that drive brain evolution. Comparison of neural cells derived from human, chimpanzee, and macaque PSCs demonstrated species-specific duration of proliferation and cumulative neuronal output (Otani et al., 2016). Of note, human neural stem and progenitor cells displayed longer periods of expansion in both 2D adherent cultures and 3D brain organoids than cells from other species (Otani et al., 2016). While the underlying mechanism for the species-specific cellular behaviors is likely multifaceted, recent studies indicate that human-specific gene duplication may be a contributing factor (Florio et al., 2015, 2018; Fiddes et al., 2018; Suzuki et al., 2018). Features of species-specific cortical development, such as the differential abundance of outer radial glial cells, could be detected in brain organoids derived from human and nonhuman primates (Pollen et al., 2019).
Interspecies comparisons also revealed that differential reliance on conserved molecular pathways modulates species-specific brain development. It has been shown that preferential utilization of the Robo or Dll1 pathways distinguishes the mode of neurogenesis in mammals versus in reptiles and birds (Cárdenas et al., 2018). In particular, in human brain organoids, genetic manipulations that modulate the strength of Robo and Dll1 signaling were sufficient to change the output of neuronal differentiation (Cárdenas et al., 2018). Another conserved signaling cascade that shows species-specific utilization is the mTOR pathway. Transcriptomic analysis of brain organoids derived from multiple primate species revealed a preferential reliance on this pathway in human, compared with chimpanzee and macaque (Pollen et al., 2019). This is consistent with the finding that activation of the mTOR signaling pathway via genetic ablation of PTEN resulted in pronounced expansion and folding in human brain organoids (Y. Li et al., 2017). In contrast, Pten deletion in mouse PSCs led to a modest increase in brain organoid size and failed to induce structural folding (Y. Li et al., 2017). Together, these studies exemplify the possibilities of using PSCs and brain organoids to investigate the conserved and nonconserved principles of brain development across multiple species. While much remains unknown about the mechanisms that drove the expansion and complexification of the human brain, the availability of these technological implements opens up exciting opportunity for exploration.
Experimental considerations for using brain organoids
Because of the many advantages brain organoids provide, it could be beneficial to incorporate them into study designs in pursuit of a pertinent biological question about neural development, disease, and evolution. However, it is important to note that the current brain organoid technology has yet to succeed in recapitulating many key aspects of human fetal brain. These limitations include incomplete cellular composition and anatomy, lack of synaptic and circuit maturation, as well as high levels of variability. These caveats are to be carefully weighed to determine whether brain organoids are suitable and advantageous in addressing a particular biological question.
The initial considerations of implementing a brain organoid experiment are common for all PSC-based models. While the ability to derive iPSCs from any individual creates an opportunity to examine the underlying biology across diverse human genetic backgrounds, it inevitably generates hurdles in establishing causal correlations between genetic variants (genotypes) and experimental readouts (phenotypes). For many of these instances, inclusion of isogenic controls is a natural solution to exclude variations caused by genetic background differences. Gene editing tools, such as TALEN and CRISPR/Cas9, have made it possible to create isogenic pairs of hPSCs that differ exclusively at the desired genetic locus. Inclusion of more than one parental hPSC line can be helpful in validating genetically penetrant findings. For other studies where a genetically diverse cohort is a necessary starting point, such as investigating polygenic risks associated with neural developmental and degenerative diseases, a sufficiently powered initial population with appropriate clinical stratification should be assembled. From these steps onward, the decision of whether and how to incorporate experiments using brain organoids should be made cognizant of their limitations and guided by the underlying biological question.
Generating the right cell types and structures
Not unlike their 2D counterparts, 3D brain organoids can be generated to assume different regional, temporal, and lineage identity. These methods use developmental cues and patterning factors that guide the differentiation of hPSCs and their primitive neuroepithelial progenies. A critical first evaluation is needed to identify the appropriate organoid derivation method for the biological question at hand. The least restrictive differentiation method relies on the neuralization propensity of hPSCs and generates organoids bearing broad cerebral cortical identity (Lancaster et al., 2013). Addition of morphogens and growth factors generates brain organoids bearing the specific identity of cortex, hippocampus, thalamus, ventral forebrain, midbrain, and hindbrain (Kadoshima et al., 2013; Muguruma et al., 2015; Paşca et al., 2015; Sakaguchi et al., 2015; Jo et al., 2016; Qian et al., 2016).
During in vivo development, morphogens and growth factors often exist in limited amounts and in gradients that instruct different signaling responses and contribute to the spatial patterning of the brain. A recent study by Cederquist et al. (2019) generated organoids with inherent positional identity in response to a gradient of sonic hedgehog. This strategy is broadly applicable to model the formation of in vivo-like topography in brain organoids and explore long-range signal propagation. As mentioned above, fusion of multiple region-specific brain organoids has been used successfully to study the directional migration of inhibitory neurons (Bagley et al., 2017; Birey et al., 2017; Xiang et al., 2017) and to model reciprocal projections between cortical and thalamic organoids (Xiang et al., 2019).
The choice of differentiation method also depends on the cell types of interest. Studies that require access to early neural precursors can rely on the ability of brain organoids to form the equivalent of early germinal zones encompassing the diverse neural stem and progenitor cells that exist in vivo. However, cell types that emerge later during fetal brain development, such as mature neurons whose identities are coupled with their physical positions and functional connections, may not be sufficiently specified within the brain organoids due to the absence of this prerequisite information. Gene expression and electrophysiological studies showed that cell types within the developing brain organoids most closely resemble first and second trimester human fetal brain (Paşca et al., 2015; Qian et al., 2016; Watanabe et al., 2017). The limitation in achieving cellular maturity levels equivalent of postnatal and adult brain is not limited to neurons. Other resident cell types, such as astrocytes and oligodendrocytes, can be generated in brain organoids (Paşca et al., 2015; Madhavan et al., 2018; Sloan et al., 2018; Marton et al., 2019). Their maturation is also protracted and may be difficult to accelerate without a cost to neuronal differentiation using existing methods developed based on 2D directed differentiation (Yang et al., 2013; Tcw et al., 2017; Lager et al., 2018; X. Li et al., 2018; Zheng et al., 2018). We also note that anatomical features, such as the development of a consistent axis, an expanded cortical plate, and its complexification into the stereotypic 6-layer cortex structure, are currently poorly recapitulated in brain organoids. These limitations are not unique to brain organoids, and are instead common to PSC-based modeling efforts where adult cell types and structures are desirable. It remains to be seen whether approaches that have been effective in promoting accelerated maturation of distinct cell types in 2D cultures can be adapted to brain organoids (Zhang et al., 2013; Qi et al., 2017; Yang et al., 2017; Tchieu et al., 2019). Last, a sometimes-overlooked consideration in the choice of organoid derivation method is the desired size and complexity of the 3D structure. Whereas large and complex self-patterned cerebral organoids can be more effective in modeling developmental and disease processes that affect neural tissue expansion and organization, can support the study of long-range signal transduction in 3D space, and permit the investigation of regional interactions, they tend to be more heterogeneous and variable (Lancaster et al., 2013; Velasco et al., 2019). This may limit their use in phenotypic discovery and high throughput assays. Protocols have been developed to generate less variable neural structures with reproducible size, cellular composition (Velasco et al., 2019; Yoon et al., 2019), and even structural symmetry (Zhu et al., 2013; Knight et al., 2018). While some of these approaches may not permit the development of complex features, they can be preferred where variability is less tolerated.
Incorporating the missing cellular and structural components
While the majority of the cells in the human brain come from the neuroectoderm lineage, an important minority population derives from the mesoderm. This includes the vascular endothelial cells and resident microglia. Directed differentiation methods used to convert hPSCs into the neural lineage do not generate these cell types. The endothelial cells and microglia migrate into the early developing brain and codevelop with the resident neural cells. Dysfunction in these cell types is involved in a diverse array of neurological conditions, including Alzheimer's disease, Parkinson's disease, autism, schizophrenia, and cerebrovascular diseases, such as cerebral cavernous malformations (Fischer et al., 2013; Li and Barres, 2018). To model these complex disorders, strategies to incorporate these missing cell types into 3D brain organoids are required.
The efforts to generate vascularized brain organoids can benefit from the recent development of hPSC-derived blood vessel organoids (Wimmer et al., 2019), potentially allowing codevelopment or fusion of organoids with brain and vasculature identities. While current iterations of blood-vessel organoids are not yet capable of supporting circulation outside a xenograft host, future engineering approaches may facilitate this process in vitro via active mechanical perfusion.
Brain organoids are also devoid of microglia, the resident immune cells of the CNS. We (Muffat et al., 2016) and others (Abud et al., 2017; Douvaras et al., 2017; Haenseler et al., 2017; Pandya et al., 2017; Takata et al., 2017; Brownjohn et al., 2018) have recently shown that hPSCs can be differentiated into primitive yolk-sac macrophages, which can then be induced to mature into microglia-like cells sharing many characteristics of developing human microglia. These cells can be cocultured with isogenic neuronal and astrocytic cells in 2D. However, their identity, fine-tuned by the neuroectodermal environment in which they reside, is remarkably labile: ex vivo microglia lose their morphology, functional characteristics, and steady-state expression profile. From an experimental standpoint, it is therefore highly beneficial to provide them with neural tissue to reside in, by reaggregating them with isogenic neurons and glia to assemble an organoid with defined cellular components, or letting them invade preformed 3D brain organoids (Muffat et al., 2016, 2018). Their microglial identity on the transcriptomic level is enhanced under these coculture conditions, and they can display physiological characteristics of in vivo microglia, including highly branched ramifications, surveying behavior, and responses to cellular damage and infectious agents (Muffat et al., 2016, 2018). It is reasonable to posit that the presence of microglia will in return affect the fate and function of neuroectodermal cells that form the bulk of the organoid. It is worth noting that rare homozygous CSF1R mutations in humans lead to loss of microglia and severe developmental malformations, such as callosal agenesis (Guo et al., 2019; Oosterhof et al., 2019). Furthermore, recent findings showed that IL1A, C1Q, and TNF-α secreted by microglia can control the cytotoxicity of astrocytes toward neurons and oligodendrocytes (Liddelow et al., 2017).
Recent studies comparing the transcriptomic profiles of human fetal brain tissues and brain organoids revealed many similarities but also distinct differences (Camp et al., 2015; Qian et al., 2016; Amiri et al., 2018; Pollen et al., 2019; Velasco et al., 2019). The absence of the aforementioned cellular and structural components figures prominently in these differences. Lack of cell types that originate from non-neuroectoderm lineage (e.g., microglia, endothelial cells), elevated metabolic stress due to nutrient and oxygen deprivation, and delayed maturation are among the most noted limitations of the current iterations of brain organoids. These findings highlight the value of continuous research on primary human cells and tissues, to understand human neurodevelopment, and to guide the implementation of more physiological organoid systems.
Navigating the variability of the assays
Brain organoids can be analyzed in a variety of phenotypic assays. They are conducive to canonical in vitro approaches that may be difficult in vivo, such as live imaging over long developmental time courses. They also have tissue-like density and anatomical features that benefit from handling techniques intended for tissues. For example, histology methods for structural analysis, organotypic slice culture for imaging and electrophysiology, and intraventricular injection of DNA for electroporation have often been used on brain organoids.
A major limitation of many current brain organoid systems is their variability, reflected in the differences in size, shape, cellular composition, and cytoarchitecture among samples generated in the same batch (Kelava and Lancaster, 2016; Heide et al., 2018; Pollen et al., 2019). Approaches that improve the homogeneity of the starting hPSCs or narrow their differentiation trajectory to a defined regional identity can reduce such variability (Velasco et al., 2019; Yoon et al., 2019). The use of Matrigel as an embedding material and media supplement may provide a supportive extracellular matrix and a concentrated source of nutrients. However, this creates batch-to-batch variability due to the undefined nature of Matrigel. In addition, the step of embedding an embryoid body into a droplet of Matrigel leads to variability and introduces organoid-to-organoid differences. Efforts to identify the mechanical properties and molecular components of Matrigel that contribute to organoid growth could help to create more defined culture protocols. A better understanding of the parameters leading to formation of ventricular structures in organoids may illuminate the role of these cerebrospinal fluid compartments during brain development while allowing generation of more reproducibly polarized features. Methods that generate smaller neural structures (Knight et al., 2018) also enjoy higher reproducibility, albeit at the cost of having fewer structural features.
Overall it may seem that the current approach of relying on the self-organizing and self-patterning properties of stem cells necessitates the tolerance of some stochasticity in the system. However, it should be remembered that in vivo development, from a single cell to an entire organism, is remarkably reproducible. It is foreseeable that further knowledge in human development, gained through studying hPSC-based models, will guide rational design of better differentiation and engineering strategies to recapitulate more in vivo-like features, thus driving iterative advances.
Navigating the scales and throughput
Brain organoid systems provide an opportunity for systematic assessment of cell types and anatomical structures that are otherwise difficult to access during fetal development. Whereas successful high-throughput chemical and genetic screens using hPSCs-derived 2D neural cultures have recently been reported (Xu et al., 2016; Zhou et al., 2017; Y. Li et al., 2019), implementing high throughput assays in organoids may encounter added hurdles. Achieving consistency and reproducibility among individual organoids will be of paramount importance for scaled up productions. Developing assays that can reliably and sensitively detect phenotypic alterations in free-floating 3D structures is another key challenge. We envision that effective strategies may emerge from new engineering solutions as well as approaches adapted from chemical and genetic screens in model organisms, such as Caenorhabditis elegans, zebrafish, and planarian.
In conclusion, organoid systems leverage the amazing self-organizing properties of stem cells to re-create complex tissue and organ development in a dish. In the span of just a few years, this emerging tool has already advanced our knowledge of human development, disease, and evolution. As for every nascent technology, the organoid system will continue to evolve. This requires the users of the technology to be cognizant of current limitations and agile in implementing adaptations. We are confident that the combination of genome engineering, uses of gradients to guide differentiation into polarized neural tissues, modular assembly of regional elements, and inclusion of relevant neural and non-neural cell types will lead to the establishment of improved brain organoid systems.
Footnotes
J.M. was supported by the Canada Research Chairs Program, and the National Sciences and Engineering Research Council of Canada. Y.L. was supported by Simons Foundation SFARI 479754 and the Ontario Institute for Regenerative Medicine. This research was part of the University of Toronto's Medicine by Design initiative, which received funding from the Canada First Research Excellence Fund. We thank members of the Y.L. and J.M. laboratories for helpful discussions.
The authors declare no competing financial interests.
References
- Abud EM, Ramirez RN, Martinez ES, Healy LM, Nguyen CH, Newman SA, Yeromin AV, Scarfone VM, Marsh SE, Fimbres C, Caraway CA, Fote GM, Madany AM, Agrawal A, Kayed R, Gylys KH, Cahalan MD, Cummings BJ, Antel JP, Mortazavi A, et al. (2017) iPSC-derived human microglia-like cells to study neurological diseases. Neuron 94:278–293.e9. 10.1016/j.neuron.2017.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiri A, Coppola G, Scuderi S, Wu F, Roychowdhury T, Liu F, Pochareddy S, Shin Y, Safi A, Song L, Zhu Y, Sousa AM, Gerstein M, Crawford GE, Sestan N, Abyzov A, Vaccarino FM (2018) Transcriptome and epigenome landscape of human cortical development modeled in organoids. Science 362:eaat6720. 10.1126/science.aat6720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagley JA, Reumann D, Bian S, Lévi-Strauss J, Knoblich JA (2017) Fused cerebral organoids model interactions between brain regions. Nat Methods 14:743–751. 10.1038/nmeth.4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bershteyn M, Nowakowski TJ, Pollen AA, Di Lullo E, Nene A, Wynshaw-Boris A, Kriegstein AR (2017) Human iPSC-derived cerebral organoids model cellular features of lissencephaly and reveal prolonged mitosis of outer radial glia. Cell Stem Cell 20:435–449.e4. 10.1016/j.stem.2016.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian S, Repic M, Guo Z, Kavirayani A, Burkard T, Bagley JA, Krauditsch C, Knoblich JA (2018) Genetically engineered cerebral organoids model brain tumor formation. Nat Methods 15:631–639. 10.1038/s41592-018-0070-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birey F, Andersen J, Makinson CD, Islam S, Wei W, Huber N, Fan HC, Metzler KR, Panagiotakos G, Thom N, O'Rourke NA, Steinmetz LM, Bernstein JA, Hallmayer J, Huguenard JR, Paşca SP (2017) Assembly of functionally integrated human forebrain spheroids. Nature 545:54–59. 10.1038/nature22330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair JD, Hockemeyer D, Bateup HS (2018) Genetically engineered human cortical spheroid models of tuberous sclerosis. Nat Med 24:1568–1578. 10.1038/s41591-018-0139-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownjohn PW, Smith J, Solanki R, Lohmann E, Houlden H, Hardy J, Dietmann S, Livesey FJ (2018) Functional studies of missense TREM2 mutations in human stem cell-derived microglia. Stem Cell Rep 10:1294–1307. 10.1016/j.stemcr.2018.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystron I, Blakemore C, Rakic P (2008) Development of the human cerebral cortex: Boulder Committee revisited. Nat Rev Neurosci 9:110–122. 10.1038/nrn2252 [DOI] [PubMed] [Google Scholar]
- Camp JG, Badsha F, Florio M, Kanton S, Gerber T, Wilsch-Bräuninger M, Lewitus E, Sykes A, Hevers W, Lancaster M, Knoblich JA, Lachmann R, Pääbo S, Huttner WB, Treutlein B (2015) Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc Natl Acad Sci U S A 112:15672–15677. 10.1073/pnas.1520760112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cárdenas A, Villalba A, de Juan Romero C, Picó E, Kyrousi C, Tzika AC, Tessier-Lavigne M, Ma L, Drukker M, Cappello S, Borrell V (2018) Evolution of cortical neurogenesis in amniotes controlled by Robo signaling levels. Cell 174:590–606.e21. 10.1016/j.cell.2018.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederquist GY, Asciolla JJ, Tchieu J, Walsh RM, Cornacchia D, Resh MD, Studer L (2019) Specification of positional identity in forebrain organoids. Nat Biotechnol 37:436–444. 10.1038/s41587-019-0085-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douvaras P, Sun B, Wang M, Kruglikov I, Lallos G, Zimmer M, Terrenoire C, Zhang B, Gandy S, Schadt E, Freytes DO, Noggle S, Fossati V (2017) Directed differentiation of human pluripotent stem cells to microglia. Stem Cell Reports 8:1516–1524. 10.1016/j.stemcr.2017.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiraku M, Watanabe K, Matsuo-Takasaki M, Kawada M, Yonemura S, Matsumura M, Wataya T, Nishiyama A, Muguruma K, Sasai Y (2008) Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 3:519–532. 10.1016/j.stem.2008.09.002 [DOI] [PubMed] [Google Scholar]
- Fiddes IT, Lodewijk GA, Mooring M, Bosworth CM, Ewing AD, Mantalas GL, Novak AM, van den Bout A, Bishara A, Rosenkrantz JL, Lorig-Roach R, Field AR, Haeussler M, Russo L, Bhaduri A, Nowakowski TJ, Pollen AA, Dougherty ML, Nuttle X, Addor MC, et al. (2018) Human-specific NOTCH2NL genes affect notch signaling and cortical neurogenesis. Cell 173:1356–1369.e22. 10.1016/j.cell.2018.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Zalvide J, Faurobert E, Albiges-Rizo C, Tournier-Lasserve E (2013) Cerebral cavernous malformations: from CCM genes to endothelial cell homeostasis. Trends Mol Med 19:302–308. 10.1016/j.molmed.2013.02.004 [DOI] [PubMed] [Google Scholar]
- Florio M, Albert M, Taverna E, Namba T, Brandl H, Lewitus E, Haffner C, Sykes A, Wong FK, Peters J, Guhr E, Klemroth S, Prüfer K, Kelso J, Naumann R, Nüsslein I, Dahl A, Lachmann R, Pääbo S, Huttner WB (2015) Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science 347:1465–1470. 10.1126/science.aaa1975 [DOI] [PubMed] [Google Scholar]
- Florio M, Heide M, Pinson A, Brandl H, Albert M, Winkler S, Wimberger P, Huttner WB, Hiller M (2018) Evolution and cell-type specificity of human-specific genes preferentially expressed in progenitors of fetal neocortex. eLife 7:e32332. 10.7554/eLife.32332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcez PP, Loiola EC, Madeiro da Costa R, Higa LM, Trindade P, Delvecchio R, Nascimento JM, Brindeiro R, Tanuri A, Rehen SK (2016) Zika virus impairs growth in human neurospheres and brain organoids. Science 352:816–818. 10.1126/science.aaf6116 [DOI] [PubMed] [Google Scholar]
- Giandomenico SL, Mierau SB, Gibbons GM, Wenger LM, Masullo L, Sit T, Sutcliffe M, Boulanger J, Tripodi M, Derivery E, Paulsen O, Lakatos A, Lancaster MA (2019) Cerebral organoids at the air–liquid interface generate diverse nerve tracts with functional output. Nat Neurosci 22:669–679. 10.1038/s41593-019-0350-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Bertola DR, Takanohashi A, Saito A, Segawa Y, Yokota T, Ishibashi S, Nishida Y, Yamamoto GL, Franco JF, Honjo RS, Kim CA, Musso CM, Timmons M, Pizzino A, Taft RJ, Lajoie B, Knight MA, Fischbeck KH, Singleton AB, et al. (2019) Bi-allelic CSF1R mutations cause skeletal dysplasia of dysosteosclerosis-Pyle disease spectrum and degenerative encephalopathy with brain malformation. Am J Hum Genet 104:925–935. 10.1016/j.ajhg.2019.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenseler W, Sansom SN, Buchrieser J, Newey SE, Moore CS, Nicholls FJ, Chintawar S, Schnell C, Antel JP, Allen ND, Cader MZ, Wade-Martins R, James WS, Cowley SA (2017) A highly efficient human pluripotent stem cell microglia model displays a neuronal-co-culture-specific expression profile and inflammatory response. Stem Cell Reports 8:1727–1742. 10.1016/j.stemcr.2017.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heide M, Huttner WB, Mora-Bermúdez F (2018) Brain organoids as models to study human neocortex development and evolution. Curr Opin Cell Biol 55:8–16. 10.1016/j.ceb.2018.06.006 [DOI] [PubMed] [Google Scholar]
- Hu WF, Chahrour MH, Walsh CA (2014) The diverse genetic landscape of neurodevelopmental disorders. Annu Rev Genomics Hum Genet 15:195–213. 10.1146/annurev-genom-090413-025600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itskovitz-Eldor J, Schuldiner M, Karsenti D, Eden A, Yanuka O, Amit M, Soreq H, Benvenisty N (2000) Differentiation of human embryonic stem cells into embryoid bodies compromising the three embryonic germ layers. Mol Med 6:88–95. 10.1007/BF03401776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M, Pomp O, Shinoda T, Toba S, Torisawa T, Furuta K, Oiwa K, Yasunaga T, Kitagawa D, Matsumura S, Miyata T, Tan TT, Reversade B, Hirotsune S (2017) Katanin p80, NuMA and cytoplasmic dynein cooperate to control microtubule dynamics. Sci Rep 7:39902. 10.1038/srep39902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo J, Xiao Y, Sun AX, Cukuroglu E, Tran HD, Göke J, Tan ZY, Saw TY, Tan CP, Lokman H, Lee Y, Kim D, Ko HS, Kim SO, Park JH, Cho NJ, Hyde TM, Kleinman JE, Shin JH, Weinberger DR (2016) Midbrain-like organoids from human pluripotent stem cells contain functional dopaminergic and neuromelanin-producing neurons. Cell Stem Cell 19:248–257. 10.1016/j.stem.2016.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoshima T, Sakaguchi H, Nakano T, Soen M, Ando S, Eiraku M, Sasai Y (2013) Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc Natl Acad Sci U S A 110:20284–20289. 10.1073/pnas.1315710110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawada J, Kaneda S, Kirihara T, Maroof A, Levi T, Eggan K, Fujii T, Ikeuchi Y (2017) Generation of a motor nerve organoid with human stem cell-derived neurons. Stem Cell Reports 9:1441–1449. 10.1016/j.stemcr.2017.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelava I, Lancaster MA (2016) Stem cell models of human brain development. Cell Stem Cell 18:736–748. 10.1016/j.stem.2016.05.022 [DOI] [PubMed] [Google Scholar]
- Kim H, Park HJ, Choi H, Chang Y, Park H, Shin J, Kim J, Lengner CJ, Lee YK, Kim J (2019) Modeling G2019S-LRRK2 sporadic Parkinson's disease in 3D midbrain organoids. Stem Cell Reports 12:518–531. 10.1016/j.stemcr.2019.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaus J, Kanton S, Kyrousi C, Ayo-Martin AC, Di Giaimo R, Riesenberg S, O'Neill AC, Camp JG, Tocco C, Santel M, Rusha E, Drukker M, Schroeder M, Götz M, Robertson SP, Treutlein B, Cappello S (2019) Altered neuronal migratory trajectories in human cerebral organoids derived from individuals with neuronal heterotopia. Nat Med 25:561–568. 10.1038/s41591-019-0371-0 [DOI] [PubMed] [Google Scholar]
- Knight GT, Lundin BF, Iyer N, Ashton LM, Sethares WA, Willett RM, Ashton RS (2018) Engineering induction of singular neural rosette emergence within hPSC-derived tissues. eLife 7:e37549. 10.7554/eLife.37549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lager AM, Corradin OG, Cregg JM, Elitt MS, Shick HE, Clayton BL, Allan KC, Olsen HE, Madhavan M, Tesar PJ (2018) Rapid functional genetics of the oligodendrocyte lineage using pluripotent stem cells. Nat Commun 9:3708. 10.1038/s41467-018-06102-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Knoblich JA (2014) Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc 9:2329–2340. 10.1038/nprot.2014.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA (2013) Cerebral organoids model human brain development and microcephaly. Nature 501:373–379. 10.1038/nature12517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Huynh M, Silhavy JL, Kim S, Dixon-Salazar T, Heiberg A, Scott E, Bafna V, Hill KJ, Collazo A, Funari V, Russ C, Gabriel SB, Mathern GW, Gleeson JG (2012) De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly. Nat Genet 44:941–945. 10.1038/ng.2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Barres BA (2018) Microglia and macrophages in brain homeostasis and disease. Nat Rev Immunol 18:225–242. 10.1038/nri.2017.125 [DOI] [PubMed] [Google Scholar]
- Li X, Tao Y, Bradley R, Du Z, Tao Y, Kong L, Dong Y, Jones J, Yan Y, Harder CR, Friedman LM, Bilal M, Hoffmann B, Zhang SC (2018) Fast generation of functional subtype astrocytes from human pluripotent stem cells. Stem Cell Reports 11:998–1008. 10.1016/j.stemcr.2018.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Muffat J, Omer A, Bosch I, Lancaster MA, Sur M, Gehrke L, Knoblich JA, Jaenisch R (2017) Induction of expansion and folding in human cerebral organoids. Cell Stem Cell 20:385–396.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Muffat J, Omer Javed A, Keys HR, Lungjangwa T, Bosch I, Khan M, Virgilio MC, Gehrke L, Sabatini DM, Jaenisch R (2019) Genome-wide CRISPR screen for Zika virus resistance in human neural cells. Proc Natl Acad Sci U S A 116:9527–9532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddelow SA, Guttenplan KA, Clarke LE, Bennett FC, Bohlen CJ, Schirmer L, Bennett ML, Münch AE, Chung WS, Peterson TC, Wilton DK, Frouin A, Napier BA, Panicker N, Kumar M, Buckwalter MS, Rowitch DH, Dawson VL, Dawson TM, Stevens B, et al. (2017) Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541:481–487. 10.1038/nature21029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YT, Seo J, Gao F, Feldman HM, Wen HL, Penney J, Cam HP, Gjoneska E, Raja WK, Cheng J, Rueda R, Kritskiy O, Abdurrob F, Peng Z, Milo B, Yu CJ, Elmsaouri S, Dey D, Ko T, Yankner BA, et al. (2018) APOE4 causes widespread molecular and cellular alterations associated with Alzheimer's disease phenotypes in human iPSC-derived brain cell types. Neuron 98:1141–1154.e7. 10.1016/j.neuron.2018.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkous A, Balamatsias D, Snuderl M, Edwards L, Miyaguchi K, Milner T, Reich B, Cohen-Gould L, Storaska A, Nakayama Y, Schenkein E, Singhania R, Cirigliano S, Magdeldin T, Lin Y, Nanjangud G, Chadalavada K, Pisapia D, Liston C, Fine HA (2019) Modeling patient-derived glioblastoma with cerebral organoids. Cell Rep 26:3203–3211.e5. 10.1016/j.celrep.2019.02.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C, Lancaster MA, Castanon R, Nery JR, Knoblich JA, Ecker JR (2016) Cerebral organoids recapitulate epigenomic signatures of the human fetal brain. Cell Rep 17:3369–3384. 10.1016/j.celrep.2016.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhavan M, Nevin ZS, Shick HE, Garrison E, Clarkson-Paredes C, Karl M, Clayton BL, Factor DC, Allan KC, Barbar L, Jain T, Douvaras P, Fossati V, Miller RH, Tesar PJ (2018) Induction of myelinating oligodendrocytes in human cortical spheroids. Nat Methods 15:700–706. 10.1038/s41592-018-0081-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani J, Coppola G, Zhang P, Abyzov A, Provini L, Tomasini L, Amenduni M, Szekely A, Palejev D, Wilson M, Gerstein M, Grigorenko EL, Chawarska K, Pelphrey KA, Howe JR, Vaccarino FM (2015) FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell 162:375–390. 10.1016/j.cell.2015.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marton RM, Miura Y, Sloan SA, Li Q, Revah O, Levy RJ, Huguenard JR, Paşca SP (2019) Differentiation and maturation of oligodendrocytes in human three-dimensional neural cultures. Nat Neurosci 22:484–491. 10.1038/s41593-018-0316-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride KL, Varga EA, Pastore MT, Prior TW, Manickam K, Atkin JF, Herman GE (2010) Confirmation study of PTEN mutations among individuals with autism or developmental delays/mental retardation and macrocephaly. Autism Res 3:137–141. 10.1002/aur.132 [DOI] [PubMed] [Google Scholar]
- Mellios N, Feldman DA, Sheridan SD, Ip JP, Kwok S, Amoah SK, Rosen B, Rodriguez BA, Crawford B, Swaminathan R, Chou S, Li Y, Ziats M, Ernst C, Jaenisch R, Haggarty SJ, Sur M (2018) MeCP2-regulated miRNAs control early human neurogenesis through differential effects on ERK and AKT signaling. Mol Psychiatry 23:1051–1065. 10.1038/mp.2017.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muffat J, Li Y, Yuan B, Mitalipova M, Omer A, Corcoran S, Bakiasi G, Tsai LH, Aubourg P, Ransohoff RM, Jaenisch R (2016) Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat Med 22:1358–1367. 10.1038/nm.4189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muffat J, Li Y, Omer A, Durbin A, Bosch I, Bakiasi G, Richards E, Meyer A, Gehrke L, Jaenisch R (2018) Human induced pluripotent stem cell-derived glial cells and neural progenitors display divergent responses to Zika and dengue infections. Proc Natl Acad Sci U S A 115:7117–7122. 10.1073/pnas.1719266115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muguruma K, Nishiyama A, Kawakami H, Hashimoto K, Sasai Y (2015) Self-organization of polarized cerebellar tissue in 3D culture of human pluripotent stem cells. Cell Rep 10:537–550. 10.1016/j.celrep.2014.12.051 [DOI] [PubMed] [Google Scholar]
- Muñoz-Sanjuán I, Brivanlou AH (2002) Neural induction, the default model and embryonic stem cells. Nat Rev Neurosci 3:271–280. 10.1038/nrn786 [DOI] [PubMed] [Google Scholar]
- Nakano T, Ando S, Takata N, Kawada M, Muguruma K, Sekiguchi K, Saito K, Yonemura S, Eiraku M, Sasai Y (2012) Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell 10:771–785. 10.1016/j.stem.2012.05.009 [DOI] [PubMed] [Google Scholar]
- Omer Javed A, Li Y, Muffat J, Su KC, Cohen MA, Lungjangwa T, Aubourg P, Cheeseman IM, Jaenisch R (2018) Microcephaly modeling of kinetochore mutation reveals a brain-specific phenotype. Cell Rep 25:368–382.e5. 10.1016/j.celrep.2018.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill AC, Kyrousi C, Klaus J, Leventer RJ, Kirk EP, Fry A, Pilz DT, Morgan T, Jenkins ZA, Drukker M, Berkovic SF, Scheffer IE, Guerrini R, Markie DM, Götz M, Cappello S, Robertson SP (2018) A primate-specific isoform of PLEKHG6 regulates neurogenesis and neuronal migration. Cell Rep 25:2729–2741.e6. 10.1016/j.celrep.2018.11.029 [DOI] [PubMed] [Google Scholar]
- Oosterhof N, Chang IJ, Karimiani EG, Kuil LE, Jensen DM, Daza R, Young E, Astle L, van der Linde HC, Shivaram GM, Demmers J, Latimer CS, Keene CD, Loter E, Maroofian R, van Ham TJ, Hevner RF, Bennett JT (2019) Homozygous mutations in CSF1R cause a pediatric-onset leukoencephalopathy and can result in congenital absence of microglia. Am J Hum Genet 104:936–947. 10.1016/j.ajhg.2019.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormel PR, Vieira de Sá R, van Bodegraven EJ, Karst H, Harschnitz O, Sneeboer MA, Johansen LE, van Dijk RE, Scheefhals N, Berdenis van Berlekom A, Ribes Martínez E, Kling S, MacGillavry HD, van den Berg LH, Kahn RS, Hol EM, de Witte LD, Pasterkamp RJ (2018) Microglia innately develop within cerebral organoids. Nat Commun 9:4167. 10.1038/s41467-018-06684-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otani T, Marchetto MC, Gage FH, Simons BD, Livesey FJ (2016) 2D and 3D stem cell models of primate cortical development identify species-specific differences in progenitor behavior contributing to brain size. Cell Stem Cell 18:467–480. 10.1016/j.stem.2016.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya H, Shen MJ, Ichikawa DM, Sedlock AB, Choi Y, Johnson KR, Kim G, Brown MA, Elkahloun AG, Maric D, Sweeney CL, Gossa S, Malech HL, McGavern DB, Park JK (2017) Differentiation of human and murine induced pluripotent stem cells to microglia-like cells. Nat Neurosci 20:753–759. 10.1038/nn.4534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paşca AM, Sloan SA, Clarke LE, Tian Y, Makinson CD, Huber N, Kim CH, Park JY, O'Rourke NA, Nguyen KD, Smith SJ, Huguenard JR, Geschwind DH, Barres BA, Paşca SP (2015) Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat Methods 12:671–678. 10.1038/nmeth.3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poduri A, Evrony GD, Cai X, Elhosary PC, Beroukhim R, Lehtinen MK, Hills LB, Heinzen EL, Hill A, Hill RS, Barry BJ, Bourgeois BF, Riviello JJ, Barkovich AJ, Black PM, Ligon KL, Walsh CA (2012) Somatic activation of AKT3 causes hemispheric developmental brain malformations. Neuron 74:41–48. 10.1016/j.neuron.2012.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen AA, Bhaduri A, Andrews MG, Nowakowski TJ, Meyerson OS, Mostajo-Radji MA, Di Lullo E, Alvarado B, Bedolli M, Dougherty ML, Fiddes IT, Kronenberg ZN, Shuga J, Leyrat AA, West JA, Bershteyn M, Lowe CB, Pavlovic BJ, Salama SR, Haussler D, et al. (2019) Establishing cerebral organoids as models of human-specific brain evolution. Cell 176:743–756.e17. 10.1016/j.cell.2019.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, Yao B, Hamersky GR, Jacob F, Zhong C, Yoon KJ, Jeang W, Lin L, Li Y, Thakor J, Berg DA, Zhang C, Kang E, Chickering M, Nauen D, et al. (2016) Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell 165:1238–1254. 10.1016/j.cell.2016.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Zhang XJ, Renier N, Wu Z, Atkin T, Sun Z, Ozair MZ, Tchieu J, Zimmer B, Fattahi F, Ganat Y, Azevedo R, Zeltner N, Brivanlou AH, Karayiorgou M, Gogos J, Tomishima M, Tessier-Lavigne M, Shi SH, Studer L (2017) Combined small-molecule inhibition accelerates the derivation of functional cortical neurons from human pluripotent stem cells. Nat Biotechnol 35:154–163. 10.1038/nbt.3777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrato G, Nguyen T, Macosko EZ, Sherwood JL, Min Yang S, Berger DR, Maria N, Scholvin J, Goldman M, Kinney JP, Boyden ES, Lichtman JW, Williams ZM, McCarroll SA, Arlotta P (2017) Cell diversity and network dynamics in photosensitive human brain organoids. Nature 545:48–53. 10.1038/nature22047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja WK, Mungenast AE, Lin YT, Ko T, Abdurrob F, Seo J, Tsai LH (2016) Self-organizing 3D human neural tissue derived from induced pluripotent stem cells recapitulate Alzheimer's disease phenotypes. PLoS One 11:e0161969. 10.1371/journal.pone.0161969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. (1974) Neurons in rhesus monkey visual cortex: systematic relation between time of origin and eventual disposition. Science 183:425–427. 10.1126/science.183.4123.425 [DOI] [PubMed] [Google Scholar]
- Rivière JB, Mirzaa GM, O'Roak BJ, Beddaoui M, Alcantara D, Conway RL, St-Onge J, Schwartzentruber JA, Gripp KW, Nikkel SM, Worthylake T, Sullivan CT, Ward TR, Butler HE, Kramer NA, Albrecht B, Armour CM, Armstrong L, Caluseriu O, Cytrynbaum C, et al. (2012) De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nat Genet 44:934–940. 10.1038/ng.2331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi H, Kadoshima T, Soen M, Narii N, Ishida Y, Ohgushi M, Takahashi J, Eiraku M, Sasai Y (2015) Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue. Nat Commun 6:8896. 10.1038/ncomms9896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Clevers H (2013) Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science 340:1190–1194. 10.1126/science.1234852 [DOI] [PubMed] [Google Scholar]
- Sloan SA, Darmanis S, Huber N, Khan TA, Birey F, Caneda C, Reimer R, Quake SR, Barres BA, Paşca SP (2017) Human astrocyte maturation captured in 3D cerebral cortical spheroids derived from pluripotent stem cells. Neuron 95:779–790.e6. 10.1016/j.neuron.2017.07.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan SA, Andersen J, Paşca AM, Birey F, Paşca SP (2018) Generation and assembly of human brain region-specific three-dimensional cultures. Nat Protoc 13:2062–2085. 10.1038/s41596-018-0032-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth P, Lagomarsino VN, Muratore CR, Ryu SC, He A, Taylor WM, Zhou C, Arellano M, Young-Pearse TL (2018) Shared effects of DISC1 disruption and elevated WNT signaling in human cerebral organoids. Transl Psychiatry 8:77. 10.1038/s41398-018-0122-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Hevner RF (2014) Growth and folding of the mammalian cerebral cortex: from molecules to malformations. Nat Rev Neurosci 15:217–232. 10.1038/nrn3707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki IK, Gacquer D, Van Heurck R, Kumar D, Wojno M, Bilheu A, Herpoel A, Lambert N, Cheron J, Polleux F, Detours V, Vanderhaeghen P (2018) Human-specific NOTCH2NL genes expand cortical neurogenesis through delta/notch regulation. Cell 173:1370–1384.e16. 10.1016/j.cell.2018.03.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata K, Kozaki T, Lee CZ, Thion MS, Otsuka M, Lim S, Utami KH, Fidan K, Park DS, Malleret B, Chakarov S, See P, Low D, Low G, Garcia-Miralles M, Zeng R, Zhang J, Goh CC, Gul A, Hubert S, et al. (2017) Induced-pluripotent-stem-cell-derived primitive macrophages provide a platform for modeling tissue-resident macrophage differentiation and function. Immunity 47:183–198.e6. 10.1016/j.immuni.2017.06.017 [DOI] [PubMed] [Google Scholar]
- Tchieu J, Calder EL, Guttikonda SR, Gutzwiller EM, Aromolaran KA, Steinbeck JA, Goldstein PA, Studer L (2019) NFIA is a gliogenic switch enabling rapid derivation of functional human astrocytes from pluripotent stem cells. Nat Biotechnol 37:267–275. 10.1038/s41587-019-0035-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcw J, Wang M, Pimenova AA, Bowles KR, Hartley BJ, Lacin E, Machlovi SI, Abdelaal R, Karch CM, Phatnani H, Slesinger PA, Zhang B, Goate AM, Brennand KJ (2017) An efficient platform for astrocyte differentiation from human induced pluripotent stem cells. Stem Cell Reports 9:600–614. 10.1016/j.stemcr.2017.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco S, Kedaigle AJ, Simmons SK, Nash A, Rocha M, Quadrato G, Paulsen B, Nguyen L, Adiconis X, Regev A, Levin JZ, Arlotta P (2019) Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature 570:523–527. 10.1038/s41586-019-1289-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CA. (1999) Genetic malformations of the human cerebral cortex. Neuron 23:19–29. 10.1016/S0896-6273(00)80749-7 [DOI] [PubMed] [Google Scholar]
- Watanabe M, Buth JE, Vishlaghi N, de la Torre-Ubieta L, Taxidis J, Khakh BS, Coppola G, Pearson CA, Yamauchi K, Gong D, Dai X, Damoiseaux R, Aliyari R, Liebscher S, Schenke-Layland K, Caneda C, Huang EJ, Zhang Y, Cheng G, Geschwind DH, et al. (2017) Self-organized cerebral organoids with human-specific features predict effective drugs to combat Zika virus infection. Cell Rep 21:517–532. 10.1016/j.celrep.2017.09.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer RA, Leopoldi A, Aichinger M, Wick N, Hantusch B, Novatchkova M, Taubenschmid J, Hämmerle M, Esk C, Bagley JA, Lindenhofer D, Chen G, Boehm M, Agu CA, Yang F, Fu B, Zuber J, Knoblich JA, Kerjaschki D, Penninger JM (2019) Human blood vessel organoids as a model of diabetic vasculopathy. Nature 565:505–510. 10.1038/s41586-018-0858-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Tanaka Y, Patterson B, Kang YJ, Govindaiah G, Roselaar N, Cakir B, Kim KY, Lombroso AP, Hwang SM, Zhong M, Stanley EG, Elefanty AG, Naegele JR, Lee SH, Weissman SM, Park IH (2017) Fusion of regionally specified hPSC-derived organoids models human brain development and interneuron migration. Cell Stem Cell 21:383–398.e7. 10.1016/j.stem.2017.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Tanaka Y, Cakir B, Patterson B, Kim KY, Sun P, Kang YJ, Zhong M, Liu X, Patra P, Lee SH, Weissman SM, Park IH (2019) hESC-derived thalamic organoids form reciprocal projections when fused with cortical organoids. Cell Stem Cell 24:487–497.e7. 10.1016/j.stem.2018.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Lee EM, Wen Z, Cheng Y, Huang WK, Qian X, Tcw J, Kouznetsova J, Ogden SC, Hammack C, Jacob F, Nguyen HN, Itkin M, Hanna C, Shinn P, Allen C, Michael SG, Simeonov A, Huang W, Christian KM, et al. (2016) Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat Med 22:1101–1107. 10.1038/nm.4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Zuchero JB, Ahlenius H, Marro S, Ng YH, Vierbuchen T, Hawkins JS, Geissler R, Barres BA, Wernig M (2013) Generation of oligodendroglial cells by direct lineage conversion. Nat Biotechnol 31:434–439. 10.1038/nbt.2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Chanda S, Marro S, Ng YH, Janas JA, Haag D, Ang CE, Tang Y, Flores Q, Mall M, Wapinski O, Li M, Ahlenius H, Rubenstein JL, Chang HY, Buylla AA, Südhof TC, Wernig M (2017) Generation of pure GABAergic neurons by transcription factor programming. Nat Methods 14:621–628. 10.1038/nmeth.4291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Kang E, Yu C, Qian X, Jacob F, Yu C, Mao M, Poon RY, Kim J, Song H, Ming GL, Zhang M (2017) DISC1 regulates neurogenesis via modulating kinetochore attachment of Ndel1/Nde1 during mitosis. Neuron 96:1041–1054.e5. 10.1016/j.neuron.2017.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SJ, Elahi LS, Paşca AM, Marton RM, Gordon A, Revah O, Miura Y, Walczak EM, Holdgate GM, Fan HC, Huguenard JR, Geschwind DH, Paşca SP (2019) Reliability of human cortical organoid generation. Nat Methods 16:75–78. 10.1038/s41592-018-0255-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SC, Wernig M, Duncan ID, Brüstle O, Thomson JA (2001) In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol 19:1129–1133. 10.1038/nbt1201-1129 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S, Marro S, Patzke C, Acuna C, Covy J, Xu W, Yang N, Danko T, Chen L, Wernig M, Südhof TC (2013) Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 78:785–798. 10.1016/j.neuron.2013.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Li Q, Zhao C, Da Y, Zhang HL, Chen Z (2018) Differentiation of glial cells from hiPSCs: potential applications in neurological diseases and cell replacement therapy. Front Cell Neurosci 12:239. 10.3389/fncel.2018.00239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Tan L, Cederquist GY, Fan Y, Hartley BJ, Mukherjee S, Tomishima M, Brennand KJ, Zhang Q, Schwartz RE, Evans T, Studer L, Chen S (2017) High-content screening in hPSC-neural progenitors identifies drug candidates that inhibit Zika virus infection in fetal-like organoids and adult brain. Cell Stem Cell 21:274–283.e5. 10.1016/j.stem.2017.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Carido M, Meinhardt A, Kurth T, Karl MO, Ader M, Tanaka EM (2013) Three-dimensional neuroepithelial culture from human embryonic stem cells and its use for quantitative conversion to retinal pigment epithelium. PLoS One 8:e54552. 10.1371/journal.pone.0054552 [DOI] [PMC free article] [PubMed] [Google Scholar]


