Abstract
Standard chemotherapy for acute myeloid leukemia (AML) targets proliferative cells and efficiently induces complete remission; however, many patients relapse and die of their disease. Relapse is caused by leukemia stem cells (LSCs), the cells with self-renewal capacity. Self-renewal and proliferation are separate functions in normal hematopoietic stem cells (HSCs) in steady state conditions. If these functions are also separate functions in LSCs, then antiproliferative therapies may fail to target self-renewal, allowing for relapse. We investigated whether proliferation and self-renewal are separate functions in LSCs as they often are in HSCs. Distinct transcriptional profiles within LSCs of Mll-AF9/NRASG12V murine AML were identified using single-cell RNA sequencing. Single-cell qPCR revealed that these genes were also differentially expressed in primary human LSCs and normal human HSPCs. A smaller subset of these genes was upregulated in LSCs relative to HSPCs; this subset of genes constitutes “LSC-specific” genes in human AML. To assess the differences between these profiles, we identified cell surface markers, CD69 and CD36, whose genes were differentially expressed between these profiles. In vivo mouse reconstitution assays resealed that only CD69High LSCs were capable of self-renewal and were poorly proliferative. In contrast, CD36High LSCs were unable to transplant leukemia but were highly proliferative. These data demonstrate that the transcriptional foundations of self-renewal and proliferation are distinct in LSCs as they often are in normal stem cells and suggest that therapeutic strategies that target self-renewal, in addition to proliferation, are critical to prevent relapse and improve survival in AML.
Keywords: acute myeloid leukemia, NRAS, self-renewal, leukemia stem cell, single-cell
Introduction
Acute myeloid leukemia (AML) is a lethal malignancy with a dismal overall survival rate. Standard AML chemotherapy induces complete remission in 60–80% of patients; however, because of high relapse rates, only 20–30% of patients survive during the following two years[1]. Relapse is caused by the ability of a small subset of cells, called leukemia stem cells (LSCs), to recapitulate disease; these leukemia cells are endowed with long-term self-renewal potential[2]. Self-renewal is a process distinct from proliferation in that it can give rise to pluripotent progenitors (stem cells) in addition to daughter cells that differentiate into effector cells. Experimentally, self-renewal is defined as the ability to transplant leukemia to secondary recipients. Most leukemia cells have high proliferative rates, but lack self-renewal capacity: they cannot transplant leukemia to secondary recipients. In contrast, LSCs are a minor leukemia cell population that can give rise to the full spectrum of leukemia subpopulations, including highly proliferative daughter cells[2]. Initial therapy for AML is efficient at eliminating rapidly dividing leukemia cells (the bulk of the leukemia population), but self-renewing LSCs that survive can cause relapse. Our work seeks to identify molecular features of self-renewal so that we can therapeutically target LSCs and prevent AML relapse.
A major challenge to studying LSCs is that the precise immunophenotypic profile of true LSCs (leukemia cells that self-renew) varies by leukemia subtype and individual patients[3–6]. The LSC-enriched compartment in AML has been identified immunophenotypically by a cell surface protein expression profile. Thus far, many studies that query the molecular features of self-renewal have relied on cell surface proteins to identify an LSC-enriched population for evaluation. Relying on immunophenotypic markers of stemness may mask true features of self-renewal. In immunophenotypically identified normal hematopoietic stem cells (HSCs), oncogenic NRAS induced either proliferation or self-renewal, but these two functions were mutually exclusive[7]. We previously demonstrated that oncogenic NRAS (NRASG12V) enforces a self-renewal gene expression profile and is required for self-renewal of LSCs in a murine model of AML (Mll-AF9/NRASG12V)[8, 9]. In this model, NRASG12V is a tetracycline repressible transgene under the control of the Vav1 promoter[10, 11]: treatment with the tetracycline analog, doxycycline, causes loss of NRASG12V expression and leads to disease remission. Moreover, in comparing the gene expression profiles of human and murine AML, we found that the NRASG12V-enforced self-renewal gene expression profile was expressed in multiple human AML self-renewal datasets[8].
This report demonstrated that NRASG12V directs self-renewal in LSCs as it does in normal HSCs[7]; however, these studies did not distinguish the gene expression profile of self-renewal within the immunophenotypically-defined LSC compartment.
If self-renewal and rapid proliferation are separate functions in LSCs as they are in normal HSCs, then therapeutically targeting mediators of rapid proliferation would fail to target self-renewing LSCs, allowing for disease relapse. In this report, we tested whether these functions are likewise separate in LSCs. We used whole transcriptome single-cell RNA sequencing of the LSC-enriched subpopulation in Mll-AF9/NRASG12V murine AML and found distinct transcriptional profiles within these LSCs. To investigate whether these transcriptional profiles are clinically relevant, we performed single-cell gene expression profiling of normal human bone marrow and primary human AML precursors. We found that the genes we identified in the murine model are expressed in a similar pattern in primary human LSCs. Importantly, we found a subset of this signature that was higher in human AML LSCs at the single-cell level; this represents a single-cell LSC-specific gene list. Finally, we identified genes that encode cell surface markers (Cd36 and Cd69) that delineate these gene expression profiles for functional assays. Using CD36 and CD69 protein expression to label and detect cellular subsets within the murine LSC-enriched compartment, we performed in vivo mouse reconstitution assays and found that these subpopulations differ in their self-renewal and proliferative capabilities. We also found that these markers delineate human AML subsets with different proliferative capacities. These findings define and validate a leukemia self-renewal gene expression profile at the single-cell level. Furthermore, these studies demonstrate that proliferation and self-renewal are separate functions in immunophenotypically-defined LSCs of this mouse model, as they are in HSCs, and suggest that curing AML requires therapeutically targeting self-renewal in addition to rapid proliferation.
Materials and Methods
Experimental Design
Our study uses single-cell RNA sequencing to identify transcriptional heterogeneity within LSCs and define the self-renewing subset within this compartment. See Experimental workflow figure (Supplementary Fig. S1A–B). We performed single-cell RNA sequencing on the LSC-enriched compartment of our murine leukemia model. As we previously demonstrated that self-renewal is dependent on NRASG12V activity in this model, we also performed single-cell RNA sequencing on the LSC-enriched compartment after turning off NRASG12V transgene expression. To identify the functional contribution of each single-cell transcriptional profile, we performed in vivo assays of proliferation (using CellTrace labeling) and self-renewal (using in vivo leukemia reconstitution assays). We also performed single-cell qPCR on primary human LSCs and normal bone marrow HSPCs to determine whether these cells express components of this single-cell self-renewal signature. Finally, we analyzed the TCGA gene expression data to determine whether elements of the single-cell self-renewal gene expression signature that we identified are associated with survival in AML.
Murine single-cell RNA sequencing analysis
FPKMs were modified by adding 0.1 to each value (to minimize the effects of dividing by zero). Genes with FPKMs >0.1 in 50% of the cells were log2 transformed. The cells were hierarchically clustered using average linkage clustering using Pearson Correlation. In Figure 1A–B, the values were mean centered prior to clustering. Cell surface markers (Cd36 and Cd69) corresponding to different groups were identified by unsupervised clustering. Groups composed of samples with high Cd36 (>16 FPKM) or high Cd69 (>16 FPKM) were compared using a two-group (Group 1: CD36LowCD69High, Group 3: CD36HighCD69Low) t-test to compare transcript levels for all genes where 4 or more non-zero abundance estimates were present. A list of differentially expressed genes in each dataset was defined as genes with an average fold change ≥ 2 between the two groups and a p-value of ≤ 0.05 in each dataset. This comparison was carried out independently in both the discovery and validation datasets; the overlap of genes with a fold change of ≥2.0 and a p value of ≤ 0.05 in both data sets was defined as the murine single-cell differentially expressed gene expression profile. All RNA sequencing data can be accessed at GSE140896.
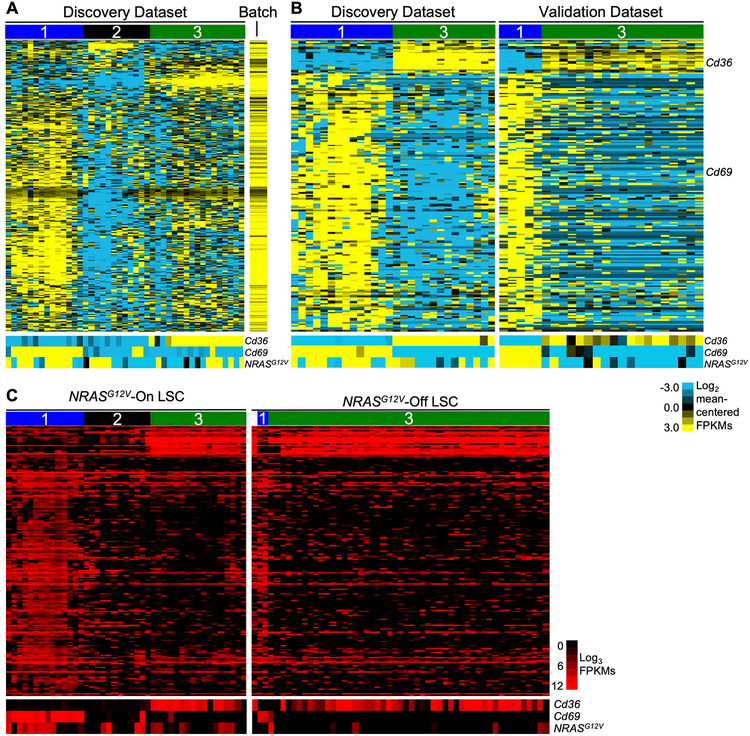
Figure 1.
Single-cell RNA sequencing identifies three distinct gene expression profiles in immunophenotypic LSCs. Primary leukemia cells harvested from the spleen of a leukemic mouse were sorted to isolate the LSC-enriched fraction (Mac1LowKit+Sca1+). This sorted population was stained for viability and captured in microfluidic chambers (1 cell per chamber, C1 Single-Cell Auto Prep System, Fluidigm, South San Francisco, California) and processed to isolate RNA. Chambers were visualized to select single, live cells for cDNA library creation and sequencing. A single cDNA library was generated per cell. An aliquot of cells was used to generate bulk RNA for sequencing as a population control. Each single-cell library was sequenced to a depth of 1–3 million reads per cell (100 bp, paired-end reads) to generate the discovery dataset. Cells with > 75% alignment were used for analysis. A, Unsupervised two-dimensional hierarchical clustering of single-cell RNA sequencing data from Mac1LowKit+Sca1+ leukemia cells in the discovery dataset (n=42). To generate this analysis, normalized expression values from each cell were mean-centered. The colors on the heatmap represent relative expression of each gene in each cell in comparison to all the cells in the dataset. Mean expressed genes per cell=3,171; range=1,491–5,387. Bottom panel: heatmap of Cd36, Cd69, and NRASG12V expression in corresponding cells. Normalized expression values (FPKMs) were mean centered (FPKM value for each gene was divided by the mean FPKM value of the gene for the entire dataset) and log2 transformed. B, Primary leukemia cells were harvested from the spleen of a second leukemic mouse and processed for single-cell capture and RNA sequencing (as described above) to generate a validation dataset (n=33, mean expressed genes per cell=2,680, range=1,392–4,409). Genes differentially expressed between Cd69High (Group 1) and Cd36High (Group 3) cells in both datasets were used to generate heatmaps. In this panel, the discovery dataset is reproduced from Fig. 1A, but Group 2 (Cd36LowCd69Low) cells are omitted from this analysis to aid in visualization. As in panel A, normalized expression values were mean-centered and log2 transformed. C, A leukemic mouse was treated with doxycycline to abolish NRASG12V transgene expression. Primary leukemia cells were harvested from the spleen of this mouse after 72 hours of treatment. These “Ras-Off” leukemia cells were sorted to isolate Mac1LowKit+Sca1+ (MKS) cells and processed for single-cell capture and RNA sequencing (as described above) to generate the Ras-Off single-cell dataset (n=51, mean expressed genes per cell=2,355, range=1,576–4,035). We used the 197 common differentially expressed genes (defined in the Ras-On datasets) to perform one-dimensional hierarchical clustering of the single-cell transcriptional data of sorted LSCs from all of these datasets. Bottom panels: heatmap of Cd36, Cd69, and NRASG12V expression in corresponding cells. Unlike the analyses in panels A and B, where the data was mean-centered to highlight genes that were differentially expressed within the dataset, these data represent FPKM expression values that were not mean-centered. As there was little cell-to-cell variation in the expression of these genes in the NRASG12V-Off dataset, mean-centering the values would have obscured differences between Group 1 and Group 3 genes in this dataset.
SingleScore: a computational approach designed to quantitate the levels of a gene expression profile within single cell RNA sequencing data.
We developed SingleScore, a single-cell transcriptional scoring system that assesses the expression of a gene expression profile in a single-cell transcriptional dataset. SingleScore is designed to overcome the sparsity and frequent gene drop-outs inherent to single-cell data. SingleScore reports a composite score that reflects each cell’s average expression of a gene list as a deviation from the median expression of those genes in the entire dataset. To account for dropout, we use the dataset’s nonzero median expression of each gene in the list. Z-scores are computed for each cell to reflect the deviation of the fold change of the gene set of interest relative to the typical deviation observed across all cells in the dataset. A positive z-score indicates that an individual cell over-expresses the genes of interest compared to other cells in that dataset; a negative z-score indicates a cell expresses few genes of interest or expresses them at a lower expression level, relative to other cells in the dataset. To calculate a SingleScore z-score:
- The L2 Norm of each cell is calculated by taking the square root of the sum of the FPKMs squared for each gene in that single-cell.
- The L2-normalized expression value of each gene in each cell is then calculated by dividing the FPKM of each gene by the L2 Norm of the cell.
- To calculate the fold change value for each gene, the nonzero median of each gene is first identified. The nonzero median of each gene is the median expression value of that gene among all of the non-zero expression values of that gene in the dataset (the median expression value among the cells that express that gene). The log2 fold change for each gene is then computed as follows:
- The average fold change for each cell is calculated by taking the mean of the log2-transformed fold change values for all of the genes in the gene set of interest
- The overall z-score for each cell (the SingleScore z-score) is calculated by subtracting the average of these average fold changes values computed across all cells from the average fold change of the single cell of interest and dividing this number by the standard deviation of the fold changes calculated for all cells in the dataset.
Human single-cell qPCR data analysis
Clustering.
The normalized data were clustered using K-means (k=2) and hierarchical agglomerative clustering, using cosine as the distant metric (k-means and clustergram functions, respectively, in MATLAB, Mathworks Inc, Natick, MA), yielding similar results. Overall mean expression of each gene was compared between clusters by calculating the difference between the mean expression vector for each gene in each cluster (by subtraction).
Enrichment analyses.
Enrichment was calculated as the expected fraction (of G1 or G3 genes) divided by the observed fraction. Housekeeping genes were omitted for these analyses. P values for enrichment per sample were calculated exactly using the Hypergeometric function (hygecdf function, Mathworks Inc.). Due to the symmetry of the Hypergeometric function, the p value for Clusters A and B are identical. The enrichment probabilities of AML samples were compared to those of the normal samples with the Wilcoxon Rank Sum test (ranksum function, Mathworks Inc.) for each cluster, then combined using the formula p1p2(1-log(p1p2)), where pi represents the p-value obtained for each cluster individually.
Determination of LSC-specific genes.
To identify LSC-specific genes, the rank-based values were assigned in the 95–0 range in each sample, the top gene in each cell was assigned rank 95. This adjustment allow direct comparison of the most highly expressed genes across all samples. Cells with the top 10% rank of each gene were selected in all samples and compared between pooled AML and pooled normal bone marrow cells. Statistical significance of the difference of expression of genes between AML and normal cells was calculated using the Wilcoxon Rank Sum test, performed using the ranksum function in MATLAB (Mathworks, Inc).
Comparison of gene expression in pooled AML relative to pooled normal bone marrow samples.
All of the cells from all of the AML samples were pooled to generate a single group of AML cells. Likewise, all of the cells from all of the normal bone marrow samples were pooled to generate a single group of normal bone marrow cells. Genes that passed quality control (as described in the quality control section, above) and that were expressed by at least one AML sample and one normal bone marrow sample were included in the analysis. The expression distribution of each gene was compared between the pooled AML samples and the pooled normal samples using a Student’s t test and Rank Sum analysis (ttests2 function, ranksum function, Mathworks, Inc).
Assessment of gene expression distribution of AML cells.
The Kolmogorov-Smirnoff test (kstest2 function, Mathworks, Inc.) was used to assess the degree of divergence of each gene in each AML sample from a reference distribution. All of the normal bone marrow samples were pooled to generate a normal reference distribution. All of the AML samples, excluding the sample being tested, were pooled to generate an AML reference distribution. Resulting values from each reference distribution were analyzed using the Rank Sum test (ranksum function, Mathworks, Inc.).
Human Samples
All human samples were obtained with written informed consent in accordance with the policies of the Institutional Review Board of the University of Minnesota (IRB study #1506E72802 and 0611M96846). They were provided as de-identified specimens.
Mice
Mice were handled in accordance with protocols and procedures approved by the University of Minnesota Institutional Animal Care and Use Committee.
Additional information, including single-cell capture and data processing, are described in Supplementary Materials.
Results
Single-cell RNA sequencing identifies three distinct gene expression profiles in immunophenotypic LSCs
We performed single-cell RNA sequencing of the Mac1LowKit+Sca1+ (MKS) subpopulation of Mll-AF9/NRASG12V AML, the LSC-enriched compartment of this leukemia[8]. We obtained MKS cells as we previously described[8]. Briefly, primary Mll-AF9/NRASG12V murine leukemia cells were transplanted into a mouse and allowed to engraft. Eighteen days after transplant, the mouse was sacrificed, and leukemia cells were harvested from the spleen and sorted to isolate the MKS cells. We previously demonstrated that self-renewing stem cells reside in the spleen in this model[8]. Sorted MKS cells were submitted for single-cell capture (C1, Fluidigm) and RNA sequencing. Unsupervised, two-dimensional hierarchical clustering of the single-cell data revealed that the MKS subpopulation expressed three distinct gene expression profiles (Fig. 1A). We defined three groups of cells based on these profiles (Group 1, 2, and 3). Next, we searched for genes that encode cell surface markers that could distinguish cells from each of these groups. We reasoned that we could use such markers to isolate cells that express each of these transcriptional groups. We found that Cd36 and Cd69 were the most differentially expressed cell surface markers between the single-cell transcriptional groups (Fig. 1A bottom panel, Supplementary Fig. S2A–B). Group 1 was Cd36LowCd69High, Group 2 was Cd36LowCd69Low, and Group 3 was Cd36HighCd69Low.
Next, we sought to define a reproducible single-cell-based gene expression signature associated with these groups. To generate a validation dataset, we repeated the experiment and performed single-cell capture (C1, Fluidigm) and RNA sequencing of the LSC-enriched subpopulation (MKS) of another leukemic mouse. As in the first dataset (the “discovery dataset”), Cd36 and Cd69 were the most differentially expressed cell surface markers. We generated a list of genes that were differentially expressed between Group 1 (Cd36LowCd69High) and Group 3 (Cd36HighCd69Low) with a p-value <0.05 separately in each dataset. There were 899 such differentially expressed genes in the discovery dataset and 916 such genes in the validation dataset. There were 198 such genes that were differentially expressed in both datasets. Of these 198 genes, 197 were concordantly differentially expressed: 174 genes were upregulated in Group 1 (Cd36LowCd69High) and 23 genes were upregulated in Group 3 (Cd36HighCd69Low) in both datasets (Fig. 1B, Supplementary Table S1). This list of 197 differentially expressed genes represents the murine LSC single-cell gene expression profile. We hypothesize that this concordance and reproducibility indicates that these differentially expressed genes likely represent relevant biology.
Because activated NRAS enforces self-renewal and increased proliferation in normal HSCs, we investigated the effect of NRASG12V-withdrawal on the expression of the LSC single-cell gene expression profile. We treated a leukemic mouse with doxycycline to abolish NRASG12V transgene expression and harvested its “Ras-Off” leukemia cells. Ras-Off MKS cells were isolated by sorting and submitted for single-cell capture and RNA sequencing. As expected, Ras-Off MKS cells expressed significantly lower levels of NRASG12V (Fig. 1C, bottom panel). Additionally, Group 2 and 3 cells within the Ras-On MKS dataset expressed lower levels of the NRASG12V transgene than Group 1 cells (Fig. 1C, bottom panel) despite equivalent levels of Vav1 (the Vav1 promoter drives NRASG12V transgene expression) among these cells (Supplementary Fig. S2C). These findings suggest that differential expression of NRASG12V at the single-cell level may account for the transcriptional differences we observe.
Using the LSC single-cell gene expression profile, we performed unsupervised hierarchical clustering of the Ras-Off single-cell gene expression dataset (Fig. 1C). After NRASG12V-withdrawl, Cd69 and the Cd69-associated gene expression profile (Group 1) was rarely detected. Likewise, the Cd36LowCd69Low-associated profile (Group 2) was not detected in these cells. In contrast, nearly all Ras-Off cells uniformly expressed Cd36 and the Cd36-associated profile (Group 3, Fig. 1C and Supplementary Fig. S2D). These analyses reveal that NRASG12V-withdrawal leads to a loss of Group 1 and 2-associated gene expression and that the Group 3-associated gene expression persists at 72 hours post doxycycline-treatment in the LSC compartment.
Discrepant LSC gene expression profiles correspond to varying self-renewal potential and differentiation stages
Next, we compared our Group 1 (Cd69High) and Group 3 (Cd36High) transcriptional profiles to previously published gene expression profiles and databases to annotate their predicted biological significance. Using Ingenuity Pathway Analysis (IPA, Redwood City, California), we found Group 1 genes to be significantly associated with repression of hematopoietic differentiation effectors (let-7, GATA1) and activation of regulators associated with hematopoietic self-renewal, including NFκB1, HIF1α (Table 1, Supplementary Table S2). In addition, Group 1 was significantly correlated with genes of leukemia and cancer relapse. Using gene set enrichment analysis (GSEA[12]), we found that Group 1 was enriched in gene sets of leukemia self-renewal and hematopoietic stem cells (Table 2[13–15]). In contrast, Group 3 was correlated with inflammatory and hematopoietic differentiation pathways (Table 1, Supplementary Table S2). These data suggest that Group 1 and 3 transcriptional profiles represent distinct functional states of LSCs.
Table 1. Ingenuity Pathway Analysis.
Representative Ingenuity Pathway Analysis of the common differentially expressed genes.
| Upstream Regulator, Pathway, or Function | z-score | p-value |
|---|---|---|
| CD69-associated genes | ||
| let-7 | −3 | 0.00000287 |
| GATA-1 | −2.546 | 3.05E-10 |
| NFKB | 1.715 | 0.0167 |
| HIF1A | 2.651 | 0.00007 |
| Recurrent hematological cancer | 0.0000004 | |
| CD36-associated genes | ||
| IL4 | 1.103 | 0.00194 |
| Tretinoin | 1.4 | 0.0219 |
| IFNG | 1.836 | 0.0000452 |
| Differentiation of myeloid leukocytes | 0.83 | 0.000027 |
Table 2. Gene Set Enrichment Analysis (GSEA): Group 1 relative to Group 3 gene expression.
Gene set enrichment analysis, comparing Group 1 and Group 3 transcriptional profiles. NES=normalized enrichment score, FDR=false discovery rate; FDR is used as a p-value which has been corrected for multiple hypothesis testing.
| Gene sets enriched in Group 1 LSCs | NES | FDR q |
|---|---|---|
| MLL-AF9-dependent self-renewal genes (Zuber et al.) | 2.01 | 0.00 |
| LSC genes (Somervaille et al.) | 1.71 | 0.01 |
| Myb-dependent self-renewal genes (Zuber et al.) | 1.84 | 0.01 |
| HSC and CMP genes (Laurenti et al.) | 1.60 | 0.04 |
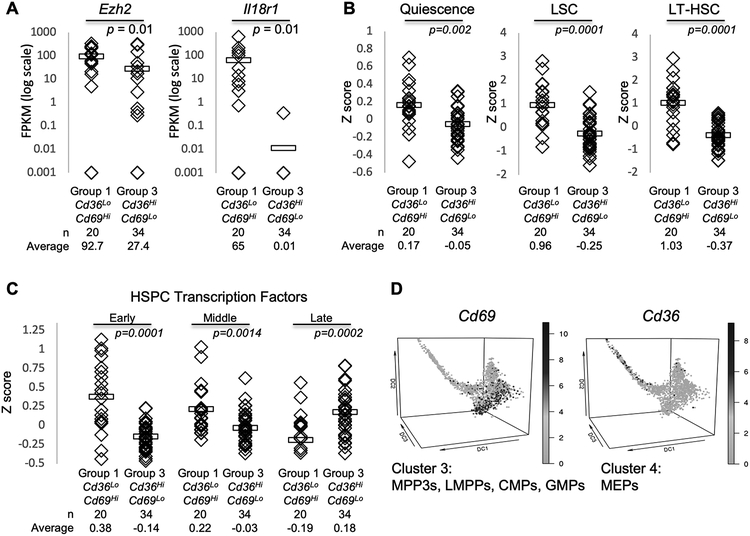
We also compared our data to a recently published single-cell qPCR analysis of a retrovirally-transduced model of MLL-AF9 AML[16]. In that study, they identified a group of genes that are associated with increased aggressiveness (reduced leukemia latency). Our Group 1 cells expressed higher levels of these aggressiveness genes, most notably, Ezh2, which is critical for maintaining the quiescence of leukemia and embryonic-like stem cells (Fig. 2A)[16–18]. Additionally, a single-cell RNA sequencing study of Flt3(ITD)/Dnmt3ainsufficient murine AML revealed that elevated levels of Il18r1 were associated with increased in vitro clonogenicity. Accordingly, this gene was higher in Group 1 (Cd69High) relative to Group 3 (Fig. 2A).
Figure 2.
Discrepant LSC gene expression profiles correspond to varying differentiation stages and self-renewal potential. A, The average FPKM values for genes in Group 1 and 3 single-cell data pooled from the discovery and validation datasets. Ezh2 is associated with aggressive leukemic behavior in the Guo et al. Cell Stem Cell 2013 dataset; Il18r1 is associated with in vitro clonogenicity in the Meyer et al. Cancer Discovery 2016 dataset. Horizontal bars indicate the mean expression value. Unpaired Student’s t-test was used to calculate p-values for each comparison indicated. B-C, SingleScore z scores were calculated to compare the levels of previously published gene expression profiles between Group 1 and Group 3 cells pooled from the discovery and validation datasets. B, Quiescence[19], LSC[20], and LT-HSC[21] gene expression profiles were defined in bulk RNA sequencing data. C, HSPCs transcription factors were curated from recently published single-cell gene expression studies of normal hematopoiesis reporting the expression of transcription factor genes, at the single-cell level, at different stages of HSPC development[22, 23]. D, Cd69 and Cd36 expression in normal mouse HSPCs and progenitors (Nestorowa et al. dataset). MPP=multipotent progenitors, LMPP=lymphoid-primed multipotent progenitors, CMP=common myeloid progenitor, GMP=granulocyte-monocyte progenitor, MEP=megakaryocyte erythrocyte progenitor. Unpaired Student’s t-test was used to calculate p-values for each comparison indicated (A-D).
We developed a computational approach, SingleScore, to directly quantitate the levels of gene profiles in single-cell transcriptional data. SingleScore is designed to overcome the sparsity of single-cell data and calculates a z-score that represents the overall expression level of a list of genes in each cell, relative to other cells in the population. We used SingleScore to compare the expression levels of gene profiles between Group 1 and Group 3 cells. We found that gene expression profiles of Quiescence[19], LSCs[20], and LT-HSCs[21] were preferentially expressed in higher levels in Group 1 cells (relative to Group 3 cells, Fig. 2B). Taken together, these data suggest that Group 1 (Cd69High) and Group 3 (Cd36High) may represent groups with varying self-renewal potential.
We interrogated our single-cell dataset for genes that encode markers of hematopoietic differentiation and found that cells from both Groups 1 and 3 express Flt3 and Cd48, but only cells from Group 1 express Cd34 (Supplementary Fig. S3A). Since AMLs express cell surface markers aberrantly, we used SingleScore to assess the levels of hematopoietic transcription factors (TFs) in our single-cell dataset as an additional assessment of the differentiation status of our transcriptional subgroups. For this analysis, we reviewed recently-published single-cell transcriptional datasets of normal hematopoiesis that defined TF expression along differentiation of LT-HSCs to more committed hematopoietic progenitors, at the single-cell level[22, 23]. We used SingleScore to compare the expression of early, middle, and late HSPC TFs between Group 1 and Group 3 cells (Fig. 2C). Group 1 cells preferentially expressed higher levels of TF of early hematopoiesis while Group 3 cells preferentially expressed higher levels of late HSPC TFs, suggesting that these two LSC subpopulations might phenocopy HSPCs at varying stages of differentiation.
Because Cd36 and Cd69 distinguish between the single-cell transcriptional subgroups, we asked whether these genes are expressed at distinct stages of normal hematopoiesis. We queried a previously published catalog of single-cell gene expression profiles of early mouse hematopoiesis[23] and found that Cd69 was most highly expressed in very early multipotent progenitor cells (Fig. 2D, Supplementary Fig. S3B–D). In contrast, high Cd36 expression was restricted to more committed megakaryocyte-erythrocyte progenitors (MEPs). As in our leukemia progenitors, high Cd36 and high Cd69 levels were generally mutually exclusive in all hematopoietic cells reported in this study, but this effect was most prominent in long-term hematopoietic stem cells (LT-HSCs), the cells with self-renewal potential (Supplementary Fig. S3E–G).
Human LSCs recapitulate the murine LSC single-cell gene expression profile at the single-cell level
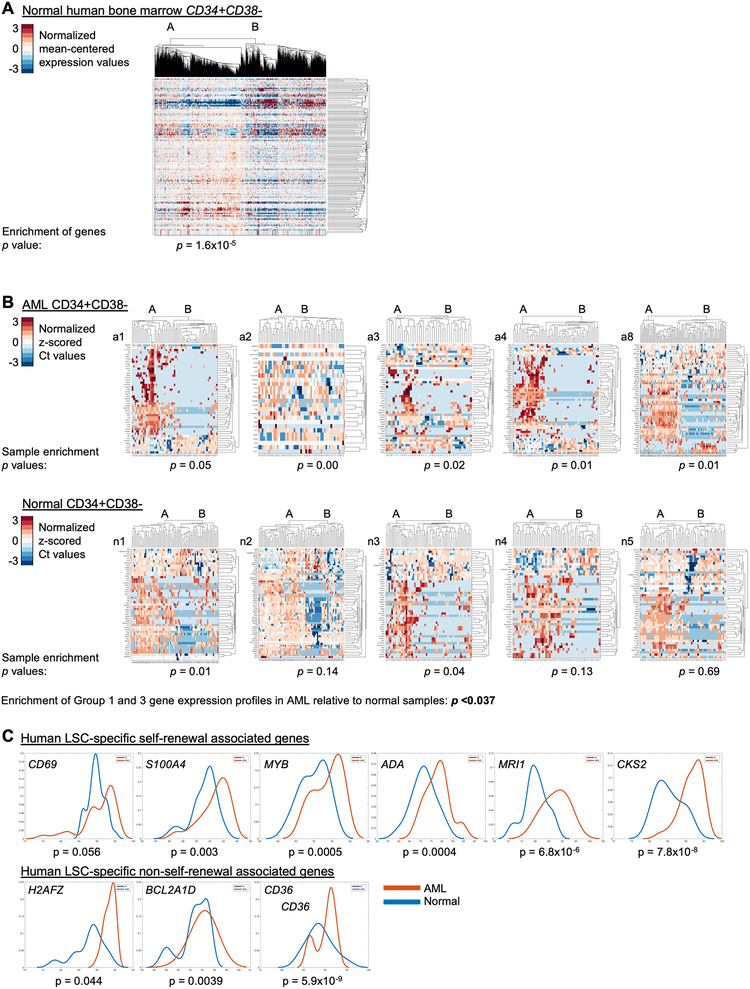
To determine whether the transcriptional behaviors we observed in our murine model can be detected in normal human HSPCs, we interrogated a recently published single-cell transcriptional dataset derived from 25 normal human bone marrow samples[24]. We selected CD34+CD38- cells based on expression of these two genes (CD34 >0, CD38≤1) and analyzed the expression of the murine LSC single-cell gene expression profile in these cells. Unsupervised, two-dimensional hierarchical clustering of this data identified 2 clusters (Fig. 3A). To investigate if the clusters in normal human HSPCs recapitulate the patterns found in murine LSCs, we compared the relative levels of Group 1 and Group 3 genes in these two clusters and found a strong enrichment of Group 1 genes in one cluster and strong enrichment of Group 3 genes in the other cluster (p value for enrichment = 1.6×10−5), indicating that these normal HSPCs differentially express Group 1 and Group 3 genes in a manner similar to the murine LSCs. These data suggest that the single-cell transcriptional patterns we identified in the murine LSCs might represent normal hematopoietic transcriptional programs that are co-opted in leukemia.
Figure 3.
Human LSCs and normal HSPCs recapitulate the murine LSC single-cell gene expression profiles at the single-cell level. A, The expression of genes from the murine LSC single-cell gene expression profile in a recently published single-cell RNA sequencing dataset of normal human bone marrow[24] is displayed in a heatmap representing an unsupervised, two-dimensional hierarchical clustering of the data. CD34+CD38- cells were selected from the data set for this analysis. Each column represents a single cell. Each row represents a gene. B-C, The murine single-cell gene expression dataset was used to develop a 96-gene PCR assay. Diagnostic bone marrow aspirates from patients with AML (n=5) or normal human bone marrow specimens (Lonza, n=5) were CD34+CD38- selected and submitted for single-cell capture and quantitative, reverse transcriptase PCR using this 96-gene PCR assay. The Ct values were normalized (as described in the methods). B, Data from each patient were clustered by two-dimensional hierarchical clustering. Each column represents a single cell. Each row represents a gene. The cluster with the highest expression of genes is labeled Cluster A and the other is labeled Cluster B. The p-value for enrichment of these genes within each cluster is displayed for each sample and directly represents the degree of enrichment seen (low p value indicates strong enrichment). C, The top 10% of cells expressing each gene from each AML or normal sample were pooled. Genes that were higher among LSCs relative to normal HSPCs are displayed. The Wilcoxon Rank Sum test was performed to compare expression values between LSCs and normal HSPCs.
To determine if these patterns are also shared with human AML, we performed single-cell whole transcriptome RNA sequencing of CD34+ cells from a bone marrow specimen obtained at AML diagnosis (Mll rearranged, Supplementary Fig. S4A–B, Supplementary Tables S3–4, sample a2). Direct assessment of the murine LSC single-cell gene expression profile could not be performed due to data sparsity: very few of the genes in our murine LSC single-cell gene expression profile were detected in this dataset. Unsupervised hierarchical clustering of the genes that were detected and differentially expressed within this dataset revealed two distinct gene expression profiles. IPA identified differential expression of pathways of hematological development and cell survival between these profiles (Supplementary Fig. S4C). Additionally, IPA Upstream Regulators analysis revealed that these CD34+ diagnostic human AML cells differ in their expression of RAS-activated pathways (Supplementary Fig. S4C). These findings suggest that features of the transcriptional heterogeneity we detect in our mouse model may be similarly heterogeneous in human AML stem cells.
Next, we sought to determine whether the differentially expressed genes identified in the murine single-cell datasets are variably expressed, at the single-cell level, in human AML stem cells. For these experiments, we developed a 96 gene qPCR panel that represents genes that differ between Group 1 and Group 3 in the validation dataset (Supplementary Table S5 for the gene panel). We sorted human LSCs (hLSCs, CD34+CD38-) from diagnostic bone marrow aspirates of five adult AML patients with intermediate or poor risk cytogenetic and molecular features (Supplementary Tables S3–4) and performed single-cell qPCR using the 96-gene panel. We clustered the data from each patient and identified two clusters of cells (using K-means and hierarchical agglomerative clustering). We characterized the expression profile of each cluster with respect to higher overall expression of each gene. In each AML sample, we found that one cluster was enriched in Group 1 genes and the other enriched in Group 3 genes (Fig. 3B, Supplementary Table S6). This enrichment pattern was significant in each of the AML samples. These data indicate that the murine LSC single-cell gene expression profile is expressed in human LSCs and that Group 1 and Group 3 genes are differentially expressed in human LSCs similarly to the murine model.
Next, we sought to determine whether this profile was also expressed in the CD34+CD38- HSPCs from normal human bone marrow. We sorted CD34+CD38- HSPCs and performed single-cell capture and qPCR analysis using the 96-gene panel. Hierarchical clustering of the data reveals two clusters in each sample. As in the AML samples, one cluster (A) is enriched in Group 1 genes and Cluster B is enriched in Group 3 genes. However, in the normal HSPCs, this enrichment was significantly reduced relative to the AML samples: enrichment in two of five normal samples was significant. The enrichment of these gene profiles was significantly stronger in the AML samples relative to the normal samples (p <0.037 for enrichment, p<0.008 for enrichment probabilities). These findings suggest that these gene expression patterns are more strongly associated with leukemic LSCs than with normal HSPCs.
Next, we investigated whether we could identify a subset of LSC-specific genes based on high expression in LSCs relative to normal HSPCs. We pooled the highest expressing 10% of cells for each gene (the cells that express the highest amounts of each gene) from all AML samples and all normal HSPC samples, and compared expression of these genes, among these top-expressing cells, between pooled AML cells and pooled HSPC cells. This analysis identified six Group 1 and three Group 3 LSC-specific genes (Group 1: CD69, S100A4, MYB, ADA, MRI1, CKS2; Group 3: H2AFZ, BCL2A1D, CD36; p-value of ≤0.05, Fig. 3C). These hLSC-specific genes included CD36 and CD69. MYB encodes a transcription factor that has been implicated in leukemia self-renewal transcriptional programming[15] and was also higher in a subset of hLSCs. Likewise, S100A4 has been implicated in glioma self-renewal[25] and fibroblast invasiveness[26]. This list of LSC-specific genes may represent mediators of aberrant self-renewal and leukemic behavior in these cells.
Next, we assessed whether Group 1 and Group 3 gene expression profiles are likely specific to AML (relative to normal HSPCS) using two approaches. First, we asked whether individual genes are expressed differently between AML and normal cells. For this analysis, we pooled all AML cells into one group and all normal bone marrow cells into a separate group. We performed both parameteric (Student’s t test) and nonparametric (Rank Sum) analyses to compare expression of each gene between AML and normal cells and found that 61% (t-test)-57% (Rank Sum) of the genes were differentially expressed between AML and normal bone marrow with a p<0.05. Second, we tested whether each AML sample appears to be drawn from the distribution of the other AML samples or from the distribution of normal bone marrow samples. Using the Kolmogorov-Smirnov test, we found that that deviation of each AML sample from the distribution of normal bone marrow samples was much greater than its deviation from the distribution of other AML samples (p<3.4197e-04, see Supplementary Fig. S4DE), indicating that each AML sample individually is unlikely to be drawn from the distribution of the normal samples. These findings suggest that the AML samples likely include very few residual normal HSPCs. Together, these analyses demonstrate that the Group 1 and Group 3 genes are differentially expressed in human AML stem cells, as they are in murine LSCs, and that these gene expression profiles appear to be largely leukemia-specific.
In vivo leukemia reconstitution assays demonstrate that Group 1 is the self-renewing subset of immunophenotypic LSCs
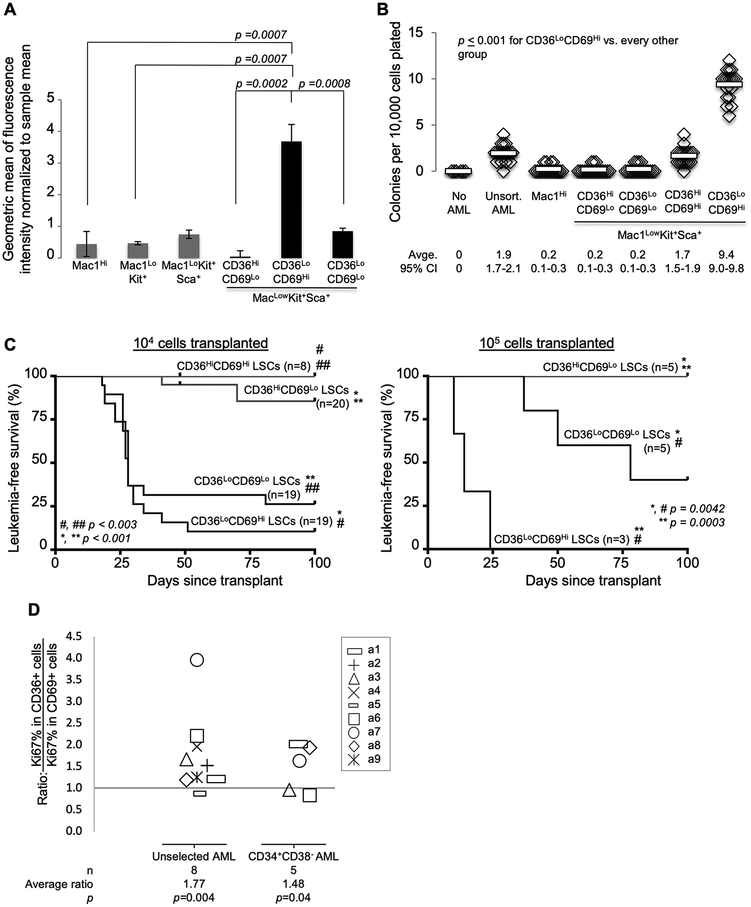
Since previous work suggest that self-renewal and proliferation can be separate functions in normal HSCs, we asked whether Group 1 and Group 3 represent cells with discrepant proliferative capacity in LSCs. We stained bulk leukemia cells from our murine model with CellTrace, transplanted these cells into mice, and harvested leukemia from these mice 14 days after transplant. Within the MKS compartment, CellTrace was retained at significantly higher levels in cells with an immunophenotype corresponding to the Group 1 gene expression profile (Mac1LowKit+Sca1+CD36LowCD69High) than any other immunophenotypic subgroup within the MKS population (Fig. 4A). In contrast, CellTrace was retained at the lowest level among MKS cells with an immunophenotype corresponding to the Group 3 gene expression profile (Mac1LowKit+Sca1+CD36HighCD69Low). Another MKS subgroup expressed low levels of both CD36 and CD69 (Group 2) and displayed intermediate levels of CellTrace. These data indicate that MKS cells with an immunophenotype consistent with Group 1 are poorly proliferative while those with the Group 3 pattern are highly proliferative.
Figure 4.
In vivo leukemia reconstitution assays demonstrate that Group 1 is the self-renewing subset of immunophenotypic LSCs. A, Primary leukemia cells were stained with CellTrace and transplanted into mice via tail vein injection. Leukemia cells were harvested 14 days later. Levels of CellTrace are shown according to immunophenotypic subgroups as assayed by flow cytometry. Error bars indicate standard error of the mean. Unpaired Student’s t-test was used to calculate p-values for each comparison indicated. B-C, The LSC-enriched compartment (Mac1LowKit+Sca1+) was sorted based on CD36 and CD69 status. B, Sorted cells were plated in methylcellulose colony forming assays. Results are representative of 4 independent experiments. Horizontal bars indicate mean value. Unpaired Student’s t-test was calculated for each comparison indicated. CI=confidence interval. C, Sorted cells were transplanted into mice via tail vein injection. Each mouse received either 104 or 105 sorted cells (as indicated). Leukemia status was assessed by weekly peripheral blood complete blood counts. MKSCD36HighCD69High were very rare and were therefore not tested at the higher cell dose. Leukemia-free survival is plotted as Kaplan-Meier curves. Log-rank tests were used to calculate p-values. D, The levels of Ki67 (a proliferation marker) were compared between CD36High and CD69High subpopulations of AML. Diagnostic bone marrow aspirates from patients with AML were stained with antibodies to CD34, CD38, CD36, CD69, and Ki67. The ratio of the percentage of cells that express Ki67 in CD36HighCD69Low cells was compared to that of CD36LowCD69High cells within the bulk sample and within the CD34+CD38- (LSC-enriched) compartment. Each symbol represents this ratio in a single sample. Not all samples had sufficient CD36HighCD69Low or CD36LowCD69High cells within the CD34+CD38- compartment for these measurements.
We next tested whether these MKS subgroups vary in their self-renewal capacity. We sorted cells based on immunophenotypic markers (sorting strategy and post-sort assessments in Supplementary Fig. S5–6) and plated them in colony forming assays. As we previously described, Mac1High leukemia cells lack colony forming capacity, while MKS (Mac1LowKit+Sca1+) cells harbor the colony forming capacity of this leukemia[8]. Among the MKS population, the CD36LowCD69HighMKS subpopulation (Group 1), which accounts for approximately 5–10% of the MKS compartment, formed the most colonies, while the CD36HighCD69LowMKS subpopulation (Group 3), which accounts for approximately 20–50% of the MKS compartment, was unable to form colonies (p = 0.00001, Fig. 4B). We assessed the viability of these leukemic subgroups using a dead-cell exclusion dye and found that Group 3 cells were highly viable (more so than Group 1 cells, Supplementary Fig. S7A). We also used these sorted cells to perform in vivo mouse leukemia reconstitution assays. CD36LowCD69High MKS (Group 1) cells induced leukemia with the highest penetrance and lowest latency of all groups: 20 of 22 mice developed leukemia with a median latency of 14 and 21 days, depending on transplanted cell dose (Fig. 4C). In contrast, CD36HighCD69Low MKS (Group 3) cells were almost uniformly unable to induce leukemia in mice: 2 of 25 mice developed leukemia at 41 and 48 days after transplant. CD36LowCD69Low MKS (Group 2) cells displayed an intermediate phenotype: 17 of 24 mice developed leukemia with a prolonged latency. Leukemias that developed in the transplanted mice matched the immunophenotype of the parent leukemia (Supplementary Fig. S7BC). These data demonstrate that Group 1 (CD36LowCD69HighMKS) cells are relatively quiescent and harbor the self-renewal capacity of this leukemia.
To assess whether CD36 and CD69 also delineate cells with discrepant proliferative capacity in human AML, we compared the levels of Ki67 (a proliferation marker) in CD36HighCD69Low and CD36LowCD69High subsets of human AML bone marrow samples, obtained at diagnosis. We used mass cytometry (CyTOF) to compare the percentage of Ki67+ cells within these compartments in CD34+CD38- and unselected leukemia cells and found that the CD36LowCD69High subset was less proliferative than the CD36HighCD69Low subset within each of these compartments (Fig. 4D, Supplementary Fig. S8, patient information in Supplementary Tables S3–4). These findings suggest that CD36 and CD69 may identify human AML subsets with differing functional capacity. As in murine LSCs, where CD69 defines a relatively quiescent, self-renewing subpopulation of LSCs, we found that CD69High human AML cells were less proliferative than CD36High cells.
Discussion
Using single-cell transcriptional profiling of an NRASG12V-driven murine model of AML, we defined distinct transcriptional profiles in immunophenotypic LSCs, delineating distinct groups of cells. In comparison to previously published gene expression studies, we found that Group 1 cells (Cd69High) expressed genes of self-renewal and relapse while Group 3 cells (Cd36High) expressed a signature consistent with hematopoietic differentiation. Genes from this murine LSC single-cell profile were also differentially expressed in primary human LSCs at the single-cell level. When comparing normal human HSPCs to human LSCs, we found a subset of these genes that were LSC-specific and included both CD69 and CD36. Finally, we used CD69 and CD36 protein expression to delineate murine LSCs from these groups. We found that Group 1 efficiently transplanted leukemia but was poorly proliferative, while Group 3 was unable to transplant leukemia and was highly proliferative. Similarly, CD69High human AML cells were more quiescent than CD36High cells, suggesting that these markers may also identify human LSCs subsets with varying self-renewal potential. These experiments define an LSC self-renewal gene expression profile at the single-cell level and demonstrate that self-renewal and proliferation are separate functions in LSCs, as they often are in HSCs. These results highlight the importance of therapeutically targeting self-renewal, in addition to rapid proliferation, to prevent relapse in AML.
Recent reports of single-cell profiling of AML and normal precursors have been instrumental in describing the architecture of molecular heterogeneity of these cells[5, 27–32]. Some studies have started to test the functional relevance of single-cell heterogeneity in leukemic behavior. Single-cell transcriptomics, coupled with functional assays, demonstrated the bimodality of proliferation and self-renewal in normal HSCs[32]. A single-cell qPCR assessment of genes encoding cell surface proteins identified Cd24 expression as a marker of decreased leukemia latency in an MLL-AF9 retrovirally transduced model of murine AML and described a set of Cd24-associated genes at the single-cell level[16]. Single-cell RNA sequencing of AML precursors from a murine model of Dnmt3amutant/Flt3ITD revealed a panel of genes that encoded cell surface markers whose expression correlated with clonogenicity in vitro[33]. Studies that have experimentally tested the single-cell gene expression behavior of AML self-renewal in vivo have not been reported to our knowledge.
Normal HSCs have been shown to acquire a proliferation or self-renewal advantage in response to NRAS activation[7]. In these experiments, we find that the gene expression profile of self-renewal (Group 1) is closely correlated with NRASG12V levels at the single-cell level, suggesting NRASG12V is enforcing this profile, consistent with our earlier analysis of bulk LSCs in this model[8]. As in normal hematopoiesis, leukemia is hierarchically organized with pluripotent stem cells that give rise to progressively more differentiated leukemia cells[3, 34, 35]. Other groups have demonstrated that leukemia stem cells are relatively quiescent compared to more differentiated leukemia cells[35–39]. However, the bimodality of proliferation versus self-renewal has not been previously demonstrated in LSCs. Our experiments demonstrate that the gene expression profiles associated with self-renewal and proliferation are indeed distinct and that these functions are separate in LSCs in our murine AML model.
The Group 1 gene expression profile described in murine LSCs was associated with self-renewal in our functional assays. This profile was also associated with signatures of previously validated mediators of AML and HSC self-renewal (such as NFκB and HIF1α) and was enriched in previously annotated gene sets of leukemia self-renewal (identified in bulk studies). Notably, CD69, a calcium dependent lectin superfamily type II transmembrane receptor best known for its role in lymphocyte activation, is an NFκB-responsive gene[40]. Recently, CD69 inhibition was shown to increase the proliferation of HSPCs[41], consistent with our finding that CD69High LSCs are poorly proliferative. Our functional validation experiments along with comparisons to previously published gene expression datasets collectively identify Group 1 as the single-cell self-renewal signature of LSCs in our murine model.
IPA analyses show that the Group 1 profile is associated with the expression of numerous profiles of hematopoietic and solid tumor relapse. Because stem cell self-renewal is a critical feature that allows these cells to cause relapse, expression of the Group 1 profile may be an important determinant of relapse. Future work could interrogate the specific contribution of genes within these profiles to self-renewal and relapse in AML models and AML patients.
Several features of the Group 3 profile are consistent with a differentiated state (relative to the Group 1 profile). IPA analysis identified myeloid differentiation pathways that are repressed in Group 1. In normal mouse hematopoiesis, Cd69 is primarily limited to early HSPCs, while Cd36 is seen only in more committed progenitors. An intermediate population of cells, displaying the Group 2 gene expression profile, was CD36LowCD69Low and displayed intermediate self-renewal and proliferative capacity; this finding suggests that these cells may represent an intermediate stage between CD69High self-renewing cells and CD36High proliferative cells. These data are consistent with a model of CD69High self-renewing stem cells (Group 1) that give rise to progressively more committed and proliferative CD36High cells (Group 3) with a CD36LowCD69Low intermediate (Group 2).
Importantly, we found evidence that components of these gene expression profiles are expressed in human LSCs and HSPCs at the single-cell level. Previous work implies that LSCs self-renew by enforcing the self-renewal gene expression profile of normal HSCs[42]. Our qPCR data support this model at the single-cell level. Additionally, our data identified a group of genes that were specifically highly expressed in human LSCs. These LSC-specific genes may provide functional insights into leukemia-specific behaviors and may be useful in identifying self-renewal-specific therapeutic targets.
Consistent with the murine data, CD69 and CD36 genes correlated with these signatures in human samples and were LSC-specific genes. While CD69 and CD36 protein levels correspond to different proliferative rates in human AML, our data do not formally establish whether these proteins can identify human LSCs with different self-renewal capacities. Future work, utilizing CD36High or CD69High sorted human LSCs in serial colony forming or leukemia xenograft assays could demonstrate these functions. Nevertheless, our data implicate CD36 or CD69 as possible subjects for antibody-based targeted therapies, which have revolutionized treatment of hematological malignancies[43–46]. CD36 expression is associated with altered leukemia cell metabolism and chemoresistance[47, 48]; this finding is consistent with a pro-leukemic effect. CD36 is a glycoprotein cell surface receptor that plays a wide variety of functions, including cell adhesion, platelet regulation, and fatty acid transport. MYB, another LSC-specific gene in our analysis, has been implicated in AML self-renewal[8, 14, 15] and has several candidate inhibitors in development[49–51].
In conclusion, our findings demonstrate that self-renewal and proliferation are mutually exclusive features of LSCs and suggest that targeting self-renewal, in addition to proliferation, may be an important strategy to prevent AML relapse.
Supplementary Material
Statement of Significance.
These findings define and functionally validate a self-renewal gene profile of leukemia stem cells at the single-cell level and demonstrate that self-renewal and proliferation are distinct in AML.
Acknowledgements
The authors thank Michael Franklin for editorial assistance. The authors thank Katy Richards-Hrdlicka and Jason McKinney from Fluidigm for helping us to assess and ensure the quality of the single-cell qPCR data.
This work utilized the resources of the University of Minnesota Hematological Malignancies Tissue Bank which is supported by the National Cancer Institute (5P30CA077598-18), Minnesota Masonic Charities, and the Killebrew-Thompson Memorial Fund through the Cancer Experimental Therapeutics Initiative (CETI); the Flow Cytometry Resource and other services of the Masonic Cancer Center (which is supported by NIH 2P30CA077598-21) at the University of Minnesota; Jerry Daniel and Darrell Johnson and the University of Minnesota Genomics Center Fluidigm C1 single-cell capture and BioMark qPCR and next-generation sequencing services; the Mass Cytometry Shared Resource at the University of Minnesota (which is supported by the Office of the Vice President for Research, U of MN) and the Minnesota Supercomputing Institute. This work and Z. Sachs were supported by the American Cancer Society, Frederick A. DeLuca Foundation, Mentored Research Scholar Grant (MRSG-16-195-01-DDC); the Clinical and Translational Science Institute at the University of Minnesota KL2 Career Development and K to R01 awards (National Institutes of Health’s National Center for Advancing Translational Sciences, grants KL2TR002492 and UL1TR002494. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health’s National Center for Advancing Translational Sciences); the Lois and Richard King Assistant Professorship in Medicine, the American Cancer Society Institutional Research Grant at the University of Minnesota (124166-IRG-58-001-55-IRG12); the Masonic Cancer Center at the University of Minnesota Translational Working Group Award and Genetic Mechanisms of Cancer Award; the University of Minnesota Department of Medicine Women’s Early Research Career Award; the University of Minnesota Genomic Center Pilot Award; the Randy Shaver Cancer Research and Community Fund, the Masonic Cancer Center, the Division of Hematology, Oncology, and Transplantation, Department of Medicine; and the University of Minnesota Foundation donors. K. Sachs was supported in part by an award from the Muscular Dystrophy Association (MDA Award #574137). D. Largaespada was supported by an American Cancer Society Research Professorship.
Footnotes
Conflict of interest: Karen Sachs had been paid consulting fees by Fluidigm in the past for analyses that predate the data in this manuscript. The authors declare no other potential conflicts of interest.
References
- 1.Kubal T and Lancet JE, The thorny issue of relapsed acute myeloid leukemia. Curr Opin Hematol, 2013. 20(2): p. 100–6. [DOI] [PubMed] [Google Scholar]
- 2.Kreso A and Dick JE, Evolution of the cancer stem cell model. Cell Stem Cell, 2014. 14(3): p. 275–91. [DOI] [PubMed] [Google Scholar]
- 3.Shlush LI, et al. , Tracing the origins of relapse in acute myeloid leukaemia to stem cells. Nature, 2017. 547(7661): p. 104–108. [DOI] [PubMed] [Google Scholar]
- 4.Ng SW, et al. , A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature, 2016. 540(7633): p. 433–437. [DOI] [PubMed] [Google Scholar]
- 5.Saito Y, et al. , Overcoming mutational complexity in acute myeloid leukemia by inhibition of critical pathways. Sci Transl Med, 2017. 9(413). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas D and Majeti R, Biology and relevance of human acute myeloid leukemia stem cells. Blood, 2017. 129(12): p. 1577–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Q, et al. , Oncogenic Nras has bimodal effects on stem cells that sustainably increase competitiveness. Nature, 2013. 504(7478): p. 143–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sachs Z, et al. , NRASG12V oncogene facilitates self-renewal in a murine model of acute myelogenous leukemia. Blood, 2014. 124(22): p. 3274–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corral J, et al. , An Mll-AF9 fusion gene made by homologous recombination causes acute leukemia in chimeric mice: a method to create fusion oncogenes. Cell, 1996. 85(6): p. 853–61. [DOI] [PubMed] [Google Scholar]
- 10.Wiesner SM, et al. , Repressible transgenic model of NRAS oncogene-driven mast cell disease in the mouse. Blood, 2005. 106(3): p. 1054–62. [DOI] [PubMed] [Google Scholar]
- 11.Kim WI, et al. , RAS oncogene suppression induces apoptosis followed by more differentiated and less myelosuppressive disease upon relapse of acute myeloid leukemia. Blood, 2009. 113(5): p. 1086–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Subramanian A, et al. , Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A, 2005. 102(43): p. 15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laurenti E, et al. , The transcriptional architecture of early human hematopoiesis identifies multilevel control of lymphoid commitment. Nat Immunol, 2013. 14(7): p. 756–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Somervaille TC, et al. , Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell, 2009. 4(2): p. 129–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zuber J, et al. , An integrated approach to dissecting oncogene addiction implicates a Myb-coordinated self-renewal program as essential for leukemia maintenance. Genes Dev, 2011. 25(15): p. 1628–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo G, et al. , Mapping cellular hierarchy by single-cell analysis of the cell surface repertoire. Cell Stem Cell, 2013. 13(4): p. 492–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujita S, et al. , Dual inhibition of EZH1/2 breaks the quiescence of leukemia stem cells in acute myeloid leukemia. Leukemia, 2018. 32(4): p. 855–864. [DOI] [PubMed] [Google Scholar]
- 18.Shin DM, et al. , Global gene expression analysis of very small embryonic-like stem cells reveals that the Ezh2-dependent bivalent domain mechanism contributes to their pluripotent state. Stem Cells Dev, 2012. 21(10): p. 1639–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graham SM, et al. , Transcriptional analysis of quiescent and proliferating CD34+ human hemopoietic cells from normal and chronic myeloid leukemia sources. Stem Cells, 2007. 25(12): p. 3111–20. [DOI] [PubMed] [Google Scholar]
- 20.Gal H, et al. , Gene expression profiles of AML derived stem cells; similarity to hematopoietic stem cells. Leukemia, 2006. 20(12): p. 2147–54. [DOI] [PubMed] [Google Scholar]
- 21.Ivanova NB, et al. , A stem cell molecular signature. Science, 2002. 298(5593): p. 601–4. [DOI] [PubMed] [Google Scholar]
- 22.Buenrostro JD, et al. , Integrated Single-Cell Analysis Maps the Continuous Regulatory Landscape of Human Hematopoietic Differentiation. Cell, 2018. 173(6): p. 1535–1548 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nestorowa S, et al. , A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood, 2016. 128(8): p. e20–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oetjen KA, et al. , Human bone marrow assessment by single-cell RNA sequencing, mass cytometry, and flow cytometry. JCI Insight, 2018. 3(23). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chow KH, et al. , S100A4 Is a Biomarker and Regulator of Glioma Stem Cells That Is Critical for Mesenchymal Transition in Glioblastoma. Cancer Res, 2017. 77(19): p. 5360–5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xia H, et al. , Calcium-binding protein S100A4 confers mesenchymal progenitor cell fibrogenicity in idiopathic pulmonary fibrosis. J Clin Invest, 2017. 127(7): p. 2586–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corces MR, et al. , Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet, 2016. 48(10): p. 1193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dai YJ, et al. , Conditional knockin of Dnmt3a R878H initiates acute myeloid leukemia with mTOR pathway involvement. Proc Natl Acad Sci U S A, 2017. 114(20): p. 5237–5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jan M, et al. , Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci Transl Med, 2012. 4(149): p. 149ra118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paguirigan AL, et al. , Single-cell genotyping demonstrates complex clonal diversity in acute myeloid leukemia. Sci Transl Med, 2015. 7(281): p. 281re2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Grandis M, et al. , JAM-C Identifies Src Family Kinase-Activated Leukemia-Initiating Cells and Predicts Poor Prognosis in Acute Myeloid Leukemia. Cancer Res, 2017. 77(23): p. 6627–6640. [DOI] [PubMed] [Google Scholar]
- 32.Wilson NK, et al. , Combined Single-Cell Functional and Gene Expression Analysis Resolves Heterogeneity within Stem Cell Populations. Cell Stem Cell, 2015. 16(6): p. 712–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyer SE, et al. , DNMT3A Haploinsufficiency Transforms FLT3ITD Myeloproliferative Disease into a Rapid, Spontaneous, and Fully Penetrant Acute Myeloid Leukemia. Cancer Discov, 2016. 6(5): p. 501–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonnet D and Dick JE, Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med, 1997. 3(7): p. 730–7. [DOI] [PubMed] [Google Scholar]
- 35.Hope KJ, Jin L, and Dick JE, Acute myeloid leukemia originates from a hierarchy of leukemic stem cell classes that differ in self-renewal capacity. Nat Immunol, 2004. 5(7): p. 738–43. [DOI] [PubMed] [Google Scholar]
- 36.Behbehani GK, et al. , Mass Cytometric Functional Profiling of Acute Myeloid Leukemia Defines Cell-Cycle and Immunophenotypic Properties That Correlate with Known Responses to Therapy. Cancer Discov, 2015. 5(9): p. 988–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guan Y, Gerhard B, and Hogge DE, Detection, isolation, and stimulation of quiescent primitive leukemic progenitor cells from patients with acute myeloid leukemia (AML). Blood, 2003. 101(8): p. 3142–9. [DOI] [PubMed] [Google Scholar]
- 38.Ishikawa F, et al. , Chemotherapy-resistant human AML stem cells home to and engraft within the bone-marrow endosteal region. Nat Biotechnol, 2007. 25(11): p. 1315–21. [DOI] [PubMed] [Google Scholar]
- 39.Terpstra W, et al. , Fluorouracil selectively spares acute myeloid leukemia cells with long-term growth abilities in immunodeficient mice and in culture. Blood, 1996. 88(6): p. 1944–50. [PubMed] [Google Scholar]
- 40.Cibrian D and Sanchez-Madrid F, CD69: from activation marker to metabolic gatekeeper. Eur J Immunol, 2017. 47(6): p. 946–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Notario L, et al. , Anti-CD69 therapy induces rapid mobilization and high proliferation of HSPCs through S1P and mTOR. Leukemia, 2018. 32(6): p. 1445–1457. [DOI] [PubMed] [Google Scholar]
- 42.Krivtsov AV, et al. , Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature, 2006. 442(7104): p. 818–22. [DOI] [PubMed] [Google Scholar]
- 43.Wei AH and Tiong IS, Midostaurin, enasidenib, CPX-351, gemtuzumab ozogamicin, and venetoclax bring new hope to AML. Blood, 2017. 130(23): p. 2469–2474. [DOI] [PubMed] [Google Scholar]
- 44.Kantarjian H, et al. , Monoclonal antibody-based therapies: a new dawn in the treatment of acute lymphoblastic leukemia. J Clin Oncol, 2012. 30(31): p. 3876–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee HC and Weber DM, Advances and practical use of monoclonal antibodies in multiple myeloma therapy. Hematology Am Soc Hematol Educ Program, 2016. 2016(1): p. 512–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maloney DG, Anti-CD20 antibody therapy for B-cell lymphomas. N Engl J Med, 2012. 366(21): p. 2008–16. [DOI] [PubMed] [Google Scholar]
- 47.Ye H, et al. , Leukemic Stem Cells Evade Chemotherapy by Metabolic Adaptation to an Adipose Tissue Niche. Cell Stem Cell, 2016. 19(1): p. 23–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Farge T, et al. , Chemotherapy-Resistant Human Acute Myeloid Leukemia Cells Are Not Enriched for Leukemic Stem Cells but Require Oxidative Metabolism. Cancer Discov, 2017. 7(7): p. 716–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pattabiraman DR and Gonda TJ, Role and potential for therapeutic targeting of MYB in leukemia. Leukemia, 2013. 27(2): p. 269–77. [DOI] [PubMed] [Google Scholar]
- 50.Ramaswamy K, et al. , Peptidomimetic blockade of MYB in acute myeloid leukemia. Nat Commun, 2018. 9(1): p. 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walf-Vorderwulbecke V, et al. , Targeting acute myeloid leukemia by drug-induced c-MYB degradation. Leukemia, 2018. 32(4): p. 882–889. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.