Adolescence is a time of extensive neural restructuring, leaving one susceptible to atypical development. Although neural maturation in humans can be measured using functional and structural MRI, the subtle patterns associated with the initial stages of abnormal change may be difficult to identify, particularly at an individual level.
Keywords: adolescence, brain age, brain development, fMRI, machine learning, sMRI
Abstract
Adolescence is a time of extensive neural restructuring, leaving one susceptible to atypical development. Although neural maturation in humans can be measured using functional and structural MRI, the subtle patterns associated with the initial stages of abnormal change may be difficult to identify, particularly at an individual level. Brain age prediction models may have utility in assessing brain development in an individualized manner, as deviations between chronological age and predicted brain age could reflect one's divergence from typical development. Here, we built a support vector regression model to summarize high-dimensional neuroimaging as an index of brain age in both sexes. Using structural and functional MRI data from two large pediatric datasets and a third clinical dataset, we produced and validated a two-dimensional neural maturation index (NMI) that characterizes typical brain maturation patterns and identifies those who deviate from this trajectory. Examination of brain signatures associated with NMI scores revealed that elevated scores were related to significantly lower gray matter volume and significantly higher white matter volume, particularly in high-order regions such as the prefrontal cortex. Additionally, those with higher NMI scores exhibited enhanced connectivity in several functional brain networks, including the default mode network. Analysis of data from a sample of male and female patients with schizophrenia revealed an association between advanced NMI scores and schizophrenia diagnosis in participants aged 16–22, confirming the NMI's utility as a marker of atypicality. Altogether, our findings support the NMI as an individualized, interpretable measure by which neural development in adolescence may be assessed.
SIGNIFICANCE STATEMENT The substantial neural restructuring that occurs during adolescence increases one's vulnerability to aberration. A brain index that is capable of capturing one's conformance with typical development will allow for individualized assessment and enhance our understanding of typical and atypical development. In this analysis, we produce a neural maturation index (NMI) using support vector regression and a large pediatric sample. This index generalizes across multiple cohorts and shows potential in the identification of clinical groups. We also implement a novel method for examining the developmental trajectory through data-driven analysis. The signatures identified by the NMI reflect key stages of the extensive neural development that occurs during adolescence and support its utility as a metric of typical brain development.
Introduction
Brain development during adolescence is associated with profound changes in brain structure and function. The synaptic pruning and increase in myelination that are hallmarks of this neurodevelopmental stage are reflected in widespread gray matter loss (Giedd et al., 1999; Gogtay et al., 2004; Giorgio et al., 2010). This loss occurs in a spatially heterogeneous manner, beginning in the sensorimotor cortex and proceeding rostrally and caudally, with regions involved in complex function (i.e., the dorsolateral prefrontal cortex and superior temporal cortex) continuing to decrease in volume through late adolescence and early adulthood (Gogtay et al., 2004). Conversely, regional gray matter density increases (Gennatas et al., 2017) and an escalation in white matter and structural connectivity throughout the brain contribute to increased functional network integration (Giorgio et al., 2008; Dennis et al., 2013). The default mode network (DMN) in particular undergoes extensive maturation, integrating into a more cohesive network (Fair et al., 2008). Structural MRI (sMRI) and resting-state functional MRI (fMRI) measure several aspects of this brain maturation process. However, capturing the complexity and subtleties of brain development necessitates the use of advanced multivariate analytic methods. Machine learning algorithms have been efficacious in capturing consistent patterns of age-related neural change over the lifespan (Dosenbach et al., 2010; Franke et al., 2010, 2012; Erus et al., 2015; Habes et al., 2016). Moreover, machine learning algorithms can encapsulate these complex features in a small number of indices that allow one to assess brain signatures at an individual level. Structural and functional brain age are two such indices.
Brain age, as estimated by neuroimaging-based age prediction models, may accurately reflect one's deviation, or lack thereof, from the typical developmental trajectory. Thus, it may be used as a comprehensive and individualized marker of brain health (Cole and Franke, 2017). A growing volume of research in adults has associated advanced brain age with a wide range of disorders, including schizophrenia (Koutsouleris et al., 2014; Schnack et al., 2016; Shahab et al., 2019). Although the extensive brain changes that occur during the critical period of adolescence are thought to confer vulnerability to emergence of neuropsychiatric symptoms (Paus et al., 2008), the relationship between brain age and psychiatric pathology has been only sparsely investigated. Existing work indicates that deviations between brain age and chronological age may capture anomalies in structural and functional brain development (Chung et al., 2018; Hajek et al., 2019). A developmental index based on one's brain age could provide a standardized measure of risk and contribute to the early detection of neuropathology at an individual level. In the current work we derive a comprehensive neural maturation index (NMI) of structure and function and test its reproducibility across two large studies of brain development.
To investigate the generalizability of our multimodal NMI, we first derived separate models for both structural and functional data from participants in the Philadelphia Neurodevelopmental Cohort (PNC; Satterthwaite et al., 2016). These models were then applied to participants from the Pediatric Imaging, Neurocognition, and Genetics (PING; Jernigan et al., 2016) study, an entirely independent study whose data were collected under different imaging conditions and locations. We hypothesized that machine learning techniques would identify an accurate and reproducible index of brain development. In this framework, we examined the association of the distance between one's predicted age and their chronologic age with the respective patterns of brain structure and function. We understood that differences would exist, as the NMI model uses brain structure and function as the basis for assigning one's position in the chart. Therefore, our intention was not to identify the existence of differences between the groups produced by our NMI but to investigate the brain patterns that the model identified as being the most effective estimators of development. Finally, we evaluated whether schizophrenia is associated with deviations from typical patterns of brain development summarized by the NMI using a third independent dataset of schizophrenia patients between the ages of 16 and 22.
Materials and Methods
Participants
Subjects were drawn from the PNC and the PING study. Participants without appropriate imaging data were excluded from the corresponding analysis. Three MRI protocols were used to collect data from PING participants; only participants following the Siemens MRI protocol were included. The final sMRI analysis included 1396 PNC participants (731 females, ages 8–23) and 314 PING participants (160 females, ages 8–21). The final fMRI analysis included 968 PNC participants (534 females, ages 8–23) and 176 PING participants (88 females, ages 8–21). Data collection methods for PNC and PING were described by Satterthwaite et al. (2014) and Jernigan et al. (2016), respectively. For computation of age-specific brain maps, we used a 4 year sliding window. With so few participants older than 22, we were unable to produce representative brain maps for ages >20. This necessitated the exclusion of participants older than 22 (n = 12) for this portion of the analysis.
To compare the NMI scores of schizophrenia patients to controls, we used participants from a multisite schizophrenia study (United States, Germany, and China). We excluded participants whose age fell outside the range of our NMI, giving us a final sample of 96 participants (ages 16–22, 25 females; 43 schizophrenia, 53 control).
Data processing
MRI preprocessing.
An automated pipeline was applied for preprocessing T1-weighted scans of subjects. This pipeline included magnetic field inhomogeneity correction (Sled et al., 1998) and extraction of the intracranial brain tissues using multi-atlas skull-stripping (Doshi et al., 2013). Anatomical regions-of-interest (ROIs) were identified using a multi-atlas label fusion method (Doshi et al., 2016). Volumes of segmented ROIs were used as input features for the NMI model. For voxelwise analyses, gray and white matter tissue density maps were computed using the RAVENS algorithm, a method extensively validated both using simulated data and on cross-sectional and longitudinal MRI images (Davatzikos et al., 2001; Resnick et al., 2003; Rozycki et al., 2018). The RAVENS algorithm involved tissue segmentation followed by nonlinear registration to atlas space for spatial normalization of volumetric images in a common atlas space. To maximize the accuracy of image registrations from subject to atlas space, we used study-specific intermediate atlases independently for PNC and PING, selected automatically as the single most-representative T1 image of each sample. RAVENS maps were normalized by individual intracranial volume to adjust for global differences in intracranial size, downsampled to 2 × 2 × 2 mm, and smoothed using a 2-mm-diameter Gaussian filter.
fMRI preprocessing.
Anatomical T1 images were registered to MNI152 Template using ANTS diffeomorphic SyN registration (Avants et al., 2008). All subjects' functional time-series were slice-time and motion corrected using MCFLIRT with six degrees of freedom (3 rotations and 3 translations). Corrected functional data were coregistered to anatomical T1 and bandpass filtered with passband frequency of 0.01–0.08 Hz. To reduce the influence of motion, we applied a validated confound regression procedure using a 36-parameter regression model (Satterthwaite et al., 2013). Participants who had a mean relative displacement value of >0.2 mm were excluded from subsequent analysis. ROIs segmented on the T1-weighted images were transferred to functional image space through linear alignment and average time series were calculated within each ROI. Connectivity matrices that represent the correlation of average time series between each ROI pair were calculated to be used as input to the functional NMI model.
Quality control protocol.
Structural image quality was verified using automated and manual quality control procedures. Images that failed the quality control protocol were removed from subsequent analyses. An image motion score was assigned to each individual based on visual inspection and agreement between two raters. Motion was corrected for in each study using a linear model with motion score, age, and sex included as covariates, so that all data included in the NMI score calculation were corrected for motion. fMRI image and processing quality was controlled using metrics that were automatically calculated during image preprocessing and registration. No associations were found between participants' mean relative displacement values and their SCP coefficients.
Data harmonization procedures.
Derived structural and functional imaging variables from PNC and PING were harmonized by correcting each variable independently for site effects using a linear model that included age, sex, and intracranial volume of the subjects as covariates. During the harmonization the model also corrected the values for motion using individual motion scores for structural and functional data. To eliminate potential site biases between the PNC data and the multisite schizophrenia dataset, we applied the ComBat harmonization procedure (Fortin et al., 2017).
Statistical analysis
NMI calculation.
We used a support vector machine (SVM) regression method for calculating the NMI (Smola and Schölkopf, 2004). The SVM model included a nonlinear radial based functions kernel. The structural model used gray and white matter region of interest volumes from each scan as input, and the functional model used connectivity matrices. Optimization of cost and kernel parameters for each model was done through grid search in a standard 10-fold nested cross-validation framework. Both structural and functional NMIs were trained using data from PNC participants. NMI scores for the PNC were derived using tenfold cross-validation. The NMI that was trained on the complete PNC set was applied to the PING and schizophrenia study cohorts to estimate NMI scores for these participants. Therefore, NMI scores for all participants were derived using a model fit to an external sample. NMI scores for each participant were computed by taking the residuals from a linear regression of predicted brain age on chronological age, described by Le et al. (2018). Model effectiveness was evaluated using the mean absolute error (MAE) and a Pearson correlation between chronological age and predicted brain age. Participants were grouped based on their estimated NMI score. Participants with NMI scores >70th percentile of the sample were placed in the high NMI group and those <30th percentile were placed in the low NMI group. The remaining participants were classified as having typical NMI.
Voxelwise analyses of structural group differences.
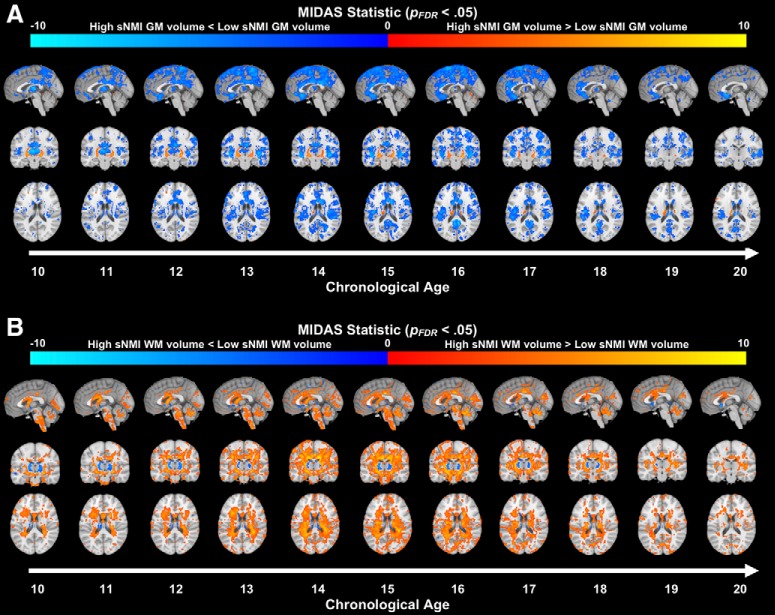
Voxelwise group comparisons were performed using RAVENS maps of the subjects. For the age-specific analyses a 4 year sliding age window was applied to the data, i.e., an age bin centered at age a included participants a ± 2. To maintain balanced sample sizes across age bins, comparisons were conducted for ages 10 through 20. Voxelwise comparisons were applied using a multivariate discriminative statistical mapping method (MIDAS; 28). MIDAS scans a voxelwise volumetric map using a set of overlapping neighborhoods and conducts regional discriminative analysis with least-squares SVM. A regional volumetric profile is identified which maximally discriminates between groups similar to searchlight analysis (Kriegeskorte et al., 2006). For each voxel, a statistic is calculated. p values corresponding to each statistic are then computed using an analytic approximation of permutation tests (Nichols and Holmes, 2002). Once p value maps were produced, FDR correction was applied. For the exploratory sNMI/fNMI multidimensional analysis, we first created a mask based on the MIDAS statistic, and then performed a standard effect size computation. Areas where the effect size was >0.3 are shown in Figure 6.
Figure 6.

Patterns of development in the typical sNMI group. Significant gray matter (A) and white matter (B) volume changes (pFDR < 0.05) across age. Each age bin centered at age p included participants of age p ± 2. Comparisons were made between participants within each age bin and those in the age bin reflecting p − 4 (e.g., those in the 13-year-old age bin were compared with those in the 9-year-old age bin). Blue/light blue overlay indicates regions where participants at age p exhibit less volume than those at age p − 4, whereas red/yellow overlay indicates regions where they exhibit more volume.
Computation of sparse connectivity patterns.
Because of the complexity of functional connectivity data, group comparisons for fMRI analysis were performed on a smaller set of variables derived using a validated method of sparsity-based dimensionality reduction (Eavani et al., 2015). This data-driven method takes as input a group of connectivity matrices and calculates a low-rank decomposition into a set of distinct functional subnetworks or “sparse connectivity patterns” (SCPs). Additionally, coefficients are produced for each participant that reflect the degree to which a respective connectivity pattern captured by an SCP is present in an individual. SCPs and their coefficients were computed using the SCPLearn toolbox v1.1.0. As with the previous analyses, SCPs were calculated using the PNC connectivity data as input. SCP coefficients were then computed for both the PNC and PING groups.
Other analyses.
R v3.5.1 was used to evaluate NMI model performance and perform group comparisons of NMI scores and SCP coefficients. Group comparison results were evaluated at p < 0.05. To control for multiple comparisons, p values were corrected using the false discovery rate (FDR).
Results
MRI data from participants who were imaged as part of the PNC were used to develop structural NMI (sNMI) and functional NMI (fNMI) models that were tested on unseen PNC subjects via 10-fold cross-validation, and then applied to participants from the PING study. Individual NMI scores were computed by taking the residuals of a linear regression of predicted age on chronological age. In this way, typical brain development would be represented by NMI scores close to 0. Brain age estimates older or younger than respective chronological age, i.e., positive and negative NMI scores, respectively, indicated deviation from typical brain development (Fig. 1). These thresholds were selected to identify atypical NMI scores while also preserving similar group sizes to maintain homogeneity of variance. Importantly, the NMIs were calculated using tenfold cross-validation, i.e., an individual's brain age score was always calculated from a model fit to a subset of the sample not including the individual.
Figure 1.
NMI group separation by NMI scores across dataset and imaging type. These scatterplots indicate the groups identified using structural NMI scores for PNC (A) and PING (B) and functional NMI scores for PNC (C) and PING (D). Those with NMI scores >70th percentile were placed in the high NMI group, those <30th percentile in the low NMI group, and the remaining in the typical NMI group. Percentile rankings were calculated separately for each sex.
sNMIs and fNMIs generalize across cohorts and sites
In the PNC, the cross-validated correlation between chronological age and sNMI score was r = 0.822 (MAE = 1.67). The sNMI model developed using the PNC as applied to the PING cohort resulted in a correlation of r = 0.872 (MAE = 1.59). To determine consistency in model performance between two separate sites, a comparison of regional volumetric patterns between sNMI groups was performed. For input, we used the RAVENS algorithm (Davatzikos et al., 2001) to produce gray and white matter tissue-density maps for each participant and applied voxelwise comparisons using MIDAS in which a regional volumetric profile is identified that maximally discriminates between groups (Varol et al., 2018). This comparison revealed that similar imaging patterns of brain maturation differentiated between those with high and low NMI scores, regardless of site (Fig. 2). These patterns were more significant in the PNC than in the PING cohort. This could be because of the pattern being identified using the PNC before being tested on PING but could also be a result of increased power afforded by the PNC's much larger sample.
Figure 2.
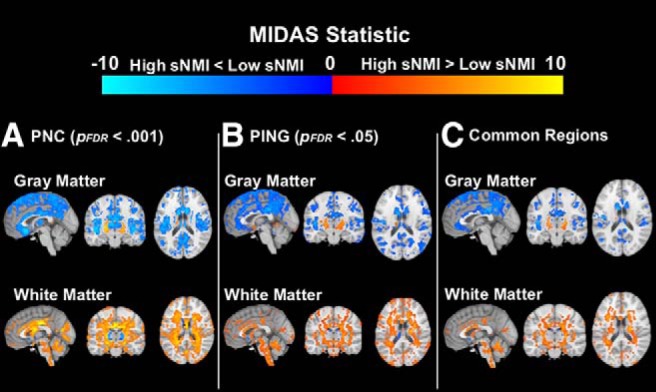
Significant regional volumetric differences between high and low sNMI groups by study. Significant regions are shown separately for the PNC (A; p < 0.001) and PING (B; p < 0.05) cohorts, along with the significant regions that were shared between the two sites (C). Blue/light blue overlay indicates regions where those with high sNMI scores exhibit less volume than those with low sNMI scores, whereas red/yellow overlay indicates regions where those with high sNMI scores exhibit more volume than those with low sNMI scores.
fMRI data were expected to produce less consistent functional development trajectories than the sMRI data due to the relatively higher between-subject variability in functional connectivity patterns. Despite the higher variability, fNMI prediction in the PING cohort was still comparable to the cross-validated results of the PNC group, indicating good generalizability of the fNMI across sites. In the PNC, the correlation between chronological age and fNMI score was r = 0.615 (MAE = 2.14). The fNMI model as applied to the PING cohort resulted in a correlation of r = 0.605 (MAE = 2.58).
To compare regional connectivity patterns of functional maturation across cohorts, we decomposed high-dimensional functional connectivity data into 10 SCPs (Eavani et al., 2015; Fig. 3). SCPs concisely capture brain connectivity in a manner that reveals heterogeneity across individuals while also identifying subsets within the data that exhibit similar patterns.
Figure 3.
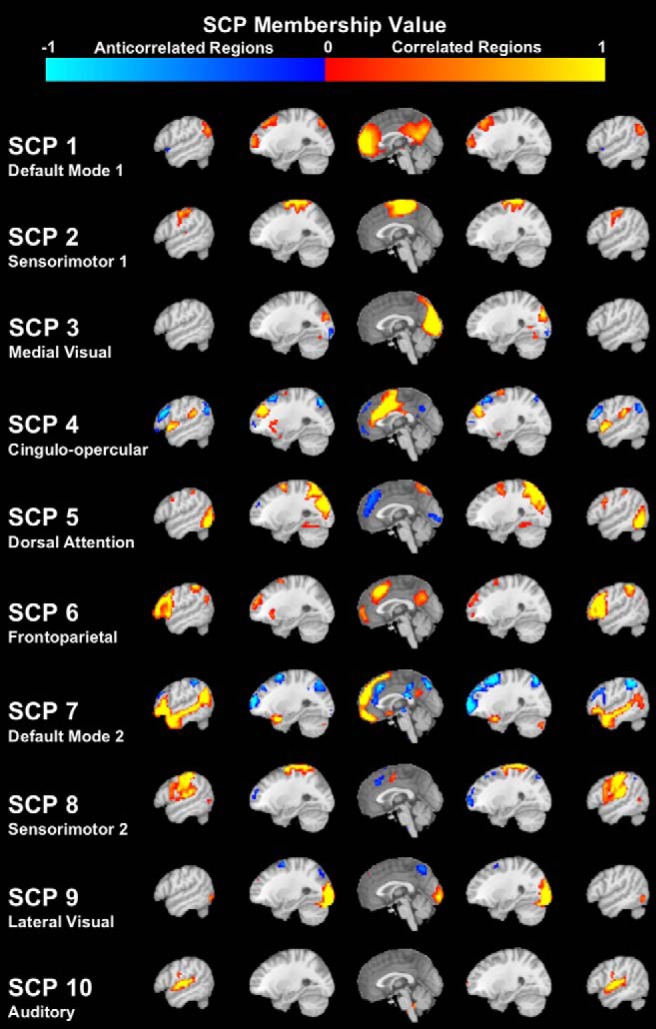
Sparse connectivity patterns extracted from the PNC resting-state fMRI data. Within each SCP, red-yellow shading indicates correlated regions within the network, while blue shading indicates anti-correlated regions within the network.
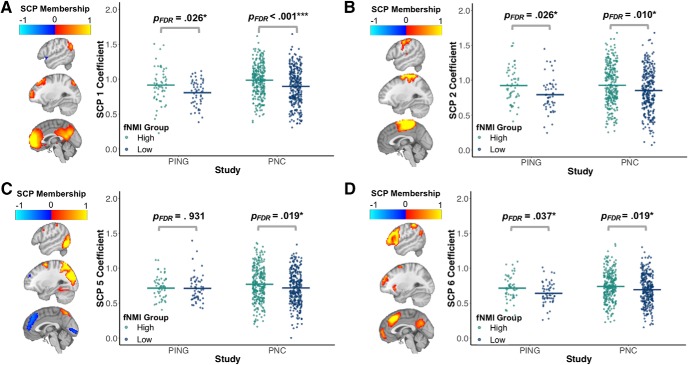
SCPs were derived from the PNC, and participants in both cohorts were assigned coefficients that indicated the degree to which each SCP is represented in that participant, with higher coefficients indicating greater component representation. Similar to the sNMI, we checked the fNMI model's consistency across sites by performing comparisons of the associations of these coefficients with the fNMI. These comparisons indicated general consistency in subnetwork presentation between the PING and PNC groups, i.e., the magnitude and direction of the difference between high and low fNMI groups was similar across sites in the majority of SCP components (Fig. 4). Although there was more between-site variance than produced by the comparison of sNMI groups, it was not unexpected given that the sNMI was highly successful in capturing brain maturation patterns, whereas the fNMI was less robust.
Figure 4.
Significant functional connectivity differences between the high and low fNMI groups in the PNC and PING cohorts. Four SCPs showed significant differences in connectivity between developmentally advanced and delayed groups. On the brain maps, red/yellow shading indicates correlated regions within the network, whereas blue shading indicates anti-correlated regions within the network. The segments on the jitterplot indicate the mean of each group. p values reflect the study-specific results of independent t tests between fNMI groups. There was relative consistency between the studies in the components that reflect the medial regions of the default mode network (A), the sensorimotor network (B), and the frontoparietal network (D). However, in the component that reflects the dorsal attention network (C), there were significant differences in the PNC, but not the PING cohort.
Both sNMIs and fNMIs characterize expected developmental trajectory
sNMI across age
To investigate the dynamic patterns that contribute to the sNMI estimation, we compared the RAVENS maps of high and low sNMI groups at each age, a. For a ∈ ℤ : 10 ≤ a ≤ 20, a 4-year sliding age window was applied to the data; each age bin centered at a included participants of age a ± 2. (for descriptive data for each age bin, see Table 1).
Table 1.
Demographic data for each NMI group by age window
| Age window | NMI group | sNMI |
fNMI |
||
|---|---|---|---|---|---|
| Age | Sex, M/F | Age | Sex, M/F | ||
| 10 | High | 10.15 ± 1.09 | 68/71 | 10.48 ± 1.09 | 28/30 |
| Low | 10.26 ± 1.08 | 68/71 | 9.92 ± 0.96 | 28/30 | |
| 11 | High | 10.99 ± 1.16 | 76/69 | 11.38 ± 1.15 | 33/36 |
| Low | 11.10 ± 1.18 | 76/70 | 10.78 ± 1.24 | 33/36 | |
| 12 | High | 12.13 ± 1.24 | 81/69 | 12.35± 1.08 | 41/39 |
| Low | 12.06 ± 1.14 | 81/69 | 12.09 ± 1.27 | 41/39 | |
| 13 | High | 13.27 ± 1.19 | 82/71 | 13.31 ± 1.08 | 44/46 |
| Low | 12.85 ± 1.15 | 82/71 | 13.13 ± 1.05 | 44/46 | |
| 14 | High | 14.35 ± 1.10 | 83/81 | 14.26 ± 1.13 | 50/57 |
| Low | 13.81 ± 1.19 | 83/81 | 14.02 ± 1.13 | 50/57 | |
| 15 | High | 15.21 ± 1.12 | 85/90 | 15.16 ± 1.06 | 58/67 |
| Low | 15.00 ± 1.18 | 85/91 | 15.12 ± 1.20 | 58/67 | |
| 16 | High | 16.09 ± 1.14 | 82/95 | 15.97 ± 1.11 | 59/75 |
| Low | 15.99 ± 1.20 | 82/95 | 16.15 ± 1.13 | 59/75 | |
| 17 | High | 16.90 ± 1.13 | 77/103 | 16.90 ± 1.22 | 63/81 |
| Low | 17.11 ± 1.24 | 77/103 | 16.95 ± 1.10 | 63/81 | |
| 18 | High | 17.68 ± 1.07 | 77/99 | 17.88 ± 1.14 | 63/81 |
| Low | 18.20 ± 1.23 | 77/99 | 17.99 ± 1.17 | 63/81 | |
| 19 | High | 18.53 ± 1.05 | 69/91 | 18.58 ± 0.93 | 56/75 |
| Low | 19.03 ± 1.08 | 69/91 | 19.06 ± 1.17 | 56/75 | |
| 20 | High | 19.25 ± 0.97 | 55/73 | 19.10 ± 0.84 | 45/59 |
| Low | 19.64 ± 1.00 | 55/73 | 19.76 ± 0.97 | 45/59 | |
Each age window centered at age a includes participants of age a ± 2.
We found a widespread decrease in volume over the course of adolescence consistent with previous developmental research (Giedd et al., 1999). Comparisons between the high and low groups by age indicated that regions with significant group differences vary across adolescence (Fig. 5) and follow the expected developmental trajectory; those with high sNMI scores exhibit reduced gray matter volume throughout the cortex compared with those with low sNMI scores. Volume differences in the posterior portion of the brain are most pronounced in young adolescents. These differences advance rostrally, with the frontal and temporal lobes exhibiting the most significant group differences during late adolescence and early adulthood.
Figure 5.
Significant volumetric differences (pFDR < 0.05) in gray matter (A) and white matter (B) between the high and low sNMI groups across adolescence. Blue/light blue overlay indicates regions where the high sNMI group exhibits less volume than the low sNMI group, whereas red/yellow overlay indicates regions where the high sNMI group exhibits more volume than the low sNMI group. Each age bin centered at age a includes participants of age a ± 2.
To determine whether this pattern of gray matter loss was consistent with traditional development, we compared gray matter volume between older and younger typical developers, i.e., those with sNMI scores between the 30th and 70th percentiles (n = 682). Comparison groups were separated in a manner consistent with the 4-year sliding window applied to the rapid and gradual development groups. Specifically, for a ∈ ℤ : 10 ≤ a ≤ 20, a 4-year sliding age window was applied to the data; each age bin centered at a included participants of age a ± 2. Starting at age 13, participants in each age bin were compared with the participants in the age a–4 (to avoid overlap between groups). We found similar neural signatures in the typical developers as those produced by the high/low sNMI comparisons, i.e., reduced gray matter volume that is noted in the posterior regions in the youngest participants and the frontal and superior temporal cortices in the oldest participants (Fig. 6A). Additionally, we see a general increase in white matter throughout the brain over the span of adolescence (Fig. 6B).
Although the spatial patterns are similar between the typical and high/low sNMI groups, the temporal pattern is shifted. Specifically, the neural signatures found in the typical group are most consistent with the high sNMI participants who are a few years older. This suggests that, generally, those with extreme sNMI scores are quantitatively precocious or delayed in their structural development rather than exhibiting a qualitatively different maturation pattern.
fNMI across age
To explore patterns of functional development and identify those that contribute most strongly to fNMI assignment, we performed additional analyses on SCPs across adolescence. In general, SCP coefficients were positively correlated with age, which indicates that connectivity within the networks represented by these SCPs increased over the course of adolescence. The SCP that captures medial regions of the DMN exhibited the strongest correlation with age, r(1142) = 0.19, reflecting enhanced within-network connectivity over the course of adolescence (Fair et al., 2008). Using the same sliding window applied to the structural data, we performed comparisons between the high and low fNMI groups across the span of adolescence (Table 1). Similar to the structural results, independent t tests indicated that networks with significant group differences vary across adolescence, with the high fNMI group exhibiting significantly stronger connectivity in the default mode, sensorimotor, dorsal attention, frontoparietal, and auditory networks (Fig. 7).
Figure 7.
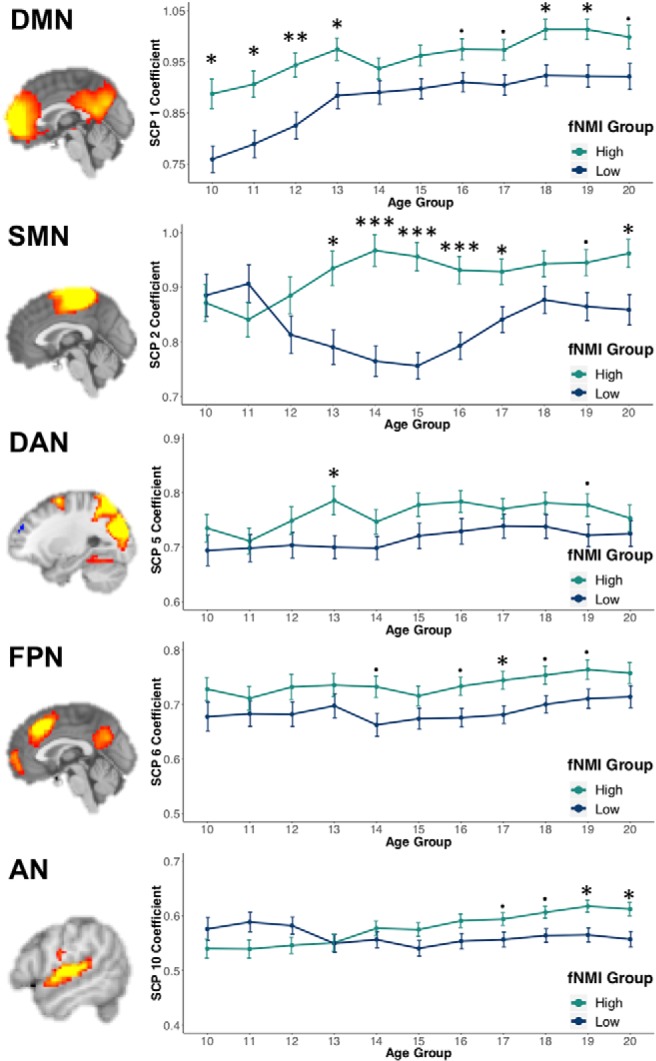
Mean SCP coefficient values for the significant networks in the high and low fNMI groups across adolescence. ***pFDR < 0.001, **pFDR < 0.01, *pFDR < 0.05. pFDR < 0.1. p values reflect the age-specific results of independent t-tests between fNMI groups. Error bars reflect the SEM. SMN, Sensorimotor network; DAN, dorsal attention network; FPN, frontoparietal network; AN, auditory network.
Multidimensionality of the neural maturation index
To further investigate the differences identified by our NMI model and illustrate the use of the brain development chart, we performed an exploratory analysis by grouping participants based upon their structural and functional NMI scores jointly. When stratified in this manner, each quadrant was found to possess unique neural signatures that separated them from those who were classified as both structurally and functionally typical (Fig. 8). Interestingly, differences captured by one modality were not necessarily associated with consistent differences in another; those who with high fNMI scores exhibited greater differences in structural development than those with low fNMI scores, suggesting an interaction between structural and functional differences.
Figure 8.
Results of the analyses between each quadrant and typical developers. All differences presented in this figure are those in which d ≥ 0.30. Quadrant membership was defined by both structural and functional NMI scores. Those with scores >70th percentile were placed in the high NMI group, those <30th percentile in the low NMI group, and the remaining in the typical NMI group. Participants from each quadrant were then compared with those who fell into both structural and functional typical groups. For the sMRI analyses, the highlighted regions reflect areas where Cohen's d ≥ 0.30. Yellow/red shading indicates regions where those in the respective quadrant exhibit greater gray matter volume than the typical group, whereas blue shading indicates regions where those in the respective quadrant exhibit less gray matter volume than the typical group. For the fMRI analyses, Sparse connectivity patterns that were significantly different between each quadrant and the typical group are shown, if significant differences exist. Yellow/red shading indicates regions that contribute to the SCP, whereas blue shading indicates regions that are anti-correlated with the SCP. The corresponding boxplots indicate the degree to which a respective connectivity pattern captured by an SCP is present in each group.
sNMI in youth with schizophrenia
We next investigated the indicative value of sNMI in identifying imaging patterns of brain change in disease. We used an independent cohort of schizophrenia patients (n = 43) and controls (n = 53) in the age range 16–22. Previous studies have reported that patients with schizophrenia exhibit accelerated brain aging compared with normal controls in their age group (Koutsouleris et al., 2014). Structural NMI scores for both schizophrenia and control groups were estimated using the original sNMI model derived from the PNC sample (MAE = 2.03) and sNMI scores were computed from a linear regression of estimated age on chronological age. An independent t test indicated that participants with schizophrenia had significantly higher sNMI scores (0.41 ± 1.82) than the controls (−0.33 ±2.06), tone-tailed(93.33) = 1.88, p = 0.031, d = 0.38 (Fig. 9A), suggesting that the schizophrenia patients had brains that looked older than the normative control group. This difference increased with subjects' age (Fig. 9B).
Figure 9.
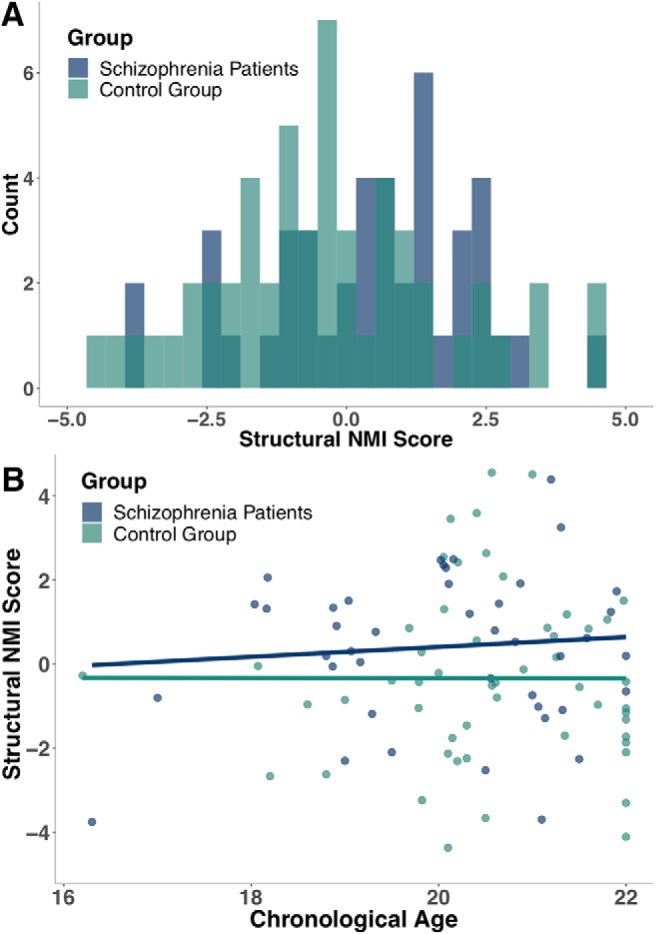
Differences in sNMI scores in schizophrenia patients and a control group. A, Schizophrenia patients were generally assigned higher NMI scores than those in the control group. B, The difference in sNMI between groups increased across adolescence, such that control participants exhibited brain patterns consistent with typical development regardless of age, whereas schizophrenia patients exhibited increasingly atypical neural development with age.
Discussion
In this study, we capitalized upon two large neurodevelopmental datasets and a multisite schizophrenia study to derive and validate a two-dimensional chart of brain development, and to investigate patterns of structure and function associated with typical and atypical brain development. Both structural and functional NMIs captured robust and reproducible development patterns, as demonstrated by within-dataset cross-validation and between-dataset application. Both general NMI performance and NMI group comparison results were highly consistent between studies, thereby bolstering our confidence that NMI scores generalize across sites, a critical factor for these imaging indices to be adopted broadly. Additionally, sNMI scores were associated with schizophrenia, which underlines the NMI's potential utility as a measure of clinical risk assessment. The results of our study establish a dimensional neuroimaging system for evaluating brain development concisely using machine learning methods. They also shed light on the spatial patterns of reduced cortical volume and concomitant functional connectivity changes.
When investigating the developmental signatures captured by the NMIs, we found a widespread reduction in gray matter, an increase in white matter, and a general increase in within-network functional connectivity. Moreover, analysis of independent age groups showed that the NMI captures age-specific patterns of change. Early adolescents displayed the greatest differences in the posterior parietal cortex, those in middle adolescence displayed the greatest differences in the parietal, frontal, and superior temporal cortex, and the oldest adolescents and young adults displayed the greatest differences in the orbitofrontal and superior temporal cortex. The posterior to anterior progression of volume loss is highly consistent with previous developmental research (Gogtay et al., 2004), was verified in the typical developers identified by our sNMI, and strongly supports the sNMI's characterization of normal development. Additionally, this result suggests that extreme developers identified by the sNMI are not generally exhibiting abnormal patterns of neural development but, rather, developing more rapidly or gradually than their peers. Notably, there were very few differences between the sNMI groups in lower-order regions such as the visual and somatosensory cortices. The most prominent differences between groups were in higher-order regions such as the prefrontal cortex, supporting the hypothesis that development occurs along an evolutionary hierarchy (Sotiras et al., 2017), with lower-order regions stabilizing during childhood and higher-order regions continuing to develop throughout adolescence. Additionally, the results suggest that higher-order regions have a greater potential for developmental variation, which emphasizes the need for individualized measures of brain maturation (Satterthwaite and Davatzikos, 2015).
To examine developmental patterns captured by the fNMI, we produced SCPs reflecting the default mode, dorsal attention, frontoparietal, salience, visual, auditory, and sensorimotor networks. The most robust correlations with age were observed in a component reflecting the DMN, which undergoes profound development during adolescence (Fair et al., 2008). Remarkably, the single component that was negatively correlated with age also reflected regions of the DMN. However, this component included the superior frontal cortex; connectivity between the hemispheres in this region decreases with age (Power et al., 2010). The high and low fNMI groups showed significant differences in 6 of the 10 networks. The DMN showed the greatest differences over the entire span of adolescence, while differences between other networks were confined to a limited age range, suggesting connectivity within the DMN as a key feature of our fNMI model, and likely one of the most consistent patterns of functional maturation during this age range. Interestingly, but not unexpectedly, the age-constrained functional differences are associated with corresponding structural differences between groups. For example, the auditory network shows no significant difference between groups until early adulthood during which we also see significant structural differences in the superior temporal lobe.
In our multidimensional NMI analysis, we found distinct differences between each quadrant and typically developing youth. In cases where differences are due to individual variance in brain maturation, we might expect structural and functional brain development to correspond, as in Quadrants 1 and 3 (Fig. 8). Less intuitive are those individuals whose structural and functional development do not align. Given that volume loss during adolescent brain development primarily reflects increased myelination (Natu et al., 2019), higher gray matter volume is presumably associated with either a less mature brain (i.e., less myelination) or an atypical source. Individuals in Quadrant 2 have more gray matter and increased functional connectivity, which could indicate normal myelination with a lack of corresponding gray matter loss. Individuals in Quadrant 4 have less gray matter and decreased functional connectivity, which might imply a decline in gray matter that is not associated with increased myelination, as is the case with patients with schizophrenia (Cannon et al., 1998). Future longitudinal studies will be able to investigate this result in greater depth.
Adolescence is a time of increased vulnerability to neural aberration (Paus et al., 2008). NMI scores reflect one's deviation from typical development and therefore could be suitable for determining an individual's need for observation or further assessment, especially if cognitive and behavioral characteristics suggest atypical development. As adolescence has been hypothesized to be a critical period in the development of schizophrenia (Selemon and Zecevic, 2015), sNMI scores obtained during this time may be useful in assessing risk. In our analysis, schizophrenia patients were assigned higher sNMI scores than the healthy control group, indicating that elevated sNMI scores could be a marker of this disorder. Notably, previous research has associated schizophrenia with particularly prominent volume loss in the superior temporal lobe and prefrontal cortex (Gur et al., 2000; Mathalon et al., 2001). In our nonclinical sample, these regions were identified by the sNMI as the primary areas of development in late adolescence and early adulthood. Indeed, the high sNMI group exhibited significantly lower volumes in these regions compared with the low group. This supports the idea that there is some overlap between typical age-related volume loss and pathological volume loss (Schnack et al., 2016), but adolescents with schizophrenia exhibit precocious development in these regions to the extent it becomes atypical. Although further research is necessary to determine the proposed NMI's effectiveness in identification of other clinical groups, previous research indicating atypical gray matter volume in other clinical cohorts is promising (Nakao et al., 2011; Zarei et al., 2011; Grieve et al., 2013).
In addition to producing an NMI that may be used to assess risk of atypical development, we also implemented a novel method for examining the developmental trajectory through the use of data-driven analysis. One criticism of brain age models is that they are a “black box” in which complex imaging data are provided as input and a single outcome is derived (Cole and Franke, 2017), with little understanding of how the input produces the outcome. In this analysis, we attempted to unpack the black box by comparing those classified as extremes (i.e., the high and low NMI groups). The goal was not to determine the existence of group differences, as we were aware that differences would inherently exist. Rather, we explored those differences that the model deemed most effective at estimating brain development. By splitting the groups into small age windows, we were able to identify age-specific patterns of deviation between high and low groups in a novel manner. Thus, we use an innovative data-driven method to identify the structural and functional signatures most indicative of typical aging.
Some limitations must be noted. First, previous research has established sex differences in both structural and functional development (Satterthwaite et al., 2015; Gennatas et al., 2017). Because of restrictions in sample size, we did not produce sex-specific NMIs and were unable to compute separate age-specific pattern analyses of group differences. We expect that a model incorporating both males and females will naturally seek common patterns of brain development or would produce a high MAE. The relatively low MAE scores produced by each of our NMI models suggest they successfully captured neural signatures shared between the sexes. Additionally, we attempted to further compensate for sex differences by (1) using sex-corrected imaging data in these analyses and (2) performing NMI group classification separately by sex. However, it is understood that future studies, if sufficiently powered, could derive sex-specific models that may yield more detailed results. Second, cross-sectional data were used to develop the NMI. However, longitudinal data would undoubtedly provide a richer picture of brain development across adolescence and would allow for the investigation of how one's NMI score changes with time and other factors, such as disease. NMI scores could potentially be used to show one's rate of decline or as an indicator of treatment effectiveness.
Together, our results delineate a dimensional imaging system of adolescent brain development based on structural and functional brain age, validated across multiple samples. Further analysis indicated that the signatures identified by the NMI for brain age prediction reflect key stages of the neural restructuring that occurs during adolescence, suggesting that the NMI is a concise summary of typical brain development. Additionally, we confirmed the NMI's efficacy as a marker for atypicality in patients diagnosed with schizophrenia. In sum, the NMI is a useful and interpretable metric by which neural development in late childhood and adolescence may be assessed.
Footnotes
This work was supported by NIH Grants R01EB022573 (C.D.), and R01MH112070 (C.D.), with additional support from R01MH120482 (T.D.S.), R01MH112847 (T.D.S.), R01MH113550 (T.D.S.), R01MH113565 (D.H.W.).
The authors declare no competing financial interests.
References
- Avants BB, Epstein CL, Grossman M, Gee JC (2008) Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal 12:26–41. 10.1016/j.media.2007.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon TD, van Erp TG, Huttunen M, Lönnqvist J, Salonen O, Valanne L, Poutanen VP, Standertskjöld-Nordenstam CG, Gur RE, Yan M (1998) Regional gray matter, white matter, and cerebrospinal fluid distributions in schizophrenic patients, their siblings, and controls. Arch Gen Psychiatry 55:1084–1091. 10.1001/archpsyc.55.12.1084 [DOI] [PubMed] [Google Scholar]
- Chung Y, Addington J, Bearden CE, Cadenhead K, Cornblatt B, Mathalon DH, McGlashan T, Perkins D, Seidman LJ, Tsuang M, Walker E, Woods SW, McEwen S, van Erp TGM, Cannon TD; North American Prodrome Longitudinal Study (NAPLS) Consortium; Pediatric Imaging, Neurocognition, and Genetics (PING) Study Consortium (2018) Use of machine learning to determine deviance in neuroanatomical maturity associated with future psychosis in youths at clinically high risk. JAMA Psychiatry 75:960–968. 10.1001/jamapsychiatry.2018.1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JH, Franke K (2017) Predicting age using neuroimaging: innovative brain ageing biomarkers. Trends Neurosci 40:681–690. 10.1016/j.tins.2017.10.001 [DOI] [PubMed] [Google Scholar]
- Davatzikos C, Genc A, Xu D, Resnick SM (2001) Voxel-based morphometry using the RAVENS maps: methods and validation using simulated longitudinal atrophy. Neuroimage 14:1361–1369. 10.1006/nimg.2001.0937 [DOI] [PubMed] [Google Scholar]
- Dennis EL, Jahanshad N, McMahon KL, de Zubicaray GI, Martin NG, Hickie IB, Toga AW, Wright MJ, Thompson PM (2013) Development of brain structural connectivity between ages 12 and 30: a 4-tesla diffusion imaging study in 439 adolescents and adults. Neuroimage 64:671–684. 10.1016/j.neuroimage.2012.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosenbach NUF, Nardos B, Cohen AL, Fair DA, Power JD, Church JA, Nelson SM, Wig GS, Vogel AC, Lessov-Schlaggar CN, Barnes KA, Dubis JW, Feczko E, Coalson RS, Pruett JR Jr, Barch DM, Petersen SE, Schlaggar BL (2010) Prediction of individual brain maturity using fMRI. Science 329:1358–1361. 10.1126/science.1194144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doshi J, Erus G, Ou Y, Gaonkar B, Davatzikos C (2013) Multi-atlas skull-stripping. Acad Radiol 20:1566–1576. 10.1016/j.acra.2013.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doshi J, Erus G, Ou Y, Resnick SM, Gur RC, Gur RE, Satterthwaite TD, Furth S, Davatzikos C; Alzheimer's Disease Neuroimaging Initiative (2016) MUSE: MUlti-atlas region segmentation utilizing ensembles of registration algorithms and parameters, and locally optimal atlas selection. Neuroimage 127:186–195. 10.1016/j.neuroimage.2015.11.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eavani H, Satterthwaite TD, Filipovych R, Gur RE, Gur RC, Davatzikos C (2015) Identifying sparse connectivity patterns in the brain using resting-state fMRI. Neuroimage 105:286–299. 10.1016/j.neuroimage.2014.09.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erus G, Battapady H, Satterthwaite TD, Hakonarson H, Gur RE, Davatzikos C, Gur RC (2015) Imaging patterns of brain development and their relationship to cognition. Cereb Cortex 25:1676–1684. 10.1093/cercor/bht425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair DA, Cohen AL, Dosenbach NU, Church JA, Miezin FM, Barch DM, Raichle ME, Petersen SE, Schlaggar BL (2008) The maturing architecture of the brain's default network. Proc Natl Acad Sci U S A 105:4028–4032. 10.1073/pnas.0800376105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin JP, Parker D, Tunç B, Watanabe T, Elliott MA, Ruparel K, Roalf DR, Satterthwaite TD, Gur RC, Gur RE, Schultz RT, Verma R, Shinohara RT (2017) Harmonization of multi-site diffusion tensor imaging data. Neuroimage 161:149–170. 10.1016/j.neuroimage.2017.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke K, Ziegler G, Klöppel S, Gaser C (2010) Estimating the age of healthy subjects from T1-weighted MRI scans using kernel methods: exploring the influence of various parameters. Neuroimage 50:883–892. 10.1016/j.neuroimage.2010.01.005 [DOI] [PubMed] [Google Scholar]
- Franke K, Luders E, May A, Wilke M, Gaser C (2012) Brain maturation: predicting individual BrainAGE in children and adolescents using structural MRI. Neuroimage 63:1305–1312. 10.1016/j.neuroimage.2012.08.001 [DOI] [PubMed] [Google Scholar]
- Gennatas ED, Avants BB, Wolf DH, Satterthwaite TD, Ruparel K, Ciric R, Hakonarson H, Gur RE, Gur RC (2017) Age-related effects and sex differences in gray matter density, volume, mass, and cortical thickness from childhood to young adulthood. J Neurosci 37:5065–5073. 10.1523/JNEUROSCI.3550-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedd JN, Blumenthal J, Jeffries NO, Castellanos FX, Liu H, Zijdenbos A, Paus T, Evans AC, Rapoport JL (1999) Brain development during childhood and adolescence: a longitudinal MRI study. Nat Neurosci 2:861–863. 10.1038/13158 [DOI] [PubMed] [Google Scholar]
- Giorgio A, Watkins KE, Douaud G, James AC, James S, De Stefano N, Matthews PM, Smith SM, Johansen-Berg H (2008) Changes in white matter microstructure during adolescence. Neuroimage 39:52–61. 10.1016/j.neuroimage.2007.07.043 [DOI] [PubMed] [Google Scholar]
- Giorgio A, Watkins KE, Chadwick M, James S, Winmill L, Douaud G, De Stefano N, Matthews PM, Smith SM, Johansen-Berg H, James AC (2010) Longitudinal changes in grey and white matter during adolescence. Neuroimage 49:94–103. 10.1016/j.neuroimage.2009.08.003 [DOI] [PubMed] [Google Scholar]
- Gogtay N, Giedd JN, Lusk L, Hayashi KM, Greenstein D, Vaituzis AC, Nugent TF 3rd, Herman DH, Clasen LS, Toga AW, Rapoport JL, Thompson PM (2004) Dynamic mapping of human cortical development during childhood through early adulthood. Proc Natl Acad Sci U S A 101:8174–8179. 10.1073/pnas.0402680101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieve SM, Korgaonkar MS, Koslow SH, Gordon E, Williams LM (2013) Widespread reductions in gray matter volume in depression. Neuroimage Clin 3:332–339. 10.1016/j.nicl.2013.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur RE, Cowell PE, Latshaw A, Turetsky BI, Grossman RI, Arnold SE, Bilker WB, Gur RC (2000) Reduced dorsal and orbital prefrontal gray matter volumes in schizophrenia. Arch Gen Psychiatry 57:761–768. 10.1001/archpsyc.57.8.761 [DOI] [PubMed] [Google Scholar]
- Habes M, Janowitz D, Erus G, Toledo JB, Resnick SM, Doshi J, Van der Auwera S, Wittfeld K, Hegenscheid K, Hosten N, Biffar R, Homuth G, Völzke H, Grabe HJ, Hoffmann W, Davatzikos C (2016) Advanced brain aging: relationship with epidemiologic and genetic risk factors and overlap with Alzheimer disease atrophy patterns. Transl Psychiatry 6:e775. 10.1038/tp.2016.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajek T, Franke K, Kolenic M, Capkova J, Matejka M, Propper L, Uher R, Stopkova P, Novak T, Paus T, Kopecek M, Spaniel F, Alda M (2019) Brain age in early stages of bipolar disorders or schizophrenia. Schizophr Bull 45:190–198. 10.1093/schbul/sbx172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jernigan TL, Brown TT, Hagler DJ Jr, Akshoomoff N, Bartsch H, Newman E, Thompson WK, Bloss CS, Murray SS, Schork N, Kennedy DN, Kuperman JM, McCabe C, Chung Y, Libiger O, Maddox M, Casey BJ, Chang L, Ernst TM, Frazier JA, et al. (2016) The pediatric imaging, neurocognition, and genetics (PING) data repository. Neuroimage 124:1149–1154. 10.1016/j.neuroimage.2015.04.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsouleris N, Davatzikos C, Borgwardt S, Gaser C, Bottlender R, Frodl T, Falkai P, Riecher-Rössler A, Möller HJ, Reiser M, Pantelis C, Meisenzahl E (2014) Accelerated brain aging in schizophrenia and beyond: a neuroanatomical marker of psychiatric disorders. Schizophr Bull 40:1140–1153. 10.1093/schbul/sbt142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegeskorte N, Goebel R, Bandettini P (2006) Information-based functional brain mapping. Proc Natl Acad Sci U S A 103:3863–3868. 10.1073/pnas.0600244103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TT, Kuplicki RT, McKinney BA, Yeh HW, Thompson WK, Paulus MP; Tulsa 1000 Investigators (2018) A nonlinear simulation framework supports adjusting for age when analyzing BrainAGE. Front Aging Neurosci 10:317. 10.3389/fnagi.2018.00317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathalon DH, Sullivan EV, Lim KO, Pfefferbaum A (2001) Progressive brain volume changes and the clinical course of schizophrenia in men. Arch Gen Psychiatry 58:148–157. 10.1001/archpsyc.58.2.148 [DOI] [PubMed] [Google Scholar]
- Nakao T, Radua J, Rubia K, Mataix-Cols D (2011) Gray matter volume abnormalities in ADHD: voxel-based meta-analysis exploring the effects of age and stimulant medication. Am J Psychiatry 168:1154–1163. 10.1176/appi.ajp.2011.11020281 [DOI] [PubMed] [Google Scholar]
- Natu VS, Gomez J, Barnett M, Jeska B, Kirilina E, Jaeger C, Zhen Z, Cox S, Weiner KS, Weiskopf N, Grill-Spector K (2019) Apparent thinning of human visual cortex during childhood is associated with myelination. Proc Natl Acad Sci U S A 116:20750–20759. 10.1073/pnas.1904931116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols TE, Holmes AP (2002) Nonparametric permutation tests for functional neuroimaging: a primer with examples. Hum Brain Mapp 15:1–25. 10.1002/hbm.1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paus T, Keshavan M, Giedd JN (2008) Why do many psychiatric disorders emerge during adolescence? Nat Rev Neurosci 9:947–957. 10.1038/nrn2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Fair DA, Schlaggar BL, Petersen SE (2010) The development of human functional brain networks. Neuron 67:735–748. 10.1016/j.neuron.2010.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick SM, Pham DL, Kraut MA, Zonderman AB, Davatzikos C (2003) Longitudinal magnetic resonance imaging studies of older adults: a shrinking brain. J Neurosci 23:3295–3301. 10.1523/JNEUROSCI.23-08-03295.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozycki M, Satterthwaite TD, Koutsouleris N, Erus G, Doshi J, Wolf DH, Fan Y, Gur RE, Gur RC, Meisenzahl EM, Zhuo C, Yin H, Yan H, Yue W, Zhang D, Davatzikos C (2018) Multisite machine learning analysis provides a robust structural imaging signature of schizophrenia detectable across diverse patient populations and within individuals. Schizophr Bull 44:1035–1044. 10.1093/schbul/sbx137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite TD, Davatzikos C (2015) Towards an individualized delineation of functional neuroanatomy. Neuron 87:471–473. 10.1016/j.neuron.2015.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite TD, Elliott MA, Gerraty RT, Ruparel K, Loughead J, Calkins ME, Eickhoff SB, Hakonarson H, Gur RC, Gur RE, Wolf DH (2013) An improved framework for confound regression and filtering for control of motion artifact in the preprocessing of resting-state functional connectivity data. Neuroimage 64:240–256. 10.1016/j.neuroimage.2012.08.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite TD, Elliott MA, Ruparel K, Loughead J, Prabhakaran K, Calkins ME, Hopson R, Jackson C, Keefe J, Riley M, Mentch FD, Sleiman P, Verma R, Davatzikos C, Hakonarson H, Gur RC, Gur RE (2014) Neuroimaging of the philadelphia neurodevelopmental cohort. Neuroimage 86:544–553. 10.1016/j.neuroimage.2013.07.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite TD, Wolf DH, Roalf DR, Ruparel K, Erus G, Vandekar S, Gennatas ED, Elliott MA, Smith A, Hakonarson H, Verma R, Davatzikos C, Gur RE, Gur RC (2015) Linked sex differences in cognition and functional connectivity in youth. Cereb Cortex 25:2383–2394. 10.1093/cercor/bhu036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite TD, Connolly JJ, Ruparel K, Calkins ME, Jackson C, Elliott MA, Roalf DR, Hopson R, Prabhakaran K, Behr M, Qiu H, Mentch FD, Chiavacci R, Sleiman PMA, Gur RC, Hakonarson H, Gur RE (2016) The Philadelphia neurodevelopmental cohort: a publicly available resource for the study of normal and abnormal brain development in youth. Neuroimage 124:1115–1119. 10.1016/j.neuroimage.2015.03.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnack HG, van Haren NE, Nieuwenhuis M, Hulshoff Pol HE, Cahn W, Kahn RS (2016) Accelerated brain aging in schizophrenia: a longitudinal pattern recognition study. Am J Psychiatry 173:607–616. 10.1176/appi.ajp.2015.15070922 [DOI] [PubMed] [Google Scholar]
- Selemon LD, Zecevic N (2015) Schizophrenia: a tale of two critical periods for prefrontal cortical development. Transl Psychiatry 5:e623. 10.1038/tp.2015.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahab S, Mulsant BH, Levesque ML, Calarco N, Nazeri A, Wheeler AL, Foussias G, Rajji TK, Voineskos AN (2019) Brain structure, cognition, and brain age in schizophrenia, bipolar disorder, and healthy controls. Neuropsychopharmacology 44:898–906. 10.1038/s41386-018-0298-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC (1998) A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging 17:87–97. 10.1109/42.668698 [DOI] [PubMed] [Google Scholar]
- Smola AJ, Schölkopf B (2004) A tutorial on support vector regression. Stat Comput 14:199–222. 10.1023/B:STCO.0000035301.49549.88 [DOI] [Google Scholar]
- Sotiras A, Toledo JB, Gur RE, Gur RC, Satterthwaite TD, Davatzikos C (2017) Patterns of coordinated cortical remodeling during adolescence and their associations with functional specialization and evolutionary expansion. Proc Natl Acad Sci U S A 114:3527–3532. 10.1073/pnas.1620928114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varol E, Sotiras A, Davatzikos C (2018) MIDAS: regionally linear multivariate discriminative statistical mapping. Neuroimage 174:111–126. 10.1016/j.neuroimage.2018.02.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarei M, Mataix-Cols D, Heyman I, Hough M, Doherty J, Burge L, Winmill L, Nijhawan S, Matthews PM, James A (2011) Changes in gray matter volume and white matter microstructure in adolescents with obsessive-compulsive disorder. Biol Psychiatry 70:1083–1090. 10.1016/j.biopsych.2011.06.032 [DOI] [PubMed] [Google Scholar]