Abstract
The RAS family of proteins are at the apex of several pathways implicated in a multitude of epithelial cancers, but have remained stubbornly resistant to the wave of targeted small molecules and antibodies that have revolutionized clinical oncology. KRAS, the most commonly mutated of the isoforms, represents an attractive target for treatment given its ubiquity, central role as a driver mutation and association with poor prognosis. This review is a comprehensive summary of the existing approaches to targeting KRAS spanning small molecule inhibitors, cancer vaccines, and with a focus on trials in adoptive cell therapy. Here we explain how the limitations of existing drugs and non-specific immune-based therapies are circumvented with techniques in modern immunotherapy. The successes outlined represent the most promising path to finally targeting the prototypical “undruggable” RAS oncogene family.
Introduction
The RAS family of proteins are small guanosine triphosphatases (GTPases) that regulate cell proliferation and survival, with abnormal function contributing to developmental disorders and cancer [1]. Mutations in RAS oncogenes are present in 20-30% of epithelial cancers, including 56% of pancreatic, 16% of lung, and 33% colorectal adenocarcinomas [2]. Of the four clinically relevant isoforms (KRAS4A, KRAS4B, NRAS, HRAS), the KRAS splice-variants, differing only by 23-24 carboxy-terminal residues, are the most frequently mutated in cancer [2]. Therapies targeting proteins downstream of activated RAS, such as PI 3’ kinase and BRAF, have been largely unsuccessful due to paradoxical activation of adjacent pathways, implying that RAS, a protein at the apex of several pathways, would be better served as a direct target. Yet this has proven to be challenging and in the four decades since its discovery, mutated RAS oncogenes have remained stubbornly resistant to the wave of targeted small molecules and antibodies that have revolutionized clinical oncology [3].
KRAS stands at the center of numerous intracellular signaling cascades, such as the mitogen-activated protein kinase (MAP-K), phosphatidylinositol 3-kinase (PI3K), and mammalian target of rapamycin (mTOR) pathways, among others, all of which promote cell growth and suppress apoptosis [3]. When functioning normally, the RAS protein acts as a molecular switch, turned on by the binding of GTP and off by cleavage to GDP. Although the protein possesses slow, intrinsic GTPase activity, this transition is catalyzed 100,000-fold by GTPase Activating Proteins (GAPs) [4]. GDP eventually makes way for new GTP, a process facilitated by guanine nucleotide exchange factors (GEFs) such as Son of Sevenless (SOS). Mutant KRAS proteins are constitutively locked in the GTP-bound, active state, due to defective interactions with GAPs, decrease in intrinsic GTPase activity, or both; this leads to chronic activation of downstream pathways and, subsequently, uncontrolled cellular proliferation. This effect has been shown with mutations in the catalytic domain of the protein (nucleotides 12, 13, and 61), which disrupt the interaction between RAS and GAPs [2, 4, 5]. In the context of a picomolar binding affinity, the high intra-cellular concentration of GTP and what amounts to a “loss of function” of GAP proteins, specific targeting of mutated RAS without affecting wild type RAS has thus far not been achievable.
Clinical Relevance of KRAS Mutations
KRAS is most commonly mutated at codon 12, though the variant amino acid substitution varies by cancer histology (Table 1)[6]. The G12D mutation, in which glycine is replaced by aspartate, is the most common overall, present in over one third of KRAS-mutated tumors. G12D (substitution of aspartate) is found at an overall frequency of 45% in pancreatic cancers and 13% in colorectal adenocarcinomas [7-10]. There is some frequency variation by histology, most notably the higher incidence of G12C in non-small cell lung cancer (Table 1) [11, 12]. From a clinical standpoint, some studies have shown KRAS-mutant tumors, particularly lung and colon cancers, are associated with poorer overall survival and resistance to treatment [13-18]. Of greatest clinical significance is the finding that patients with KRAS-mutant colorectal cancers are resistant to targeted inhibition of EGFR [15-17, 19-21].
Table 1.
Breakdown of KRAS mutations across various histologies and annual incidences as reported in the COSMIC Database.
| Histology | % KRAS Mutated |
% G12C* |
% G12D* |
% G12V* |
% G13D* |
% Other* |
Annual Incidence |
Est # KRAS Mut/yr |
|---|---|---|---|---|---|---|---|---|
| Large Intestine | 33.0 | 2.6 | 11.4 | 7.4 | 6.2 | 5.4 | 140,250 | 46,319 |
| Lung | 16.0 | 6.1 | 3.2 | 3.7 | 0.5 | 2.5 | 234,030 | 37,440 |
| Pancreas | 55.6 | 1.5 | 26.7 | 17.6 | 0.9 | 8.9 | 55,440 | 30,846 |
| Biliary | 20.8 | 1.4 | 9.9 | 4.1 | 0.1 | 5.3 | 12,360 | 2,572 |
| Endometrial | 14.9 | 1.3 | 5.4 | 3.4 | 1.7 | 3.1 | 61,880 | 9,237 |
Percentage of total KRAS mutations: G12C (11.7%), G12D (34.5%), G12V (24%), G13D (12.6%) and others (i.e. G12A, G12S, G12R, and Q61H) total to 15%.
The ubiquity of KRAS mutations, coupled with its central role as a driver mutation in a variety of epithelial cancers and its association with poor prognosis, makes it a prime target for treatment. This review will summarize the efforts in this field, with special attention paid to the success and promise of immunotherapy-based treatments.
Small Molecule Inhibitors and Non-Immune Approaches
Constitutive activation of RAS paradoxically results from the equivalent of a loss-of-function mutation in that the interactions with GAP family proteins are lost. This has made the development of drugs specific for mutant RAS isoforms particularly difficult. Efforts at KRAS inhibition have targeted interactions with downstream effectors, reduction of RAS in the GTP-bound state, stabilization of the inactive form (i.e. GEF inhibition), or an overall reduction in the protein’s presence at the membrane [5].. Another challenge in targeting a key protein such as KRAS, however, lies in avoiding wild-type reactivity; the risks of this were clearly highlighted in KRAS-knockout mice, which die at 12-14 weeks of gestation [22].
The lack of a sufficiently large and hydrophobic pocket for small-molecule binding limits drug-development for inhibiting interactions with downstream effector molecules [23]. Nonetheless, some groups have demonstrated pre-clinical success in this endeavor. Various small molecule inhibitors (SMIs) of the Ras-GTP/c-Raf and RAS-GTP interactions block downstream effectors and have shown in vivo success in a xenograft model [24, 25], while others have been crippled by off-target toxicities [23].
Inhibition of GEF function stabilizes the inactive form of KRAS proteins by increasing the RAS-GDP fraction within the cell. Maurer et al [26] and Lito et al have demonstrated in vitro inhibition of tumor growth using small molecule inhibitors that stabilize the GDP-bound form of G12C mutated KRAS [12]. This mechanism, however, is limited in its application by the pharmacokinetic limitations of the drugs, and given their dependence on residual GTPase function within the mutant protein. Efforts to identify molecules better suited to occupy this domain in-vivo are still underway [27].
To date, no SMI targeting mutant-KRAS/effector interactions, GEF-inhibition, or RAS membrane localization has been able to translate success from the animal model to clinical application in humans; though there are many promising approaches, avoidance of wild-type reactivity remains the greatest challenge to making this transition.
Immunotherapy -- Vaccine Approaches
The idea of targeting mutated RAS isoforms as antigens is not new. Jung and Schluesener in 1991 described Class II-restricted proliferative responses in the PBL of normal volunteers in response to a G12V mutated RAS peptide but not the wild type after multiple in vitro stimulations. Researchers in 1995 vaccinated patients with pancreatic cancer with autologous antigen presenting cells loaded with synthetic mutated RAS peptides. From the blood of one of the patients, they demonstrated transient antigen-specific proliferative CD4 responses and cloned a CD8+ T-cell specific for the G12V mutation in RAS. This clone was HLA-B35 restricted and specifically lysed a tumor line from that patient’s ascites [28]. When these findings were pursued with phase I and II clinical trials, 25/43 patients (58%) with both surgically resectable and locally advanced pancreatic adenocarcinoma demonstrated peptide-specific immunity; an immune response to the mutated peptide was also associated with a statistically significant survival advantage (141 vs 61 days, P = 0.0002) [29, 30]. Of 20 surgically resected and vaccinated patients who received long-term follow up by this group, 4 were alive at the 10 year mark, and 3 demonstrated ongoing T-cell memory response to RAS peptides. Yet little can be concluded from this limited uncontrolled clinical experience given the non-specific associations between immune competence and overall survival of cancer patients [31].
Subsequently, others reported Class II restricted proliferative responses and Class I restricted T-cell clones in the blood of cancer patients immunized with mutated RAS peptides [32]. Yet despite documenting these immune responses [1], the RAS peptide vaccines evaluated in clinical trials (GI-4000, TG-01 and TG-02) in patients with RAS mutated cancers did not show clear clinical benefit [33, 34]. In general, vaccine approaches to tumor-associated antigens have been disappointing, though they may be best applied in the adjuvant setting [35]. The optimal method of generating T-cell responses in humans remains unclear, and all of these approaches attempt to generate these responses in the continued presence of a hostile tumor microenvironment. Even murine models typically fail against realistic tumors when vaccines are applied against tumors that are established and vascularized. Certain commonly RAS-mutated histologies have shown good responses to checkpoint inhibitors (i.e. lung), however, these encompass only a small portion of RAS-mutated tumors. This highlights why other approaches to treating cancers with T-cells are needed. [35].
Adoptive Cell Therapy
Overview/History
Adoptive cell therapy (ACT) is the use of ex-vivo expanded tumor-reactive T-cells administered to a properly prepared recipient. Early experience used naturally occurring populations found in the tumor-infiltrating lymphocytes (TIL) of some cancers, such as melanoma, which frequently contain T-cells recognizing antigens displayed by the autochthonous tumor [36, 37]. Demonstrating reactivity against tumor-specific mutated proteins led to the hypothesis that immune attack against these mutated neoantigens by TIL was the ‘final common pathway’ of human cancer immunotherapy [38]. To transfer T-cells to a patient effectively, it was found that the recipient had to undergo preparative lymphodepletion, using a non-myeloablative combination of cyclophosphamide and fludarabine. This transiently removed T-regulatory cells, induced lymphotrophic cytokines such as IL-7 and IL-15 and removed irrelevant host lymphocytes that could compete with the infused T-cells for homeostatic cytokines [39, 40]. The regimen of lymphodepletion, melanoma TIL transfer and brief in vivo support with systemic IL-2 led to complete, durable regression of metastatic disease in 24% of patients in two sequential trials (total of 194 patients) [39, 41, 42]; similar results were replicated in studies at other centers [43-45]. Subsequent studies showed that tumor-reactive T-cells generated by retrovirally introducing tumor-reactive T-cell receptors into autologous peripheral blood lymphocytes (PBL) could be similarly effective [46-48]. The strategy of separating the T-cell response from the patient tumor microenvironment by in vitro expansion and allowing the independent manipulation of the recipient appears to accomplish meaningful tumor regressions where vaccines have not. The major challenge at this point is identifying safe and effective antigen targets, especially for the common epithelial cancers. It has become clear in the last few years that nearly all effective T-cell based immunotherapies, including checkpoint inhibitors, rely heavily on immune responses to tumor-specific mutated antigens [49-52].
In retrospect, neoantigens represent the ideal tumor-associated antigen in that they are completely tumor specific and may be more immunogenic because they are “foreign” or “non-self” and thus have not been selected against during thymic development. Furthermore, targeting them is much less likely to induce autoimmunity compared to unmutated melanocyte antigens (such as gp100 or MART-1) or overexpressed proteins (such as CEA), which have led to significant “on-target, off-tumor” activity [53, 54]. Despite this, prior ACT trials have also seen “off-target, off-tumor” toxicities due to the unpredictable cross-reactivity of the transduced TCRs [55]. Early data on TIL selected for mutation reactivity from epithelial cancers has demonstrated that such T-cells can be highly effective in some patients when used for ACT [56-59]. Given the difficulties of establishing patient-specific cell lines from epithelial cancers, these tumor-specific TIL are identified by co-culture with autologous antigen presenting cells which display a patient’s tumor specific mutations via transfection with multiple minigenes or loading of synthetic mutated peptides [38, 58]. Unfortunately, nearly all of these TIL are recognizing completely private antigens requiring patient-specific T-cell products for every patient. In our initial cohort of 53 patients, only two shared an immunogenic non-synonymous mutation based on our TIL-screen (Unpublished Data). This shared neoantigen was KRAS G12D, reviving interest in targeting recurring mutations in common cancers using off-the-shelf reagents.
RAS-Specific ACT in Humans
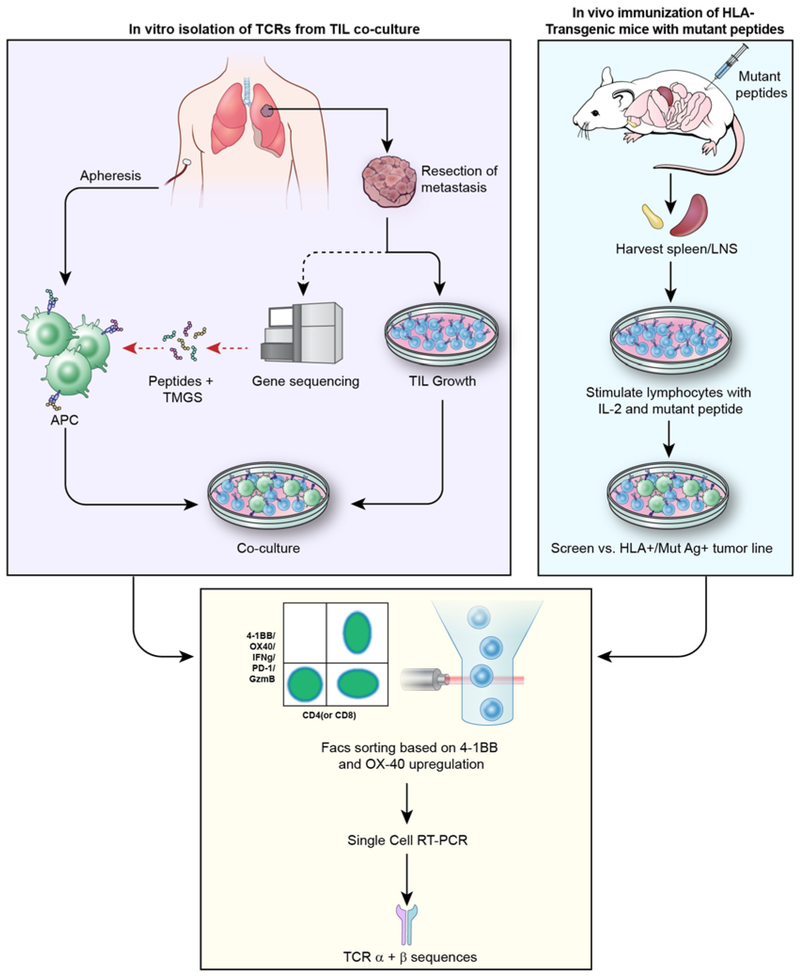
Two avenues of investigation have demonstrated that mutated RAS proteins are immunogenic and could produce reagents that might be clinically effective. The first involves the analysis of patient TIL to isolate naturally-occurring T-cells populations recognizing that patient’s RAS mutation and if present in sufficient numbers, potentially administering them clinically. The second (discussed below) uses in vivo immunization and in-vitro sensitization of lymphocytes from HLA-transgenic mice to generate HLA-restricted murine TCRs reactive with specific candidate RAS mutations. Such TCRs can be used in clinical studies by retrovirally introducing these TCRs (either murine or human) into autologous PBL for administration. A summary of the methods used to isolate or generate mutation-reactive T-cells and clone their TCRs is shown in Figure 1.
Figure 1.
Available approaches to isolating mutation-specific TCRs. For in vitro isolation of TCRs, a metastatic lesion is resected for whole exome sequencing and TIL growth; mutant peptides are presented on autologous APCs and co-cultured with TIL to identify mutant-reactive T-cells. In vivo immunization of HLA-transgenic mice with mutant peptides yields sensitized lymphocytes that can be re-stimulated and co-cultured with mutant antigen-bearing tumor cells in vitro. In both pathways, T-cells activated in co-culture are sorted on 4-1BB/OX40 upregulation and subjected to single-cell RT-PCR for identification of unique TCR α & β sequences.
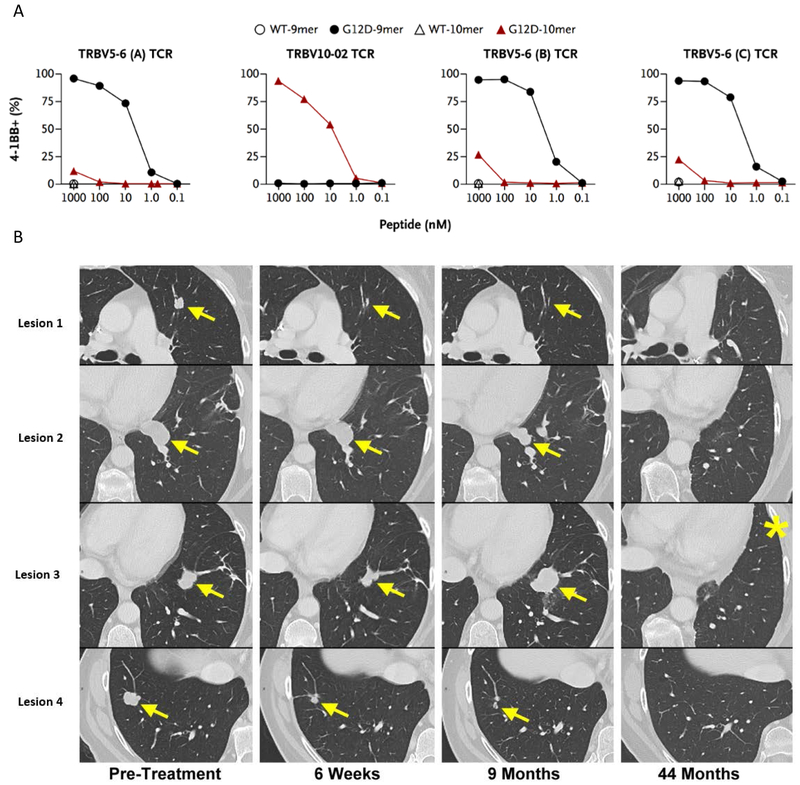
In 2016, Tran et al found polyclonal CD8+ TIL reactivity against KRAS G12D in the TIL of a patient with metastatic colorectal cancer and isolated 4 separate HLA-C*08:02 restricted TCRs recognizing the patient’s KRAS G12D mutation [53]. TIL grown from a resected metastatic lesion were co-cultured with autologous APCs electroporated with minigenes (in the form of RNA) encoding all the patient’s tumor specific mutations to present them in the context of all MHC Class I alleles. TIL reactivity was assessed by secretion of IFN-γ on ELISPOT assay and flow cytometric analysis of T-cell activation markers 4-1BB and OX-40. The mutated minimal determinant epitopes were determined using synthetic peptides and reactivity to both the KRAS G12D 9-mer (GADGVGKSA) and 10-mer (GADGVGKSAL) peptides were found. TCR α and β chains were cloned from reactive TIL and retrovirally reintroduced into PBL to verify the reactivities (Figure 2a). Furthermore, these TCR-transduced PBL also recognized allogeneic HLA-C*08:02+, KRAS G12D-mutated pancreatic adenocarcinoma cell lines.
Figure 2.
Panel A: PBL transfected with KRAS mutation-specific TCRs (isolated and cloned from reactive TIL) upregulate the activation marker 4-1BB when co-incubated with PBMC pulsed with titrated amounts of KRAS G12D mutant peptides (both 9-mer and 10-mer). There is no reactivity seen when co-cultured with PBMC pulsed with the corresponding KRAS wild-type peptides. Panel B: In this patient with metastatic colon cancer, adoptive transfer of 1.1×1011 autologous TIL, reactive against KRAS G12D presented by HLA-C*08:02, mediated durable regression (ongoing at 44 months) of all but one lesion (#3). This sole relapse was excised (*) and found to have lost the C*08:02 MHC Class I allele. Figure adapted from Tran et al. [57] from The New England Journal of Medicine, “T-Cell Transfer Therapy Targeting Mutant KRAS in Cancer,” volume 375, page 2255. Copyright © 2016 Massachusetts Medical Society. Reprinted with permission from Massachusetts Medical Society.
Adoptive transfer of 1.1×1011 of these G12D-reactive T-cells mediated significant regression of all seven pulmonary metastases at just 40 days post-transfer (Figure 2b). This partial response was maintained for 9 months, at which time one lesion progressed and was resected; genomic analysis of this lesion demonstrated loss of the chromosome 6 haplotype encoding HLA-C*08:02, compatible with immune escape from MHC loss. This escape mechanism could be addressed by treating with combinations of CD8+ and CD4+ reactive T-cells to counter this possibility. Nonetheless, this patient remains free of disease almost 4 years post-cell transfer and three years after resection of the solitary relapse.
Interestingly, at 40 days post-cell transfer, the dominant infused T-cell clone (49.5% of infusion bag) was no longer detectable in the patient’s blood, however, the three clones administered at lower frequencies (19.1%, 6.9%, and 0.04%, respectively) persisted in vivo. Murine data would predict that this could be due to the epigenetic state of differentiation of the clones [60] and retrospective flow cytometric analysis of the infusion bag was supportive of this hypothesis. This discovery of KRAS-reactive TCRs is not an isolated occurrence; Veatch et al recently published the isolation of a KRAS G12V-rective TCR from a CD4+ T-cell in the peripheral blood of a patient with non-small cell lung cancer [61]. The availability of these TCRs encoded in highly efficient retroviruses for introduction into the PBL of other patients would allow them to be treated with PBL selected or manipulated to enhance persistence of transferred cells.
RAS-Specific ACT in the Murine Model
As previously mentioned, targeted generation of T-cells recognizing mutated RAS isoforms can also be accomplished with in vivo vaccination of HLA-transgenic mice [57]. Previous in vitro work as well as clinical ACT protocols have shown that unmodified murine TCRs can be retrovirally inserted into human PBL and mediate tumor recognition and clinical regressions [48, 54, 62]. Humoral and cellular immune responses to these murine receptors can develop over time but have not consistently affected their efficacy. Therefore, mice transgenic for HLA alleles predicted to bind and present epitopes containing common codon 12 mutations in RAS were immunized with mutated RAS peptides, their lymphocytes harvested and stimulated in vitro.
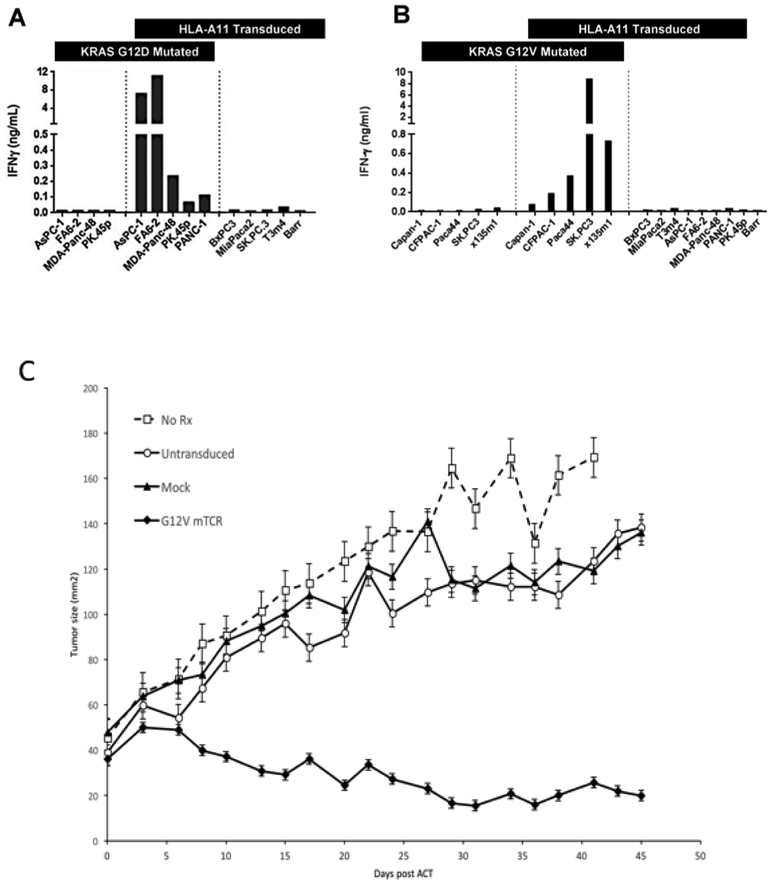
Wang et al generated HLA-A*11:01 restricted mouse T-cells against KRAS G12D and G12V mutations by immunizing HLA-A*11:01 transgenic mice with 9-mer (D: VVGADGVGK, V: VVGAVGVGK) and 10-mer (D: VVVGADGVGK, V: VVVGAVGVGK) mutant RAS peptides and performing in vitro sensitizations [63]. These T-cells demonstrated reactivity via upregulation of IFN-γ and CD107a when co-cultured with appropriate KRAS-mutant cell lines, verifying their cytolytic potential and specificity. Multiple TCR α and β chain cDNA combinations against KRAS G12D and G12V were subsequently identified and retrovirally transfected into peripheral blood lymphocytes. All TCRs were mutation-specific, HLA-A*11:01 restricted, and led to upregulation of T-cell activation markers when co-cultured with titrated amounts of the relevant 9-mer and 10-mer peptides (Figure 3a/b). In a subsequent in vivo experiment, immunodeficient mice bearing large established human KRAS-mutant pancreatic adenocarcinomas were treated with PBL transduced with G12D- or G12V-reactive TCRs. In some cases, significant growth inhibition was seen [63] and in others complete regression was seen in some mice (Figure 3c). Although developed from a focus on KRAS, these mutations occur in the completely conserved region of KRAS, HRAS, and NRAS, and all of those mutated isoforms can be recognized by these KRAS-specific TCRs.
Figure 3.
Panel A/B: T-cells transduced with KRAS G12D/V TCR were cocultured with pancreatic tumor lines with or without KRAS G12D/V mutation and HLA-A*11:01 expression to demonstrate the mutant epitope specificity and HLA-restriction of the receptors. Figure adapted from paper by Wang et al [63]. Panel C: Treatment of human pancreatic cancer xenograft in NSG mice using adoptive cell transfer. Pancreatic adenocarcinoma line TC4177-HLA*A11:01 (1E6 cells) was injected subcutaneously into NSG mice. After 26 days when tumors had grown to approximately 6mm in diameter, they were 1) left untreated, 2) given PBL stimulated with OKT3 but not transduced, 3) stimulated with OKT and transduced with empty retrovirus or 4) OKT stimulated and transduced with the retrovirus encoding the anti-G12V KRAS T-cell receptor. The cell dose was 5e6 T-cells and all cell-treated mice were also given a daily i.p. dose of IL-2 (aldesleukin - 180,000 IU) for 3 days. There were 5 mice per treatment group and tumors were measured by a blinded investigator. All mice treated with TCR transduced PBL showed tumor regression and two of the five mice achieved complete regressions.
Expanding the RAS-Reactive T-Cell Repertoire
The HLA-A*11:01 allele occurs in 14% of US Caucasians, 23% of Asian Americans and up to 50% of Han Chinese [63], while the HLA-C*08:02 is found in 8% of US Caucasians [57], implying a significant number of beneficiaries from these receptors alone, but also speaking to the need for a more diverse collection of TCR reagents [64]. To broaden this library, Yossef et al recently demonstrated that FACS enrichment for CD134+, CD137+ and PD-1+ T-cells followed by limiting dilution cloning could reveal Class II restricted anti-G12V T-cell not detected by conventional screening [65]. Cafri et al have also isolated KRAS-mutant reactive TCRs in populations of in-vitro stimulated memory T cells (both CD4 and CD8) collected from the peripheral blood of patients with known reactivity against the shared antigen [66]. This has the potential to eliminate the TIL harvest process entirely. Table 2 shows the various TCRs which have been isolated from patients with KRAS mutated cancers in the Surgery Branch at the NCI, using either conventional tandem mini-gene (TMG) screening or in vitro stimulation of peripherally circulating memory T cells, as described above. With multiple methods of TCR detection and generation, a library of reactive TCRs that covers a wider breadth of KRAS mutations and HLA alleles can be developed.
Table 2.
Anti-KRAS T-cell receptors isolated from patients with metastatic cancer treated in the Surgery Branch at the NCI in Bethesda, MD.
|
KRAS Mutation |
Patient # | Cancer Diagnosis |
CD4/CD8 | HLA- Restriction |
Method |
|---|---|---|---|---|---|
| G12D | 4095 4238 |
Colon Colon |
CD8 CD4 |
C*08:02 (8%) Class II (DR) |
TMG IVS |
| G12V | 4148 | Endometrial | CD4 CD8 |
DRB1*07:01 (25%) A*11:01 (14%) |
TMG IVS |
| G12C | 4173 | Ovarian | CD4 | DRB1*11:01 (10%) | IVS |
| G12R | 4268 | Colon | CD4 | Class II | TMG |
Future Directions & Challenges
While over 20 years have elapsed since the immunogenicity of KRAS mutations was demonstrated with peptide vaccination in vitro, clinical translation of this to human trials is just now being achieved. Direct targeting of the KRAS protein using small molecule inhibitors remains a daunting task and, in general, small molecule inhibitors have not been proven to have curative potential.
ACT represents a promising medium through which to attack RAS-mutated cancers, however, while its efficacy in hematologic malignancies and melanoma is better established, its utility in epithelial cancers remains in the early phases of investigation.
Proof of principle in one patient using a KRAS-reactive TIL transfer is encouraging, but methods to more frequently and quickly derive T-cells reactive with mutated RAS are needed, as in vivo discovery is simply too rare an event. HLA-transgenic murine models, when used in tandem with HLA-prediction algorithms, have demonstrated utility in broadening the anti-RAS T-cell repertoire, however, much work remains in building this library of receptors. The engineering of these cells using existing off-the-shelf TCRs is available to an increasing number of patients; as this library of receptors expands, off-the-shelf combination therapies may be a future effort. Two trials using this approach for HLA-A11+ patients with either G12V or G12D mutations are now open and accruing: and (https://clinicaltrials.gov/).
Conclusions
As driver mutations present in a significant percentage of epithelial tumors, RAS mutations represent ideal targets for immune-based treatments. Adoptive cell transfer of TIL or PBL engineered to attack KRAS-mutations represents a unique therapy that overcomes the many limitations of existing small molecule inhibitors, non-specific immune-based therapies, and passive vaccination trials. For some patients, it may represent the most promising path to finally targeting the prototypical “undruggable” RAS oncogene family.
Footnotes
Potential conflicts of interest: J.C. Yang may receive royalties from the T-cell receptors described in this article, and reports receiving other commercial research support from Kite/Gilead. No potential conflicts of interest were disclosed by the other author.
Sources
- 1.Pant S, Hubbard J, Martinelli E, and Bekaii-Saab T, Clinical update on K-Ras targeted therapy in gastrointestinal cancers. Crit Rev Oncol Hematol, 2018. 130: p. 78–91. [DOI] [PubMed] [Google Scholar]
- 2.Singh H, Longo DL, and Chabner BA, Improving Prospects for Targeting RAS. J Clin Oncol, 2015. 33(31): p. 3650–9. [DOI] [PubMed] [Google Scholar]
- 3.McCormick F, KRAS as a Therapeutic Target. Clin Cancer Res, 2015. 21(8): p. 1797–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCormick F, Targeting KRAS Directly. 2018. 2(1): p. 81–90. [DOI] [PubMed] [Google Scholar]
- 5.Ostrem JM and Shokat KM, Direct small-molecule inhibitors of KRAS: from structural insights to mechanism-based design. Nat Rev Drug Discov, 2016. 15(11): p. 771–785. [DOI] [PubMed] [Google Scholar]
- 6.Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. , COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Research, 2018. 47(D1): p. D941–D947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vaughn CP, Zobell SD, Furtado LV, Baker CL, and Samowitz WS, Frequency of KRAS, BRAF, and NRAS mutations in colorectal cancer. Genes Chromosomes Cancer, 2011. 50(5): p. 307–12. [DOI] [PubMed] [Google Scholar]
- 8.Bryant KL, Mancias JD, Kimmelman AC, and Der CJ, KRAS: feeding pancreatic cancer proliferation. Trends Biochem Sci, 2014. 39(2): p. 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andreyev HJ, Norman AR, Cunningham D, Oates J, Dix BR, Iacopetta BJ, et al. , Kirsten ras mutations in patients with colorectal cancer: the "RASCAL II" study. British journal of cancer, 2001. 85(5): p. 692–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esteller M, González S, Risques RA, Marcuello E, Mangues R, Germà JR, et al. , K-ras and p16 Aberrations Confer Poor Prognosis in Human Colorectal Cancer. 2001. 19(2): p. 299–304. [DOI] [PubMed] [Google Scholar]
- 11.Stephen AG, Esposito D, Bagni RK, and McCormick F, Dragging ras back in the ring. Cancer Cell, 2014. 25(3): p. 272–81. [DOI] [PubMed] [Google Scholar]
- 12.Lito P, Solomon M, Li LS, Hansen R, and Rosen N, Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science, 2016. 351(6273): p. 604–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zocche DM, Ramirez C, Fontao FM, Costa LD, and Redal MA, Global impact of KRAS mutation patterns in FOLFOX treated metastatic colorectal cancer. Frontiers in genetics, 2015. 6: p. 116–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marabese M, Ganzinelli M, Garassino MC, Shepherd FA, Piva S, Caiola E, et al. , KRAS mutations affect prognosis of non-small-cell lung cancer patients treated with first-line platinum containing chemotherapy. Oncotarget, 2015. 6(32): p. 34014–34022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amado RG, Wolf M, Peeters M, Cutsem EV, Siena S, Freeman DJ, et al. , Wild-Type KRAS Is Required for Panitumumab Efficacy in Patients With Metastatic Colorectal Cancer. 2008. 26(10): p. 1626–1634. [DOI] [PubMed] [Google Scholar]
- 16.Benvenuti S, Sartore-Bianchi A, Di Nicolantonio F, Zanon C, Moroni M, Veronese S, et al. , Oncogenic Activation of the RAS/RAF Signaling Pathway Impairs the Response of Metastatic Colorectal Cancers to Anti–Epidermal Growth Factor Receptor Antibody Therapies. 2007. 67(6): p. 2643–2648. [DOI] [PubMed] [Google Scholar]
- 17.Di Fiore F, Blanchard F, Charbonnier F, Le Pessot F, Lamy A, Galais MP, et al. , Clinical relevance of KRAS mutation detection in metastatic colorectal cancer treated by Cetuximab plus chemotherapy. British journal of cancer, 2007. 96(8): p. 1166–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pao W, Wang TY, Riely GJ, Miller VA, Pan Q, Ladanyi M, et al. , KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS medicine, 2005. 2(1): p. e17–e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, Tebbutt NC, et al. , K-ras Mutations and Benefit from Cetuximab in Advanced Colorectal Cancer. 2008. 359(17): p. 1757–1765. [DOI] [PubMed] [Google Scholar]
- 20.Loupakis F, Ruzzo A, Cremolini C, Vincenzi B, Salvatore L, Santini D, et al. , KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. British journal of cancer, 2009. 101(4): p. 715–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Douillard J-Y, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, et al. , Panitumumab–FOLFOX4 Treatment and RAS Mutations in Colorectal Cancer. 2013. 369(11): p. 1023–1034. [DOI] [PubMed] [Google Scholar]
- 22.Johnson L, Greenbaum D, Cichowski K, Mercer K, Murphy E, Schmitt E, et al. , K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes Dev, 1997. 11(19): p. 2468–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Welsch ME, Kaplan A, Chambers JM, Stokes ME, Bos PH, Zask A, et al. , Multivalent Small-Molecule Pan-RAS Inhibitors. Cell, 2017. 168(5): p. 878–889 e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shin SM, Choi DK, Jung K, Bae J, Kim JS, Park SW, et al. , Antibody targeting intracellular oncogenic Ras mutants exerts anti-tumour effects after systemic administration. Nat Commun, 2017. 8: p. 15090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shima F, Yoshikawa Y, Ye M, Araki M, Matsumoto S, Liao J, et al. , In silico discovery of small-molecule Ras inhibitors that display antitumor activity by blocking the Ras-effector interaction. Proc Natl Acad Sci U S A, 2013. 110(20): p. 8182–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maurer T, Garrenton LS, Oh A, Pitts K, Anderson DJ, Skelton NJ, et al. , Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc Natl Acad Sci U S A, 2012. 109(14): p. 5299–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spencer-Smith R, Koide A, Zhou Y, Eguchi RR, Sha F, Gajwani P, et al. , Inhibition of RAS function through targeting an allosteric regulatory site. Nat Chem Biol, 2017. 13(1): p. 62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gjertsen MK, Bjorheim J, Saeterdal I, Myklebust J, and Gaudernack G, Cytotoxic CD4+ and CD8+ T lymphocytes, generated by mutant p21-ras (12Val) peptide vaccination of a patient, recognize 12Val-dependent nested epitopes present within the vaccine peptide and kill autologous tumour cells carrying this mutation. Int J Cancer, 1997. 72(5): p. 784–90. [DOI] [PubMed] [Google Scholar]
- 29.Gjertsen MK, Buanes T, Rosseland AR, Bakka A, Gladhaug I, Soreide O, et al. , Intradermal ras peptide vaccination with granulocyte-macrophage colony-stimulating factor as adjuvant: Clinical and immunological responses in patients with pancreatic adenocarcinoma. Int J Cancer, 2001. 92(3): p. 441–50. [DOI] [PubMed] [Google Scholar]
- 30.Gjertsen MK, Saeterdal I, Saeboe-Larssen S, and Gaudernack G, HLA-A3 restricted mutant ras specific cytotoxic T-lymphocytes induced by vaccination with T-helper epitopes. J Mol Med (Berl), 2003. 81(1): p. 43–50. [DOI] [PubMed] [Google Scholar]
- 31.Weden S, Klemp M, Gladhaug IP, Moller M, Eriksen JA, Gaudernack G, et al. , Long-term follow-up of patients with resected pancreatic cancer following vaccination against mutant K-ras. Int J Cancer, 2011. 128(5): p. 1120–8. [DOI] [PubMed] [Google Scholar]
- 32.Abrams SI, Khleif SN, Bergmann-Leitner ES, Kantor JA, Chung Y, Hamilton JM, et al. , Generation of stable CD4+ and CD8+ T cell lines from patients immunized with ras oncogene-derived peptides reflecting codon 12 mutations. Cell Immunol, 1997. 182(2): p. 137–51. [DOI] [PubMed] [Google Scholar]
- 33.Richards D. e.a., Oral presentations. Annals of Oncology, 2012. 23(suppl_4): p. iv5–iv18.22774231 [Google Scholar]
- 34.Coeshott C, Holmes T, Mattson A, King T, Guo Z, Cohn A, et al. , Abstract A28: Immune responses to mutated Ras - development of a yeast-based immunotherapeutic. 2014. 12(12 Supplement): p. A28–A28. [Google Scholar]
- 35.Rosenberg SA, Yang JC, and Restifo NP, Cancer immunotherapy: moving beyond current vaccines. Nat Med, 2004. 10(9): p. 909–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosenberg SA, Packard BS, Aebersold PM, Solomon D, Topalian SL, Toy ST, et al. , Use of Tumor-Infiltrating Lymphocytes and Interleukin-2 in the Immunotherapy of Patients with Metastatic Melanoma. 1988. 319(25): p. 1676–1680. [DOI] [PubMed] [Google Scholar]
- 37.Robbins PF, El-Gamil M, Li YF, Kawakami Y, Loftus D, Appella E, et al. , A mutated beta-catenin gene encodes a melanoma-specific antigen recognized by tumor infiltrating lymphocytes. The Journal of experimental medicine, 1996. 183(3): p. 1185–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tran E, Robbins PF, and Rosenberg SA, 'Final common pathway' of human cancer immunotherapy: targeting random somatic mutations. Nat Immunol, 2017. 18(3): p. 255–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goff SL, Dudley ME, Citrin DE, Somerville RP, Wunderlich JR, Danforth DN, et al. , Randomized, Prospective Evaluation Comparing Intensity of Lymphodepletion Before Adoptive Transfer of Tumor-Infiltrating Lymphocytes for Patients With Metastatic Melanoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology, 2016. 34(20): p. 2389–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dudley ME, Wunderlich JR, Robbins PF, Yang JC, Hwu P, Schwartzentruber DJ, et al. , Cancer regression and autoimmunity in patients after clonal repopulation with antitumor lymphocytes. Science, 2002. 298(5594): p. 850–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenberg SA and Restifo NP, Adoptive cell transfer as personalized immunotherapy for human cancer. Science, 2015. 348(6230): p. 62–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosenberg SA, Yang JC, Sherry RM, Kammula US, Hughes MS, Phan GQ, et al. , Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin Cancer Res, 2011. 17(13): p. 4550–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pilon-Thomas S, Kuhn L, Ellwanger S, Janssen W, Royster E, Marzban S, et al. , Efficacy of adoptive cell transfer of tumor-infiltrating lymphocytes after lymphopenia induction for metastatic melanoma. Journal of immunotherapy (Hagerstown, Md. : 1997), 2012. 35(8): p. 615–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Radvanyi LG, Bernatchez C, Zhang M, Fox PS, Miller P, Chacon J, et al. , Specific lymphocyte subsets predict response to adoptive cell therapy using expanded autologous tumor-infiltrating lymphocytes in metastatic melanoma patients. Clin Cancer Res, 2012. 18(24): p. 6758–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Besser MJ, Shapira-Frommer R, Itzhaki O, Treves AJ, Zippel DB, Levy D, et al. , Adoptive transfer of tumor-infiltrating lymphocytes in patients with metastatic melanoma: intent-to-treat analysis and efficacy after failure to prior immunotherapies. Clin Cancer Res, 2013. 19(17): p. 4792–800. [DOI] [PubMed] [Google Scholar]
- 46.Lu Y-C, Parker LL, Lu T, Zheng Z, Toomey MA, White DE, et al. , Treatment of Patients With Metastatic Cancer Using a Major Histocompatibility Complex Class II-Restricted T-Cell Receptor Targeting the Cancer Germline Antigen MAGE-A3. Journal of clinical oncology : official journal of the American Society of Clinical Oncology, 2017. 35(29): p. 3322–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robbins PF, Kassim SH, Tran TL, Crystal JS, Morgan RA, Feldman SA, et al. , A pilot trial using lymphocytes genetically engineered with an NY-ESO-1-reactive T-cell receptor: long-term follow-up and correlates with response. Clin Cancer Res, 2015. 21(5): p. 1019–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robbins PF, Morgan RA, Feldman SA, Yang JC, Sherry RM, Dudley ME, et al. , Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. Journal of clinical oncology : official journal of the American Society of Clinical Oncology, 2011. 29(7): p. 917–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schumacher TN and Schreiber RD, Neoantigens in cancer immunotherapy. Science, 2015. 348(6230): p. 69–74. [DOI] [PubMed] [Google Scholar]
- 50.Colli LM, Machiela MJ, Myers TA, Jessop L, Yu K, and Chanock SJ, Burden of Nonsynonymous Mutations among TCGA Cancers and Candidate Immune Checkpoint Inhibitor Responses. Cancer Res, 2016. 76(13): p. 3767–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, et al. , Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science, 2015. 350(6257): p. 207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. , Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med, 2014. 371(23): p. 2189–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parkhurst MR, Yang JC, Langan RC, Dudley ME, Nathan DA, Feldman SA, et al. , T cells targeting carcinoembryonic antigen can mediate regression of metastatic colorectal cancer but induce severe transient colitis. Mol Ther, 2011. 19(3): p. 620–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson LA, Morgan RA, Dudley ME, Cassard L, Yang JC, Hughes MS, et al. , Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood, 2009. 114(3): p. 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morgan RA, Chinnasamy N, Abate-Daga D, Gros A, Robbins PF, Zheng Z, et al. , Cancer regression and neurological toxicity following anti-MAGE-A3 TCR gene therapy. J Immunother, 2013. 36(2): p. 133–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zacharakis N, Chinnasamy H, Black M, Xu H, Lu YC, Zheng Z, et al. , Immune recognition of somatic mutations leading to complete durable regression in metastatic breast cancer. Nat Med, 2018. 24(6): p. 724–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tran E, Robbins PF, Lu YC, Prickett TD, Gartner JJ, Jia L, et al. , T-Cell Transfer Therapy Targeting Mutant KRAS in Cancer. N Engl J Med, 2016. 375(23): p. 2255–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tran E, Ahmadzadeh M, Lu YC, Gros A, Turcotte S, Robbins PF, et al. , Immunogenicity of somatic mutations in human gastrointestinal cancers. Science, 2015. 350(6266): p. 1387–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tran E, Turcotte S, Gros A, Robbins PF, Lu Y-C, Dudley ME, et al. , Cancer Immunotherapy Based on Mutation-Specific CD4+ T Cells in a Patient with Epithelial Cancer. 2014. 344(6184): p. 641–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang J, Khong HT, Dudley ME, El-Gamil M, Li YF, Rosenberg SA, et al. , Survival, persistence, and progressive differentiation of adoptively transferred tumor-reactive T cells associated with tumor regression. J Immunother, 2005. 28(3): p. 258–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Veatch JR, Jesernig BL, Kargl J, Fitzgibbon M, Lee SM, Baik C, et al. , Endogenous CD4(+) T Cells Recognize Neoantigens in Lung Cancer Patients, Including Recurrent Oncogenic KRAS and ERBB2 (Her2) Driver Mutations. Cancer Immunol Res, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sommermeyer D, Conrad H, Kronig H, Gelfort H, Bernhard H, and Uckert W, NY-ESO-1 antigen-reactive T cell receptors exhibit diverse therapeutic capability. Int J Cancer, 2013. 132(6): p. 1360–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang QJ, Yu Z, Griffith K, Hanada K, Restifo NP, and Yang JC, Identification of T-cell Receptors Targeting KRAS-Mutated Human Tumors. Cancer Immunol Res, 2016. 4(3): p. 204–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gonzalez-Galarza FF, Takeshita LY, Santos EJ, Kempson F, Maia MH, da Silva AL, et al. , Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res, 2015. 43(Database issue): p. D784–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yossef R, Tran E, Deniger DC, Gros A, Pasetto A, Parkhurst MR, et al. , Enhanced detection of neoantigen-reactive T cells targeting unique and shared oncogenes for personalized cancer immunotherapy. JCI Insight, 2018. 3(19). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cafri G, Yossef R, Pasetto A, Deniger DC, Lu YC, Parkhurst M, et al. , Memory T cells targeting oncogenic mutations detected in peripheral blood of epithelial cancer patients. Nat Commun, 2019. 10(1): p. 449. [DOI] [PMC free article] [PubMed] [Google Scholar]