Abstract
Semiconductor quantum dots (QDs) have demonstrated utility in long-term single particle tracking of membrane proteins in live cells in culture. To extend the superior optical properties of QDs to more physiologically relevant cell platforms, such as acute brain slices, we examine the photophysics of compact ligand-conjugated CdSe/CdS QDs using both ensemble and single particle analysis in brain tissue media. We find that symmetric core passivation is critical for both photostability in oxygenated media and for prolonged single particle imaging in brain slices. We then demonstrate the utility of these QDs by imaging single dopamine transporters in acute brain slices, achieving 20 nm localization precision at 10 Hz frame rates. These findings detail design requirements needed for new QD probes in complex living environments, and open the door to physiologically relevant studies that capture the utility of QD probes in acute brain slices.
Introduction
Fluorescence microscopy has long served as a cornerstone technique in biology for addressing many of the fundamental processes of life. Consequent to the dynamic nature of biomolecules, single molecule imaging approaches have been developed to achieve finer spatiotemporal resolution sufficient for dynamic molecular imaging in live cells.1,2 Single particle tracking (SPT) has been used to investigate protein localization and dynamics in mammalian cells and established the basis of protein dynamics such as membrane protein trafficking and clustering in detail.3–5 Neuronal membrane protein membrane dynamics have been investigated by SPT image analysis such as glycine,5 GABA, and glutamate-gated receptors,6–9 as well as epidermal growth factor receptors,10–12 and G-protein coupled receptors.13 SPT approaches like these require bright probes such as semiconductor quantum dots (QDs) to achieve the high signal-to-background ratios (SBRs) needed to track individual proteins. In 2002, ligand-conjugated QDs were introduced as probes in order to specifically label serotonin transporters (SERT).14 Ligand conjugation of QDs have since been adapted to image single SERT and dopamine transporter (DAT) proteins, finding alterations in diffusion patterns associated with neuropsychiatric diseases.15–21
Many single molecule imaging studies rely on heterologous expression systems and cultured neurons, systems in which large SBRs arise from having imaging planes close to the glass substrate. In acute brain slices and other physiologically relevant environments, single molecule imaging has remained challenging and is a necessary next step to link mammalian physiology to protein dynamics. A key challenge is the development of specific, bright, and stable probes that can be imaged deep in tissue at millisecond SPT frame rates. While some organic fluorophores and fluorescent proteins exhibit high fluorescence quantum yields, the high excitation powers needed for SPT have been shown to cause significant photodamage to cells and the probes themselves.22–24 QDs may be tracked at lower fluences because of their large absorption cross-sections, which lead to enhanced emission, reduced photobleaching, and lower phototoxicity compared to conventional probes.25
While various tissues have been imaged with QD labelling for ensemble analysis,26–28 few examples of QD tracking in brain tissue have been reported, leaving native 3D neuronal architectures largely unexplored.29–31 In this study, we provide a structural and photophysical basis for the importance of shell geometry of high quality CdSe/CdS QDs in single molecule imaging deep in living brain slices. These pseudo type II core/shell structures have been reported to have enhanced photon output,32 stability in common aqueous buffer, as well as much smaller hydrodynamic diameters compared to streptavidin-coated QDs frequently used in SPT experiments (Fig. S1†).24,33 In the interest of transitioning single particle studies from in vitro to ex vivo platforms, we demonstrate here that these nanoparticles (i) maintain their photostability in oxygenated brain slice media (i.e., artificial cerebrospinal fluid, aCSF) and (ii) show significantly great stability and less blinking than the widely-used QD655 probes (ThermoFisher) via a detailed ensemble and single particle investigation. Using energy-dispersive X-ray (EDS) chemical mapping, we show that thick, symmetric CdS shells are required for prolonged photostability in brain slice media. We substantiate our motivation for this study by successful subcellular localization imaging of dopamine transporters and SPT experiments of these probes in acute brain slices (20 nm localization precision, 10 Hz frame rates) using a conventional spinning disk confocal microscope. To address commonly available QDs fall short of meeting the photophysical criteria for SPT of membrane proteins in oxygenated brain slice media, we introduce the blueprint of critical considerations in probe design. This study paves the way to development of probes for long-term monitoring of targeted protein dynamics in their native environments and sets the course for direct observation of these dynamics deep in the brain tissue of neuropsychiatric disease models.
Results and discussion
Chemically mapping structural differences in core/shell aqueous probes
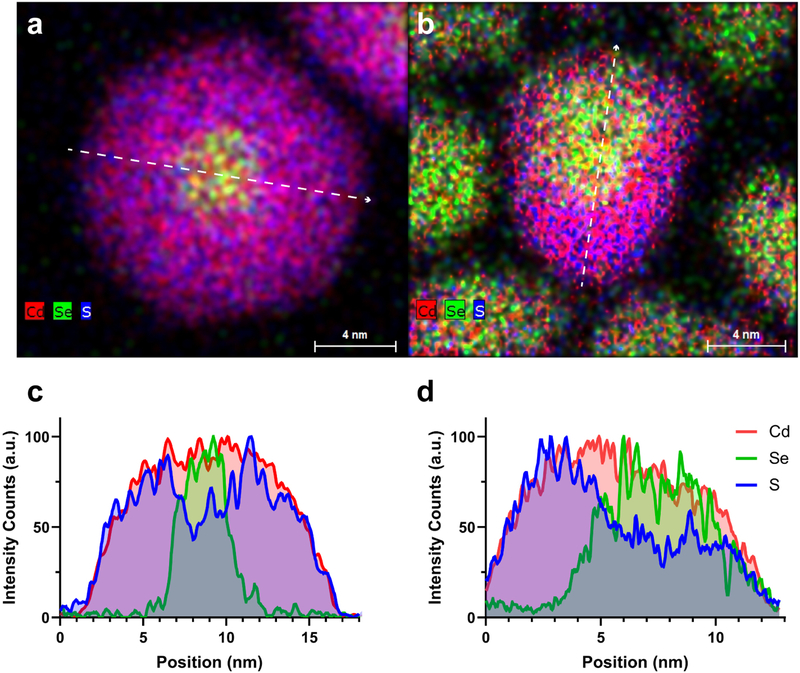
The fluorescence efficiency and stability of a QD is exceedingly dependent on how well the shell passivates the core surface. Proper passivation relies on both the degree of lattice mismatch, surface coverage, as well as by how many layers of shell are grown.34 Although a thin shell can significantly enhance the QD fluorescence, these particles are far from robust and will photobleach rapidly in all but the most benign environments.35 Conversely, too thick of a shell, as in those for ‘giant’ shelled quantum dots, leads to significant charge state emission that caps the ensemble fluorescence to a maximum of roughly 50%.35–38 Ou Chen demonstrated that ∼8 monolayers of a uniform CdS shell leads to a balance of high quantum yield and improved stability.32 Ultimately, in a chemically and physically challenging environment where a minimum QD hydrodynamic radius is desired, the amount and location of shell is critical for long-term performance. The clearest way to observe shell coverage is through STEM-EDS imaging.37,39 Fig. 1 shows STEM-EDS maps of the QDs engineered for brain slice imaging and QD655s (High-resolution TEM images also provided in Fig. S2†). While both exhibit thick CdS shells, the QD655s dissimilar to our symmetrically shelled QDs show preferential shell growth along the c-axis leading to asymmetric shell coverage, as evident in the line scan (Fig. 1c, d and S3–S5†). The close proximity of the core to the surface likely enhances electron and hole overlap with trap sites that form during illumination. Notably, the native ligands on both types of QDs are encapsulated with an amphiphilic polymer which enables water solubility, while maintaining a similar particle size and colloidal stability (Fig. S6†). Although ligand shells are needed for solution stability and chemical functionality, long term photostability relies on inorganic shell passivation. It is also worth noting that although these QD655s are quoted as having a ZnS shell, only trace amounts of Zn signal were detected for this particular batch (Fig. S7 and S8†).40
Fig. 1.
Elemental characterization of CdSe/CdS QD architectures. Combined Cd (red), Se (green), and S (blue) elemental maps of (a) symmetrically shelled QDs and (b) QD655s show core/shell structures. Linescans of the elemental maps (white arrows) Se distribution relative to Cd and S in both (c) symmetrically shelled QDs and (d) QD655s.
Ensemble photophysical investigation
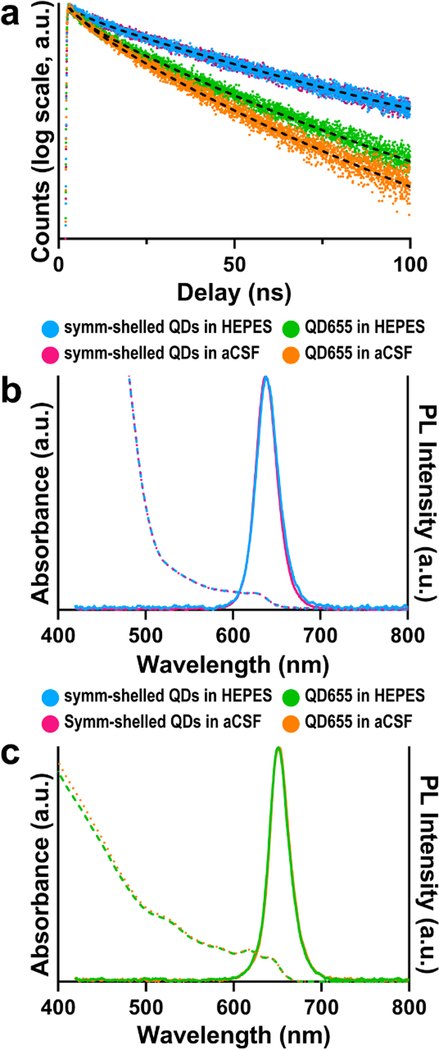
In order to compare the photostability of both these QD constructs in brain slice media, we performed time-resolved photoluminescence measurements on dilute concentrations of each QD type suspended in both 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer and oxygenated cerebrospinal fluid. We note that HEPES is a commonly used buffer for storing and performing conjugation reactions on QDs as well as for coupling techniques such as diimide/N-hydroxysuccinimide (e.g. EDC/NHS), making it a useful control. The samples were excited at low fluences (∼1 μJ cm−2). At these low fluences, we expect the majority of photo-excited QDs to contain a single electron–hole pair.39 Interestingly, the symmetrically shelled QDs displayed similar lifetimes in both HEPES and oxygenated cerebrospinal fluid (τavg ∼46.0 ± 0.2 ns), indicating that the QDs retain their photostability on exchange in the brain slice media (Fig. 2a). In comparison, the lifetime of QD655s was notably shortened upon exchange into the brain slice media (τavg = 27.0 ± 0.2 ns in HEPES, τavg = 22.0 ± 0.1 ns in oxygenated cerebrospinal fluid) (Table S1†), suggesting symmetric shells are required to suppress O2-mediated quenching.
Fig. 2.
Ensemble photophysical profiles of symmetrically shelled (symm-shelled) CdSe/CdS QDs and QD655s in HEPES buffer or oxygenated cerebrospinal fluid (aCSF). (a) Transient PL dynamics of symm-shelled QDs and QD655s in HEPES and oxygenated aCSF. (b) Absorbance (dotted) and emission (solid) spectra of (b) symm-shelled QDs and (c) QD655s in HEPES and oxygenated aCSF.
To examine whether this apparent quenching is due to spectral-shifting processes such as etching or aggregation, UV-visible and PL spectroscopy in various buffers were performed. Interestingly, the differences observed in PL lifetimes of the commercial QD655s are not observed in the absorbance and photoluminescence spectra (Fig. 2b and c). Considering oxygenated environments have been shown in general to quench the photoluminescence of QDs by creating defects at the nanocrystal surface that introduce nonradiative recombination centres, a process that is accelerated under high flux,41–43 it is likely that O2-rich media further quenches QD655s. In combination with our structural results that the cores of the QD655 probes are asymmetrically passivated (Fig. 1b), these findings support the possibility of the cores being vulnerable to their environment. More generally, our results emphasize the need to investigate photoluminescence lifetimes of probes in their intended environments (e.g. oxygenated tissue media) during development and optimization.
Single QD analysis in biological media
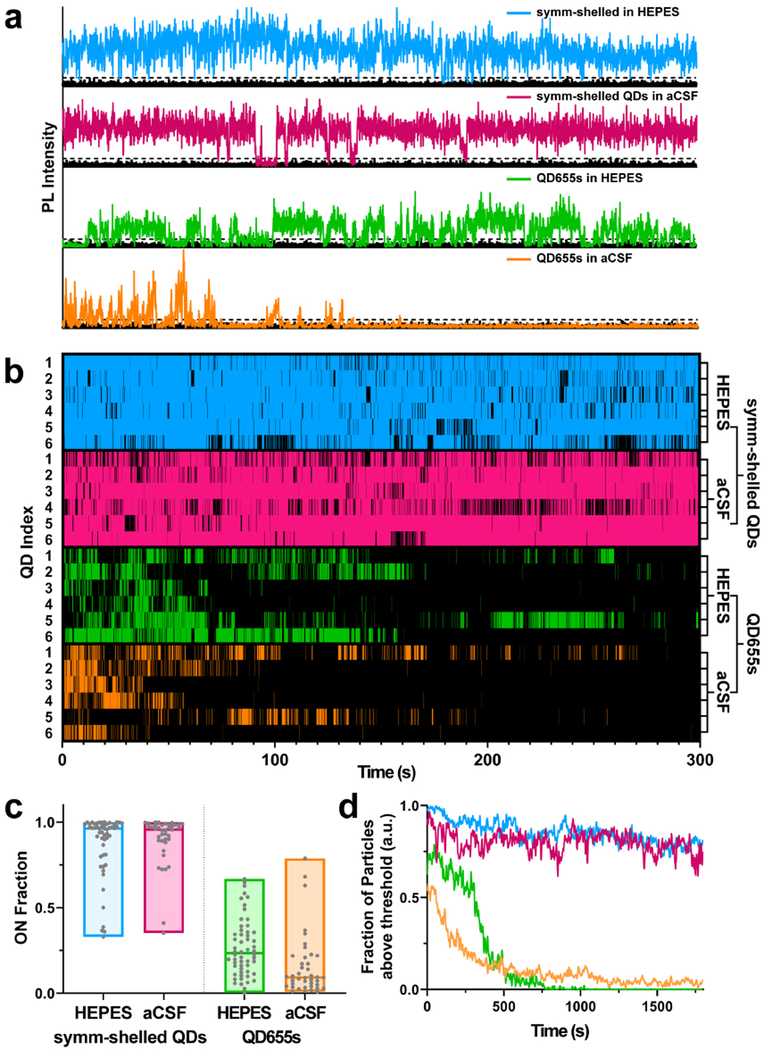
Further photophysical differences between QDs systems were measured using single-molecule imaging modalities. We first set out to test whether QD blinking is altered in brain slice media. Using a spinning disk confocal microscope system (100 ms exposure time at 51 W cm−2), PL intensity traces of single QDs incubated in oxygenated cerebrospinal fluid were compared to QDs incubated in HEPES as a negative control. Representative PL intensity time traces (Fig. 3a) of the two QD in both HEPES and oxygenated cerebrospinal fluid show QD classic PL fluctuations between high (ON) and low (OFF) emission intensity values. Binarization of the intensity traces by assigning time bins with intensity above 6σ of the background level (Fig. 3b) clearly reveals strong blinking suppression of the symmetrically shelled QDs in both HEPES and cerebrospinal fluid. Furthermore, distributions in ON-fractions—the fraction of time a QD spends in the emissive state over the course of the experiment—display no significant difference between symmetrically shelled QD populations (Fig. 3c) diluted in HEPES (ON-fraction = 0.87 ± 0.03) and oxygenated cerebrospinal fluid (ON-fraction = 0.89 ± 0.02). In contrast, QD655s are characterized by strong blinking with low ON-times in HEPES (ON-fraction = 0.27 ± 0.02) and even lower ON-times (ON-fraction = 0.16 ± 0.03) were observed for QDs in oxygenated cerebrospinal fluid (see ESI† for statistics using data analysis with bootstrap-coupled estimation,45 Fig. S9 and Table S2†). These low ON-times can also be attributed to incomplete passivation of the CdSe core in QD655s, resulting in greater overlap of excited charge carriers with the nanocrystal surface where they can become trapped and render the particles non-emissive.46–49 CdSe/CdS heterostructures display type II exciton behaviour, in which holes are confined to the CdSe core whereas electrons may reside in core or CdS shell. The presence of thin CdS shell domains then opens the possibility of O2 quenching of electrons that venture to the QD surface.50–52 Exposure of the poorly passivated QDs on exchange in oxygenated cerebrospinal fluid additionally increases the number of available trap sites and further enhances blinking, concomitant with the shortening of the PL lifetimes observed for QD655s in ensemble.
Fig. 3.
Time series and blinking behaviour of single QDs. (a) Representative intensity trajectories for symmetrically shelled (symm-shelled) QDs and QD655s in both HEPES and oxygenated artificial cerebrospinal fluid (aCSF). (b) Subsets of blinking dynamics for symm-shelled QDs and QD655s in HEPES and aCSF. A total of 6 particles per condition were populated to display blinking behaviour. Colours represent times that the particle was in the ON state. (c) Comparison of ON fraction populations under each condition (N ≥ 40 QDs). (d) Comparison of photobleaching profiles for symm-shelled QDs vs. QD655s under each condition (N > 40 QDs).
To further investigate the photophysical properties of the two QD types under imaging conditions in brain slices, we examined the photobleaching behaviour of the samples over 30 minutes under continuous photo-illumination. Using the same microscope and imaging parameters as the blinking analysis but increasing the bin time from 100 ms to 1 s, we compared the intensity traces of single QDs incubated in oxygenated cerebrospinal fluid versus control HEPES buffer. For the symmetrically shelled QDs, the majority of particles remain emissive (Fig. 3d) in both HEPES (80%) and oxygenated cerebrospinal fluid (78%) over the course of 30 minutes, whereas nearly all QD655 particles completely photobleach during the first ∼8 minutes of the experiment. Intriguingly, a small fraction (<5%) of QD655s remain luminescent in oxygenated cerebrospinal fluid, but completely bleach in HEPES, exhibiting no additional decline over the measured time. A similar effect has been observed in studies of O2 sensing53 and highlights the possibility that a small fraction of QDs are sufficiently passivated. Nonetheless, the degrees of depreciation in ON-fractions and photostability are pronounced in QD655 populations, which imposes major experimental limitations for single particle brain slice imaging. Taken together, the blinking and photobleaching studies suggest out symmetrically shelled QDs should provide superior photon output under brain slice imaging conditions and should enable the capture of membrane diffusion dynamics of neuronal proteins with higher fidelity over extended periods of time compared to QD655s.
Imaging ligand-conjugated SPT probes in acute brain slices
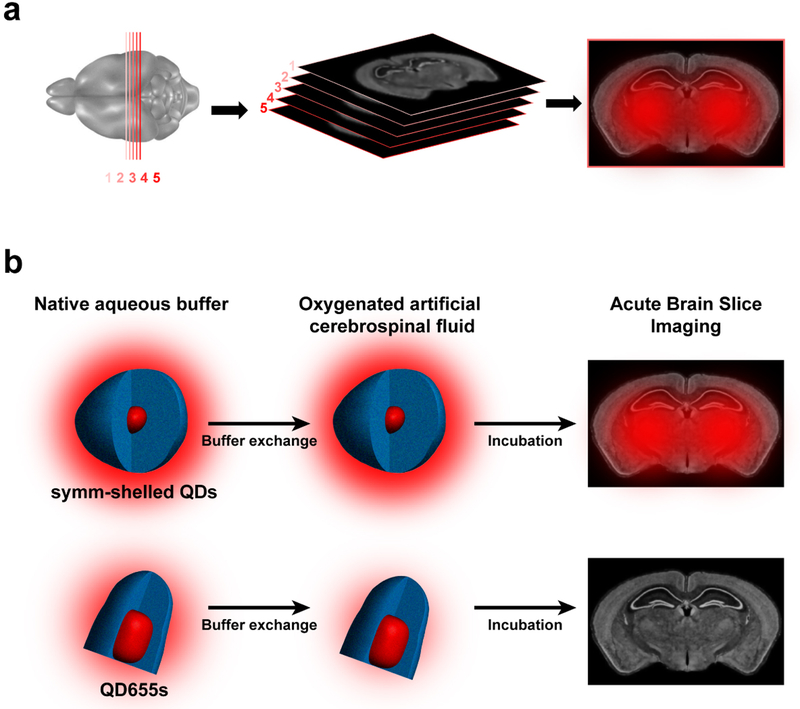
Acute brain slices are prepared such that slices undergo little change in physiological conditions (e.g. pH, oxygen concentration, glucose levels) from the time of dissection to image acquisition at the microscope (Fig. 4a). During this time prior to mounting samples to the microscope stage, specimens are incubated with QDs diluted in oxygenated cerebrospinal fluid, allowing for fluorophores to reach depths of interest. Taking into account the ensemble and single particle characterizations included in this study, we propose the use of QDs with the symmetrically shelled composition reported here will greatly facilitate practical single target imaging in living brain tissue. Since the onset of any environmental effects on QD655s occurs at the exchange into oxygenated cerebrospinal fluid, we expect fluorescence to diminish mostly by the time of brain slice incubation (Fig. 4b). Thus, imaging QDs in acute brain slices is dependent on the fate of emission efficiency in cerebrospinal fluid.
Fig. 4.
(a) Acute brain slice imaging with CdSe/CdS QDs. Mouse brain slices (1–5 300 μm slices) are cut by vibratome and incubated with ligand-conjugated QDs prior to imaging. (b) Schemes outlining buffer exchange of QDs (drawn to scale) into brain slice media (oxygenated artificial cerebrospinal fluid, aCSF) for symmetrically shelled QDs and commercial QD655s. The schemes illustrate comparison of symmetrically shelled QD and QD655 performance in oxygenated aCSF and their photoluminescence fate in tissue specimens. The auras surrounding QD structures illustrate relative photoluminescence and the fate of diminished performance of QD655 in slice media. Whole brain slice representations are provided by the Allen Institute.44
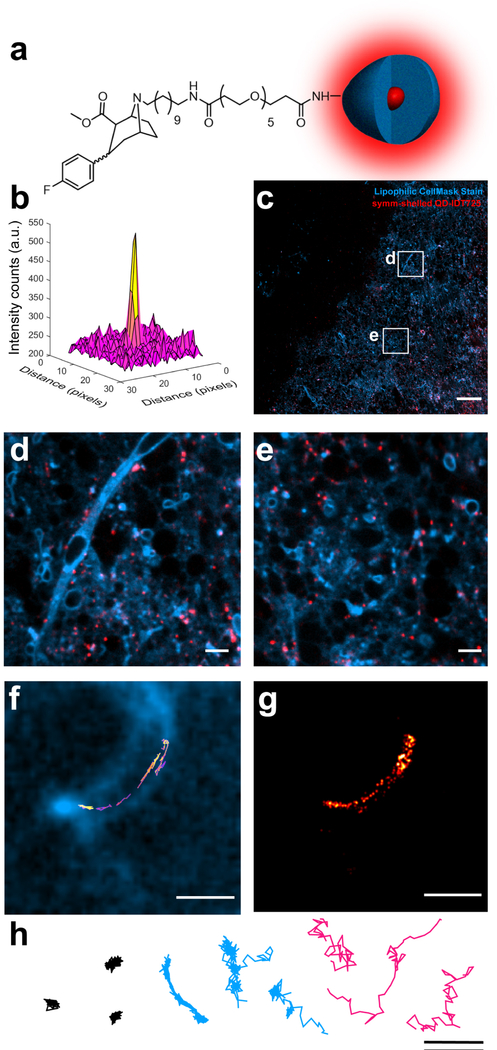
Using a simple spinning disk confocal microscope and a dedicated 405 nm continuous-wave excitation source, we observed the symmetrically shelled QDs are detectable 50 μm deep in a brain slice (Fig. 5). Prior to imaging, these QDs were PEGylated and conjugated with IDT725 (Fig. 5a), which is furnished with a terminal cocaine analogue previously used to label DAT proteins.54 Accordingly, imaging was performed on striatal regions known to be rich in available DAT proteins. To show QD localization with SBRs suitable for tracking experiments (Fig. 5b), 25 images (255 025 μm2) of the QDs dispersed in a striatal slice were stitched together with a 100 ms exposure time for each image acquired (Fig. 5c). Studies that include brain slice imaging typically outline network morphology by using genetic expression of fluorescent proteins, which only display a small fraction of neurons.55,56 To illuminate the whole slice, a lipophilic stain was used to outline the complex morphologies of a native neuronal architecture. Fig. 5d shows that the QDs can be detected along axonal regions of neurons as well as regions that lie just outside of focal plane (Fig. 5d and e). Excitingly, time series on the order of minutes could be acquired such that we could generate high quality trajectories of QDs along a neuron at focal planes reaching 50 μm into brain tissue (Fig. 5f and Movie S1 in ESI†). The localization map provides detailed hot spots where the QDs show extended residence times during the course of the tracking experiment (Fig. 5g). A challenge to overcome in these experiments is washing unbound QDs from the slice. To circumvent this, QD diffusion patterns were characterized in accordance to the physical nature (Fig. 5h), whereby immobilized and unbound diffusion patterns are filtered from trajectory sets with anomalous diffusion characteristic of QDs specifically bound to the respective target.
Fig. 5.
Detection and tracking analysis of QD probes in acute brain slices. (a) Structure of symmetrically shelled (symm-shelled) QD-IDT725 conjugate (not to scale). (b) Surface plot of a QD point spread function imaged 50 μm into a live brain slice. (c) Stitched image of symm-shelled QD conjugates dispersed in a brain tissue (scale bar = 50 μm). (d and e) 10× magnification of various regions captured in the stitched image in panel c (scale bar = 5 μm). (f) QD trajectory with non-Brownian diffusion dynamics along a neuron 50 μm into the slice (scale bar = 2 μm). (g) Localization map of the QD trajectory in panel e (scale bar = 2 μm). (h) Representative trajectories displaying diffusion of immobilized (black), neuron-bound (blue), and unbound (magenta) symm-shelled QD conjugates (scale bar = 2 μm). All images and tracking data were collected in striatal brain slices incubating in oxygenated aCSF. Lipophilic CellMask stain was used to outline the cell morphology.
Given the photostability and photon output requirements of SPT analysis, QD655s have been used in the few examples of SPT in acute brain slices reported, rather than organic or protein-based fluorophores.29,30 For example, antibody-conjugated QD655s have been successful in imaging organotypic slices at depth with multi-photon imaging,29 although organotypic slices do not require oxygenated cerebrospinal fluid. One example of SPT in acute brain slices incubated in oxygenated cerebrospinal fluid show successful detection of QD655s, but trajectories generated in these studies are limited to under 30 seconds.30 The limited number of reports is likely a result of the difficulty in overcoming the O2-dependent instability of conventional QDs. Since more than 80% of the QDs are photobleached 10 minutes after exchange into brain slice media (Fig. 3), and a subpopulation (∼8%) of QDs are expected to be non-emissive prior to excitation,34 the possibility of crowding the tissue sample with undetectable QDs should be of concern. In this regard, the images (Fig. 5) display QD dispersal that accurately represents the concentration of QDs used in our imaging experiments.
It is important to consider that Cd-containing QDs can induce dose-dependent cytotoxicity.57 The QDs in this study are passivated with oleic acid and encapsulated within a PEGylated amphiphilic polymer layer, which forms a low dielectric layer that limits escape of metal ions.33 Toxicology studies have shown that addition of PEG compounds to QD surfaces significantly reduces cytotoxicity effects.58–60 Our labelling concentration is far below concentrations that induce any considerable cytotoxicity seen for PEGylated CdSe/CdS QDs in ex vivo slices,60 and we observe no apparent toxicity on live slices based on neuronal function. Additional experiments are needed to determine precise toxicity levels for our symmetrically shelled QDs.
Conclusions
This work introduced QD probes tailored for dynamic imaging of protein targets in native brain tissue. With ensemble and single-particle analysis, we compared QD performances in oxygenated brain slice media. Our findings show commercially available QDs are prone to fouling due to asymmetric shell coverage. On the other hand, our thick symmetrically shelled QDs feature superior photon output that is resilient to oxygenated cerebrospinal fluid. We conclude that not just shell thickness, but geometry of QD shells impact performance in physiologically relevant environments. Moreover, the effects of these architectural differences are silent in classic UV-visible/PL analysis. The considerations we reported here can be applied to all QD imaging platforms that involve chemically challenging systems. With our ligand-conjugated QD constructs, we also show that neuronal proteins can be imaged at the nanoscale for long time periods in live brain tissue. This is the critical step forward for future pursuits investigating molecular underpinnings of neurological diseases.
Supplementary Material
Acknowledgements
The authors would like to thank Dr Brian Bachmann for assistance with reverse-phase HPLC purification of IDT725; Vanderbilt Institute of Chemical Biology and Chemical Synthesis Core for assistance with Gilson purification of IDT725-SE. LBT was supported by the CBI Training Grant (NIH T32GM065086-14). This work was supported by CAL-BRAIN award 350327 (B.E.C.) and National Institutes of Health award R01NS096317 (B.E.C.). Work at the Molecular Foundry was supported by the Director, Office of Science, Office of Basic Energy Sciences, Division of Materials Sciences and Engineering, of the U.S. Department of Energy under Contract No. DE-AC02- 05CH11231. D. S. and D. G. M. were supported by the National institute of Mental Health (NIH 5R01MH108562). Spinning disk confocal microscopy experiments were performed in part through the use of the Vanderbilt Cell Imaging Shared Resource (supported by NIH grants CA68485, DK20593, DK58404, DK59637 and EY08126). We also acknowledge support from Vanderbilt University.
Footnotes
Conflicts of interest
There are no conflicts to declare.
References
- 1.Patterson G, Davidson M, Manley S and LippincottSchwartz J, Annu. Rev. Phys. Chem., 2010, 61, 345–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Z, Lavis LD and Betzig E, Mol. Cell, 2015, 58, 644–659. [DOI] [PubMed] [Google Scholar]
- 3.Choquet D and Triller A, Neuron, 2013, 80, 691–703. [DOI] [PubMed] [Google Scholar]
- 4.Triller A and Choquet D, Neuron, 2008, 59, 359–374. [DOI] [PubMed] [Google Scholar]
- 5.Dahan M, Lévi S, Luccardini C, Rostaing P, Riveau B and Triller A, Science, 2003, 302, 442–445. [DOI] [PubMed] [Google Scholar]
- 6.Bouzigues C, Morel M, Triller A and Dahan M, Proc. Natl. Acad. Sci. U. S. A, 2007, 104, 11251–11256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jézéquel J, Johansson EM, Dupuis JP, Rogemond V, Gréa H, Kellermayer B, Hamdani N, Le Guen E, Rabu C, Lepleux M, Spatola M, Mathias E, Bouchet D, Ramsey AJ, Yolken RH, Tamouza R, Dalmau J, Honnorat J, Leboyer M and Groc L, Nat. Commun., 2017, 8, 1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai E, Ge P, Lee SH, Jeyifous O, Wang Y, Liu Y, Wilson KM, Lim SJ, Baird MA, Stone JE, Lee KY, Davidson MW, Chung HJ, Schulten K, Smith AM, Green WN and Selvin PR, Angew. Chem., Int. Ed., 2014, 53, 12484–12488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee SH, Le P, Youn Y, Smith AM and Selvin PR, Biophys. J., 2018, 114, 668a. [Google Scholar]
- 10.Kawashima N, Nakayama K, Itoh K, Itoh T, Ishikawa M and Biju V, Chemistry, 2010, 16, 1186–1192. [DOI] [PubMed] [Google Scholar]
- 11.Freed DM, Bessman NJ, Kiyatkin A, Salazar-Cavazos E, Byrne PO, Moore JO, Valley CC, Ferguson KM, Leahy DJ, Lidke DS and Lemmon MA, Cell, 2017, 171, 683–695.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lidke DS, Lidke KA, Rieger B, Jovin TM and Arndt-Jovin DJ, J. Cell Biol., 2005, 170, 619–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yanagawa M, Hiroshima M, Togashi Y, Abe M, Yamashita T, Shichida Y, Murata M, Ueda M and Sako Y, Sci. Signaling, 2018, 11, eaao1917. [DOI] [PubMed] [Google Scholar]
- 14.Rosenthal SJ, Tomlinson I, Adkins EM, Schroeter S, Adams S, Swafford L, McBride J, Wang Y, DeFelice LJ and Blakely RD, J. Am. Chem. Soc., 2002, 124, 4586–4594. [DOI] [PubMed] [Google Scholar]
- 15.Kovtun O, Tomlinson ID, Sakrikar DS, Chang JC, Blakely RD and Rosenthal SJ, ACS Chem. Neurosci., 2011, 2, 370–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang JC, Tomlinson ID, Warnement MR, Ustione A, Carneiro AMD, Piston DW, Blakely RD and Rosenthal SJ, J. Neurosci., 2012, 32, 8919–8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kovtun O, Sakrikar D, Tomlinson ID, Chang JC, Arzeta-Ferrer X, Blakely RD and Rosenthal SJ, ACS Chem. Neurosci., 2015, 6, 526–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bailey DM, Catron MA, Kovtun O, Macdonald RL, Zhang Q and Rosenthal SJ, ACS Chem. Neurosci., 2018, 9, 2534–2541. [DOI] [PubMed] [Google Scholar]
- 19.Thal LB, Tomlinson ID, Quinlan MA, Kovtun O, Blakely RD and Rosenthal SJ, ACS Chem. Neurosci., 2019, 10, 460–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenthal SJ, ACS Chem. Neurosci., 2019, 10, 30–32. [DOI] [PubMed] [Google Scholar]
- 21.Thal LB, Bailey DM, Kovtun O and Rosenthal SJ, in Chemical and Synthetic Approaches in Membrane Biology, ed. Shukla AK, Springer New York, New York, NY, 2017, pp. 219–230, DOI: 10.1007/8623_2016_12. [DOI] [Google Scholar]
- 22.Ji N, Magee JC and Betzig E, Nat. Methods, 2008, 5, 197–202. [DOI] [PubMed] [Google Scholar]
- 23.Laissue PP, Alghamdi RA, Tomancak P, Reynaud EG and Shroff H, Nat. Methods, 2017, 14, 657–661. [DOI] [PubMed] [Google Scholar]
- 24.Wichner SM, Mann VR, Powers AS, Segal MA, Mir M, Bandaria JN, DeWitt MA, Darzacq X, Yildiz A and Cohen BE, ACS Nano, 2017, 11, 6773–6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenthal SJ, Chang JC, Kovtun O, McBride JR and Tomlinson ID, Chem. Biol., 2011, 18, 10–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larson DR, Zipfel WR, Williams RM, Clark SW, Bruchez MP, Wise FW and Webb WW, Science, 2003, 300, 1434–1436. [DOI] [PubMed] [Google Scholar]
- 27.So MK, Xu C, Loening AM, Gambhir SS and Rao J, Nat. Biotechnol., 2006, 24, 339–343. [DOI] [PubMed] [Google Scholar]
- 28.Orndorff RL, Warnement MR, Mason JN, Blakely RD and Rosenthal SJ, Nano Lett., 2008, 8, 780–785. [DOI] [PubMed] [Google Scholar]
- 29.Biermann B, Sokoll S, Klueva J, Missler M, Wiegert J, Sibarita JB and Heine M, Nat. Commun., 2014, 5, 3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Varela JA, Dupuis JP, Etchepare L, Espana A, Cognet L and Groc L, Nat. Commun., 2016, 7, 10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Varela JA, Ferreira JS, Dupuis JP, Durand P, Bouchet-Tessier D and Groc L, Neurophotonics, 2016, 3, 041808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen O, Zhao J, Chauhan VP, Cui J, Wong C, Harris DK, Wei H, Han HS, Fukumura D, Jain RK and Bawendi MG, Nat. Mater., 2013, 12, 445–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mann VR, Powers AS, Tilley DC, Sack JT and Cohen BE, ACS Nano, 2018, 12, 4469–4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orfield NJ, McBride JR, Keene JD, Davis LM and Rosenthal SJ, ACS Nano, 2015, 9, 831–839. [DOI] [PubMed] [Google Scholar]
- 35.van Embden J, Jasieniak J and Mulvaney P, J. Am. Chem. Soc., 2009, 131, 14299–14309. [DOI] [PubMed] [Google Scholar]
- 36.Chen Y, Vela J, Htoon H, Casson JL, Werder DJ, Bussian DA, Klimov VI and Hollingsworth JA, J. Am. Chem. Soc., 2008, 130, 5026–5027. [DOI] [PubMed] [Google Scholar]
- 37.Orfield NJ, McBride JR, Wang F, Buck MR, Keene JD, Reid KR, Htoon H, Hollingsworth JA and Rosenthal SJ, ACS Nano, 2016, 10, 1960–1968. [DOI] [PubMed] [Google Scholar]
- 38.McBride J, Treadway J, Feldman LC, Pennycook SJ and Rosenthal SJ, Nano Lett., 2006, 6, 1496–1501. [DOI] [PubMed] [Google Scholar]
- 39.Reid KR, McBride JR, Freymeyer NJ, Thal LB and Rosenthal SJ, Nano Lett., 2018, 18, 709–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qdot Nanocrystals – Section 6.6, https://www.thermofisher.com/us/en/home/references/molecular-probes-the-handbook/ultrasensitive-detection-technology/qdot-nanocrystal-technology.html.
- 41.van Sark WGJHM, Frederix PLTM, Van den Heuvel DJ, Gerritsen HC, Bol AA, van Lingen JNJ, de Mello Donegá C and Meijerink A, J. Phys. Chem. B, 2001, 105, 8281–8284. [Google Scholar]
- 42.van Sark WGJHM, Frederix PLTM, Bol AA, Gerritsen HC and Meijerink A, ChemPhysChem, 2002, 3, 871–879. [Google Scholar]
- 43.Lee SF and Osborne MA, ChemPhysChem, 2009, 10, 2174–2191. [DOI] [PubMed] [Google Scholar]
- 44.Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen TM, Chi Chin M, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, Desaki AL, Desta T, Diep E, Dolbeare TA, Donelan MJ, Dong HW, Dougherty JG, Duncan BJ, Ebbert AJ, Eichele G, Estin LK, Faber C, Facer BA, Fields R, Fischer SR, Fliss TP, Frensley C, Gates SN, Glattfelder KJ, Halverson KR, Hart MR, Hohmann JG, Howell MP, Jeung DP, Johnson RA, Karr PT, Kawal R, Kidney JM, Knapik RH, Kuan CL, Lake JH, Laramee AR, Larsen KD, Lau C, Lemon TA, Liang AJ, Liu Y, Luong LT, Michaels J, Morgan JJ, Morgan RJ, Mortrud MT, Mosqueda NF, Ng LL, Ng R, Orta GJ, Overly CC, Pak TH, Parry SE, Pathak SD, Pearson OC, Puchalski RB, Riley ZL, Rockett HR, Rowland SA, Royall JJ, Ruiz MJ, Sarno NR, Schaffnit K, Shapovalova NV, Sivisay T, Slaughterbeck CR, Smith SC, Smith KA, Smith BI, Sodt AJ, Stewart NN, Stumpf KR, Sunkin SM, Sutram M, Tam A, Teemer CD, Thaller C, Thompson CL, Varnam LR, Visel A, Whitlock RM, Wohnoutka PE, Wolkey CK, Wong VY, Wood M, Yaylaoglu MB, Young RC, Youngstrom BL, Feng Yuan X, Zhang B, Zwingman TA and Jones AR, Nature, 2006, 445, 168–176. [DOI] [PubMed] [Google Scholar]
- 45.Ho J, Tumkaya T, Aryal S, Choi H and Claridge-Chang A, Nat. Methods, 2019, 16, 565–566. [DOI] [PubMed] [Google Scholar]
- 46.Efros AL and Rosen M, Phys. Rev. Lett., 1997, 78, 1110–1113. [Google Scholar]
- 47.Frantsuzov PA, Volkán-Kacsó S and Jankó B, Phys. Rev. Lett., 2009, 103, 207402. [DOI] [PubMed] [Google Scholar]
- 48.Qin W and Guyot-Sionnest P, ACS Nano, 2012, 6, 9125–9132. [DOI] [PubMed] [Google Scholar]
- 49.Lim SJ, Ma L, Schleife A and Smith AM, Coord. Chem. Rev., 2016, 320–321, 216–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Talapin DV, Nelson JH, Shevchenko EV, Aloni S, Sadtler B and Alivisatos AP, Nano Lett., 2007, 7, 2951–2959. [DOI] [PubMed] [Google Scholar]
- 51.Luo Y and Wang LW, ACS Nano, 2010, 4, 91–98. [DOI] [PubMed] [Google Scholar]
- 52.Rainò G, Stöferle T, Moreels I, Gomes R, Kamal JS, Hens Z and Mahrt RF, ACS Nano, 2011, 5, 4031–4036. [DOI] [PubMed] [Google Scholar]
- 53.Lorenzon M, Christodoulou S, Vaccaro G, Pedrini J, Meinardi F, Moreels I and Brovelli S, Nat. Commun., 2015, 6, 6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kovtun O, Tomlinson ID, Bailey DM, Thal LB, Ross EJ, Harris L, Frankland MP, Ferguson RS, Glaser Z, Greer J and Rosenthal SJ, Chem. Phys. Lett., 2018, 706, 741–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW and Sanes JR, Neuron, 2000, 28, 41–51. [DOI] [PubMed] [Google Scholar]
- 56.Karra D and Dahm R, J. Neurosci., 2010, 30, 6171–6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oh E, Liu R, Nel A, Gemill KB, Bilal M, Cohen Y and Medintz IL, Nat. Nanotechnol., 2016, 11, 479. [DOI] [PubMed] [Google Scholar]
- 58.Selim KMK, Xing Z-C, Choi M-J, Chang Y, Guo H and Kang I-K, Nanoscale Res. Lett., 2011, 6, 528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Clift MJD, Varet J, Hankin SM, Brownlee B, Davidson AM, Brandenberger C, Rothen-Rutishauser B, Brown DM and Stone V, Nanotoxicology, 2011, 5, 664–674. [DOI] [PubMed] [Google Scholar]
- 60.Zhang M, Bishop BP, Thompson NL, Hildahl K, Dang B, Mironchuk O, Chen N, Aoki R, Holmberg VC and Nance E, Nanoscale Adv., 2019, 1, 3424–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.