Abstract
Acquired resistance to HER2-targeted therapies occurs frequently in HER2+ breast tumors and new strategies for overcoming resistance are needed. Here we report that resistance to trastuzumab is reversible, as resistant cells regained sensitivity to the drug after being cultured in drug-free media. RNA-sequencing analysis showed that cells resistant to trastuzumab or trastuzumab + pertuzumab in combination increased expression of oxidative phosphorylation pathway genes. Despite minimal changes in mitochondrial respiration, these cells exhibited increased expression of ATP synthase genes and selective dependency on ATP synthase function. Resistant cells were sensitive to inhibition of ATP synthase by oligomycin A, and knockdown of ATP5J or ATP5B, components of ATP synthase complex, rendered resistant cells responsive to a low dose of trastuzumab. Furthermore, combining ATP synthase inhibitor oligomycin A with trastuzumab led to regression of trastuzumab-resistant tumors in vivo. In conclusion, we identify a novel vulnerability of cells with acquired resistance to HER2-targeted antibody therapies and reveal a new therapeutic strategy to overcome resistance.
Keywords: HER2+ breast cancer, trastuzumab, pertuzumab, acquired resistance, ATP synthase, oxidative phosphorylation
Introduction
About 20–30% of breast cancers exhibit gene amplification and/or overexpression of human epidermal growth factor receptor 2 (HER2/gene name ERBB2) and are classified as HER2-positive (HER2+) (1). HER2 is a receptor tyrosine kinase that forms dimers with itself or other family members, including epidermal growth factor receptor (EGFR) and human epidermal growth factor receptor 3 (HER3), to activate downstream signaling. Signaling from HER family kinases controls several important processes that are often deregulated in cancers such as cell growth, proliferation, motility, and survival (reviewed in (2)). The standard of care for HER2+ breast cancer includes a HER2-targeted agent in addition to chemotherapy and surgery. The first and most widely used HER2-targeted therapy is a humanized monoclonal antibody called trastuzumab (trade name Herceptin; Genentech/Roche). Trastuzumab functions by HER2 downregulation (3) and degradation (4), blocking ligand-independent HER2 dimerization and downstream signaling (5–9), and through antibody dependent cellular cytotoxicity (ADCC) (10,11). The addition of trastuzumab to a chemotherapy regimen was shown to significantly increase median progression-free survival (PFS) and overall survival (OS) in metastatic breast cancers by 3 and 5 months, respectively (12). Rates of disease-free survival (DFS) and OS were also significantly improved in early stage breast cancer patients treated with trastuzumab (13–15). Pertuzumab (trade name Perjeta; Genentech/Roche) is a humanized monoclonal antibody that can be used in combination with trastuzumab and chemotherapy for treatment of HER2+ breast cancers at early (16) and late (17) stages. It targets the dimerization domain of HER2 (18) to block ligand-induced dimerization between HER2 and other family members, reducing activation of important downstream signaling pathways such as the phosphoinositide 3-kinase (PI3K) pathway (19). Addition of pertuzumab to a docetaxel and trastuzumab regimen in metastatic breast cancer patients increased median PFS by 6 months and median OS by 16 months (16). Furthermore, addition of pertuzumab to trastuzumab-based adjuvant chemotherapy increased rates of invasive disease-free survival in operable, early stage HER2+ breast cancer patients (17).
Unfortunately, HER2+ breast cancer patients do not respond uniformly to HER2-targeted antibodies. Fewer than half of HER2+ metastatic breast cancer patients initially respond to trastuzumab (20). Additionally, while trastuzumab significantly extends OS in a fraction of HER2+ metastatic breast cancer patients, the majority of those patients will experience tumor progression within only one year (12). Many early stage HER2+ patients treated with trastuzumab also experience recurrence (13,15,21). Several strategies to overcome primary and acquired trastuzumab resistance have been proposed, but at this time, with the exception of pertuzumab, few have translated into clinical improvements (22). Patients with HER2+ breast cancer treated with the combination of chemotherapy, trastuzumab, and pertuzumab may also experience tumor recurrence after adjuvant therapy for early stage breast cancer and with metastatic breast cancer nearly all inevitably ultimately progress (16,17). To our knowledge, there are currently no published studies investigating mechanisms of acquired resistance to this combination therapy. These will become increasingly important, as this regimen is becoming the new standard of care for HER2+ breast cancer patients diagnosed at any stage.
Several recent studies suggest that drug resistance to anti-cancer therapies develops initially through a reversible phase. For example, it has been shown in several cancer cell line models that drug-tolerant persister cells can become re-sensitized to the drug after being cultured without it for some time (23–26). In line with this model of plastic resistance, we show here that trastuzumab resistance was reversed when resistant cells were cultured for several passages in drug-free media. Thus, we set out to uncover novel features of trastuzumab and trastuzumab + pertuzumab resistance by narrowing our focus on changes in gene expression programs. We identified a selective dependency of HER2-antibody resistant breast cancer cells on mitochondrial oxidative phosphorylation (OXPHOS). This is a major process by which adenosine triphosphate (ATP) is produced in cells. The flow of electrons through the mitochondrial electron transport chain creates an electrochemical proton gradient across the inner mitochondrial membrane. The energy from this gradient is harnessed by the passage of protons through ATP synthase (also called complex V) to phosphorylate adenosine diphosphate (ADP) to produce ATP in the mitochondrial matrix.
Metabolic rewiring to shift reliance towards OXPHOS or glycolysis has been shown to contribute to tumor recurrence in many settings (reviewed in (27)). Trastuzumab was shown to hinder glycolysis in breast cancer cells, and the combination of trastuzumab with an inhibitor of glycolysis synergistically decreased tumor cell growth (28). Trastuzumab resistance was also linked to an increase in glycolysis (28,29). Thus, the glycolytic contribution to trastuzumab resistance has been well characterized in HER2+ breast cancer, but to our knowledge, the contribution from OXPHOS has never been considered. Here, we demonstrate that resistant cells upregulated OXPHOS-related genes and were more sensitive to OXPHOS blockade through inhibition of ATP synthase, even though sensitive and resistant cells respired at a similar rate. Combining trastuzumab with an ATP synthase inhibitor led to regression of trastuzumab-resistant tumors in vivo. We also show that expression of the ATP synthase gene ATP5B correlated with poor outcome in HER2+ breast cancer patients. These novel findings may provide a new avenue of treatment for patients with trastuzumab- or trastuzumab + pertuzumab- resistant disease.
Materials and Methods
Chemicals and antibodies
DMSO was purchased from Sigma-Aldrich. Oligomycin A (11342) and rotenone (13995) were purchased from Cayman Chemical. 2-Deoxy-d-glucose (2-DG) (202010) was purchased from Santa Cruz Biotechnology, Inc. Antimycin A (A8674) was purchased from Sigma-Aldrich. Trastuzumab and pertuzumab were provided or purchased from Genentech/Roche. Anti-phospho-EGFR (D7A5), anti-EGFR (4267), anti-phospho-HER2 (2241), anti-HER2 (4290), anti-phospho-HER3 (2842), anti-HER3 (12708), anti-phospho-AKT S473 (4060), anti-AKT (4691), anti-phospho-ERK (4376), anti-ERK (46955), anti-ß-actin (3700), and anti-vinculin (13901) antibodies were purchased from Cell Signaling Technologies. Anti-ATP5B (ab14730) and anti-ATP5J (ab224139) antibodies were purchased from Abcam. Anti-cytokeratin (Z0622) and Envision + System HRP-labelled polymer anti-mouse (K4001) antibodies were purchased from Agilent Dako. Goat anti-rabbit Alexa-546 (A11010) and DAPI (D1306) were purchased from Life Technologies. Cyanine-5 tyramide (SAT705A001EA) was purchased from Perkin-Elmer.
Cell culture
BT474 and SKBR3 cells were obtained from the American Type Culture Collection. BT474 cells were cultured in RPMI 1640. SKBR3 and HEK293T cells were cultured in DMEM. All media were supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. Resistant cell pools were gradually made resistant to trastuzumab or trastuzumab + pertuzumab by being cultured in 10 μg/ml up to 100 μg/ml over the course of ~3 months. Cells were subsequently continuously maintained in 10 μg/ml trastuzumab or 10 μg/ml trastuzumab + 10 μg/ml pertuzumab. Trastuzumab and pertuzumab were used at 10 μg/ml unless explicitly stated otherwise. To allow for direct comparison of parental cells and drug resistant cells without the influence of drug(s), resistant cells were seeded into media without drug(s), for 2–7 days unless indicated otherwise. MCF10A cells were cultured as previously described (30). Cells were periodically tested for mycoplasma contamination and authenticated using short tandem repeat profiling.
RNA-sequencing
RNA was isolated from BT474 cells treated for three days with 10 μg/ml trastuzumab, 10 μg/ml trastuzumab + 10 μg/ml pertuzumab, or no treatment. Resistant cells were cultured for 7 days without drugs and then RNA was harvested. RNA was harvested from three replicate 10 cm plates for each cell line using the RNeasy Plus Mini Kit (Qiagen 74136). Samples were evaluated using the Agilent Bioanalyzer before mRNA library preparation. Libraries were prepared according to standard Illumina protocols by the Yale Stem Cell Center Genomics and Bioinformatics Core and run on Illumina HiSeq2000. Bioinformatic analyses were performed as previously described (31). In brief, reads were mapped to the human genome (hg38) with Bowtie2 (32), differential gene expression was determined with DESeq2 (33), and Gene Set Enrichment Analysis (GSEA) software (34) was utilized for pathway analysis. RNA-seq data were deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus database under accession number GSE121105.
Oxygen consumption measurements
Agilent Seahorse XF96 plate wells (101085–004) were coated with Cell-Tak (Corning C354240) according to the manufacturer’s protocol. BT474 cells ± 10 μg/m1 trastuzumab for three days, BT-TR cells ± 10 μg/m1 trastuzumab for seven days, and BT-TPR cells ± 10 μg/m1 trastuzumab and 10 μg/m1 pertuzumab for seven days were seeded at 25,000 cells/well. Each cell line was seeded into 4–8 wells. The next day, oxygen consumption measurements were obtained on the Agilent Seahorse Bioscience XF96 analyzer with injection ports loaded to attain final concentrations of 1.5 μM oligomycin A, 1 μM carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP), and 1 μM rotenone + 1 μM antimycin A. Afterwards, cells were lysed in high salt lysis buffer and protein content was measured by Bradford assay as the loading control. This assay was performed twice with four or more replicates per condition.
Mitochondrial membrane potential measurements
BT474, BT-TR1 and BT-TR2 cells were seeded into 12 well plates at 250,000 cells/well after being kept out of drug(s) for 7 days. The next day, cells were treated with DMSO or 20 μM FCCP for 10 minutes at 37°C. Subsequently, cells were exposed to 50 nM solution of tetramethylrhodamine ethyl ester (TMRE) (Abcam 113852) for 30 minutes at 37°C. Cells were washed with PBS, trypsinized, resuspended and pushed through a mesh lid to separate cell clumps. Phycoerythrin (PE) signal was measured by flow cytometry. Membrane potential was calculated as the ratio of PE signal of DMSO-treated cells/ FCCP-treated cells, as FCCP-treated cells represent staining associated with mitochondrial mass.
Knockdown of ATP5J and ATP5B
pLKO.1 plasmids containing short hairpins against ATP5J (TRCN0000038219-shATP5J-1, TRCN0000038220-shATP5J-2) and ATP5B (TRCN0000300454- shATP5B-1, TRCN0000300455- shATP5B-2) were obtained from Sigma-Aldrich. Scrambled shRNA plasmid was used as control (35). Insert sequences are listed in Supplementary Table 1. HEK293T cells were transfected with the pLKO.1 plasmids and second generation psPAX2 and pMD2.G packaging system using Lipofectamine 2000 (ThermoFisher 12566014). Virus was sterile filtered and applied to target cells for 16 hours with 8 μg/ml polybrene (hexadimethrine bromide, Sigma H9268). Target cells were selected with 1 μg/ml puromycin and utilized without delay in downstream applications. The knockdown procedure was repeated 3 times independently to capture differences in cell proliferation and trastuzumab sensitivity before any cellular adaptations.
Animal studies
17β-estradiol pellets (17 mg) were purchased from Huntsman Cancer Institute and implanted subcutaneously into the loose skin at the back of the neck of female NOD scid gamma mice (4–5 weeks old). Five days later, each mouse received two cell injections into the fourth mammary fat pads on opposite flanks. The injections were comprised of 5 × 106 viable cells resuspended in sterile phosphate-buffered saline mixed at 1:1 with Matrigel (Corning 356231) in a total volume of 0.1 ml. BT474 cells were injected into the right flank and BT-TR2 cells were injected into the left flank of each animal. After 20 days, mice were randomly assigned to four treatment groups: saline control, trastuzumab [5 mg/kg intraperitoneal (i.p.) every 3 days], oligomycin A [0.5 mg/kg i.p. daily for 3 days followed by 0.25 mg/kg i.p. daily], or the combination of trastuzumab + oligomycin A (each at the same dosage as the monotherapy). Mice in the saline and single treatment arms received control saline injections matching their counterparts receiving drug treatments (1–2 injections per day per mouse). Tumors were measured daily using digital calipers and volume calculated as 0.5 × length × width2. After 13 days of treatment, mice were euthanized and tumors were collected for histological analyses. This work was performed according to NIH guidelines and approved by the Yale University Institutional Animal Care and Use Committee.
Tissue microarray staining and analysis
The optimal titer of ATP5B antibody was determined by staining the breast cancer standardization TMA YTMA263 at 5 dilutions of primary antibody as previously described (36). ATP5B antibody was used at 0.5 μg/ml for subsequent staining. A tissue microarray was constructed from BT474 cell pellets expressing shControl, shATP5B-1, and shATP5B-2 by the Yale Cancer Center/Pathology Tissue Microarray facility to evaluate ATP5B antibody specificity. Tissue microarrays containing tumor tissue from the Hellenic Oncology Group (HeCOG) 10/05 clinical trial (37) were stained, Automated Quantitative Analysis (AQUA) scores were calculated, and Kaplan-Meier estimates of disease-free and overall survival were calculated as previously described (36). All tissue samples were used in accordance with U.S. Common Rule after approval by the Yale Human Investigation Committee. Informed written consent was obtained from all the patients.
Statistical analysis
Unless indicated otherwise, unpaired, two-tailed student’s t-test was used to determine significance using GraphPad Prism 7.0 software.
Results
Acquired resistance to trastuzumab is reversible.
To study resistance to HER2-targeted antibodies, we used trastuzumab sensitive BT474 and SKBR3 HER2+ breast cancer cells to generate several derivative pools of BT474 and SKBR3 cells resistant to trastuzumab or trastuzumab + pertuzumab (Figure 1A, Supplementary Figure 1A–B). We treated parental BT474 and SKBR3 cells with increasing doses of trastuzumab or both antibodies, escalating the dose when the cells appeared to be proliferating normally despite the presence of drug(s) as determined by colony formation assays (Supplementary Figure 1C). The pools were named with the first two letters of their parental cell line (BT or SK), followed by the designation of trastuzumab-resistant (TR) or trastuzumab + pertuzumab-resistant (TPR) and a number to differentiate between pools generated separately. We first characterized the expression and phosphorylation level of HER-family signaling pathway kinases in BT474 and BT-TR2 cells. We found that BT-TR1 and BT-TR2 cells have increased EGFR, HER3 and phosphorylated HER3 compared to parental cells, while they did not show a marked increase in phosphorylation of HER2, EGFR, and ERK (Supplementary Figure 1D). Short term trastuzumab treatment decreased phosphorylation of HER3 and ERK while inducing phosphorylation of HER2 and EGFR for both parental and drug resistant cells (Supplementary Figure 1D), indicating that these resistant cells respond to trastuzumab similarly as the parental cells for these phosphorylation-mediated signaling events. In contrast to gatekeeper mutations accounting for the majority of acquired resistance cases to targeted small molecule ATP analog tyrosine kinase inhibitors, no specific mutation or genetic event has been shown to account for the majority of acquired resistance cases to trastuzumab (38). In line with this, the trastuzumab- and trastuzumab + pertuzumab- resistant pools we generated showed no significant increases in copy number of key receptor tyrosine kinase genes EGFR, ERBB2 (HER2), ERBB3 (HER3), MET, or IGF1R compared to parental cells (Supplementary Figure 2).
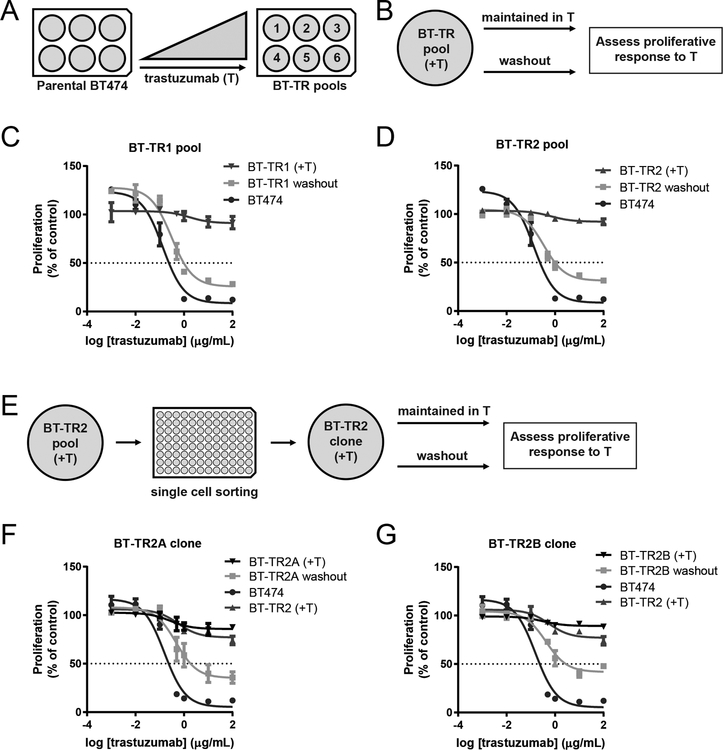
Figure 1: Trastuzumab resistance is reversible.
(A) Trastuzumab-resistant pools were generated by exposing parental BT474 cells to increasing doses of trastuzumab over the course of 3+ months. (B) Schematic of resistance reversal experiment for BT-TR cells. (C-D) Pools of BT474 cells made resistant to trastuzumab were cultured in trastuzumab (+T; triangle) or without drugs (washout; square) for 20 doublings (9 passages) and their proliferation after ten days of trastuzumab treatment was measured by WST-1 assays. BT474 cells (circle) were included as a control. Proliferation is shown as a percentage of no treatment control growth. (E) Schematic of resistance reversal experiment for BT-TR2-derived clones. (F-G) Clones of BT-TR2 cells were cultured in trastuzumab (+T; down triangle) or without drugs (washout; square) for 23 doublings. Proliferation after ten days of trastuzumab treatment was measured by WST-1 assays. BT474 cells (circle) and BT-TR2 cells cultured continuously in trastuzumab (up triangle) were included as controls. Proliferation is shown as a percentage of no treatment control growth. Data points in C, D, F and G represent means of three replicate wells ± SEM.
We passaged BT474-derived resistant pools side by side in trastuzumab or drug-free media (referred to as “washout”) and examined their sensitivity to trastuzumab periodically. After twenty doublings (nine passages) in drug-free media, all pools became more sensitive to the drug, and three out of four pools of resistant cells tested regained sensitivity to trastuzumab (Figure 1B–D, Supplementary Figure 3A–B). To decipher if the pools regained sensitivity due to clonal selection or flexibility of individual clones, we generated single cell clones from the resistant pool BT-TR2 and repeated the assay (Figure 1E). After 23 doublings, two of three resistant clones tested regained sensitivity to trastuzumab (Figure 1F–G). The third clone demonstrated increased sensitivity after 34 doublings (Supplementary Figure 3C). Taken together, these results suggested that non-genetic changes may mediate resistance to trastuzumab.
The oxidative phosphorylation gene signature is enriched in resistant cells.
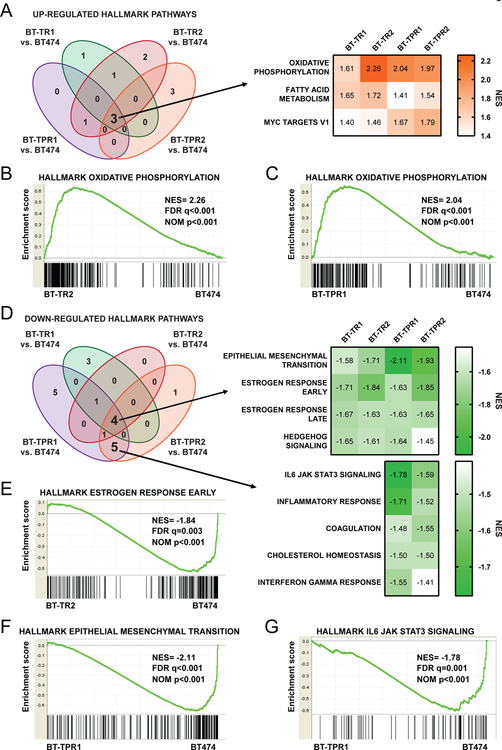
We hypothesized that alterations in gene expression programs could be the major contributors to resistance. Thus, RNA-sequencing was performed for sensitive BT474 cells, two pools of BT-TR cells and two pools of BT-TPR cells cultured in the absence of drug(s) for seven days in order to exclude gene expression changes induced by the drug(s) (Supplementary Tables 2–5). We utilized GSEA to identify differences between resistant pools and BT474 parental cells (Supplementary Tables 6–13). Several hallmark pathways were positively enriched with nominal p-value <0.05 and FDR q-value <0.1 in each resistant pool compared to BT474 cells. Only one hallmark pathway, protein secretion, was common to both BT-TR pools, but not BT-TPR pools (Figure 2A). Surprisingly, no pathways were common to both BT-TPR pools without also being enriched in BT-TR pools, highlighting similarities in pools resistant to single and combination therapies. Three GSEA hallmark pathways were positively enriched in all four resistant pools compared to BT474 cells: oxidative phosphorylation, fatty acid metabolism, and MYC targets V1 (Figure 2A). Oxidative phosphorylation (OXPHOS) was the top positively enriched pathway in BT-TR2, BT-TPR1, and BT-TPR2 cells, and third for BT-TR1 (Figure 2B–C, Supplementary Tables 6–9).
Figure 2: The oxidative phosphorylation gene program is elevated in resistant cells.
(A) GSEA hallmark pathways positively enriched with nominal p-value<0.05 and FDR q-value<0.1 in resistant pools versus BT474 parental cells (left). NES scores of each resistant pool for pathways enriched in all pools compared to BT474 cells (right). (B-C) GSEA enrichment plots of the hallmark oxidative phosphorylation pathway for BT-TR2 (B) and BT-TPR1 (C) versus BT474 parental cells. (D) GSEA hallmark pathways negatively enriched with nominal p-value<0.05 and FDR q-value<0.1 in resistant pools versus BT474 parental cells (left). NES scores of each resistant pool for pathways enriched in all pools compared to BT474 cells (right, top) or in BT-TPR pools only (right, bottom). (E) GSEA enrichment plots of the hallmark estrogen response early pathway for BT-TR2 versus BT474 parental cells. (F) GSEA enrichment plots of the hallmark epithelial mesenchymal transition pathway for BT-TPR1 versus BT474 parental cells. (G) GSEA enrichment plots of the hallmark IL6 JAK STAT3 signaling pathway for BT-TPR1 versus BT474 parental cells.
Among the pathways down-regulated in resistant pools compared to BT474 cells with nominal p-value <0.05 and FDR q-value <0.1, four pathways were negatively enriched in all resistant cell pools compared to parental cells: estrogen response early, epithelial mesenchymal transition, estrogen response late, and Hedgehog signaling (Figure 2D–F). The hallmark gene set for estrogen response early was the top down-regulated pathway for both BT-TR pools, while epithelial mesenchymal transition was the top down-regulated pathway for both BT-TPR pools (Figure 2E–F, Supplementary Tables 10–13). No down-regulated gene set was common to BT-TR pools without also being down-regulated in a BT-TPR pool (Figure 2D, Supplementary Tables 10–13). Interestingly, five hallmark pathways were negatively enriched in both BT-TPR pools, without significant enrichment in BT-TR pools (Figure 2D). These BT-TPR-specific down-regulated pathways include IL6 JAK STAT3 signaling (Figure 2G), inflammatory response, interferon gamma response, cholesterol homeostasis, and coagulation (Supplementary Tables 12–13).
We decided to investigate the role of OXPHOS in resistance to HER2-targeted antibodies as the OXPHOS pathway gene set was highly enriched in all profiled resistant pools compared to parental cells and because OXPHOS is targetable with small molecule inhibitors, facilitating potential translation to the clinic.
Resistant cells exhibit increased sensitivity to inhibition of oxidative phosphorylation, but not increased oxidative phosphorylation.
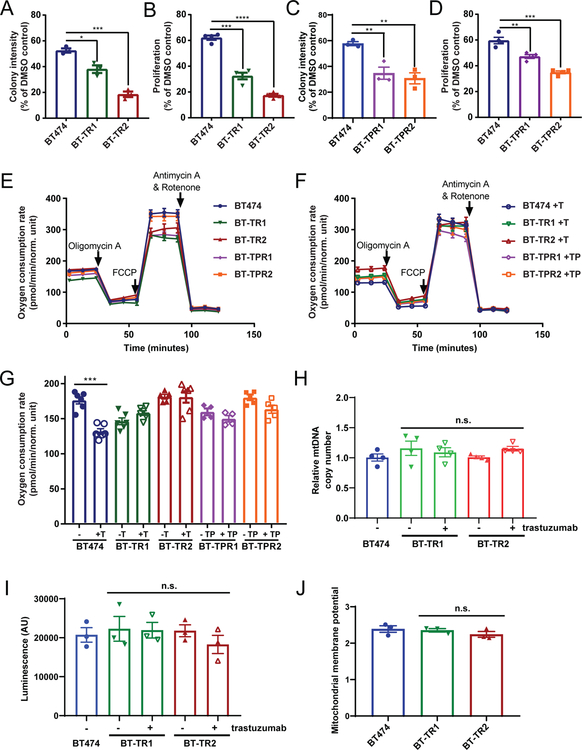
To determine if the increased expression of OXPHOS genes in resistant cells indicated an increased reliance on this pathway, we treated sensitive and resistant cells with oligomycin A, an OXPHOS inhibitor targeting the F0 subunit of ATP synthase. Although BT474 and BT-TR cells were all sensitive to 24-hour exposure to oligomycin A at low nanomolar concentrations, BT-TR1 and BT-TR2 cells were more sensitive than the parental BT474 cells in colony formation and proliferation assays (Figure 3A–B, Supplementary Figure 4A). Similarly, BT-TPR1 and BT-TPR2 cells showed more growth inhibition by oligomycin A treatment than BT474 cells in proliferation assays (Figure 3C–D, Supplementary Figure 4B). One out of two tested SK-TR cell lines also responded more strongly to oligomycin A than parental SKBR3 cells (Supplementary Figure 4C). In contrast, MCF10A normal-like mammary epithelial cells did not show any growth inhibition at doses several fold greater than those required to inhibit the breast cancer cell lines (Supplementary Figure 4D). We then asked whether trastuzumab-resistant cells were also more sensitive to other OXPHOS inhibitors. BT-TR1, BT-TR2, BT-TPR1 and BT-TPR2 cells all showed greater sensitivity to antimycin A, an inhibitor of mitochondrial complex III, and rotenone, an inhibitor of mitochondrial complex I than the parental cells (Supplementary Figure 4E–H).
Figure 3: Resistant cells are particularly sensitive to OXPHOS inhibition, but metabolically similar to parental cells.
(A) Quantification of colony formation after two weeks for BT474, BT-TR1, and BT-TR2 cells exposed to 2 nM of oligomycin A for 24 hours. (B) Proliferation after six days of BT474, BT-TR1, and BT-TR2 cells exposed to 3 nM oligomycin A for 24 hours normalized to respective DMSO controls. (C) Quantification of colony formation after three weeks for BT474, BT-TPR1, and BT-TPR2 cells exposed to 4 nM of oligomycin A for 24 hours. (D) Proliferation after six days of BT474, BT-TPR1, and BT-TPR2 cells exposed to 3 nM oligomycin A for 24 hours normalized to respective DMSO controls. Data in A-D is representative of three or more independent experiments with three or more replicates. (E-F) Oxygen consumption rate during Seahorse metabolic flux assay for designated cells without drug(s) (E) or with drug(s) (F). BT474 cells were treated with 10 μg/ml trastuzumab for 3 days. Resistant cells were cultured with drug(s) or without drug(s) for 7 days prior to the measurement. (G) Basal oxygen consumption rate (measurement 3 from E-F) of designated cells with or without drugs. T; 10 μg/ml trastuzumab. TP; 10 μg/ml trastuzumab + 10 μg/ml pertuzumab. Data in E-G is representative of two independent experiments with 4–8 replicates per sample. (H) Relative mitochondrial DNA (mtDNA) copy number in BT474 cells, BT-TR1 and BT-TR2 cells kept in trastuzumab or cultured without for seven days. All samples were normalized to BT474 copy number. Data is representative of three experiments performed in triplicate. (I) ATP level in BT474, BT-TR1, BT-TR2 cells measured using CellTiter-Glo assay. Data represents three biological replicates performed in triplicate. (J) Mitochondrial membrane potential measured by TMRE signal/ TMRE signal in the presence of uncoupler FCCP. Data is representative of two experiments performed in triplicate. Data points in A-J represent mean ± SEM. n.s., not significant. *, p< 0.05. **, p< 0.01. ***, p<0.001. ****, p< 0.0001.
Given the resistant cells’ enrichment in OXPHOS and several other metabolic gene programs compared to BT474 cells (Supplementary Tables 6–13), it was expected that they were operating at a higher metabolic rate. However, using Seahorse metabolic flux assays, we found that the overall metabolic features of sensitive and resistant cells were very similar, consistent with a previous report (29). While three-day treatment with trastuzumab lowered cellular oxygen consumption rate (OCR) compared to untreated BT474 cells, BT-TR and BT-TPR cells demonstrated basal OCRs similar to or slightly less than parental cells, whether they were kept in drug or cultured in drug-free media for one week beforehand (Figure 3E–G). Mitochondrial DNA copy number remained very similar between parental BT474 cells and BT-TR cells with or without drug treatments (Figure 3H). While BT-TPR cells have either decreased or increased mitochondrial DNA copy number compared to parental BT474 cells (Supplementary Figure 5A), there was no difference in mitochondrial DNA copy number between parental SKBR3 cells and SK-TR cells (Supplementary Figure 5B). Likewise, ATP production was comparable between BT474 and BT-TR or BT-TPR cells (Figure 3I, Supplementary Figure 5C), and between SKBR3 and SK-TR cells (Supplementary Figure 5D). Furthermore, we did not observe significant difference of mitochondrial membrane potential between parental BT474 and BT-TR cells as determined by TMRE signal normalized to the basal signal in the presence of the uncoupler FCCP (Figure 3J).
Trastuzumab was reported to shut down glycolysis in HER2+ breast cancer cells (28). Consistent with that, the gene signature for glycolysis is negatively enriched in BT474 cells after three-day trastuzumab treatment or three-day trastuzumab + pertuzumab treatment (Supplementary Figures 6A–B). Resistance to trastuzumab has been associated with increased glycolytic capacity (28,29). In line with this, one of the two BT-TR cell lines profiled with RNA sequencing, BT-TR2, showed marginal enrichment (NES=1.16, nominal p= 0.102, FDR q=0.237) of the glycolysis gene program (Supplementary Figure 6C, Supplementary Table 7). This resistant cell line also displayed enhanced ability to increase glycolysis upon inhibition of mitochondrial respiration, as measured by the extracellular acidification rate (ECAR) during the Seahorse metabolic flux assay (Supplementary Figure 6D–E). However, similar to BT-TR1 cells, BT-TR2 cells were less sensitive to 2-Deoxy-D-glucose (2-DG), a glucose analog that inhibits glycolysis, compared to parental BT474 cells, indicating that they do not harbor a selective dependence on glycolysis (Supplementary Figure 6F).
ATP synthase expression is increased in resistant cells, correlates with poor survival of HER2+ breast cancer patients and is necessary for maintenance of resistance.
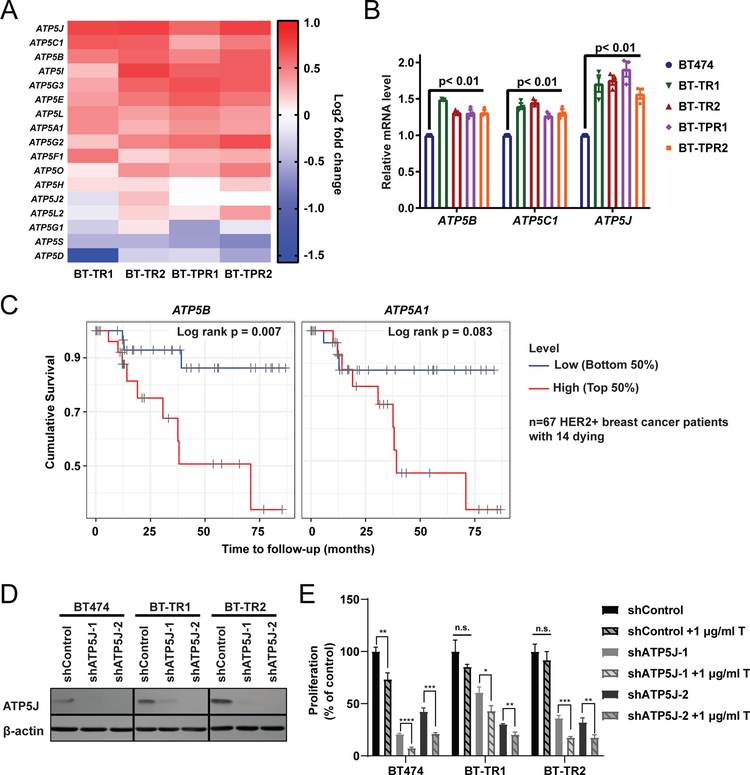
To better understand the resistant cells’ sensitivity to OXPHOS pathway inhibitors, we looked closer into the genes that comprised the OXPHOS signature and found that several ATP synthase transcripts were increased ~1.5 fold in BT-TR and BT-TPR resistant pools compared to sensitive cells (Figure 4A). This was validated by RT-qPCR (Figure 4B). Similar increases were observed in two single cell clones derived from the resistant pool BT-TR2 cells (Supplementary Figure 7A). SK-TR2 also demonstrated a moderate increase in ATP5J and ATP5B gene expression compared to parental SKBR3 cells, correlating with its increased sensitivity to oligomycin A (Supplementary Figure 7B).
Figure 4: ATP synthase expression is increased in resistant cells, correlates with poor survival of HER2+ breast cancer patients, and is required for maintenance of resistance.
(A) Heatmap showing relative ATP synthase complex V gene expression from RNA-sequencing in resistant cells compared to BT474 parental cells. (B) Relative mRNA levels of ATP5B, ATP5C1, and ATP5J measured by RT-qPCR in resistant cells normalized to BT474. Each symbol represents the mean of three technical replicates. Bars represent mean ± SEM of biological replicates. (C) Cumulative survival of HER2+ breast cancer patients with patients divided in half by ATP5B (left) and ATP5A1 (right) mRNA expression level. (D) Western blot analysis of BT474, BT-TR1 and BT-TR2 cells expressing shControl, shATP5J-1 and shATP5J-2. (E) WST-1 proliferation assay of BT474, BT-TR1 and BT-TR2 cells expressing the designated shRNAs ± 1 μg/ml trastuzumab for 7 days. Bars represent mean ± SEM. n.s., not significant. *, p< 0.05. **, p< 0.01. ***, p<0.001. ****, p< 0.0001.
In order to assess the clinical relevance of increased ATP synthase expression in resistant cells, mRNA expression data of tumors collected from HER2+ breast cancer patients (n=67) with no prior treatment in The Cancer Genome Atlas (TCGA) dataset were analyzed using the online tool TIMER (39). The majority of these patients were treated with HER2-targeted therapies. We found that patients with high mRNA levels of ATP5B, one of the core components of ATP synthase, had significantly poorer survival compared to the low expression group, despite the small cohort size and limited number of events (Figure 4C). mRNA levels of ATP5A1, another core component of the complex, followed a similar pattern, though this was not statistically significant by log-rank test due to the limited number of events (Figure 4C). In contrast, ATP5B and ATPA1 were not significantly associated with survival in basal or luminal subtypes of breast cancer, or in the combined cohorts with all subtypes of breast cancers (Supplementary Figure 8). The importance of ATP5B protein was explored in HER2+ breast cancer patients from a clinical trial of adjuvant chemotherapy followed by trastuzumab (HeCOG 10/05) (37). In this unique clinical trial, HER2+ patients were given trastuzumab after (rather than concurrent with) their chemotherapy. Although we observed a trend of association between expression of ATP5B protein with poor overall survival, the associations between expression of ATP5B protein and treatment outcome in this cohort were not statistically significant (Supplementary Figure 9). However, the analysis was underpowered with only 11 and 7 events for disease-free and overall survival, respectively, for a total of 117 patients included in the analysis (Supplementary Figure 9).
To examine the relationship between ATP synthase function and trastuzumab sensitivity, we knocked down expression of complex component ATP5J with two independent shRNAs in BT474 and BT-TR cells (Figure 4D). Notably, knockdown of ATP5J allowed a low dose of trastuzumab to inhibit growth of BT-TR cells, while control BT-TR cells did not respond to trastuzumab at this dose (Figure 4E). Similarly, ATP5B downregulation with two independent shRNAs also led to reduced growth of BT-TR2 cells by a low dose of trastuzumab (Supplementary Figure 10). These results confirm a role for ATP synthase in trastuzumab tolerance.
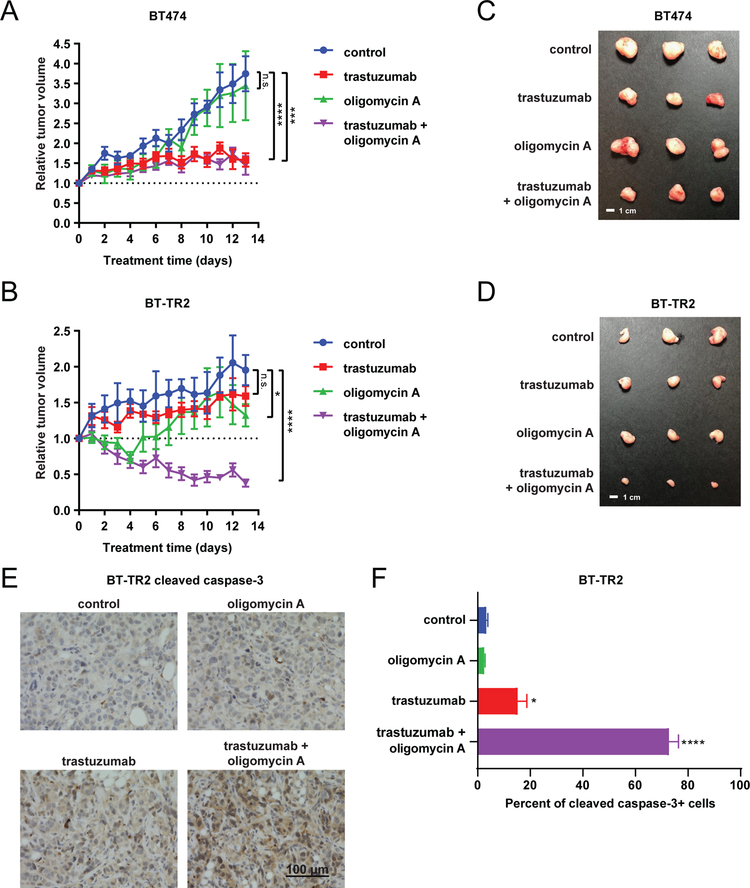
To interrogate these concepts in vivo, we injected BT474 and BT-TR2 cells into the mammary fat pads of female nude mice implanted with pellets to release β-estradiol. After tumor formation, mice were split into four treatment groups: control, trastuzumab, oligomycin A, and trastuzumab + oligomycin A. Despite similar in vitro growth rates, BT474 tumors grew faster than BT-TR2 tumors in vivo with control tumors more than tripling in size for BT474 over ~2 weeks, but only doubling for BT-TR2 (Figures 5A–D). This may be because BT-TR2 tumors have a diminished response to estrogen compared to BT474 tumors, as we noted that the hallmark pathways for estrogen response were significantly down-regulated in BT-TR2 cells compared to parental cells (Figure 2D–E). We analyzed the effect of treatment on each tumor by comparing tumor volume relative to the pre-treatment tumor volume. BT474 tumors responded to trastuzumab, but not to oligomycin A. Trastuzumab significantly stagnated tumor growth, while oligomycin A had negligible effects on growth. The combination of the two did not offer further benefit beyond the effects of trastuzumab alone (Figure 5A). In contrast, BT-TR2 tumors exhibited only small benefit from trastuzumab or oligomycin A as single agents. The loading dose of oligomycin A appeared to decrease tumor volume for the first days of the experiment, but tumors recovered during the maintenance dose phase. Notably, the combination of the two incited significant tumor regression (Figure 5B). BT-TR2 tumors treated with trastuzumab + oligomycin A shrank by an average of 60% compared to their starting volumes, demonstrating the potential of this combination for treating trastuzumab-resistant tumors. This combination treatment led to significant apoptosis of tumor cells as measured by cleaved caspase-3 staining (Figure 5E–F), without affecting overall tumor morphology or tumor cell proliferation as measured by Ki67 staining (Supplementary Figure 11).
Figure 5: Trastuzumab-resistant tumors regress with oligomycin A and trastuzumab combination therapy.
Tumor volume relative to pre-treatment tumor volume of BT474 (A) and BT-TR2 (B) tumors treated with trastuzumab (5 mg/kg i.p. every 3 days), oligomycin A (0.5 mg/kg i.p. daily for 3 days followed by 0.25 mg/kg i.p. daily), the combination, or saline as control. n=6 for control group, n=10 for trastuzumab group in panel A, n=8 for trastuzumab group in panel B, n=5 for oligomycin A group, and n=9 for combination treatment group. Representative BT474 (C) and BT-TR2 (D) tumors at endpoint for each treatment group. (E-F) Representative images (E) and quantification (F) of cleaved caspase 3 immunohistochemical staining in BT-TR2 tumors (n=3) at endpoint for each treatment group. 3 representative fields were quantified per tumor. Data points in A, B and F represent mean ± SEM. n.s., not significant. *, p<0.05. ***, p<0.001. ****, p<0.0001.
Discussion
In this study, we showed that trastuzumab resistance is reversible, suggesting that non- mutational changes contribute considerably to acquired trastuzumab resistance. Studies in other types of cancer have demonstrated the flexibility of drug tolerance within persister cell populations (23,25). Here, we demonstrated that fully developed trastuzumab-resistant cell pools and clones are able to revert back to a sensitive state if cultured in drug-free media. This provides rationale for considering a clinical model which includes a drug-free hiatus for patients, followed by re-treatment with trastuzumab. It also provides support for identifying new therapeutic targets based on transcriptional reprogramming in resistant cells.
Through transcriptional profiling, we identified several gene expression programs significantly up or down-regulated in resistant pools compared to parental cells. Our analysis revealed similarities and differences between the single or dual therapy acquired resistance settings. For instance, the protein secretion pathway was exclusively up-regulated in trastuzumab-resistant pools, while five pathways were exclusively down-regulated in trastuzumab + pertuzumab-resistant pools. Several of the pathways down-regulated only in dual-resistant pools were related to inflammation, revealing an interesting finding for future exploration. Four pathways were significantly down-regulated by all resistant pools, while three were significantly up-regulated. Of note, we found elevated expression of genes involved in oxidative phosphorylation, particularly ATP synthase genes, in all four tested trastuzumab- and trastuzumab + pertuzumab-resistant pools. These increases could be due to increased Myc activity (Figure 2A), which was shown to regulate expression of complex V genes (40), or other posttranscriptional regulatory mechanisms of ATP synthase components that can lead to change of mRNA levels (41). Furthermore, we found that higher expression of ATP synthase component ATP5B significantly correlated with poor overall survival of HER2+ breast cancer patients. These findings indicate that expression of ATP synthase complex genes, especially ATP5B, may be a good biomarker for predicting response to trastuzumab and clinical outcomes of HER2+ breast cancer patients.
Treatment with a small molecule tyrosine kinase inhibitor was previously shown to sensitize leukemia cancer cells to ATP synthase inhibition (42), but it was not known whether antibody-based targeted therapies had a similar consequence or if this occurred in alternate cancer settings. We demonstrated here that long-term HER2-targeted antibody treatment to generate resistant breast cancer cells also created sensitivity to ATP synthase inhibition. Resistant cells that demonstrated increased gene expression of ATP synthase demonstrated increased sensitivity to pharmacological inhibition of ATP synthase compared to parental cells. This was the case for both single and dual-resistant cells, broadening the potential clinical relevance of these findings to a wider population of patients. Knockdown of ATP5J or ATP5B was sufficient to make trastuzumab-resistant cells respond to a low dose of trastuzumab not effective on the corresponding control cells. Trastuzumab had a cytostatic effect on BT474 tumors in vivo and very little effect on BT-TR2 tumors. However, the combination of oligomycin A and trastuzumab induced regression of trastuzumab resistant tumors in vivo.
These data suggest that ATP synthase function is required for the maintenance of resistance and propose interference with ATP synthase activity as a method for overcoming resistance. Consistent with our findings, oligomycin A treatment prevented outgrowth of a therapy-resistant melanoma cell subpopulation (43) and prevented tumor recurrence in a mouse model of KRAS-mutant pancreatic cancer (44). However, while the oligomycin A-sensitive cell populations demonstrated increased OCR or ATP production in other models, the resistant cells in this study had similar OCR and ATP production compared to parental cells. These results suggest that ATP synthase in these cells is less efficient in producing ATP and that these cells functionally compensate for this defect by inducing expression of ATP synthase, therefore creating vulnerability to ATP synthase inhibition. The resistant cells showed less sensitivity to 2-DG (Supplementary Figure 6F) and greater sensitivity to OXPHOS inhibitors than the parental cells (Figure 3 and Supplementary Figure 4). Interestingly, Myc-positive cells also showed similar trends in sensitivity to these drugs compared to Myc-deficient cells (45). The phenotypic similarity described above is consistent with the idea that increased Myc activity leads to increased expression of OXPHOS genes in drug-resistant cells. In fact, it was reported that breast cancer cells resistant to lapatinib, an inhibitor of HER2 and EGFR, are sensitive to Myc inhibition (46).
Overall, the data presented here suggests that cells resistant to HER2-targeted antibodies have shifted their metabolism in such a way as to make them vulnerable to disruption of ATP synthase function. This approach as a cancer therapy is nuanced because all cells require mitochondrial respiration, but our data suggests that a therapeutic window may exist. The low nanomolar doses of oligomycin A used in vitro selectively slowed the growth of breast cancer cells, but not normal-like breast epithelial cells. Daily oligomycin A injection in vivo did not significantly affect blood counts or markers of renal and hepatic toxicities as assessed by Alvarez-Calderon et al. (42) and it was tolerated in experiments performed by us and others (44). Alternative inhibitors of ATP synthase may demonstrate stronger safety profiles and should be explored. For example, FDA-approved drugs such as bedaquiline (47), simvastatin (48), paroxetine (48) and tamoxifen (48) were recently identified as inhibitors of ATP synthase activity and may be better tolerated for this purpose. Taken together, our study revealed that ATP synthase inhibition is a new avenue to defeat resistance to HER2-targeted antibodies in breast cancers.
Supplementary Material
Significance:
These findings implicate ATP synthase as a novel potential target for tumors resistant to HER2-targeted therapies.
Acknowledgements
We thank the members of the Yan, Shadel, Rimm, Stern, Wajapeyee, and Nguyen laboratories at Yale School of Medicine for their helpful discussions and support. The Illumina sequencing service was conducted by Dr. Mei Zhong at Yale Stem Cell Center Genomics Core facility, which was supported by the Connecticut Regenerative Medicine Research Fund and the Li Ka Shing Foundation. We thank Dr. Wei Wei at Yale School of Public Health for his biostatistical support. This work was supported in part by the National Institutes of Health (R21 CA187862 to Q.Y., R01 CA216101 to G.S.S., F32 AG052995 to M.A.H., P30 CA016359 to Yale Comprehensive Cancer Center), the Natural Science Foundation of China (Major International Joint Research Program 81620108024 to X.C.), a National Science Foundation Graduate Research Fellowship (DGE-1122492 to M.G.), a James Hudson Brown – Alexander Brown Coxe Postdoctoral Fellowship (to M.Z.) and a Leslie H. Warner Postdoctoral Fellowship (to L.W.).
Footnotes
Disclosure of Potential Conflict of Interest
M. P. DiGiovanna is a consultant for Merck and Immunogen, receives royalties from DAKO and NeoMarkers, had speaking engagement with Total Health Information Services. No potential conflicts of interest were disclosed by the other authors.
References
- 1.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 1987;235:177–82 [DOI] [PubMed] [Google Scholar]
- 2.Arteaga CL, Engelman JA. ERBB receptors: from oncogene discovery to basic science to mechanism-based cancer therapeutics. Cancer cell 2014;25:282–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cuello M, Ettenberg SA, Clark AS, Keane MM, Posner RH, Nau MM, et al. Down-regulation of the erbB-2 receptor by trastuzumab (herceptin) enhances tumor necrosis factor-related apoptosis-inducing ligand-mediated apoptosis in breast and ovarian cancer cell lines that overexpress erbB-2. Cancer research 2001;61:4892–900 [PubMed] [Google Scholar]
- 4.Klapper LN, Waterman H, Sela M, Yarden Y. Tumor-inhibitory antibodies to HER-2/ErbB-2 may act by recruiting c-Cbl and enhancing ubiquitination of HER-2. Cancer research 2000;60:3384–8 [PubMed] [Google Scholar]
- 5.Nagata Y, Lan KH, Zhou X, Tan M, Esteva FJ, Sahin AA, et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer cell 2004;6:117–27 [DOI] [PubMed] [Google Scholar]
- 6.Junttila TT, Akita RW, Parsons K, Fields C, Lewis Phillips GD, Friedman LS, et al. Ligand-independent HER2/HER3/PI3K complex is disrupted by trastuzumab and is effectively inhibited by the PI3K inhibitor GDC-0941. Cancer cell 2009;15:429–40 [DOI] [PubMed] [Google Scholar]
- 7.Zhang S, Huang WC, Li P, Guo H, Poh SB, Brady SW, et al. Combating trastuzumab resistance by targeting SRC, a common node downstream of multiple resistance pathways. Nature medicine 2011;17:461–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghosh R, Narasanna A, Wang SE, Liu S, Chakrabarty A, Balko JM, et al. Trastuzumab has preferential activity against breast cancers driven by HER2 homodimers. Cancer research 2011;71:1871–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Molina MA, Codony-Servat J, Albanell J, Rojo F, Arribas J, Baselga J. Trastuzumab (herceptin), a humanized anti-Her2 receptor monoclonal antibody, inhibits basal and activated Her2 ectodomain cleavage in breast cancer cells. Cancer research 2001;61:4744–9 [PubMed] [Google Scholar]
- 10.Arnould L, Gelly M, Penault-Llorca F, Benoit L, Bonnetain F, Migeon C, et al. Trastuzumab-based treatment of HER2-positive breast cancer: an antibody-dependent cellular cytotoxicity mechanism? British journal of cancer 2006;94:259–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clynes RA, Towers TL, Presta LG, Ravetch JV. Inhibitory Fc receptors modulate in vivo cytotoxicity against tumor targets. Nature medicine 2000;6:443–6 [DOI] [PubMed] [Google Scholar]
- 12.Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med 2001;344:783–92 [DOI] [PubMed] [Google Scholar]
- 13.Slamon D, Eiermann W, Robert N, Pienkowski T, Martin M, Press M, et al. Adjuvant trastuzumab in HER2-positive breast cancer. N Engl J Med 2011;365:1273–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perez EA, Romond EH, Suman VJ, Jeong JH, Sledge G, Geyer CE Jr., et al. Trastuzumab plus adjuvant chemotherapy for human epidermal growth factor receptor 2-positive breast cancer: planned joint analysis of overall survival from NSABP B-31 and NCCTG N9831. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2014;32:3744–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Piccart-Gebhart MJ, Procter M, Leyland-Jones B, Goldhirsch A, Untch M, Smith I, et al. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med 2005;353:1659–72 [DOI] [PubMed] [Google Scholar]
- 16.Swain SM, Baselga J, Kim SB, Ro J, Semiglazov V, Campone M, et al. Pertuzumab, trastuzumab, and docetaxel in HER2-positive metastatic breast cancer. N Engl J Med 2015;372:724–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von Minckwitz G, Procter M, de Azambuja E, Zardavas D, Benyunes M, Viale G, et al. Adjuvant Pertuzumab and Trastuzumab in Early HER2-Positive Breast Cancer. N Engl J Med 2017;377:122–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franklin MC, Carey KD, Vajdos FF, Leahy DJ, de Vos AM, Sliwkowski MX. Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex. Cancer cell 2004;5:317–28 [DOI] [PubMed] [Google Scholar]
- 19.Agus DB, Akita RW, Fox WD, Lewis GD, Higgins B, Pisacane PI, et al. Targeting ligand-activated ErbB2 signaling inhibits breast and prostate tumor growth. Cancer cell 2002;2:127–37 [DOI] [PubMed] [Google Scholar]
- 20.Vogel CL, Cobleigh MA, Tripathy D, Gutheil JC, Harris LN, Fehrenbacher L, et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol 2002;20:719–26 [DOI] [PubMed] [Google Scholar]
- 21.Romond EH, Perez EA, Bryant J, Suman VJ, Geyer CE, Davidson NE, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med 2005;353:1673–84 [DOI] [PubMed] [Google Scholar]
- 22.Vu T, Sliwkowski MX, Claret FX. Personalized drug combinations to overcome trastuzumab resistance in HER2-positive breast cancer. Biochimica et biophysica acta 2014;1846:353–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hangauer MJ, Viswanathan VS, Ryan MJ, Bole D, Eaton JK, Matov A, et al. Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature 2017;551:247–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knoechel B, Roderick JE, Williamson KE, Zhu J, Lohr JG, Cotton MJ, et al. An epigenetic mechanism of resistance to targeted therapy in T cell acute lymphoblastic leukemia. Nat Genet 2014;46:364–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 2010;141:69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liau BB, Sievers C, Donohue LK, Gillespie SM, Flavahan WA, Miller TE, et al. Adaptive Chromatin Remodeling Drives Glioblastoma Stem Cell Plasticity and Drug Tolerance. Cell stem cell 2017;20:233–46 e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morandi A, Indraccolo S. Linking metabolic reprogramming to therapy resistance in cancer. Biochimica et biophysica acta 2017;1868:1–6 [DOI] [PubMed] [Google Scholar]
- 28.Zhao Y, Liu H, Liu Z, Ding Y, Ledoux SP, Wilson GL, et al. Overcoming trastuzumab resistance in breast cancer by targeting dysregulated glucose metabolism. Cancer research 2011;71:4585–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lenz G, Hamilton A, Geng S, Hong T, Kalkum M, Momand J, et al. t-Darpp Activates IGF-1R Signaling to Regulate Glucose Metabolism in Trastuzumab-Resistant Breast Cancer Cells. Clinical cancer research : an official journal of the American Association for Cancer Research 2018;24:1216–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gale M, Sayegh J, Cao J, Norcia M, Gareiss P, Hoyer D, et al. Screen-identified selective inhibitor of lysine demethylase 5A blocks cancer cell growth and drug resistance. Oncotarget 2016;7:39931–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu L, Cao J, Cai WL, Lang SM, Horton JR, Jansen DJ, et al. KDM5 histone demethylases repress immune response via suppression of STING. PLoS Biol 2018;16:e2006134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang H, Minamishima YA, Yan Q, Schlisio S, Ebert BL, Zhang X, et al. pVHL acts as an adaptor to promote the inhibitory phosphorylation of the NF-kappaB agonist Card9 by CK2. Mol Cell 2007;28:15–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carvajal-Hausdorf DE, Schalper KA, Pusztai L, Psyrri A, Kalogeras KT, Kotoula V, et al. Measurement of Domain-Specific HER2 (ERBB2) Expression May Classify Benefit From Trastuzumab in Breast Cancer. J Natl Cancer Inst 2015;107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fountzilas G, Dafni U, Papadimitriou C, Timotheadou E, Gogas H, Eleftheraki AG, et al. Dose-dense sequential adjuvant chemotherapy followed, as indicated, by trastuzumab for one year in patients with early breast cancer: first report at 5-year median follow-up of a Hellenic Cooperative Oncology Group randomized phase III trial. BMC cancer 2014;14:515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luque-Cabal M, Garcia-Teijido P, Fernandez-Perez Y, Sanchez-Lorenzo L, Palacio-Vazquez I. Mechanisms Behind the Resistance to Trastuzumab in HER2-Amplified Breast Cancer and Strategies to Overcome It. Clin Med Insights Oncol 2016;10:21–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108–e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Graves JA, Wang Y, Sims-Lucas S, Cherok E, Rothermund K, Branca MF, et al. Mitochondrial structure, function and dynamics are temporally controlled by c-Myc. PloS one 2012;7:e37699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Willers IM, Cuezva JM. Post-transcriptional regulation of the mitochondrial H(+)-ATP synthase: a key regulator of the metabolic phenotype in cancer. Biochimica et biophysica acta 2011;1807:543–51 [DOI] [PubMed] [Google Scholar]
- 42.Alvarez-Calderon F, Gregory MA, Pham-Danis C, DeRyckere D, Stevens BM, Zaberezhnyy V, et al. Tyrosine kinase inhibition in leukemia induces an altered metabolic state sensitive to mitochondrial perturbations. Clinical cancer research : an official journal of the American Association for Cancer Research 2015;21:1360–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roesch A, Vultur A, Bogeski I, Wang H, Zimmermann KM, Speicher D, et al. Overcoming Intrinsic Multidrug Resistance in Melanoma by Blocking the Mitochondrial Respiratory Chain of Slow-Cycling JARID1B(high) Cells. Cancer Cell 2013;23:811–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sanchez N, Marchesini M, et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature 2014;514:628–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morrish F, Neretti N, Sedivy JM, Hockenbery DM. The oncogene c-Myc coordinates regulation of metabolic networks to enable rapid cell cycle entry. Cell Cycle 2008;7:1054–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matkar S, Sharma P, Gao S, Gurung B, Katona BW, Liao J, et al. An Epigenetic Pathway Regulates Sensitivity of Breast Cancer Cells to HER2 Inhibition via FOXO/c-Myc Axis. Cancer cell 2015;28:472–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fiorillo M, Lamb R, Tanowitz HB, Cappello AR, Martinez-Outschoorn UE, Sotgia F, et al. Bedaquiline, an FDA-approved antibiotic, inhibits mitochondrial function and potently blocks the proliferative expansion of stem-like cancer cells (CSCs). Aging (Albany NY) 2016;8:1593–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nadanaciva S, Bernal A, Aggeler R, Capaldi R, Will Y. Target identification of drug induced mitochondrial toxicity using immunocapture based OXPHOS activity assays. Toxicol In Vitro 2007;21:902–11 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.