Summary
A mismatch at HLA-DPB1 locus is associated with higher acute GVHD and lower relapse rate after myeloablative (MAC) allogeneic hematopoietic cell transplantation (alloHCT). Also, in MAC setting, mismatch permissiveness and expression level impact alloHCT outcomes. However, in reduced intensity (RIC), DP mismatch effect on transplant outcomes is unknown. We retrospectively evaluated DP mismatch influence (number, permissiveness, and expression) on HCT outcomes in 310 patients with high-resolution typing (HLA-A, -B, -C, -DRB1, -DQB1 and -DPB1), who underwent RIC-HCT. By multivariable analysis, 11/12 had better overall survival (OS) and relapse vs. 12/12 (HR=1.61 and 2.02; p=0.04 and 0.01, respectively) and better OS vs. 10/12 (HR=1.68; p=0.02). Within the 11/12, non-permissive mismatch (NoPR) was associated with higher risk of grade II-IV acute GVHD (HR=1.97; p=0.005) and non-relapse mortality (HR=2.13; p=0.02) vs. permissive (PR). Grouping 11/12 based on the DP expression conferred higher mortality (HR=3.78; p= 0.003) when low expressers received a graft from high expressers (AG) vs. low expressers (AA). Better OS was achieved in PR 11/12, when expression was low in patient and donor (AA) vs. all other combinations. Therefore, in RIC HCT, a single DP mismatch has a protective role, especially in permissive setting, when donor and recipient are low expressers.
Introduction
It is well-established that outcomes of allogenic hematopoietic cell transplantation (alloHCT) from a matched unrelated donor (MUD) are directly influenced by the degree of human leukocyte antigen (HLA) compatibility between the unrelated donor and the recipient, with donor-recipient HLA disparity increasing the risk of post-transplant complications such as graft rejection, graft-versus-host disease (GvHD), and mortality.1, 2 Currently, using high-resolution 10/10 matched donor pair (donor-recipient match at HLA-A, -B, -C, -DRB1, and DQB1 alleles) is accepted as the standard practice at most centers. HLA-DPB1 is the sixth classic HLA molecule, located in the class II region of chromosome 6p21.3.3 While this locus is not routinely typed for or included in donor selection, multiple clinical studies have indicated that an HLA mismatch at this locus is associated with higher incidence of GvHD and lower relapse rate in patients undergoing myeloablative 10/10 MUD-HCT.4-6
A single-nucleotide polymorphism in HLA-DPB1 regulatory region variant rs9277534 is associated with HLA-DPB1 cell-surface expression,7, 8 and Petersdorf et al, recently reported that the risk of GvHD is higher when a patient with low HLA-DPB1 expression receives a graft from a high-expression allele donor.8, 9 Furthermore, HLA-DPB1 mismatches can induce alloreactive T-cell response and classification of HLA-DPB1 mismatches based on the T-cell epitope (TCE) grouping has identified permissive (PR) and nonpermissive (NoPR) mismatches relevant to acute GvHD after alloHCT.4, 10 Petersdorf et al, also described a linkage between the HLA-DPB1 expression level and TCE permissiveness,8, 11 by which when both donor and recipient were low expressers, mismatches were mostly PR, whereas mismatches in the HLA-DPB1 high-expresser pairs were mainly NoPR.
HLA-DPB1 mismatch number, DP allele expression level and TCE permissiveness are interdependent factors, and a combination of these factors dictates transplant outcomes. It is important to note that, majority of patients included in the above studies undergone myeloablative conditioning regimens. However, to our knowledge, there are currently no reports on the impact of HLA-DPB1 on transplant outcomes after alloHCT with reduced intensity conditioning (RIC). With the increased use of RIC regimens within the last decade, specifically for patients not suitable for myeloablative conditioning including older population, the impact of HLA-DPB1 mismatch on transplant outcomes with this less intensive regimen becomes increasingly important. Therefore, the main objective of this single center retrospective study was to analyze the effect of different combinations of HLA-DPB1 mismatches on the outcome of patients who underwent alloHCT with RIC regimen.
Methods
Study population
From January of 2006 to December of 2012, 310 patients with hematological disease underwent RIC- MUD alloHCT (high-resolution 10/10 HLA typing: HLA-A, -B, -C, -DRB1, and -DQB1) at City of Hope. All patients were eligible for inclusion in the study regardless of age, conditioning regimen, GvHD prophylaxis, or stem cell source. All research samples and data were collected according to institutional review board-approved guidelines and protocols.
HLA typing
High-resolution HLA typing was performed using a combination of the following methods (resolving common ambiguous genotypes): sequencing-based typing, PCR-SSOP (sequence-specific oligonucleotide probes) and/or PCR-SSP (sequence-specific primers). Different alleles from the same G groups were considered matched. If patient and/or donor were homozygous for HLA-DPB1, we assumed the presence of two copies from the same allele. PR and NoPR mismatches were assigned according to Crivello et al 201512 using tools available at IPD-IMGT/HLA database.13
HLA-DPB1 alleles were divided into high and low expressers based on the linkage disequilibrium pattern between HLA-DPB1 alleles and single nucleotide polymorphism (SNP) rs9277534 as previously described by Petersdorf et al,8 and updated by Schone et al.14 (Brief description of this technique is provided in the supplementary material)
Endpoint definitions
Overall survival (OS) was defined as the time interval from stem cell infusion to date of death from any cause or date of last contact; Event free survival (EFS) was defined as the time interval from stem cell infusion to date of first documented disease relapse, progression, death from any cause, or date of last contact. For cumulative incidence of relapse (CIR), non-relapse mortality (NRM), acute and chronic GvHD competing risks models were used.15 Time to event was measured from time interval from stem cell infusion to event of interest, competing risk, or date of last follow up. CIR event was defined as the first documented relapse or progression, while death from anything other than disease progression was treated as a competing event. NRM was death from any cause other than relapse/progression and relapse/progression was treated as a competing event. Grades II-IV and III-IV acute GvHD were defined by the Glucksberg scale,16 and chronic GvHD was defined as limited or extensive according to the Seattle criteria.17 Death and relapse/progression were treated as a competing risk for both acute and chronic.
Statistical methods
Descriptive statistics included medians and ranges for continuous variables and frequencies/percentages for categorical variables. Each endpoint was assessed by HLA match number, TCE classification, and HLA-DPB1 expression using Cox regression. An interaction term between recipient and donor HLA-DPB1 expression was used to determine whether the effect of donor expression was similar, regardless of recipient expression. In order to determine whether a cumulative effect of TCE classification and HLA-DPB1 expression was present we ran Cox regression analysis with an interaction term between TCE and HLA-DPB1 expression. Finally, we ran a subset analysis with only 11/12-PR patients, in order to asses which patient/donor expression combinations within this subset were better than fully matched transplants. All multivariable models were adjusted for age, disease risk, and conditioning, while models testing TCE classification were also adjusted for stem cell source due to potential confounding. All calculations were performed using SAS v9.4 (SAS Institute, Cary NC).
Results
Patient and Transplant characteristics
Transplantations were performed between 2006 and 2012. When grouped based on the number of HLA-DPB1 mismatches, of the 310 patients, 65 cases were 10/12, 172 were 11/12, and 73 were 12/12 matched. Patient and transplant characteristics are presented in Table 1. The median age of patients and donors were 58 (range: 2–73), and 30 (range: 18–57), respectively. The study population was 58% (n=179) male. Of all patients, 36% (n=112) had acute myeloid leukemia, 24% (n=84) had lymphoproliferative disorders (non-Hodgkin lymphoma, Hodgkin lymphoma, chronic lymphocytic leukemia, and multiple myeloma), and 24% (n=74) had myelodysplastic syndromes/myeloproliferative disorders/chronic myelomonocytic leukemia. Majority of patients (89%, n=275) received a peripheral blood stem cell as graft source, and tacrolimus/sirolimus-based GVHD prophylaxis was used in 93% (n=289). Median follow-up for surviving patients was of 33.3 months (range: 2.0–85.1). When grouped according to the TCE grouping,4, 10 44% (n=105) of the mismatches were non-permissive (NoPR). Assessment of baseline characteristics across DPB1 mismatched groups showed no statistically significant differences between patient and donor age, gender, CMV risk per serology, performance status, HCT comorbidity index, disease risk, conditioning and GVHD prophylaxis regimens, and stem cell source.
Table 1.
Patient, disease, and transplantation characteristics based on number of HLA-DPB1 mismatches
| Variable | 10/12 | 11/12 | 12/12 | Total |
|---|---|---|---|---|
| (N=65) | (N=172) | (N=73) | (N=310) | |
| Diagnosis at transplant | ||||
| AML | 35 (54%) | 47 (27%) | 30(41%) | 112 (36%) |
| Lymphoproliferative discordes | 12 (18%) | 55 (32%) | 17 (23%) | 84 (27%) |
| MDS/MPN/CMML | 12 (18%) | 39 (23%) | 23 (32%) | 74 (24%) |
| ALL | 4 (6%) | 9 (5%) | 3 (4%) | 16 (5%) |
| Non-malignant | 2 (3%) | 14 (8%) | 0 | 16 (5%) |
| Leukemia | 0 | 6 (3%) | 0 | 6 (2%) |
| Other | 0 | 2 (1%) | 0 | 2 (1%) |
| Disease risk | ||||
| Low | 26 (40%) | 57 (33%) | 25 (34%) | 108 (35%) |
| Intermediate | 8 (12%) | 21 (12%) | 9 (12%) | 38 (12%) |
| High | 26 (40%) | 65 (38%) | 32 (44%) | 123 (40%) |
| Non-Malignant | 5 (8%) | 23 (13%) | 7 (10%) | 35 (11%) |
| Other | 0 | 6 (3%) | 0 | 6 (2%) |
| Gender | ||||
| Female | 29 (45%) | 70 (41%) | 32 (44%) | 131 (42%) |
| Male | 36 (55%) | 102 (59%) | 41 (56%) | 179 (58%) |
| Female donor, male recipient | ||||
| Yes | 10 (15%) | 20 (12%) | 10 (14%) | 40 (13%) |
| No | 55 (85%) | 152 (88%) | 63 (86%) | 270 (87%) |
| CMV status(recipient/donor) | ||||
| −/− | 12 (19%) | 21 (12%) | 7 (10%) | 40 (13%) |
| −/+ | 6 (9%) | 19 (11%) | 5 (7%) | 30 (9%) |
| +/− | 26 (40%) | 88 (51%) | 37 (50%) | 151 (49%) |
| +/+ | 21 (32%) | 44 (26%) | 24 (33%) | 89 (29%) |
| Patient age (median, range) | 58 (3-71) | 57 (2-72) | 59 (5-73) | 58 (2-73) |
| <58 | 31 (48) | 89 (52) | 34 (47) | 154 (50) |
| ≥58 | 34 (52) | 83 (48) | 39 (53) | 156 (50) |
| Donor age (median, range) | 31 (20-52) | 31 (19-57) | 28 (18-55) | 30 (18-57) |
| <30 | 27 (42) | 78 (46) | 39 (53) | 144 (47) |
| ≥30 | 38 (58) | 93 (54) | 34 (47) | 165 (53) |
| Graft source | ||||
| Bone marrow | 5 (8%) | 21 (12%) | 9 (12%) | 35 (11%) |
| Peripheral blood stem cells | 60 (92%) | 151 (88%) | 64 (88%) | 275 (89%) |
| GvHD prophylaxis | ||||
| Tacro/Siro based | 60 (92%) | 158 (92%) | 71 (97%) | 289 (93%) |
| Other | 5 (8%) | 14 (8%) | 2 (3%) | 21 (7%) |
| Conditioning regimen* | ||||
| Fludarabine/Melphalan based | 57 (88%) | 155 (90%) | 64 (88%) | 276 (89%) |
| Fludarabine/Cyclophosphamide based | 2 (3%) | 11 (6%) | 3 (4%) | 16 (5%) |
| Clofarabine/Melphalan based | 5 (8%) | 6 (3%) | 6 (8%) | 17 (5%) |
| Fludarabine/TBI | 1 (2%) | 0 | 0 | 1 (1%) |
| KPS | ||||
| ≤80 | 11 (17%) | 33 (19%) | 22 (30%) | 66 (22%) |
| 90+ | 37 (57%) | 106 (62%) | 38 (52%) | 181 (58%) |
| Unknown | 17 (26%) | 33 (19%) | 13 (18%) | 63 (20%) |
| Comorbidity – HCT CI | ||||
| 0 | 22 (34%) | 80 (47%) | 28 (38%) | 130 (42%) |
| 1 | 11 (17%) | 23 (13%) | 9 (12%) | 43 (14%) |
| 2 | 3 (5%) | 5 (3%) | 6 (8%) | 14 (5%) |
| ≥3 | 10 (15%) | 30 (17%) | 17 (23%) | 57 (18%) |
| Unknown | 19 (29%) | 34 (20%) | 13 (18%) | 66 (21%) |
AML: Acute Myeloid Leukemia; Lymphoproliferative Disorders includes: Non-Hodgkin Lymphoma, Hodgkin Lymphoma, Multiple Myeloma and Chronic Lymphocytic Leukemia; MDS/MPN/CMMoL: Myelodysplastic Syndrome/Myeloproliferative Neoplasms/Chronic Myelomonocytic Leukemia; ALL: Acute Lymphocytic Leukemia; Non-Malignant includes: Aplastic Anemia, Sickle Cell Disease, and Thalassemia; Other Leukemia includes: Acute Biophenotypic Leukemia, Leukemia, general, Chronic Myeloid Leukemia, Histiocytic Syndrome, and Other Hematopoietic; Other includes: Other and Kidney, Renal Cell; TBI: Total Body Irradiation; KPS: Karnofsky Performance Status; HCT CI: Hematopoietic Cell Transplant Specific Co-morbidity Index.
Main conditioning regimen consisted of Fludarabine (25 mg/m2 for 5 days) with Melphalan (100 or 140 mg/m2)-based regimen was the main condition regimen in our cohort.
Effect of number of HLA-DPB1 locus mismatches
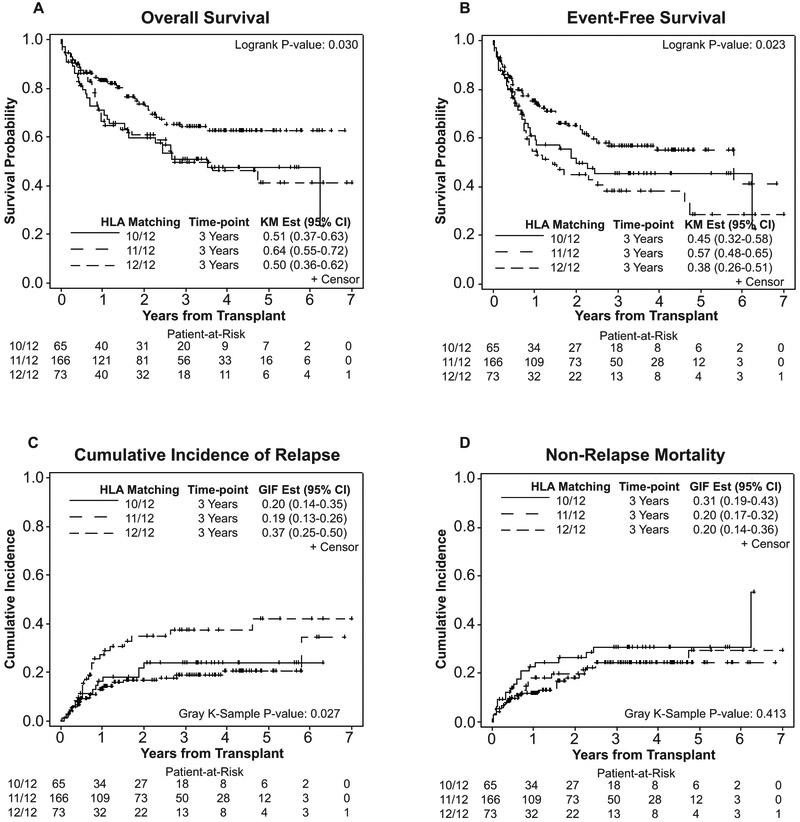
Results of our primary multivariable analysis investigating the effect of HLA-DPB1 mismatch number on Transplant outcomes (Table 2) indicated significantly better OS (HR= 1.61; 95% CI: 1.03–2.51; p=0.04), EFS (HR= 1.66; 95% CI: 1.12–2.47; p=0.01), and lower CIR (HR=2.02; 95% CI: 1.18–3.47; p=0.01) when patients received HCT from a single DP mismatched (11/12) compared to a fully matched (12/12) donor. However, statistical significance was not achieved for NRM (Figure 1). Lower OS was achieved when patients receiving HCT from double DP mismatched URD (10/12) were compared to single DP mismatched URD (HR=1.68; 95% CI: 1.07–2.64; p=0.02), but there were no differences in the EFS and CIR among these groups (Figure 1 and Table 2).
Table 2.
Effect of HLA match on transplant outcomes (N=304)*
| Clinical endpoint | HLA Match | N | Events | Hazard ratio | 95% CI | p |
|---|---|---|---|---|---|---|
| OS | 11/12 | 166 | 50 | |||
| 10/12 | 65 | 30 | 1.68 | 1.07 -2.64 | 0.02 | |
| 12/12 | 73 | 32 | 1.61 | 1.03 -2.51 | 0.04 | |
| EFS | 11/12 | 166 | 64 | |||
| 10/12 | 65 | 33 | 1.47 | 0.96-2.24 | 0.07 | |
| 12/12 | 73 | 40 | 1.66 | 1.12-2.47 | 0.01 | |
| CIR | 11/12 | 166 | 30 | |||
| 10/12 | 65 | 14 | 1.25 | 0.66-2.35 | 0.50 | |
| 12/12 | 73 | 24 | 2.02 | 1.18-3.47 | 0.01 | |
| NRM | 11/12 | 166 | 34 | |||
| 10/12 | 65 | 19 | 1.48 | 0.84-2.59 | 0.17 | |
| 12/12 | 73 | 16 | 1.09 | 0.61-1.96 | 0.76 | |
| Acute GvHD II-IV | 11/12 | 165 | 66 | |||
| 10/12 | 65 | 35 | 1.43 | 0.95-2.15 | 0.09 | |
| 12/12 | 72 | 27 | 0.92 | 0.59-1.44 | 0.72 | |
| Acute GvHD III-IV | 11/12 | 165 | 22 | |||
| 10/12 | 65 | 14 | 1.57 | 0.79-3.13 | 0.20 | |
| 12/12 | 73 | 11 | 1.14 | 0.55-2.36 | 0.73 | |
| Chronic GvHD | 11/12 | 164 | 119 | |||
| 10/12 | 65 | 48 | 1.19 | 0.84-1.69 | 0.32 | |
| 12/12 | 73 | 47 | 0.84 | 0.60-1.19 | 0.33 |
All models are multivariable and adjusted for age (<58 vs. ≥58) and disease risk (high, intermediate, low/non-malignant). Patients with incomplete clinical data or with disease risk of other were excluded.
Figure 1.
Effect of number of HLA-DPB1 locus mismatches on transplant outcomes in patients receiving transplant form 12/12, 11/12, and 10/12 matched donors. (A) Overall survival, (B) Event-free survival, (C) Cumulative incidence of relapse, and (D) Non-relapse mortality.
Effect of HLA-DPB1 mismatch according to T-cell epitope classification
Patient, disease and transplant characteristics of patients characteristics based on TCE classification is summarized in supplemental Table 1. Analysis of the impact of TCE classification on transplant outcomes after RIC alloHCT from in 11/12 HLA match (n=166). (Table 3) showed that while OS was not adversely by mismatch permissiveness, PR (n=132) or NoPR (n=105) MUD HCT, incidence of acute GvHD grades II-IV (HR= 1.97; 95% CI: 1.22–3.18; p=0.005) and NRM (HR=2.13; 95% CI: 1.10–4.12; p=0.02) were significantly higher in patients with NoPR mismatches (n=105) compared to PR mismatches (n=132). No statistical significance was detected in the incidence of chronic GvHD, acute GvHD grades III-IV or CIR when PR and NoPR mismatches were compared. When 11/12 and 10/12 were included in the analysis (Supplemental Table 2) OS (HR=1.62; P= 0.04) and acute GvHD grades II-IV (HR= 1.62; P=0.02) were significantly affected.
Table 3.
Effect of TCE on transplant outcomes for patients with 11/12 HLA match (N=166)*
| Clinical endpoint | TCE | N | Events | Hazard Ratio | 95% CI | P |
|---|---|---|---|---|---|---|
| OS | PR | 105 | 28 | |||
| NoPR | 61 | 22 | 1.38 | 0.79-2.42 | 0.26 | |
| EFS | PR | 105 | 36 | |||
| NoPR | 61 | 28 | 1.41 | 0.85-2.32 | 0.18 | |
| CIR | PR | 105 | 20 | |||
| NoPR | 61 | 10 | 0.75 | 0.34-1.66 | 0.48 | |
| NRM | PR | 105 | 16 | |||
| NoPR | 61 | 18 | 2.13 | 1.10-4.12 | 0.02 | |
| Acute GvHD II-IV | PR | 104 | 34 | |||
| NoPR | 61 | 32 | 1.97 | 1.22 -3.18 | 0.005 | |
| Acute GvHD III-IV | PR | 104 | 12 | |||
| NoPR | 61 | 10 | 1.42 | 0.62-3.23 | 0.40 | |
| Chronic GVHD | PR | 103 | 75 | |||
| NoPR | 61 | 44 | 1.02 | 0.69-1.50 | 0.93 |
All models are multivariable and adjusted for age (<58 vs. ≥58) and disease risk (high, intermediate, low/non-malignant). Patients with incomplete clinical data or with disease risk of other were excluded.
Effect of HLA-DPB1 expression level
When we grouped 11/12 pairs (n=166) according to HLA-DPB1 expression level, 31% (n=51) were low expresser patient and donor (AA or LoDP/LoDP), 24% (n=40) were low expresser patients with high expresser donors (AG or LoDP/HiDP), 29% (n=47) were high expresser patients with low expresser donors (GA or HiDP/LoDP), and 16% (n=28) were high expresser patient and donor (GG or HiDP/HiDP) combinations. On multivariable analysis (Table 4), OS was almost 4 times higher in the AG (LoDP patients with HiDP donors) compared to the AA combination (HR=3.78; 95% CI: 1.56–9.16; p=0.003) (Supplementary Figure 1A). There was also a more than 5-fold increase in the NRM risk when AG pairs (LoDP/HiDP) when compared with AA (LoDP/LoDP) (HR=5.33; 95% CI: 1.81–15.6; p=0.002) (Supplementary Figure 1B). The same detrimental effect was not seen in the GG pairs (HiDP/HiDP) were compared with GA combination (HiDP/LoDP). (Table 4) This finding was also true when we restricted the analysis to patients who received HCT from donors with mismatched DP in the graft-vs.-host (GVH) vector (Supplementary Table 3). Incidences of acute and chronic GvHD or CIR were not affected by HLA-DPB1 expression in our cohort (Table 4).
Table 4:
Effect of HLA-DPB1 expression on transplant outcomes for patients with 11/12 HLA match (N=166)*
| Clinical endpoint | Recipient | Donor | N | Events | Hazard Ratio | 95% CI | P | P-value for interaction |
|---|---|---|---|---|---|---|---|---|
| OS | A | G | 40 | 19 | 3.78 | 1.56-9.16 | 0.003 | |
| A | 51 | 7 | 0.01 | |||||
| G | G | 28 | 7 | 0.86 | 0.35-2.14 | 0.75 | ||
| A | 47 | 17 | ||||||
| EFS | A | G | 40 | 19 | 1.91 | 0.93-3.94 | 0.08 | |
| A | 51 | 13 | 0.13 | |||||
| G | G | 28 | 11 | 0.95 | 0.45-2.02 | 0.89 | ||
| A | 47 | 21 | ||||||
| CIR | A | G | 40 | 5 | 0.56 | 0.20-1.58 | 0.27 | |
| A | 51 | 9 | 0.52 | |||||
| G | G | 28 | 6 | 1.01 | 0.40-2.50 | 0.99 | ||
| A | 47 | 10 | ||||||
| NRM | A | G | 40 | 14 | 5.33 | 1.81-15.68 | 0.002 | |
| A | 51 | 4 | 0.03 | |||||
| G | G | 28 | 5 | 0.98 | 0.32-3.03 | 0.97 | ||
| A | 47 | 11 | ||||||
| Acute GvHD II-IV | A | G | 40 | 14 | 0.99 | 0.48-2.06 | 0.99 | |
| A | 51 | 18 | 0.91 | |||||
| G | G | 28 | 12 | 0.99 | 0.48-2.04 | 0.98 | ||
| A | 46 | 22 | ||||||
| Acute GvHD III-IV | A | G | 40 | 4 | 1.06 | 0.29-3.86 | 0.93 | |
| A | 51 | 5 | 0.49 | |||||
| G | G | 28 | 3 | 0.49 | 0.13-1.87 | 0.29 | ||
| A | 46 | 10 | ||||||
| Chronic GvHD | A | G | 39 | 29 | 1.02 | 0.62-1.67 | 0.95 | |
| A | 50 | 39 | 0.49 | |||||
| G | G | 28 | 17 | 0.78 | 0.43-1.41 | 0.40 | ||
| A | 47 | 34 |
All models are multivariable and adjusted for age (<58 vs. ≥58) and disease risk (high, intermediate, low/non-malignant). Patients with incomplete clinical data or with disease risk of other were excluded.
Cumulative effect of a combination of DP mismatches
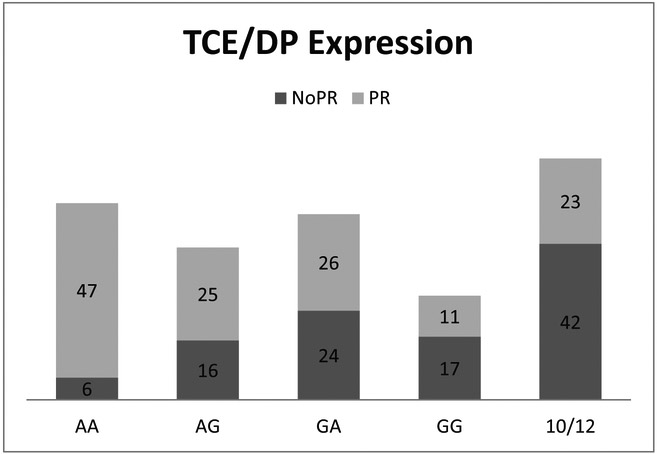
To answer the question if a combination of DP mismatches have a cumulative effect on transplant outcomes after RIC alloHCT, we analyzed and compared 11/12 pairs (n=172) based on both mismatch permissiveness and DP expression level, and found that AA patients-donor combination (LoDP/LoDP) were more likely to have PR mismatches compared to the GG (HiDP/HiDP) pairs (89% vs. 39%, p=<0.0001) as observed by others.8 (Figure2) Analysis of 10/12 pairs (n=65) based on TCE status showed that majority of mismatches (65%) were NoPR. (Figure 2).
Figure 2.
Patient distribution based on mismatch permissiveness and DP expression level in 11/12 and 10/12 pairs.
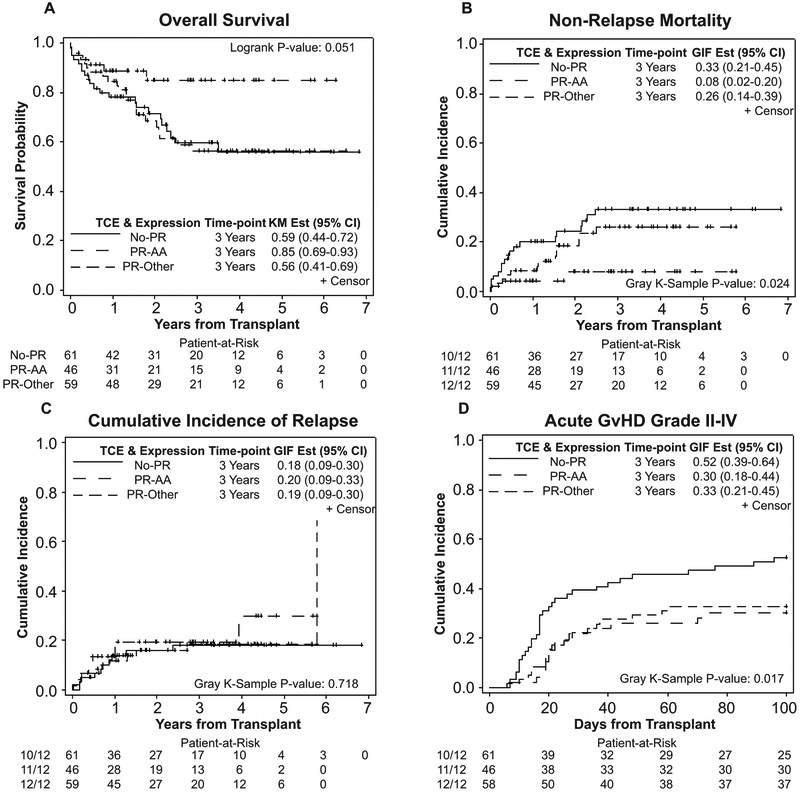
To further determine the simultaneous impact of a combination of HLA-DPB1 mismatches, we ran models using a composite of permissiveness and DP expression level, limiting our analysis to 11/12 pairs (Table 5). Results of our analysis showed that AA pairs (LoDP/LoDP) with PR mismatches had significantly lower NRM and consequently better OS compared to other combinations (AG,GA, and GG pairs) with PR mismatches (HR=3.47; 95% CI: 1.00–12.09; p=0.05 and HR=2.82; 95% CI: 1.13–7.09; p=0.03) or patients with non-PR mismatches (HR=5.08; 95% CI: 1.51–17.08; p=0.01 and HR=2.82; 95% CI: 1.13–7.03; p=0.03).(Figure 3) AA pairs with PR mismatches also had lower grade II-IV acute GvHD compared with patients with NoPR mismatches (HR=2.02; 95% CI: 1.11–3.68; p=0.02). (Figure 3 and Table 5)
Table 5.
Cumulative effect of HLA match, TCE, and DP expression, model restricted to 11/12 permissive (N=166)*
| Clinical Endpoint | TCE*DP Expression | N | Events | Hazard Ratio | 95% CI | P |
|---|---|---|---|---|---|---|
| OS | PR-AA | 46 | 6 | |||
| PR-Other | 59 | 22 | 2.82 | 1.13-7.09 | 0.03 | |
| NoPR | 61 | 22 | 2.82 | 1.13-7.03 | 0.03 | |
| EFS | PR-AA | 46 | 12 | |||
| PR-Other | 59 | 24 | 1.47 | 0.74-2.95 | 0.27 | |
| NoPR | 61 | 28 | 1.80 | 0.90-3.58 | 0.10 | |
| CIR | PR-AA | 46 | 9 | |||
| PR-Other | 59 | 11 | 0.78 | 0.33-1.85 | 0.57 | |
| NoPR | 61 | 10 | 0.65 | 0.25-1.70 | 0.38 | |
| NRM | PR-AA | 46 | 3 | |||
| PR-Other | 59 | 13 | 3.47 | 1.00-12.09 | 0.05 | |
| NoPR | 61 | 18 | 5.08 | 1.51-17.08 | 0.01 | |
| Acute GvHD II-IV | PR-AA | 46 | 15 | |||
| PR-Other | 58 | 19 | 1.04 | 0.53-2.04 | 0.91 | |
| NoPR | 61 | 32 | 2.02 | 1.11 -3.68 | 0.02 | |
| Acute GvHD III-IV | PR-AA | 46 | 4 | |||
| PR-Other | 58 | 8 | 1.73 | 0.50-5.96 | 0.38 | |
| NoPR | 61 | 10 | 1.98 | 0.63-6.25 | 0.25 | |
| Chronic GvHD | PR-AA | 45 | 35 | |||
| PR-Other | 58 | 40 | 0.93 | 0.59-1.46 | 0.75 | |
| NoPR | 61 | 44 | 0.98 | 0.62-1.54 | 0.92 |
All models are multivariable and adjusted for age (<58 vs. ≥58) and disease risk (high, intermediate, low/non-malignant). Patients with incomplete clinical data or with disease risk of other were excluded.
Figure 3.
Cumulative effect of mismatch permissiveness and DP expression level on transplant outcomes. (A) Overall survival, (B) Non-relapse mortality, (C) Cumulative incidence of relapse, and (D) Acute graft-versus-host disease, grades II-IV.
Discussion
To date, multiple investigators have studied the impact of HLA-DPB1 mismatches on transplant outcomes demonstrating that number of mismatches (single versus double), mismatch permissiveness based on TCE grouping, and expression levels of HLA-DPB1 alleles can impact transplant outcomes after myeloablative alloHCT.4, 8-10, 12 While these variables are not independent and have overlapping effects (for instance, most of the 10/12 pairs are also NoPR, and mismatch for low expression is linked to permissiveness), there are no reports of examining whether a combination of all three different types of DP mismatches have the same impact on transplant outcomes after alloHCT with RIC. Our single center retrospective study, to our knowledge, is the first to analyze the impact of DP mismatch impact on a homogenous patient population undergoing RIC alloHCT for malignant hematological diseases.
While our results confirms the importance of including HLA-DPB1 typing during the donor selection process by demonstrating the impact of DP mismatches on RIC alloHCT outcomes, in contrast to previous reports describing alloHCT outcomes with myeloablative conditioning, in our study a single DP mismatch (11/12) resulted in significantly lower disease relapse compared to fully matched pairs (12/12), without significantly increasing the incidence of acute GvHD. These outcomes could possibly be explained by the stronger graft-versus-tumor (GVT) effect in 11/12 mismatches, resulting in lower relapse rate. In accordance to previously published studies, 4, 8, 10, 12 results of our study also prove that NoPR HLA-DPB1 mismatches can increase the incidence of acute GvHD and decrease the incidence of disease relapse in the reduced-intensity setting, confirming that NoPR mismatches should be avoided, if possible, regardless of the conditioning regimen.
Analysis of transplant outcomes based on DP expression levels confirmed the immunogenic potential of HLA-DPB1 high expression allele (G), especially in a low expresser recipient (A),8 in the RIC setting. In our cohort, AA pairs (LoDP/LoDP) were mostly PR and had better OS and lower NRM compared with AG pairs (LoDP/HiDP). However, statistical significance was not met for any of the transplant outcomes when high expresser patients were grouped solely based on the expression level without including the permissiveness variable in the analysis (Table 4), most probably because majority (70%) of the GG pairs (HiDP/HiDP) in our cohort carried NoPR mismatches (Figure 2).
Furthermore, in a recent study by Morishima et al, patients mismatched for DP5 group (low DP expression) were at higher risk for acute GVHD compared with DP2 (high DP expression), regardless of donors’ DP status.9 Similar to our findings, Morishima et al, reported that the combination of permissiveness and expression was associated with transplant outcomes. Our data is also in agreement with Morishima et al, findings that DP2 group is mostly permissive, contrary to DP5 group.
In this study, we aimed to provide clinical evidence for the innovative concept of combining different types of DP mismatches (number, permissiveness and expression level) for alloHCT from a matched unrelated donor after reduced intensity conditioning. Thus, we grouped patients in our cohort based on all three DP mismatch variables, then restricted our analysis to patient/donor pairs with best transplant outcomes per variable. In other words, 11/12 and PR status were chosen over 10/12 and NoPR due to lower relapse and GvHD, respectively. Our data demonstrated significantly better OS and NRM with 11/12 PR mismatches, when a low expresser donor was matched with a low expresser patient (11/12, AA-PR)
Based on the results of this retrospective study, the beneficial effect of typing for DP expression-linked SNP in donor selection becomes more evident, specifically when the overlapping effects of DP expression and permissiveness (i.e. majority of AA pairs were PR and majority of GG pairs were NoPR) are taken into consideration. Therefore, based on our algorithm choosing 11/12 AA donor with PR mismatch is preferred over 12/12 HLA matched donor and could result in better transplant outcome in the setting of RIC MUD HCT. We recognize that based on our older method of HLA typing our G group assignment is not specific to HLA-DP and some patients might have been misclassified into matched or mismatched groups. Unfortunately, NGS assays that are more appropriate for higher resolution HLA typing were not available at the time of typing for this study. However, based on the data described by Schone et al, while a stronger linkage exists between exon 3 and the rs9277534 SNP, a prediction of rs9277534 based on exon 2 sequence alone yields an error probability of only 0.9%.14
In conclusion, although our study carries the inherent limitations of a single center retrospective analysis, it includes a large number of homogeneously treated patients with RIC HCT. Our data provide new information on the impact of a combination of HLA-DPB1 mismatches on transplant outcomes in RIC setting, which could be used to substantially reduce post-transplant complications by avoiding high-risk mismatches. Results of this retrospective analysis further demonstrates the feasibility and influence of donor selection in MUD HCT setting based on a combination of DP mismatches and set the stage for prospective studies in which a combination of DP mismatches will be used in the donor selection process.
Supplementary Material
Acknowledgments:
The authors would like to thank Dr. Joycelynne Palmer for assistance with biostatistical data analysis and reviewing this manuscript. This study was partially funded by NIH P30 CA033572 (Biostatistics Core).
Footnotes
Conflict of Interests
Authors declare no relevant conflict of interest.
References
- 1.Farhadfar N, Hogan WJ. Overview of the progress on haploidentical hematopoietic transplantation. World journal of transplantation 2016; 6(4): 665–674. e-pub ahead of print 2017/01/07; doi: 10.5500/wjt.v6.i4.665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fabricius WA, Ramanathan M. Review on Haploidentical Hematopoietic Cell Transplantation in Patients with Hematologic Malignancies. Advances in hematology 2016; 2016: 5726132. e-pub ahead of print 2016/04/02; doi: 10.1155/2016/5726132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choo SY. The HLA system: genetics, immunology, clinical testing, and clinical implications. Yonsei medical journal 2007; 48(1): 11–23. e-pub ahead of print 2007/02/28; doi: 10.3349/ymj.2007.48.1.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pidala J, Lee SJ, Ahn KW, Spellman S, Wang HL, Aljurf M et al. Nonpermissive HLA-DPB1 mismatch increases mortality after myeloablative unrelated allogeneic hematopoietic cell transplantation. Blood 2014; 124(16): 2596–2606. doi: 10.1182/blood-2014-05-576041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shaw BE, Gooley TA, Malkki M, Madrigal JA, Begovich AB, Horowitz MM et al. The importance of HLA-DPB1 in unrelated donor hematopoietic cell transplantation. Blood 2007; 110(13): 4560–4566. e-pub ahead of print 2007/08/30; doi: 10.1182/blood-2007-06-095265 [DOI] [PubMed] [Google Scholar]
- 6.Lee SJ, Klein J, Haagenson M, Baxter-Lowe LA, Confer DL, Eapen M et al. High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood 2007; 110(13): 4576–4583. e-pub ahead of print 2007/09/06; doi: 10.1182/blood-2007-06-097386 [DOI] [PubMed] [Google Scholar]
- 7.Thomas R, Thio CL, Apps R, Qi Y, Gao X, Marti D et al. A novel variant marking HLA-DP expression levels predicts recovery from hepatitis B virus infection. Journal of virology 2012; 86(12): 6979–6985. e-pub ahead of print 2012/04/13; doi: 10.1128/jvi.00406-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petersdorf EW, Malkki M, O’HUigin C, Carrington M, Gooley T, Haagenson MD et al. High HLA-DP Expression and Graft-versus-Host Disease. The New England journal of medicine 2015; 373(7): 599–609. doi: 10.1056/NEJMoa1500140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morishima S, Shiina T, Suzuki S, Ogawa S, Sato-Otsubo A, Kashiwase K et al. Evolutionary basis of HLA-DPB1 alleles affects acute GVHD in unrelated donor stem cell transplantation. Blood 2018; 131(7): 808–817. e-pub ahead of print 2017/12/17; doi: 10.1182/blood-2017-08-801449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fleischhauer K, Shaw BE, Gooley T, Malkki M, Bardy P, Bignon JD et al. Effect of T-cell-epitope matching at HLA-DPB1 in recipients of unrelated-donor haemopoietic-cell transplantation: a retrospective study. The Lancet. Oncology 2012; 13(4): 366–374. doi: 10.1016/S1470-2045(12)70004-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fleischhauer K Immunogenetics of HLA-DP--A New View of Permissible Mismatches. The New England journal of medicine 2015; 373(7): 669–672. e-pub ahead of print 2015/08/13; doi: 10.1056/NEJMe1505539 [DOI] [PubMed] [Google Scholar]
- 12.Crivello P, Zito L, Sizzano F, Zino E, Maiers M, Mulder A et al. The impact of amino acid variability on alloreactivity defines a functional distance predictive of permissive HLA-DPB1 mismatches in hematopoietic stem cell transplantation. Biol Blood Marrow Transplant 2015; 21(2): 233–241. doi: 10.1016/j.bbmt.2014.10.017 [DOI] [PubMed] [Google Scholar]
- 13.Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh SG. The IPD and IMGT/HLA database: allele variant databases. Nucleic acids research 2015; 43(Database issue): D423–431. e-pub ahead of print 2014/11/22; doi: 10.1093/nar/gku1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schone B, Bergmann S, Lang K, Wagner I, Schmidt AH, Petersdorf EW et al. Predicting an HLA-DPB1 expression marker based on standard DPB1 genotyping: Linkage analysis of over 32,000 samples. Human immunology 2018; 79(1): 20–27. e-pub ahead of print 2017/11/12; doi: 10.1016/j.humimm.2017.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fine JP, Gray RJ. A Proportional Hazards Model for the Subdistribution of a Competing Risk. Journal of the American Statistical Association 1999; 94(446): 496–509. doi: 10.1080/01621459.1999.10474144 [DOI] [Google Scholar]
- 16.Glucksberg H, Storb R, Fefer A, Buckner CD, Neiman PE, Clift RA et al. Clinical manifestations of graft-versus-host disease in human recipients of marrow from HL-A-matched sibling donors. Transplantation 1974; 18(4): 295–304. [DOI] [PubMed] [Google Scholar]
- 17.Shulman HM, Sullivan KM, Weiden PL, McDonald GB, Striker GE, Sale GE et al. Chronic graft-versus-host syndrome in man. A long-term clinicopathologic study of 20 Seattle patients. The American journal of medicine 1980; 69(2): 204–217. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.