Abstract
Genome-wide association studies (GWAS) have identified more than 250 loci associated with body mass index (BMI) and obesity. However, post-GWAS functional genomic investigations have been inadequate for understanding how these genetic loci physiologically impact disease development. We performed a PCR-free expression assay targeting genes located nearby the GWAS-identified SNPs associated with BMI/obesity in a large panel of human tissues. Furthermore, we analyzed several genetic risk scores (GRS) summing GWAS-identified alleles associated with increased BMI in 4,236 individuals. We found that the expression of BMI/obesity susceptibility genes is strongly enriched in the brain, especially in the insula (p = 4.7×10-9) and substantia nigra (p = 6.8×10-7), which are two brain regions involved in addiction and reward. Inversely, we found that top obesity/BMI-associated loci, including FTO, showed the strongest gene expression enrichment in the two brain regions. Our data suggest for the first time that the susceptibility genes for common obesity may have an effect on eating addiction and reward behaviors through their high expression in substantia nigra and insula, i.e. a different pattern from monogenic obesity genes that act in the hypothalamus and cause hyperphagia. Further epidemiological studies with relevant food behavior phenotypes are necessary to confirm these findings.
Common obesity is a polygenic disorder, resulting from a complex interplay between genetics and environmental factors. Since 2007, genome-wide association studies (GWAS) utilized DNA to assess millions of single nucleotide polymorphisms (SNPs) in large cohorts and have identified >250 loci associated with body mass index (BMI)1. However, elucidating the functional link between the causal variant and BMI has been a challenge.
Recently, we found that the expression of genes located in closed proximity to type 2 diabetes GWAS SNPs was significantly enriched in insulin-secreting beta cells, but not in insulin sensitive tissues2. This was performed using PCR-free technology in various human tissues, which has the advantage of avoiding genetic material amplification2. This study confirmed that the pathophysiology of common type 2 diabetes caused by the genetic variants acts through insulin-secreting cells2. In obesity, by using public gene expression microarray data, a recent study has demonstrated that the expression of genes, located nearby GWAS-identified BMI-associated SNPs, is significantly enriched in the central nervous system3. These findings were consistent with studies from monogenic forms of obesity, as the implicated genes, like MC4R, POMC, and LEPR, play a key role in the central nervous system, regulating appetite and body weight. Here, using the same strategy as previously implemented2, we aimed to confirm and refine this enrichment of BMI-associated gene expression in the central nervous system. In addition, we used the French D.E.S.I.R. study to assess the association between BMI and several genetic risk scores (GRS), including alleles increasing BMI closest to the genes with the highest expression level and specificity in two brain regions of interest.
Methods
Expression study of obesity/BMI susceptibility genes
The expression study of obesity/BMI susceptibility genes was performed using the NanoString technology (Seattle, WA, USA) which is a multiplex digital quantification of nucleic acids, in a large panel of human tissues, as previously described2. Our panel included human RNA from colon, small intestine, liver, kidney, adipose tissue, primary pre-adipocytes, mature adipocytes, lung, skeletal muscle, heart, brain, substantia nigra, hippocampus, dorsal root ganglion, insula, hypothalamus, pituitary gland, caudate nucleus, frontal lobe, pancreatic islets, pancreatic beta cells, exocrine pancreas and the pancreatic beta-cell line EndoC-βH12. The probes were designed to target 111 genes, including five housekeeping genes for normalization2 and 106 susceptibility genes for obesity/BMI previously published in GWAS (Supplementary Table 1)3–11. Notably, as the probes were designed in 2016, we were unable to assess novel susceptibility genes for obesity/BMI that were identified in the latest GWAS1,12. The expression profiles of obesity/BMI susceptibility genes were analyzed using heat maps, as previously described2. We established a threshold to classify the expression enrichment of obesity/BMI susceptibility genes in each tissue of the panel2. This threshold was defined as the average gene expression across all tissues, plus one standard deviation (SD). A Fisher's exact test was applied to test the expression enrichment of the gene set in each tissue of the panel2.
Calculation of GRS
Five GRS were calculated as the sum of five alleles increasing BMI closest to the five genes with the highest expression level and specificity in human insula and substantia nigra (top-1→top-5 genes, top-6→top-10 genes, top-11→top-15 genes, top-16→top-20 genes and top-21→top-25 genes; Supplementary Table 2).
Study population
We used genetic and clinical data from the D.E.S.I.R. cohort13. We analyzed 4,236 European-Caucasian individuals (mean±SD age of 47±10 years; mean±SD BMI of 24.7±3.8 kg/m2). The ethnicity of the participants was assessed as previously described13. All participants provided written informed consent and the study protocol was approved by the Ethics Committee for the Protection of Subjects for Biomedical Research of Bicêtre Hospital (France).
Genotyping
The genotyping of the 25 SNPs included in our GRS was performed using Metabochip DNA arrays (Illumina), as previously described13. The genotyping success rate in the study was >99% for the 25 SNPs, and no departures from Hardy-Weinberg equilibrium were observed. We excluded from the analysis the individuals with ≥2 missing SNP calls. For individuals with only one missing SNP call, the missing alleles were assigned to the most common allele.
Statistical analyses
The association between each of the five GRS and BMI was assessed through a linear regression adjusted for age and sex, using the IBM SPSS software (version 22).
Results
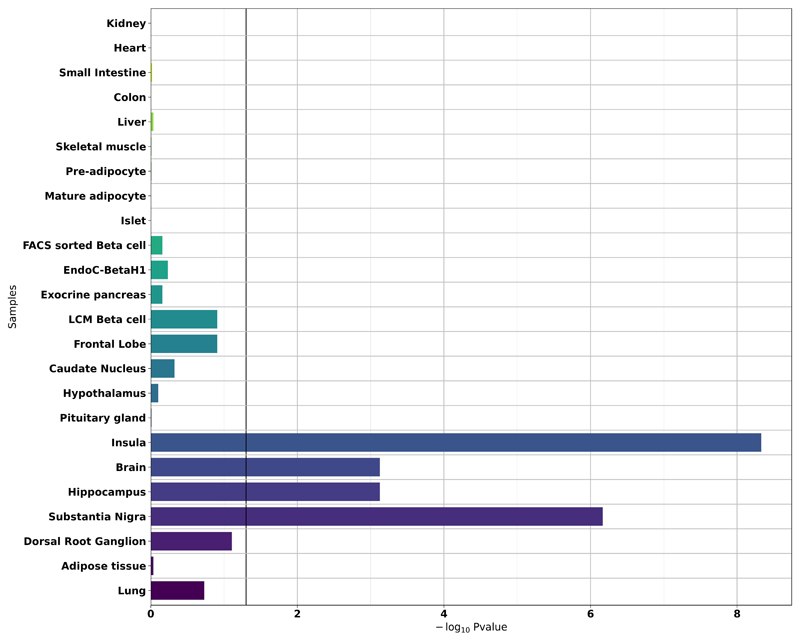
We assessed the expression of 106 genes (Supplementary Table 1) located close to GWAS-identified SNPs associated with obesity/BMI in our panel of human tissues using the PCR-free NanoString technology. The heat map in Supplementary Figure 1 shows the expression profile of the 106 susceptibility genes for obesity and/or BMI in the panel. The expression of the 106 genes was strongly enriched in the insula (p=4.7×10-9) and substantia nigra (p=6.8×10-7) (Figure 1). We identified two other tissues with a less significant expression enrichment pattern for BMI/obesity susceptibility genes, namely the whole brain (p=7.5×10-4) and hippocampus (p=7.5×10-4) (Figure 1).
Figure 1. Enrichment analysis of the expression of 106 susceptibility genes for obesity/BMI in the panel of human organs, tissues and cells.
The black vertical line denotes a p-value of 0.05.
EndoC-βH1, human pancreatic beta-cell line; FACS, fluorescence-activated cell sorting; LCM, laser capture microdissection.
In order to correlate our enrichment expression analysis and genetic association data, we next generated five GRS, each summing five obesity/BMI associated alleles (found by GWAS) closest to the five genes with the highest expression level and specificity in human insula and substantia nigra. Namely, GRS-1 included alleles nearby top-1 to top-5 insula/substantia nigra enriched genes, GRS-2 included alleles nearby top-6 to top-10 genes, GRS-3 included alleles nearby top-11 to top-15 genes, GRS-4 included alleles nearby top-16 to top-20 genes and GRS-5 included alleles nearby top-21 to top-25 genes (Supplementary Table 2). We assessed the association of each of these GRS with BMI in 4,236 European individuals. We found that GRS-1 (including effect alleles close to CADM2, NUDT3, FTO, FAIM2 and NRXN3) had the strongest association with an increased BMI (per additional risk allele: β=0.188±0.039 kg/m2, p=1.3×10-6; Table 1). This was followed by GRS-3 (including effect alleles close to NAV1, NEGR1, RALYL, LRP1B and STXBP6; β=0.154±0.040 kg/m2, p=1.5×10-4; Table 1) and GRS-2 (including effect alleles close to C6orf106, ETV5, ERBB4, GRID1 and HIP1; β=0.138±0.044 kg/m2, p=0.0016; Table 1). GRS-4 and GRS-5 did not associate with BMI in our study (p>0.01; Table 1). Of note, FTO SNP alone had the following effect on BMI in our study: β=0.265±0.081 kg/m2, p=0.0010.
Table 1. Association between several GRS and BMI in the D.E.S.I.R. study, adjusted for age and sex.
| SNPs increasing BMI | Closest genes | n | Per allele effect size ± SE | p-value | |
|---|---|---|---|---|---|
| GRS-1 | rs13078960 | CADM2 | 4,236 | 0.188 ± 0.039 | 1.3 × 10-6 |
| rs206936 | NUDT3 | ||||
| rs1558902 | FTO | ||||
| rs7138803 | FAIM2 | ||||
| rs7141420 | NRXN3 | ||||
| GRS-2 | rs205262 | C6orf106 | 4,236 | 0.138 ± 0.044 | 1.6 × 10-3 |
| rs1516725 | ETV5 | ||||
| rs7599312 | ERBB4 | ||||
| rs7899106 | GRID1 | ||||
| rs1167827 | HIP1 | ||||
| GRS-3 | rs2820292 | NAV1 | 4,235 | 0.154 ± 0.040 | 1.5 × 10-4 |
| rs3101336 | NEGR1 | ||||
| rs2033732 | RALYL | ||||
| rs2121279 | LRP1B | ||||
| rs10132280 | STXBP6 | ||||
| GRS-4 | rs11727676 | HHIP | 4,236 | 0.055 ± 0.048 | 0.25 |
| rs11191560 | NT5C2 | ||||
| rs2075650 | TOMM40 | ||||
| rs1000940 | RABEP1 | ||||
| rs1528435 | UBE2E3 | ||||
| GRS-5 | rs9400239 | FOXO3 | 4,235 | 0.072 ± 0.034 | 0.037 |
| rs657452 | AGBL4 | ||||
| rs11583200 | ELAVL4 | ||||
| rs887912 | FANCL | ||||
| rs16951275 | MAP2K5 |
BMI, body mass index; GRS, genetic risk score; SE, standard error; SNP, single nucleotide polymorphism.
Discussion
The physiopathology of obesity is still an area of immense controversy. Although there is a general agreement that the heritability of obesity is substantial, its contribution to energy imbalance remains largely elusive. Since the 1990s, considerable progress has been made in the elucidation of the most extreme forms of early-onset obesity, which are often monofactorial. These monofactorial forms of obesity are primarily caused by defects in appetite regulation, as most of the monogenic obesity genes are part of the leptin-melanocortin pathway that mainly acts within the hypothalamus. However, it remains unknown whether these mechanisms of disease are relevant to common obesity. Of note, under clinical protocols, unlike patients with monogenic obesity who display constant hyperphagia, obese individuals do not exhibit free-running behavior14. Conversely, it has been demonstrated that eating addiction strongly contributes to obesity15,16.
Our results have unambiguously shown that the expression of obesity/BMI susceptibility genes is strongly enriched in two regions of the brain, the insula and substantia nigra, which are related to addiction and reward. Moreover, the strongest gene enrichment in those two brain regions concerns genes within loci that have the strongest impact on BMI, including FTO, which is the most potent locus in common obesity. The insula (or insular cortex) is located within the cerebral cortex and is subdivided into two main groups, the granular and agranular insula17. The agranular insula likely plays a key role in emotion and motivation, in addition to addiction and the conscious urges to take drugs17. This is due to the strong dopaminergic innervation within the agranular insula and its high density of D1 dopamine receptors, endogenous opioids and µ-opioid receptors17. Moreover, the substantia nigra, which is part of the basal ganglia located in the midbrain, also plays a role in addiction, motivation and reward-seeking behavior and displays a high density of dopaminergic neurons18,19. Previous studies showed that the obesity/BMI-associated locus in FTO (i.e. the top 3 gene enriched in insula and substantia nigra in the present study) impacts eating disorders20,21, including binge eating, which was found to disrupt dopaminergic signaling in the human brain22.
The main limitation of our study is that we predominantly focused on genes closest to the BMI-associated SNPs found by GWAS. This approach excluded all genes located in the same topologically associating domains (TADs) as the BMI-associated SNPs, namely the genes that may in theory be regulated by these SNPs, and expression quantitative trait loci (eQTL) particularly within brain regions. However, using publicly available data from the Genotype-Tissue Expression (GTEx) project, in 1,497 human brain tissue samples, out of the 25 BMI-associated SNPs listed in the five GRS, only four SNPs had a significant effect on mRNA expression, thus, limiting the second option. It is important to note that when we analyzed all available tissues (i.e. 10,294 human tissue samples with donor genotype), we found no significant eQTLs for 15 of the 25 BMI-associated SNPs listed in the five GRS. Of the remaining 10 BMI-associated SNPs, eight SNPs (rs206936, rs1558902, rs205262, rs7599312, rs3101336, rs11191560, rs1000940, rs16951275) had a significant effect on the expression of their nearest gene (NUDT2, FTO, C6orf106, ERBB4, NEGR1, NT5C2, RABEP1, MAP2K5, respectively) in at least one of the assessed human tissues. These results were in line with our strategy, which focused on investigating the expression of genes closest to BMI-associated SNPs. Assessing the expression of all genes located in the same TAD as the BMI-associated SNPs would have reduced or eliminated the signal of expression enrichment in brain regions. Of note, our library also included IRX5 located in the same TAD as FTO, which was suggested to play a role in weight regulation through adipocyte thermogenesis23. The highest expression of IRX5 was actually found in pancreatic beta cells, exocrine pancreas, lung and heart, and to a lesser extent in the brain areas (Supplementary Figure 2).
In conclusion, the present study suggests that the genetic component of common obesity has an impact on eating addiction and reward behaviors in the central nervous system. Further epidemiological studies with relevant food behavior phenotypes are necessary to confirm these findings.
Supplementary Material
Acknowledgements
We thank Endocells for providing the pancreatic beta cell line, EndoC-βH1. The GTEx Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from the GTEx Portal on 11/23/2018. This work was supported by grants from the French National Research Agency (ANR-10-LABX-46 [European Genomics Institute for Diabetes] and ANR-10-EQPX-07-01 [LIGAN-PM], to PF), from the European Research Council (ERC GEPIDIAB – 294785, to PF; ERC Reg-Seq – 715575, to AB), from FEDER (to PF) and from the ‘Région Nord Pas-de-Calais’ (to PF and to FKN). AB was supported by Inserm. The D.E.S.I.R. study has been funded by Inserm contracts with Caisse nationale de l'assurance maladie des travailleurs salariés (CNAMTS), Lilly, Novartis Pharma, and Sanofi-Aventis; Inserm (Réseaux en Santé Publique, Interactions entre les déterminants de la santé, Cohortes Santé TGIR 2008); the Association Diabète Risque Vasculaire; the Fédération Française de Cardiologie; La Fondation de France; the Association de Langue Française pour l'Etude du Diabète et des Maladies Métaboliques (ALFEDIAM)/Société Francophone de Diabétologie (SFD); the Office national interprofessionnel des vins (ONIVINS); Ardix Medical; Bayer Diagnostics; Becton Dickinson; Cardionics; Merck Santé; Novo Nordisk; Pierre Fabre; Roche; Topcon. The D.E.S.I.R. Study Group includes: Inserm U1018: B. Balkau, P. Ducimetière, E. Eschwège; Inserm U367: F. Alhenc-Gelas; CHU D’Angers: Y Gallois, A. Girault; Centre de Recherche des Cordeliers, Inserm U1138, Bichat Hospital: F. Fumeron, M. Marre, R. Roussel; CHU de Rennes: F. Bonnet; CNRS UMR8199, Lille: A. Bonnefond, P. Froguel; Centres d’Examens de Santé: Alençon, Angers, Blois, Caen, Chateauroux, Chartres, Cholet, Le Mans, Orléans, Tours; Institute de Recherche Médecine Générale: J. Cogneau; General practitioners of the region; Institute Inter-Regional pour la Santé: C. Born, E. Caces, M. Cailleau, O Lantieri, J.G. Moreau, F. Rakotozafy, J. Tichet, S. Vol.
Footnotes
Conflict of Interest
The authors declare to have no conflict of interest.
References
- 1.Turcot V, Lu Y, Highland HM, Schurmann C, Justice AE, Fine RS, et al. Protein-altering variants associated with body mass index implicate pathways that control energy intake and expenditure in obesity. Nat Genet. 2018;50:26–41. doi: 10.1038/s41588-017-0011-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ndiaye FK, Ortalli A, Canouil M, Huyvaert M, Salazar-Cardozo C, Lecoeur C, et al. Expression and functional assessment of candidate type 2 diabetes susceptibility genes identify four new genes contributing to human insulin secretion. Mol Metab. 2017;6:459–470. doi: 10.1016/j.molmet.2017.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berndt SI, Gustafsson S, Mägi R, Ganna A, Wheeler E, Feitosa MF, et al. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat Genet. 2013;45:501–512. doi: 10.1038/ng.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wen W, Cho Y-S, Zheng W, Dorajoo R, Kato N, Qi L, et al. Meta-analysis identifies common variants associated with body mass index in east Asians. Nat Genet. 2012;44:307–311. doi: 10.1038/ng.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradfield JP, Taal HR, Timpson NJ, Scherag A, Lecoeur C, Warrington NM, et al. A genome-wide association meta-analysis identifies new childhood obesity loci. Nat Genet. 2012;44:526–531. doi: 10.1038/ng.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wheeler E, Huang N, Bochukova EG, Keogh JM, Lindsay S, Garg S, et al. Genome-wide SNP and CNV analysis identifies common and low-frequency variants associated with severe early-onset obesity. Nat Genet. 2013;45:513–517. doi: 10.1038/ng.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monda KL, Chen GK, Taylor KC, Palmer C, Edwards TL, Lange LA, et al. A meta-analysis identifies new loci associated with body mass index in individuals of African ancestry. Nat Genet. 2013;45:690–696. doi: 10.1038/ng.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiao H, Arner P, Hoffstedt J, Brodin D, Dubern B, Czernichow S, et al. Genome wide association study identifies KCNMA1 contributing to human obesity. BMC Med Genomics. 2011;4:51. doi: 10.1186/1755-8794-4-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson D, Cordell HJ, Fakiola M, Francis RW, Syn G, Scaman ESH, et al. First genome-wide association study in an Australian aboriginal population provides insights into genetic risk factors for body mass index and type 2 diabetes. PloS One. 2015;10:e0119333. doi: 10.1371/journal.pone.0119333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yengo L, Sidorenko J, Kemper KE, Zheng Z, Wood AR, Weedon MN, et al. Meta-analysis of genome-wide association studies for height and body mass index in ~700000 individuals of European ancestry. Hum Mol Genet. 2018;27:3641–3649. doi: 10.1093/hmg/ddy271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaxillaire M, Yengo L, Lobbens S, Rocheleau G, Eury E, Lantieri O, et al. Type 2 diabetes-related genetic risk scores associated with variations in fasting plasma glucose and development of impaired glucose homeostasis in the prospective DESIR study. Diabetologia. 2014;57:1601–1610. doi: 10.1007/s00125-014-3277-x. [DOI] [PubMed] [Google Scholar]
- 14.Blundell JE, Gillett A. Control of food intake in the obese. Obes Res. 2001;9(Suppl 4):263S–270S. doi: 10.1038/oby.2001.129. [DOI] [PubMed] [Google Scholar]
- 15.Flint AJ, Gearhardt AN, Corbin WR, Brownell KD, Field AE, Rimm EB. Food-addiction scale measurement in 2 cohorts of middle-aged and older women. Am J Clin Nutr. 2014;99:578–586. doi: 10.3945/ajcn.113.068965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hebebrand J, Albayrak Ö, Adan R, Antel J, Dieguez C, de Jong J, et al. ‘Eating addiction’, rather than ‘food addiction’, better captures addictive-like eating behavior. Neurosci Biobehav Rev. 2014;47:295–306. doi: 10.1016/j.neubiorev.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 17.Naqvi NH, Bechara A. The hidden island of addiction: the insula. Trends Neurosci. 2009;32:56–67. doi: 10.1016/j.tins.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zaghloul KA, Blanco JA, Weidemann CT, McGill K, Jaggi JL, Baltuch GH, et al. Human substantia nigra neurons encode unexpected financial rewards. Science. 2009;323:1496–1499. doi: 10.1126/science.1167342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo SX, Huang EJ. Dopaminergic Neurons and Brain Reward Pathways: From Neurogenesis to Circuit Assembly. Am J Pathol. 2016;186:478–488. doi: 10.1016/j.ajpath.2015.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Micali N, Field AE, Treasure JL, Evans DM. Are obesity risk genes associated with binge eating in adolescence? Obes Silver Spring Md. 2015;23:1729–1736. doi: 10.1002/oby.21147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Castellini G, Franzago M, Bagnoli S, Lelli L, Balsamo M, Mancini M, et al. Fat mass and obesity-associated gene (FTO) is associated to eating disorders susceptibility and moderates the expression of psychopathological traits. PloS One. 2017;12:e0173560. doi: 10.1371/journal.pone.0173560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corwin RLW, Wojnicki FHE, Zimmer DJ, Babbs RK, McGrath LE, Olivos DR, et al. Binge-type eating disrupts dopaminergic and GABAergic signaling in the prefrontal cortex and ventral tegmental area. Obes Silver Spring Md. 2016;24:2118–2125. doi: 10.1002/oby.21626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Claussnitzer M, Dankel SN, Kim K-H, Quon G, Meuleman W, Haugen C, et al. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N Engl J Med. 2015;373:895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.