Abstract
The transcription factor BHLHE40 is an emerging regulator of the immune system. Recent studies suggest that BHLHE40 regulates type 2 immunity, but this has not been demonstrated in vivo. We found that BHLHE40 is required in T cells for a protective TH2 cell response in mice infected with the helminth Heligmosomoides polygyrus bakeri (H. polygyrus). H. polygyrus elicited changes in gene and cytokine expression by lamina propria CD4+ T cells, many of which were BHLHE40-dependent, including production of the common beta (βC, CSF2RB) chain family cytokines GM-CSF and IL-5. In contrast to deficiency in GM-CSF or IL-5 alone, loss of both GM-CSF and IL-5 signaling impaired protection against H. polygyrus. Overall, we show that BHLHE40 regulates the TH2 cell transcriptional program during helminth infection to support normal expression of Csf2, Il5, and other genes required for protection and reveal unexpected redundancy of βC chain-dependent cytokines previously thought to possess substantially divergent functions.
Introduction
The transcription factor BHLHE40 has recently been characterized as a key regulator of the hematopoietic system (1–8). We and others have discovered that BHLHE40 modulates TH1 and TH17 cell cytokine production in infection and autoimmunity, particularly by controlling GM-CSF and IL-10 (1–3, 5, 6, 8). Several studies have also suggested a role for BHLHE40 as a regulator of type 2 immunity, possibly by a TH2 cell- (9, 10) or myeloid cell-intrinsic role (7).
Helminthic worms manipulate the immune system to establish chronic infections in diverse sites, often resulting in significant tissue damage (11, 12). Helminths elicit production of alarmins including IL-25, IL-33, and thymic stromal lymphopoietin (TSLP) and classic type 2 cytokines such as IL-4, IL-5, and IL-13 (11, 12). The helminth Heligmosomoides polygyrus bakeri (H. polygyrus) is a natural pathogen of mice and an oft-employed experimental model (12, 13). Several cytokines contribute to protective immunity against H. polygyrus, including IL-4 and IL-25, while IL-5, IL-33, and TSLP are considered dispensable for control of this infection (14–17).
In many other instances of type 2 immunity, IL-5 plays a crucial role (11, 18, 19). Controversy previously existed as to whether IL-5 was protective against helminths, but it is now appreciated that deficiency in IL-5 only affects immunity to select parasites and sometimes in a life cycle stage-dependent manner (11, 18). IL-5 binds to the unique IL-5Rα chain paired with the common beta (βC) chain, which is shared with the receptors for GM-CSF and IL-3 (20). While IL-3 signaling is maintained in mice in the absence of the βC chain (Csf2rb) through a unique beta chain (βIL3, Csf2rb2), GM-CSF and IL-5 signaling is abrogated in Csf2rb−/− mice (20). Despite absolute dependence on this shared receptor chain, which is largely responsible for downstream signaling (20), known roles for GM-CSF and IL-5 are divergent. GM-CSF has been proposed to be a central regulator of inflammation driven by TH1 and TH17 cells and affects both autoimmunity and infection (21–26). GM-CSF also contributes to pathological TH2 cell responses in allergy and asthma (27, 28). However, little is known regarding GM-CSF during type 2 infections, though it is thought to be dispensable during infection with the helminth Nippostrongylus brasiliensis (29).
We have found that BHLHE40 and βC chain-dependent cytokine signaling are essential to protective memory to secondary H. polygyrus infection. We defined a T cell-intrinsic role for BHLHE40 to support the helminth-induced gene signature in lamina propria CD4+ T cells, one of the first such analyses in helminth-infected mice. In vitro and in vivo, TH2 cells exhibited BHLHE40-dependent production of GM-CSF, IL-5, and other cytokines. Secondary infection of mice deficient in both GM-CSF and IL-5 signaling resulted in severely impaired protective immunity. Overall, BHLHE40 serves as a pivotal regulator of the TH2 cell transcriptional response to helminth infection, in part by modulating GM-CSF and IL-5, and reveals redundant roles for these cytokines not observed during deficiency of either factor alone.
Materials and Methods
Mice and infections
Il10−/−, Csf2−/−, Csf2rb−/−, Cd4-Cre, and LysM-Cre mice were obtained from Jackson. C57BL/6 mice were obtained from Jackson or Taconic. Bhlhe40−/− (2, 30), Bhlhe40GFP+ (3), and Bhlhe40fl/fl (5) mice have been reported. Mice were maintained in our specific pathogen free facility. Sex-matched littermates were used for experiments whenever possible. Animal experiments were approved by the Animal Studies Committee of Washington University.
Infective Heligmosomoides polygyrus bakeri third-stage larvae (L3) were prepared as described (13) and 200 L3 were given using a 20-gauge gavage needle. Fecal eggs were counted on a McMaster counting chamber (Chalex LLC) (13). For rechallenge experiments, infection was cleared with pyrantel pamoate p.o. (2 mg, Columbia Laboratories) on d14 and 15 after primary infection. Mice were rested for 3–4 weeks before reinfection. Blood was collected at d21 post-rechallenge to assess antibody titers. Mice were sacrificed on d17 or 22 of secondary infection for assessment of cellular responses, macroscopic intestinal granulomas, and intestinal worm burden.
For in vivo antibody blockade, mice were injected i.p. with 300 μg of αGM-CSF (Leinco, MP1–22E9), αIL-5 (BioXCell, TRFK5), or control polyclonal rat IgG (Sigma Aldrich I4131) every other day from the day of reinfection.
Cell culture
The EasySep mouse naïve CD4+ T cell isolation kit (Stemcell Technologies) was used to isolate splenic T cells. Cells were cultured in complete IMDM (cIMDM, with added 10% FBS, L-glutamine, sodium pyruvate, non-essential amino acids, penicillin/streptomycin, and 2-ME) with plate-bound anti-CD3 antibody (2 μg/mL) and anti-CD28 antibody (2 μg/ml). IL-4 (10 ng/mL) and anti-IFN-γ (5 μg/mL) were added at the start of culture to generate TH2 cells. Cells were split at d3. On d4, cells were counted and stimulated for 24 hours with plate-bound anti-CD3 and anti-CD28 for assessment of secreted cytokines. In some experiments, naïve CD4+ T cells were labelled with 40 μM CFSE (Tonbo) for 10 min at room temperature, followed by culture under TH2 polarizing conditions as above and assessment of CFSE dilution by flow cytometry on d3.
ELISA
Nunc Maxisorp plates were coated with capture antibodies (for cytokine ELISAs) or H. polygyrus lysate (for H. polygyrus-specific IgG1 ELISA) overnight. Following blocking, culture supernatants (cytokine ELISAs) or serum (IgG1 ELISA) were added, followed by incubation with cytokine-detection biotinylated antibodies or horseradish peroxidase (HRP)-conjugated anti-mouse IgG1 antibody. For cytokine ELISAs, streptavidin-conjugated horseradish peroxidase (HRP) was added. Substrate solution (BD OptEIA 555214) was added, reactions were stopped with 1 M H3PO4, and absorbance was read at OD450. For cytokine ELISAs, standard curves were generated with purified cytokines. For the IgG1 ELISA, serum was diluted between 10−2 and 10−8 and the last well with an OD450 above 0.100 represented the titer. The Mouse IL-4 and IL-5 ELISA Max Standard Sets (BioLegend), Mouse IL-13 DuoSet ELISA (R&D), and C57BL/6 Mouse Immunoglobulin Panel (SouthernBiotech) were used. For detection of GM-CSF, biotinylated αGM-CSF (BioLegend, MP1–31G6), unconjugated αGM-CSF (BioLegend, MP1–22E9), and murine GM-CSF (Peprotech) were used.
To generate H. polygyrus lysate, adult worms were washed in PBS and ground in 1 mL of PBS. Debris was pelleted by centrifugation at 16,000 g for 20 minutes at 4 °C. The supernatant was passed through a 0.2 μm filter and stored at −80 °C.
Microscopy
The proximal 6 cm of the small intestine were formed into a swiss roll, frozen in OCT media, and cryosectioned. For H&E staining, sections were fixed in 4% paraformaldehyde (PFA). For immunofluorescent staining, sections were fixed in acetone and blocked with CAS-Block (Invitrogen). Staining was performed with Cy3-anti-α-smooth muscle actin (Sigma, 1A4) and AlexaFluor647-anti-F4/80 (BioLegend, BM8) diluted in CAS-Block. Sections were mounted with Abcam Fluoroshield Mounting Medium with DAPI. Images were captured with a Nikon Eclipse E800 microscope and MicroPublisher 5.0 RTV or EXi Blue cameras (QImaging) using QCapture software. Fluorescence images were merged and levelled in Adobe Photoshop.
Cell isolation
Peritoneal cells were collected by lavage. Spleens were homogenized through a 70 μm filter. SILP cells were isolated from 4 cm of the proximal small intestine, cleaned of fecal contents and cut into 2–3 pieces. Tissues were shaken twice in 1x HBSS (with added 10% FBS, HEPES, EDTA, and DTT) for 20 minutes, vigorous vortexed, and then shaken at 220 rpm at 37 °C for 1 hour in an Innova 4330 shaker (New Brunswick Scientific) with Collagenase IV (0.8 mg/mL, Sigma C5138) in RPMI 1640 (with added 10% FBS, L-glutamine, penicillin/streptomycin, and 2-ME). RBC were lysed when needed.
Flow cytometry
Cells were blocked with αCD16/32 (BioXCell, 2.4G2) and stained with antibodies before flow cytometry on LSRFortessa X20 and FACSCanto II instruments (BD). Analysis was performed with FlowJo software (Treestar). The following antibodies were purchased from BioLegend: αCD3 APC/Cy7 (17A2), αCD4 PE/Cy7 or BV510 (RM4–5), αCD11b PE/Cy7 or APC/Cy7 (M1/70), αCD45.2 FITC (104), αCD64 APC (X54–5/7.1), αCD102 FITC or AF647 (3C4), αCD115 BV421 (AFS98), αI-A/I-E BV510 or PB (M5/114.15.2), αIL-4 APC (11B11), αIL-5 BV421 (TRFK5), αLy6C APC/Cy7 or BV510 (HK1.4), and αTCRβ PE (H57–597). The following antibodies were purchased from Tonbo: αCD8 PerCP/Cy5.5 or APC/Cy7 (53–6.7), αCD11b PE/Cy7 or APC/Cy7 (M1/70), αCD19 PerCP/Cy5.5 (1D3), αCD45 RF710 (30-F11), αCD115 PE (AFS98), αF4/80 PerCP/Cy5.5 (BM8.1), and αI-A/I-E RF710 (M5/114.15.2). The following antibodies were purchased from Becton Dickinson: αCD45 BV510 (30-F11), αGM-CSF BV421 (MP1–22E9), and αSiglec-F PE (E50–2440). αIL-5 FITC (TRFK5) was purchased from Leinco. Unconjugated αRELMα was purchased from Peprotech (500-P214).
SILP macrophages were gated as CD45+F4/80+CD64+MHC-II+Ly6C−, while GMMs were gated as CD45+F4/80+CD64+MHC-IIloLy6Clo. SILP eosinophils were gated as CD45+F4/80+CD64−CD11b+SSC-Ahi (validated by Siglec-F staining in some experiments). SILP CD3+ T cells were gated as CD45+F4/80−CD64−CD19−CD3+. SILP CD4+ T cells were gated as CD19−F4/80−TCRβ+CD4+CD8−. LPMs were gated as CD115+CD11b+ICAM2+MHC-IIlo. Peritoneal eosinophils were gated as Siglec-F+ICAM2−. Peritoneal CD4+ T cells were gated as ICAM2−TCRβ+CD4+CD8−.
For intracellular cytokine staining of T cells, total SILP and peritoneal cells were cultured in cIMDM for 3–4 hours at 37 °C in the presence of PMA (50 ng/ml), ionomycin (1 μM), and brefeldin A (1 μg/ml). Cells were then surface stained and fixed with 4% PFA. To permeabilize the cells, samples were washed with 1x Perm/Wash buffer (BD 554714) and stained for 20 minutes at 4 °C. For RELMα staining, the True-Nuclear Transcription Factor Buffer set (BioLegend 424401) was used.
Microarrays
CD4+ SILP T cells were sorted using a FACSAria II (BD). RNA was purified with E.Z.N.A. MicroElute Total RNA kits (Omega Bio-Tek). cDNA was synthesized from RNA (NuGen Pico SL) followed by analysis on Affymetrix Mouse Gene 1.0 ST microarrays. CEL files were normalized with the DNASTAR ArrayStar program. Genes with an expression value of >5 (in log 2 scale) in at least one replicate were considered expressed. Lists of differentially expressed genes required a p-value significance of ≤0.01 by the moderated t-test. Morpheus was used to generate heatmaps (Broad). GSEA software (Broad) was used to analyze enrichment of C5 database gene sets. Microarray data have been deposited in the Gene Expression Omnibus under accession number GSE135182 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE135182).
Chromatin immunoprecipitation sequencing (ChIP-Seq)
Samples GSM3094815 and GSM3094816 from GSE113054 (5) were reanalyzed for BHLHE40 binding at the Csf2 and Il5 loci. Previously published peak calls were downloaded from the Gene Expression Omnibus.
Statistics
Statistical analysis was performed with Prism 7 (GraphPad) as described in the figure legends.
Results
BHLHE40 is required for control of H. polygyrus rechallenge
Primary oral challenge of C57BL/6 mice with H. polygyrus results in chronic infection, but rechallenge evokes a protective recall response which limits infection (31). To assess whether BHLHE40 was required for immunity to H. polygyrus, we challenged C57BL/6 Bhlhe40+/+ and Bhlhe40−/− mice with infective H. polygyrus larvae (L3), cured them by treatment with pyrantel pamoate, and rechallenged them with infective L3. Mature H. polygyrus worms mate in the intestinal lumen, resulting in females laying eggs which are then shed in the feces (13). While Bhlhe40+/+ and Bhlhe40−/− mice exhibited similar parasite fecal egg burdens during primary infection, Bhlhe40−/− mice had a much higher egg burden during secondary infection as compared to Bhlhe40+/+ mice (Fig. 1A). Increased production of eggs could result from impaired expulsion of the parasites or enhanced parasite fitness. While there was a trend towards increased adult worm burden in Bhlhe40−/− as compared to Bhlhe40+/+ mice after rechallenge (Supplemental Fig. 1A), this did not reach statistical significance, indicating that their increased egg burden was primarily due to increased worm fecundity. H. polygyrus-rechallenged Bhlhe40+/+ and Bhlhe40−/− mice had dramatically different pathology. Type 2 granulomas can form around developing parasites and have been correlated with protective immunity (31). Small intestines from Bhlhe40+/+ mice exhibited many granulomas, while those from Bhlhe40−/− mice resembled healthy tissue (Fig. 1B, 1C). Histological analysis and immunostaining showed reduced immune infiltration and damage to the smooth muscle layer in H. polygyrus-rechallenged Bhlhe40−/− as compared to Bhlhe40+/+ mice (Fig. 1D, 1E).
Figure 1. BHLHE40 is required for a protective recall response to H. polygyrus.
(A and B) H. polygyrus-infected Bhlhe40+/+ and Bhlhe40−/− mice were analyzed for (A) quantitation of H. polygyrus eggs/gram feces and (B) small intestine morphology after secondary infection. Arrows point to granulomas. Scale bar, 1 cm. (C) Quantitation of intestinal granulomas. (D and E) H. polygyrus-rechallenged Bhlhe40+/+ and Bhlhe40−/− mice were analyzed histologically on adjacent sections by (D) hematoxylin and eosin staining and (E) immunostaining for F4/80, α-SMA, and DAPI on swiss rolls of the proximal small intestine (2 representative lesions from each genotype). G, granuloma. M, muscle layers. P, peritoneal space. V, villi. Scale bar, 200 μm. (F and G) Naïve and H. polygyrus-rechallenged Bhlhe40+/+ and Bhlhe40−/− mice were analyzed by flow cytometry for (F) SILP myeloid cells and (G) quantitation as in (F). Data are representative of or pooled from at least 3 independent experiments. Data are mean ± s.e.m. Significance calculated with an unpaired Student’s t-test.
Flow cytometry of the small intestine from helminth-infected mice is challenging because of extensive tissue remodeling and enhanced mucus production. Therefore, we adapted a previously published protocol for intestinal flow cytometry for this purpose (32). When we explored the cellular composition of the small intestine lamina propria (SILP) following H. polygyrus rechallenge of Bhlhe40+/+ mice by flow cytometry, we found both CD45+CD64+F4/80+MHC-II+Ly6C− resident macrophages (33, 34) and another CD45+F4/80+CD64+MHC-IIloLy6Clo autofluorescent population, which we termed granuloma-associated monocytes/macrophages (GMMs) (Fig. 1F, 1G). This latter population may correspond to previously described clodronate-sensitive alternatively activated macrophages seen by immunostaining during H. polygyrus infection (35). SILP CD45+F4/80+CD64−CD11b+SSC-Ahi eosinophils were also significantly increased after secondary H. polygyrus infection (Fig. 1F, 1G, Supplemental Fig. 1B). However, GMMs and eosinophils were greatly reduced in H. polygyrus-rechallenged Bhlhe40−/− as compared to Bhlhe40+/+ mice, in contrast to SILP CD3+ T cells (Fig. 1F, 1G, Supplemental Fig. 1C). Because H. polygyrus infection is known to change the cellular composition of the peritoneal cavity (11, 36), we assessed accumulation of LPMs and peritoneal eosinophils, and found severe reductions in both populations in H. polygyrus-rechallenged Bhlhe40−/− as compared to Bhlhe40+/+ mice, as well as impaired LPM polarization as assessed by resistin-like molecule alpha (RELMα) staining (Supplemental Fig. 1D–H). In contrast, serum H. polygyrus-specific IgG1 titers were not affected by loss of BHLHE40 (Supplemental Fig. 1I). These data indicated impaired responses by multiple myeloid cell lineages to H. polygyrus rechallenge in Bhlhe40−/− mice.
BHLHE40 is required in T cells for a normal immune response to H. polygyrus
We next employed Cd4-Cre+ Bhlhe40fl/fl and LysM-Cre+ Bhlhe40fl/fl mice to address whether loss of BHLHE40 specifically in T cells or myeloid cells recapitulated the phenotype of Bhlhe40−/− mice during secondary H. polygyrus infection. Cd4-Cre+ Bhlhe40fl/fl mice were severely impaired in controlling H. polygyrus rechallenge and lacked intestinal granulomas as compared to Cd4-Cre− Bhlhe40fl/fl mice (Fig. 2A, 2B). When we assessed the cellular response to secondary infection, we found that loss of BHLHE40 selectively in T cells was sufficient to perturb both the SILP and peritoneal myeloid cell responses, as well as to reduce SILP T cells in some animals (Fig. 2C, Supplemental Fig. 1J–M). In contrast, LysM-Cre+ Bhlhe40fl/fl mice were able to control infection comparably to LysM-Cre− Bhlhe40fl/fl mice (Fig. 2D–F). Therefore, BHLHE40 expression was required in T cells to control secondary H. polygyrus infection, but was dispensable in LysM-expressing myeloid cells.
Figure 2. BHLHE40 is required in T cells for a normal memory response to H. polygyrus.
(A) H. polygyrus-rechallenged Cd4-Cre− Bhlhe40fl/fl and Cd4-Cre+ Bhlhe40fl/fl mice were analyzed for quantitation of H. polygyrus eggs/gram feces over time. (B) Naïve and H. polygyrus-rechallenged Cd4-Cre− Bhlhe40fl/fl and Cd4-Cre+ Bhlhe40fl/fl mice were analyzed for quantitation of intestinal granulomas. (C) Naïve and H. polygyrus-rechallenged Cd4-Cre− Bhlhe40fl/fl and Cd4-Cre+ Bhlhe40fl/fl mice were analyzed by flow cytometry for quantitation of SILP myeloid cells. (D) H. polygyrus-rechallenged LysM-Cre− Bhlhe40fl/fl and LysM-Cre+ Bhlhe40fl/fl mice were analyzed for quantitation of H. polygyrus eggs/gram feces over time. (E) Naïve and H. polygyrus-rechallenged LysM-Cre− Bhlhe40fl/fl and LysM-Cre+ Bhlhe40fl/fl mice were analyzed for quantitation of intestinal granulomas. (F) Naïve and H. polygyrus-rechallenged LysM-Cre− Bhlhe40fl/fl and LysM-Cre+ Bhlhe40fl/fl mice were analyzed by flow cytometry for quantitation of SILP myeloid cells. Data are representative of or pooled from at least 2 independent experiments. Data are mean ± s.e.m. Significance calculated with an unpaired Student’s t-test.
BHLHE40 is required for a normal CD4+ T cell transcriptional response to H. polygyrus
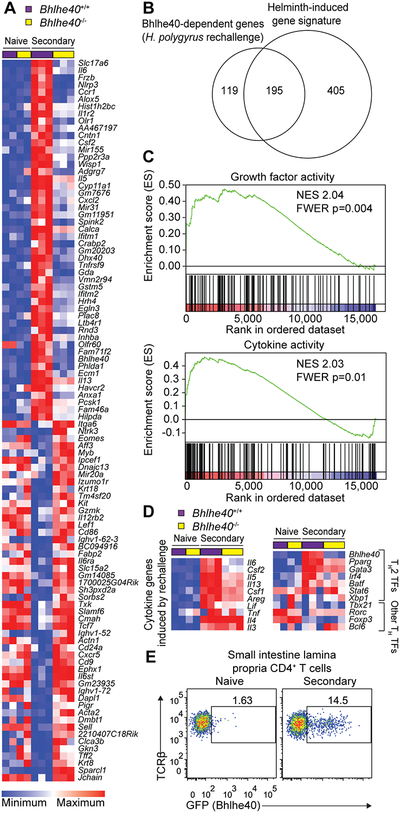
We next asked whether loss of BHLHE40 dysregulated CD4+ T cell gene expression in response to H. polygyrus rechallenge. To address this, we sorted CD4+ T cells from the SILP of naïve and rechallenged Bhlhe40+/+ and Bhlhe40−/− mice for gene expression microarrays. By comparing CD4+ T cells from naïve and H. polygyrus-rechallenged Bhlhe40+/+ mice, we defined a helminth-induced signature which included transcripts for cytokines (including Areg, Il3, Il4, Il5, Il6, Il13, Csf1, Csf2, Lif, Tnf, Tnfsf11), cytokine receptors (including Il1rl1, Il1r2, Il17rb), and transcription factors (including Atf3, Bhlhe40, Gata3, Nfil3, Pparg, Rbpj, Vdr, and Zeb2) (Fig. 3A, Supplemental Fig. 2A). When we compared differentially expressed transcripts between H. polygyrus-elicited and naive SILP CD4+ T cells, we found a considerable overlap with the recently described transcriptional profile of HDM-elicited airway TH2 cells (including Bhlhe40, Cd200r1, Il6, Plac8, and Igfbp7) (Fig. 3A, Supplemental Fig. 2A) (10), demonstrating significant conservation of the TH2 cell transcriptional program independent of tissue environment and stimulus. When we assessed BHLHE40-dependent genes after H. polygyrus rechallenge, we found that a significant majority were part of the helminth-induced signature and that BHLHE40-dependent genes were distinct in CD4+ T cells from naïve and H. polygyrus-rechallenged mice (Fig. 3A, 3B, Supplemental Fig. 2B, 2C).
Figure 3. Loss of BHLHE40 dysregulates the CD4+ T cell transcriptional response to H. polygyrus rechallenge.
(A) Gene expression microarray data were analyzed for the 100 most differentially expressed probe sets in SILP CD4+ T cells from H. polygyrus-rechallenged Bhlhe40+/+ and Bhlhe40−/− mice. (B) Gene expression microarray data were analyzed for shared and unique Bhlhe40-dependent genes (≥2-fold differentially expressed between SILP CD4+ T cells from H. polygyrus-rechallenged Bhlhe40+/+ and Bhlhe40−/− mice) with the helminth-induced signature (≥2-fold differentially expressed between SILP CD4+ T cells from naïve and H. polygyrus-rechallenged Bhlhe40+/+ mice), depicted as a Venn diagram. (C) GSEA of gene expression microarray data for selected C5 gene sets enriched in Bhlhe40+/+ versus Bhlhe40−/− SILP CD4+ T cells from H. polygyrus-rechallenged mice. NES, normalized enrichment score. FWER, family-wise error rate. (D) Gene expression microarray data were analyzed for (left) expression of cytokines induced by H. polygyrus rechallenge and (right) expression of TH2 and T helper cell lineage-specifying transcription factors in SILP CD4+ T cells from naïve and H. polygyrus-rechallenged Bhlhe40+/+ and Bhlhe40−/− mice. TF, transcription factor. (E) Flow cytometry of Bhlhe40GFP transgene reporter expression in SILP CD4+ T cells from Bhlhe40GFP+ mice. Data in (E) are representative of 2 independent experiments. Microarray data are from 1 experiment.
When we used gene set enrichment analysis (GSEA) to look for BHLHE40-dependent gene modules, we noted that two of the most enriched sets in Bhlhe40+/+ as compared to Bhlhe40−/− CD4+ T cells after H. polygyrus rechallenge were “growth factor activity” and “cytokine activity,” reflecting altered expression of cytokine genes including Areg, Il5, Il6, Il13, Csf1, Csf2, and Lif, but not Il3, Il4, or Tnf (Fig. 3C, 3D). Furthermore, when we assessed differential expression of the lineage-specifying transcription factors of each TH cell subset as well as a recently defined set of transcriptional regulators of in vitro-polarized TH2 cells (9), we observed reduced expression of Pparg in Bhlhe40−/− as compared to Bhlhe40+/+ CD4+ T cells from H. polygyrus-rechallenged mice (2.4-fold reduced), but only subtle changes in other factors including Gata3 (1.4-fold reduced) (Fig. 3D). As PPARγ is required for normal TH2 cell responses and protective immunity to H. polygyrus, this may indicate that some effects of BHLHE40 deficiency are indirect (37, 38). Bhlhe40 expression was induced in mice experiencing secondary infection as compared to naïve mice (Fig. 3D). Using Bhlhe40GFP bacterial artificial chromosome reporter mice (3), we observed a marked increase in SILP GFP+ CD4+ T cells after H. polygyrus rechallenge (Fig. 3E). Taken together, these data showed that BHLHE40 was induced by SILP CD4+ T cells in response to H. polygyrus rechallenge and was critical for their normal transcriptional program.
Loss of BHLHE40 impairs CD4+ T cell cytokine production in response to H. polygyrus
Next, we restimulated SILP cells from naïve and rechallenged Cd4-Cre− Bhlhe40fl/fl and Cd4-Cre+ Bhlhe40fl/fl mice ex vivo with PMA and ionomycin to assess cytokine production. Rechallenge with H. polygyrus induced production of IL-4, IL-5, and IL-13 from Cd4-Cre− Bhlhe40fl/fl and Cd4-Cre+ Bhlhe40fl/fl CD4+ T cells; however, BHLHE40 was required for normal frequencies of single- and multi-cytokine-producing cells, largely due to reductions in IL-5+ and IL-13+ CD4+ T cells, consistent with the results of our microarray gene expression analysis (Fig. 4A–C). Because Csf2 was upregulated by H. polygyrus rechallenge in a BHLHE40-dependent fashion, we also assessed GM-CSF and found that it was markedly induced by rechallenge (~35% of Cd4-Cre− Bhlhe40fl/fl CD4+ T cells) and that this required BHLHE40 (~10% of Cd4-Cre+ Bhlhe40fl/fl CD4+ T cells) (Fig. 4D, 4E). SILP CD4+ T cells could coproduce GM-CSF and IL-5 (Supplemental Fig. 3A). TH2 cells disseminate widely in mice infected with H. polygyrus (36). CD4+ T cell cytokine responses in the peritoneal cavity and mesenteric lymph nodes from H. polygyrus-rechallenged Cd4-Cre− Bhlhe40fl/fl and Cd4-Cre+ Bhlhe40fl/fl mice were generally consistent with those in the SILP (Supplemental Fig. 3B and data not shown). To assess whether BHLHE40 was also required for normal cytokine production by a more homogenous population of TH2 cells, we differentiated naïve splenic CD4+ T cells into TH2 cells and restimulated them to assess cytokine production. We found that BHLHE40 was essential for production of GM-CSF and that loss of BHLHE40 also impaired in vitro production of IL-4, IL-5, and IL-13 (Supplemental Fig. 3C). As published literature is unclear as to whether Bhlhe40 controls TH cell proliferation (1, 6), we assessed this in vitro with Bhlhe40+/+ and Bhlhe40−/− TH2 cells and found that BHLHE40 was dispensable for TH2 cell proliferation (Supplemental Fig. 3D, 3E). As we have previously published BHLHE40 ChIP-Seq for in vitro-polarized TH1 cells (5), we also assessed whether BHLHE40 was bound near the Csf2 and Il5 loci in this TH subset. BHLHE40 was bound to the Csf2 locus, while the nearest peaks to the Il5 locus were more proximal to Rad50 and Irf1 (Supplemental Fig. 3F, 3G). Taken together, these data indicated that BHLHE40 was required in vitro and in vivo for normal TH2 cell function.
Figure 4. BHLHE40 is required for normal CD4+ T cell production of βC chain-dependent cytokines.
(A-C) Naïve and H. polygyrus-rechallenged Cd4-Cre− Bhlhe40fl/fl and Cd4-Cre+ Bhlhe40fl/fl mice were analyzed by flow cytometry for (A) IL-4-, IL-5-, and IL-13-producing CD4+ T cells, (B) quantitation of the frequency of IL-4+, IL-5+, and IL-13+ CD4+ T cells, and (C) quantitation of the frequency of CD4+ T cells producing one or more cytokines after in vitro PMA/ionomycin stimulation of SILP cells. (D and E) Naïve and H. polygyrus-rechallenged Cd4-Cre− Bhlhe40fl/fl and Cd4-Cre+ Bhlhe40fl/fl mice were analyzed by flow cytometry for (D) GM-CSF-producing CD4+ T cells and (E) quantitation as in (D) after in vitro PMA/ionomycin stimulation of SILP cells. Data are representative of or pooled from 2 independent experiments. Data are mean ± s.e.m. Significance calculated with an unpaired Student’s t-test.
As BHLHE40 is a known repressor of IL-10 (3, 5, 6), we also assessed IL-10 production from SILP CD4+ T cells. Indeed, SILP CD4+ T cells lacking BHLHE40 produced significantly higher levels of IL-10 after H. polygyrus rechallenge (Supplemental Fig. 4A). Nevertheless, genetic deletion of IL-10 in Bhlhe40−/− Il10−/− mice was not sufficient to restore control of infection or normal intestinal granulomatous pathology (Supplemental Fig. 4B, 4C). These data demonstrated a key role for BHLHE40 in CD4+ T cell cytokine responses to helminth infection, notably controlling production of the βC chain family cytokines GM-CSF and IL-5.
Loss of the βC chain impairs protective memory to H. polygyrus
As protective memory responses to H. polygyrus rechallenge are unaffected by IL-5 blockade (14), we first asked whether loss of GM-CSF signaling was sufficient to impair control of a secondary infection with H. polygyrus. Genetic deletion of GM-CSF (Csf2−/− mice) did not substantially affect control of H. polygyrus infection as compared to Csf2+/+ mice (Supplemental Fig. 4D–F). As GM-CSF and IL-5 were individually dispensable, we then rechallenged Csf2rb+/+ and Csf2rb−/− mice. Csf2rb−/− mice had a severe defect in control of H. polygyrus as compared to Csf2rb+/+ mice and did not form intestinal granulomas or develop normal SILP and peritoneal myeloid cell responses (Fig. 5A–C, Supplemental Fig. 4G–I). To exclude a role for βC chain-dependent IL-3 signaling, we singly blocked GM-CSF or IL-5 signaling with neutralizing antibodies or blocked both cytokines together. Double, but not single, blockade resulted in impaired protective immunity (Fig. 5D, 5E). Taken together, these data indicated that BHLHE40 was required for normal production of GM-CSF and IL-5 from CD4+ T cells and that these cytokines were collectively, but not individually, critical for control of H. polygyrus rechallenge.
Figure 5. βC chain signaling is required for control of H. polygyrus rechallenge.
(A) H. polygyrus-rechallenged Csf2rb+/+ and Csf2rb−/− mice were analyzed for quantitation of H. polygyrus eggs/gram feces over time. (B) Naïve and H. polygyrus-rechallenged Csf2rb+/+ and Csf2rb−/− mice were analyzed for quantitation of intestinal granulomas. (C) Naïve and H. polygyrus-rechallenged Csf2rb+/+ and Csf2rb−/− mice were analyzed by flow cytometry for quantitation of SILP myeloid cells. (D) Wild type mice were rechallenged with H. polygyrus concurrent with control IgG, αGM-CSF, αIL-5, or αGM-CSF plus αIL-5 antibody treatment and were analyzed for quantitation of H. polygyrus eggs/gram feces over time. (E) Naïve (open circles) and H. polygyrus-rechallenged mice (filled triangles) treated as in (D) were analyzed for quantitation of intestinal granulomas. Data are pooled from 2 independent experiments (A-C) or are from 1 experiment (D and E). Data are mean ± s.e.m. Significance calculated with an unpaired Student’s t-test.
Discussion
Our findings establish BHLHE40 as an essential, T cell-intrinsic regulator of type 2 immune responses. Mechanistically, we found that SILP CD4+ T cells activated by H. polygyrus rechallenge required BHLHE40 for normal expression of many helminth-induced genes, most notably Csf2 and Il5, and that these and potentially other factors were necessary for normal myeloid cell-mediated responses to H. polygyrus. In the absence of BHLHE40 in T cells, we observed a near-complete loss of the myeloid cell-mediated response, which encapsulates developing parasites in a type 2 granuloma structure and directly targets them through effector mechanisms including arginase-1 activity (35). These data also demonstrate the severity of type 2 immunopathology in the H. polygyrus model, as an impaired immune response in the absence of BHLHE40 protected mice from severe damage to the intestinal wall and smooth muscle layers. While our data suggest that BHLHE40 uniquely regulates some TH2-specific genes, our work and that of others has now established a broader role for BHLHE40 in TH1, TH2, and TH17 cells as a pivotal regulator of GM-CSF, IL-10, and other cytokines, acting as both a direct and indirect transcriptional regulator of these loci (1–3, 5, 6). These data and recent work on c-Maf (39) suggest that T cell production of many cytokines may be controlled by multi-lineage transcription factors in addition to lineage-restricted factors. In light of the critical role for BHLHE40 in multiple CD4+ T cell subsets, clinical targeting of BHLHE40 may possess therapeutic potential.
A significant effect of BHLHE40 deficiency on CD4+ T cells was reduced production of the βC chain family members GM-CSF and IL-5. Using our previously published TH1 cell ChIP-Seq data set (5), we found that BHLHE40 bound to the Csf2 locus, while Il5-proximal peaks were more closely associated with other genes. As Bhlhe40−/− SILP CD4+ T cells exhibited decreased expression of Pparg, a known regulator of type 2 cytokines (37, 38), BHLHE40 likely regulates Csf2 and Il5 via a combination of direct and indirect effects. While these cytokines have long been known to share common signaling through the βC chain, their described functions are largely distinct, with GM-CSF contributing to TH1 and TH17 cell-driven inflammation and IL-5 contributing to type 2 immunity (19, 26). We have demonstrated that protective memory to a helminth infection unaffected by IL-5 blockade and also insensitive to GM-CSF deficiency is nonetheless dependent on the combination of GM-CSF and IL-5 signaling through the βC chain. It remains to be determined how GM-CSF and IL-5 compensate for each other, whether by direct substitution or via effects on distinct arms of the type 2 response. While redundancy between βC chain family cytokines is not well described, it is known that these cytokines can regulate eosinophils in a complementary manner (40). Future studies should establish whether GM-CSF and IL-5 are collectively involved in protective immunity to other helminth infections. As βIL3 chain-dependent IL-3 signaling is preserved in Csf2rb−/− mice (20), it is also of interest to establish whether additionally blocking IL-3 results in a more severe defect in immunity to H. polygyrus than is observed in Csf2rb−/− mice. IL-3 regulates basophilia in response to H. polygyrus and basophils help control infection with this helminth (41, 42).
Our findings reveal novel transcriptional and cytokine regulation of the immune response to a helminth infection, elucidating a critical function for BHLHE40 during in vivo TH2 cell immunity and an unexpected role for βC chain-dependent signaling. Our data indicate that the importance of βC chain family cytokines may have been underestimated in type 2 diseases because of redundancy and suggest that combinatorial targeting of these factors could yield improved clinical outcomes in diseases of pathological type 2 immunity.
Supplementary Material
Key points.
BHLHE40 is required for protective immunity to the helminth H. polygyrus.
BHLHE40 is required for normal TH2 gene transcription and cytokine production.
CSF2RB-dependent cytokines can compensate for each other during type 2 immunity.
Acknowledgements
We thank N. Zhang, B. Saunders, and G. Randolph for help with microscopy and technical advice, E. Lantelme, D. Brinja, and A. Mitra for help with FACS, and J. Bando for help with flow cytometry of the gut.
This work was supported by National Institutes of Health Grants R01AI113118 and R01AI132653 (to B.T.E.), T32AI007163 (to N.N.J.), and P30DK097948 (to S.C.-C. H); a Burroughs Wellcome Fund Career Award for Medical Scientists (to B.T.E.); National Science Foundation Grant DGE-1745038 (to M.E.C.); and American Cancer Society Grant IRG-16–186-21 (to S.C.-C.H.). Research reported in this publication was supported by the Washington University Institute of Clinical and Translational Sciences grant UL1TR002345 from the National Center for Advancing Translational Sciences of the National Institutes of Health.
Abbreviations used in this article:
- CSF2RB and βC
beta common
- BHLHE40
basic helix-loop-helix, member e40
- GMM
granuloma-associated monocyte/macrophage
- GSEA
gene set enrichment analysis
- H. polygyrus
Heligmosomoides polygyrus bakeri
- L3
infective stage H. polygyrus larvae
- LPM
large peritoneal macrophage
- RELMα
resistin-like molecule alpha
- SILP
small intestine lamina propria
Footnotes
The online version of this article contains supplemental material.
Disclosures
The authors have no financial conflicts of interest
References
- 1.Martínez-Llordella M, Esensten JH, Bailey-Bucktrout SL, Lipsky RH, Marini A, Chen J, Mughal M, Mattson MP, Taub DD, and Bluestone JA. 2013. CD28-inducible transcription factor DEC1 is required for efficient autoreactive CD4+ T cell response. J. Exp. Med 210: 1603–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin CC, Bradstreet TR, Schwarzkopf EA, Sim J, Carrero JA, Chou C, Cook LE, Egawa T, Taneja R, Murphy TL, Russell JH, and Edelson BT. 2014. Bhlhe40 controls cytokine production by T cells and is essential for pathogenicity in autoimmune neuroinflammation. Nat. Commun 5: 3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin CC, Bradstreet TR, Schwarzkopf EA, Jarjour NN, Chou C, Archambault AS, Sim J, Zinselmeyer BH, Carrero JA, Wu GF, Taneja R, Artyomov MN, Russell JH, and Edelson BT. 2016. IL-1-induced Bhlhe40 identifies pathogenic T helper cells in a model of autoimmune neuroinflammation. J Exp Med 213: 251–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanda M, Yamanaka H, Kojo S, Usui Y, Honda H, Sotomaru Y, Harada M, Taniguchi M, Suzuki N, Atsumi T, Wada H, Baghdadi M, and Seino K. 2016. Transcriptional regulator Bhlhe40 works as a cofactor of T-bet in the regulation of IFN-γ production in iNKT cells. Proc Natl Acad Sci U S A 113: E3394–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huynh JP, Lin CC, Kimmey JM, Jarjour NN, Schwarzkopf EA, Bradstreet TR, Shchukina I, Shpynov O, Weaver CT, Taneja R, Artyomov MN, Edelson BT, and Stallings CL. 2018. Bhlhe40 is an essential repressor of IL-10 during Mycobacterium tuberculosis infection. J Exp Med 215: 1823–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu F, Sharma S, Jankovic D, Gurram RK, Su P, Hu G, Li R, Rieder S, Zhao K, Sun B, and Zhu J. 2018. The transcription factor Bhlhe40 is a switch of inflammatory versus antiinflammatory Th1 cell fate determination. J Exp Med 215: 1813–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jarjour NN, Schwarzkopf EA, Bradstreet TR, Shchukina I, Lin CC, Huang SC, Lai CW, Cook ME, Taneja R, Stappenbeck TS, Randolph GJ, Artyomov MN, Urban JF Jr., and Edelson BT. 2019. Bhlhe40 mediates tissue-specific control of macrophage proliferation in homeostasis and type 2 immunity. Nat. Immunol 20: 687–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ow JR, Tan YH, Jin Y, Bahirvani AG, and Taneja R. 2014. Stra13 and Sharp-1, the non-grouchy regulators of development and disease. Curr. Top. Dev. Biol 110: 317–338. [DOI] [PubMed] [Google Scholar]
- 9.Henriksson J, Chen X, Gomes T, Ullah U, Meyer KB, Miragaia R, Duddy G, Pramanik J, Yusa K, Lahesmaa R, and Teichmann SA. 2019. Genome-wide CRISPR Screens in T Helper Cells Reveal Pervasive Crosstalk between Activation and Differentiation. Cell 176: 882–896.e818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tibbit CA, Stark JM, Martens L, Ma J, Mold JE, Deswarte K, Oliynyk G, Feng X, Lambrecht BN, De Bleser P, Nylen S, Hammad H, Henriksson MA, Saeys Y, and Coquet JM. 2019. Single-Cell RNA Sequencing of the T Helper Cell Response to House Dust Mites Defines a Distinct Gene Expression Signature in Airway Th2 Cells. Immunity 51: 1–16. [DOI] [PubMed] [Google Scholar]
- 11.Allen JE, and Sutherland TE. 2014. Host protective roles of type 2 immunity: parasite killing and tissue repair, flip sides of the same coin. Semin. Immunol 26: 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maizels RM, Smits HH, and McSorley HJ. 2018. Modulation of Host Immunity by Helminths: The Expanding Repertoire of Parasite Effector Molecules. Immunity 49: 801–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camberis M, Le Gros G, and Urban J Jr. 2003. Animal model of Nippostrongylus brasiliensis and Heligmosomoides polygyrus. Curr Protoc Immunol Chapter 19: Unit 19.12. [DOI] [PubMed] [Google Scholar]
- 14.Urban JF Jr., Katona IM, Paul WE, and Finkelman FD. 1991. Interleukin 4 is important in protective immunity to a gastrointestinal nematode infection in mice. Proc Natl Acad Sci U S A 88: 5513–5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Massacand JC, Stettler RC, Meier R, Humphreys NE, Grencis RK, Marsland BJ, and Harris NL. 2009. Helminth products bypass the need for TSLP in Th2 immune responses by directly modulating dendritic cell function. Proc Natl Acad Sci U S A 106: 13968–13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zaiss MM, Maslowski KM, Mosconi I, Guenat N, Marsland BJ, and Harris NL. 2013. IL-1beta suppresses innate IL-25 and IL-33 production and maintains helminth chronicity. PLoS Pathog 9: e1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pei C, Zhao C, Wang AJ, Fan AX, Grinchuk V, Smith A, Sun R, Xie Y, Lu N, Urban JF Jr., Shea-Donohue T, Zhao A, and Yang Z. 2016. Critical Role for Interleukin-25 in Host Protective Th2 Memory Response against Heligmosomoides polygyrus bakeri. Infect Immun 84: 3328–3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maizels RM, Balic A. 2004. Resistance to Helminth Infection: The Case for Interleukin-5-Dependent Mechanisms. J. Infect. Dis 190: 427–429. [DOI] [PubMed] [Google Scholar]
- 19.Roufosse F 2018. Targeting the Interleukin-5 Pathway for Treatment of Eosinophilic Conditions Other than Asthma. Front. Med. (Lausanne) 5: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dougan M, Dranoff G, and Dougan SK. 2019. GM-CSF, IL-3, and IL-5 Family of Cytokines: Regulators of Inflammation. Immunity 50: 796–811. [DOI] [PubMed] [Google Scholar]
- 21.Mandujano JF, D’Souza NB, Nelson S, Summer WR, Beckerman RC, and Shellito JE. 1995. Granulocyte-macrophage colony stimulating factor and Pneumocystis carinii pneumonia in mice. Am J Respir Crit Care Med 151: 1233–1238. [DOI] [PubMed] [Google Scholar]
- 22.Zhan Y, Lieschke GJ, Grail D, Dunn AR, and Cheers C. 1998. Essential roles for granulocyte-macrophage colony-stimulating factor (GM-CSF) and G-CSF in the sustained hematopoietic response of Listeria monocytogenes-infected mice. Blood 91: 863–869. [PubMed] [Google Scholar]
- 23.LeVine AM, Reed JA, Kurak KE, Cianciolo E, and Whitsett JA. 1999. GM-CSF–deficient mice are susceptible to pulmonary group B streptococcal infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deepe GS Jr., Gibbons R, and Woodward E. 1999. Neutralization of endogenous granulocyte-macrophage colony-stimulating factor subverts the protective immune response to Histoplasma capsulatum. J Immunol 163: 4985–4993. [PubMed] [Google Scholar]
- 25.Hirata Y, Egea L, Dann SM, Eckmann L, and Kagnoff MF. 2010. GM-CSF-facilitated dendritic cell recruitment and survival govern the intestinal mucosal response to a mouse enteric bacterial pathogen. Cell Host Microbe 7: 151–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Becher B, Tugues S, and Greter M. 2016. GM-CSF: From Growth Factor to Central Mediator of Tissue Inflammation. Immunity 45: 963–973. [DOI] [PubMed] [Google Scholar]
- 27.Yamashita N, Tashimo H, Ishida H, Kaneko F, Nakano J, Kato H, Hirai K, Horiuchi T, and Ohta K. 2002. Attenuation of airway hyperresponsiveness in a murine asthma model by neutralization of granulocyte-macrophage colony-stimulating factor (GM-CSF). Cell Immunol 219: 92–97. [DOI] [PubMed] [Google Scholar]
- 28. Cates EC, Fattouh R, Wattie J, Inman MD, Goncharova S, Coyle AJ, Gutierrez-Ramos JC, and Jordana M. 2004. Intranasal exposure of mice to house dust mite elicits allergic airway inflammation via a GM-CSF-mediated mechanism. J Immunol 173: 6384–6392. [DOI] [PubMed] [Google Scholar]
- 29.Shim DS, Schilter HC, Knott ML, Almeida RA, Short RP, Mackay CR, Dent LA, and Sewell WA. 2012. Protection against Nippostrongylus brasiliensis infection in mice is independent of GM-CSF. Immunol Cell Biol 90: 553–558. [DOI] [PubMed] [Google Scholar]
- 30.Sun H, Lu B, Li RQ, Flavell RA, and Taneja R. 2001. Defective T cell activation and autoimmune disorder in Stra13-deficient mice. Nat Immunol 2: 1040–1047. [DOI] [PubMed] [Google Scholar]
- 31.Filbey KJ, Grainger JR, Smith KA, Boon L, van Rooijen N, Harcus Y, Jenkins S, Hewitson JP, and Maizels RM. 2014. Innate and adaptive type 2 immune cell responses in genetically controlled resistance to intestinal helminth infection. Immunol Cell Biol 92: 436–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bando JK, Gilfillan S, Song C, McDonald KG, Huang SC-C, Newberry RD, Kobayashi Y, Allan DSJ, Carlyle JR, Cella M, and Colonna M The Tumor Necrosis Factor Superfamily Member RANKL Suppresses Effector Cytokine Production in Group 3 Innate lymphoid Cells. Immunity 48: 1208–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bain CC, Bravo-Blas A, Scott CL, Perdiguero EG, Geissmann F, Henri S, Malissen B, Osborne LC, Artis D, and Mowat AM. 2014. Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat Immunol 15: 929–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scott CL, Bain CC, and Mowat AM. 2017. Isolation and Identification of Intestinal Myeloid Cells. Methods Mol Biol 1559: 223–239. [DOI] [PubMed] [Google Scholar]
- 35.Anthony RM, Urban JF, Alem F, Hamed HA, Rozo CT, Boucher JL, Van Rooijen N, and Gause WC. 2006. Memory T(H)2 cells induce alternatively activated macrophages to mediate protection against nematode parasites. Nat Med 12: 955–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohrs K, Harris DP, Lund FE, and Mohrs M. 2005. Systemic dissemination and persistence of Th2 and type 2 cells in response to infection with a strictly enteric nematode parasite. J. Immunol 175: 5306–5313. [DOI] [PubMed] [Google Scholar]
- 37.Nobs SP, Natali S, Pohlmeier L, Okreglicka K, Schneider C, Kurrer M, Sallusto F, and Kopf M. 2017. PPARgamma in dendritic cells and T cells drives pathogenic type-2 effector responses in lung inflammation. J Exp Med 214: 3015–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen T, Tibbitt CA, Feng X, Stark JM, Rohrbeck L, Rausch L, Sedimbi SK, Karlsson MCI, Lambrecht BN, Karlsson Hedestam GB, Hendriks RW, Chambers BJ, Nylen S, and Coquet JM. 2017. PPAR-gamma promotes type 2 immune responses in allergy and nematode infection. Sci Immunol 2. [DOI] [PubMed] [Google Scholar]
- 39.Gabrysova L, Alvarez-Martinez M, Luisier R, Cox LS, Sodenkamp J, Hosking C, Perez-Mazliah D, Whicher C, Kannan Y, Potempa K, Wu X, Bhaw L, Wende H, Sieweke MH, Elgar G, Wilson M, Briscoe J, Metzis V, Langhorne J, Luscombe NM, and O’Garra A. 2018. c-Maf controls immune responses by regulating disease-specific gene networks and repressing IL-2 in CD4(+) T cells. Nat. Immunol 19: 497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Esnault S, and Kelly EA. 2016. Essential Mechanisms of Differential Activation of Eosinophils by IL-3 Compared to GM-CSF and IL-5. Crit Rev Immunol 36: 429–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herbst T, Esser J, Prati M, Kulagin M, Stettler R, Zaiss MM, Hewitson JP, Merky P, Verbeek JS, Bourquin C, Camberis M, Prout M, Maizels RM, Le Gros G, and Harris NL. 2012. Antibodies and IL-3 support helminth-induced basophil expansion. Proc Natl Acad Sci U S A 109: 14954–14959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwartz C, Turqueti-Neves A, Hartmann S, Yu P, Nimmerjahn F, and Voehringer D. 2014. Basophil-mediated protection against gastrointestinal helminths requires IgE-induced cytokine secretion. Proc Natl Acad Sci U S A 111: E5169–5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.