Abstract
The APOE4 protein affects the primary neuropathological markers of Alzheimer’s disease (AD): amyloid plaques, neurofibrillary tangles, and gliosis. These interactions have been investigated to understand the strong effect of APOE genotype on risk of AD. However, APOE genotype has strong effects on processes in normal brains, in the absence of the hallmarks of AD. We propose that CNS APOE is involved in processes in the normal brains that in later years apply specifically to processes of AD pathogenesis. We review the differences of the APOE protein found in the CNS compared to the plasma, including post-translational modifications (glycosylation, lipidation, multimer formation), focusing on ways that the common APOE isoforms differ from each other. We also review structural and functional studies of young human brains and control APOE knock-in mouse brains. These approaches demonstrate the effects of APOE genotype on microscopic neuron structure, gross brain structure, and behavior, primarily related to the hippocampal areas. By focusing on the effects of APOE genotype on normal brain function, approaches can be pursued to identify biomarkers of APOE dysfunction, to promote normal functions of the APOE4 isoform, and to prevent the accumulation of the pathologic hallmarks of AD with aging.
Keywords: APOE, apolipoprotein, lipoprotein, inflammation, glycosylation, mouse model, functional MRI
Introduction
For the past 25 years, a great deal of research has examined APOE genotype in the context of its profound effect on the risk of Alzheimer’s Disease (AD) (Strittmatter et al., 1993). In this time, a literature has also developed on APOE genotype in the context of normal brain function (Di Battista et al., 2016; Iacono and Feltis, 2019; Wisdom et al., 2011). Knowledge of the effects of APOE genotype prior to AD could provide insight into normal cognitive strengths and weaknesses of individuals based on their APOE genotypes as well as their later risks of cognitive dysfunctions. As more people make use of commercial DNA sequencing tools (Campion et al., 2019) and discover their APOE genotypes at young ages, public interest in the effects of APOE genotype throughout life will increase.
Knowledge of the effects of APOE genotype in the brain is based in part on its effects on neural dysfunction. In addition to the long history with late onset AD (Raber et al., 2004), APOE associations been observed in other conditions, such as risk of Diffuse Lewy Body Disease (Hansen et al., 2019), recovery from traumatic brain injuries (TBI) (Kassam et al., 2016; Merritt et al., 2018), recovery from stroke (Cramer et al., 2012; Wagle et al., 2009), and risk of cognitive impairment after chemotherapy (Buskbjerg et al., 2019; Mandelblatt et al., 2018) or HIV infection (Chang et al., 2014). These various findings are supported in preclinical studies, including mouse models of TBI (Main et al., 2018), stroke recovery (Lei et al., 2012), and chemotherapy-induced cognitive impairment (Speidell et al., 2019). These conditions support a model in which APOE genotype effects in normal brain create conditions that make adverse responses to injury more likely (Mahley and Huang, 2012). In this context, aging can be considered a condition of accumulating brain damages that are affected by APOE genotype: populations of the oldest old show increased prevalence of the APOE2 allele and decreased prevalence of the APOE4 allele (Garatachea et al., 2015; Rebeck et al., 1994; Revelas et al., 2018; Schachter et al., 1994; Sebastiani et al., 2019).
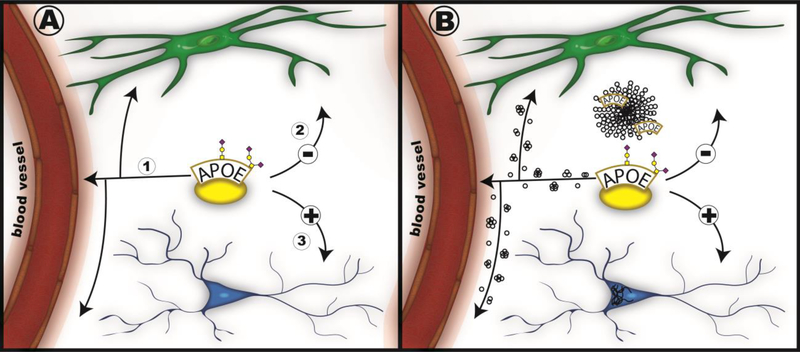
Our overall hypothesis is that CNS APOE is involved in processes in the normal brain that in later years apply to processes of AD pathogenesis. In normal brain, these processes are related to clearance of debris for homeostasis, inhibition of inflammation, and promotion of neuronal network resilience (Figure 1A). In AD brain, these processes are related to clearance of Aβ oligomers, glial activation in response to protein aggregates, and neuronal dysfunction and death (Figure 1B).
Figure 1. APOE functions in normal brain are reflected in functions in AD brain.
Secreted lipoproteins containing modified APOE are indicated as a yellow disk holding the APOE protein with two representative glycans. This CNS lipoprotein interacts with a variety of CNS cells to 1) clear debris through binding to molecules at the surface of the endothelial cells and basement membrane along CNS blood vessels (in red) and to CNS glia (in green); 2) inhibit activation of glia through signaling through cell surface receptors; and 3) promote neurite outgrowth and dendritic spine formation on neurons (in blue). In the AD brain, these functions act to promote clearance of Aβ monomers and oligomers (small collections of circles), to promote anti-inflammatory processes in response to Aβ plaques (round collection of circles containing APOE molecules), and to slow intracellular neurofibrillary tangle formation (black curved lines) and propagation.
In this review, we will first consider the APOE protein that is present in the central nervous system (CNS); this form differs in important ways from APOE found in the periphery. We will then synthesize data on the effects of APOE genotype on brain structure and function in the absence of signs of AD pathogenesis. Finally, we will speculate on ways that the structure of CNS APOE could be related to some of the observed effects of APOE genotype on CNS structure and function. These observations of how APOE genotype predisposes brains to damage are particularly important because they will direct the development of prevention methods for conditions such as AD. Furthermore, they will help in the targeted identification of biomarkers that can be used to test prevention approaches that do not rely on the phenotypes observed in the later stages of AD, such as cognitive impairment and the accumulations of the pathogenic proteins Aβ and phospho-tau.
CNS APOE protein structure
APOE is present both in the CNS and in the periphery, although the structure of the protein is different in these two systems. The mature APOE protein is 299 amino acids with a single amino acid substitution defining each of the three common isoforms: APOE2 (Cys112, Cys158), APOE3 (Cys112, Arg158) and APOE4 (Arg112, Arg158) (Rall et al., 1982) (Figure 2). The rare Christchurch variant consists of a Ser136 variant (Wardell et al., 1987). APOE has three main domains: an N-terminal, four helix, receptor binding domain; a C-terminal, triple helix, lipid binding domain; and an intervening flexible hinge region (Chen et al., 2011; Lalazar et al., 1988; Nguyen et al., 2010; Sakamoto et al., 2008). In the periphery, APOE is synthesized and secreted by hepatocytes (Mahley, 1988) and macrophages (Kockx et al., 2008), and is involved in the HDL, exogenous, and endogenous cholesterol metabolism pathways. It associates with a wide array of varied lipoproteins, ranging from small (7–14 nm) plasma HDL particles (Otvos, 2002) to the larger (30–100 nm) and polyhedral VLDL particles (Yu et al., 2016), to the very large (75–1200 nm) chylomicrons (Dawson and Rudel, 1999; Mahley and Ji, 1999; Patsch, 1998). It functions in the transport of lipoproteins and regulation of plasma lipid levels, with additional functions such as immune modulation (Bennet et al., 2007; Mahley, 1988; Sing and Davignon, 1985; Tenger and Zhou, 2003; Vitek et al., 2009).
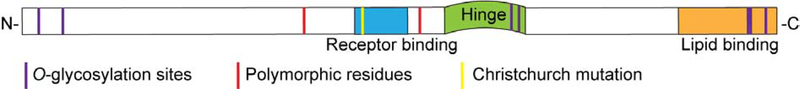
Figure 2. Structural components of APOE isoforms.
The 299 amino acid APOE protein consists of an N-terminal receptor-binding domain and a C-terminal lipid-binding domain (light orange) with an intervening flexible hinge region (green). The schematic also shows amino acids 112 and 158 (red) which determine APOE2, APOE3 and APOE4 status, and the rare Christchurch mutation at amino acid 136 (yellow). Glycosylation sites are in purple.
APOE CNS lipoproteins
APOE is the most abundant apolipoprotein in the brain, although other apolipoproteins are also present, including abundant APOA-I and less abundant apolipoproteins including APOA-II, APOA-IV, APOJ, APOD and APOH (Roher et al., 2009; Wang and Eckel, 2014). In the CNS, APOE is primarily expressed by astrocytes and to a lesser extent by pericytes, oligodendrocytes, choroid plexus, and neurons under stressed physiological conditions (Achariyar et al., 2016; Bruinsma et al., 2010; Nelissen et al., 2012; Pitas et al., 1987a; Xu et al., 2006). APOE is secreted by glia associated with lipids forming small (8–15 nm) discoid particles, which increase in size, becoming spherical as they accumulate lipids and flow into the CSF (12–20 nm with a fraction up to 30 nm) (Koch et al., 2001; LaDu et al., 1998; Pitas et al., 1987b). APOE lipoproteins produced in the choroid plexus are secreted directly into the CSF (Achariyar et al., 2016).
CNS APOE secretion and lipidation occurs in conjunction with the ATP binding cassette (ABC) proteins, ABCA1 and ABCG1 (Courtney and Landreth, 2016). These proteins are embedded in the cell membrane and act to pump lipid molecules into the extracellular space, where they bind apolipoproteins such as APOE and APOA-I (Tall, 2018). Like APOE, the expression of ABCA1 and ABCG1 is increased by the transcription factor LXR (either directly (Xu et al., 2013) or indirectly (Fan et al., 2018)) to promote APOE and lipid efflux (Courtney and Landreth, 2016). ABCA1 activity can also be increased through binding of specific peptides based on the sequences of APOA-I (Sherman et al., 2010) or APOE (Bielicki, 2016), leading to increased lipidation of APOE (Boehm-Cagan et al., 2016a; Chernick et al., 2018).
The three APOE isoforms, APOE2, APOE3, and APOE4, have different levels of lipidation and related functions. In CSF from both middle aged and older cognitively normal individuals, those who carried an APOE4 allele had significantly smaller APOE containing particle distributions compared to those without an APOE4 allele, and APOE2.3 individuals had significantly larger APOE particle distributions (Heinsinger et al., 2016). In a study of viral construct expression of APOE2, APOE3 or APOE4, the APOE4 protein promoted the development of less lipidated APOE particles while APOE2 was more highly lipidated (Hu et al., 2015). A consistent isoform effect on lipidation is apparent in the related function of lipid efflux: APOE2 promoted significantly more lipid efflux from both astrocytes and neurons compared to APOE3, which promoted more lipid efflux than APOE4 (Michikawa et al., 2000; Minagawa et al., 2009). Complete APOE lipidation alters the hinge region movement for access to the N-terminal receptor binding domain, according to APOE-lipid binding models (Chen et al., 2011). Thus, the level of lipidation of each APOE isoform is essential, not only for efficiency of lipid transport, but also for downstream effects involving receptor-binding interactions.
The C-terminal of APOE, from amino acid 244 onwards (from middle of Helix C2 (Chen et al., 2011)), critically affects any lipoprotein binding, driving APOE isoform specificity (Minagawa et al., 2009; Nguyen et al., 2010; Sakamoto et al., 2008; Westerlund and Weisgraber, 1993). Lipid binding differences of APOE isoforms are observed in the periphery, with APOE3 binding preferentially with the more protein-rich HDL while APOE4 binds more effectively to the lipid-rich VLDL particles (Minagawa et al., 2009; Nguyen et al., 2010; Sakamoto et al., 2008; Weisgraber, 1990). Isoform-dependent lipoprotein binding preference is due to the APOE4 protein being more dependent on the C-terminal region (273–299) for binding than APOE3 (Nguyen et al., 2010; Sakamoto et al., 2008). Variations within the most C-terminal region of the APOE4 molecule are therefore more likely to have an impact on its lipid binding properties.
APOE self-association may be another important binding-related property for CSF lipoproteins. Unlipidated APOE monomers form multimers including dimers and tetramers, and APOE can further aggregate to form fibrils. Although there is no major difference in the overall quaternary structure or stability of APOE tetramers between isoforms using a range of techniques (Garai and Frieden, 2010; Raulin et al., 2019; Wang et al., 2019), APOE4 has slight differences in two helical regions (amino acids 12–20 and 204–210) which may result in the reduced formation of tetramers (Chetty et al., 2017). These data indicate that isoform differences in tetramers are more associated with number rather than structure. There are also differences in binding domains and larger structures. The APOE4 molecule has been shown to again rely on the C-terminal domain for self-association: when amino acids 273–299 are removed self-association is lost in APOE4 but not APOE3, and APOE4 creates more homo-isomers than APOE3 (Sakamoto et al., 2008). Purified recombinant unlipidated APOE4 forms large oligomers that create fibril-like structures over time; APOE2 and APOE3 make these structures to a lesser degree over the same timeframe (Hatters et al., 2006; Raulin et al., 2019). The reduced HDL binding affinity of APOE4 may result in a larger proportion of unlipidated APOE that is more likely to aggregate (Hatters et al., 2006). The APOE4 large aggregates are more toxic to neurons than APOE2 and APOE3 aggregates (Hatters et al., 2006). Normal self-association up to tetramers, however, may be important for the construction of large complexes with lipoprotein particles able to hold at least two APOE proteins (Chen et al., 2011; Minagawa et al., 2009; Raussens et al., 2005). Consequently, changes in the C-terminal region of APOE4 may not only impact lipid-binding but also healthy oligomer formation.
APOE dimers
The APOE isoform differences at positions 112 and 158 in the N-terminal domain. They are cysteine-arginine substitutions, altering both the charge of the protein and its ability to form cysteine-cysteine dimers (Mahley, 1988). Indeed, APOE4 contains no cysteine residues throughout the protein. Through the cysteine 112 residue, APOE can form disulfide bonds with other APOE proteins and with APOA-II proteins (Weisgraber and Shinto, 1991). As expected, CNS APOE3 isoforms in brain and CSF form APOE-APOE and APOE-APOA-II dimers, while APOE4 isoforms do not (Elliott et al., 2010; Rebeck et al., 1998), although the levels in CSF are much lower than in plasma (Weisgraber and Shinto, 1991). Dimerization at the cysteine 112 site in APOE3 negatively affects its interaction with HDL (Weisgraber, 1990), consistent with the existence of only HDL-like particles in the CSF. Levels of plasma APOE3 homo- and heterodimers correlate with HDL levels (Yamauchi et al., 2017).
APOE protein levels in the CNS
Individuals that express APOE4 have lower levels of APOE in the CNS than those that express APOE3. Some of these data derive from the study of APOE targeted replacement (APOE TR) mice (Riddell et al., 2008; Sullivan et al., 2011; Vitek et al., 2009). These mice express APOE alleles from the endogenous mouse APOE promoter (Sullivan et al., 1997), with the expected glial expression of APOE isoforms (Sullivan et al., 2004). This glial expression pattern is consistent with the observations from a mouse model of GFP expression under the mouse APOE promoter (Xu et al., 2006). APOE4 TR mice have the lowest levels of APOE and APOE2 TR mice the highest APOE levels in: frontal cortex brain extracts (Riddell et al., 2008); hippocampus brain extracts (which had overall more APOE than the frontal cortex (Riddell et al., 2008)); CSF (Fryer et al., 2005; Riddell et al., 2008), and interstitial fluid (Ulrich et al., 2013). Primary astrocytes grown alone show these same trends with APOE4 astrocytes exhibiting reduced APOE secretion compared to APOE3 astrocytes (Riddell et al., 2008).
Findings in the APOE TR mouse model are supported by studies in humans, with APOE2 alleles having a positive impact on APOE protein concentration in the CSF and APOE4 alleles having a negative impact. The CSF from APOE2.3 individuals had the highest levels of APOE, and APOE3.4 and APOE4.4 individuals had the lowest levels (Cruchaga et al., 2012). This same trend has been found in other analyses of CSF APOE concentration (Castellano et al., 2011). A genome wide association study has shown that of the APOE genotype has a strong (p=6.9×10−13) association with CSF protein level and no other SNP reached genome wide significance (Cruchaga et al., 2012). Finally, astrocytes derived from lines of inducible pluripotent stem cells also demonstrated higher levels of cellular and secreted APOE3 than APOE4 (Lin et al., 2018).
APOE glycosylation
APOE is an O-glycoprotein that was initially shown to hold glycosylation at a site in the hinge region (Thr194) (Wernette-Hammond et al., 1989), but has since been shown to also hold glycosylation at sites within the N-terminus (Thr8 and Thr18), the C-terminus (Thr289, Ser290 and Ser296), and at a second site (at low abundance) within the hinge region, Ser197 (Flowers et al., 2019; Halim et al., 2013; Lee et al., 2010; Nilsson et al., 2009; Steentoft et al., 2011). Although identification of the attached glycan is a more technical challenge, APOE holds predominately monosialylated (Neu5Acα2–3Galβ1–3GalNAcα1-) and disialylated (Neu5Acα2–3Galβ1–3(Neu5Acα2–6)GalNAcα1-) core 1 O-glycan structures (Flowers et al., 2019). APOE in the cell is more heavily glycosylated than the secreted forms (Lee et al., 2010; Zannis et al., 1986) and APOE from the CSF is more highly glycosylated compared to APOE isolated from the plasma (Flowers et al., 2019; Pitas et al., 1987c; Rebeck et al., 1998). Normal human CSF holds ten times more abundant glycosylation within the C-terminal lipid-binding domain (CSF 37.8%, Plasma 3.7%), and also holds a higher proportion of larger disialylated core 1 glycans compared to plasma derived APOE (Flowers et al., 2019). Plasma APOE, on the other hand, holds greater glycosylation on the N-terminal domain sites (CSF 0.2%, Plasma 15.8%). Finally, while the hinge domain glycosylation was more similar for both plasma and CSF derived APOE, the CSF APOE held more abundant glycosylation (CSF 26.8%, 11.4% plasma) (Flowers et al., 2019). These analyses have important implications for the binding properties of the APOE from these two compartments, with plasma APOE holding little glycosylation in the C-terminal lipid domain and having a more diverse lipoprotein binding profile. The CSF APOE, on the other hand, binds only the small HDL particles and has higher abundance of C-terminal glycosylation. These observations suggest that C-terminal APOE glycosylation may tailor the disparate lipoprotein binding requirements in the two compartments. In support of this hypothesis, when sialylation was removed from APOE with a neuraminidase that removes α2–3 linked and α2–6 linked Neu5Ac (the linkages since confirmed to be common on APOE (Flowers et al., 2019)), the de-sialylated APOE binding to HDL was more detrimentally impacted than VLDL binding (Marmillot et al., 1999). This binding deficit was then rescued by the re-addition of sialic acid, confirming the importance of complete normal glycosylation including sialylation to effective HDL binding (Marmillot et al., 1999).
Glycosylation differences have been shown by two-dimensional electrophoresis under certain physiological conditions and with APOE genotype. Cells stably expressing APOE under a CMV promoter when loaded with cholesterol showed decreased APOE secretion, and decreased APOE sialylation (Kockx et al., 2012). It is unknown whether the APOE2, APOE3, or APOE4 variants differ in specific aspects of glycosylation in the brain. Brain samples solubilized sequentially in Tris-buffered saline (TBS) and then 1% Triton X-100, to separate soluble and membrane associated fractions, showed that APOE4 brains, both mouse and human, held more soluble higher molecular weight APOE compared to the APOE3 brain samples (DiBattista et al., 2016). Isoelectric focusing of the two fractions showed differences in a series of post-translation modifications, indicating that APOE O-glycosylation is associated with the more soluble forms of APOE in the brain (DiBattista et al., 2016). Interestingly, these modifications were also linked to neuron health: when APOE4 TR mice were treated with an non-steroidal anti-inflammatory drug, soluble, glycosylated APOE decreased and neuronal dendritic spine density increased (DiBattista et al., 2016).
The structural differences in APOE isoforms are outlined in Figure 2, highlighting the regions that are affected by genetic variation, the lipid- and receptor-binding domains, and the glycosylation sites.
CNS APOE genotype effects
The functional consequences of the different APOE isoforms in the CNS can be inferred from the effects of APOE genotype on cognition and behavior before the onset of AD. However, the APOE4 allele is associated with an earlier appearance of amyloid as determined by amyloid PET scans (Jansen et al., 2015) and more amyloid as defined in post-mortem studies (Rebeck et al., 1993; Schmechel et al., 1993), consistent with its correlation with an earlier age of onset of AD (Corder et al., 1993). A meta-analysis of Alzheimer’s Disease Neuroimaging studies showed that very many APOE4-positive control individuals have positive amyloid PET scans by age 60 (Jansen et al., 2015). Thus, it is likely that studies of control individuals middle-aged or older include the effects of both amyloid and APOE4 genotype on brain structures and function. Thus, in this review, we will focus on studies of young human populations, and on mouse models with normal brain APOE regulation and without engineered AD pathological processes.
Human studies
Brain structure
There is a mixed literature on whether APOE genotype affects normal grey matter structure in younger individuals as evaluated by Magnetic Resonance Imaging (MRI) (Alexopoulos et al., 2011; Dennis et al., 2010; DiBattista et al., 2014; Filippini et al., 2009b; Matura et al., 2014; O’Dwyer et al., 2012b). Several recent studies show no APOE genotype-dependent effects in very young populations (Bussy et al., 2019; Lyall et al., 2019; Lyall et al., 2013; Wisdom et al., 2011; Zheng et al., 2017). Effects may be limited to specific hippocampal substructures, e.g., entorhinal cortex, or they may change substantially with normal development. Different effects associated with the APOE4 allele have been reported in small medial temporal lobe structures in infants, children, and young adults (Chang et al., 2016; Dean et al., 2014; Knickmeyer et al., 2014; O’Dwyer et al., 2012a; Shaw et al., 2007). White matter microstructure, as measured by fractional anisotropy and white matter intensities, is impaired in APOE4 carriers compared to non-carriers (Heise et al., 2011; Lyall et al., 2019; Westlye et al., 2012), consistent with potential APOE4-related problems with brain connectivity and activity.
Brain activity
Blood Oxygen Level Dependent (BOLD) contrast imaging in functional MRI is a measure of brain activity. Resting brain activity, analyzed through co-activation of the default mode networks (DMN), showed that young APOE4 individuals have higher co-activations that include the medial temporal lobe than young APOE3 individuals (Filippini et al., 2009a; Shen et al., 2017). APOE genotype effects on the DMN are not only related to APOE4, but include effects of APOE2 as well (Trachtenberg et al., 2012). These effects on the DMN may be related to differences in spontaneous brain activity (Zheng et al., 2017) or lower functional connectivity (Su et al., 2017). During active encoding tasks, APOE genotype is also associated with altered medial temporal lobe activity with increased BOLD signal in young carriers of the APOE4 allele compared to non-carriers (Dennis et al., 2010; Evans et al., 2017; Filippini et al., 2009a). An increased hippocampal activity in APOE4 carriers compared to non-carriers occurred in the cognitive generation of grid-cell-like representations (Kunz et al., 2015), this signal correlates with cerebrovascular reactivity to CO2 (Suri et al., 2015). In contrast to increased activity during encoding tasks, APOE4-positive individuals showed decreased medial temporal lobe activity during executive attention compared to APOE4-negative individuals, which is a task dependent on frontal lobe activation (Green et al., 2014). Thus, in the unimpaired brain, APOE4 is associated with higher levels of medial temporal lobe activity during resting state as well as during functions that depend on its efficient function.
Several studies have identified differences in measures of brain utilization of glucose and oxygen dependent on APOE genotype, supporting a model with APOE4-positive individuals are unable to efficiently regulate cerebral metabolism compared to APOE4-negative individuals (Brandon et al., 2018). The FDG PET measure of glucose uptake was lower in APOE4 individuals in posterior cingulate, parietal, temporal and prefrontal cortex (Reiman et al., 2004). Post-mortem analysis of brains from young individuals show APOE genotype had several effects on levels of brain glucose and lactate transporters, and on mitochondrial electron transport proteins (Perkins et al., 2016). The lower glucose metabolism associated with APOE4 may cause alterations in specific brain activities, or the lower energy metabolism may be caused by alterations in brain activities from other APOE4-related effects.
Behavior
Differences in brain activity and connectivity, particularly related to structures in the medial temporal lobe, may affect behaviors. However, there is a lack of consensus on behavioral effects of APOE genotype in young individuals. Compared to non-APOE4 carriers, young APOE4 carriers perform better in tasks of executive function, verbal fluency and memory (Jochemsen et al., 2012; Mondadori et al., 2007; Rusted et al., 2013). APOE4 individuals have altered navigational behavior, consistent with differences in grid cell-like activity (Kunz et al., 2015) and decreased associative memory (Bussy et al., 2019), compared to individuals without APOE4. A meta-analysis from 2011 concluded that APOE4 was associated with worse measures of episodic memory and global cognitive ability (Wisdom et al., 2011), although many of those studies included older individuals, when APOE4-related impairments increase (Rusted and Carare, 2015; Wisdom et al., 2011), thus potentially lessening the magnitude of direct cognitive effects of APOE4 (Reinvang et al., 2013).
Mouse APOE knock-in model studies
Studies of mouse models of APOE complement human studies, allowing more genetically and environmentally controlled experiments (although in the absence of important factors relevant to human disease). Normal expression of specific human APOE isoforms in AD mouse models (e.g., EFAD mice (Youmans et al., 2012)), are useful in understanding how APOE genotype affects processes such as the deposition of Aβ (Youmans et al., 2012) or inflammatory responses to its accumulation (Rodriguez et al., 2014), and how APOE-related treatments alter AD pathologies (Safieh et al., 2019). However, here we will consider the effects of APOE genotype in mice lacking overt AD pathological changes, comparing mice expressing only APOE4 with those expressing only APOE3.
Brain structure
The ease of collection of mouse brain tissue has allowed detailed studies of microscopic neuronal structures. These studies have consistently revealed that APOE4 mice have simpler structures compared to APOE3 mice. As demonstrated with Golgi stain analyses, APOE4 mice had simpler neuronal dendritic arborization in the amygdala (Wang et al., 2005), cortex (Dumanis et al., 2009; Neustadtl et al., 2017), and hippocampus (Maezawa et al., 2006), including less branching or reduced spine densities. Decreased complexity of neurons in APOE4 brains is also seen in the entorhinal cortex (DiBattista et al., 2016; Rodriguez et al., 2013), consistent with the altered function of that brain region in humans (Kunz et al., 2015). In older mice, APOE4 is associated with fewer inhibitory neurons in the hippocampus (Andrews-Zwilling et al., 2010). APOE4 brains have a lower vascular density, associated with white matter damage (Koizumi et al., 2018) and smaller hippocampal regions (Speidell et al., 2019). Importantly, some structural effects can be modified in ways that make APOE4 mice more like APOE3 mice in terms of neuronal complexity, for example with the anti-inflammatory agent ibuprofen (DiBattista et al., 2016).
Brain activity
The ease of collection of mouse brain tissue has also allowed cellular studies of neuronal activity and synaptic measures. The electrophysiology of amygdala neurons showed reduced excitatory transmission in APOE4 mouse brain (Wang et al., 2005). There are lower levels of inhibitory tone of the APOE4 entorhinal cortex (Nuriel et al., 2017) and hippocampal hilus (Andrews-Zwilling et al., 2010). Evoked release of acetylcholine of hippocampal neurons is lower in older APOE4 mice (Dolejsi et al., 2016). Effects of APOE genotype on hippocampal neurotransmission could account for fewer short wave ripples and reduced slow gamma wave activity in aged APOE4 mice (Gillespie et al., 2016). Thus, there are changes to neuronal activity concomitant with the changes to neuronal structures seen in APOE4 mice.
The molecular processes behind these effects of APOE genotype on neuronal activities remain to be defined, but there are many effects of the endogenous APOE protein on intracellular signaling processes (Hoe et al., 2005; Huang et al., 2017; Lane-Donovan and Herz, 2017). Presynaptically, APOE4 mice show lower glutaminase levels (Dumanis et al., 2013) and altered levels of the vesicular glutamate transporter 1 (Boehm-Cagan and Michaelson, 2014; Dumanis et al., 2013) compared to APOE3 mice. Effects of APOE4 on neuronal activity could be mediated by its effects on the family of low density lipoprotein receptors, such as ApoER2 (Beffert et al., 2004; Weeber et al., 2002). APOE4 mice have lower levels of ApoER2 in the CA1 and CA3 neurons of the hippocampus (Boehm-Cagan et al., 2016b; Gilat-Frenkel et al., 2014). These in vitro and in vivo studies combine to demonstrate that APOE isoforms differentially affect neuronal cell signaling.
Behavior
Effects of APOE genotype on mouse brain structure and activity are reflected in numerous behavioral assays. It is important to reiterate that the APOE-driven differences in behavior reviewed here occur in the absence of pathological changes introduced by transgenes or exogenous agents, and thus do not reflect the effects of gross AD pathological changes. Compared to APOE3 mice, APOE4 mice are impaired in spatial learning as measured in the Barnes maze (Rodriguez et al., 2013; Speidell et al., 2019), the Morris Water Maze (Boehm-Cagan and Michaelson, 2014; Bour et al., 2008; Knoferle et al., 2014; Salomon-Zimri et al., 2014), and Novel Place Recognition (Grootendorst et al., 2005). They are impaired in other memory related pathways, as evidenced by Novel Object Recognition (Boehm-Cagan and Michaelson, 2014; Salomon-Zimri et al., 2014), Contextual Fear Conditioning (Boehm-Cagan and Michaelson, 2014; Salomon-Zimri et al., 2014; Segev et al., 2013) and Y-maze active avoidance (Bour et al., 2008). Several studies demonstrated that deficits were particularly observed in older APOE4 mice (Andrews-Zwilling et al., 2010; Bour et al., 2008), which would be consistent with the increased risk of AD in older individuals. These behaviors present opportunities to alter APOE4-associated phenotypes in the absence of AD pathological changes, relevant for the generation of early prevention approaches. For example GABA potentiation alleviated APOE4-related behavioral deficits in the Morris Water Maze (Andrews-Zwilling et al., 2012) and deficits in Morris Water Maze and Novel Object Recognition were alleviated by bexarotene (Boehm-Cagan and Michaelson, 2014) and an ABCA1 agonist (Boehm-Cagan et al., 2016b).
Thus, human and mouse studies are consistent in their findings that the APOE4 genotype affects the activity and function of the hippocampus, reflected in behavioral differences. These effects may lead to, or be exacerbated by, the presence of the various pathological changes later in life.
CNS APOE structure-function relationships
The effects of APOE genotype on APOE protein, APOE levels, brain structure, and brain function in normal brains are logical targets for studies on the prevention of brain dysfunction in AD (Yamazaki et al., 2016). However, linking measures in the normal brain to prevention of later AD-associated symptoms is a difficult task (Gomez-Isla and Frosch, 2019).
Increasing APOE levels could aid in the clearance of debris, inhibition of inflammation, and delivery of lipids to neurons for increased resilience (Figure 1). APOE levels are increased through activation of various transcription factors related to lipid homeostasis (Cao et al., 2007). APOE levels are further affected by recycling through neuronal endocytic pathways, with deficits in this recycling evidenced with APOE4 (Heeren et al., 2004; Xian et al., 2018). Through interactions with APOE receptors, APOE can promote neural complexity (Lane-Donovan and Herz, 2017), reduce inflammation (Pocivavsek et al., 2009), and promote debris clearance (Rasmussen et al., 2018). Importantly, APOE and APOE-derived peptides have anti-inflammatory effects (Laskowitz et al., 2017; Vitek et al., 2012) through interactions with the family of lipoprotein receptors; increasing APOE functionality could address the connections of pathological changes (Perez-Nievas et al., 2013) and genetics (Malik et al., 2015) with inflammation.
Increasing APOE lipidation in brain-specific HDL can be accomplished using ABCA1 agonists (Boehm-Cagan et al., 2016b) and perhaps through altering C-terminal glycosylation events specific to the CNS (Flowers et al., 2019). Chemical and thermal denaturation studies demonstrate that the APOE4 monomer tertiary structure is less stable and less structured than APOE3 and APOE2, prone to a molten globule state (Morrow et al., 2002; Ray et al., 2017). The altered folding of APOE4 can be targeted with small molecules that stabilize APOE4 (Petros et al., 2019) or prevent APOE domain interactions (Wang et al., 2018). More stable and lipidated forms of APOE have conformations that also promote receptor interactions (Frieden et al., 2017).
Conclusions
The effects of APOE genotype on APOE modification, lipidation, or levels could influence neuronal resilience, the time course or intensity of neuroinflammation, and the homeostasis of extracellular hydrophobic molecules (Figure 1A). These functions are the same ones that are hypothesized to contribute to AD pathogenesis (Figure 1B). Approaches to address these properties of the APOE4 protein are being pursued to treat or prevent the symptoms of AD (Safieh et al., 2019). These potential treatments to address deficiencies in APOE4 positive AD patients could be developed using assays in preclinical studies of normal mice and humans. Some approaches may depend on beginning treatments in advance of marked amyloid accumulation, since there may be adverse effects of the form of APOE4, which is bound chronically to plaques (Wisniewski and Frangione, 1992). Thus, assays need to be developed to monitor characteristics of APOE and its effects in normal brain, such as state of lipidation, basal inflammation, or ability to transport hydrophobic molecules. Ideally, these measures could be based on APOE analyzed in the peripheral circulation, perhaps including studies of glycosylated APOE isoforms that may pass from the CNS to the periphery if the blood brain barrier is impaired. Overall, APOE-directed studies under non-pathological conditions are necessary for testing preventative approaches in this large population genetically at risk for AD.
Acknowledgements
Support for work on the analysis of the published data and the writing of the manuscript was provided by the National Institutes of Health, NIA R56AG062305 (SF) and NINDS R01NS100704 (GWR). We thank Christi Anne Ng for design and production of the Figure 1.
Footnotes
The authors have no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Achariyar TM, et al. , 2016. Glymphatic distribution of CSF-derived apoE into brain is isoform specific and suppressed during sleep deprivation. Mol Neurodegener. 11, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexopoulos P, et al. , 2011. Hippocampal volume differences between healthy young apolipoprotein E epsilon2 and epsilon4 carriers. J Alzheimers Dis. 26, 207–10. [DOI] [PubMed] [Google Scholar]
- Andrews-Zwilling Y, et al. , 2010. Apolipoprotein E4 causes age- and Tau-dependent impairment of GABAergic interneurons, leading to learning and memory deficits in mice. J Neurosci. 30, 13707–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews-Zwilling Y, et al. , 2012. Hilar GABAergic interneuron activity controls spatial learning and memory retrieval. PLoS One. 7, e40555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beffert U, et al. , 2004. Functions of lipoprotein receptors in neurons. J Lipid Res. 45, 403–9. [DOI] [PubMed] [Google Scholar]
- Bennet AM, et al. , 2007. Association of apolipoprotein E genotypes with lipid levels and coronary risk. JAMA. 298, 1300–11. [DOI] [PubMed] [Google Scholar]
- Bielicki JK, 2016. ABCA1 agonist peptides for the treatment of disease. Curr Opin Lipidol. 27, 40–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm-Cagan A, et al. , 2016a. Differential Effects of apoE4 and Activation of ABCA1 on Brain and Plasma Lipoproteins. PLoS One. 11, e0166195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm-Cagan A, et al. , 2016b. ABCA1 Agonist Reverses the ApoE4-Driven Cognitive and Brain Pathologies. J Alzheimers Dis. 54, 1219–1233. [DOI] [PubMed] [Google Scholar]
- Boehm-Cagan A, Michaelson DM, 2014. Reversal of apoE4-driven brain pathology and behavioral deficits by bexarotene. J Neurosci. 34, 7293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bour A, et al. , 2008. Middle-aged human apoE4 targeted-replacement mice show retention deficits on a wide range of spatial memory tasks. Behav Brain Res. 193, 174–82. [DOI] [PubMed] [Google Scholar]
- Brandon JA, et al. , 2018. APOE and Alzheimer’s Disease: Neuroimaging of Metabolic and Cerebrovascular Dysfunction. Front Aging Neurosci. 10, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruinsma IB, et al. , 2010. Apolipoprotein E protects cultured pericytes and astrocytes from D-Abeta(1–40)-mediated cell death. Brain Res. 1315, 169–80. [DOI] [PubMed] [Google Scholar]
- Buskbjerg CDR, et al. , 2019. Genetic risk factors for cancer-related cognitive impairment: a systematic review. Acta Oncol. 58, 537–547. [DOI] [PubMed] [Google Scholar]
- Bussy A, et al. , 2019. Effect of apolipoprotein E4 on clinical, neuroimaging, and biomarker measures in noncarrier participants in the Dominantly Inherited Alzheimer Network. Neurobiol Aging. 75, 42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campion M, et al. , 2019. Genomic education for the next generation of health-care providers. Genet Med. [DOI] [PubMed] [Google Scholar]
- Cao G, et al. , 2007. Liver X receptor-mediated gene regulation and cholesterol homeostasis in brain: relevance to Alzheimer’s disease therapeutics. Curr Alzheimer Res. 4, 179–84. [DOI] [PubMed] [Google Scholar]
- Castellano JM, et al. , 2011. Human apoE isoforms differentially regulate brain amyloid-beta peptide clearance. Sci Transl Med. 3, 89ra57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, et al. , 2016. Gray matter maturation and cognition in children with different APOE epsilon genotypes. Neurology. 87, 585–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, et al. , 2014. Effects of APOE epsilon4, age, and HIV on glial metabolites and cognitive deficits. Neurology. 82, 2213–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, et al. , 2011. Topology of human apolipoprotein E3 uniquely regulates its diverse biological functions. Proceedings of the National Academy of Sciences of the United States of America. 108, 14813–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernick D, et al. , 2018. High-density lipoprotein mimetic peptide 4F mitigates amyloid-beta-induced inhibition of apolipoprotein E secretion and lipidation in primary astrocytes and microglia. J Neurochem. 147, 647–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetty PS, et al. , 2017. Helical structure, stability, and dynamics in human apolipoprotein E3 and E4 by hydrogen exchange and mass spectrometry. Proc Natl Acad Sci U S A. 114, 968–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corder EH, et al. , 1993. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 261, 921–3. [DOI] [PubMed] [Google Scholar]
- Courtney R, Landreth GE, 2016. LXR Regulation of Brain Cholesterol: From Development to Disease. Trends Endocrinol Metab. 27, 404–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer SC, et al. , 2012. Correlation between genetic polymorphisms and stroke recovery: analysis of the GAIN Americas and GAIN International Studies. Eur J Neurol. 19, 718–24. [DOI] [PubMed] [Google Scholar]
- Cruchaga C, et al. , 2012. Cerebrospinal fluid APOE levels: an endophenotype for genetic studies for Alzheimer’s disease. Hum Mol Genet. 21, 4558–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson PA, Rudel LL, 1999. Intestinal cholesterol absorption. Curr Opin Lipidol. 10, 315–20. [DOI] [PubMed] [Google Scholar]
- Dean DC 3rd, et al. , 2014. Brain differences in infants at differential genetic risk for late-onset Alzheimer disease: a cross-sectional imaging study. JAMA Neurol. 71, 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis NA, et al. , 2010. Temporal lobe functional activity and connectivity in young adult APOE varepsilon4 carriers. Alzheimers Dement. 6, 303–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Battista AM, et al. , 2016. Alzheimer’s Disease Genetic Risk Factor APOE-epsilon4 Also Affects Normal Brain Function. Curr Alzheimer Res. 13, 1200–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiBattista AM, et al. , 2016. Identification and modification of amyloid-independent phenotypes of APOE4 mice. Exp Neurol. 280, 97–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiBattista AM, et al. , 2014. Two Alzheimer’s disease risk genes increase entorhinal cortex volume in young adults. Front Hum Neurosci. 8, 779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolejsi E, et al. , 2016. Apolipoprotein E4 reduces evoked hippocampal acetylcholine release in adult mice. J Neurochem. 136, 503–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumanis SB, et al. , 2013. APOE genotype affects the pre-synaptic compartment of glutamatergic nerve terminals. J Neurochem. 124, 4–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumanis SB, et al. , 2009. ApoE4 decreases spine density and dendritic complexity in cortical neurons in vivo. J Neurosci. 29, 15317–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott DA, et al. , 2010. Apolipoprotein-E forms dimers in human frontal cortex and hippocampus. BMC Neurosci. 11, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans S, et al. , 2017. Disrupted neural activity patterns to novelty and effort in young adult APOE-e4 carriers performing a subsequent memory task. Brain Behav. 7, e00612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, et al. , 2018. Small molecule inducers of ABCA1 and apoE that act through indirect activation of the LXR pathway. J Lipid Res. 59, 830–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippini N, et al. , 2009a. Distinct patterns of brain activity in young carriers of the APOE-epsilon 4 allele. Proceedings of the National Academy of Sciences of the United States of America. 106, 7209–7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippini N, et al. , 2009b. Anatomically-distinct genetic associations of APOE epsilon4 allele load with regional cortical atrophy in Alzheimer’s disease. Neuroimage. 44, 724–8. [DOI] [PubMed] [Google Scholar]
- Flowers SA, et al. , 2019. O-glycosylation on cerebrospinal fluid and plasma Apolipoprotein E differs in the lipid binding domain. Glycobiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieden C, et al. , 2017. A mechanism for lipid binding to apoE and the role of intrinsically disordered regions coupled to domain-domain interactions. Proc Natl Acad Sci U S A. 114, 6292–6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer JD, et al. , 2005. The low density lipoprotein receptor regulates the level of central nervous system human and murine apolipoprotein E but does not modify amyloid plaque pathology in PDAPP mice. J Biol Chem. 280, 25754–9. [DOI] [PubMed] [Google Scholar]
- Garai K, Frieden C, 2010. The association-dissociation behavior of the ApoE proteins: kinetic and equilibrium studies. Biochemistry. 49, 9533–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garatachea N, et al. , 2015. The ApoE gene is related with exceptional longevity: a systematic review and meta-analysis. Rejuvenation Res. 18, 3–13. [DOI] [PubMed] [Google Scholar]
- Gilat-Frenkel M, et al. , 2014. Involvement of the Apoer2 and Lrp1 receptors in mediating the pathological effects of ApoE4 in vivo. Curr Alzheimer Res. 11, 549–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie AK, et al. , 2016. Apolipoprotein E4 Causes Age-Dependent Disruption of Slow Gamma Oscillations during Hippocampal Sharp-Wave Ripples. Neuron. 90, 740–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Isla T, Frosch MP, 2019. The Challenge of Defining Alzheimer Disease Based on Biomarkers in the Absence of Symptoms. JAMA Neurol. [DOI] [PubMed] [Google Scholar]
- Green AE, et al. , 2014. A combined effect of two Alzheimer’s risk genes on medial temporal activity during executive attention in young adults. Neuropsychologia. 56, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grootendorst J, et al. , 2005. Human apoE targeted replacement mouse lines: h-apoE4 and h-apoE3 mice differ on spatial memory performance and avoidance behavior. Behav Brain Res. 159, 1–14. [DOI] [PubMed] [Google Scholar]
- Halim A, et al. , 2013. LC-MS/MS characterization of O-glycosylation sites and glycan structures of human cerebrospinal fluid glycoproteins. Journal of Proteome Research. 12, 573–84. [DOI] [PubMed] [Google Scholar]
- Hansen D, et al. , 2019. Review: Clinical, neuropathological and genetic features of Lewy body dementias. Neuropathol Appl Neurobiol. [DOI] [PubMed] [Google Scholar]
- Hatters DM, et al. , 2006. Amino-terminal domain stability mediates apolipoprotein E aggregation into neurotoxic fibrils. J Mol Biol. 361, 932–44. [DOI] [PubMed] [Google Scholar]
- Heeren J, et al. , 2004. Impaired recycling of apolipoprotein E4 is associated with intracellular cholesterol accumulation. J Biol Chem. 279, 55483–92. [DOI] [PubMed] [Google Scholar]
- Heinsinger NM, et al. , 2016. Apolipoprotein E Genotype Affects Size of ApoE Complexes in Cerebrospinal Fluid. J Neuropathol Exp Neurol. 75, 918–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heise V, et al. , 2011. The APOE varepsilon4 allele modulates brain white matter integrity in healthy adults. Mol Psychiatry. 16, 908–16. [DOI] [PubMed] [Google Scholar]
- Hoe HS, et al. , 2005. Multiple pathways of apolipoprotein E signaling in primary neurons. J Neurochem. 93, 145–55. [DOI] [PubMed] [Google Scholar]
- Hu J, et al. , 2015. Opposing effects of viral mediated brain expression of apolipoprotein E2 (apoE2) and apoE4 on apoE lipidation and Abeta metabolism in apoE4-targeted replacement mice. Mol Neurodegener. 10, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YA, et al. , 2017. ApoE2, ApoE3, and ApoE4 Differentially Stimulate APP Transcription and Abeta Secretion. Cell. 168, 427–441 e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono D, Feltis GC, 2019. Impact of Apolipoprotein E gene polymorphism during normal and pathological conditions of the brain across the lifespan. Aging (Albany NY). 11, 787–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen WJ, et al. , 2015. Prevalence of cerebral amyloid pathology in persons without dementia: a meta-analysis. JAMA. 313, 1924–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jochemsen HM, et al. , 2012. APOE epsilon4 differentially influences change in memory performance depending on age. The SMART-MR study. Neurobiol Aging. 33, 832 e15–22. [DOI] [PubMed] [Google Scholar]
- Kassam I, et al. , 2016. Association of the APOE-epsilon4 allele with outcome of traumatic brain injury in children and youth: a meta-analysis and meta-regression. J Neurol Neurosurg Psychiatry. 87, 433–40. [DOI] [PubMed] [Google Scholar]
- Knickmeyer RC, et al. , 2014. Common variants in psychiatric risk genes predict brain structure at birth. Cereb Cortex. 24, 1230–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoferle J, et al. , 2014. Apolipoprotein E4 produced in GABAergic interneurons causes learning and memory deficits in mice. J Neurosci. 34, 14069–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch S, et al. , 2001. Characterization of four lipoprotein classes in human cerebrospinal fluid. J Lipid Res. 42, 1143–51. [PubMed] [Google Scholar]
- Kockx M, et al. , 2012. Cholesterol accumulation inhibits ER to Golgi transport and protein secretion: studies of apolipoprotein E and VSVGt. Biochem J. 447, 51–60. [DOI] [PubMed] [Google Scholar]
- Kockx M, et al. , 2008. Regulation of endogenous apolipoprotein E secretion by macrophages. Arterioscler Thromb Vasc Biol. 28, 1060–7. [DOI] [PubMed] [Google Scholar]
- Koizumi K, et al. , 2018. Apoepsilon4 disrupts neurovascular regulation and undermines white matter integrity and cognitive function. Nat Commun. 9, 3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz L, et al. , 2015. Reduced grid-cell-like representations in adults at genetic risk for Alzheimer’s disease. Science. 350, 430–3. [DOI] [PubMed] [Google Scholar]
- LaDu MJ, et al. , 1998. Nascent astrocyte particles differ from lipoproteins in CSF. J Neurochem. 70, 2070–81. [DOI] [PubMed] [Google Scholar]
- Lalazar A, et al. , 1988. Site-specific mutagenesis of human apolipoprotein E. Receptor binding activity of variants with single amino acid substitutions. Journal of Biological Chemistry. 263, 3542–5. [PubMed] [Google Scholar]
- Lane-Donovan C, Herz J, 2017. The ApoE receptors Vldlr and Apoer2 in central nervous system function and disease. J Lipid Res. 58, 1036–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowitz DT, et al. , 2017. Neuroprotective pentapeptide CN-105 is associated with reduced sterile inflammation and improved functional outcomes in a traumatic brain injury murine model. Sci Rep. 7, 46461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, et al. , 2010. Glycosylation and sialylation of macrophage-derived human apolipoprotein E analyzed by SDS-PAGE and mass spectrometry: evidence for a novel site of glycosylation on Ser290. Molecular & Cellular Proteomics. 9, 1968–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei B, et al. , 2012. Interaction between sex and apolipoprotein e genetic background in a murine model of intracerebral hemorrhage. Transl Stroke Res. 3, 94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YT, et al. , 2018. APOE4 Causes Widespread Molecular and Cellular Alterations Associated with Alzheimer’s Disease Phenotypes in Human iPSC-Derived Brain Cell Types. Neuron. 98, 1141–1154 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyall DM, et al. , 2019. Association between APOE e4 and white matter hyperintensity volume, but not total brain volume or white matter integrity. Brain Imaging Behav. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyall DM, et al. , 2013. Alzheimer’s disease susceptibility genes APOE and TOMM40, and hippocampal volumes in the Lothian birth cohort 1936. PLoS One. 8, e80513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maezawa I, et al. , 2006. Apolipoprotein E isoform-dependent dendritic recovery of hippocampal neurons following activation of innate immunity. J Neuroinflammation. 3, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahley RW, 1988. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 240, 622–30. [DOI] [PubMed] [Google Scholar]
- Mahley RW, Huang Y, 2012. Apolipoprotein e sets the stage: response to injury triggers neuropathology. Neuron. 76, 871–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahley RW, Ji ZS, 1999. Remnant lipoprotein metabolism: key pathways involving cell-surface heparan sulfate proteoglycans and apolipoprotein E. J Lipid Res. 40, 1–16. [PubMed] [Google Scholar]
- Main BS, et al. , 2018. Apolipoprotein E4 impairs spontaneous blood brain barrier repair following traumatic brain injury. Mol Neurodegener. 13, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik M, et al. , 2015. Genetics ignite focus on microglial inflammation in Alzheimer’s disease. Mol Neurodegener. 10, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelblatt JS, et al. , 2018. Cancer-Related Cognitive Outcomes Among Older Breast Cancer Survivors in the Thinking and Living With Cancer Study. J Clin Oncol. JCO1800140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmillot P, et al. , 1999. Desialylation of human apolipoprotein E decreases its binding to human high-density lipoprotein and its ability to deliver esterified cholesterol to the liver. Metabolism. 48, 1184–92. [DOI] [PubMed] [Google Scholar]
- Matura S, et al. , 2014. Differential effects of the ApoE4 genotype on brain structure and function. Neuroimage. 89, 81–91. [DOI] [PubMed] [Google Scholar]
- Merritt VC, et al. , 2018. APOE-epsilon4 Genotype is Associated with Elevated Post-Concussion Symptoms in Military Veterans with a Remote History of Mild Traumatic Brain Injury. Arch Clin Neuropsychol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michikawa M, et al. , 2000. Apolipoprotein E exhibits isoform-specific promotion of lipid efflux from astrocytes and neurons in culture. J Neurochem. 74, 1008–16. [DOI] [PubMed] [Google Scholar]
- Minagawa H, et al. , 2009. Mechanism underlying apolipoprotein E (ApoE) isoform-dependent lipid efflux from neural cells in culture. J Neurosci Res. 87, 2498–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondadori CR, et al. , 2007. Better memory and neural efficiency in young apolipoprotein E epsilon4 carriers. Cereb Cortex. 17, 1934–47. [DOI] [PubMed] [Google Scholar]
- Morrow JA, et al. , 2002. Apolipoprotein E4 forms a molten globule. A potential basis for its association with disease. J Biol Chem. 277, 50380–5. [DOI] [PubMed] [Google Scholar]
- Nelissen K, et al. , 2012. Liver X receptors regulate cholesterol homeostasis in oligodendrocytes. J Neurosci Res. 90, 60–71. [DOI] [PubMed] [Google Scholar]
- Neustadtl AL, et al. , 2017. Reduced cortical excitatory synapse number in APOE4 mice is associated with increased calcineurin activity. Neuroreport. 28, 618–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D, et al. , 2010. Molecular basis for the differences in lipid and lipoprotein binding properties of human apolipoproteins E3 and E4. Biochemistry. 49, 10881–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson J, et al. , 2009. Enrichment of glycopeptides for glycan structure and attachment site identification. Nat Methods. 6, 809–11. [DOI] [PubMed] [Google Scholar]
- Nuriel T, et al. , 2017. Neuronal hyperactivity due to loss of inhibitory tone in APOE4 mice lacking Alzheimer’s disease-like pathology. Nat Commun. 8, 1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Dwyer L, et al. , 2012a. White matter differences between healthy young ApoE4 carriers and non-carriers identified with tractography and support vector machines. PLoS One. 7, e36024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Dwyer L, et al. , 2012b. Reduced hippocampal volume in healthy young ApoE4 carriers: an MRI study. PLoS One. 7, e48895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otvos JD, 2002. Measurement of lipoprotein subclass profiles by nuclear magnetic resonance spectroscopy. Clin Lab. 48, 171–80. [PubMed] [Google Scholar]
- Patsch J, 1998. Influence of lipolysis on chylomicron clearance and HDL cholesterol levels. Eur Heart J. 19 Suppl H, H2–6. [PubMed] [Google Scholar]
- Perez-Nievas BG, et al. , 2013. Dissecting phenotypic traits linked to human resilience to Alzheimer’s pathology. Brain. 136, 2510–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins M, et al. , 2016. Altered Energy Metabolism Pathways in the Posterior Cingulate in Young Adult Apolipoprotein E varepsilon4 Carriers. J Alzheimers Dis. 53, 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petros AM, et al. , 2019. Fragment-Based Discovery of an Apolipoprotein E4 (apoE4) Stabilizer. J Med Chem. 62, 4120–4130. [DOI] [PubMed] [Google Scholar]
- Pitas RE, et al. , 1987a. Astrocytes synthesize apolipoprotein E and metabolize apolipoprotein E-containing lipoproteins. Biochimica et Biophysica Acta. 917, 148–61. [DOI] [PubMed] [Google Scholar]
- Pitas RE, et al. , 1987b. Lipoproteins and their receptors in the central nervous system. Characterization of the lipoproteins in cerebrospinal fluid and identification of apolipoprotein B,E(LDL) receptors in the brain. Journal of Biological Chemistry. 262, 14352–60. [PubMed] [Google Scholar]
- Pitas RE, et al. , 1987c. Lipoproteins and their receptors in the central nervous system. Characterization of the lipoproteins in cerebrospinal fluid and identification of apolipoprotein B,E(LDL) receptors in the brain. J Biol Chem. 262, 14352–60. [PubMed] [Google Scholar]
- Pocivavsek A, et al. , 2009. Low-density lipoprotein receptors regulate microglial inflammation through c-Jun N-terminal kinase. Glia. 57, 444–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raber J, et al. , 2004. ApoE genotype accounts for the vast majority of AD risk and AD pathology. Neurobiol Aging. 25, 641–50. [DOI] [PubMed] [Google Scholar]
- Rall SC Jr., et al. , 1982. Human apolipoprotein E. The complete amino acid sequence. Journal of Biological Chemistry. 257, 4171–8. [PubMed] [Google Scholar]
- Rasmussen MK, et al. , 2018. The glymphatic pathway in neurological disorders. Lancet Neurol. 17, 1016–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raulin AC, et al. , 2019. The Molecular Basis for Apolipoprotein E4 as the Major Risk Factor for Late-Onset Alzheimer’s Disease. J Mol Biol. 431, 2248–2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raussens V, et al. , 2005. Orientation and mode of lipid-binding interaction of human apolipoprotein E C-terminal domain. Biochem J. 387, 747–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, et al. , 2017. Atomistic Insights into Structural Differences between E3 and E4 Isoforms of Apolipoprotein E. Biophys J. 113, 2682–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeck GW, et al. , 1998. Structure and functions of human cerebrospinal fluid lipoproteins from individuals of different APOE genotypes. Exp Neurol. 149, 175–82. [DOI] [PubMed] [Google Scholar]
- Rebeck GW, et al. , 1994. Reduced apolipoprotein epsilon 4 allele frequency in the oldest old Alzheimer’s patients and cognitively normal individuals. Neurology. 44, 1513–6. [DOI] [PubMed] [Google Scholar]
- Rebeck GW, et al. , 1993. Apolipoprotein E in sporadic Alzheimer’s disease: allelic variation and receptor interactions. Neuron. 11, 575–80. [DOI] [PubMed] [Google Scholar]
- Reiman EM, et al. , 2004. Functional brain abnormalities in young adults at genetic risk for late-onset Alzheimer’s dementia. Proc Natl Acad Sci U S A. 101, 284–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinvang I, et al. , 2013. APOE-related biomarker profiles in non-pathological aging and early phases of Alzheimer’s disease. Neurosci Biobehav Rev. 37, 1322–35. [DOI] [PubMed] [Google Scholar]
- Revelas M, et al. , 2018. Review and meta-analysis of genetic polymorphisms associated with exceptional human longevity. Mech Ageing Dev. 175, 24–34. [DOI] [PubMed] [Google Scholar]
- Riddell DR, et al. , 2008. Impact of apolipoprotein E (ApoE) polymorphism on brain ApoE levels. J Neurosci. 28, 11445–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez GA, et al. , 2013. Young APOE4 targeted replacement mice exhibit poor spatial learning and memory, with reduced dendritic spine density in the medial entorhinal cortex. Learning & Memory. 20, 256–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez GA, et al. , 2014. Human APOE4 increases microglia reactivity at Abeta plaques in a mouse model of Abeta deposition. J Neuroinflammation. 11, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roher AE, et al. , 2009. Proteomics-derived cerebrospinal fluid markers of autopsy-confirmed Alzheimer’s disease. Biomarkers. 14, 493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusted J, Carare RO, 2015. Are the effects of APOE 4 on cognitive function in nonclinical populations age- and gender-dependent? Neurodegener Dis Manag. 5, 37–48. [DOI] [PubMed] [Google Scholar]
- Rusted JM, et al. , 2013. APOE e4 polymorphism in young adults is associated with improved attention and indexed by distinct neural signatures. Neuroimage. 65, 364–73. [DOI] [PubMed] [Google Scholar]
- Safieh M, et al. , 2019. ApoE4: an emerging therapeutic target for Alzheimer’s disease. BMC Med. 17, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, et al. , 2008. Contributions of the carboxyl-terminal helical segment to the self-association and lipoprotein preferences of human apolipoprotein E3 and E4 isoforms. Biochemistry. 47, 2968–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon-Zimri S, et al. , 2014. Hippocampus-related cognitive impairments in young apoE4 targeted replacement mice. Neurodegener Dis. 13, 86–92. [DOI] [PubMed] [Google Scholar]
- Schachter F, et al. , 1994. Genetic associations with human longevity at the APOE and ACE loci. Nat Genet. 6, 29–32. [DOI] [PubMed] [Google Scholar]
- Schmechel DE, et al. , 1993. Increased amyloid beta-peptide deposition in cerebral cortex as a consequence of apolipoprotein E genotype in late-onset Alzheimer disease. Proc Natl Acad Sci U S A. 90, 9649–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiani P, et al. , 2019. APOE Alleles and Extreme Human Longevity. J Gerontol A Biol Sci Med Sci. 74, 44–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segev Y, et al. , 2013. ApoE epsilon4 is associated with eIF2alpha phosphorylation and impaired learning in young mice. Neurobiol Aging. 34, 863–72. [DOI] [PubMed] [Google Scholar]
- Shaw P, et al. , 2007. Cortical morphology in children and adolescents with different apolipoprotein E gene polymorphisms: an observational study. Lancet Neurol. 6, 494–500. [DOI] [PubMed] [Google Scholar]
- Shen J, et al. , 2017. Modulation of APOE and SORL1 genes on hippocampal functional connectivity in healthy young adults. Brain Struct Funct. 222, 2877–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman CB, et al. , 2010. Apolipoprotein A-I mimetic peptides: a potential new therapy for the prevention of atherosclerosis. Cardiol Rev. 18, 141–7. [DOI] [PubMed] [Google Scholar]
- Sing CF, Davignon J, 1985. Role of the apolipoprotein E polymorphism in determining normal plasma lipid and lipoprotein variation. Am J Hum Genet. 37, 268–85. [PMC free article] [PubMed] [Google Scholar]
- Speidell AP, et al. , 2019. Development of a Human APOE Knock-in Mouse Model for Study of Cognitive Function After Cancer Chemotherapy. Neurotox Res. 35, 291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steentoft C, et al. , 2011. Mining the O-glycoproteome using zinc-finger nuclease-glycoengineered SimpleCell lines. Nat Methods. 8, 977–82. [DOI] [PubMed] [Google Scholar]
- Strittmatter WJ, et al. , 1993. Binding of human apolipoprotein E to synthetic amyloid beta peptide: isoform-specific effects and implications for late-onset Alzheimer disease. Proc Natl Acad Sci U S A. 90, 8098–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YY, et al. , 2017. Lower functional connectivity of default mode network in cognitively normal young adults with mutation of APP, presenilins and APOE epsilon4. Brain Imaging Behav. 11, 818–828. [DOI] [PubMed] [Google Scholar]
- Sullivan PM, et al. , 2011. Reduced levels of human apoE4 protein in an animal model of cognitive impairment. Neurobiol Aging. 32, 791–801. [DOI] [PubMed] [Google Scholar]
- Sullivan PM, et al. , 2004. Marked regional differences of brain human apolipoprotein E expression in targeted replacement mice. Neuroscience. 124, 725–33. [DOI] [PubMed] [Google Scholar]
- Sullivan PM, et al. , 1997. Targeted replacement of the mouse apolipoprotein E gene with the common human APOE3 allele enhances diet-induced hypercholesterolemia and atherosclerosis. J Biol Chem. 272, 17972–80. [DOI] [PubMed] [Google Scholar]
- Suri S, et al. , 2015. Reduced cerebrovascular reactivity in young adults carrying the APOE epsilon4 allele. Alzheimers Dement. 11, 648–57 e1. [DOI] [PubMed] [Google Scholar]
- Tall AR, 2018. Plasma high density lipoproteins: Therapeutic targeting and links to atherogenic inflammation. Atherosclerosis. 276, 39–43. [DOI] [PubMed] [Google Scholar]
- Tenger C, Zhou X, 2003. Apolipoprotein E modulates immune activation by acting on the antigen-presenting cell. Immunology. 109, 392–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachtenberg AJ, et al. , 2012. The effects of APOE on the functional architecture of the resting brain. Neuroimage. 59, 565–72. [DOI] [PubMed] [Google Scholar]
- Ulrich JD, et al. , 2013. In vivo measurement of apolipoprotein E from the brain interstitial fluid using microdialysis. Mol Neurodegener. 8, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitek MP, et al. , 2009. APOE genotype-specific differences in the innate immune response. Neurobiol Aging. 30, 1350–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitek MP, et al. , 2012. APOE-mimetic peptides reduce behavioral deficits, plaques and tangles in Alzheimer’s disease transgenics. Neurodegener Dis. 10, 122–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagle J, et al. , 2009. Association between ApoE epsilon4 and cognitive impairment after stroke. Dement Geriatr Cogn Disord. 27, 525–33. [DOI] [PubMed] [Google Scholar]
- Wang C, et al. , 2018. Gain of toxic apolipoprotein E4 effects in human iPSC-derived neurons is ameliorated by a small-molecule structure corrector. Nat Med. 24, 647–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, et al. , 2005. Human apoE4-targeted replacement mice display synaptic deficits in the absence of neuropathology. Neurobiol Dis. 18, 390–8. [DOI] [PubMed] [Google Scholar]
- Wang H, Eckel RH, 2014. What are lipoproteins doing in the brain? Trends Endocrinol Metab. 25, 8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, et al. , 2019. Native Mass Spectrometry, Ion Mobility, Electron-Capture Dissociation, and Modeling Provide Structural Information for Gas-Phase Apolipoprotein E Oligomers. J Am Soc Mass Spectrom. 30, 876–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardell MR, et al. , 1987. Apolipoprotein E2-Christchurch (136 Arg----Ser). New variant of human apolipoprotein E in a patient with type III hyperlipoproteinemia. J Clin Invest. 80, 483–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeber EJ, et al. , 2002. Reelin and ApoE receptors cooperate to enhance hippocampal synaptic plasticity and learning. J Biol Chem. 277, 39944–52. [DOI] [PubMed] [Google Scholar]
- Weisgraber KH, 1990. Apolipoprotein E distribution among human plasma lipoproteins: role of the cysteine-arginine interchange at residue 112. J Lipid Res. 31, 1503–11. [PubMed] [Google Scholar]
- Weisgraber KH, Shinto LH, 1991. Identification of the disulfide-linked homodimer of apolipoprotein E3 in plasma. Impact on receptor binding activity. J Biol Chem. 266, 12029–34. [PubMed] [Google Scholar]
- Wernette-Hammond ME, et al. , 1989. Glycosylation of human apolipoprotein E. The carbohydrate attachment site is threonine 194. Journal of Biological Chemistry. 264, 9094–101. [PubMed] [Google Scholar]
- Westerlund JA, Weisgraber KH, 1993. Discrete carboxyl-terminal segments of apolipoprotein E mediate lipoprotein association and protein oligomerization. Journal of Biological Chemistry. 268, 15745–50. [PubMed] [Google Scholar]
- Westlye LT, et al. , 2012. Effects of APOE on brain white matter microstructure in healthy adults. Neurology. 79, 1961–9. [DOI] [PubMed] [Google Scholar]
- Wisdom NM, et al. , 2011. The effects of apolipoprotein E on non-impaired cognitive functioning: a meta-analysis. Neurobiol Aging. 32, 63–74. [DOI] [PubMed] [Google Scholar]
- Wisniewski T, Frangione B, 1992. Apolipoprotein E: a pathological chaperone protein in patients with cerebral and systemic amyloid. Neurosci Lett. 135, 235–8. [DOI] [PubMed] [Google Scholar]
- Xian X, et al. , 2018. Reversal of ApoE4-induced recycling block as a novel prevention approach for Alzheimer’s disease. Elife. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, et al. , 2013. LXR agonists: new potential therapeutic drug for neurodegenerative diseases. Mol Neurobiol. 48, 715–28. [DOI] [PubMed] [Google Scholar]
- Xu Q, et al. , 2006. Profile and regulation of apolipoprotein E (ApoE) expression in the CNS in mice with targeting of green fluorescent protein gene to the ApoE locus. J Neurosci. 26, 4985–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi K, et al. , 2017. Redox status of serum apolipoprotein E and its impact on HDL cholesterol levels. Clin Biochem. 50, 777–783. [DOI] [PubMed] [Google Scholar]
- Yamazaki Y, et al. , 2016. Apolipoprotein E as a Therapeutic Target in Alzheimer’s Disease: A Review of Basic Research and Clinical Evidence. CNS Drugs. 30, 773–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youmans KL, et al. , 2012. APOE4-specific changes in Abeta accumulation in a new transgenic mouse model of Alzheimer disease. J Biol Chem. 287, 41774–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, et al. , 2016. Polyhedral 3D structure of human plasma very low density lipoproteins by individual particle cryo-electron tomography1. J Lipid Res. 57, 1879–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannis VI, et al. , 1986. Intracellular modifications of human apolipoprotein E. Journal of Biological Chemistry. 261, 13415–21. [PubMed] [Google Scholar]
- Zheng LJ, et al. , 2017. Altered spontaneous brain activity pattern in cognitively normal young adults carrying mutations of APP, presenilin-1/2 and APOE epsilon4. Eur J Radiol. 95, 18–23. [DOI] [PubMed] [Google Scholar]