Abstract
daf-2 is one of the most studied mutants in C.elegans: it contains a deletion in the gene orthologue of the insulin/insulin-like growth factor (IGF) receptor. Using high resolution 1H NMR spectroscopy, metabolomics has helped to dissect the metabolic consequences of altered daf-2 signalling. Here, we present a detailed metabolomic analysis of daf-2, using NMR spectroscopy, gas chromatographymass spectrometry (GC-MS) and liquid chromatography mass spectrometry (LC-MS) to integrate information from different pathways. We have then used Pearson and partial correlation analysis to build networks to explore the central role of daf-2 in regulating fatty acid and amino acid metabolism. The results show the tight link between these two parts of the metabolome.
Introduction
Caenorhabditis elegans has become a versatile model organism. The reasons for this can be found in its rapid life cycle, the ease of maintenance, mode of reproduction (it can be either hermaphrodite or male), small genome size (it was the first multicellular organism to have its genome completely sequenced [1]), transparent body and invariable cell number [2]. Originally, it was introduced as a model organism to study the genetic basis of nervous system function and embryonic development, but it is now widely used in many other fields, including aging, microRNA research, pathogenesis and fat metabolism. The milestone in terms of fat metabolism research is the publication by Ashrafi and co-workers which identified 112 genes associated with increased fat accumulation and 305 genes associated with reduced concentrations using a systematic RNAi screen of the C. elegans genome using the vital dye Nile Red [3]. This was the first whole-genome, systematic study of the genes involved in fat metabolism. Since this publication, the interest in the field has increased rapidly, with a search on Pubmed for “C. elegans AND fat” giving 272 manuscripts as of 2012).
One of the most studied mutants in the field of lipid research in C. elegans contains a deletion in the gene daf-2, the orthologue of the insulin/insulin-like growth factor (IGF) receptor, which negatively regulates the activity of DAF-16, a Forkhead-related transcription factor, by inducing its phosphorylation and nuclear exclusion, and generates extreme changes on the animal phenotype, including increased lifespan and lipid content, and severe alteration to the organism, such as adult size and morphology, pumping rate and gonad morphology [4 ; 5 ; 6]. The role of daf-2 in metabolism has been demonstrated previously, with adult mutants having a higher metabolic rate than wild type animals as measured using a chemiluminescent assay to estimate the maximal metabolic performance and the expression of a range of enzymes were increased including catalase, superoxide dismutase, isocitrate lyase and malate synthase [7]. The interest in the daf-2 signalling pathway in part reflects the fact that the gene is involved in fat and carbohydrate metabolism, being an orthologue to insulin, but is also involved in ageing and has a pivotal role in reproduction in C.elegans. The results of the research performed on daf-2 demonstrated a significant contribution to the theory about the impact of ROS on aging.
Metabolomics has previously been used to examine the metabolic consequences of altered daf-2 signalling. Fuchs and co-workers [8] investigated metabolic changes associated with long-lived worm mutants, including daf-2, using high resolution 1H NMR spectroscopy, suggesting the existence of a common metabolic signature for long life. From this work, regulatory pathways associated with longevity, previously considered independent, may, in fact, regulate the same regions of the metabolic network. Some of the metabolites that comprise this “metabolic signature of long life”, such as those involved in carbohydrate metabolism, were expected from studies of global gene expression, while others, such as those involving amino acid metabolism, were new and were confirmed in a subsequent study [9]. Using again a 1H NMR spectroscopy approach, both profound and subtle metabolic alterations induced by differential nutrient sensing and insulin-IGF signalling pathways in C. elegans were mapped. In particular, altered amino acid profiles were a key feature of the daf-2 phenotype.
Previously, we used a range of metabolomics approaches to characterise the phenotype of C. elegans mutants with altered fatty acids contents, following deletions in the genes expressing the delta-9 fatty acid desaturase enzymes [10]. We have also investigated the role microRNAs play in regulating fatty acid and aqueous metabolism [11]. In this manuscript our aim is to reconstruct the metabolic interactions regulated by daf-2 in C.elegans following a detailed application of metabolomics in conjunction with the creation of partial correlation networks [12; 13] from wild type and daf-2 mutants. First, we present a detailed metabolomic analysis of daf-2, using NMR spectroscopy, Gas Chromatography-Mass Spectrometry (GC-MS) and liquid chromatography mass spectrometry (LC-MS) to integrate information coming from different parts of the metabolome, and compare this with previous literature. We have then used Pearson and partial correlation analysis to build networks to explore the central role of daf-2 in regulating fatty acid and amino acid metabolism. This highlights the central role of the gene for the regulation and the connection of these two parts of the metabolome.
Results
The strain CB1370 is a temperature-sensitive mutant with a deletion in the allele e1370 of the daf-2 gene. As a result of this the phenotype of the animal changes according to the temperature it is kept at. In the specific case of daf-2(e1370), its phenotype at 15° C is virtually identical to the wild type, becoming more like the mutant with increasing temperature: at 20° C the expression of daf-2 is altered around 15%, and the gene is completely disrupted at 25° C [14]. We took advantage of this characteristic analysing the metabolome at two different temperatures and hence different levels of daf-2 expression.
1. Metabolomic analysis of daf-2(e1370) at 20° C
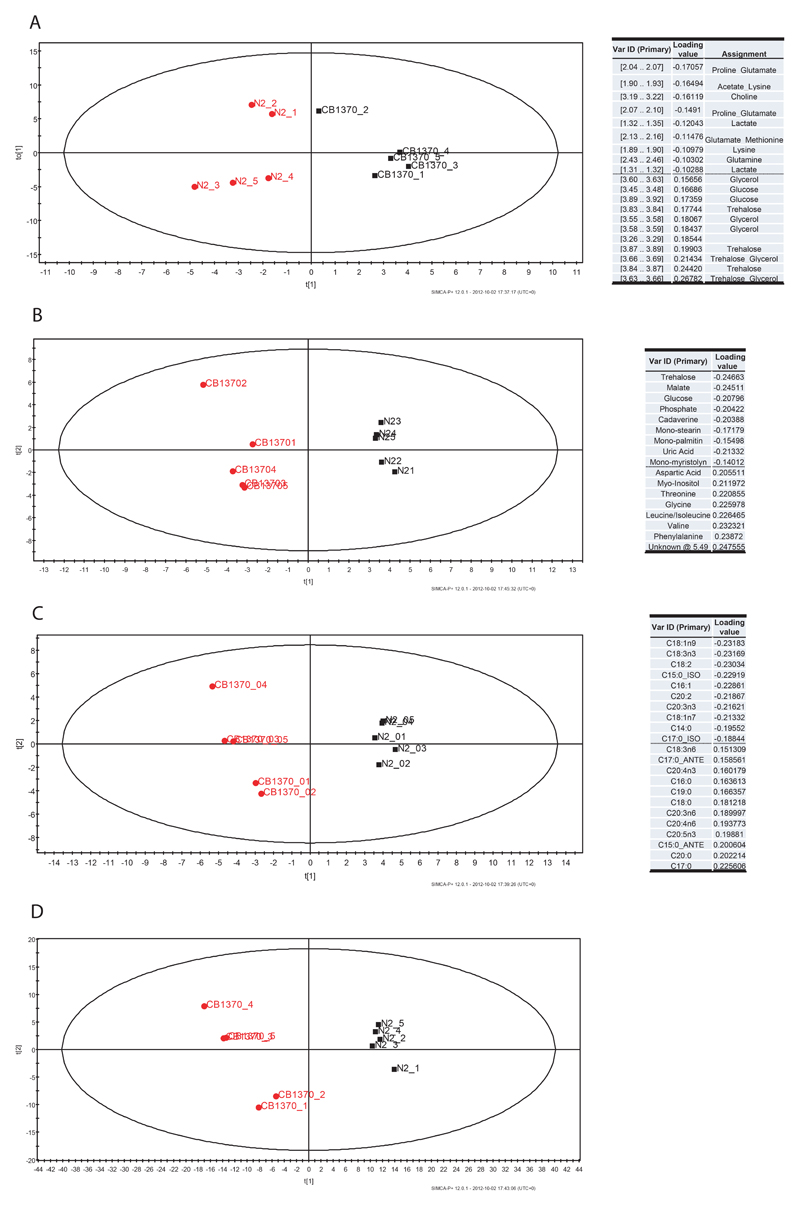
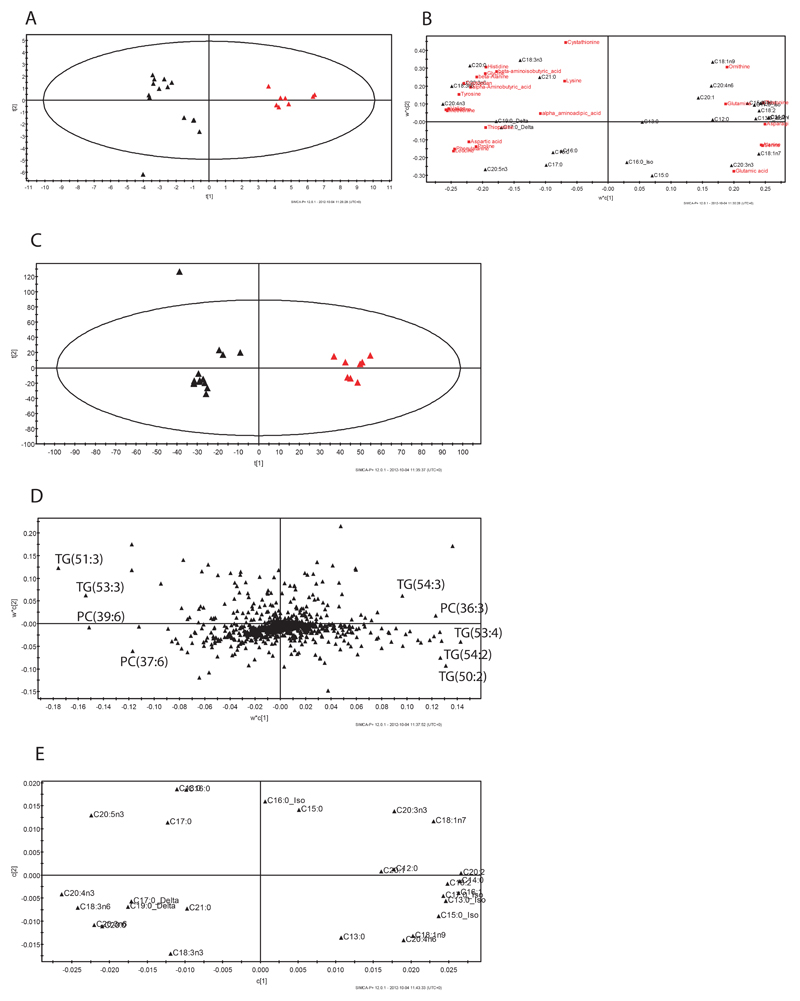
Proton-NMR spectroscopy, gas chromatography-mass spectrometry and liquid chromatography mass spectrometry were used to profile a range of aqueous and lipid metabolites in samples belonging to the strains daf-2(e1370) and wild type, kept at 20° C. For the aqueous fraction, application of Partial Least Squares-Discriminate Analysis (PLS-DA) to the NMR spectroscopy dataset produced 2 latent variables by cross-validation (model parameters: R2(X)=73.3%, R2(Y)=83.4%, Q2=60.4%) and separated the mutants from the wild types (Figure 1A). Analysis of the loadings plot associated with the PLS-DA model identified a range of metabolic changes - trehalose, glycerol, and glucose had increased concentrations in the mutant, while a number of amino acids, including proline, lysine and glutamine, branched chain amino acids and choline were decreased.
Figure 1.
Metabolomic analysis of daf-2(e1370) mutants at 20°. Score plots and loading tables showing the clustering pattern according to genotype and the metabolites responsible for separation in the profiles obtained by A. NMR spectroscopy, B. aqueous fraction and GC-MS, C. fatty acids in GC-MS, D. LC-MS in positive mode.
A more robust PLS-DA model was formed from the GC-MS dataset of the aqueous fraction. This PLS-DA model had 2 latent variables by cross-validation (model parameters: R2(X)=61.4%, R2(Y)=98.8%, Q2=89.4%) (Figure 1B). Analysis of the loadings plot again identified a number of metabolites with altered concentrations (Figure 1B) - trehalose, malate, glucose, mono-stearin, mono-palmitin, cadaverine and uric acid had increased concentrations in the mutant, while a number of amino acids, including phenylalanine, valine, leucine/isoleucine, glycine, threonine, aspartate, glutamate and serine in addition to myo-Inositol, were decreased.
The lipid fraction was analysed by both GC-MS to examine total fatty acid content and LC-MS to examine intact lipids directly. The PLS-DA model obtained from the GC-MS analysis of total fatty acids (FA) also demonstrated clear separation between the mutant and wild type samples (Figure 1C) using 5 latent variables by cross validation (model parameters: R2X=86.3%, R2Y=99% and Q2=97.9%). From an examination of the loadings plot (Figure 1E) the mutant strain had higher concentrations of monounsaturated fats C16:1 and C18:1n9 as well as C18:3n3, C18:2, and C20:2. There was also an increase in the branched fatty acids (C15:0_Iso, C17:0_Iso). The wild type, in contrast, had higher concentrations of saturated (C17:0, C20:0, C18:0, C16:0) and polyunsaturated fatty acids (C20:5n3, C20:4n6, C20:3 n6).
Finally, the PLS-DA model obtained from the LC-MS of the lipid fraction also showed excellent discrimination between the two strains (Figure 1D; model validation parameters: R2X=61.1%, R2Y=99.1% and Q2=95%). The loadings of this plot demonstrated that a higher content of triglycerides containing branched and monounsaturated FA were found in the mutants (with lipid species such as TAG(18:1/17:0Δ/15:0), TAG(18:1/19:0Δ/15:0), TAG(16:0/18:2/18:1), TAG(18:2/18:1/17:0Δ), TAG(18:2/17:0Δ/17:0Δ) and TAG(18:1/18:1/18:1) increased) and saturated FA in the wild type (with lipid species such as TAG (18:0/16:0/18:0), TAG(18:0/18:0/18:0), TAG(16:0/16:0/18:0) increased in the wild type). In relative proportion there was also higher concentrations of phospholipids in the wild type (such as PC(20:5/20:5)) and diglycerides (such as DG(18:0/0/18:0) and DG(18:0/0/16:0).
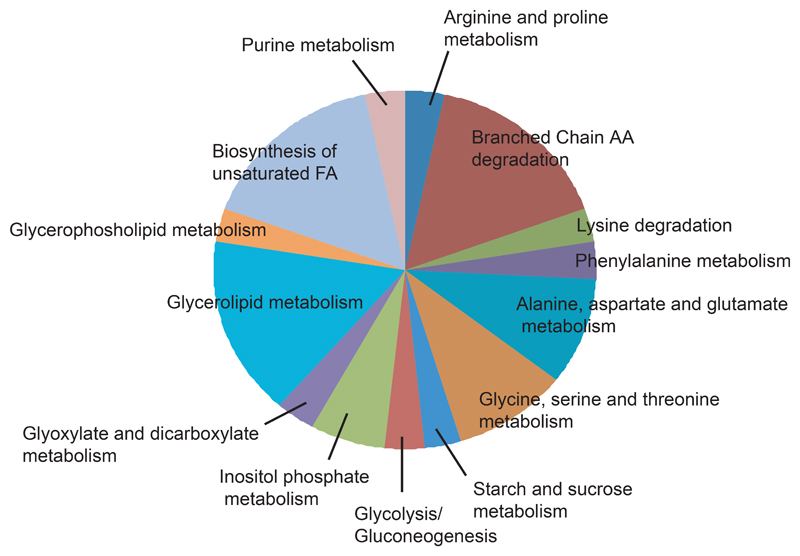
Figure 2 summarizes together all the results obtained from the different techniques, showing the pathways involved in the changes.
Figure 2.
Metabolic pathways changing for daf-2(e1370) mutants at 20°.
2. Correlation networks for daf-2(e1370) at 25° C
As already stated, the strain is temperature-sensitive, and its characteristics change according to the temperature. At 20° C, it has increased longevity, but it feeds at a similar rate to the wildtype control and produces progeny. However, the metabolic changes compared to the wild type are significant. To explore further the changes in the metabolome of CB1370, the strain was grown at 15° C until L4 stage (to avoid dauer formation) and shifted at 25° C for 24 h, at which temperature 100% of the gene was deleted. NMR spectroscopy has already been used to characterise this mutant under these conditions [8; 9]. In the following, a series of correlation networks for the more extreme conditions were built to explore the consequences on selected classes of metabolites, which have been shown to change significantly.
2.1. Amino Acid metabolism
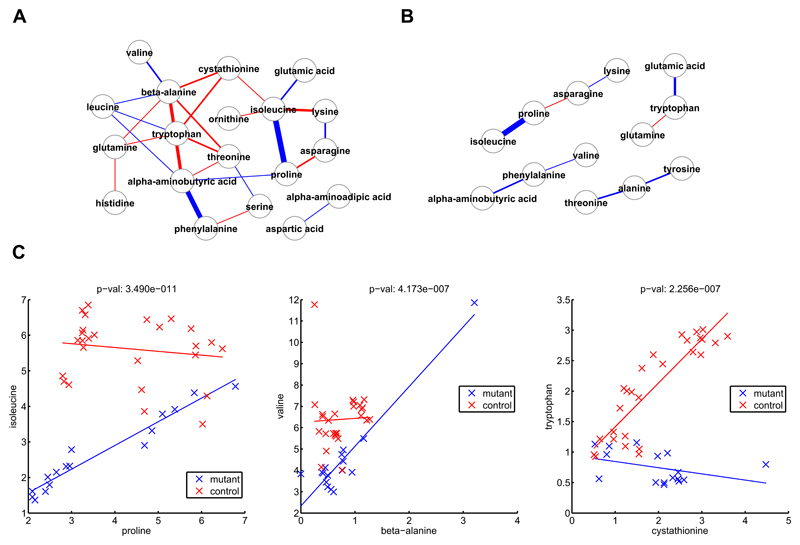
In Figure 3A, the differential correlation network obtained considering the amino acid profiles as measured by GC-MS shows the changes in the Pearson correlation structures considering the two genotypes. Blue edges indicate a higher correlation in mutants, red a higher correlation in controls. This network shows the changes associated with metabolism of a high number of key amino acids, including threonine, branched-chain amino acids and proline. In Figure 3B, the differential network calculating the partial correlation coefficient for the control only and the mutant only is shown. Partial correlation coefficients aim to separate the contribution of the direct dependency between couples of metabolites from the indirect one. The number of edges is reduced in this kind of analysis underlying how the network in its entirety changes more than specific pathways.
Figure 3.
Correlation analysis for metabolites from the Amino Acid kit for daf-2(e1370) and the control at 25°. A. Differential network obtained considering Pearson correlation coefficients and a Bonferroni cut-off of 3.33e-5. Blue edges indicate a higher correlation in mutants, red a higher correlation in controls. B. Differential Gaussian graphical model obtained considering partial correlation coefficients. Again, blue edges indicate a higher correlation in mutants, red a higher correlation in controls. C. Biplots for Pearson correlation coefficients between couples of amino acids with significant changes between mutants and controls.
In Figure 3C, example biplots of the change in the correlation coefficients between mutants and controls are shown: a number of pairs of amino acids are non-correlated in one strain but are highly correlated in the other, including the pairs proline and isoleucine correlated in the wild type, beta-alanine and valine correlated in the wild type, phenylalanine and alpha-aminobutyric acid in daf-2 and cystathionine and tryptophan in daf-2, demonstrating again the reorganization in the network of amino acid metabolism following a reduction in the expression of daf-2.
2.2. Total Fatty Acids
The differential correlation network obtained considering the total fatty acid profile as measured by GC-MS for daf-2(e1370) and the control strain was calculated. The common features between the two networks refer to basic characteristics for C. elegans metabolism. For example, we detect indications of the origin of the fatty acids, as represented by the high correlation coefficient for C17:0_delta- and C19:0_delta for both genotypes, which are both being derived from diet. Moreover, we observe signatures of the common synthesis pathway, e.g. iso-C17:0 (branch chain C17:0 fatty acid) which is correlated with_iso-C15:0, iso-C15:0 which is correlated with iso-C13:0, and iso-C17:0 which is correlated with iso-C13:0, demonstrating the pairing between fatty acids that are synthesized from one another.
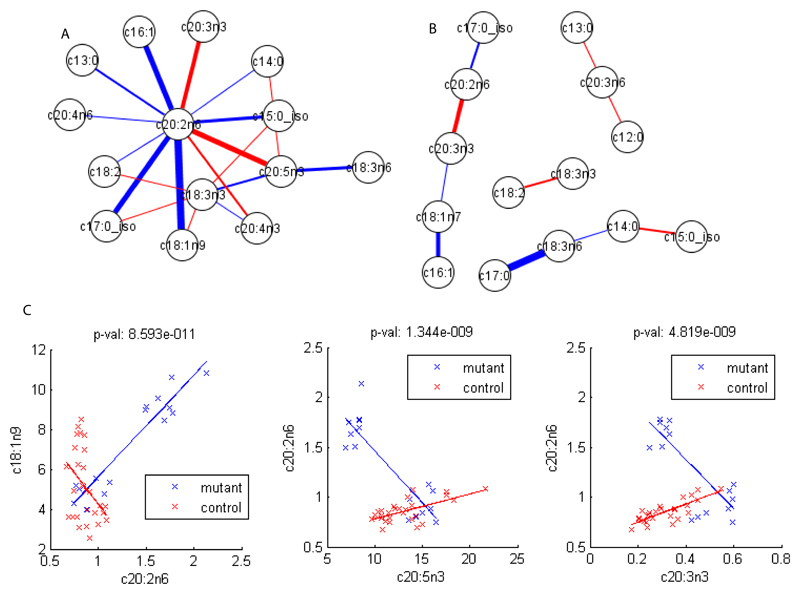
In Figure 4A, the changes in the correlation structures of the fatty acids considering the two genotypes are shown. For both genotypes there is a central role for C20:2 in the networks. The fatty acid C20:2 displays higher correlation coefficients with the fatty acids C16:1, C18:1n9, C17:0_iso in the daf-2 mutant, which from the LC-MS analysis are major constituents of the triglycerides detected in daf-2. However, in the wild type C20:2 has higher correlation coefficients with fatty acids C20:5n3, C20:4n3, C20:3n3. These fatty acids are key constituents of the phospholipid species as detected by LC-MS, for their crucial role in maintaining the fluidity of the membranes. This result suggests a different utilization of C20:2 dependent on daf-2 expression in the two strains, a distinct switch between the partitioning of C20:2 and its central role in regulating fatty acid metabolism. When a partial correlation networks is built the number of significant edges again decreases, compared with the network built using Pearson correlation coefficients (Figure 4B), similarly to what we observed for the amino acids. These results suggest daf-2 to be responsible for a shift in metabolism, in part reflecting the balance between triglyceride and phospholipid metabolism. This is reflected in a profound remodelling of the network about C20:2 away from correlations associated with phospholipid metabolism, towards a phenotype here the production of triglycerides is readily apparent. Plots with the most significantly affected correlation coefficients are shown in Figure 4C.
Figure 4.
Correlation analysis for metabolites from the GC-MS for daf-2(e1370) and the control at 25°. A. Differential network obtained considering Pearson correlation coefficients and a Bonferroni cut-off of 2.64e-6. Blue edges indicate a higher correlation in mutants, red a higher correlation in controls. B. Differential Gaussian graphical model obtained considering partial correlation coefficients. Again, blue edges indicate a higher correlation in mutants, red a higher correlation in controls. C. Biplots for Pearson correlation coefficients between couples of fatty acids with significant changes between mutants and controls.
3. Correlation between amino acids and fatty acids for daf-2(e1370) at 25° C
3.1. Metabolic Correlations between amino acid and fatty acid metabolism
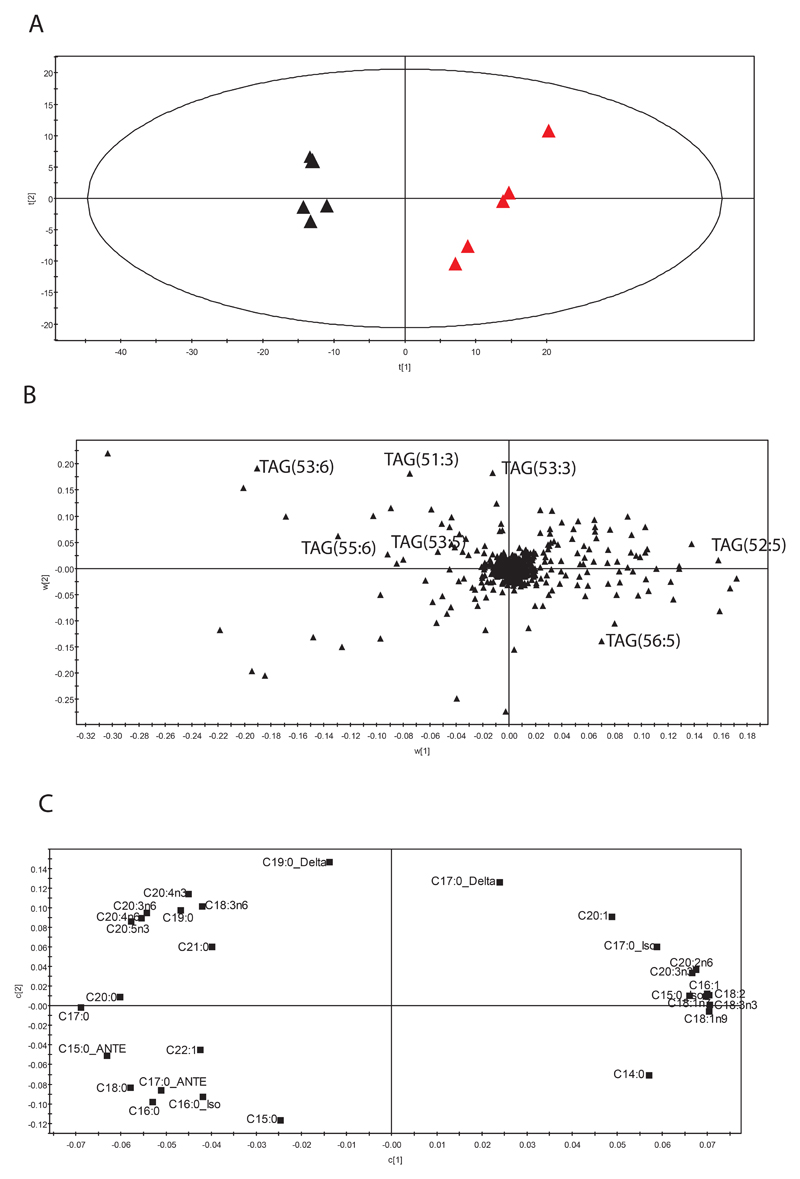
To examine the interactions between amino acid and fatty acid metabolism a PLS regression model was built between the amino acid dataset as detected by GC-MS and total fatty acid analysis by GC-MS. This model had 4 components (Figure 5A; model parameters: R2X=84.5%, R2Y=85% and Q2=70.2%). The score plot demonstrated a good discrimination between control and modified strains. The loading analysis (Figure 5B) shows results in agreement with the previous analysis, but it underlines the link between the amino acid and fatty acid metabolism: the increase in the content of branched-chain amino acids in the control is parallel to the increase in the branched fatty acids in the mutants (this class of fatty acids are synthesized from branched chain amino acids).
Figure 5.
Multi-platform analysis for daf-2(e1370) and the control at 25°. A. Score plot of the PLS model obtained considering the fatty acids as X matrix and the amino acids as Y. Black triangles represent samples belonging to the control, red triangles samples belonging to the mutant strain. The subgroups among samples belonging to the same strain refers to “different days of bleaching”. B. Loading plot for the score in plot A shows the amino acids and fatty acids responsible for the model. C. Score plot of the PLS model obtained considering the lipids in positive mode as X matrix and the fatty acids as Y. Again, the black triangles represent samples belonging to the control, the red triangles samples belonging to the mutant strain. C. Loading plot associated to the X matrix for the score in C shows the lipids responsible for the model. D. Loading plot associated to the Y matrix for the score in C shows the fatty acids responsible for the model.
In addition, the correlation of α-aminobutyric acid with leucine, iso-C15:0 and iso-C17:0 produced very strong correlation coefficients (negative for the first metabolite, positive for the other two), but these correlations are very close to zero in the mutant.
3.2. Correlation between the intact lipid data as collected by LC-MS and total fatty acid analysis
In order to understand better why C20:2 appears to have such a pivotal role in the network of fatty acid metabolism generated for the daf-2 mutant using total fatty acid data, we performed a correlation analysis between the total fatty acids as measured by GC-FID and the intact lipids as measured by LC-MS. The concentration of total fatty acid C20:2 was highly negatively correlated with the following phosphatidylcholines: PC(39:6), PC(39:5), PC(35:2) and PC(39:4), as well as with TG(53:3); and high positive correlation coefficients were found instead with PC(38:5) and PC(34:3), plus with an large number of triglycerides. Correlation coefficients were also investigated for the other fatty acids: in the cases of C18:1n9 and C18:1n7, high correlation coefficients were found with triglycerides, underlying the importance of these fatty acids in the synthesis of these lipids.
To further investigate the correlations between the total fatty acid analysis and the intact lipids we used partial least squares. A PLS regression model was formed between the LC-MS data of intact lipids and the GC-MS total fatty acid analysis, obtaining a model with 7 components (Figure 5C; R2X=81.3%, R2Y=89.8% and Q2=58%). The scores plot demonstrated a good discrimination between control and modified strains. The loading plot identified the most altered lipids (X-block data, Figure 5D) and fatty acids (Y-block data, Figure 5E) across the dataset. Again a higher content of triglycerides containing branched and monounsaturated FA was found in the mutants, and saturated fatty acids in the control, with correlations between monounsaturated FAs and branched fatty acids, with both increasing in the controls. In addition, we performed multivariate analysis on a dataset from C.elegans maintained at 20° C. A PLS regression between the intact lipid data and the total fatty acid pool was built (Figure 6A; 2 latent variables R2X=63.9%, R2Y=86.5% and Q2=72.8%). The score plot showed a very good discrimination between control and the daf-2 mutant. The loading plot identified the lipids (X-block, Figure 6B) and fatty acids (Y-block, Figure 6C) with the largest variance. Again, higher concentrations of triglycerides containing branched and monounsaturated FAs were found in the mutants, and saturated fatty acids increased in the control animals, with a correlated increase in monounsaturated FAs and branched fatty acids in the control.
Figure 6.
Multi-platform analysis for daf-2(e1370) and the control at 20°. A. Score plot of the PLS model obtained considering the lipids in positive mode as X matrix and the fatty acids as Y. Again, the black triangles represent samples belonging to the control, the red triangles samples belonging to the mutant strain. B. Loading plot associated to the X matrix for the score in A shows the lipids responsible for the model. C. Loading plot associated to the Y matrix for the score in A shows the fatty acids responsible for the model.
Discussion
The results obtained from our metabolomics approach are in agreement with reported previous studies: at 20°C the phenotype is mild, although significantly present [9]. The higher concentration of trehalose detected by both NMR spectroscopy and GC-MS in the mutants has been extensively discussed previously from a transcriptional [15] and metabolic [8] perspective. Both techniques also demonstrated an increase in glucose, as expected from an upregulation of transcripts in the gluconeogenesis pathway previously associated with a failure to express daf-2 [16], while the increase of malate in the mutants detected by GC-MS is associated with an increase in activity of the glyoxylate shunt [16; 7], to increase the synthesis of sugars from lipids. Alterations in amino acid metabolism are evident from both the aqueous profiles detected by NMR spectroscopy and GC-MS. The total content of amino acids in the wild type is higher than that in the mutant, which is in good agreement with a shift towards an higher catabolism of amino acids in daf-2 as previously reported [8]. The central role of proline in the correlation network and its decrease in the mutants has been discussed by Zarse and co-workers [17]. The activation of mitochondrial L-proline catabolism and an associated transiently increased ROS levels induce an adaptive response that culminates in increased stress resistance and extends life span in C.elegans. Finally, the increase in monoglycerides (monomyristolin, monopalmitin, monostearin) in the mutants is in good agreement with the overall increase in lipid synthesis in daf-2. Parallel to this there is a significant decrease in the expression of the class of protein vitallogenin, that can be linked to a reduction in the mobility of the lipids in this mutant [16] and an increase towards the catabolism of amino acids, again demonstrating the tight link between the two metabolic classes.
The central role of daf-2 in the regulation of lipid metabolism was further studied using GC-MS and LC-MS analysis of the lipid fraction. The higher content of 16:1, 18:1n7 and 18:1n9 in the mutants is explained by the previously reported upregulation of the transcripts of fat-5 and fat-6, two of the three genes expressing Δ9 desaturase enzymes [16]. Several studies have been conducted on the importance of monounsaturated fatty acids in storage of fat, showing how the deletion of stearoyl-CoA desaturase (a Δ9 desaturase) decrease the content of the triglycerides both in mice [18] and worms [19; 20]. This reorganization is also the reason for the lower proportion of phospholipids in the LC-MS dataset and of 20:5n3 detected by GC-MS in the mutants. The fatty acid 20:5n3 is a major constituent of phosphocholines. The LC-MS data reported also an increase of triglycerides containing monounsaturated fatty acids in the mutants, demonstrating the switch induced by daf-2 between sequestration of fatty acids into phospholipids in cell membranes and the storage of fatty acids in stores such as triglycerides.
Finally, a genetic interaction between the genes daf-2 and elo-5 has been found: elo-5 is the gene that specifically elongates branched chain fatty acids. It has been shown that its role is important for the extension of lifespan [21]. Furthermore, elo-5 has been found among the genes functioning within the daf-2 pathway to shorten lifespan. The correlation between the branched-chain fatty acids of different length does not change, but the flux through the pathway increases in the mutants.
The results of the correlation networks help in defining better the metabolic consequences of a failure to express daf-2, suggesting a different use of fatty acids (and in particular of C20:2) in the lipid synthesis, in agreement with the phenotype of the animal, and a wide reorganization of the amino acids network as shown in previous papers.
A-aminobutyric acid can be produced from threonine, as a minor catabolic pathway of this amino acid and enter in the TCA through succinyl-CoA. An alternative pathway in the utilisation of threonine is the conversion to serine. In the mutants, lower correlation coefficients between α-aminobutyric acid and threonine, with a parallel increase in the correlation coefficients between threonine and serine, suggesting an alteration in the major pathway which consumes this essential amino acid. This change is also correlated with the catabolic metabolism of the branched chain amino acids, as shown by the parallel changes in the correlation coefficients of threonine and leucine, α-aminobutyric acid and leucine, and α-aminobutyric acid and C17:0_iso.
Experimental
Culture of nematodes
C. elegans were grown under standard conditions on Nematode Growth Media (NGM) with HB101 [32] as a food source. The wild-type strain used was the Bristol N2 [2]. Mutant strains were obtained from the Caenorhabditis Genetics Center, University of Minnesota, Twin Cities, MN and included: CB1370 daf-2(e1370) For each experiment, early embryos were isolated by alkaline hypochlorite treatment [2] and plated after synchronisation on NGM plates seeded with bacteria. Two different experimental protocols were used at this stage: in one case the samples were kept at 20° C, while in the other they were kept at 15° C until they reached the L4 stage and then shifted to 25° C. This provided two levels of reduced expression for daf-2 to test our approach. In the first experiment, 5 samples for each strain were collected at the late L4 stage of development, while in the second one, eight samples per strain were collected both before and 24 h after the shift of temperature. For each biological replicate, ~10000 animals were harvested and washed. All the samples were stored at -80° C until extraction.
Extraction procedure for metabolites
Metabolites from whole nematodes were extracted using a methanol–chloroform procedure [10]. 600 μl of methanol–chloroform (2:1 v:v) was added to the frozen nematodes and samples were sonicated for 15 min. 200 μl each of chloroform and water were added, the samples centrifuged and the aqueous layer separated from the lipid layer. The procedure was repeated twice to form a double extract and maximise the recovery of metabolites from the protein pellet. The aqueous layer was dried overnight in an evacuated centrifuge, while the lipid fraction was dried under a stream of nitrogen gas in a fumehood.
Analysis of aqueous extracts
NMR spectroscopy
The dried extracts were rehydrated in 550 μl D2O, containing 0.05 mM (sodium-3-(tri-methylsilyl)-2,2,3,3-tetradeuteriopropionate (TSP) (Cambridge Isotope Laboratories, MA, USA) as an internal standard. The samples were analysed using an AVANCE II+ NMR spectrometer operating at 500.13 MHz for the 1H frequency (Bruker, Germany) using a 5 mm TXI probe. Spectra were collected using a solvent suppression pulse sequence based on a one-dimensional NOESY pulse sequence to saturate the residual 1H water signal (relaxation delay = 2 s, t1 increment = 3 us, mixing time = 150 ms, solvent presaturation applied during the relaxation time and the mixing time). Two hundred and fifty-six transients were collected into 16 K data points over a spectral width of 12 ppm at 27 °C.
Amino Acid analysis
The samples analysed by NMR spectroscopy were dried overnight in an evacuated centrifuge, rehydrated in 100 μl of water and analysed by the EZ:faast Free (Physiological) Amino Acid Analysis by GC-MS kit (Phenomenex, USA). The samples were treated according to the procedure described by the manufacturer. In brief, 100 μl of norvaline was added as internal standard, then the sample got through a solid phase extraction step, followed by a derivatization step, aiming to form chloroformate-derivates of the amino acids.
GC-MS
For the worms grown at 20° C, the samples analysed by NMR spectroscopy were subsequently dried overnight in a centrifuge. 50 μl of methoxyamine hydrochloride (20 mg/ml in pyridine) were added to the dried sample and left for 17 hours at room temperature. 50 μl of N-methyl-N-trimethylsilyltrifluoroacetamide were added and the samples left at room temperature for another hour. 600 μl of hexane were added: 100 μl of this solution were mixed with 400 μl of hexane and the sample analysed by GC-MS Experiments were collected using the ISQ (ThermoScientific, Hemel Hempstead, UK) on 2 μl per injection, in a splitless mode. The injector temperature was 230°C and helium carrier gas was used at a flow rate of 1.2 ml/minute. The column was 30 m × 0.25 mm 5% phenylpolysilphenylene-siloxane one with a 0.25 μm ZB-5 ms stationary phase (Phenomenex, Macclesfield, Cheshire, UK). The initial column temperature of 70°C was increased by 5°C/minute to 230°C and then increased at a rate of 20°C/minute to 310°C (transfer line temperature = 250°C; ion source = 250°C; electron ionization = 70 eV). The detector was turned on after 240 s and full-scan spectra were collected using three scans/s over a range of 50 to 650 m/z.
Analysis of lipid extracts
Lipid extracts were dissolved in 200 μl of chloroform/methanol (1:1 v/v): half was used for GC-FID analysis of total fatty acids and half for LC-MS analysis of intact lipids. For the GC-FID analysis 50 μl of D-25 tridecanoic acid (200 μM in chloroform), 650 μl of chloroform/methanol (1:1 v/v) and 250 μl BF3/methanol (Sigma-Aldrich) was added to the extract and the vials were incubated at 80° C for 90 min. 500 μl H2O and 1 ml hexane were added and each vial vortex mixed. The organic layer was evaporated to dryness before reconstitution in 100 μl hexane for analysis. The derivatised organic metabolites were injected onto a TR-fatty acid methyl ester (FAME) stationary phase column (Thermo Electron; 30 m × 0.25 mm ID × 0.25 μm; 70% cyanopropyl polysilphenylene-siloxane) with a split ratio of 20. The injector temperature was 230°C and the helium carrier gas flow rate was 1.2 ml/min. The column temperature was 60°C for 2 min, increased by 15°C/min to 150°C, and then increased at a rate of 4°C/min to 230°C (transfer line = 240°C; ion source = 250°C, EI = 70 eV). The detector was turned on after 240 s, and full-scan spectra were collected using 3 scans/s over a range of 50–650 m/z. Peaks were assigned using the Food Industry FAME Mix (Restek 6098) and the Bacterial Acid Methyl Ester (BAME) Mix solution (Supelco 47080).
For LC-MS, half of the lipid fraction was resuspended in 100 μl methanol:chloroform 1:1, then 10 μl of it were added to 0.5 ml of IPA:Methanol:Water (2:1:1). Samples were analyzed using a Waters Q-ToF Xevo (Waters Corporation, Manchester, UK) combined with an Acquity Ultra Performance Liquid Chromatogram (UPLC). Ten μl of the sample was injected onto a 1.7 μm bridged ethyl hybrid C8 column (2.1 × 100 mm; Waters Corporation, Manchester, UK) held at 55°C. The binary solvent system (flow rate 0.400 ml/min) consisted of solvent A containing HPLC grade acetonitrile:water 60:40 with 10 mM ammonium formate and solvent B consisting of LC-MS grade acetonitrile:isopropanol 10:90 and 10 mM ammonium formate. The gradient started from 60% A/40% B, reached 99% B in 18 min, then returned back to the starting condition, and remained there for the next 2 min. The data was collected over the mass range of m/z 100–1200 with a scan duration of 0.2 sec and an interscan delay of 0.014 s. The source temperature was set at 120°C and nitrogen was used as the desolvation gas (600 L/h) at 280°C. The voltages of the sampling cone and capillary were 30 V and 3 kV, respectively and collision energy 4.6 V. As lockmass, a solution of 2 ng/μl (50:50 acetonitrile:water) leucine enkephaline (m/z 556.2771) was infused into the instrument at 3 μl/min.
Data processing
NMR spectra were processed using ACD one-dimensional NMR processor (version 12, ACD, Toronto, Canada). Free induction decays were Fourier transformed following multiplication by a line broadening of 1 Hz, and referenced to TSP at 0.0 ppm. Spectra were phased and baseline corrected manually. Each spectrum was integrated using 0.02 ppm integral regions between 0.5 and 4.5, and 5.2–9.5 ppm. Each spectral region was normalised to a total integral value of 1000. GC–FID chromatograms were analysed using Xcalibur, version 2.0 (Thermo Fisher), integrating each peak individually. Each integrated peak was normalised so that the total sum of peaks was set to 1000. LC-MS chromatograms were analysed using Micromass Markerlynx Applications Manager Version 4.1 (Waters Corporation). The ion intensities for each lipid identified were normalized so that the sum of the peak within each sample was set to 1000.
Multivariate analysis of metabolic profiles
Each set of metabolic profiles obtained was analysed by multivariate statistical analysis. Datasets were imported into SIMCA-P 12.0 (Umetrics, Umea, Sweden) for processing using PCA and PLS-DA (a regression extension of PCA used for supervised classification). Proton NMR and LC-MS data were Pareto scaled, in which each variable was centred and multiplied by 1/(Sk)1/2 where Sk is the standard deviation of the variable. GC–MS data were scaled to unit variance by dividing each variable by Sk
Differential correlation networks
We sought to detect pathway steps that are specifically altered between two conditions by applying a differential correlation network approach. Let be the Pearson product-moment correlation coefficients between metabolites A and B in condition 1 and 2, respectively. Then the significance of difference between these two correlation values can be computed using the following test statistic:
where is the Fisher transformation [22] and n1 and n2 represent the sample sizes of condition 1 and 2. If the difference between correlations is zero, TAB will be standard normally distributed. Thus, a two-sided statistical test for a non-zero correlation difference can be constructed by calculating p-values p(TAB) ≔ ϕ(TAB) ⋅ 2, where ϕ(⋅) represents the standard normal cumulative distribution function. An edge was drawn in the differential correlation network if the correlation difference p-value was below α=0.01 following a Bonferroni correction. For the partial correlation networks, we employed a permutation-based significance test of differences.
The amino acid and fatty acid datasets, obtained considering mutants and controls after the shift at 25° C, were used for the correlation analysis: data were imported into Matlab R2011a and investigated with in-house developed code.
References
- 1.The C. elegans sequencing consortium. Genome Sequence of the Nematode C. elegans: A Platform for Investigating Biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 2.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashrafi K, Chang FY, Watts JL, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature. 2003;421(6920):268–272. doi: 10.1038/nature01279. [DOI] [PubMed] [Google Scholar]
- 4.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 5.Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 6.Gems D, Sutton AJ, Sundermeyer ML, Albert PS, King KV, Edgley ML, Larsen PL, Riddle DL. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics. 1998;150:129–55. doi: 10.1093/genetics/150.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vanfleteren JR, De Vreese A. The gerontogenes age-1 and daf-2 determine metabolic rate potential in aging caenorhabditis elegans. FASEB Journal. 1995;9:1355–1361. doi: 10.1096/fasebj.9.13.7557026. [DOI] [PubMed] [Google Scholar]
- 8.Fuchs S, B J, Davies SK, Viney JM, Swire JS, Leroi AM. A metabolic signature of long life in Caenorhabditis elegans. BMC Biology. 2010;8:14. doi: 10.1186/1741-7007-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin FJ, Spanier B, Collino S, Montoliu I, Kolmeder C, Giesbertz P, Affolter M, Kussmann M, Daniel H, Kochhar S, Rezzi S. Metabotyping of Caenorhabditis elegans and their culture media revealed unique metabolic phenotypes associated to amino acid deficiency and insulin-like signalling. J Proteome Res. 2011;10:990–1003. doi: 10.1021/pr100703a. [DOI] [PubMed] [Google Scholar]
- 10.Castro C, Sar F, Shaw WR, Mishima M, Miska EA, Griffin JL. A metabolomic strategy defines the regulation of lipid content and global metabolism by Δ9 desaturases in Caenorhabditis elegans. BMC Genomics. 2012;13:36. doi: 10.1186/1471-2164-13-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lehrbach NJ, Castro C, Murfitt KJ, Abreu-Goodger C, Griffin JL, Miska EA. Post-developmental microRNA expression is required for normal physiology, and regulates ageing in parallel to Insulin/IGF-1 signaling in C. elegans. RNA. 2012 doi: 10.1261/rna.035402.112. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krumsiek J, Suhre K, Illig T, Adamski J, Theis FJ. Gaussian graphical modeling reconstructs pathway reactions from high-throughput metabolomics data. BMC Syst Biol. 2011;5:21. doi: 10.1186/1752-0509-5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valcárcel B, Würtz P, Seich al Basatena N-K, Tukiainen T, Kangas AJ, Soininen P, Järvelin M, Ala-Korpela M, Ebbels TM, de Iorio M. A Differential Network Approach to Exploring Differences between Biological States: An Application to Prediabetes. PLoS ONE. 2011;6:e24702. doi: 10.1371/journal.pone.0024702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rudner A, Tabtiang R, Kenyon CJ. A Mutation Which Doubles C. elegans Life Span. Worm Breeder's Gazette. 1993;12:94. [Google Scholar]
- 15.Lamitina ST, Strange K. Transcriptional targets of DAF-16 insulin signaling pathway protect C. elegans from extreme hypertonic stress. Am J Physiol Cell Physiol. 2005;288:C467–C474. doi: 10.1152/ajpcell.00451.2004. [DOI] [PubMed] [Google Scholar]
- 16.McElwee JJ, Schuster E, Blanc E, Thornton J, Gems D. Diapause-associated metabolic traits reiterated in long-lived daf-2 mutants in the nematode Caenorhabditis elegans. Mechanisms of Ageing and Development. 2006;127:458–472. doi: 10.1016/j.mad.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 17.Zarse K, Schmeisser S, Groth M, Priebe S, Beuster G, Kuhlow D, Guthke R, Platzer M, Kahn CR, Ristow M. Impaired Insulin/IGF1 signaling extends life span by promoting mitochondrial L-proline catabolism to induce a transient ROS signal. Cell Metabolism. 2012;15:451–465. doi: 10.1016/j.cmet.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dobrzyn A, Ntambi JM. The role of stearoyl-CoA desaturase in the control of metabolism. Prostaglandins Leukot Essent Fatty Acids. 2005;73:35–41. doi: 10.1016/j.plefa.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 19.Brock TJ, Browse J, Watts JL. Fatty acid desaturation and the regulation of adiposity in Caenorhabditis elegans. Genetics. 2007;176:865–875. doi: 10.1534/genetics.107.071860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le TT, Duren HM, Slipchenko MN, Hu CD, Cheng JX. Label-free quantitative analysis of lipid metabolism in living Caenorhabditis elegans. J Lipid Res. 2010;51:672–677. doi: 10.1194/jlr.D000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samuelson AV, Carr CE, Ruvkun G. Gene activities that mediate increased life span of C. elegans insulin-like signaling mutants. Genes Dev. 2007;21:2976–94. doi: 10.1101/gad.1588907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fisher RA. On the `probable error' of a coefficient of correlation deduced from a small sample. Metron. 1921;1:3–32. [Google Scholar]