Abstract
Activation of the epithelial-mesenchymal transition (EMT) program is a critical mechanism for initiating cancer progression and migration. Colorectal cancers (CRCs) contain many genetic and epigenetic alterations that can contribute to EMT. Mutations activating the PI3K/AKT signaling pathway are observed in >40% of patients with CRC contributing to increased invasion and metastasis. Little is known about how oncogenic signaling pathways such as PI3K/AKT synergize with chromatin modifiers to activate the EMT program. Lysine Specific Demethylase 1 (LSD1) is a chromatin-modifying enzyme that is overexpressed in colorectal cancer (CRC) and enhances cell migration. In this study we determine that LSD1 expression is significantly elevated in CRC patients with mutation of the catalytic subunit of PI3K, PIK3CA, compared to CRC patients with WT PIK3CA. LSD1 enhances activation of the AKT kinase in CRC cells through a non-catalytic mechanism, acting as a scaffolding protein for the transcription-repressing CoREST complex. Additionally, growth of PIK3CA mutant CRC cells is uniquely dependent on LSD1. Knockdown or CRISPR knockout of LSD1 blocks AKT-mediated stabilization of the EMT-promoting transcription factor Snail and effectively blocks AKT-mediated EMT and migration. Overall we uniquely demonstrate that LSD1 mediates AKT activation in response to growth factors and oxidative stress, and LSD1-regulated AKT activity promotes EMT-like characteristics in a subset of PIK3CA mutant cells.
Implications
Our data supports the hypothesis that inhibitors targeting the CoREST complex may be clinically effective in CRC patients harboring PIK3CA mutations.
Keywords: Epigenetics, PIK3CA, LSD1, CoREST, EMT, pAKT
Introduction
Cancer cells have numerous genetic and epigenetic alterations that contribute to tumor formation, progression, and therapy resistance. One role of chromatin-modifying complexes is to maintain cellular identity by chemically modifying amino acid residues on histone and non-histone substrates. Lineage-specific transcriptional networks maintained by these complexes provide context for specific mutations which determines the phenotypic outcome of the mutation(1). This information can be leveraged to identify how chromatin modifiers synergize with specific driver mutations in tissue-specific tumor models and discover synthetic-lethal relationships.
Mutations in the PI3K/AKT pathway are critical for invasive properties and malignant transformation in colorectal cancer (CRC)(2). The two most common genetic PI3K/AKT pathway alterations in CRC are activating mutations in the PI3K catalytic subunit gene PIK3CA or loss of the pathway suppressor PTEN(3). Mutations in PIK3CA occur in roughly 25% of CRC patients(4) and have been functionally implicated in epithelial-to-mesenchymal transition (EMT), migration and chemoresistance(5). While aberrant activation of the PI3K/AKT pathway has been implicated in CRC progression, single nucleotide PIK3CA mutations that activate the PI3K/AKT pathway are not significantly associated with alterations in patient survival(6). These findings indicate that PI3K-pathway activating mutations may require additional factors for full activation of the pathway. Recently, the lysine demethylase JMJD2A was found to be critical for steps involved in activation of AKT, including the recruitment of AKT to the cell membrane and phosphorylation of AKT at threonine 308(7, 8). These studies suggest that aberrant overexpression of chromatin-modifying proteins can further activate the PI3K/AKT pathway and therefore may work synergistically with PIK3CA mutations. Little is known with regard to how chromatin modifiers function in the context of PIK3CA mutation to mediate tumorigenic processes in the gut.
The chromatin modifier lysine specific demethylase 1 (LSD1) is overexpressed in CRC and positively correlates with advanced tumor staging(9). LSD1 is functionally linked to EMT-like changes and invasion in CRC(10–12). LSD1 is a member of the RE1 silencing transcription factor corepressor (CoREST) complex(13), which also contains the scaffolding protein RCOR1 and other chromatin-modifying subunits, including histone deacetylase 1 and 2 (HDAC1/2)(14, 15). LSD1 and HDAC1/2 within CoREST demethylate and deacetylate active chromatin, respectively, to maintain a repressive chromatin state. In some cellular contexts, LSD1, as a member of CoREST, demethylates di-methyl Histone H3 Lysine 4 (H3K4me2) at the promoter of epithelial genes to drive CRC(10–12). Recent studies, however, have highlighted catalysis-independent functions for LSD1, where it instead acts as a scaffold for the CoREST complex to maintain transcriptional repression of lineage-specific genes(16, 17). For example, RE1 silencing transcription factor (REST) can confine expression of neuronal genes to neuronal cells by mediating their silencing in non-neuronal cell types through the recruitment of CoREST(14, 15, 18). Furthermore, mechanistic studies of LSD1 catalytic inhibitors in SCLC(19), AML(20, 21) and erythroleukemia(22) demonstrate that these inhibitors reactivate gene expression and alter processes such as survival, proliferation and differentiation by disrupting the recruitment of CoREST to chromatin by SNAG domain transcription factors as opposed to inhibiting LSD1 demethylase activity. These studies further support the notion that non-catalytic LSD1 functions are critical for tumorigenesis.
We hypothesize that LSD1 overexpression synergizes with PIK3CA mutation to enhance invasive phenotypes in CRC. In this study, we demonstrate that LSD1 is significantly overexpressed in patients harboring PIK3CA mutations in the gut, but not in cancers arising from other tissues. This observation is functionally significant as we demonstrate that PIK3CA mutant colorectal and stomach cancer cells exhibit reduced growth after perturbation of LSD1. We further find that LSD1 regulates activation of AKT at the level of phosphorylation at serine 473 and EMT characteristics downstream of active AKT through a non-catalytic scaffolding role in the CoREST complex. Altogether we illustrate a paradigm wherein LSD1 synergizes with a specific PIK3CA mutation to enhance EMT characteristics and migration.
Materials and Methods
Cell Culture and Treatments
All cell lines were maintained in a humidified atmosphere with 5% CO2. Our study included five colon cell lines (HT29, SW480, HCT116, LoVo and RKO) and one stomach cell line (AGS). HT29, SW480, HCT116 and LoVo cells were cultured in McCoys 5A media (Corning), RKO and AGS were cultured in RPMI 1640 media (Corning) supplemented with 10% FBS (Gibco). All cell lines were purchased from the ATCC and authenticated and tested for Mycoplasma by IDEXX on 6/20/2019. All cells used in experiments were passaged fewer than 15 times with most being passaged fewer than 10 times. For H2O2 treatments, 30% H2O2 (Sigma) was diluted in PBS immediately prior to treatment at 250 μM for 1H at 37°C. For EGF treatments, cells were starved in media lacking serum for 48H prior to treatment. Cells were then treated with 100 ng/ml recombinant EGF (R&D Systems: 236-EG) for 48H. GSK-LSD1 (Sigma, SML1072), GSK690693 (Sigma, SML0428) and corin (generously provided by Dr. Philip Cole and Dr. Jay Kalin) were solubilized in DMSO (Sigma) prior to treatment. Treatment dosages and durations are defined in the figure legends.
Knockdown, Knockout and Transient Transfections
LSD1 (KDM1A) (TRCN0000327856), RCOR1 (TRCN0000128570) and HDAC1 (TRCN0000195467, TRCN0000195103) knockdown constructs were purchased from Sigma Aldrich mission shRNA; empty plasmid was used as a vector control. Lentiviral-mediated knockdowns were performed as previously described(23). CRISPR/Cas9 LSD1 KO plasmid (sc-430289) and LSD1 HDR plasmid (sc-430289-HDR) were purchased from Santa Cruz and knockout was performed according to manufacturer’s protocol. Individual LSD1 KO clones were isolated for experiments, or mixed population KO’s were used, as defined in the figure legends. Cells were transfected with Lipofectamine 3000 (Invitrogen) per the manufacturer’s protocol. N-terminus HA tagged LSD1 plasmid was purchased from Sino Biological (HG13721-NY) with pCMV3-N-HA used as a negative control vector.
Site-directed Mutagenesis
Mutagenesis was performed according to manufacturer’s protocol (NEB, E0554) using HA-LSD1 plasmid as template, and confirmed via Sanger sequencing by Eurofins Scientific. LSD1 K661A substitution primers were generated using NEBaseChanger.
Primer sequences:
forward, CAACCTTAACgcGGTGGTGTTGTG
reverse, CCAAATCCCATCCTTTGG
Annealing temperature: 58°C
Chromatin Immunoprecipitation (ChIP)-sequencing
ChIP was performed using Diagenode iDeal ChIP-seq kit (C01010055 - for Transcription factors) (C01010057 - for histone modifications) per manufacturer’s protocol. Libraries were generated for sequencing using the NEBNext Ultra DNA Library Prep Kit for Illumina (NEB) per manufacturer’s protocol.
Chromatin Bound Fraction and Whole Cell Isolation
Chromatin bound (or tight chromatin) fractions were isolated as previously described(23). For protein isolation, cell pellets were lysed in 4% SDS buffer using a Qiashredder (Qiagen). Relative densitometry for western blots were determined using ImageJ software, and normalized to density of loading controls Lamin-B, β-Actin or Histone H3. All antibodies used in this study are included in the Supplementary Materials and Methods.
Immunofluorescence and Imaging
2 × 105 HT29 cells were grown on coverslips at 37°C. After 48 hours, they were treated with 250μM H2O2 for 1 hour. The cells were then fixed with 4% paraformaldehyde in PBS for 15 minutes at RT. Cells were permeabilized with 0.5% Triton-X in PBS for 10 minutes at RT and then fixed with 1% BSA in PBST (PBS + 0.2% tween 20) for 30 minutes. They were then incubated with anti-LSD1 (1:100) and anti-pAKT (1:100) in 1% BSA in PBST for 1 hour at RT. This was followed by incubation with Alexa flour anti-rabbit 594 (1:500) and Alexa flour anti-mouse 488 (1:1000) for 1 hour at RT. Coverslips were mounted using prolong gold antifade with DAPI molecular probes (CST #8961). Images were acquired using Leica SP8 confocal microscope at a magnification of 63X. The NA of 63X objective used is 1.4. Images were processed using Image J.
Proliferation Assays
Assays were performed using the CellTiter-Glo Luminescent Cell Viability Assay (Promega #G7572) per manufacturer’s protocol. Briefly, 1×10^3 cells were plated in 96 well plates and allowed to incubate under standard growth conditions. Luminescent signals were detected on a SYNERGY H1 microplate reader (BioTek) using Gen 5 software (v2.09). All plate readings were normalized to a control plate to account for variance in plating density.
Clonogenic Growth and Migration Assays
500 (HT29) or 1500 (SW480) single cells were plated and allowed to culture at 37°C. After 10 days, cells were fixed with 10% formalin and stained with crystal violet. Crystal violet stained cells were imaged using a SYNGENE G:BOX and quantified using the GeneSys and GeneTools programs. For migration, 7.5 × 10^4 cells in serum free media were plated into transwell in 24 well plates (corning #40578) for 48H with media containing 10% FBS at the bottom. Transwell inserts were stained using Hema 3 Stat Pack (Fisher #123–869). Migration inserts were randomized prior to manual quantification and the outer 5% of the inserts were not included during quantification to reduce edge-effect bias. All images were taken on an EVOS FL Auto microscope.
RNA Isolation for RNA-sequencing and Quantitative PCR
Total RNA was isolated using the RNeasy mini kit (Qiagen, 74104). For RNA-sequencing of empty vector and LSD1 knockdown cells, libraries were generated using Illumina TruSeq Stranded mRNA (Illumina, 20020594), or as previously described for RNA-sequencing of DMSO and GSK-LSD1 treated cells(24). For qPCR, RNA was used to generate cDNA via reverse transcription (Thermo, K1642). cDNA was amplified using gene-specific primers and FastStart Essential DNA Green Master (Roche, 06402712001). Cq values of non-housekeeping genes were normalized to GAPDH expression. qPCR Primer sequences listed below:
LSD1, forward, GGTGAGCTCTTCCTCTTCTGG
LSD1, reverse, TCGGCCAACAATCACATCGT
SNAI1, forward, CTAGGCCCTGGCTGCTACA
SNAI1, reverse, TGGCACTGGTACTTCTTGACA
GAPDH, forward, GAAGGTCGGAGTCAACGGATTT
GAPDH, reverse, ATGGGTGGAATCATATTGGAC
Sequencing, TCGA and Statistical Analyses
Detailed description included in the Supplementary Materials and Methods. All experiments were performed in biological triplicate unless otherwise specified, with representative results displayed. All quantitative plots depict mean ± SD unless otherwise stated.
Data availability
RNA-sequencing and ChIP-sequencing data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE139927.
Results
Knockdown of LSD1 reduces phosphorylation of AKT S473
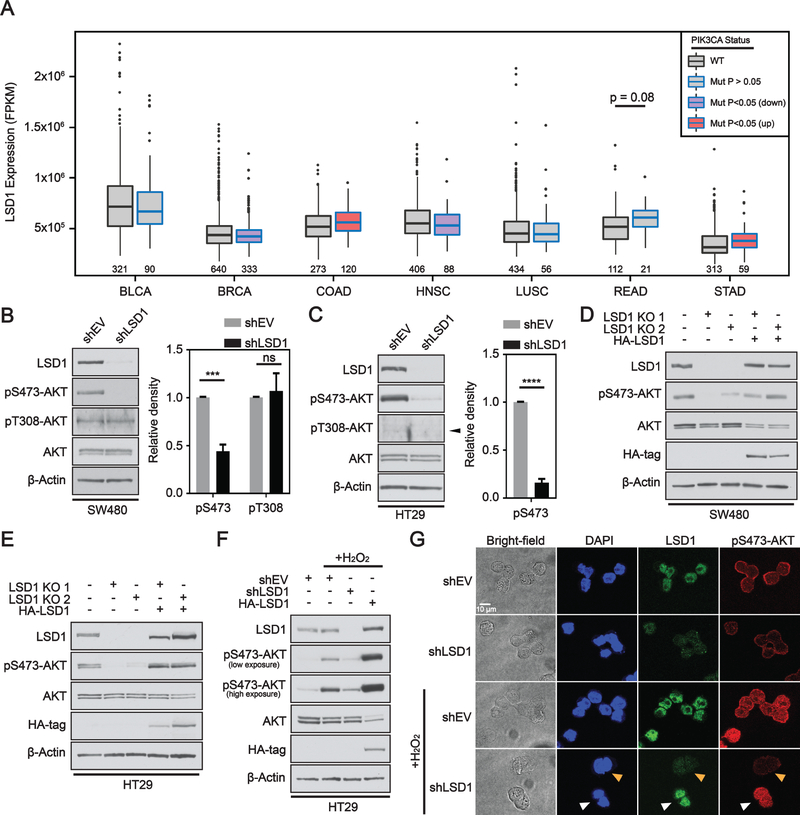
To initially determine if LSD1 works synergistically with PIK3CA, we analyzed The Cancer Genome Atlas (TCGA) patient data from 7 different cancer types where PIK3CA mutations are common. LSD1 expression was significantly higher in PIK3CA mutant versus WT tumors for gastrointestinal cancers colon adenocarcinoma (COAD) and stomach adenocarcinoma (STAD) (Figure 1A). There was a trend toward increased LSD1 expression in PIK3CA mutant rectal adenocarcinomas (READ) in the modest number of cases analyzed. All other tumor types either had no significant change (BLCA, LUSC) or a significant decrease (BRCA, HNSC) in LSD1 expression in the presence of a PIK3CA mutation compared to PIK3CA WT tumors.
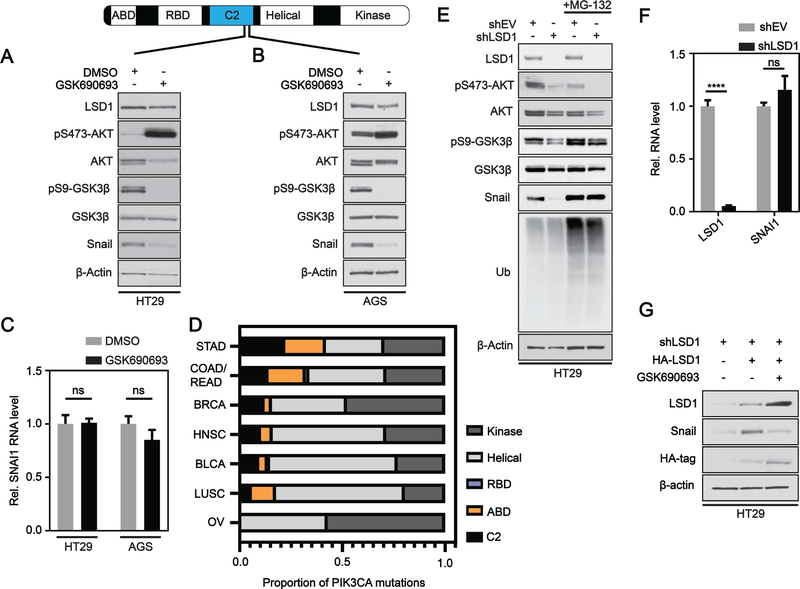
Figure 1: LSD1 regulates phosphorylation of AKT in CRC.
(A) Box and whisker plot of fragments per kilobase of transcript per million mapped reads (FPKM) expression values for LSD1 across different TCGA datasets. Box limits are set at the third and first quartile range with central line at the median, with whiskers depicting 1.5 times the interquartile range. Datapoints outside this range are represented at outliers (black dots). Black and blue outline indicates data for WT and PIK3CA mutant tumors, respectively. Red and purple fill represent significant increase and decrease in LSD1 expression, respectively, between PIK3CA mutant and PIK3CA WT tumors. The numbers under the box plots are the number of samples used to generate the plots. Western blot analysis of empty vector (shEV) or LSD1 KD in (B) SW480 or (C) HT29 cells. Arrow head indicates correct position of pT308-AKT band. Western blot quantified by densitometric analysis and normalized to β-actin and shEV. Results are represented as mean ± SD. (N=3). Significance was determined by two-tailed Student’s t-test. LSD1 CRISPR KO clones with or without LSD1 OE plasmid (HA-LSD1) in (D) SW480 or (E) HT29 cells analyzed by western blot. (F) EV, LSD1 KD or LSD1 OE cells treated w/ 250 μM H2O2 for 1H. (G) Brightfield and immunofluorescence images of EV or LSD1 KD HT29 cells under untreated or H2O2 treated conditions. A field was selected in the H2O2 treated shLSD1 cells to facilitate direct comparison of LSD1 deficient and LSD1 proficient cells. White arrow indicates cells with LSD1 expression and orange arrow indicates cells deficient in LSD1. (p-value; ***<.001, ****<.0001, ns - not significant).
The PI3K pathway has been extensively shown to mediate signaling through the activation of AKT. AKT is synergistically activated(25) by phosphorylation of threonine 308 by PDK1(26), and serine 473 mediated by mTORC2 as part of the mTORC/Rictor complex(27). Chromatin modifiers, specifically histone demethylating enzymes, have recently been linked to the regulation of AKT phosphorylation(7, 8). Therefore, we wanted to test the hypothesis that LSD1 may be a mediator of complete AKT activation. shRNA-mediated knockdown (KD) of LSD1 resulted in a significant reduction in pS473-AKT relative to EV cells in both HT29 and SW480 CRC cell lines (Figure 1B,C). There was no significant change in pT308-AKT in LSD1 KD SW480 cells and we were unable to detect pT308-AKT in HT29 cells, potentially due to low sensitivity. To confirm the effect of LSD1 on pS473-AKT, we used CRISPR to generate two LSD1 knockout (KO) clones each in the SW480 and HT29 cell lines. KO of LSD1 also resulted in reduction of pS473-AKT (Figure 1D, E). Importantly, re-introduction of LSD1 was sufficient to rescue pS473-AKT levels in both cell lines (Figure 1D, E). To test the function of LSD1 in de-novo activation of AKT, we stimulated pS473-AKT using hydrogen peroxide (H2O2)(28). LSD1 KD blocked H2O2-dependent pS473-AKT and, conversely, overexpression of LSD1 enhanced H2O2-dependent pS473-AKT (Figure 1F). Overexpression of LSD1 in combination with H2O2 reduced total AKT, potentially due to a feedback mechanism associated with hyper-phosphorylation of AKT, as has been shown with some AKT inhibitors(29). LSD1-mediated regulation of pS473-AKT observed by western blot was confirmed by immunofluorescence under both basal and H2O2-treated conditions (Figure 1G). pS473-AKT was reduced in LSD1 KD cells compared to empty vector. LSD1 deficient cells exhibited no change in pS473-AKT after H2O2 treatment, while LSD1 proficient cells exhibited H2O2-induced pS473-AKT. To test the alternative hypothesis that mutant PIK3CA was directly causing increased LSD1 expression, we inhibited PI3K in PIK3CA mutant CRC cells, but saw no change in LSD1 levels (Supplementary Figure S1). Together these results demonstrate that LSD1 is more highly expressed in PIK3CA mutant CRC compared to WT PIK3CA CRC and that LSD1 levels positively correlate with pS473-AKT, highlighting a molecular connection between LSD1 levels and activation of the PI3K/AKT pathway.
LSD1 catalytic activity is not required for its regulation of pS473-AKT
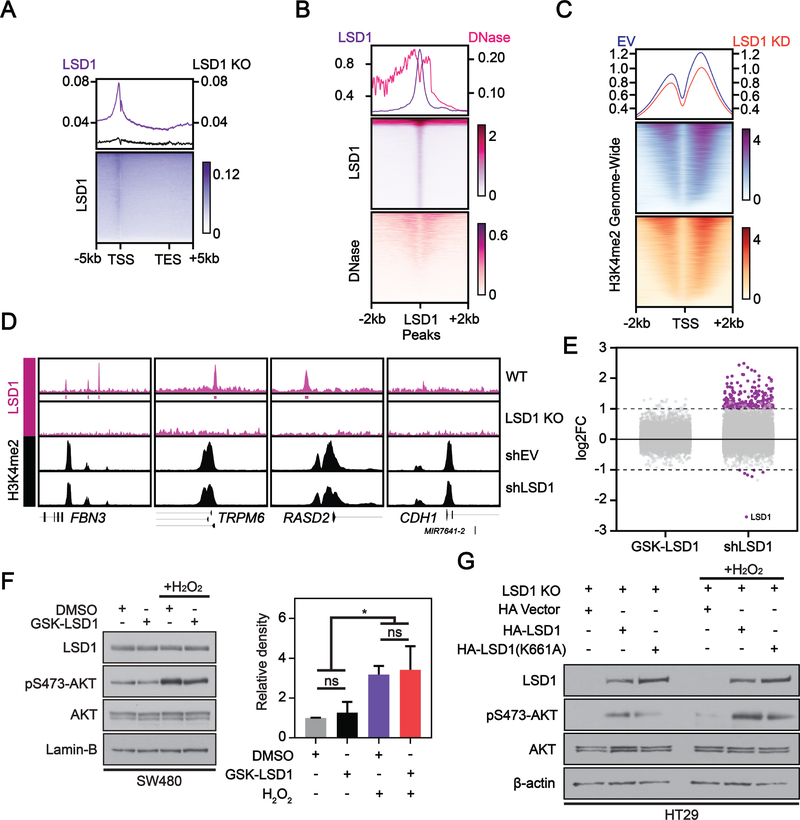
LSD1 catalyzes demethylation of mono- and di-methylated lysine residues on histone(13) and non-histone substrates(30). The oncogenic function of LSD1 in CRC has previously been attributed to its ability to demethylate H3K4me2 at the promoter of epithelial genes, causing their repression and promoting more aggressive cellular phenotypes via enhanced EMT(11, 12). In contrast, newly emerging research has documented non-catalytic oncogenic functions of LSD1(16, 17, 19–22) and suggests we need to consider LSD1 enzymatic activity in the regulation of AKT activation. To address this, we first performed ChIP-seq of LSD1 in SW480 cells to identify direct binding targets genome-wide, generating a catalog of potential catalytic target loci. Consistent with other studies, LSD1 enrichment at genes was primarily detected near transcription start sites (TSS)(31) (Figure 2A). To generate a list of potential LSD1 targets de novo and confirm the specificity of our antibody, LSD1 peaks were called by normalizing LSD1 enrichment in parental cells to our LSD1 CRISPR KO. We detected 6,221 LSD1 peaks shared between replicates. Using DNase sequencing data from the SW480 cell line, we observed depletion of DNase cleavage centered at LSD1 peaks genome-wide (Figure 2B), confirming occupancy of these regions. This DNase footprint is not an artifact of LSD1 enrichment at TSSs, as we do not observe an overall depletion of DNase cleavages over TSSs (Supplementary Figure S2A). To determine if LSD1 KD alters genome-wide histone methylation status, we performed ChIP-seq for the LSD1 substrate H3K4me2 in control and LSD1 KD SW480 cells. There was a slight decrease in H3K4me2 enrichment genome-wide after LSD1 KD, in contrast to the increase that would be expected based on LSD1’s known histone demethylase activity (Figure 2C). There was no significant change in bulk H3K4me2 after LSD1 KD suggesting the slight change in ChIP-seq may not be biologically relevant (Supplementary Figure S2B). To identify potential direct transcriptional targets of LSD1, we performed RNA-seq after LSD1 KD in SW480 cells. We combined this transcriptional data with our ChIP-seq datasets to assess H3K4me2 levels at genes with LSD1 enrichment near their promoters and significantly increased expression after LSD1 KD. The promoter regions of genes FBN3, TRPM6, and RASD2 are depicted as representative loci for this analysis. There were no significant changes in H3K4me2 enrichment at the promoter of FBN3, TRPM6, or RASD2 (Figure 2D), nor at the promoter of any gene following LSD1 KD (data not shown). CDH1, a published target of LSD1, showed significantly increased gene expression (log2FC=1.070, FDR=0.035) after LSD1 KD. However, there was no change in H3K4me2 at the CDH1 promoter after LSD1 KD, nor did we detect enrichment of LSD1. A recent study in PC3 prostate cancer cells demonstrated similar results, wherein levels of H3K4me2 following LSD1 KD only changed at the promoter of 2 genes(16). Together, these data suggest that LSD1 impacts gene expression in SW480 colon cells without demethylating H3K4me2.
Figure 2: LSD1 catalytic activity is dispensable for regulation of gene expression and activation of AKT.
(A) Metageneplot and heatmap depicting ChIP-seq of LSD1 in WT (N=2) or LSD1 KO (N=1) SW480 cells at gene enrichment sites genome-wide. Average plots and heatmaps depicting: (B) LSD1 enrichment peak overlap with DNase-seq peaks in SW480, and (C) H3K4me2 ChIP-seq signal at TSS enrichment sites genome-wide in shEV and shLSD1 SW480 cells (N=3). (A-C) Values are derived from CPM (counts per million) normalized reads. (E) Differentially expressed genes (DEGs) from RNA-seq (log2FC>=1 & FDR<=0.05 = purple) after 40 nM GSK-LSD1 for 48H vs. DMSO or shLSD1 vs shEV in SW480 cells (N=3). (D) ChIP-seq gene tracks of representative DEGs in LSD1 versus EV KD SW480 cells with or without LSD1 promoter enrichment. (F) Cells pretreated with DMSO or 40 nM GSK-LSD1 for 48H then treated with 250 μM H2O2 for 1H. Western blots were quantified by densitometric analysis and normalized to loading control and DMSO. Graph represents mean ± SD. ns – not significant. Significance determined using one-way ANOVA with Tukey’s multiple comparisons test (N=3). (G) Mixed population LSD1 KO cells were transfected with vector control, HA-LSD1, or HA-LSD1(K661A) for 48H. Whole cell extract from untreated and cells treated with 250 μM H2O2 for 1H were analyzed by western blot.
To further characterize LSD1’s role in SW480 cells, we treated these cells with a highly potent and selective inhibitor, GSK-LSD1(31) and performed RNA-sequencing. Treatment with this small molecule had a minimal effect on gene expression (0 genes with expression significantly altered >Log2FC=1), whereas LSD1 KD caused robust activation of gene expression, consistent with the role of LSD1 as a transcriptional repressor (Figure 2E). Additionally, while H2O2 treatment induced the expected increases in pS473-AKT, inhibiting LSD1 did not significantly alter basal or H2O2-induced pS473-AKT levels (Figure 2F), with similar results in HT29 cells (Supplementary Figure S2C). Moreover, catalytically defective K661A mutant LSD1 was able to rescue pS473-AKT levels in LSD1 KO HT29 cells similarly to WT LSD1, both basally and in response to H2O2 (Figure 2G). Together these studies strongly support the concept that LSD1-mediated transcriptional repression and regulation of AKT-activation are independent of LSD1 catalytic activity in these CRC cell lines.
LSD1 regulates pS473-AKT by scaffolding the CoREST complex on chromatin
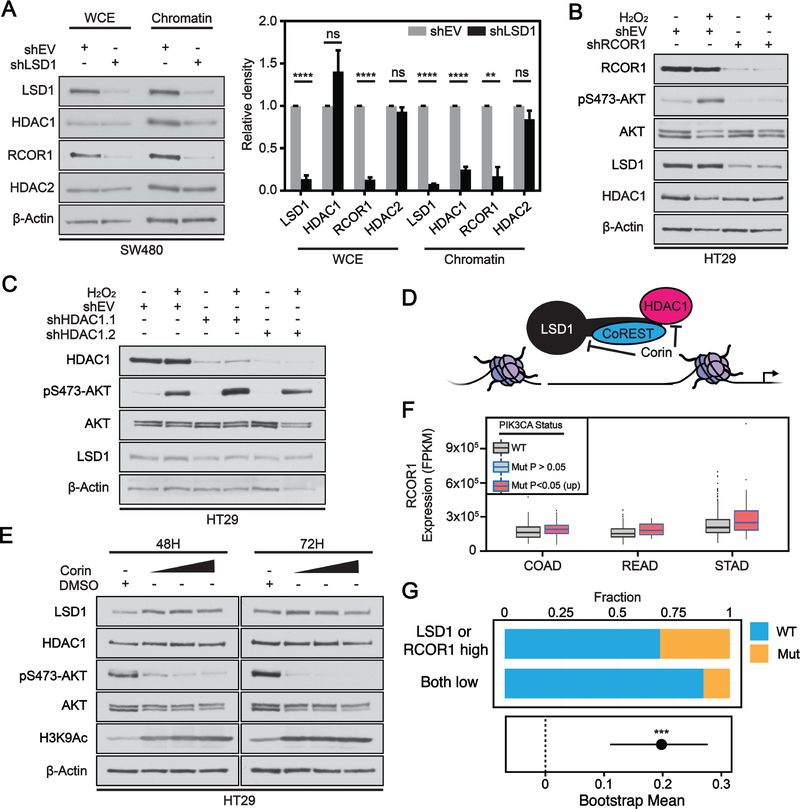
Our RNA- and ChIP-seq data suggest that LSD1-mediated regulation of AKT-activation is independent of demethylase activity; we therefore hypothesized that this regulation may instead occur through LSD1’s stabilizing role in the repressive CoREST complex. To address this, we performed a chromatin fractionation assay. KD of LSD1 in SW480 cells led to a significant decrease in whole cell and chromatin bound levels of the CoREST scaffold protein RCOR1 (Figure 3A). HDAC1 and HDAC2 can be recruited to assist in the repressive functions of CoREST complexes(32). While whole cell levels of HDAC1/HDAC2 remain unchanged after LSD1 KD, there is a significant reduction in the chromatin binding of HDAC1 (Figure 3A). This result supports the notion that loss of LSD1 reduces formation of the CoREST complex and therefore reduces HDAC1 recruitment to chromatin. This is consistent with a recent study, where loss of LSD1 decreased HDAC1 binding at LSD1-enriched enhancers in leukemia cells(20). There was a slight but insignificant decrease in the recruitment of HDAC2 to chromatin (Figure 3A). It is possible that a smaller proportion of cellular HDAC2 is recruited to CoREST complexes in comparison to HDAC1. RCOR1 is one of three CoREST isoforms that interact with LSD1(33). RCOR contains a SANT domain which enhances the complex’s interaction with histones and therefore plays an important scaffolding role for the CoREST complex(34). To determine whether the CoREST complex functions in the regulation of AKT activation, we performed KD of RCOR1 using shRNA and induced pS473-AKT using H2O2. KD of RCOR1 in HT29 cells reduced basal levels of pS473-AKT and blocked H2O2-induced pS473-AKT (Figure 3B). Additionally, there was a decrease in whole cell levels of LSD1 protein, consistent with the scaffolding function ascribed to RCOR1 (Figure 3B).
Figure 3: LSD1 regulates AKT activation via scaffolding of the CoREST complex on chromatin.
(A) Chromatin affinity assay performed in shEV or shLSD1 with whole cell extract (WCE) or chromatin bound fraction. Western blots were quantified by densitometric analysis and normalized to loading control and shEV fraction. Significance determined by 2way ANOVA with Sidak’s multiple comparisons test (N=3). Graphs depict mean ± SD (adj. p-value; **<.01, ****<.0001, ns – not significant). (B) shEV or shRCOR1 cells treated with 250 μM H2O2 for 1H, and analyzed by western blot. (C) shEV or shHDAC1 cells treated as in B. (D) Model for corin inhibitor mode of action. (E) Cells treated with DMSO or 3, 5 or 7 μM corin over time course, and analyzed by western blot. (F) Box and whisker plot of fragments per kilobase of transcript per million mapped reads (FPKM) expression values for RCOR1 across different TCGA datasets. Box limits are set at the third and first quartile range with central line at the median, with whiskers depicting 1.5 times the interquartile range. Datapoints outside this range are represented as outliers (black dots). Black and blue outline indicates data for WT and PIK3CA mutant tumors, respectively. Red fill represent significant increase in LSD1 expression, respectively, between PIK3CA mutant and PIK3CA WT tumors. (G) Fraction of patients with PIK3CA mutation separated by high expression of LSD1 or RCOR1 vs low expression of both LSD1 and RCOR1. Plotted below is bootstrapped 90% confidence interval for the difference in fraction for patients with the PIK3CA mutation between the two groups. Significance determined by permutation test (p-value; ***< 0.001).
To our knowledge, HDAC1 does not function as a scaffold in the CoREST complex but instead contributes to the formation of a repressive chromatin environment by catalyzing the removal of acetyl groups from histone lysine residues. To determine if loss of HDAC1 alone is sufficient to block AKT activation, we knocked down HDAC1 using two independent shRNAs and stimulated AKT using H2O2. HDAC1 KD had no effect on H2O2-induced pS473-AKT or whole cell levels of LSD1 in HT29 (Figure 3C) or SW480 cells (Supplementary Figure S3A). Overall, knockdowns of proteins that stabilize the CoREST complex (LSD1 or RCOR1) were sufficient to perturb AKT activation while knockdown of a protein (HDAC1) that interacts with the core CoREST complex had no effect on AKT activation, indicating that an intact, chromatin-associated core CoREST complex is required for full activation of AKT.
Recently, a single molecule hybrid inhibitor targeting multiple chromatin modifiers within the CoREST complex was synthesized. The compound corin contains an LSD1 inhibitor (tranylcypromine analog) fused to a class 1 HDAC inhibitor (MS-275) and exhibits near irreversible inhibition of the CoREST complex(35) (Figure 3D). In vitro and cellular studies demonstrated this compound is selective for complexes containing HDAC1 and LSD1 over complexes that only contain HDAC1. Corin treatment led to a dose-dependent reduction in pS473-AKT, with the effect increasing over time (Figure 3E). Consistent with HDAC1 inhibition, global H3K9Ac levels increased after treatment with corin. At similar concentrations, MS-275 alone does not reduce pS473-AKT(36).
To test whether these observations mirror the patient data available through the TCGA, we investigated RCOR1 expression in PIK3CA WT and mutant tumors. Like LSD1, RCOR1 showed significantly higher levels of expression in PIK3CA mutant compared to WT in GI cancer types COAD, READ and STAD (Figure 3F). Generally, there was no significant association between RCOR1 expression and PIK3CA mutation in non-GI cancer types (Supplementary Figure S3B). However, there was no significant correlation observed between LSD1 and RCOR1 expression in COAD and READ datasets but there was a significant correlation in the STAD dataset (Supplementary Figure S3C). Consistent with the idea that only LSD1 or RCOR1 need to be overexpressed to stabilize the other, high expression of LSD1 or RCOR1 in COADREAD patient data was significantly associated with PIK3CA mutation compared to patients with low expression of both LSD1 and RCOR1, and the magnitude of change was significantly greater than expected by random chance from permutation testing (Figure 3G). Altogether these data suggest that LSD1 regulates AKT activation through a scaffolding function for the CoREST complex. An intact chromatin-bound CoREST complex is required to mediate positive regulation of AKT activation via regulation of gene expression. Furthermore, high expression of CoREST core members LSD1 or RCOR1 is significantly associated with the presence of PIK3CA mutation.
LSD1 regulates EMT-associated gene programs in PIK3CA mutant cells
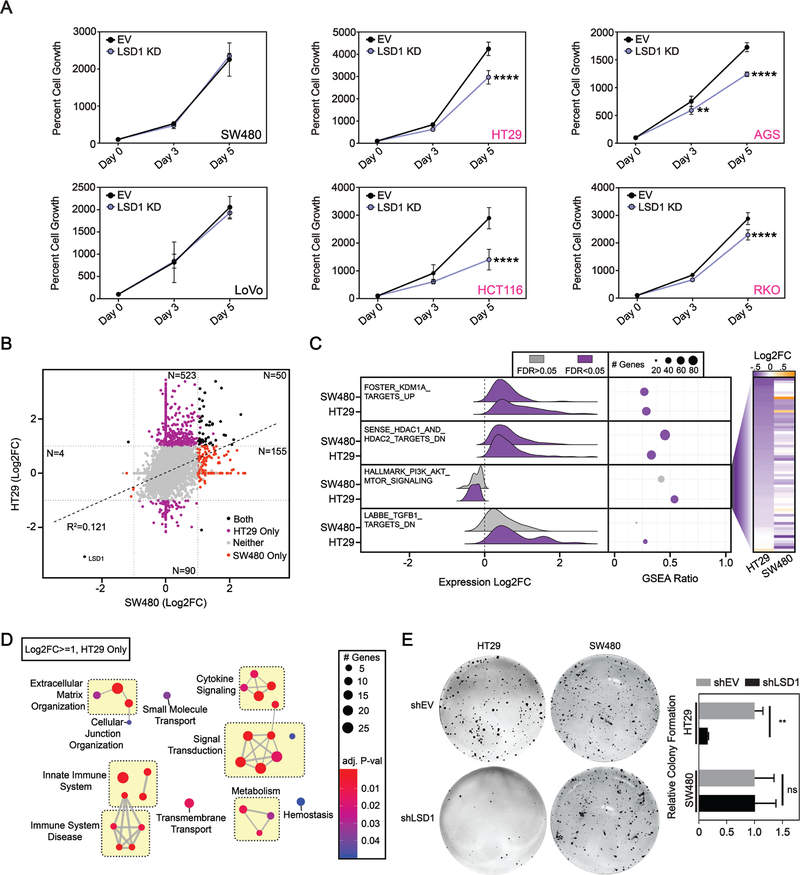
Based on our observations from clinical datasets (Figure 1A), we tested a phenotypic link between PIK3CA mutational status and cellular levels of LSD1. We selected six cell lines for this analysis: five colon cancer lines (SW480, HT29, LoVo, HCT116, and RKO) and one stomach cancer line (AGS). SW480 and LoVo are PIK3CA WT, whereas HT29, AGS, HCT116, and RKO all carry PI3K-activating PIK3CA mutation(s). The cell lines selected have several other common CRC mutations but none are unique to the PIK3CA mutant or WT cells (Supplementary Table S1)(37). LSD1 KD had no effect on the growth of PIK3CA WT cell lines (Figure 4A). Interestingly, LSD1 KD caused a significant reduction in growth in all PIK3CA mutant cell lines. Similar results were obtained with LSD1 CRISPR knockout, where LSD1 KO significantly reduced growth in PIK3CA mutant HT29 cells, but had no effect on the growth of PIK3CA WT SW480 cells (Supplementary Figure S4A–D). These data suggest that PIK3CA mutant cancers may be sensitive to targeting of LSD1, and raise the unique possibility that LSD1 may be important for the tumorigenic activity of PIK3CA mutations.
Figure 4: Gastrointestinal cell lines with mutant PIK3CA are sensitive to LSD1 KD.
(A) SW480, LoVo, HT29, AGS, HCT116 and RKO cell growth over a five-day time course determined by the CellTiter-Glo Luminescent Cell Viability Assay. N=4. Graph depicts mean + SD. Statistical analyses are performed using 2way ANOVA and Sidaks multiple comparisons test with all statistically significant comparisons shown. adj. p-value; **<.01, ****<.0001. (B) Correlation plot of RNA-seq data from LSD1 versus EV KD in SW480 and HT29 cells. Significant data points are defined as abs(Log2FC)>=1 & FDR<=0.05 for LSD1 compared to EV KD for each cell line, including those unique to HT29 (purple), unique to SW480 (red), and shared between HT29 and SW480 (black). N=3. (C) Ridge plot depicts LSD1 versus EV KD expression changes of genes contributing to max enrichment score of hallmark and curated gene sets accessed from the molecular Signatures Database (v6.2). GSEA Ratio shows the ratio of genes contributing to max enrichment score, to the total number of genes in the gene set. Heatmap on right shows Log2FC value for each gene enriched in HALLMARK_PI3K_AKT_MTOR_SIGNALING in either HT29 or SW480 cells sorted by HT29 Log2FC. (D) Network analysis of uniquely upregulated genes in HT29 after LSD1 KD with significantly enriched processes from the Reactome database. Similar terms were manually grouped, and size and color of circles were set to indicate number of genes and the p-value, respectively. (E) Clonogenic growth assay. Significance determined by 2way ANOVA with Sidak’s multiple comparisons test. N = 3. Results are represented as mean ± SD. adj. p-value; **<.01, ns – not significant.
Since perturbing LSD1 abrogates activation of AKT in both HT29 (PIK3CA mutant) and SW480 (PIK3CA WT) cells but only reduces the viability of HT29 cells, we hypothesized that AKT may be uniquely targeting pathways associated with cell survival in HT29 but not SW480 cells. To test this hypothesis, we performed RNA-seq in the EV and LSD1 KD HT29 cells and generated datasets that consisted of genes with significantly altered expression in LSD1 KD versus EV for each cell line. When comparing these two datasets, there was very little overlap between genes upregulated by LSD1 KD, exemplified by a weak positive correlation (R2=0.121), suggesting that the majority of significant changes were unique between the two cell lines (Figure 4B). The largest set of genes with significantly altered expression after LSD KD compared to EV by >3-fold was genes uniquely upregulated in HT29 cells. The only commonly downregulated gene between the datasets was LSD1, further validating the specificity of our KD. To identify conserved and divergent characteristics between the two datasets, we performed gene set enrichment analysis (Figure 4C). As validation, genes upregulated after LSD1 KD in an independent dataset were enriched and upregulated by LSD1 KD in both HT29 and SW480 cells as determined by RNA-seq. In agreement with our finding that LSD1 KD reduces recruitment of HDACs to chromatin, genes downregulated after HDAC1 and HDAC2 overexpression were also significantly enriched and upregulated in our data. In support of our finding that LSD1 is critical for AKT activation, genes upregulated during active PI3K/AKT signaling were significantly enriched in our HT29 dataset, and trended towards downregulated after LSD1 KD in both SW480 and HT29 RNA-seq datasets, indicating reduced active AKT signaling. 65 PI3K/AKT signaling genes were downregulated between the two cell lines. While 51% were commonly downregulated, 69% of the remaining genes were uniquely downregulated in HT29 cells indicating LSD1 KD may have a more significant effect on gene expression in this pathway in HT29 cells. Further, by gene ontology analysis of genes uniquely upregulated after LSD1 KD in SW480 cells no enriched genesets were associated with proliferation indicating that there is likely not a compensatory pathway activated in SW480 cells to buffer loss of AKT activation (Supplementary Figure S4E). Additionally, genes downregulated by TGFβ1 were enriched and upregulated in HT29 cells after LSD1 KD (Figure 4C). This is interesting since cross-talk between the PI3K/AKT and TGF-β signaling networks plays a critical role in tumor progression(38).
We next narrowed our focus to genes that were uniquely upregulated in the HT29 cells after LSD1 KD (N=523) and performed hypergeometric enrichment analysis using the Reactome database. In agreement with another study in CRC cells, ontologies associated with immune response were enriched(39). We additionally identified pathways associated with extracellular matrix organization and cellular junction organization, one example being the formation of hemidesmosomes, which play a role in epithelial cell adherence to extracellular matrices and are lost during EMT(40) (Figure 4D). Among genes uniquely downregulated after LSD1 KD in HT29 cells, were mediators of EMT-associated elastic fibre formation, TGFB3 and LOXL1 (data not shown). Together these data suggest that LSD1 regulates EMT characteristics in HT29 cells. Clonogenic assays test the ability of a tumor cell to survive and grow in isolation, an important characteristic of EMT and stem-like cells, which may be a critical attribute for metastasis(41, 42). HT29 cells exhibited > 7-fold reduction in clonogenic survival after LSD1 KD, while there was no significant loss of clonogenic survival in PIK3CA WT SW480 cells (Figure 4E). Together this data demonstrates that LSD1 KD causes a significant reduction in growth and survival as well as alterations in gene expression programs related to EMT in PIK3CA mutant but not PIK3CA WT CRC cell lines.
LSD1 regulates protein stability of Snail through regulation of AKT
AKT has been extensively implicated in the EMT process through protein stabilization of the EMT-promoting transcription factor Snail family Transcriptional Repressor 1 (SNAI1 or Snail). Snail can be phosphorylated by GSK3β leading to its ubiquitination and subsequent proteasomal degradation(43). Active AKT can phosphorylate GSK3β at serine 9(44), inhibiting its kinase activity toward Snail, and increasing the protein half-life of Snail. However, the cellular contexts that support the AKT-GSK3β-Snail axis are not well understood. While the downstream functional outcomes of PIK3CA mutations in different domains are poorly characterized, one study suggests they may alter different downstream pathways, and require unique co-factors to exert their oncogenic effects(45). Further, PIK3CA mutations in the p85, C2, helical and kinase domains all increase PIK3CA lipid kinase activity, but only mutations in C2, helical and kinase domains are sufficient to robustly increase pS473-AKT and transform cells(46). The exact functions of C2 domain mutations in driving tumorigenesis have remained largely uncharacterized, as researchers have focused primarily on more commonly occurring kinase and helical domain mutations. We hypothesize that the activity of the AKT-GSK3β-Snail axis in CRC cells is dependent on the PIK3CA mutational status.
To test for the effect of PIK3CA mutational status on AKT-mediated stabilization of Snail protein, we inhibited AKT across our cell line panel using a potent and selective competitive pan-AKT inhibitor, GSK690693(29). Treating cells with this inhibitor generates a feedback that causes hyper-phosphorylation of S473-AKT, reduction of AKT protein levels, and loss of pS9-GSK3β. Inhibiting AKT did not change or caused an increase in Snail protein levels in PIK3CA WT (SW480/LoVo) or PIK3CA kinase mutant (HCT116/RKO) cells, respectively (Supplementary Figure S5A–D). Inhibiting AKT in PIK3CA C2 (HT29) or C2 and helical (AGS) mutant cells led to a marked decrease in the protein levels of Snail (Figure 5A, B) independent of significant changes in the mRNA levels of Snail (Figure 5C). Using the TCGA pancancer datasets, we stratified seven different cancer types based on the domain frequency of PIK3CA mutations (Figure 5D). While mutations in the C2 domain represented on average 10.6% of PIK3CA mutations across the different cancer types, mutations in GI cancers of the stomach and colon/rectum were disproportionately higher at 22.2% and 14%, respectively. Together these results suggest that activation of the AKT-GSK3β-Snail axis may depend on PIK3CA mutational status and correlates with mutations in the C2 domain which are more prevalent in some GI cancers.
Figure 5: LSD1 regulates Snail stability via AKT in PIK3CA C2 domain mutant cancer cells.
(A) Western blot analysis of HT29 or (B) AGS cells treated with DMSO or 10 μM GSK690693 for 48H. (C) Real-time PCR analysis of SNAI1 RNA expression levels after 48H DMSO or 10 μM GSK690693 treatment in HT29 and AGS cells. Expression was normalized to Gapdh and DMSO. Results are represented as mean ± SD. Significance determined by 2way ANOVA with Sidak’s multiple comparisons test. ns – not significant. (D) Proportion of total PIK3CA mutations occurring in the different domains indicated across various cancer types in the TCGA pancancer datasets. (E) shEV and shLSD1 HT29 cells treated with DMSO or 10 μM MG-132 for 4H and analyzed by western blot. (F) Real-time PCR analysis of LSD1 and SNAI1 RNA expression levels in shEV and shLSD1 HT29 cells as in (C). adj. p-value; ****<.0001. (G) HT29 shLSD1 cells were transfected with HA-LSD1 alone or in combination with 10 μM GSK690693.
We next sought to determine if LSD1 enhances EMT-like changes in HT29 cells through positive regulation of the AKT-GSK3β-Snail axis. LSD1 KD resulted in a strong reduction in Snail protein levels (Figure 5E) that was independent of significant changes in SNAI1 mRNA (Figure 5F). As expected, changes in Snail protein level were accompanied by decreases in pS473-AKT and pS9-GSK3β. Treatment of cells with proteasome inhibitor MG-132 was sufficient to rescue Snail protein levels after LSD1 KD (Figure 5E). While re-expression of LSD1 in LSD1 KD cells was sufficient to rescue protein levels of Snail, re-expressing LSD1 in the context of AKT inhibition was not sufficient to rescue Snail protein levels, further supporting our hypothesis that LSD1 is acting upstream of AKT in the stabilization of Snail (Figure 5G). LSD1 did not significantly effect AKT activation in AGS cells nor did LSD1 KD significantly reduce Snail protein level, supporting the notion that LSD1-mediated changes in Snail protein rely on LSD1 regulation of AKT (Supplementary Figure S5E). While LSD1 KD reduced pS473-AKT in SW480 cells, LSD1 KD had no significant effect on Snail protein level (Supplementary Figure S5F). This result is consistent with our finding that inhibiting AKT has no effect on Snail protein in this cell line (Supplementary Figure S5A). In additional cell lines with WT (LoVo) or kinase domain (RKO, HCT116) mutant PIK3CA where AKT inhibition did not alter Snail levels (Supplementary Figure S5B–D), LSD1 KD did not alter pS473-AKT levels, further suggesting this axis is mutation-dependent (Supplementary Figure S5G–I). Together these data indicate PIK3CA C2 domain mutations may function in the activation of the AKT-GSK3β-Snail axis. Furthermore, our molecular studies establish LSD1 as a context-dependent upstream regulator of this pathway.
The CRC Subtyping Consortium has previously developed a classifier that can be used to subtype cancers into 4 groups CMS1 (MSI/immune), CMS2 (canonical), CMS3 (metabolic) and CMS4 (mesenchymal) (47). While there was no significant difference in LSD1 expression between CMS1-CMS4 subtyped COADREAD datasets, LSD1 was uniquely upregulated (P < 0.05) in PIK3CA mutant CMS4 mesenchymal subtype tumors compared to WT (Supplementary Figure S6A, B). This finding suggests our observation that LSD1 is overexpressed in PIK3CA mutant vs. WT CRC tumors (Figure 1A) is at least in part explained by significant differences in CMS4 subtyped tumors. Further, we found that PIK3CA C2 domain mutations were most common in CMS4 subtyped tumors (Supplementary Figure S6C). This observation in addition to our AKT inhibitor studies established a connection between PIK3CA C2 domain mutations and mesenchymal phenotypes.
LSD1 is required for EGF-induced migration in HT29 cells
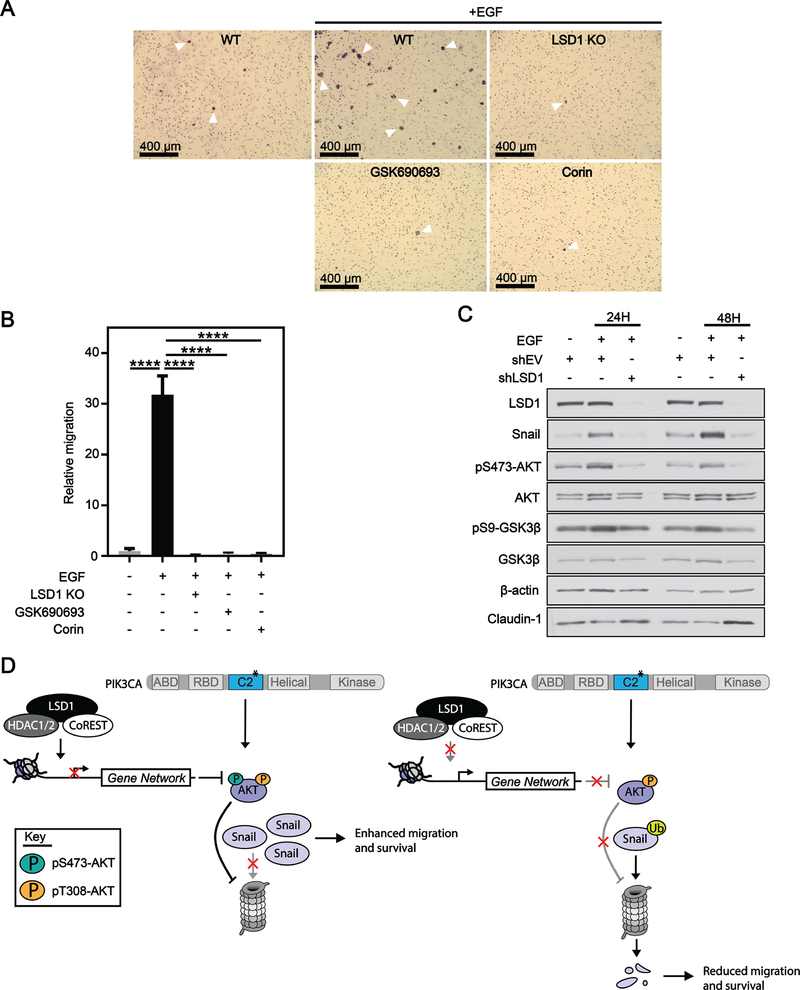
EGF has previously been used to enhance migratory phenotypes through the AKT-GSK3β-Snail axis(48). HT29 cells exhibited little to no basal migration, but upon stimulation with EGF, exhibited a significant >30 fold increase in migration (Figure 6A, B). Importantly, loss of LSD1, inhibition of AKT or inhibition of the CoREST complex completely blocked EGF-induced migration (Figure 6A, B). Treatment with EGF altered HT29 cellular morphology, with cells becoming elongated and scattered; however, LSD1 KO abrogated EGF-mediated changes in cellular morphology (Supplementary Figure S7). Treatment of cells with EGF led to an increase in levels of pS473-AKT, pS9-GSK3β and Snail at 24H and 48H (Figure 6C). LSD1 KD completely blocked EGF-induced activation of pS473-AKT, pS9-GSK3β and increase in Snail protein. LSD1 KD also increased levels of the epithelial marker Claudin-1. Together these data suggest that LSD1 is required for EGF-induced migration mediated by the AKT-GSK3β-Snail pathway.
Figure 6: LSD1 is required for EGF-induced migration of cells with an active AKT-GSK3β-Snail axis.
(A) 10x bright field images of crystal violet stained HT29 cells after 48H migration through transwell insert. WT, LSD1 KO, 10 μM GSK690693 or 3 μM corin cells were co-treated with 100 ng/mL EGF for 48H. (B) Quantification of migration normalized to migration counts for untreated cells. Results are represented as mean ± SD. Significance was determined by one-way ANOVA with Tukey’s multiple comparisons test. All significant comparisons are shown. adj. p-value; ****<.0001. (C) shEV or shLSD1 cells were treated with 100 ng/mL EGF for 24H or 48H and analyzed by western blot. (D) Model depicting CoREST complex in the regulation of AKT.
Discussion
In this study, we implicate LSD1 in the regulation of AKT activity. We find that PIK3CA mutant CRC cell lines are sensitive to LSD1 KD whereas WT cells are not. Using TCGA data, we provide evidence that this connection between LSD1 and PIK3CA mutation status may be unique to gastrointestinal tumor types. PI3K inhibitors, which have been successful in gynecological and breast cancers with mutant PIK3CA, have failed in CRC clinical trials, suggesting that additional factors influence the PI3K pathway in CRC. We demonstrate for the first time that LSD1 regulates basal AKT activation as well as activation in response to both exogenous oxidative stress and growth factors and, in a subset of PIK3CA mutant cells, LSD1-mediated AKT activity promotes EMT-like characteristics, including migration.
Our findings suggest that LSD1 may play a role in the full activation of AKT. We highlight, for the first time, an oncogenic function for LSD1 in the regulation of PI3K/AKT signaling via catalytically independent regulation of gene expression. We hypothesize that the CoREST complex represses gene(s) that normally function to regulate S473-AKT phosphorylation such as regulators of cell receptors, negative regulators of upstream kinases and/or phosphatases. When the complex is disrupted, these gene(s) are expressed and perturb AKT phosphorylation. Our RNA-seq data in HT29 cells confirms that LSD1 KD decreases PI3K/AKT pathway activity. However, it is unclear from these datasets how S473-AKT phosphorylation is regulated. Future studies will determine the mechanisms by which CoREST complexes influence pS473-AKT.
The canonical function of LSD1 is to catalyze the demethylation of histones to repress gene expression(13). Previously, a study has shown using endometrial cancer cells that during estrogen signaling, PI3K/AKT upregulated LSD1 protein levels and LSD1 sustained this process through the demethylation of histones to activate gene expression, thereby creating a positive feedback loop(49). Our study presents a model that is in contrast to a classical enzymatic role for LSD1 as we establish a catalytically independent oncogenic function for LSD1 both in the regulation of PI3K/AKT signaling and in the regulation of gene expression without the requirement of nuclear hormone signaling. Furthermore, according to our gene expression data obtained from RNA-seq of LSD1 KD cells, we detected significant increase in CDH1 gene expression after LSD1 KD. The change in CDH1 expression after LSD1 KD is consistent with previous studies, including studies performed in HCT116 cells, that demonstrate LSD1 is recruited by Snail to the promoter of E-cadherin (CDH1) to demethylate H3K4me2 to repress CDH1 gene expression and drive EMT and migration(10–12). However, there was no change in H3K4me2 at the CDH1 promoter after LSD1 KD or enrichment of LSD1 in our SW480 cells. This result suggests that regulation of CDH1 gene expression occurs through an alternative mechanism that is independent of LSD1 enzymatic activity in some CRC cell lines. Our findings are unique from studies where LSD1 interacts with Snail to promote migration through its demethylase activity in that we demonstrate LSD1 promotes survival and migration through stabilization of Snail by a non-catalytic activity. This is a critical distinction because as inhibitors of LSD1 advance in clinical trials, it is crucial to establish the importance of the enzymatic versus scaffolding activity of LSD1, as its tumorigenicity may be dynamic between CRC tumors or even within tumor cell populations. This information will enable us to determine how LSD1 may be acting in a particular tumor and inform patient enrollment in future clinical studies.
Our study demonstrates that the activation of AKT in CRC cells depends on the scaffolding function of LSD1 in the CoREST complex. In agreement with our findings, a recent study demonstrated that a demethylase dead mutant LSD1 was able to rescue viability defects in lymphoma cells where LSD1 had been knocked down (17). This study further demonstrated that an LSD1 mutant unable to interact with CoREST was not sufficient to rescue viability defects induced by LSD1 KD. In our study, we also demonstrate that the dual LSD1/HDAC1 inhibitor corin was sufficient to ablate pS473-AKT, mimicking our KD and CRISPR studies. It is tempting to speculate that corin, rather than acting through inhibiting enzymatic activity, co-binds to LSD1 and HDAC1, disrupts the complex’s histone-interacting surface, and reduces the complex’s interaction with chromatin, as has been shown with other LSD1 inhibitors(19–22). The finding that LSD1 and RCOR1 correlate in STAD, but not COAD or READ may reflect differences in how these genes are transcriptionally regulated between the large intestine and stomach. LSD1 and RCOR1 are stabilized through their protein-protein interactions with one another, (50) consistent with our observation that loss of LSD1 decreases RCOR1 protein and vice versa (Figure 3A, B). This association also suggests that only LSD1 or RCOR1 needs to be overexpressed to increase protein levels of the other. Our finding that HDAC1 KD did not phenocopy loss of LSD1 or RCOR1 in the regulation of pS473-AKT supports this conjecture, but the possibility remains that corin may function through enhanced repression of HDAC1 in CoREST complexes.
PI3K/AKT signaling is essential for gut development due to its role in the activation of EMT and extracellular matrix (ECM)-modifications that lead to the specification of foregut precursors(51). Interestingly, we demonstrate that LSD1 plays a significant role in the regulation of ECM organization and EMT-associated gene expression programs. It is possible that LSD1 and PI3K signaling may cooperate during specification of foregut precursors. This hypothesis may also explain why there is a positive association between CoREST expression in PIK3CA mutant GI cancers but not in other cancer types, suggesting that this interaction may be lineage dependent. It is also consistent with our observation of EMT-associated gene expression programs in a CRC cell line harboring PIK3CA mutation within the C2 domain.
In our TCGA analyses of PIK3CA mutation frequencies, mutations in the helical and kinase domains were most frequent across the different cancer types while less common mutations such as those in the C2 domain, RBD and ABD appeared more cancer specific. Mutations in the C2 domain were disproportionately more common in GI cancers of the stomach and colon/rectum. The exact nature of the relationship between PIK3CA mutation and LSD1 expression is currently unclear, and further studies are required. It is possible that LSD1 expression is more closely linked to specific PIK3CA mutations, and that the prevalence of the specific mutation in a given cancer type is driving the significance of the association between LSD1 expression and PIK3CA mutational status in the TCGA data. Our study also suggests that activation of the AKT-GSK3β-Snail axis to promote EMT may be dependent on PIK3CA C2 domain mutation. This mechanism for upregulation of Snail is different from a previously described mechanism by which PIK3CA with the H1047R kinase mutation upregulates SNAI1 mRNA expression(52). Further, our finding of AKT-signaling dependent Snail stabilization by LSD1 is a distinct mechanism from stabilization of Snail through interaction with the CoREST complex(10). An important caveat to our PIK3CA mutational studies is that AGS cells contain mutations in both the C2 and helical domains of PIK3CA. Our study does not dissect whether helical domain mutations alone are associated with the AKT-GSK3β-Snail axis. Future studies are required to untangle the functional role of different PIK3CA mutations and to establish a mechanistic understanding of their differential roles in mediating the AKT-GSK3β-Snail axis. While we focus on the interaction between LSD1 and PIK3CA C2 domain mutation, we also demonstrated that LSD1 KD reduced proliferation of cell lines with PIK3CA kinase domain mutations. It will be important to study LSD1 under the context of different PIK3CA mutations in order to potentially identify a synthetic lethal relationship and establish prognostic value in GI cancers.
Herein, we used EGF as tool to induce migration of HT29 cells. The tumor microenvironment plays a critical role in promoting EMT via autocrine and paracrine signaling by growth factors such as EGF, which stimulates EMT and migratory phenotypes in tumor cells (53). Further in vivo studies are required to understand how LSD1 may regulate signaling events between tumors and the surrounding microenvironment.
Overall we propose a model where the CoREST complex can act synergistically with C2 domain PIK3CA mutations and growth factors to fully active the PI3K/AKT pathway and stabilize Snail protein to enhance cell migration and survival (Figure 6D). Genetic or therapeutic perturbation of the CoREST complex is sufficient to block cancer cell migration and reduce survival. This work suggests that PIK3CA mutant CRC may be particularly sensitive to LSD1 inhibitors that block the interaction of CoREST with transcription factors.
Supplementary Material
Acknowledgements
We thank the Indiana University Center for Genomics and Bioinformatics and the Indiana University Light Microscopy Imaging Center for their assistance. This work was supported by the National Institute of Environmental Health Sciences Grant (R01ES023183 to HM O’HAGAN); and the National Institutes of Health, National Center for Advancing Translational Sciences, Clinical and Translational Sciences Award (TL1 TR001107 and UL1 TR001108 (A Shekhar, PI) to SA MILLER) as well as the National Institutes of Health Grant (GM62437 to PA COLE).
Footnotes
Conflict of Interest Statement: The authors declare no potential conflicts of interest.
References
- 1.Haigis KM, Cichowski K, Elledge SJ, Tissue-specificity in cancer: The rule, not the exception. Science 363, 1150–1151 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B et al. , Cancer genome landscapes. Science 339, 1546–1558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas N, Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samuels Y et al. , High frequency of mutations of the PIK3CA gene in human cancers. Science 304, 554 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Wang Q et al. , PIK3CA mutations confer resistance to first-line chemotherapy in colorectal cancer. Cell Death Dis 9, 739 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liao X et al. , Prognostic role of PIK3CA mutation in colorectal cancer: cohort study and literature review. Clin Cancer Res 18, 2257–2268 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang G et al. , SETDB1-mediated methylation of Akt promotes its K63-linked ubiquitination and activation leading to tumorigenesis. Nat Cell Biol 21, 214–225 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo J et al. , AKT methylation by SETDB1 promotes AKT kinase activity and oncogenic functions. Nat Cell Biol 21, 226–237 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsu HC et al. , CBB1003, a lysine-specific demethylase 1 inhibitor, suppresses colorectal cancer cells growth through down-regulation of leucine-rich repeat-containing G-protein-coupled receptor 5 expression. J Cancer Res Clin Oncol 141, 11–21 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Lin Y et al. , The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J 29, 1803–1816 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding J et al. , LSD1-mediated epigenetic modification contributes to proliferation and metastasis of colon cancer. Br J Cancer 109, 994–1003 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrari-Amorotti G et al. , Inhibiting interactions of lysine demethylase LSD1 with snail/slug blocks cancer cell invasion. Cancer Res 73, 235–245 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi Y et al. , Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Iwase S et al. , Characterization of BHC80 in BRAF-HDAC complex, involved in neuron-specific gene repression. Biochem Biophys Res Commun 322, 601–608 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Ballas N et al. , Regulation of neuronal traits by a novel transcriptional complex. Neuron 31, 353–365 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Sehrawat A et al. , LSD1 activates a lethal prostate cancer gene network independently of its demethylase function. Proc Natl Acad Sci U S A 115, E4179–E4188 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hatzi K et al. , Histone demethylase LSD1 is required for germinal center formation and BCL6-driven lymphomagenesis. Nat Immunol 20, 86–96 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andres ME et al. , CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc Natl Acad Sci U S A 96, 9873–9878 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takagi S et al. , LSD1 Inhibitor T-3775440 Inhibits SCLC Cell Proliferation by Disrupting LSD1 Interactions with SNAG Domain Proteins INSM1 and GFI1B. Cancer Res 77, 4652–4662 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Maiques-Diaz A et al. , Enhancer Activation by Pharmacologic Displacement of LSD1 from GFI1 Induces Differentiation in Acute Myeloid Leukemia. Cell Rep 22, 3641–3659 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vinyard ME et al. , CRISPR-suppressor scanning reveals a nonenzymatic role of LSD1 in AML. Nat Chem Biol 15, 529–539 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamamoto R et al. , Selective dissociation between LSD1 and GFI1B by a LSD1 inhibitor NCD38 induces the activation of ERG super-enhancer in erythroleukemia cells. Oncotarget 9, 21007–21021 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding N, Miller SA, Savant SS, O’Hagan HM, JAK2 regulates mismatch repair protein-mediated epigenetic alterations in response to oxidative damage. Environ Mol Mutagen 60, 308–319 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plotnik JP, Hollenhorst PC, Genome-Wide Analysis of RAS/ERK Signaling Targets. Methods Mol Biol 1487, 277–288 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Chu N et al. , Akt Kinase Activation Mechanisms Revealed Using Protein Semisynthesis. Cell 174, 897–907 e814 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alessi DR et al. , Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr Biol 7, 261–269 (1997). [DOI] [PubMed] [Google Scholar]
- 27.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM, Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Ushio-Fukai M et al. , Reactive oxygen species mediate the activation of Akt/protein kinase B by angiotensin II in vascular smooth muscle cells. J Biol Chem 274, 22699–22704 (1999). [DOI] [PubMed] [Google Scholar]
- 29.Rhodes N et al. , Characterization of an Akt kinase inhibitor with potent pharmacodynamic and antitumor activity. Cancer Res 68, 2366–2374 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Nicholson TB, Chen T, LSD1 demethylates histone and non-histone proteins. Epigenetics 4, 129–132 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Mohammad HP et al. , A DNA Hypomethylation Signature Predicts Antitumor Activity of LSD1 Inhibitors in SCLC. Cancer Cell 28, 57–69 (2015). [DOI] [PubMed] [Google Scholar]
- 32.You A, Tong JK, Grozinger CM, Schreiber SL, CoREST is an integral component of the CoREST- human histone deacetylase complex. Proc Natl Acad Sci U S A 98, 1454–1458 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Upadhyay G, Chowdhury AH, Vaidyanathan B, Kim D, Saleque S, Antagonistic actions of Rcor proteins regulate LSD1 activity and cellular differentiation. Proc Natl Acad Sci U S A 111, 8071–8076 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee MG, Wynder C, Cooch N, Shiekhattar R, An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 437, 432–435 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Kalin JH et al. , Targeting the CoREST complex with dual histone deacetylase and demethylase inhibitors. Nat Commun 9, 53 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen CS, Weng SC, Tseng PH, Lin HP, Chen CS, Histone acetylation-independent effect of histone deacetylase inhibitors on Akt through the reshuffling of protein phosphatase 1 complexes. J Biol Chem 280, 38879–38887 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Barretina J et al. , The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang L, Zhou F, ten Dijke P, Signaling interplay between transforming growth factor-beta receptor and PI3K/AKT pathways in cancer. Trends Biochem Sci 38, 612–620 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Jin L et al. , Loss of LSD1 (lysine-specific demethylase 1) suppresses growth and alters gene expression of human colon cancer cells in a p53- and DNMT1(DNA methyltransferase 1)-independent manner. Biochem J 449, 459–468 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borradori L, Sonnenberg A, Structure and function of hemidesmosomes: more than simple adhesion complexes. J Invest Dermatol 112, 411–418 (1999). [DOI] [PubMed] [Google Scholar]
- 41.Mani SA et al. , The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133, 704–715 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nieto MA, Huang RY, Jackson RA, Thiery JP, Emt: 2016. Cell 166, 21–45 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Zhou BP et al. , Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat Cell Biol 6, 931–940 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA, Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature 378, 785–789 (1995). [DOI] [PubMed] [Google Scholar]
- 45.Zhao L, Vogt PK, Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc Natl Acad Sci U S A 105, 2652–2657 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ikenoue T et al. , Functional analysis of PIK3CA gene mutations in human colorectal cancer. Cancer Res 65, 4562–4567 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Guinney J et al. , The consensus molecular subtypes of colorectal cancer. Nat Med 21, 1350–1356 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gan Y et al. , Differential roles of ERK and Akt pathways in regulation of EGFR-mediated signaling and motility in prostate cancer cells. Oncogene 29, 4947–4958 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Chen C et al. , LSD1 sustains estrogen-driven endometrial carcinoma cell proliferation through the PI3K/AKT pathway via di-demethylating H3K9 of cyclin D1. Int J Oncol 50, 942–952 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Shi YJ et al. , Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell 19, 857–864 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Villegas SN et al. , PI3K/Akt1 signalling specifies foregut precursors by generating regionalized extra-cellular matrix. Elife 2, e00806 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wallin JJ et al. , Active PI3K pathway causes an invasive phenotype which can be reversed or promoted by blocking the pathway at divergent nodes. PLoS One 7, e36402 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maheshwari G, Wiley HS, Lauffenburger DA, Autocrine epidermal growth factor signaling stimulates directionally persistent mammary epithelial cell migration. J Cell Biol 155, 1123–1128 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-sequencing and ChIP-sequencing data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE139927.