Age-related memory loss is observed across multiple mammalian species and preferentially affects hippocampus-dependent memory. Memory impairments are characterized by accelerated decay of spatial memories. Nevertheless, the molecular mechanisms underlying these deficits are still largely unknown. Here, we investigated the expression and function of the growth arrest DNA damage (Gadd45) family during aging and cognition, respectively.
Keywords: Gadd45g, synapse-to-nucleus communication, aging-dependent cognitive decline, activity-regulated transcription, MAPK signaling
Abstract
Age-related memory loss is observed across multiple mammalian species and preferentially affects hippocampus-dependent memory. Memory impairments are characterized by accelerated decay of spatial memories. Nevertheless, the molecular mechanisms underlying these deficits are still largely unknown. Here, we investigated the expression and function of the growth arrest DNA damage (Gadd45) family during aging and cognition, respectively. We report that aging impairs the expression of Gadd45γ in the hippocampus of cognitively impaired male mice. Mimicking this decrease in young adult male mice led to age-like memory deficits in hippocampus-dependent memory tasks. Gadd45γ reduction impaired the activity of key components of the mitogen-activated protein kinase (MAPK) pathway (p38 and JNK) in mouse hippocampal cultures. Furthermore, we found that activation of downstream targets, such as ATF-2, c-Jun, and CREB (cAMP response element-binding protein), was disrupted. Finally, we showed that Gadd45γ is required for induction of key early- and late-response genes that have been associated with aging. Together, these findings indicate that Gadd45γ expression regulates cognitive abilities and synapse-to-nucleus communication and suggest Gadd45γ dysfunction as a potential mechanism contributing to age-related cognitive impairments.
SIGNIFICANCE STATEMENT A high percentage of subjects experience age-related memory loss that burdens daily performance. Although many advances have been made, the precise changes in the brain governing these deficits are unclear. Identifying molecular processes that are required for cognition and are altered during old age is crucial to develop preventive or therapeutic strategies. Here, we show that baseline and learning-induced expression of the growth arrest DNA damage (Gadd45) γ is selectively impaired in the hippocampus of aged mice with cognitive deficits. Next, we show that modeling this impairment in young adult mice with normal cognitive performance disrupts long- and short-term memories in an age-like manner. Finally, we demonstrate that Gadd45γ regulates synapse-to-nucleus communication processes that are needed for plasticity-associated gene expression.
Introduction
Age-related cognitive decline is observed across multiple species, including rodents and humans (Erickson and Barnes, 2003). In humans, ∼40% of individuals aged 65 years or older experience some form of memory loss (Aigbogun et al., 2017), even though deterioration of cognitive functions may start earlier (Singh-Manoux et al., 2012). Aged rodents, monkeys, and humans exhibit similar age-dependent memory impairments (Erickson and Barnes, 2003; Burke et al., 2012). Hippocampus-dependent forms of memory, such as spatial memory, are particularly vulnerable to aging (Burke and Barnes, 2006) such that, in aged subjects, memories are retained for shorter periods of time (Kennard and Woodruff-Pak, 2011). Despite these conserved age-related cognitive impairments, little is known about the molecular mechanisms underlying these changes.
Memory formation requires activity-dependent changes in the function and structure of neurons. Short-term storage depends on post-translational mechanisms at the level of the synapse, whereas long-term memory requires activity-regulated gene transcription (Alberini, 2009). Learning-dependent genomic responses are disrupted in the aged hippocampus (Stefanelli et al., 2018). Therefore, dysregulation of the coupling between neuronal activity impinging on synapses and gene transcription in the nucleus likely underlies age-related cognitive deficits. The activity of transcription factors is a crucial layer of regulatory control in synapse-to-nucleus communication. The transcription factors cAMP response element-binding protein (CREB) and activating protein 1 (AP-1) are required for plasticity and memory (Alberini, 2009). Age-associated changes in CREB activity and expression have been reported, and there is compelling evidence indicating that upregulation of CREB levels or activity ameliorates age-related memory deficits (Bach et al., 1999; Yu et al., 2017b). Recently, comparison of the hippocampal transcriptome of young adult and aged mice revealed that AP-1-associated gene expression is also affected during aging (Stilling et al., 2014). Hence, these changes suggest that molecular mechanisms that regulate CREB and AP-1 transcriptional activity might be disrupted during aging.
The growth arrest DNA damage (Gadd45) family is composed of three family members—Gadd45α, Gadd45β, and Gadd45γ—which are small proteins (∼18 kDa) with both nuclear and cytoplasmic expression. Gadd45 family members regulate signaling cascades such as the mitogen-activated protein kinase (MAPK) cascade, cell cycle control, and DNA repair mechanisms and have been extensively studied during tumorigenesis and cellular stress responses, which are hallmarks of aging (Moskalev et al., 2012). In contrast, the current knowledge of their functions in the brain is limited to that gained by a few studies. Gadd45β and Gadd45γ mRNA expression is induced upon neuronal activity and learning (Ma et al., 2009). Gadd45β has been shown to be required for adult neurogenesis (Ma et al., 2009), and aberrant function of Gadd45β was implicated in psychiatric disorders (Gavin et al., 2012). Two studies have suggested that Gadd45β regulates memory formation; however, the findings in those studies were conflicting (Leach et al., 2012; Sultan et al., 2012). Gadd45γ in the prelimbic prefrontal cortex has been recently proposed to be required for memory consolidation (Li et al., 2019). The known functions of Gadd45 members led us to hypothesize that aging would be associated with dysfunction of Gadd45 family members in the brain. Here, we investigated this possibility and the functions of Gadd45β and Gadd45γ in the regulation of neuronal signaling, gene expression, and memory formation. We found that aging reduces Gadd45γ expression in the mouse hippocampus, and that mimicking this reduction in young adult mice induces age-like memory impairments. Furthermore, we showed that Gadd45γ regulates MAPK signaling as well as CREB- and AP-1-dependent transcription. The expression of genes that have been previously shown to be impaired during aging (Ryan et al., 2019), such as Arc and Nptx2/Narp, was disrupted.
Overall, this work uncovered a novel function for Gadd45γ in gene expression regulation and cognitive abilities. Furthermore, these findings may implicate a dysfunction in Gadd45γ expression as a contributing mechanism to age-related cognitive impairments.
Materials and Methods
Subjects.
We used 3-month-old and 18-month-old C57BL/6J male mice (Janvier) or male C57BL/6N mice (Charles River Wiga) that were 3 months old at the time of behavior experiments. Mice were always group housed (3–4 mice/cage), unless severe fighting occurred, and were housed on a 12 h light/dark cycle with ad libitum access to water and food (22 ± 1°C, 55 ± 10% relative humidity). All behavioral experiments took place during the light phase. Sick and/or injured mice from cage-mate fighting were excluded from the study. Animals were randomly assigned to experimental groups and blinded analysis was performed. All procedures were performed in accordance with German guidelines for the care and use of laboratory animals and with the European Community Council Directive 86/609/EEC.
Primary hippocampal cultures.
Hippocampal cultures from newborn C57BL/6N mice (Charles River Wiga) were prepared and maintained as previously described (Bading and Greenberg, 1991), except that growth medium was supplemented with Invitrogen B27 (Thermo Fisher Scientific) and 1% rat serum (v/v). Recombinant adeno-associated virus (rAAV) infection of cultures occurred on day in vitro (DIV) 4. Infection rates, determined by analyzing the proportion of GFP+ cells, ranged from 80 to 90%. Experiments were performed on DIVs 10–11. To induce action potential bursting, cultures were treated with 50 μm bicuculline (Enzo Life Sciences) or 100 μm gabazine (Biotrend). DNA cotransfection was performed after a culturing period of 8 DIVs using Invitrogen Lipofectamine 2000 (Thermo Fisher Scientific) as described previously (Pruunsild et al., 2011, 2017). N numbers represent independent cell preparations.
rAAVs.
Viral particles were produced and purified as described previously (Zhang et al., 2007). For expression of short-hairpin RNAs (shRNAs), we used a vector containing the U6 promoter upstream of the shRNA sequence (control, Gadd45β, or Gadd45γ specific). The control–shRNA, Gadd45β–shRNA, Gadd45γ–shRNA1, and Gadd45γ–shRNA2 sequences are 5′ACTACCGTTGTTATAGGTGCG′3, 5′GCAGATTCACTTCACCCTGAT′3, 5′TTGAAAGAGCAGTGCAGTCG′3, and 5′ATAGCGCTGCAGATCCATTTC′3, respectively. Both the control–shRNA and Gadd45γ–shRNA1 sequences have been previously described and validated (Zhang et al., 2009; Oliveira et al., 2012). For each virus batch produced, the infection rate, toxicity, viral titer, and effectiveness of knockdown were checked before experiments were started. Viral titers obtained after production of all viruses were similar, and the volumes were matched to obtain final working concentrations of 1010 viral particles/ml. At DIV 10, infected hippocampal cultures were imaged using identical microscope settings for infection rate and toxicity analysis. The quantification was performed using Fiji (National Institutes of Health; Schindelin et al., 2012). None of the viral batches used induced cell death.
Stereotaxic surgery.
rAAVs were injected into the dorsal hippocampus (dHPC) at the following coordinates relative to the bregma: −2 mm anteroposterior; ±1.5 mm mediolateral; −1.7, −1.9, and −2.1 mm dorsoventral. A total volume of 1.5 μl was injected per hemisphere at 200 nl/min; a 2:1 mixture of viral solution and 20% mannitol was used. Following injections at each individual site, the needle was left in place for 60 s. At the time of behavioral experiments, the experimenter was blind to the identity of the virus injected into each mouse. Behavioral experiments started 2 weeks after rAAV delivery. After behavior experiments, histological analysis was performed to confirm correct targeting and tissue and cellular integrity. Mice that showed absence or missed targeting of viral expression were excluded. For histological analysis, mice were perfused with 4% paraformaldehyde (Sigma-Aldrich). The brains were collected and postfixed in the same solution overnight, then placed into a 30% sucrose solution. Brain slices were cut at a thickness of 30 μm, incubated in Hoechst 33258 (2 μg/ml; Serva Feinbiochemica) for 5 min, and mounted on glass slides. Slices were imaged with a 10× objective mounted on a fluorescence microscope (Leica Microsystems). Identical microscope settings were used between different experimental groups.
Behavioral testing.
Before behavioral testing started, mice were habituated to the experimenter and behavioral room by handling for 3 consecutive days, 1.5 min/mouse. Object–location test and contextual fear conditioning were performed as previously described (Oliveira et al., 2012, 2016). Different mouse cohorts were used to test long-term memory (24 h), short-term memory (5 min), or short-term memory (1 h). After handling sessions, mice underwent a habituation session (6 min) where they were placed in the training arena in the absence of objects. During training, the animals were exposed to two objects and allowed to explore for 6 min during three trials with 3 min intertrial intervals. During the testing session, which occurred 24 h, 1 h, or 5 min later, one object was moved to a new location and exploration of objects was scored for 6 min. During contextual fear conditioning training, mice were allowed to explore the conditioning chamber for 2 min and 28 s until a 0.5 mA footshock was administered for 2 s; then, animals remained for 30 s before returning to their home cage (HC). The testing session consisted of exposing the animals to the conditioning chamber for 5 min. Mice first underwent the object–location test, and contextual fear conditioning was performed 1 week later. If control animals did not show a preference in one experimental batch during the object–location test due to day effects, the whole set of animals was excluded from this analysis but was still included in contextual fear conditioning data. The open-field test was performed within the first session of the object–place recognition training as previously described (Gulmez Karaca et al., 2018).
Luciferase reporter assays.
Assays were performed as previously described, with alterations (Pruunsild et al., 2017). We used the following Firefly luciferase (FFluc) expression vectors: pGL4.29[luc2P/CRE/Hygro] (Promega), which contains four CRE cis-elements and a minimal promoter (4×CRE-pmin), or pGL4.44[luc2P/AP1 RE/Hygro] (Promega), which contains six copies of an AP-1 response element and a minimal promoter (AP-1 RE-pmin). Additionally, the plasmid pGL4.83 h[RlucP/Puro] (Promega), which contains the human EF1a promoter regulating the expression of Renilla luciferase (Rluc), was used for normalization. On DIV 8, mouse primary hippocampal cultures in 48-well plates were changed to transfection medium. Lipofectamine 2000 (Thermo Fisher Scientific) was used for transfection according to the manufacturer's instructions. Neurons were cotransfected with one of the following constructs: control–shRNA, Gadd45β–shRNA, Gadd45γ–shRNA1, Gadd45γ–shRNA2, and 4×CRE-pmin or AP-1 RE-pmin (all 1 μg/well), together with Rluc (75 ng/well). The DNA (μg):Lipofectamine 2000 (μl) ratio was 1:2 in a total of 25 μl/well. On DIV 10, Dual-Glo Luciferase Assay System (Promega) was used to measure FFluc and Rluc activity levels. Background signal measured from nontransfected cells was subtracted, and FFluc levels were normalized to Rluc levels. Each condition was done in triplicate. Data are presented as fold change relative to the control–shRNA condition treated with bicuculline.
Quantitative reverse-transcription PCR.
RNA was isolated using the RNeasy Plus Mini Kit (Qiagen) with additional on-column DNase I digestion, according to the manufacturer's instructions. For RNA isolation from mouse hippocampal tissue, the tissue was rapidly dissected and placed in RNAlater (Sigma-Aldrich). For the generation of complementary DNA, RNA was reverse transcribed with the Applied Biosystems High Capacity Complementary DNA Reverse Transcription Kit (Thermo Fisher Scientific). Quantitative reverse-transcription PCR (qRT-PCR) was performed on an Applied Biosystems StepOnePlus Real-Time PCR System (Thermo Fisher Scientific) using Applied Biosystems TaqMan gene expression assays (Thermo Fisher Scientific) for the following mouse genes: Arc (Mm00479619_g1), c-Fos (Mm00487425_m1), Gadd45α (Mm00432802_m1), Gadd45β (Mm00345123_m1), Gadd45γ (Mm00442225_m1), Nptx2/Narp (Mm00479438_m1), and Tgfb2 (Mm00436955_m1). Expression levels of target genes were normalized to the expression of the housekeeping gene GusB (Mm00446953_m1). Results were further normalized to uninfected conditions. In the case of early-response genes (Arc and c-Fos) or late-response genes (Nptx2/Narp and Tgfb2), data were normalized to uninfected conditions at the expression peak, 2 or 6 h of bicuculline treatment, respectively. Controls were used to exclude the possibility of DNA or RNA contaminations.
Western blotting.
Hippocampal cultures infected on DIV 4 were lysed on DIV 10 in boiling SDS sample buffer [160 mm Tris-HCl, pH 6.8, 4% SDS, 30% glycerol, 10 mm dithiothreitol, and 0.02% bromophenol blue]. The cell lysates were loaded into a 12% acrylamide gel and blotted onto a nitrocellulose membrane (GE Healthcare). The membranes were blocked in 5% milk in PBS with 0.01% Tween (PBS-T) and probed with the following antibodies: phospho-ATF-2 (1:2500; #9221, Cell Signaling Technology), total ATF-2 (1:250; sc-242, Santa Cruz Biotechnology), phospho-c-Jun (1:1000; #9261s, Cell Signaling Technology), total-c-Jun (1:250; sc-74543, Santa Cruz Biotechnology), phospho-CREB (1:10,000; #05-667, Millipore), total-CREB (1:5000; #4820, Cell Signaling Technology), phospho-Erk (1:2500; #9106, Cell Signaling Technology), total-Erk (1:5000; #9102, Cell Signaling Technology), phospho-JNK (1:1000; #4671, Cell Signaling Technology), total-JNK (1:5000; #9258, Cell Signaling Technology), phospho-p38 (1:750; #612288, BD Biosciences), total-p38 (1:3000; #92125s, Cell Signaling Technology). Antibodies were diluted in 5% milk in PBS-T or in 5% bovine serum albumin in PBS-T, if the antibodies targeted a phospho variant. Next, blots were incubated with horseradish peroxidase-conjugated secondary antibodies. Finally, blots were incubated in enhanced chemiluminescence Western blotting detection reagent (GE Healthcare or Bio-Rad) and exposed to Hyperfilm (GE Healthcare). Data are presented as the ratio of phosphorylated/total protein normalized internally to the respective uninfected condition. The conditions at baseline served as a control to confirm the phosphorylation of the various kinases in response to bicuculline treatment.
Calcium imaging.
For calcium imaging experiments, we used a viral vector that contained the human synapsin promoter upstream of the recombinant calcium indicator jRGECO1a (Dana et al., 2016). On DIV 4, primary hippocampal cultures were infected with jRGECO1a alone or coinfected with control–shRNA, Gadd45β–shRNA, Gadd45γ–shRNA1, or Gadd45γ–shRNA2. On DIVs 10–11, individual coverslips were transferred to a dish containing CO2-independent culture medium (CICM) at room temperature. Fluorescence was detected using a cooled charge-coupled device camera (iXon, Andor) through a 20× water immersion objective (LUMPlanFl/IR, Olympus) on an upright microscope (BX51W1, Olympus). jRGECO1a-infected cells were identified by the presence of a strongly red fluorescent soma (excitation ∼560 nm). Data were collected using proprietary software (CellR, Olympus) and analyzed using ImageJ (National Institutes of Health) and Igor Pro (WaveMetrics). During imaging, cells on coverslips were transferred to an imaging chamber containing room temperature CICM, and the GABAA receptor antagonist gabazine (100 μm) was applied to induce reliable action potential bursting and associated intracellular calcium rises. Fmax was measured in CICM containing 10 μm ionomycin, and Fmin was measured in CICM for 2 min before gabazine treatment. GFP+ cells were used to draw, with confidence, regions of interest for the analysis of somatic calcium responses. Calcium responses were quantified as a fraction of the binding affinity: [Ca2+]/Kd = (F − Fmin)/(Fmax − F); Mauceri et al., 2015). A total of 120 coverslips from six independent preparations were analyzed.
Experimental design and statistical analysis.
For normally distributed data sets, two-tailed unpaired Student's t tests were used to compare two groups. If more than two groups were analyzed simultaneously, a one- or two-way ANOVA was used followed by appropriate multiple-comparisons post hoc tests to control for multiple comparisons as specified. Normally distributed significant data were marked with *. In the case of a non-Gaussian distribution, two-tailed Mann–Whitney tests were used to compare two distinct groups or a Kruskal–Wallis (KW) test followed by Dunn's post hoc test to compare more than two groups. Non-normally distributed significant data were marked with #. The sample size was determined based on similar experiments performed in the past and in the literature. All plotted data represent the mean ± SEM. Statistical analysis was performed using GraphPad Prism for Mac OS X, version 7 (GraphPad Software). For behavioral experiments, the investigators were blind to group allocation during data collection and analysis. All behavioral sessions were video recorded and manually scored by an experimenter blind to the group identity to determine the exploration of objects during training and testing phases or freezing behavior. Animal locomotion during the open-field test and mean velocity during fear conditioning training were scored using Smart Video Tracking Software (Harvard Apparatus) and the TSE Systems Fear Conditioning software, respectively. For in vitro experiments, no blinding was performed because the outcome was dependent on software analysis and not manual scoring. All statistical details of experiments can be found in the Results section.
Results
Aging reduces Gadd45γ expression in the mouse hippocampus
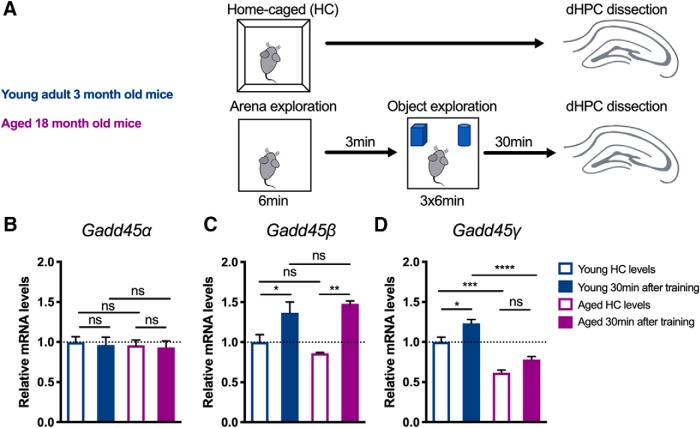
To investigate a possible link between Gadd45 family members and aging-related cognitive decline, we examined whether aging induces alterations in the expression of Gadd45 family members. We compared Gadd45α, Gadd45β, and Gadd45γ mRNA expression in the dHPC of young adult mice to that in aged mice which were previously shown to exhibit impairments in hippocampal forms of memory—namely, object–place recognition and fear conditioning (Oliveira et al., 2012). Specifically, we performed qRT-PCR analysis from isolated dHPC tissue of young and aged mice that underwent training in an object recognition test or remained in HC conditions (Fig. 1A). Young adult mice presented similar Gadd45α baseline and trained levels. In contrast, training induced the expression of Gadd45β and Gadd45γ in young adult mice (Fig. 1B–D; one-way ANOVA followed by Bonferroni's multiple-comparisons test; Gadd45α: F(3,13) = 0.104, p = 0.9563; Gadd45β: F(3,13) = 7.837, p = 0.0031; Gadd45γ: F(3,13) = 26.69, p < 0.0001). These findings are in agreement with previous studies showing that the expression of Gadd45β and Gadd45γ is regulated by neuronal activity (Ma et al., 2009; Leach et al., 2012; Sultan et al., 2012). Interestingly, we found that dHPC Gadd45γ levels were reduced in aged mice, both in HC and trained conditions (Fig. 1D). This impairment was Gadd45γ specific, as Gadd45α and Gadd45β levels were similar in young adult and aged mice. Both groups presented comparable total object exploration times (young vs aged: two-tailed Student's t test, t(7) = 1.405, p = 0.2028). Thus, our findings show that Gadd45γ expression is compromised in the dorsal hippocampus of aged mice and suggest a role in age-related cognitive decline.
Figure 1.
Gadd45γ expression is decreased in the hippocampus of aged mice. A, Schematic representation of the experimental design. B–D, qRT-PCR analysis of Gadd45α (B), Gadd45β (C), and Gadd45γ (D) expression in the dorsal hippocampus of young adult and aged mice in HC conditions or 30 min after training in the object location test. Young HC, n = 5 mice; young trained, n = 5 mice; aged HC, n = 3 mice; aged trained, n = 4 mice. Expression levels are normalized to young HC levels (dashed lines). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 by one-way ANOVA test followed by Bonferroni post hoc test with pairwise comparisons. ns, Not significant. Error bars represent SEM.
Reduced Gadd45γ levels impair memory formation in young adult mice
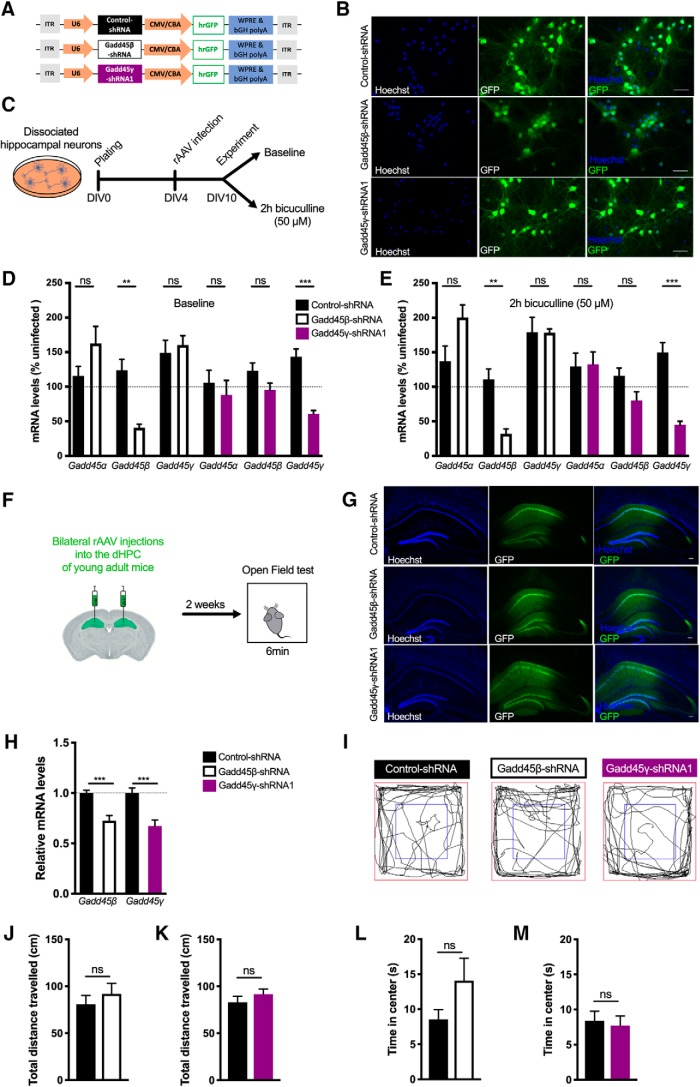
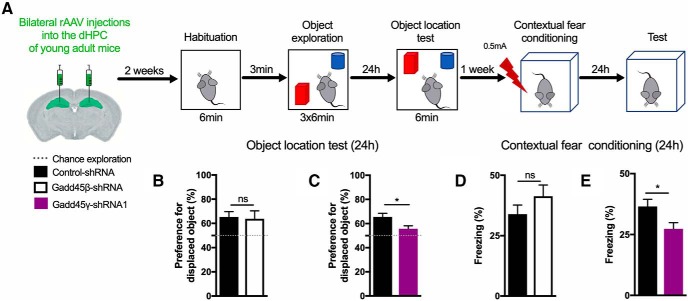
Given our finding that Gadd45γ expression is reduced in the hippocampus of cognitively impaired aged mice, we hypothesized that reducing Gadd45γ expression in the hippocampus of young adult mice would promote memory deficits. To investigate this hypothesis, we generated rAVVs to deliver a control shRNA (control–shRNA) or a previously characterized Gadd45γ-specific shRNA (Gadd45γ–shRNA1). We used the rAAV1/2 serotype to preferentially knock down Gadd45γ in neurons given its established neuronal tropism (Xu et al., 2001). Additionally, a Gadd45β-specific shRNA sequence (Gadd45β–shRNA) was designed to compare the functions of the two family members. The viral constructs also contained an expression cassette for GFP under control of the chicken β-actin promoter that served as an infection marker (Fig. 2A). Infection rates in primary hippocampal cultures were determined by analyzing the percentage of GFP+ cells, which ranged from 80 to 90% (Fig. 2B). qRT-PCR analysis revealed that both Gadd45β–shRNA- and Gadd45γ–shRNA1-infected neurons showed a significant and specific decrease in Gadd45β and Gadd45γ mRNA expression compared with control conditions. This effect was present both in baseline conditions and upon induction of action potential bursting by treatment with the GABAA receptor antagonist bicuculline (Fig. 2D,E; two-tailed Student's t test; baseline: t(4) = 1.653, p = 0.1737; t(4) = 4.994, p = 0.0075; t(4) = 0.5047, p = 0.6403; t(6) = 0.6404, p = 0.5456; t(6) = 1.843, p = 0.1149; t(8) = 6.609, p = 0.0002; 2 h bicuculline: t(4) = 2.205, p = 0.0922; t(4) = 4.767, p = 00089; t(4) = 0.03639, p = 0.9727; t(6) = 0.1194, p = 0.9088; t(6) = 2.102, p = 0.0802; t(6) = 6.975, p = 0.0004). Having established the effectiveness of our knock-down tools, we sought to test whether Gadd45β or Gadd45γ knockdown affects locomotor activity or anxiety-like behavior (tendency to thigmotaxis). We used stereotaxic surgery to deliver rAAV-control–shRNA, rAAV-Gadd45β–shRNA, or Gadd45γ–shRNA1 into the dHPC of young adult mice and performed an open-field test 2 weeks later (Fig. 2F). We confirmed robust viral-mediated expression in the dHPC of injected animals by assessing GFP expression and knock-down efficiency by qRT-PCR analysis (Fig. 2G,H; control–shRNA vs Gadd45β–shRNA: two-tailed Student's t test, t(15) = 4.940, p = 0.0002; control–shRNA vs Gadd45γ–shRNA1: two-tailed Student's t test, t(17) = 4.275, p = 0.0005). No gross anatomical or histological brain changes were observed in Gadd45β or Gadd45γ knock-down conditions. We did not find a significant difference between the three groups in the total distance traveled or the percentage of time spent in the central zone [Fig. 2I–M; two-tailed Student's t test; total distance traveled: t(21) = 0.7425, p = 0.4660 (Fig. 2J); t(26) = 1.033, p = 0.3109 (Fig. 2K); time in center: t(21) = 1.532, p = 0.1404 (Fig. 2L); t(26) = 0.3536, p = 0.7265 (Fig. 2M)]. Thus, our model allows the evaluation of cognitive functions without confounds originating from impaired locomotion or altered anxiety-like behavior. Because aging is associated with hippocampus-dependent memory impairments (Erickson and Barnes, 2003; Weber et al., 2015), we subjected mice to hippocampus-dependent memory tasks (Fig. 3A). Decreasing Gadd45β levels did not change the preference for the displaced object 24 h after training in the object–place recognition test (Fig. 3B; two-tailed Student's t test, t(14) = 0.1916, p = 0.8508). In contrast, Gadd45γ–shRNA1 animals showed an impaired preference for the displaced object 24 h after training (Fig. 3C; two-tailed Student's t test, t(21) = 2.545, p = 0.0189). This impairment was not due to differences in object exploratory behavior (one-way ANOVA followed by Bonferroni's multiple-comparisons test, F(2,21) = 0.3681, p = 0.6964). Next, we tested these animals in contextual fear conditioning. Similar to what we observed in the object–place recognition test, the Gadd45β–shRNA group did not show long-term fear memory impairments (Fig. 3D; t(14) = 1.23, p = 0.2398). Knock down of Gadd45γ in the dHPC of young adult mice, however, promoted long-term memory deficits in contextual fear memory 24 h after training (Fig. 3E; two-tailed Student's t test, t(29) = 2.334). This impairment was not a result of distinct responsiveness to the shock administration between the groups during the training session (one-way ANOVA followed by Bonferroni's multiple-comparisons test, F(5,42) = 57.66, p < 0.0001). Hence, this set of experiments shows that Gadd45γ is required for hippocampus-dependent long-term memory formation.
Figure 2.
Characterization of rAAV constructs for shRNA expression. A, Schematic representation of the viral constructs used. The viral vector contains a U6 promoter driving either Gadd45β-specific (Gadd45β–shRNA), Gadd45γ-specific (Gadd45γ–shRNA1), or control (control–shRNA) shRNA sequences and a cytomegalovirus (CMV)/chicken β-actin (CBA) hybrid promoter driving GFP as an infection marker. bGH polyA, Bovine growth hormone polyadenylation signal; ITR, inverted terminal repeat; WPRE, woodchuck hepatitis virus post-transcriptional regulatory element. B, Representative images of GFP+-cultured hippocampal cells infected with control–shRNA, Gadd45β–shRNA, or Gadd45γ–shRNA1. Scale bars, 40 μm. C, Schematic representation of the in vitro experimental design. D, E, qRT-PCR analysis of Gadd45α, Gadd45β, and Gadd45γ expression under baseline conditions (n = 3–5 independent neuronal cultures; D) or after 2 h bicuculline treatment (n = 3–4 independent neuronal cultures; E). Expression levels were normalized to the corresponding baseline or bicuculline-treated uninfected controls (dashed lines). **p < 0.01, ***p < 0.001 by two-tailed unpaired Student's t test. F, Schematic representation of the in vivo experimental design. G, Representative images of the dorsal hippocampus of control–shRNA, Gadd45β–shRNA, or Gadd45γ–shRNA1 groups 4 weeks after stereotaxic surgery. Scale bars, 100 μm. H, qRT-PCR analysis of Gadd45β and Gadd45γ expression in the dorsal hippocampus of Gadd45β–shRNA-injected mice (n = 8 mice) or Gadd45γ–shRNA1-injected mice (n = 9 mice) 30 min after object–location training normalized to control–shRNA [n = 9 or 10 mice (dashed line)]. I, Representative exploration traces of all groups during the open-field test. J, K, Locomotion analysis of control–shRNA (n = 11 mice) versus Gadd45β–shRNA groups (n = 12 mice; J) and control–shRNA (n = 14 mice) versus Gadd45γ–shRNA groups (n = 14 mice; K), quantified as the total distance traveled during the open-field test. L, M, Anxiety-like behavior, quantified as the percentage of time spent in the center of the arena during the open-field test of control–shRNA (n = 12 mice) versus Gadd45β–shRNA groups (n = 12 mice; L) and control–shRNA (n = 14 mice) versus Gadd45γ–shRNA groups (n = 14 mice; M). Two-tailed unpaired Student's t test. ns, Not significant. Error bars represent SEM.
Figure 3.
Reducing hippocampal Gadd45γ levels in young adult mice impairs long-term memory formation. A, Schematic representation of the experimental design for long-term memory tests. B, C, Twenty-four hour object–location memory test of young adult mice expressing control–shRNA (n = 8 mice), Gadd45β–shRNA (n = 8 mice; B), control–shRNA (n = 12 mice), or Gadd45γ–shRNA1 (n = 13 mice; C) in the dHPC. The dashed lines represent equal preference for either object (chance preference). D, E, Twenty-four hour contextual fear memory test of young adult mice expressing control–shRNA (n = 8 mice), Gadd45β–shRNA (n = 8 mice; D), or control–shRNA (n = 16 mice), Gadd45γ–shRNA1 (n = 15 mice; E) in the dHPC. *p < 0.05 by two-tailed unpaired Student's t test. ns, Not significant. Error bars represent SEM.
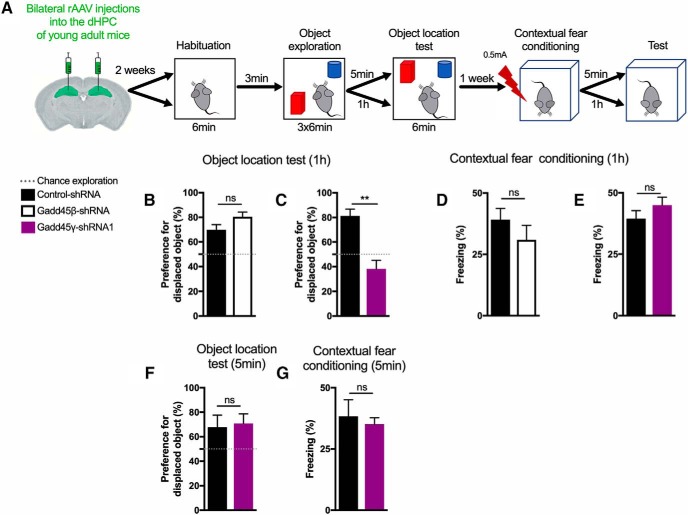
We next subjected mice to the same hippocampus-dependent memory tasks as above, but this time we used shorter time delays between training and testing to assess short-term memory (1 h or 5 min delay after training; Fig. 4A). In line with the aforementioned findings, Gadd45β knock-down mice showed intact memory compared with control animals in both tasks (Fig. 4B,D; two-tailed Student's t test, t(12) = 1.845, p = 0.0899 and t(13) = 1.086, p = 0.2971). In contrast, mice transduced with Gadd45γ–shRNA1 showed short-term memory impairments in locating the displaced object in the object–location test 1 h after training (Fig. 4C; two-tailed Student's t test, t(8) = 4.838, p = 0.0013). These effects were specific to object–location memory because no deficits were observed in contextual fear conditioning (Fig. 4E; two-tailed Student's t test, t(14) = 1.192, p = 0.2530). Knocking down Gadd45γ did not promote memory impairments when a recall session was performed 5 min after object–place recognition training (Fig. 4F; two-tailed Student's t test, t(8) = 2388, p = 0.8172) or contextual fear conditioning (Fig. 4G; two-tailed Student's t test, t(10) = 0.4399, p = 0.6694), showing that memory acquisition and early short-term memory are not compromised in Gadd45γ–shRNA1 animals. Altogether, this set of experiments indicates that hippocampal Gadd45γ is required for long-term memory and late stages of short-term recognition memory.
Figure 4.
Reducing hippocampal Gadd45γ levels in young adult mice impairs short-term recognition memory formation in a delay-specific manner. A, Schematic representation of the experimental design for short-term memory tests. B, C, One hour object location memory test of young adult mice expressing control–shRNA (n = 7 mice), Gadd45β–shRNA (n = 7 mice; B), or control–shRNA (n = 5 mice), Gadd45γ–shRNA1 (n = 5 mice; C) in the dHPC. D, E, One hour contextual fear memory test of young adult mice expressing control–shRNA (n = 7 mice), Gadd45β–shRNA (n = 8 mice; D), or control–shRNA (n = 8 mice), Gadd45γ–shRNA1 (n = 8 mice; E) in the dHPC. F, G, Five minute object–location memory test (F) and contextual fear conditioning test (G) of young adult mice expressing control–shRNA (n = 6 mice) or Gadd45γ–shRNA1 (n = 6 mice) in the dHPC. **p < 0.01 by two-tailed unpaired Student's t test. ns, Not significant. Error bars represent SEM.
Gadd45γ regulates MAPK signaling and downstream transcription factor activation
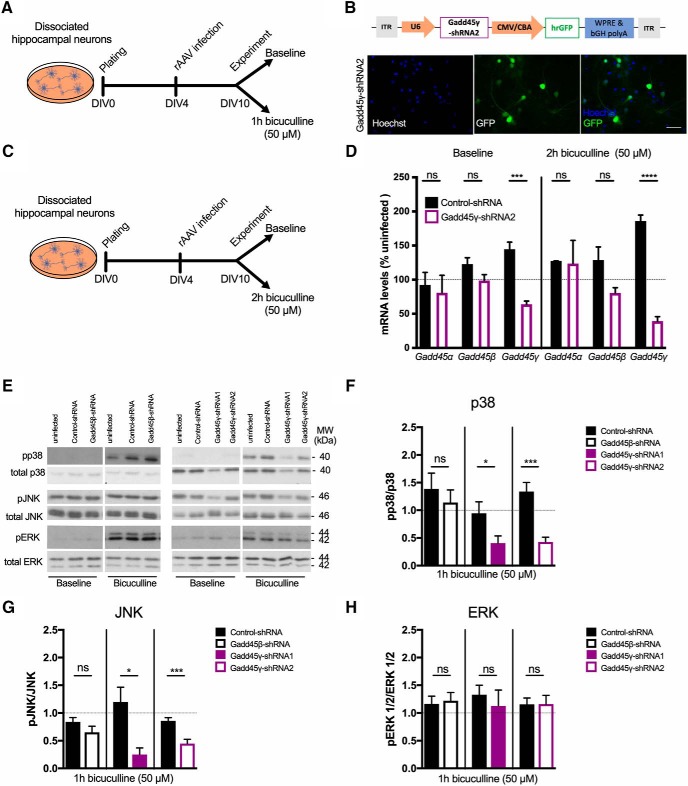
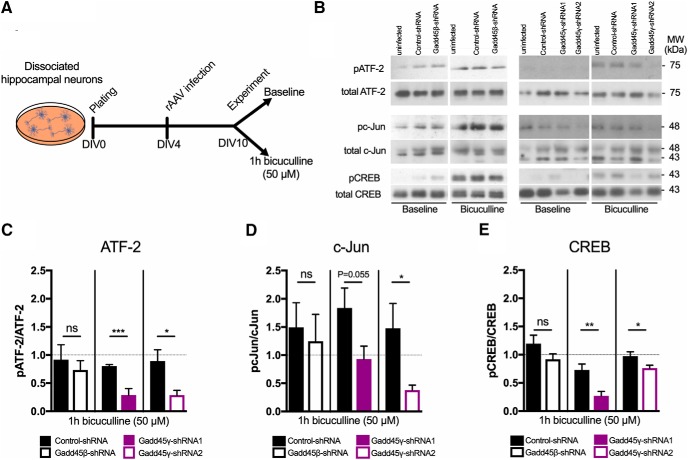
Gadd45 family members are known to regulate MAPK signaling cascades in several tissues outside of the nervous system through the interaction with kinases upstream of JNK and p38 MAPKs, but not ERK (Takekawa and Saito, 1998; Tornatore et al., 2008). Considering that both short- and long-lasting neuronal adaptive responses have been suggested to require JNK/p38 activity (Alonso et al., 2003; Bevilaqua et al., 2003; Guan et al., 2003; Sherrin et al., 2010; Giese and Mizuno, 2013; Morel et al., 2018), and Gadd45γ regulates these pathways, we next asked whether Gadd45 regulates MAPK signaling in hippocampal neurons. We addressed this by knocking down Gadd45β or Gadd45γ expression in primary hippocampal cultures and by measuring the phosphorylation levels of JNK, p38, and ERK in response to increased neuronal activity (Fig. 5A). To rule out possible off-target effects, we confirmed our findings with a second Gadd45γ-specific shRNA sequence (Gadd45γ–shRNA2). Infection rates and functional knock down of this rAAV were similar to previously validated shRNAs (Fig. 5B–D; two-tailed Student's t test; baseline: t(2) = 0.3629, p = 0.7515; t(8) = 1.901, p = 0.0938; t(8) = 7.046, p = 0.0001; bicuculline: t(2) = 0.1101, p = 0.9224; t(6) = 2.08, p = 0.0827; t(8) = 13.44, p < 0.0001). Reduction of Gadd45β expression did not affect JNK, p38, or ERK phosphorylation levels compared with control conditions (Fig. 5E–H; two-tailed Student's t test; p38: t(8) = 0.6737, p = 0.5195; JNK: t(7) = 0.9099, p = 0.3931; ERK: t(6) = 0.2913, p = 0.7806). In contrast, reduced Gadd45γ resulted in impaired JNK and p38 phosphorylation (Fig. 5E–G; two-tailed Student's t test; p38: t(18) = 2.345, p = 0.0307; t(14) = 4.885, p = 0.0002; and JNK: t(8) = 3.257, p = 0.0116; t(14) = 4.31, p = 0.0007), whereas ERK phosphorylation was not affected (Fig. 5E,H; two-tailed Student's t test; t(10) = 0.6112, p = 0.5547; t(12) = 0.01475, p = 0.9885). To further explore the functional consequences of Gadd45γ reduction, we analyzed the phosphorylation of transcription factors downstream of JNK and p38, such as ATF-2/CREB2, c-Jun, and CREB (Fig. 6A). We observed that, in Gadd45β–shRNA conditions, the phosphorylated forms of these transcription factors were not different from controls (Fig. 6B–E; two-tailed Student's t test; ATF-2: t(8) = 0.6737, p = 0.5806; c-Jun: t(8) = 0.3787, p = 0.7148; CREB: t(6) = 1.556, p = 0.1706). However, the phosphorylation of c-Jun, ATF-2, and CREB was significantly diminished when Gadd45γ levels were reduced (Fig. 6B–E; two-tailed Student's t test; ATF-2: t(10) = 5.266, p = 0.0004; t(10) = 2.792, p = 0.0191; c-Jun: t(10) = 2.167, p = 0.0554; t(8) = 2.429, p = 0.0413; CREB: t(12) = 3.57, p = 0.0039; t(12) = 2.285, p = 0.0413). These findings provide evidence that Gadd45γ regulates MAPK signaling in neurons.
Figure 5.
Gadd45γ regulates MAPK signaling in mouse hippocampal cultures. A, Schematic representation of the experimental design. B, Schematic representation of Gadd45γ–shRNA2 viral construct. The viral vector contains a U6 promoter driving Gadd45γ-specific shRNA sequence (Gadd45γ–shRNA2) and a cytomegalovirus (CMV)/chicken β-actin (CBA) hybrid promoter driving GFP as an infection marker. Representative images of cultured hippocampal cells infected with this virus are shown. Scale bar, 40 μm. bGH polyA, Bovine growth hormone polyadenylation signal; ITR, inverted terminal repeat; WPRE, woodchuck hepatitis virus post-transcriptional regulatory element. C, Schematic representation of the experimental design. D, qRT-PCR analysis of Gadd45α (n = 2 independent neuronal cultures), Gadd45β (n = 5 independent neuronal cultures), and Gadd45γ (n = 5 independent neuronal cultures) expression levels in hippocampal-cultured cells infected with control–shRNA or Gadd45γ–shRNA2 under basal conditions (left) or after 2 h bicuculline treatment (right). Expression levels were normalized to the corresponding uninfected controls in baseline conditions or upon bicuculline treatment (dashed line). E, Representative immunoblot scans using phosphospecific and total protein antibodies in hippocampal cultures infected with rAAV expressing control–shRNA, Gadd45β–shRNA, Gadd45γ–shRNA1, or Gadd45γ–shRNA2. Hippocampal cultures were harvested at baseline conditions or after 1 h of bicuculline treatment. F–H, Immunoblot quantification of p38 (n = 5–8 independent neuronal cultures; F), JNK (n = 5–8 independent neuronal cultures; G), and ERK (n = 4–7 independent neuronal cultures; H) presented in ratios of phosphorylated/total protein normalized to corresponding uninfected 1 h bicuculline condition (dashed lines). *p < 0.05, ***p < 0.001, ****p < 0.0001 by two-tailed unpaired Student's t test. ns, Not significant. Error bars represent SEM.
Figure 6.
Gadd45γ regulates the phosphorylation of downstream transcription factors in hippocampal mouse neuronal cultures. A, Schematic representation of the experimental design. B, Representative immunoblot scans using phosphospecific and total protein antibodies in hippocampal cultures infected with rAAV expressing control–shRNA, Gadd45β–shRNA, Gadd45γ–shRNA1, or Gadd45γ–shRNA2. Hippocampal cultures were harvested at baseline conditions or after 1 h of bicuculline treatment. C–E, Immunoblot quantification of ATF-2 (n = 4–6 independent neuronal cultures; C), c-Jun (n = 5–6 independent neuronal cultures; D), and CREB (n = 5–7 independent neuronal cultures; E) presented in ratios of phosphorylated/total protein normalized to uninfected 1 h bicuculline condition (dashed lines). *p < 0.05, **p < 0.01, ***p < 0.001 by two-tailed unpaired Student's t test. ns, Not significant. Error bars represent SEM.
Gadd45γ regulates CRE and AP-1 transcriptional activity and downstream activity-dependent gene transcription
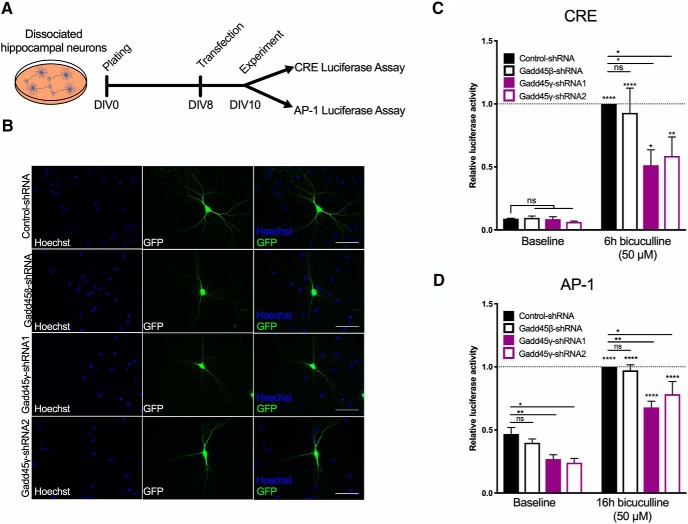
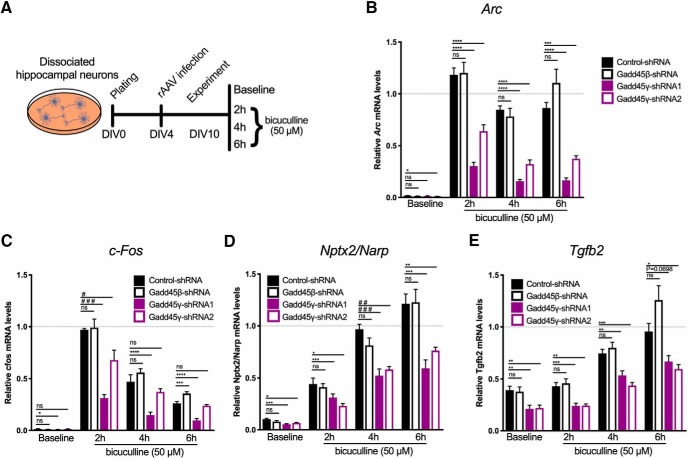
Given the disruption in the phosphorylation of CREB and in the components of the AP-1 complex [c-Jun, ATF, c-Fos (Alberini, 2009)], we next aimed at testing whether Gadd45γ reduction would impair AP-1- and CREB-dependent transcriptional activity. This analysis was further motivated by the established roles of CREB and AP-1 in memory formation (Alberini, 2009) and by studies linking CREB dysfunction and aging (Benito and Barco, 2010; Oliveira and Bading, 2011; Yu et al., 2017a,b). We performed luciferase reporter assays using a reporter plasmid containing the Firefly luciferase under the control of CRE or AP-1 binding sites in primary hippocampal cultures (Fig. 7A). To this end, we cotransfected constructs encoding CRE-luciferase or AP-1-luciferase and control–shRNA, Gadd45β–shRNA, Gadd45γ–shRNA1, or Gadd45γ–shRNA2 (Fig. 7B). Luciferase activity was assessed both under basal conditions and after induction of neuronal activity (Fig. 7C,D). In agreement with our data showing regulation of MAPK signaling specifically by Gadd45γ, Gadd45β reduction did not affect CRE or AP-1 transcriptional activity (Fig. 7C,D). In contrast, reducing Gadd45γ expression significantly impaired luciferase expression in both assays (Fig. 7C,D; one-way ANOVA followed by Sidak's multiple-comparisons test; CRE: F(7,32) = 16.23, p < 0.0001; AP-1: F(7,47) = 36.22, p < 0.0001). These results revealed that Gadd45γ expression is required for baseline AP-1 and activity-induced AP-1 and CRE transcriptional activity. To determine whether Gadd45γ regulates the expression of endogenous CRE and AP-1 target genes, we monitored the expression of CREB-dependent genes (Impey et al., 2004; Arc and c-Fos) and AP-1-dependent genes (Malik et al., 2014; Nptx2/Narp and Tgfb2) that are regulated by synaptic activity and whose expression has been shown to be dysregulated with aging (Arc and Nptx2/Narp; Ryan et al., 2019; Fig. 8A). CREB- and AP-1-dependent gene expression was disrupted in response to increased neuronal activity in Gadd45γ knock-down conditions (Fig. 8B–E; Arc: one-way ANOVA followed by Dunnett's multiple-comparisons test; baseline: F(3,28) = 2.471, p = 0.0825; 2 h: F(3,28) = 38.33, p < 0.0001; 4 h: F(3,27) = 48,85, p < 0.0001; 6 h: F(3,28) = 34,76, p < 0.0001; c-fos: one-way ANOVA followed by Dunnett's multiple-comparisons test or Kruskal–Wallis; baseline: F(3,28) = 2.974, p = 0.0486; 2 h: KW statistic = 21.55, p < 0.0001; 4 h: F(3,27) = 16.3, p < 0.0001; 6 h: F(3,28) = 45.89, p < 0.0001; Nptx2/Narp: one-way ANOVA followed by Dunnett's multiple-comparisons test or Kruskal–Wallis followed by Dunn's multiple-comparisons test; baseline: F(3,24) = 6.684, p = 0.0019; 2 h: F(3,24) = 6.369, p = 0.0025; 4 h: KW statistic = 16.95, p = 0.0007; 6 h: F(3,24) = 12.73, p < 0.0001; Tgfb2: one-way ANOVA followed by Dunnett's multiple-comparisons test; baseline: F(3,28) = 2.471, p = 0.0015; 2 h: F(3,28) = 38.33, p < 0.0001; 4 h: F(3,27) = 48.85, p < 0.0001; 6 h: F(3,28) = 34.76, p < 0.0001). Moreover, in line with luciferase assays, CREB target genes were predominantly decreased in an activity-dependent manner, whereas AP-1-target genes showed consistent baseline and activity-dependent decreases when Gadd45γ was reduced. Gadd45β knockdown did not promote any changes in gene expression compared with control conditions. These results show that Gadd45γ regulates CREB- and AP-1-dependent gene expression in neurons. It should be noted that cross talk between both signaling pathways may occur because we detected deficits in c-Fos expression, a component of the AP-1 complex (Alberini, 2009).
Figure 7.
Gadd45γ regulates CREB and AP-1 activity in mouse hippocampal cultures. A, Schematic representation of the experimental design. B, Representative images of hippocampal-cultured cells cotransfected with control–shRNA, Gadd45β–shRNA, Gadd45γ–shRNA1, or Gadd45γ–shRNA2 and luciferase reporter plasmids. Scale bars, 40 μm. C, D, Luciferase reporter assays were performed using dissociated hippocampal cultures cotransfected with control–shRNA, Gadd45β–shRNA, Gadd45γ–shRNA1, or Gadd45γ–shRNA2 constructs, and reporter plasmids expressing FFluc under the control of CRE (n = 5 independent neuronal cultures; C) or AP-1 (n = 7 independent neuronal cultures; D) transcriptional activity. C, D, Shown is luciferase activity as measured from untreated hippocampal cultures and cultures treated for 6 or 18 h with bicuculline. Values are normalized to control–shRNA condition treated with bicuculline for the respective time duration (6 or 18 h; dashed lines). *p < 0.05, **p < 0.01, and ****p < 0.0001 by one-way ANOVA test followed by Sidak's or Dunnett's post hoc test with pairwise comparisons. ns, Not significant. Error bars represent SEM.
Figure 8.
Gadd45γ regulates CREB- and AP-1-dependent transcription in mouse hippocampal cultures. A–E, Schematic representation of the experimental design for qRT-PCR analysis (A) of the expression of the CREB-dependent genes Arc (n = 7–8 independent neuronal cultures; B) and c-Fos (n = 6–8 independent neuronal cultures; C) or the AP-1-dependent genes Nptx2/Narp (n = 6–7 independent neuronal cultures; D) and Tgfb2 (n = 6–7 independent neuronal cultures; E). Hippocampal cultures were harvested at baseline conditions or after 2, 4, or 6 h of bicuculline treatment. Uninfected controls treated with bicuculline were used for normalization at the corresponding peak of gene expression: 2 h (Arc and c-Fos) or 6 h (Nptx2/Narp and Tgfb2; dashed lines). *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 by one-way ANOVA test followed by Sidak's or Dunnett's post hoc test with pairwise comparisons; #p < 0.05, ##p < 0.01, and ###p < 0.001, Kruskal–Wallis test followed by Dunn's post hoc test for non-Gaussian distribution. ns, Not significant. Error bars represent SEM.
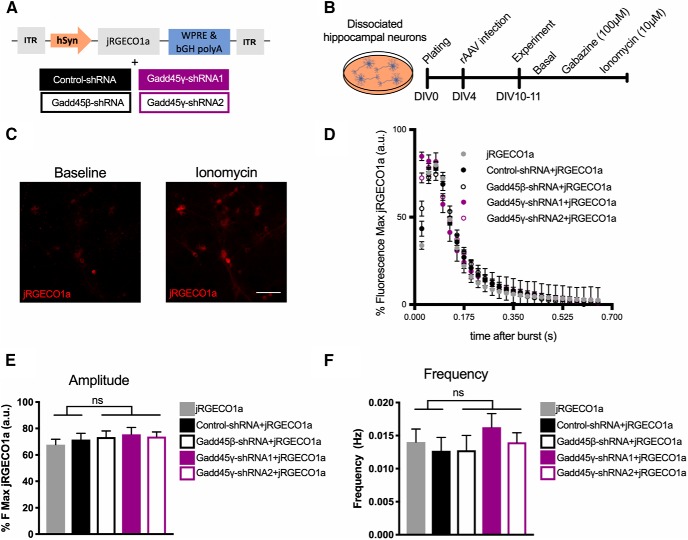
To confirm that the effects seen in MAPK signaling and downstream transcription factor activity were directly dependent on Gadd45γ function and not due to an overall disruption of neuronal responses and/or calcium-dependent signaling pathways, we assessed calcium rises triggered by neuronal activity. We measured intracellular calcium dynamics using the genetically encoded calcium indicator jRGECO1a (Fig. 9A), which has high sensitivity in dissociated neurons, in primary hippocampal cultures coinfected with the above-described shRNA constructs. Coexpression of shRNA constructs and jRGECO1a was reliably determined using GFP/mCherry fluorescence, and only GFP+ cells were analyzed. Neurons were treated with the GABAA antagonist gabazine to induce action potential bursting (Fig. 9B–D). No differences in the amplitude (Fig. 9E; one-way ANOVA followed by Sidak's multiple-comparisons test: F(4,25) = 0.435, p = 0.7820) or frequency (Fig. 9F; one-way ANOVA followed by Sidak's multiple-comparisons test: F(4,25) = 0.5321) of action potential bursts were detected in Gadd45β—shRNA-, Gadd45γ–shRNA1-, or Gadd45γ–shRNA2-infected neurons. This set of experiments indicates that deficits in MAPK signaling and activity-dependent gene expression promoted by Gadd45γ reduction are not explained by altered calcium responses to neuronal activity. This supports the hypothesis that, similar to its function in other cell types (Takekawa and Saito, 1998; Tornatore et al., 2008), Gadd45γ directly regulates MAPK signaling in neurons.
Figure 9.
Gadd45γ reduction does not alter calcium dynamics in response to neuronal activity. A, Schematic representation of the viral constructs used. The viral vector driving jRGECO1a expression does so under control of the human synapsin (hSyn) promoter. Neuronal cultures were either singly infected with jRGECO1a or coinfected with jRGECO1a and control–shRNA, Gadd45β–shRNA, Gadd45γ–shRNA1, or Gadd45γ–shRNA2. bGH polyA, Bovine growth hormone polyadenylation signal; ITR, inverted terminal repeat; WPRE, woodchuck hepatitis virus post-transcriptional regulatory element. B, Schematic representation of the experimental design. C, Images of representative cultured hippocampal cells infected with jRGECO1a at baseline or after ionomycin treatment. Scale bar, 40 μm. D, Representative traces showing the first gabazine-evoked burst of all groups. E, F, Amplitude (n = 6 independent neuronal cultures; E) and frequency (n = 6 independent neuronal cultures; F) of evoked action potential bursts during gabazine treatment in all groups. One-way ANOVA test followed by Sidak's multiple-comparisons test with pairwise comparisons was used. ns, Not significant. Error bars represent SEM.
Discussion
This study reveals that aging is associated with reduction in Gadd45γ expression, and that reducing hippocampal Gadd45γ levels in young adult mice negatively impacts cognitive function. Furthermore, our results show that Gadd45γ regulates synapse-to-nucleus signaling and AP-1- and CREB-regulated genomic programs. These findings provide insight into the functions of Gadd45 family members in the hippocampus.
Previous studies on the functions of Gadd45 family members in the brain focused mainly on Gadd45β. Knocking out Gadd45β promoted enhanced long-term memory in one study (Sultan et al., 2012) and deficient hippocampus-dependent long-term memory in another (Leach et al., 2012). Considering that both reports applied a global knock-out strategy, possible confounds originating from developmental compensatory mechanisms and/or differential functions in different brain regions that bias conclusions about the function of Gadd45β in hippocampus-dependent memory may have been present. Here, we acutely decreased the expression of Gadd45β in the dorsal hippocampus of adult mice and found no differences in long- or short-term memory. Moreover, knockdown of Gadd45β in primary hippocampal cultures did not affect stimulus-induced MAPK signaling, transcription factor activity, or gene expression. This result suggests that previously reported effects of Gadd45β knockout may not be a direct consequence of Gadd45β-associated functions in the hippocampus.
Here, we describe that, analogous to its role in other cell types (Tamura et al., 2012), Gadd45γ functions as a regulator of MAPK signaling pathways in hippocampal neurons. MAPK signaling cascades regulate the activity of downstream transcription factors with established roles in memory formation (Alberini, 2009). Here, we found that Gadd45γ regulates the transcriptional activity of CREB and AP-1 and downstream gene expression. Thus, our data suggest that Gadd45γ may mediate the formation of long-term memory through the regulation of CREB- and AP-1-dependent memory genes. However, this putative role of Gadd45γ should be tested in vivo in future studies.
Recently, it has been suggested that Gadd45γ can act as a DNA demethylase (Grassi et al., 2017; Li et al., 2019) and in this way can regulate gene expression. Our findings show a novel mechanism by which Gadd45γ regulates gene expression in neurons. This is supported by our study using luciferase assays that demonstrate the ability of Gadd45γ to regulate CREB- and AP-1-dependent transcription independently of the epigenome. Moreover, we observed a striking similarity in the magnitude of the effects obtained in the luciferase assays and the analysis of expression of CREB- and AP-1-endogenous target genes. In both assays, AP-1-dependent transcription required Gadd45γ in both basal and induced conditions, whereas CREB-dependent gene expression required Gadd45γ only after stimulation, thus suggesting that the regulation of CREB and AP-1 target genes depends on Gadd45γ-mediated synapse-to-nucleus communication. It is plausible that Gadd45γ-dependent regulation of MAPK signaling and its reported DNA demethylase activity work in conjunction in the fine tuning of genomic responses underlying memory formation. It was shown that, in the prelimbic prefrontal cortex, nuclear Gadd45γ regulates a late transcriptional response by DNA demethylation activity. Together with our data, this study suggests a mechanistic framework in which cytoplasmic Gadd45γ regulates the immediate transcriptional response to learning and nuclear Gadd45γ facilitates, through DNA demethylation, later phases of transcription activation. However, this remains to be investigated.
We found that Gadd45γ reduction impairs short-term object–place recognition memory. Short- and long-term forms of plasticity have been associated with JNK and p38 activity. JNK targets synaptic proteins that regulate the insertion of AMPA receptors (Coffey, 2014). Similarly, p38 is required for AMPA receptor trafficking associated with mGluR-induced LTD and NMDAR-induced LTD (Corrêa and Eales, 2012). In vivo studies also suggest that both JNK and p38 are activated upon learning stimuli (Giese and Mizuno, 2013). Pharmacological inhibition of p38 immediately after learning impaired both short- and long-term memory formation (Alonso et al., 2003). Therefore, impairments in the activation of p38 and/or JNK may underlie the short-term memory deficits identified in our study.
Several studies demonstrated that altered CREB activity during aging has been linked to dysregulated calcium homeostasis (Oliveira and Bading, 2011). We found, however, that Gadd45γ knockdown, which mimics age-associated changes, impacts CREB phosphorylation independently of changes in calcium dynamics. This suggests that, during aging, dysregulation of Gadd45γ-dependent regulatory mechanisms may be another factor contributing to altered CREB function. Intriguingly, the memory phenotypes identified in our study mimic well established aging-dependent cognitive deficits observed across species. We observed a delay-specific short-term recognition memory impairment, which is also observed across different species during aging. Aged animals only present short-term memory impairments when longer delays between training and recall are used (Erickson and Barnes, 2003; Burke et al., 2012). Moreover, a disruption of contextual fear memory was limited to long-term memory impairments in the case of Gadd45γ reduction. This finding is in line with rodent studies showing that associative memory—in particular, contextual fear memory—is less vulnerable to aging (Foster et al., 2012). In light of these findings, it is tempting to speculate that aging-associated Gadd45γ expression dysregulation is associated with these phenotypes. However, in addition to reduction in Gadd45γ expression, substantial alterations in signaling and gene expression accompany aging. This could potentially mask/alter Gadd45γ functions in this context compared with young adult brain. This should be addressed in future experiments by directly analyzing Gadd45γ functions in the altered setting of the aged hippocampus.
In conclusion, this study demonstrates that Gadd45γ is a critical regulator of synapse-to-nucleus communication and cognitive functions in the young adult mouse hippocampus. Our results further suggest Gadd45γ as a molecular candidate that may contribute to cognitive impairments in aging-associated pathological conditions. Identifying the underlying causes that lead to Gadd45γ dysfunction could potentially allow for the design of strategies that may prevent or delay the onset of age-associated memory deficits across species. Furthermore, Gadd45γ expression is altered in the prefrontal cortex of patients suffering from alcohol dependence (Kapoor et al., 2019) and in the hippocampus of mice subjected to unpredictable chronic mild stress, an established model for depression (Grassi et al., 2017). Therefore, our study further suggests molecular mechanisms that may be present in the pathophysiological development of psychiatric conditions and/or associated cognitive deficits.
Footnotes
This work was supported by the Sonderforschungsbereich 1134 and an Emmy Noether Grant of the Deutsche Forchungsgemeinschaft to A.M.M.O. A.M.M.O. is a member of the Excellence Cluster CellNetworks at Heidelberg University. We thank I. Bünzli-Ehret for the preparation of primary hippocampal cultures, and Dr. Anna Hagenston Hertle for providing the calcium indicator used in the study, for helpful guidance during the calcium imaging analysis, and for critical comments to the manuscript. We also thank Dr. Priit Pruunsild for the luciferase reporter constructs and guidance with the assays.
The authors declare no competing financial interests.
References
- Aigbogun MS, Stellhorn R, Krasa H, Kostic D (2017) Severity of memory impairment in the elderly: association with health care resource use and functional limitations in the United States. Alzheimers Dement (Amst) 8:51–59. 10.1016/j.dadm.2017.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberini CM. (2009) Transcription factors in long-term memory and synaptic plasticity. Physiol Rev 89:121–145. 10.1152/physrev.00017.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso M, Bevilaqua LR, Izquierdo I, Medina JH, Cammarota M (2003) Memory formation requires p38MAPK activity in the rat hippocampus. Neuroreport 14:1989–1992. 10.1097/00001756-200310270-00022 [DOI] [PubMed] [Google Scholar]
- Bach ME, Barad M, Son H, Zhuo M, Lu YF, Shih R, Mansuy I, Hawkins RD, Kandel ER (1999) Age-related defects in spatial memory are correlated with defects in the late phase of hippocampal long-term potentiation in vitro and are attenuated by drugs that enhance the cAMP signaling pathway. Proc Natl Acad Sci U S A 96:5280–5285. 10.1073/pnas.96.9.5280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bading H, Greenberg ME (1991) Stimulation of protein tyrosine phosphorylation by NMDA receptor activation. Science 253:912–914. 10.1126/science.1715095 [DOI] [PubMed] [Google Scholar]
- Benito E, Barco A (2010) CREB's control of intrinsic and synaptic plasticity: implications for CREB-dependent memory models. Trends Neurosci 33:230–240. 10.1016/j.tins.2010.02.001 [DOI] [PubMed] [Google Scholar]
- Bevilaqua LR, Kerr DS, Medina JH, Izquierdo I, Cammarota M (2003) Inhibition of hippocampal jun N-terminal kinase enhances short-term memory but blocks long-term memory formation and retrieval of an inhibitory avoidance task. Eur J Neurosci 17:897–902. 10.1046/j.1460-9568.2003.02524.x [DOI] [PubMed] [Google Scholar]
- Burke SN, Barnes CA (2006) Neural plasticity in the ageing brain. Nat Rev Neurosci 7:30–40. 10.1038/nrn1809 [DOI] [PubMed] [Google Scholar]
- Burke SN, Ryan L, Barnes CA (2012) Characterizing cognitive aging of recognition memory and related processes in animal models and in humans. Front Aging Neurosci 4:15. 10.3389/fnagi.2012.00015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey ET. (2014) Nuclear and cytosolic JNK signalling in neurons. Nat Rev Neurosci 15:285–299. 10.1038/nrn3729 [DOI] [PubMed] [Google Scholar]
- Corrêa SA, Eales KL (2012) The role of p38 MAPK and its substrates in neuronal plasticity and neurodegenerative disease. J Signal Transduct 2012:649079. 10.1155/2012/649079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana H, Mohar B, Sun Y, Narayan S, Gordus A, Hasseman JP, Tsegaye G, Holt GT, Hu A, Walpita D, Patel R, Macklin JJ, Bargmann CI, Ahrens MB, Schreiter ER, Jayaraman V, Looger LL, Svoboda K, Kim DS (2016) Sensitive red protein calcium indicators for imaging neural activity. Elife 5. 10.7554/eLife.12727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson CA, Barnes CA (2003) The neurobiology of memory changes in normal aging. Exp Gerontol 38:61–69. 10.1016/S0531-5565(02)00160-2 [DOI] [PubMed] [Google Scholar]
- Foster TC, Defazio RA, Bizon JL (2012) Characterizing cognitive aging of spatial and contextual memory in animal models. Front Aging Neurosci 4:12. 10.3389/fnagi.2012.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin DP, Sharma RP, Chase KA, Matrisciano F, Dong E, Guidotti A (2012) Growth arrest and DNA-damage-inducible, beta (GADD45b)-mediated DNA demethylation in major psychosis. Neuropsychopharmacology 37:531–542. 10.1038/npp.2011.221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giese KP, Mizuno K (2013) The roles of protein kinases in learning and memory. Learn Mem 20:540–552. 10.1101/lm.028449.112 [DOI] [PubMed] [Google Scholar]
- Grassi D, Franz H, Vezzali R, Bovio P, Heidrich S, Dehghanian F, Lagunas N, Belzung C, Krieglstein K, Vogel T (2017) Neuronal activity, TGFβ-signaling and unpredictable chronic stress modulate transcription of Gadd45 family members and DNA methylation in the hippocampus. Cereb Cortex 27:4166–4181. 10.1093/cercor/bhx095 [DOI] [PubMed] [Google Scholar]
- Guan Z, Kim JH, Lomvardas S, Holick K, Xu S, Kandel ER, Schwartz JH (2003) p38 MAP kinase mediates both short-term and long-term synaptic depression in aplysia. J Neurosci 23:7317–7325. 10.1523/JNEUROSCI.23-19-07317.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulmez Karaca K, Brito DVC, Zeuch B, Oliveira AMM (2018) Adult hippocampal MeCP2 preserves the genomic responsiveness to learning required for long-term memory formation. Neurobiol Learn Mem 149:84–97. 10.1016/j.nlm.2018.02.010 [DOI] [PubMed] [Google Scholar]
- Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, McWeeney S, Dunn JJ, Mandel G, Goodman RH (2004) Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell 119:1041–1054. 10.1016/j.cell.2004.10.032 [DOI] [PubMed] [Google Scholar]
- Kapoor M, Wang JC, Farris SP, Liu Y, McClintick J, Gupta I, Meyers JL, Bertelsen S, Chao M, Nurnberger J, Tischfield J, Harari O, Zeran L, Hesselbrock V, Bauer L, Raj T, Porjesz B, Agrawal A, Foroud T, Edenberg HJ, et al. (2019) Analysis of whole genome-transcriptomic organization in brain to identify genes associated with alcoholism. Transl Psychiatry 9:89. 10.1038/s41398-019-0384-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennard JA, Woodruff-Pak DS (2011) Age sensitivity of behavioral tests and brain substrates of normal aging in mice. Front Aging Neurosci 3:9. 10.3389/fnagi.2011.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach PT, Poplawski SG, Kenney JW, Hoffman B, Liebermann DA, Abel T, Gould TJ (2012) Gadd45b knockout mice exhibit selective deficits in hippocampus-dependent long-term memory. Learn Mem 19:319–324. 10.1101/lm.024984.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Marshall PR, Leighton LJ, Zajaczkowski EL, Wang Z, Madugalle SU, Yin J, Bredy TW, Wei W (2019) The DNA repair associated protein Gadd45γ regulates the temporal coding of immediate early gene expression within the prelimbic prefrontal cortex and is required for the consolidation of associative fear memory. J Neurosci 39:970–983. 10.1523/JNEUROSCI.2024-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, Flavell RA, Lu B, Ming GL, Song H (2009) Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science 323:1074–1077. 10.1126/science.1166859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik AN, Vierbuchen T, Hemberg M, Rubin AA, Ling E, Couch CH, Stroud H, Spiegel I, Farh KK, Harmin DA, Greenberg ME (2014) Genome-wide identification and characterization of functional neuronal activity-dependent enhancers. Nat Neurosci 17:1330–1339. 10.1038/nn.3808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauceri D, Hagenston AM, Schramm K, Weiss U, Bading H (2015) Nuclear calcium buffering capacity shapes neuronal architecture. J Biol Chem 290:23039–23049. 10.1074/jbc.M115.654962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel C, Sherrin T, Kennedy NJ, Forest KH, Avcioglu Barutcu S, Robles M, Carpenter-Hyland E, Alfulaij N, Standen CL, Nichols RA, Benveniste M, Davis RJ, Todorovic C (2018) JIP1-mediated JNK activation negatively regulates synaptic plasticity and spatial memory. J Neurosci 38:3708–3728. 10.1523/JNEUROSCI.1913-17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskalev AA, Smit-McBride Z, Shaposhnikov MV, Plyusnina EN, Zhavoronkov A, Budovsky A, Tacutu R, Fraifeld VE (2012) Gadd45 proteins: relevance to aging, longevity and age-related pathologies. Ageing Res Rev 11:51–66. 10.1016/j.arr.2011.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira AM, Bading H (2011) Calcium signaling in cognition and aging-dependent cognitive decline. Biofactors 37:168–174. 10.1002/biof.148 [DOI] [PubMed] [Google Scholar]
- Oliveira AM, Hemstedt TJ, Bading H (2012) Rescue of aging-associated decline in Dnmt3a2 expression restores cognitive abilities. Nat Neurosci 15:1111–1113. 10.1038/nn.3151 [DOI] [PubMed] [Google Scholar]
- Oliveira AM, Hemstedt TJ, Freitag HE, Bading H (2016) Dnmt3a2: a hub for enhancing cognitive functions. Mol Psychiatry 21:1130–1136. 10.1038/mp.2015.175 [DOI] [PubMed] [Google Scholar]
- Pruunsild P, Sepp M, Orav E, Koppel I, Timmusk T (2011) Identification of cis-elements and transcription factors regulating neuronal activity-dependent transcription of human BDNF gene. J Neurosci 31:3295–3308. 10.1523/JNEUROSCI.4540-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruunsild P, Bengtson CP, Bading H (2017) Networks of cultured iPSC-derived neurons reveal the human synaptic activity-regulated adaptive gene program. Cell Rep 18:122–135. 10.1016/j.celrep.2016.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan L, Hay M, Huentelman MJ, Duarte A, Rundek T, Levin B, Soldan A, Pettigrew C, Mehl MR, Barnes CA (2019) Precision aging: applying precision medicine to the field of cognitive aging. Front Aging Neurosci 11:128. 10.3389/fnagi.2019.00128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrin T, Blank T, Hippel C, Rayner M, Davis RJ, Todorovic C (2010) Hippocampal c-jun-N-terminal kinases serve as negative regulators of associative learning. J Neurosci 30:13348–13361. 10.1523/JNEUROSCI.3492-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh-Manoux A, Kivimaki M, Glymour MM, Elbaz A, Berr C, Ebmeier KP, Ferrie JE, Dugravot A (2012) Timing of onset of cognitive decline: results from whitehall II prospective cohort study. BMJ 344:d7622. 10.1136/bmj.d7622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanelli G, Azam AB, Walters BJ, Brimble MA, Gettens CP, Bouchard-Cannon P, Cheng HM, Davidoff AM, Narkaj K, Day JJ, Kennedy AJ, Zovkic IB (2018) Learning and age-related changes in genome-wide H2A.Z binding in the mouse hippocampus. Cell Rep 22:1124–1131. 10.1016/j.celrep.2018.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stilling RM, Benito E, Gertig M, Barth J, Capece V, Burkhardt S, Bonn S, Fischer A (2014) De-regulation of gene expression and alternative splicing affects distinct cellular pathways in the aging hippocampus. Front Cellular Neurosci 8:373. 10.3389/fncel.2014.00373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan FA, Wang J, Tront J, Liebermann DA, Sweatt JD (2012) Genetic deletion of Gadd45b, a regulator of active DNA demethylation, enhances long-term memory and synaptic plasticity. J Neurosci 32:17059–17066. 10.1523/JNEUROSCI.1747-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takekawa M, Saito H (1998) A family of stress-inducible GADD45-like proteins mediate activation of the stress-responsive MTK1/MEKK4 MAPKKK. Cell 95:521–530. 10.1016/S0092-8674(00)81619-0 [DOI] [PubMed] [Google Scholar]
- Tamura RE, de Vasconcellos JF, Sarkar D, Libermann TA, Fisher PB, Zerbini LF (2012) GADD45 proteins: central players in tumorigenesis. Curr Mol Med 12:634–651. 10.2174/156652412800619978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornatore L, Marasco D, Dathan N, Vitale RM, Benedetti E, Papa S, Franzoso G, Ruvo M, Monti SM (2008) Gadd45 beta forms a homodimeric complex that binds tightly to MKK7. J Mol Biol 378:97–111. 10.1016/j.jmb.2008.01.074 [DOI] [PubMed] [Google Scholar]
- Weber M, Wu T, Hanson JE, Alam NM, Solanoy H, Ngu H, Lauffer BE, Lin HH, Dominguez SL, Reeder J, Tom J, Steiner P, Foreman O, Prusky GT, Scearce-Levie K (2015) Cognitive deficits, changes in synaptic function, and brain pathology in a mouse model of normal aging(1,2,3). eNeuro 2:ENEURO.0047-15.2015 10.1523/ENEURO.0047-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Janson CG, Mastakov M, Lawlor P, Young D, Mouravlev A, Fitzsimons H, Choi KL, Ma H, Dragunow M, Leone P, Chen Q, Dicker B, During MJ (2001) Quantitative comparison of expression with adeno-associated virus (AAV-2) brain-specific gene cassettes. Gene Ther 8:1323–1332. 10.1038/sj.gt.3301529 [DOI] [PubMed] [Google Scholar]
- Yu XW, Oh MM, Disterhoft JF (2017a) CREB, cellular excitability, and cognition: implications for aging. Behav Brain Res 322:206–211. 10.1016/j.bbr.2016.07.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XW, Curlik DM, Oh MM, Yin JC, Disterhoft JF (2017b) CREB overexpression in dorsal CA1 ameliorates long-term memory deficits in aged rats. Elife 6. e19358. 10.7554/eLife.19358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SJ, Steijaert MN, Lau D, Schütz G, Delucinge-Vivier C, Descombes P, Bading H (2007) Decoding NMDA receptor signaling: identification of genomic programs specifying neuronal survival and death. Neuron 53:549–562. 10.1016/j.neuron.2007.01.025 [DOI] [PubMed] [Google Scholar]
- Zhang SJ, Zou M, Lu L, Lau D, Ditzel DA, Delucinge-Vivier C, Aso Y, Descombes P, Bading H (2009) Nuclear calcium signaling controls expression of a large gene pool: identification of a gene program for acquired neuroprotection induced by synaptic activity. PLoS Genet 5:e1000604. 10.1371/journal.pgen.1000604 [DOI] [PMC free article] [PubMed] [Google Scholar]