Abstract
Purpose:
A cryptic inv(16)(p13.3q24.3) encoding the CBFA2T3-GLIS2 fusion is associated with poor outcome in infants with acute megakaryocytic leukemia. We aimed to broaden our understanding of the pathogenesis of this fusion through transcriptome profiling.
Experimental Design:
Available RNA from children and young adults with de novo AML (N=1,049) underwent transcriptome sequencing (mRNA and miRNA). Transcriptome profiles for those with the CBFA2T3-GLIS2 fusion (N=24) and without (N=1,025) were contrasted to define fusion-specific miRNAs, genes, and pathways. Clinical annotations defined distinct fusion-associated disease characteristics and outcomes.
Results:
The CBFA2T3-GLIS2 fusion was restricted to infants < 3 years-old (p<0.001) and presence of this fusion was highly associated with adverse outcome (p<0.001) across all morphological classifications. Further, there was a striking paucity of recurrent cooperating mutations and transduction of cord blood stem cells with this fusion was sufficient for malignant transformation. CBFA2T3-GLIS2 positive cases displayed marked up-regulation of genes with cell membrane/extracellular matrix localization potential, including NCAM1 and GABRE. Additionally, miRNA profiling revealed significant over-expression of mature miR-224 and miR-452, which are intronic miRNAs transcribed from the GABRE locus. Gene-set enrichment identified dysregulated Hippo, TGFβ, and hedgehog signaling, as well as NCAM1 (CD56) Interaction pathways. Therapeutic targeting of fusion-positive leukemic cells with CD56-directed ADC caused significant cytotoxicity in leukemic blasts.
Conclusions:
The CBFA2T3-GLIS2 fusion defines a highly refractory entity limited to infants that appears to be sufficient for malignant transformation. Transcriptome profiling elucidated several highly targetable genes and pathways, including the identification of CD56, providing a highly plausible target for therapeutic intervention.
Keywords: Acute Myeloid Leukemia, Pediatric, CBFA2T3-GLIS2, Transcriptional Profiling, RNA-seq
Introduction
Pediatric acute myeloid leukemia (AML) is characterized by significant cytogenetic and molecular heterogeneity. However, the number of patients with AML having clinically actionable variants is limited (1). Although the majority of patients have identifiable chromosomal abnormalities, up to 20% of children have no cytogenetic abnormalities (1,2). Genomic profiling of megakaryocytic AML (French-American-British classification M7) identified a cryptic CBFA2T3-GLIS2 fusion that was associated with adverse outcome (3,4). While CBFA2T3-GLIS2 was subsequently found to be enriched in children with normal cytogenetics (CN-AML), and later, not exclusive to either CN or M7 AML (3,5–7), the prevalence and prognostic significance of CBFA2T3-GLIS2 in a large heterogeneous cohort of patients has not been fully evaluated.
CBFA2T3 was initially identified as a fusion partner with RUNX1 in therapy-related AML and shown to facilitate transcriptional repression, as well as being implicated in hematopoietic stem cell quiescence (3). GLIS2 is closely related to the GLI subfamily and likely functions in modulating the hedgehog signaling pathway (8,9). The fusion of GLIS2 to CBFA2T3 as a result of inv(16)(p13.3q24.3) leads to increased expression of the GLIS2 DNA-binding domain (3). A number of important features of CBFA2T3-GLIS2 AML have been identified through molecular experiments and next generation sequencing (NGS) analyses. Acute megakaryoblastic leukemia (AMKL) with CBFA2T3-GLIS2 displayed altered expression and activation of hedgehog and bone morphogenic signaling pathways, thereby representing potential therapeutic targets (3,10,11). Further genomic interrogation of fusion-positive AMKL cell-lines identified over-expression of ERG transcripts and a synergistic relationship between the CBFA2T3-GLIS2 oncoprotein and the transcription factor ERG, which primes the leukemic phenotype and blocks differentiation (12). In addition, CBFA2T3-GLIS2 positive cases were found to over-express the cell surface marker CD56 (NCAM1) and chromatin immunoprecipitation demonstrated that the fusion protein binds to the proximal promoter, inducing its over-expression (4,13).
However, previous genomic and transcriptomic studies of CBFA2T3-GLIS2 have limited the investigation to AMKL and CN-AML patients in their characterization of fusion-positive cohorts, in part due to the low frequency of the fusion in unselected populations (~2%). Our study expands on the known features of fusion-positive AML by including a diverse cohort of patients encompassing all morphological categories, in addition to normal cytogenetics and megakaryoblastic, to provide increased scope and detail in understanding the pathogenesis of this aggressive subtype. We identified 39 CBFA2T3-GLIS2 fusion-positive cases from an unselected cohort of 2,027 pediatric AML patients and all fusion-positive cases were experimentally validated using reverse-transcription (RT-PCR) based methods. Available clinical annotations and flow cytometry data were interrogated, and finally, transcriptional profiling of mRNA and miRNA sequencing data for 24/39 fusion-positive and 1,025 fusion negative cases were employed to identify fusion-specific molecular biomarkers from aberrantly expressed miRNAs, genes, and pathways. The specificity of these immunophenotypic and transcriptional markers can be leveraged to define a high-risk group of pediatric AML patients, who will need immediate and specialized treatment.
Materials and Methods
Biological Specimens and Clinical Annotations.
Pediatric patients with de novo AML enrolled on Children’s Oncology Group (COG) trials CCG-2961, AAML03P1, AAML0531, and AAML1031, with written informed consent and available clinical and/or molecular annotations were included for biological study. Trials were conducted in accordance with the Declaration of Helsinki. The Fred Hutchinson Cancer Research Center Institutional Review Board and the COG Myeloid Biology Committee approved the study. Details of treatment protocols have been previously published (14–17). Analyses of outcome and clinical characteristics were limited to patients from AAML0531 and AAML1031. Data were current as of March 31, 2018 for outcome analyses and cohort characteristics are summarized in Supplemental Table 1 and Supplemental Table 2. Patients with FLT3-ITD high allelic ratio from AAML1031 were excluded from survival and clinical associations due to enrollment on the phase I sorafenib treatment arm, since a proportion of patients were still on therapy with data under DSMC review. Risk groups were defined as follows: patients with t(8;21), inv(16), NPM1 or CEBPA mutations were considered low risk, while patients with monosomy 7, monosomy 5/del5q or high allelic ratio FLT3-ITD (ITD-AR > 0.4) were considered high risk (16). Remaining patients were considered standard risk. As a comparison, we screened 299 patients enrolled in the SWOG adult AML trial S0106.
Flow Cytometry, Cytotoxicity Assays, and CBFA2T3-GLIS2 Transduction.
Myeloid immune marker flow cytometry data were provided by Hematologics (Seattle, WA) for 437 pediatric AML patients enrolled on AAML0531. Methods for flow cytometry were previously reported (18). Cytotoxicity assays were carried out on primary AML samples by Notable Labs translational drug discovery platform (Foster City, CA) and utilized the CD56 antibody clone m906 as previously described (19). The antibody controls included chKTi-SPP-DM1 (20) and human IgG-SAP and goat IgG-ZAP (Advanced Targeting Systems, San Diego, CA, #IT-36, #IT-35). Human cord blood samples were obtained with informed consent under Swedish Medical Center Institutional Review Board (Seattle, WA). CD34+ cord blood cells were transduced with a pRRL lentivirus encoding the CBFA2T3-GLIS2 fusion transcript and GFP (21). Cell surface antigen expression was assessed by flow cytometry (18).
RNA-sequencing Library Construction.
RNA from primary patient samples were purified using the QIAcube system with AllPrep DNA/RNA/miRNA Universal Kits (QIAGEN, Valencia, CA, #80224). The mRNA libraries were prepared for 75-bp strand-specific paired-end sequencing using the ribodepletion 2.0 protocol by the British Columbia Genome Sciences Center (BCGSC, Vancouver, BC). Sequenced libraries were aligned to GRCh37 and gene level coverage was quantified using the BCGSC pipeline v1.1 with Ensembl v69 annotations. The microRNA libraries were produced using the miRNA 3.0 protocol by BCGSC. Sequenced reads (31-bp) were trimmed to remove adapter sequences and aligned to GRCh37. Perfect alignments with no mismatches were retained for mature miRNA quantification using miRBase v20 annotations. Transcriptomic data is available through the dbGaP TARGET: Acute Myeloid Leukemia study (Accession: phs000465.v19.p8).
Screening of CBFA2T3-GLIS2 Fusion.
The CBFA2T3-GLIS2 fusion transcript was detected by RNA-sequencing for AAML1031 using STAR-fusion v1.1.0 and TransAbyss v1.4.10 fusion detection software (22,23) and experimentally verified using RT-PCR. Patients enrolled on prior studies were screened by fragment length analysis (FLA) using the Applied Biosystems 3730xl DNA Analyzer. Primers for RT-PCR verification and FLA are listed in Supplemental Methods.
Differential Expression, Hierarchical Clustering, Gene-Set Enrichment Analysis, and miRNA-mRNA Interactions.
All analyses were completed in the R statistical environment. Differentially expressed genes and miRNAs were identified using Limma v.3.36.1 and those with absolute log2 fold change > 1 and false discovery rate (FDR)< 0.05 were retained. Gene counts were TMM normalized and converted to log2 scale for unsupervised hierarchical clustering. Gene-set enrichment analysis was completed using GAGE v2.30.0 (24). Interactions between miRNA-mRNA were investigated by selecting pairs of differentially expressed miRNAs and genes with significant anti-correlation using Spearman’s rho (FDR<0.05). The miRNA-miRNA pairs were investigated for both predicted and validated interactions using anamiR v1.10.0 and multiMiR v1.4.0 packages. LNA Inhibition of miR-224 and miR-452 in CBFA2T3-GLIS2 M07E cells are described in Supplemental methods.
Results
Fusion Transcript Detection.
Primary samples from children and young adults enrolled on COG AAML1031 were utilized for RNA-sequencing and the CBFA2T3-GLIS2 fusion was identified using fusion detection algorithms (22,23), and experimental validation by RT-PCR. Specimens from earlier studies were screened using RT-PCR based methods (2). In total, we identified 39 fusion-positive cases from 2,027 patients screened (1.9%) with 3 distinct chromosomal breakpoints. The majority of fusion-positive patients (80%) had a breakpoint at exon 11 of CBFA2T3 and exon 3 of GLIS2 (Figure 1A, B)(25). The remaining had breakpoints between CBFA2T3 exon 10 and GLIS2 exon 2, and one patient was found to have a fusion breakpoint in CBFA2T3 exon 9 and GLIS2 exon 3 (Supplemental Figure 1).
Figure 1. The cryptic fusion CBFA2T3-GLIS2 is most prevalent in patients less than 3 years old and is associated with poor outcomes.
A) Representation of the most common breakpoint in in pediatric AML (Zhou et al. 2015) and B) resulting in the CBA2T3-GLIS2 fusion transcript. C) Frequency of CBFA2T3-GLIS2 occurrence by age group. D) Oncoprint with CBFA2T3-GLIS2 cohort. Columns are patients and genetic abnormalities are rows. *One patient (purple) was identified as inv(16) through FISH, though no RNA-seq or karyotype evidence was detected. E) Kaplan-Meier estimates for CBFA2T3-GLIS2 AML compared to fusion-negative cohorts.
CBFA2T3-GLIS2 AML Characteristics and Clinical Outcome.
Complete clinical annotations were available for 37 fusion-positive and 1,724 fusion-negative cases from AAML0531 and AAML1031. All fusion-positive patients were < 3 years old and the median age was 1.5 years (range: 0.75 – 2.96 years) compared to a median of 10.0 years for fusion-negative AML (range: 0.01 – 29.8 years, p<0.001). In patients < 3 years of age, CBFA2T3-GLIS2 fusion was seen in 8.4% (37/441) of cases, and the majority of CBFA2T3-GLIS2 AML were diagnosed at 1-year old, accounting for 11.7% of all patients diagnosed in this age group (Figure 1C). In contrast, the CBFA2T3-GLIS2 fusion was not detected in 299 adult patients with AML (ages 20–60 years, p<0.001). There appears to be an ethnic predisposition for this fusion with nearly a third (29.4%) of CBFA2T3-GLIS2 patients being black or African American, compared with 12.8% in the fusion-negative population (p=0.009). Comparison of disease characteristics showed no significant difference in presenting white blood cell count at diagnosis between fusion-positive and negative patients.
All but one fusion-positive patient (97%) were classified as standard-risk based on conventional cytogenetic or molecular characteristics (16). One patient was classified as favorable risk based on positive FISH for Inv(16) without corresponding karyotype. Previous detailed genomic evaluation of 9 fusion-positive cases by whole genome, whole exome, or targeted exome sequencing failed to show any recurrent somatic mutations (2).
Evaluation of the karyotype of CBFA2T3-GLIS2 cases demonstrated presence of rare structural events in 63.9% of cases without any of the common AML associated recurrent fusions. A single patient with CBFA2T3-GLIS2 fusion was reported with inv(16) based on FISH but lacked the karyotype and one patient was identified with t(8;16) by karyotype, without the supporting evidence of this translocation by RNA-seq fusion calls, leading one to consider this a potential genomic rearrangement that may not produce a translated fusion product. The genomic variant trisomy 3 was detected in 7/36 cases (19.4%) vs 13 (0.8%) cases among fusion-negative patients. A minority of fusion-positive cases had trisomy 21 (4/36, 11.1%, p= 0.022) and one with trisomy 8. The CBFA2T3-GLIS2 fusion was also not seen in those with adverse karyotype including monosomy 7, monosomy 5, or deletion 5q. There was a modest enrichment of cytogenetically normal fusion-positive patients (33.3% CN-AML) compared to 22.6% CN for fusion-negative patients overall (Figure 1D). Slightly greater than half of the fusion-positive cohort (18/33) had an M7 megakaryoblastic morphology (p<0.001). The next most common FAB classification was M1 minimal differentiated morphology (6/33, 18.2%), while the remaining cases encompassed a variety of FAB types. The CBFA2T3-GLIS2 fusion was also mutually exclusive of the RBM15-MKL1 and NUP98-KDM5A fusions, despite both being prevalent in M7 megakaryoblastic and younger AML patients.
Morphologic response to induction chemotherapy was assessed for fusion-positive patients, revealing 50.0% morphologic CR rate at the end of induction 1 (EOI1) compared to 76.1% for fusion-negative (p<0.001). Fusion-positive AML had a higher median percentage of residual disease (MRD) at the end of induction 1 and 2 compared to the fusion-negative population; 80% were MRD-positive at the end of induction 1 (vs 26.7%, p<0.001) and 35.5% at the end of induction 2 (vs 13.7%, p=0.005). In fact, 94.3% of CBFA2T3-GLIS2 patients had detectable residual disease after induction 1 (median: 0.7%, range: 0.0–41.0%). Overall survival (OS) at 5-years after study entry for CBFA2T3-GLIS2 AML was 22.0% vs. that of 63.0% in fusion-negative cases (HR = 2.8, p<0.001, Figure 1E). CBFA2T3-GLIS2 also associated with a higher probability of an event (death, relapse, or induction failure) with an event-free survival (EFS) at 5-years from study entry 18.9% versus 46.9% for fusion-negative patients (HR = 2.0, p<0.001). Associated adverse prognostic impact was observed regardless of whether patients had megakaryocytic phenotype or not. A multivariable analysis including CBFA2T3-GLIS2 status, cytogenetic/molecular risk classification and M7 morphology, demonstrated that CBFA2T3-GLIS2 was an independent prognostic factor for both OS (HR = 2.3, p<0.001) and EFS (HR = 2.0, p=0.005, Supplemental Table 3). In addition, the CBFA2T3-GLIS2 fusion has prognostic value independent of age (Supplemental Figures 2 and 3). Infant AML patients ≤ 1-year old with the CBFA2T3-GLIS2 fusion (N=30) had adverse overall survival compared to the fusion-negative (N=308) population (27.6% versus 61.3% at 5-years post enrollment, p=0.001). Infant CBFA2T3-GLIS2 patients with FAB M7 morphology (N=17) remained at greater risk of death (OS 25%) compared to other FAB M7 infant AML cases (N=44, OS 58.9%, p=0.025) 5-years after study entry.
CBFA2T3-GLIS2 Immunophenotype and Targeted Therapy.
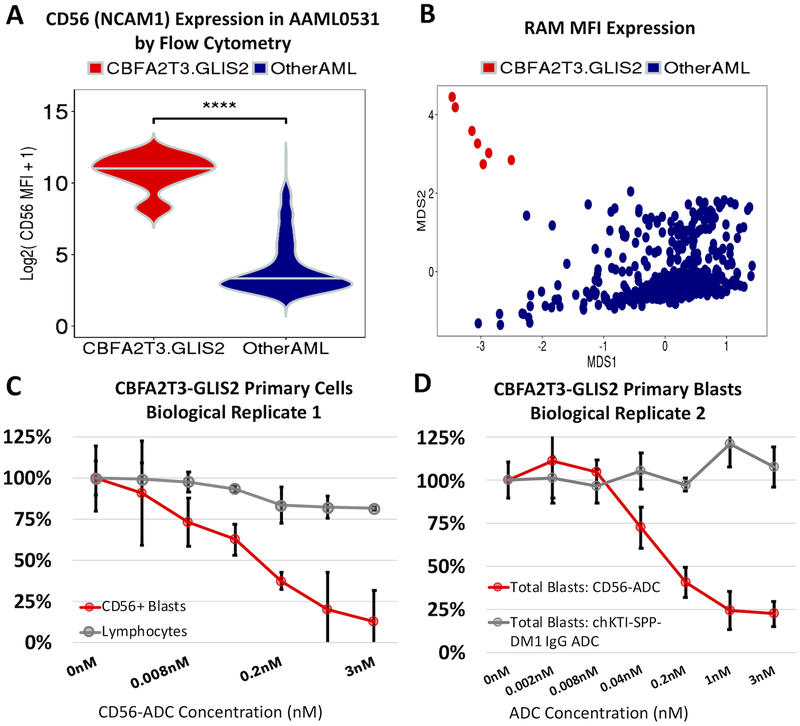
Evaluation of leukemic blast by multi-dimensional flow cytometry (MDF) demonstrated that leukemias with CBFA2T3-GLIS2 had a highly elevated CD56 expression, that in combination with dim or absent expression of HLA-DR, CD38, and CD45 can be characterized as previously described RAM phenotype (18). All patients with CBFA2T3-GLIS2 fusions were evaluated by MDF and were found to have RAM phenotype (100%, p<0.001). Expression values for 13 cell-surface antigens (myeloid marker panel) by MDF were available for a subset of patients from AAML0531 (N=437). Unsupervised hierarchical clustering demonstrated that fusion-positive tumors (7/7) clustered together by mean fluorescence intensity (MFI) of the 13 antigens, indicating that CBFA2T3-GLIS2 tumors have highly distinctive immunophenotypes, with extremely elevated surface CD56 expression (p<0.001, Figure 2A, Supplemental Figure 4). We also evaluated the available diagnostic immunophenotype data to determine whether fusion-positive cases could be identified using the characteristic RAM phenotype antigens: CD56, CD45, CD38, and HLA-DR. Unconstrained classical metric multidimensional scaling (MDS) was employed to cluster the 437 patients using MFI values of the four RAM antigens, and observed 100% (7/7) of fusion-positive AML patients clustered away from the fusion-negative cohort, providing compelling evidence that CBFA2T3-GLIS2 is associated with a unique cell surface expression profile that can be characterized by the RAM antigens alone (Figure 2B).
Figure 2. Fusion-positive cases highly over-express surface CD56 (NCAM1) and targeting CD56 with an antibody drug conjugate results in greater than 75% cytotoxicity.
A) CD56 (NCAM1) surface expression by flow cytometry in mean fluorescence intensity values (MFI) in CBFA2T3-GLIS2 patient samples (N=7) from AAML0531. B) Classical metric multidimensional scaling using expression of CD56, HLA-DR, CD38, and CD45 antigens MFI values. C) Cytotoxicity assay with CBFA2T3-GLIS2 primary cells from biological replicate #1. Blasts and normal lymphocytes from the patient were exposed in vitro to varying concentrations of the CD56 antibody drug conjugate (ADC) in triplicate. D) Cytotoxicity assay with CBFA2T3-GLIS2 blasts exposed to either the non-targeting conjugate control, chKTi-SPP-DM1 IgG ADC or CD56-ADC. Each condition and concentration were completed in triplicate.
Next, considering that a majority of CBFA2T3-GLIS2 patients express the RAM immunophenotype, we investigated whether transcript level expression would support the observations of low-to-dim expression of CD45, CD38, and HLA-DR. Using rRNA-depleted RNA-sequencing data from 24 fusion-positive cases, it was found both CD38 and CD45 transcripts were significantly down-regulated > 2-fold compared to fusion-negative AML (N=1,025), as well as the HLA-DR genes: HLA-DRA, HLA-DRB1, HLA-DRB5, and HLA-DRB6 (FDR<0.001). Investigation of miRNA-mRNA interactions using paired miRNA-sequencing data revealed CD38 mRNA had significant anti-correlation (Spearman’s rho) and predicted target binding-sites for the over-expressed miR-203a-3p (rs ≤ −0.62, FDR=0.02). For CD45, there were significant associations with highly expressed mature miR-452–3p, miR-425–5p, and both miR-224 species (rs ≤ −0.74, FDR≤0.003). HLA-DR genes also had relationships with over-expressed miRNAs: HLA-DRA had predicted target sites for miR-135a-3p (rs = −0.62, FDR=0.02) and HLA-DRB5 with miR-5683 (rs = −0.53, FDR<0.05), thus implicating miRNAs in negative regulation of the cell-surface markers that define the RAM phenotype.
We next evaluated CD56 as a potential therapeutic target using an anti-CD56 antibody-drug conjugate (m906-PBD-ADC) developed by the National Cancer Institute (19,20). Leukemic blasts and control lymphocytes from a patient with relapsed fusion-positive AML were incubated with varying doses of m906 and cytotoxicity was assessed after 72 hours. The CD56-ADC exhibited a dose-dependent and CD56-specific toxicity on leukemic blasts. There was 87% cytotoxicity of CD56+ blasts at 3nM concentration, suggesting that CD56 might prove to be a therapeutic target in this high-risk cohort of patients (p<0.001, Figure 2C). The CD56-ADC was further evaluated in an additional 3 independent CBFA2T3-GLIS2 patient samples; the dose-dependent toxicity was observed in all samples. In total, 3 of 4 CBFA2T3-GLIS2 primary samples showed > 75% cytotoxicity of leukemic blasts at 3nM concentration of the CD56-ADC (Figure 2D, Supplemental Figure 5).
Malignant transformation of cord blood stem cells by CBFA2T3-GLIS2 fusion.
Given the paucity of cooperating events in a highly aggressive disease in early life, we questioned whether this fusion may be sufficient for malignant transformation. To this end, we transduced human CD34+ cord blood stem cells (CBSC) with a lentivirus encoding the CBFA2T3-GLIS2 fusion transcript and GFP or mock control; expression of the fusion was confirmed by RT-PCR. Fusion-transduced CBSCs were more proliferative compared mock-transduced CBSCs (Supplemental Figure 6, p=0.21). After 12 weeks in culture, we evaluated immunophenotypic and morphologic features of CBFA2T3-GLIS2 fusion-transduced cells. CBFA2T3-GLIS2+ cells had immature morphology with high CD117 expression and absence of CD11b, CD36 and CD64 (Figure 3A–C). Of significance, fusion-transduced cells had remarkably high CD56 expression (3,556 MFI versus 43 MFI for the control), similar to that observed in patients with fusion-positive RAM phenotype (2,040 MFI), suggesting that CD56 expression is causally linked to the expression of the fusion transcript. In addition, morphological evaluation of CBFA2T3-GLIS2+ cells revealed multinucleated cells, prominent nucleoli, and abundant focally basophilic and vacuolated cytoplasm with cytoplasmic blebs, which are morphologic features suggestive of megakaryocytic differentiation. Megakaryocytic lineage was confirmed by demonstration of high expression of CD41 and CD61 on the surface of transduced cells (Figure 3D,E).
Figure 3. CBFA2T3-GLIS2 induces over-expression of CD56 (NCAM1) in healthy normal cord blood cells.
A) Surface immunophenotype of mock-transduced cord blood cells cultured for 6 weeks (green), B) CBFA2T3-GLIS2 transduced cord blood cells cultured for 10 weeks (purple), and C) patient with CBFA2T3-GLIS2 and RAM phenotype (red). Patient data for CD64 and CD36 not available. Morphology of CBFA2T3-GLIS2 transduced cells includes D) megakaryocytic features, such as cytoplasmic blebs, noted with arrow and E) cells express megakaryocytic surface markers CD41 and CD61.
Gene Expression Profiling of CBFA2T3-GLIS2 Patients.
Of 39 pediatric CBFA2T3-GLIS2 AML patients identified, 24/39 fusion-positive and 1,025 fusion-negative cases had material available for high depth ribodepleted mRNA-sequencing and paired miRNA-sequencing. Transcriptome data from CBFA2T3-GLIS2 positive and negative tumors were contrasted to identify fusion-specific pathways, genes, and miRNAs (Figure 4A). Gene-set enrichment analysis (24) identified a substantial activation of Hippo (FDR<0.001) and TNF signaling pathways (FDR<0.001), which have been implicated in various cancers by promoting cellular functions that enhance tumor growth, migration, and proliferation (26,27). In addition, TGFB/BMP and Hedgehog signaling pathways showed significant activation, as previously identified (3,6) (FDR≤0.001, Figure 4B,C). Furthermore, the cell-surface NCAM1 (CD56) interaction pathway was highly positively enriched (FDR<0.001), while gene-ontology (GO) term analysis revealed significant up-regulation of numerous cell-adhesion and cell-surface marker genes, including extracellular matrix binding, cell-adhesion molecule binding, and integrin binding genes (FDR<0.001).
Figure 4. A cohort of 24 CBFA2T3-GLIS2 AML patient samples were employed in transcriptional profiling for mRNA and miRNA differential expression analysis and gene-set enrichment analysis.
A) Schematic representation of the data types and transcriptomic analyses in this study. B) Heatmap of positively and negatively (blue) enriched pathways from gene-set enrichment analysis of the CBFA2T3-GLIS2 cohort. Pathways (rows) have Benjamini-Hochberg adjusted p-values ≤ 0.001. The enrichment statistic for the gene-set enrichment analysis are denoted in each tile and were derived from a two-sample t-test for differential expression of gene sets. C) Differentially expressed genes involved in Hedgehog signaling or its cross-talk found to be up-regulated in CBFA2T3-GLIS2 tumors.
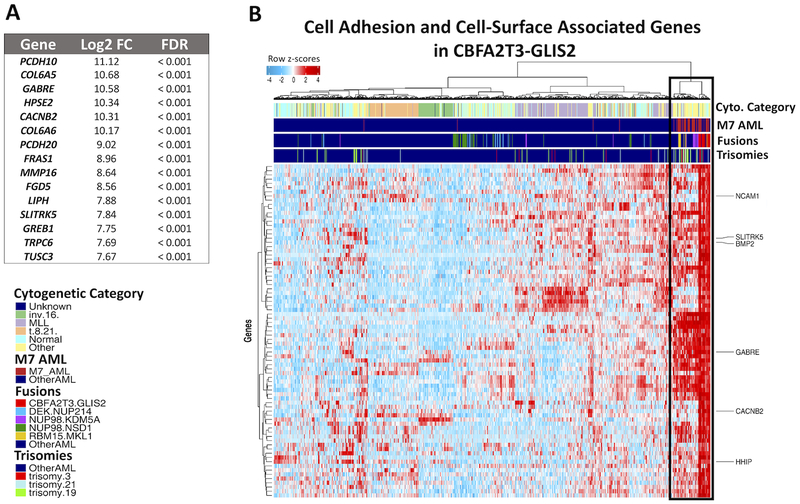
Considering that a characteristic feature of CBFA2T3-GLIS2 AML was positive regulation of cell adhesion and extracellular matrix binding (ECM), over-expressed genes whose protein products have demonstrated localization to the plasma membrane and ECM were examined (28). Of 3,711 differentially expressed genes (DEGs), 789 genes had cell adhesion, plasma membrane, and ECM associations (FDR<0.001, Figure 5A, Supplemental Figure 7). Plasma membrane and ECM genes included 10 NCAM1 interaction pathway genes, over-expressed hematopoietic lineage adhesion molecules CD44, ITGA2, and ITGA2B, as well as, numerous receptor tyrosine kinases. CD44, expressed at a level more than 2-fold greater than fusion-negative patients, is aberrantly expressed on leukemic stem cells and a marker of HSCs (29,30). Similarly, ITGA2 is a cell adhesion molecule on hematopoietic cells and used to classify subsets of HSCs (29). Upregulated receptor tyrosine kinases (RTKs) that localize to the cell periphery and cell membrane included ROR1, MET, NTRK1, and two of the TAM family kinases, TYRO3 and AXL, were noted (log2 fold-change (LFC) ≥ 2.0, FDR<0.001). The TAM family of RTKs has been shown to increase tumor cell survival, migration, and chemoresistance (31), while others have known roles in cancer progression (32,33). In addition, genes affected by BMP and hedgehog signaling crosstalk (10) with demonstrated ECM localization were upregulated, including BMP2, WNT9A, and WNT11. A concomitant overexpression of WNT11 and ERG transcripts (LFC ≥ 1.5, FDR<0.001) was observed, and it has been shown the ERG transcription factor directly binds the WNT11 locus to promote cancerous transformation in AML (34).
Figure 5. CBFA2T3-GLIS2 cases over-express a number of cell adhesion, cell-surface, and extracellular matrix associated genes.
A) Top 15 Significantly upregulated cell adhesion, extracellular matrix, and cell-surface associated genes in CBFA2T3-GLIS2 AML. B) Unsupervised hierarchal clustering of 1,049 pediatric AML patients using the 90th percentile of highest upregulated cell-surface genes (FDR < 0.001). Color bars indicate primary cytogenetic code, M7 AML classification, fusion status (CBFA2T3-GLIS2 in red), and trisomies associated with CBFA2T3-GLIS2. The patients with similar cell-adhesion/extracellular matrix associated gene expression profiles are highlighted by the black box.
Unsupervised hierarchical clustering based on the most highly expressed (90th percentile LFC, N=79 genes) cell-surface associated genes found CBFA2T3-GLIS2 AML exhibits a unique expression profile with a distinct set of dysregulated genes: NCAM1 (CD56), the NCAM1-interacting partner CACNB2, and GABRE receptor gene (Figure 5B). Hierarchical clustering on the cell adhesion/ECM associated markers also revealed a number of fusion-negative AML patients (N=66) that clustered closely with the fusion-positive cohort. This cohort shared similar characteristics with fusion-positive patients: they tend to be very young, with a median age of 1.8 years (range: 0.09 – 17.6 yrs) and 85% of cases are < 3 years-old. They were enriched in M7 AML (N=37/54, 68%), including the majority of NUP98-KDM5A (N=12/17, 70.5%) and RBM15-MKL1 (N=10/10, 100%) cases. Patients with similar cell-surface associated gene expression also had poor outcomes compared to the fusion-negative population for OS (47.5% vs 64.8%, p<0.001), though not EFS (p=0.17). Identification of similar patients allows the inclusion of many AML patients that otherwise would be missed in a targeted investigation of therapeutic biomarkers. However, these patients have some distinct differences, sharing 57/79 (72%) of the most upregulated cell-surface associated genes in common with CBFA2T3-GLIS2 (Supplemental Table 4).
MicroRNA Profiling of CBFA2T3-GLIS2 Patients.
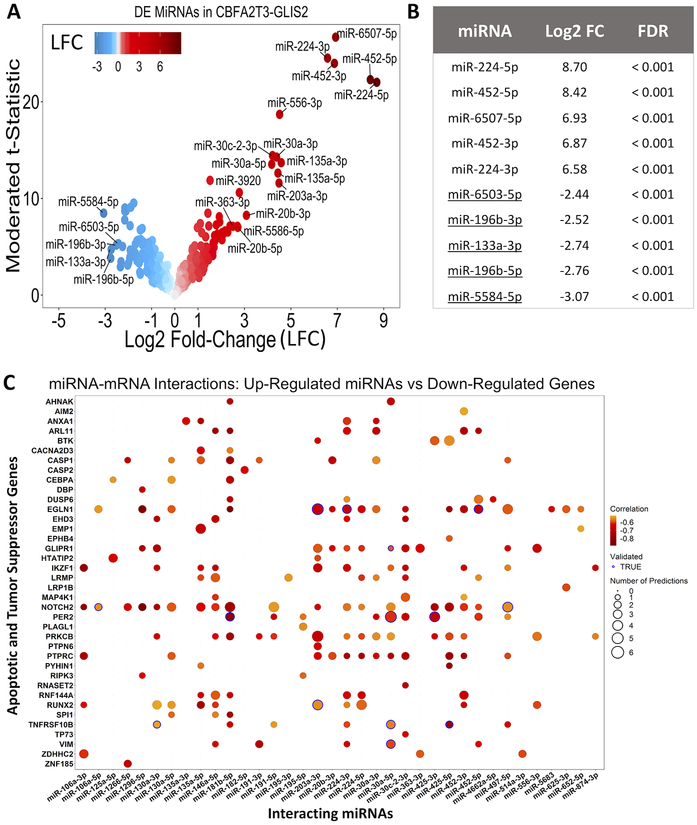
The 1,049 patients with whole transcriptome data had paired miRNA-sequencing completed. Contrasting CBFA2T3-GLIS2 AML (N=24) against the fusion-negative cohort (N=1,025) revealed 134 differentially expressed mature miRNAs, with the magnitude of differential expression being much greater for the upregulated miRNAs (Figure 6A). The most highly expressed miRNAs were miR-224–5p, miR-224–3p, miR-452–5p, miR-452–3p, and miR-6507–5p (Figure 6B). Human miR-224 and miR-452 have both been implicated as oncomirs in malignant melanoma, as well as colorectal cancers, and are intronic miRNAs transcribed from the GABRE locus, one of the most highly aberrantly expressed cell-membrane associated genes in CBFA2T3-GLIS2 (35,36). Conversely, the most down-regulated miRNAs were miR-6503–5p, miR-196b-3p, miR-196b-5p, miR-133a-3p, and miR-5584–5p. The mature miRNA miR-196b functions as a tumor suppressor during lung carcinogenesis (37), and well-characterized miR-133a has been found to function in a tumor suppressive manner in as many as 5 cancers, including breast and colorectal cancer (38).
Figure 6. CBFA2T3-GLIS2 AML have a distinct miRNA expression profile compared to the fusion-negative population and up-regulated miRNAs may inhibit tumor suppressor and apoptotic genes.
A) Volcano plot illustrating the over and under expressed miRNAs in CBFA23-GLIS2 compared to fusion-negative patients. B) The 5 most up and down regulated miRNAs ranked by absolute log2 fold-change. C) Predicted and validated miRNA-mRNA interactions with apoptosis and tumor suppressor genes found to be significantly down-regulated in CBFA2T3-GLIS2 cases.
To examine miRNA regulatory networks in CBFA2T3-GLIS2, miRNA-mRNA interactions were investigated by selecting significantly anti-correlated differentially expressed miRNA-gene pairs (Spearman’s rho, FDR<0.05). The miRNA-gene pairs were queried across independent target-prediction and experimental evidence databases, identifying 1,727 miRNA-mRNA interacting partners. Predicted interactions were only considered if two or more algorithms identified the miRNA binding-site homology or the interaction was supported by experimental evidence. The most highly over-expressed miRNAs in CBFA2T3-GLIS2 tumors, miR-224 and miR-452, were associated with numerous mRNA targets. Gene-Ontology (GO) enrichment analysis of miR-224 target genes (N=107 genes) revealed that these were involved in immune response (FDR=0.002) and leukocyte activation (FDR=0.005), indicating these pathways are inhibited in fusion-positive AML and high expression of miR-224 potentially contributes to the pathways’ modulation. The over-expressed mature miR-452 associated with 127 target genes, which are involved in leukocyte differentiation (FDR=0.001), and immune system processes (FDR=0.001), as well as myeloid leukocyte differentiation (FDR=0.02). A perturbation experiment using LNA miRNA inhibitors for 1) hsa-miR-224–5p, 2) hsa-miR-224–3p, 3) hsa-miR-452–5p, or 4) hsa-miR-452–3p was performed. The CBFA2T3-GLIS2 cell line MO7E was exposed to inhibitor for 72-hours, resulting in a knock-down efficiency > 90% for each miRNA (Supplemental Figure 8). Proliferation rates were assessed for the following 4 days (7 days of continuous LNA inhibitor exposure). The inhibition of each miRNA alone produced minimal to no morphologic or immunophenotypic alterations and led to subtle increase in proliferation compared to negative control inhibitors (Supplemental Figure 9, p≤0.01).
In contrast to gaining proliferative ability, tumors’ modulation of apoptotic genes is often involved in cancer progression (39). Two such genes, CASP1 and TRAIL-R2 were significantly down-regulated, as well as, the tumor-suppressors GLIPR1 and PER2 (39–41). CASP1 was significantly anti-correlated with and a putative target of the overexpressed miR-181b-5p (rs = −0.76, FDR=0.002, Figure 6C). TRAIL-R2 associated with upregulated mature miR-425–5p, and the regulatory interaction had experimentally validated evidence (rs = −0.80, FDR<0.001). GLIPR1 mRNA decreased with increasing miR-130a-3p transcript levels (rs = −0.72, FDR=0.003) and PER2 displayed significantly anti-correlated expression levels with miR-181b-5p (rs = 0.82, FDR<0.001) and possessed experimental evidence of the interaction.
Finally, we investigated regulatory interactions among the most significantly down-regulated miRNAs in CBFA2T3-GLIS2 AML. Mature miR-196b-5p had an identified regulatory interaction with the cell adhesion molecule ITGA2, whose expression was 22-fold greater in fusion-positive AML (rs = −0.75, FDR = 0.03). ITGA2 is expressed on HSCs (29) and silencing of miR-196b appears to induce abnormal regulation of this marker. In addition, miR-196–5p had target binding-sites identified within DNA methyltransferase DNMT3B transcripts (rs = −0.73, FDR=0.03). DNMT3B was upregulated in CBFA2T3-GLIS2 tumors and has been associated with adverse outcome in pediatric AML (42). The miR-199a/b family members, characterized as tumor suppressors in multiple cancers including AML (43,44), had a predicted regulatory relationship with the massively over-expressed plasma-membrane SLITRK5 gene (over 200-fold more highly expressed in fusion-positive cases, FDR<0.001). SLITRK5, expressed on both HSCs and aberrantly on CD34+ leukemic cells (45), had a predicted binding site for miR-199b-3p and miR-199a-3p (rs ≤ −0.73, FDR≤0.03), thus providing one putative mechanism contributing to the over-expression of this AML associated gene transcript (Supplemental Figure 10).
Discussion
This comprehensive study represents a large uniformly treated cohort of patients to be evaluated by transcriptome sequencing, in order to define the biologic implications of the CBFA2T3-GLIS2 fusion in children with de novo AML. We established that although the prevalence of CBFA2T3-GLIS2 is ~2% across an unselected pediatric cohort, it is highly enriched in younger AML populations (5,46,47), with nearly 12% of 1 year-old infants harboring this fusion. In fact, the CBFA2T3-GLIS2 fusion was not detected in those older than 3 years of age. We also confirmed the CBFA2T3-GLIS2 fusion is not limited to those with AMKL or normal karyotypes (3–7), with predilection to patients with African ancestry. Further, a majority of CBFA2T3-GLIS2 positive patients have non-recurrent karyotypic alterations with unexplained enrichment of trisomy 3, and the overwhelming majority express the unique RAM immunophenotype, which provides a seamless mechanism for rapid identification of this high risk population (18).
Furthermore, the lack of recurrent fusions or somatic mutations in a very young population suggests that this cryptic fusion may be sufficient for leukemic transformation without requirement for additional cooperating events. We confirmed this hypothesis by demonstrating that transduction of CBSC leads to malignant transformation that recapitulates infant AML with enhanced proliferation, maturation arrest, as well as morphologic and immunophenotypic alterations consistent with megakaryocytic AML. This in contrast to the disease in older patients where cooperation between class I and class II variants is required for malignant transformation (48). The fact that transduction of CBSC led to induction of NCAM1 (CD56) expression, in combination with the previously published data that the fusion protein directly binds to the NCAM1 promoter (4,13), provides substantial evidence that CD56 expression is causally and directly related to the fusion and suggests that effective targeting of CD56 may be curative.
In addition, this subgroup displayed unique transcriptional dysregulation compared to other AML groups. Similar to findings reported in AMKL, significant activation of hedgehog (HH) and TGFb/BMP signaling pathways was observed in CBFA2T3-GLIS2 AML across all FAB and cytogenetic classifications, and exhibited upregulation of the canonical HH genes PTCH1, HHIP, and GLI1, and the downstream target BMP2 (3–6). The HH and BMP pathways are known to interact with each other, and their cross talk have been implicated in multiple cancers (10,26,27). This finding provides further evidence that the CBFA2T3-GLIS2 fusion is accompanied by dysregulation of hedgehog signaling, and that this pathway may contribute to the proliferative capacity of leukemic cells in AML. Gene expression profiling also revealed a cohort of fusion-negative patients who cluster closely with CBFA2T3-GLIS2 true positive cases, and this signature identifies a larger cohort of patients with a shared expression profile of cell-surface associated genes. This may be especially important for other high-risk fusions such as NUP98-KDM5A.
In order to unravel the complex regulatory networks that are involved in the transcriptional dysregulation of CBFA2T3-GLIS2, we investigated the expression of mature miRNAs in fusion-positive AML. The most highly upregulated miRNAs were miR-224 and miR-452, which have been shown to be associated with poor outcomes across six independent colorectal cancer cohorts, and both are over-expressed in malignant melanoma (35,36). Interestingly, target genes of miR-224 and miR-452 were associated with GO enrichment of immune system processes and leukocyte differentiation, suggesting a possible mechanism of an immunosuppressive environment in fusion-positive AML. It was also demonstrated that miR-224/miR-452 genes are located in a miRNA cluster, and are induced by transactivation of GABRE, which was one of the most highly upregulated genes in CBFA2T3-GLIS2 (LFC=10.6, FDR<0.001) (35). However, the evidence suggested over-expression of E2F1 drove transactivation, yet CBFA2T3-GLIS2 AML lacked an over-expression of E2F1, suggesting an alternative mechanism of regulation. Transcription factor binding sites in K562 ChIP-seq from the ENCODE project showed that TAL1 and RBFOX2, both upregulated > 2-fold in CBFA2T3-GLIS2 patients, bind to the promoter region of GABRE, indicating alternative avenues for the observed over-expression (49). The results of the knock-down experiments are more difficult to interpret, and may indicate that these miRNAs may not be limited to enhancing cell-division/mitotic potential, but offer an alternative clonal advantage, possibly related to cell quiescence considering their apparent negative regulation of proliferation. In addition, it cannot be discounted that inhibition of these closely related miRNAs individually does may not account for potential synergistic effects of the miRNAs. Combinatorial inhibition experiments may provide improved elucidation of the regulatory mechanisms to which the four miRNAs contribute.
Alternatively, many upregulated miRNAs in fusion-positive AML were found to target apoptotic and tumor suppressor genes; miR-181b-5p was highly elevated in CBFA2T3-GLIS2 tumors and had predicted target sites on the down-regulated apoptotic genes CASP1 and TRAIL-R2, and the circadian rhythm gene PER2. Meanwhile, tumor suppressive miRNAs, including miR-196a/b, miR-133a, and miR-199a/b, had significantly reduced expression in fusion-positive AML, suggesting another mode for tumor progression. Upregulation of ITGA2 and DNMT3B, which have been shown to be independent prognostic indicators associated with poor outcomes in AML when highly expressed, were associated with the loss of miR-196b expression (42,50). Taken together, this suggests another means for leukemic tumor cells to deregulate normal signals for cell growth arrest.
In summary, CBFA2T3-GLIS2 fusion is a highly potent fusion oncogene that is sufficient for malignant transformation and induction of highly refractory leukemia. Identification of this fusion early at diagnosis would allow more appropriate risk adaptive therapeutic intervention. Although at present targeted therapies are not available for these highly refractory patients, identification of causal association of the fusion oncogene and surface CD56 expression provides a viable target that may be curative. The promising results of the CD56-ADC across 4 independent primary samples demonstrated the potency and specificity of this therapeutic strategy. Targeting the CD56 antigen offers the most immediately actionable target in this cohort of high-risk patients, and an effective CD56-ADC with limited off-target toxicity is available for continued refinement toward this goal. Additionally, this study provides comprehensive transcriptome profiling of CBFA2T3-GLIS2 patients that defines an expression signature and dysregulated pathways and networks in this distinct and highly lethal AML of infancy. The vast body of data generated defining fusion oncoprotein mediated alterations that lead to therapeutic resistance can be used for development of precisely directed targeted therapies.
Supplementary Material
Translational Relevance.
The CBFA2T3-GLIS2 fusion is a potent fusion oncogene that leads to a highly refractory phenotype in pediatric AML. Transcriptome profiling of 1,049 pediatric AML patients defined a unique expression profile for this fusion and identified a number of altered pathways and potential therapeutic targets, including NCAM1 (CD56). We further demonstrated that the CBFA2T3-GLIS2 fusion is sufficient for malignant transformation, causally associated with expression of CD56, and its induction in cord blood stem cells recapitulates human disease. The causal association between the fusion oncogene and surface CD56 expression, as well as the promising results of the CD56-ADC (antibody drug conjugate) across 4 independent primary samples, provides a viable target that may be curative for these highly refractory patients. Although at present targeted therapies are not clinically available, the vast body of data generated defines fusion oncoprotein-mediated alterations that can be used for the development of precisely directed targeted therapies.
Acknowledgements
This work utilized the computational resources of Fred Hutchinson Cancer Research Center (FHCRC) Scientific Computing and the NIH HPC Biowulf cluster (http://hpc.nih.gov). Sequencing of the AAML1031 cohort was supported by Target Pediatric AML (TpAML, https://targetpediatricaml.org/). Additional funding was provided by St. Baldrick’s Consortium Grant, Bayer HealthCare Pharmaceuticals, Inc., and the Children’s Oncology Group Foundation.
Funding: R01CA 114563-10 (SM), COG Chair’s Grant (U10CA098543), NCTN Statistics & Data Center (U10CA180899), NCTN Operations Center Grant (U10CA180886), Andrew McDonough B+ Foundation (AMBF), St. Baldrick’s Foundation (SBF), Target Pediatric AML (TpAML), COG Foundation, HHSN261200800001E (SM), Project Stella (SM), and Hyundai Hope on Wheels (SM).
Footnotes
Conflicts of Interest: Santaguida: Notable Labs: Employment. Kolb: Roche- Genentech: Membership on an entity’s Board of Directors or advisory committees; Servier: Membership on an entity’s Board of Directors or advisory committees. Eidenschink Brodersen: Hematologics, Inc: Employment. Pardo: Hematologics, Inc: Employment. Loken: Hematologics, Inc: Employment, Equity Ownership.
References
- 1.Schuback HL, Arceci RJ, Meshinchi S. Somatic characterization of pediatric acute myeloid leukemia using next-generation sequencing. Semin Hematol 2013;50(4):325–32 doi 10.1053/j.seminhematol.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 2.Bolouri H, Farrar JE, Triche T Jr., RE Ries, EL Lim, TA Alonzo, et al. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat Med 2018;24(1):103–12 doi 10.1038/nm.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gruber TA, Larson Gedman A, Zhang J, Koss CS, Marada S, Ta HQ, et al. An Inv(16)(p13.3q24.3)-encoded CBFA2T3-GLIS2 fusion protein defines an aggressive subtype of pediatric acute megakaryoblastic leukemia. Cancer Cell 2012;22(5):683–97 doi 10.1016/j.ccr.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thiollier C, Lopez CK, Gerby B, Ignacimouttou C, Poglio S, Duffourd Y, et al. Characterization of novel genomic alterations and therapeutic approaches using acute megakaryoblastic leukemia xenograft models. J Exp Med 2012;209(11):2017–31 doi 10.1084/jem.20121343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Masetti R, Pigazzi M, Togni M, Astolfi A, Indio V, Manara E, et al. CBFA2T3-GLIS2 fusion transcript is a novel common feature in pediatric, cytogenetically normal AML, not restricted to FAB M7 subtype. Blood 2013;121(17):3469–72 doi 10.1182/blood-2012-11-469825. [DOI] [PubMed] [Google Scholar]
- 6.de Rooij JD, Branstetter C, Ma J, Li Y, Walsh MP, Cheng J, et al. Pediatric non-Down syndrome acute megakaryoblastic leukemia is characterized by distinct genomic subsets with varying outcomes. Nat Genet 2017;49(3):451–6 doi 10.1038/ng.3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hara Y, Shiba N, Ohki K, Tabuchi K, Yamato G, Park MJ, et al. Prognostic impact of specific molecular profiles in pediatric acute megakaryoblastic leukemia in non-Down syndrome. Genes Chromosomes Cancer 2017;56(5):394–404 doi 10.1002/gcc.22444. [DOI] [PubMed] [Google Scholar]
- 8.Kim YS, Kang HS, Jetten AM. The Kruppel-like zinc finger protein Glis2 functions as a negative modulator of the Wnt/beta-catenin signaling pathway. FEBS Lett 2007;581(5):858–64 doi 10.1016/j.febslet.2007.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lamar E, Kintner C, Goulding M. Identification of NKL, a novel Gli-Kruppel zinc-finger protein that promotes neuronal differentiation. Development 2001;128(8):1335–46. [DOI] [PubMed] [Google Scholar]
- 10.Bertrand FE, Angus CW, Partis WJ, Sigounas G. Developmental pathways in colon cancer: crosstalk between WNT, BMP, Hedgehog and Notch. Cell Cycle 2012;11(23):4344–51 doi 10.4161/cc.22134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev 2001;15(23):3059–87 doi 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 12.Thirant C, Ignacimouttou C, Lopez CK, Diop M, Le Mouel L, Thiollier C, et al. ETO2-GLIS2 Hijacks Transcriptional Complexes to Drive Cellular Identity and Self-Renewal in Pediatric Acute Megakaryoblastic Leukemia. Cancer Cell 2017;31(3):452–65 doi 10.1016/j.ccell.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 13.Masetti R, Bertuccio SN, Astolfi A, Chiarini F, Lonetti A, Indio V, et al. Hh/Gli antagonist in acute myeloid leukemia with CBFA2T3-GLIS2 fusion gene. J Hematol Oncol 2017;10(1):26 doi 10.1186/s13045-017-0396-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aplenc R, Meshinchi S, Sung L, Alonzo TA, Pollard J, Gerbing R, et al. The Addition of Bortezomib to Standard Chemotherapy for Pediatric Acute Myeloid Leukemia Has Increased Toxicity without Therapeutic Benefit: A Report from the Children’s Oncology Group. Blood 2016;128:899-.27827828 [Google Scholar]
- 15.Cooper TM, Franklin J, Gerbing RB, Alonzo TA, Hurwitz C, Raimondi SC, et al. AAML03P1, a pilot study of the safety of gemtuzumab ozogamicin in combination with chemotherapy for newly diagnosed childhood acute myeloid leukemia: a report from the Children’s Oncology Group. Cancer 2012;118(3):761–9 doi 10.1002/cncr.26190. [DOI] [PubMed] [Google Scholar]
- 16.Gamis AS, Alonzo TA, Meshinchi S, Sung L, Gerbing RB, Raimondi SC, et al. Gemtuzumab ozogamicin in children and adolescents with de novo acute myeloid leukemia improves event-free survival by reducing relapse risk: results from the randomized phase III Children’s Oncology Group trial AAML0531. J Clin Oncol 2014;32(27):3021–32 doi 10.1200/JCO.2014.55.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lange BJ, Smith FO, Feusner J, Barnard DR, Dinndorf P, Feig S, et al. Outcomes in CCG-2961, a children’s oncology group phase 3 trial for untreated pediatric acute myeloid leukemia: a report from the children’s oncology group. Blood 2008;111(3):1044–53 doi 10.1182/blood-2007-04-084293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eidenschink Brodersen L, Alonzo TA, Menssen AJ, Gerbing RB, Pardo L, Voigt AP, et al. A recurrent immunophenotype at diagnosis independently identifies high-risk pediatric acute myeloid leukemia: a report from Children’s Oncology Group. Leukemia 2016;30(10):2077–80 doi 10.1038/leu.2016.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng Y, Wang Y, Zhu Z, Li W, Sussman RT, Randall M, et al. Differential killing of CD56-expressing cells by drug-conjugated human antibodies targeting membrane-distal and membrane-proximal non-overlapping epitopes. MAbs 2016;8(4):799–810 doi 10.1080/19420862.2016.1155014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whiteman KR, Johnson HA, Mayo MF, Audette CA, Carrigan CN, LaBelle A, et al. Lorvotuzumab mertansine, a CD56-targeting antibody-drug conjugate with potent antitumor activity against small cell lung cancer in human xenograft models. MAbs 2014;6(2):556–66 doi 10.4161/mabs.27756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, et al. A third-generation lentivirus vector with a conditional packaging system. J Virol 1998;72(11):8463–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haas BJ, Dobin A, Stransky N, Li B, Yang X, Tickle T, et al. STAR-Fusion: Fast and Accurate Fusion Transcript Detection from RNA-Seq. bioRxiv 2017. [Google Scholar]
- 23.Robertson G, Schein J, Chiu R, Corbett R, Field M, Jackman SD, et al. De novo assembly and analysis of RNA-seq data. Nat Methods 2010;7(11):909–12 doi 10.1038/nmeth.1517. [DOI] [PubMed] [Google Scholar]
- 24.Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics 2009;10:161 doi 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou X, Edmonson MN, Wilkinson MR, Patel A, Wu G, Liu Y, et al. Exploring genomic alteration in pediatric cancer using ProteinPaint. Nature Genetics 2015;48:4 doi 10.1038/ng.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neapolitan R, Horvath CM, Jiang X. Pan-cancer analysis of TCGA data reveals notable signaling pathways. BMC Cancer 2015;15:516 doi 10.1186/s12885-015-1484-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sever R, Brugge JS. Signal transduction in cancer. Cold Spring Harb Perspect Med 2015;5(4) doi 10.1101/cshperspect.a006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Binder JX, Pletscher-Frankild S, Tsafou K, Stolte C, O’Donoghue SI, Schneider R, et al. COMPARTMENTS: unification and visualization of protein subcellular localization evidence. Database (Oxford) 2014;2014:bau012 doi 10.1093/database/bau012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schroeder T Hematopoietic stem cell heterogeneity: subtypes, not unpredictable behavior. Cell Stem Cell 2010;6(3):203–7 doi 10.1016/j.stem.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 30.Schepers K, Campbell TB, Passegue E. Normal and leukemic stem cell niches: insights and therapeutic opportunities. Cell Stem Cell 2015;16(3):254–67 doi 10.1016/j.stem.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graham DK, DeRyckere D, Davies KD, Earp HS. The TAM family: phosphatidylserine sensing receptor tyrosine kinases gone awry in cancer. Nat Rev Cancer 2014;14(12):769–85 doi 10.1038/nrc3847. [DOI] [PubMed] [Google Scholar]
- 32.Lagadec C, Meignan S, Adriaenssens E, Foveau B, Vanhecke E, Romon R, et al. TrkA overexpression enhances growth and metastasis of breast cancer cells. Oncogene 2009;28(18):1960–70 doi 10.1038/onc.2009.61. [DOI] [PubMed] [Google Scholar]
- 33.Salgia R MET in Lung Cancer: Biomarker Selection Based on Scientific Rationale. Mol Cancer Ther 2017;16(4):555–65 doi 10.1158/1535-7163.MCT-16-0472. [DOI] [PubMed] [Google Scholar]
- 34.Mochmann LH, Bock J, Ortiz-Tanchez J, Schlee C, Bohne A, Neumann K, et al. Genome-wide screen reveals WNT11, a non-canonical WNT gene, as a direct target of ETS transcription factor ERG. Oncogene 2011;30(17):2044–56 doi 10.1038/onc.2010.582. [DOI] [PubMed] [Google Scholar]
- 35.Knoll S, Furst K, Kowtharapu B, Schmitz U, Marquardt S, Wolkenhauer O, et al. E2F1 induces miR-224/452 expression to drive EMT through TXNIP downregulation. EMBO Rep 2014;15(12):1315–29 doi 10.15252/embr.201439392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ling H, Pickard K, Ivan C, Isella C, Ikuo M, Mitter R, et al. The clinical and biological significance of MIR-224 expression in colorectal cancer metastasis. Gut 2016;65(6):977–89 doi 10.1136/gutjnl-2015-309372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tellez CS, Juri DE, Do K, Picchi MA, Wang T, Liu G, et al. miR-196b Is Epigenetically Silenced during the Premalignant Stage of Lung Carcinogenesis. Cancer Res 2016;76(16):4741–51 doi 10.1158/0008-5472.CAN-15-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu PY, Lopez G, Braggio D, Koller D, Bill KLJ, Prudner BC, et al. miR-133a function in the pathogenesis of dedifferentiated liposarcoma. Cancer Cell International 2018;18(1):89 doi 10.1186/s12935-018-0583-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar R, Raghava GPS. ApoCanD: Database of human apoptotic proteins in the context of cancer. Scientific Reports 2016;6:20797 doi 10.1038/srep20797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Su X, Chen D, Yang K, Zhao Q, Zhao D, Lv X, et al. The circadian clock gene PER2 plays an important role in tumor suppression through regulating tumor-associated genes in human oral squamous cell carcinoma. Oncol Rep 2017;38(1):472–80 doi 10.3892/or.2017.5653. [DOI] [PubMed] [Google Scholar]
- 41.Thompson TC. Glioma pathogenesis-related protein 1: tumor-suppressor activities and therapeutic potential. Yonsei Med J 2010;51(4):479–83 doi 10.3349/ymj.2010.51.4.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lamba JK, Cao X, Raimondi SC, Rafiee R, Downing JR, Lei S, et al. Integrated epigenetic and genetic analysis identifies markers of prognostic significance in pediatric acute myeloid leukemia. Oncotarget 2018;9(42):26711–23 doi 10.18632/oncotarget.25475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Favreau AJ, McGlauflin RE, Duarte CW, Sathyanarayana P. miR-199b, a novel tumor suppressor miRNA in acute myeloid leukemia with prognostic implications. Exp Hematol Oncol 2015;5:4 doi 10.1186/s40164-016-0033-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koshizuka K, Hanazawa T, Kikkawa N, Arai T, Okato A, Kurozumi A, et al. Regulation of ITGA3 by the antitumor miR-199 family inhibits cancer cell migration and invasion in head and neck cancer. Cancer Sci 2017;108(8):1681–92 doi 10.1111/cas.13298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Milde T, Shmelkov SV, Jensen KK, Zlotchenko G, Petit I, Rafii S. A novel family of slitrk genes is expressed on hematopoietic stem cells and leukemias. Leukemia 2007;21(4):824–7 doi 10.1038/sj.leu.2404525. [DOI] [PubMed] [Google Scholar]
- 46.de Rooij JD, Hollink IH, Arentsen-Peters ST, van Galen JF, Berna Beverloo H, Baruchel A, et al. NUP98/JARID1A is a novel recurrent abnormality in pediatric acute megakaryoblastic leukemia with a distinct HOX gene expression pattern. Leukemia 2013;27(12):2280–8 doi 10.1038/leu.2013.87. [DOI] [PubMed] [Google Scholar]
- 47.de Rooij JD, Masetti R, van den Heuvel-Eibrink MM, Cayuela JM, Trka J, Reinhardt D, et al. Recurrent abnormalities can be used for risk group stratification in pediatric AMKL: a retrospective intergroup study. Blood 2016;127(26):3424–30 doi 10.1182/blood-2016-01-695551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gilliland DG. Molecular genetics of human leukemias: new insights into therapy. Semin Hematol 2002;39(4 Suppl 3):6–11 doi 10.1053/shem.2002.36921. [DOI] [PubMed] [Google Scholar]
- 49.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489(7414):57–74 doi 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lian XY, Zhang W, Wu DH, Ma JC, Zhou JD, Zhang ZH, et al. Methylation-independent ITGA2 overexpression is associated with poor prognosis in de novo acute myeloid leukemia. J Cell Physiol 2018;233(12):9584–93 doi 10.1002/jcp.26866. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.