Abstract
As sessile organisms, plants face a variety of environmental challenges. Their reproduction and survival depend on their ability to adapt to these stressors, which include water, heat stress, high salinity, and pathogen infection. Failure to adapt to these stressors results in programmed cell death and decreased viability, as well as reduced productivity in the case of crop plants. The growth and development of plants are maintained by meiosis and mitosis as well as endoreduplication, during which DNA replicates without cytokinesis, leading to polyploidy. As in other eukaryotes, the cell cycle in plants consists of four stages (G1, S, G2, and M) with two major check points, namely, the G1/S check point and G2/M check point, that ensure normal cell division. Progression through these checkpoints involves the activity of cyclin-dependent kinases and their regulatory subunits known as cyclins. In order for plants to survive, cell cycle control must be balanced with adaption to dynamic environmental conditions. In this review, we summarize recent advances in our understanding of cell cycle regulation in plants, with a focus on the molecular interactions of cell cycle machinery in the context of stress tolerance.
Keywords: stress response, cell cycle, cyclin, cyclin-dependent kinase, immunity
Introduction
All organisms have evolved mechanisms to cope with a broad range of biotic and abiotic stresses (Hou et al., 2012; Liu et al., 2012; Tian et al., 2017; Chang et al., 2018). The former is caused by external organisms such as bacteria, virus, fungi, and animals (Liang et al., 2010; Huang et al., 2013; Zhang et al., 2014; Zhang F. et al., 2015; Xing et al., 2017), whereas the latter is attributable to environmental conditions, such as drought (Zhao S. et al., 2016; Tang et al., 2017), cold temperature (Sui, 2015), changes in salinity (Cui et al., 2018), and so on. Unlike animals, plants are sessile and have therefore developed unique stress responses involving many types of sensor that ensure their survival.
In nature, plants are vulnerable to pathogens and predators. The immune system in animals has innate and adaptive components; immune signaling cascades have been widely studied in fish (Liu et al., 2011; Sun et al., 2012; Li et al., 2013; Liu T. et al., 2017), cow (He C. et al., 2016; Hou et al., 2017; Hou et al., 2018), pig (Zheng S. et al., 2017; Zheng et al., 2018), and mouse (Cui et al., 2011; Yoshida et al., 2019). In contrast, plants lack the adaptive component and rely primarily on two layers of innate immunity in their response to pathogen infection. Plants sense pathogens through recognition of conserved microbe-associated molecular patterns and host-derived damage-associated molecular patterns (Boller and Felix, 2009). Additionally, disease resistance proteins recognize effector molecules secreted by pathogens and induce a more robust and rapid immune response that usually leads to programmed cell death (Zhang et al., 2018). However, immune activation perturbs the cell cycle, which controls plant growth; recent evidence suggests a close connection between cell cycle kinetics and immunity in plants (Bao and Hua, 2015).
Plants can detect various environmental signals such as high salt concentration (Shen et al., 2014; Wang S. et al., 2014; Liu S. et al., 2017; Sui et al., 2017), cold temperature (Tang et al., 2012; Liu et al., 2013; Zhao X. et al., 2014; He Y. et al., 2016), reactive oxygen species levels (Li K. et al., 2012), waterlogging (Song et al., 2011; Chen et al., 2016; Zhang Y. et al., 2017), phosphate scarcity (Sui et al., 2017), and ultraviolet (UV) radiation (Chen et al., 2013; Deng et al., 2015). Failure to appropriately respond to these stressors decreases plant productivity and affects human food and animal feed supplies. As such, considerable effort has been invested in developing methods to improve stress resistance in crop plants such as wheat (Gong et al., 2017; Kong et al., 2017), peanut (Hou et al., 2014; An et al., 2015), rice (Yang et al., 2013; Bai et al., 2016), tobacco (Guo et al., 2013; Cao et al., 2017), tomato (Sun et al., 2010; Meng et al., 2015), maize (Zhao K. et al., 2010; Wang S. et al., 2014), cotton (Zhao et al., 2012; Wang et al., 2016), and bean (Weng et al., 2011; Zhao B. et al., 2016).
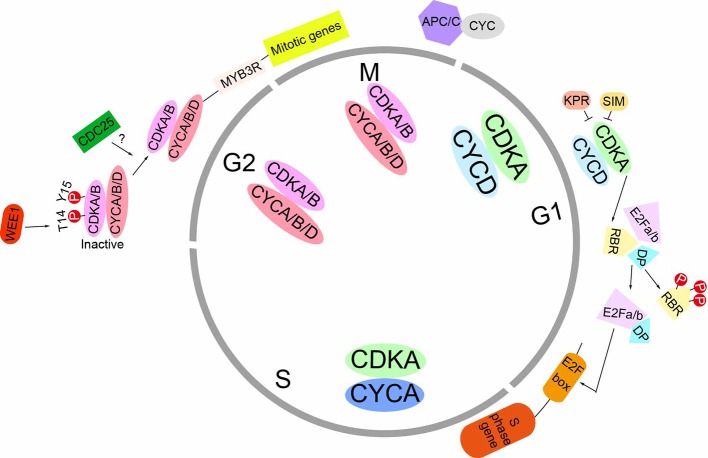
Plants growth results from the coordinated interaction of mitotic cell cycle and cell expansion. As in animals, the plant cell cycle consists of four distinct phases: G1 (postmitotic interphase), S (DNA synthesis phase), G2 (premitotic interphase), and M (mitosis/cytokinesis). Cell cycle progression is driven by cyclin-dependent kinases (CDKs) which cooperated with cyclins. There are seven classes of cyclins in plants which consists of approximately 60 cyclin genes. Among them, we know more about the A, B, and D classes. In general, D-type cyclins are considered as the regulators of G1-to-S transition, A-type cyclins are thought to control S-to-M phase, and B-type cyclins control G2-to-M transition. At G1-to-S transition, CDKA/CYCD complex phosphorylates the retinoblastoma-related (RBR) protein which activates the S-phase transcription factor, “E2F”. E2Fs promotes the G1–S transition by modulating the expression of genes involved in DNA replication, cell-cycle progression and chromatin dynamics (del Pozo et al., 2006). The negative regulation of G1/S transition is the KRPs (Kip-related proteins) and SIM (SIAMESE) which play roles as the inhibitors of CDKA/CYCD complex. At G2/M transition, CDKA and CDKB bind to CYCA, CYCB, or CYCD and drive cells into division. Meanwhile, the activities of CDKA and CDKB are negatively regulated by the WEE1 kinase. The CDC25 homologous kinase, which dephosphorylates the inhibitory phosphorylated site in CDK, still needs to be identified. Once the CDK/CYC complexes are activated, they trigger the G2-to-M transition through phosphorylating numerous of different substrates. Mitotic exit requires the proteolytic degradation of the cyclin subunits which mediated by the anaphase-promoting complex (APC) (Capron et al., 2003) ( Figure 1 ). Although the basic mechanisms which control mitotic cell cycle progression are conserved from plants to vertebrate, plants have unique molecules orchestrating the cell cycle. Moreover, plants have usually a variety of cyclins even though the function of many of which is unclear. At the same time, plants don’t have Cyclin E, which is a major target for organ regulators in animals (Halder and Johnson, 2011).
Figure 1.
Schematic representation of the mitotic cell cycle in plants. At the G1 phase, D-type cyclins (CYCD) interact with the A-type CDK (CDKA), forming the CDKA/CYCD complex. The activity of CDKA/CYCD complex can be negatively regulated by KPR and SIM proteins. Once activation, this complex phosphorylates RBR to release the transcript factor E2Fa/b-DP. This E2Fa/b-DP complex binds to the E2F box and activate the transcription of S phase genes. At the G2 and M phase S, CYCA and CYCB are strongly expressed and their gene products assemble with CDKA and CDKB. The CYCD can also associate with CDKs. At the beginning of G2 phase, CDK activity are inhibited because of the phosphorylation of Y14 and T15 site by WEEI kinase. The CDC25-related kinase, which removes the inhibitory phosphate groups, still needs to be identified. Once the CDK/CYC complex are active, they phosphorylate MYB3R transcription factors and activate mitotic genes’ transcription. Mitotic exit requires anaphase-promoting complex (APC), which degrades cyclins through ubiquitin-proteasome pathway.
In addition to mitotic cell cycle, the endocycle plays pivotal role in plant growth. Endocycle means that DNA replication in the absence of cytokinesis and leads to polyploid. The switch from the mitotic cell cycle to the endocycle involves changes in the regulation and abundance of a wide broad of cyclin-dependent kinases (CDKs), cyclins (CYC), and regulatory proteins/transcription factors. Endopolyploidy is common in reproductive tissues of plants, for example the nutritive tissue of the endosperm in seeds. The onset of endopolyploidy is induced in some cell tissues in response to stress. In this review, we summarize the molecular link between environmental signals and cell cycle progression and the stress response in plants.
Cell Cycle Regulation and Immunity
All animals and plants are vulnerable to infection with viruses and bacteria. For example, cattle and pigs are easily infected with bovine ephemeral fever virus and porcine circovirus, respectively (Yu et al., 2012; Wang X. et al., 2014), and rely on innate and adaptive immunity for protection. The bacterial strain BGI-1, isolated from the gut of the German cockroach (Blattella germanica), contributes to anti-entomopathogenic fungal infection (Huang et al., 2013). Additionally, pathogenic microorganism invasion induces the innate immune responses in zebrafish (Yang H. et al., 2017). Toll-like receptor (TLR)22 (Li H. et al., 2017), TLR18 (Shan et al., 2018), X box-binding protein 1 (Li T. et al., 2017), and interferon regulatory factor 1 (Shan et al., 2016; Zhu et al., 2016a; Zhu et al., 2016b) have been identified as critical molecules involved in immunity in the common carp. However, unlike animals, plants lack a somatic adaptive immune system and instead rely on the innate immunity of each cell to detect signals originating from sites of infection (Ausubel, 2005; Chisholm et al., 2006). Plants detect the extracellular pathogens through plasma membrane-localized pattern recognition receptors (PRRs) that recognize conserved microbe-associated molecular patterns (MAMPs), such as bacterial flg22 and elf18 (active epitopes of bacterial flagellin and elongation factor-Tu, respectively), and then drive patterns-triggered immunity (PTI) (Petutschnig et al., 2010). In addition, plants possess a variety of resistance (R) genes which encode proteins containing nucleotide binding (NB) and leucine rich repeat (LRR) domains. NB-LRR proteins induce effector-triggered immunity (ETI) after specific perception of pathogenic T3SEs.
Interaction with pathogens influences cell cycle progression in plants. In Arabidopsis thaliana, Cabbage leaf curl virus infection alters the expression of cell cycle-related factors; overexpression of CYCD3;1 or E2FB, both of which promote mitosis, increases polyploidy and strongly inhibits Cabbage leaf curl virus infection (Ascencio-Ibanez et al., 2008). Powdery mildew (PM), which is the main pathogen of cereal crops, stimulates the endocycle and increases calcium signaling at the infection site in Arabidopsis. The loss-of-function mutant of MYB3R4, a transcription factor required for endocycle initiation, suppresses PM growth (Chandran et al., 2009). However, it’s currently unclear how cell immunity response is communicated to cell cycle progression.
In a broad sense, PTI functions through several pathways such as the hormone pathways or the phosphorylation of mitogen-activated protein kinases (MAPKs) or the rapid production of reactive oxygen species (ROS) and Ca2+-mediated activation of Ca2+-dependent kinases (CDPKs) that activate transcription factors to induce the expression of down-stream genes. Immunity-triggered growth inhibition might be partly related to jasmonates (JA)-mediated inhibition of the cell cycle. Increased JA level is a positive defense response to pathogen infection. Chen et al. (2011) indicated that JA suppressed CDKA;1 and CYCB1;1. Recent studies have further revealed that JA signaling stabilizes DELLA proteins which induce the CDKA and CDKB inhibitors KRP2, SIM, and SMR (Achard et al., 2009). Therefore, inhibited cell cycle progression may be a consequence of hormone. Harnessing the toxic properties of reactive oxygen species (ROS) to fight against invading pathogens might be another reason for prolonged cell cycle. ROS in adequate concentration directs the invaded plant cells towards apoptosis so as to restrict the fungal infection spread (Bastas, 2014). In mammalian cells, ROS influence cell cycle progression via phosphorylation and ubiquitination of CDKs and cell cycle regulatory molecules, such as CKI and Cdc25. ROS exert their effect on Cdc25 activity via enhancing phosphorylation of Cdc25 or alternatively inactivation of Cdc25 by sulfonation of cysteine in the active site (Verbon et al., 2012). Therefore, it’s reasonable to speculate plant ROS play roles in the similar ways as in mammalian. Regardless of pathogen type, ETI is commonly accompanied by programmed cell death (PCD) at the infection site to stop pathogen spread (Zebell and Dong, 2015).
It’s of significant importance to investigate whether cell cycle regulatory genes promote immunity positively. The Arabidopsis genes OMISSION OF THE SECOND DIVISION (OSD)1 and its homolog UV-B-INSENSITIVE (UVI)4 negatively regulate the activities of anaphase-promoting complex (APC)/cyclosome. Overexpression of both OSD1 and UVI4, or APC6 deficiency, enhances immunity to pathogens by stimulating the expression of disease resistance genes, such as SNC1, which is associated with inhibition of the endocycle (Bao et al., 2013). However, the mechanisms by which OSD1 and UVI4 enhance immunity still need to be explored. In fact, recent studies indicated a direct link of cell cycle regulators in immunity, for example, TEOSINTE BRANCHED1, CYCLOIDEA, PCF1 (TCP)15, and MODIFIER OF SNC1 (MOS1). As a transcription factor, TCP15 regulates the transcription of CYCA2;3 (Li Z. et al., 2012) and SUPPRESSOR OF rps4-RLD1, which encodes a protein that negatively regulates plant immunity (Kim et al., 2014). In Arabidopsis, MOS1 directly interacts with TCP15, and affect the expression of CYCD3;1 and the immune receptor gene SUPRESSOR OF npr1-1, CONSTITUTIVE (SNC)1. Additionally, CYCD3;1 overexpression enhances immune responses and SNC1 protein levels. Moreover, G1-S phase checkpoint proteins Rb and E2F engaged in immune-related PCD (Chandran et al., 2014).
Taken together, these findings indicate that plants balance cell cycle regulation and immune response through manipulating the level of cyclins or the activation of CDKs. Plants respond to pathogen attack via arrested cell cycle progression depend on the state at which plants are attacked. At mitosis, inhibition of APC/C activation will contribute to immunity. At G1, S, or G2 phase, operating the expression of cyclins or check point proteins, such as E2Fs and MYB3R, are of good benefit to immunity. It’s reasonable to speculate that the biological significance of using cell cycle components as regulators of immunity is to turn cells from endoreduplication, which favors pathogen infection, to cell cycle arrest or cell death, and therefore prevent pathogen infection spread. Though recent studies have broadened our knowledge of immunity-cell cycle trade-off, future efforts are needed to identify the immunity-growth pathway and the detailed molecular mechanisms. Single-cell RNA sequence technology, RNA-Seq approach and proteomic analysis will help to underscore molecular connections that ensure antagonistic regulation of growth and immunity in plants under natural or abiotic stress conditions.
Cell Cycle Regulation and Abiotic Stress Adaptation
Salinity, temperature, moisture, and light are abiotic factors that influence plant growth and crop yield, which is shown to be a consequence of cell proliferation and the cell expansion. A rapid response to changing conditions is critical for successful adaptation. Plants respond to different stresses through a variety of mechanisms. In the following sections, we highlight the role of protein kinases that regulate cell cycle progression in the response to abiotic stressors.
Salt Stress
In general, high salinity leads to ion toxicity, water deficit, and oxidative stress. Osmotic stress also reduces cell division rates and cell numbers in leaves, roots, or the shoot meristem. Calcium ion (Ca2+) responses, reactive oxygen species (ROS) burst and abscisic acid (ABA)-dependent protein kinases are involved in response to high salinity. However, recent evidence has begun to underline the cell cycle regulator’s importance in stress response, especially cyclins and CDKs. A. thaliana, a non–salt-tolerant plant, is widely used as a model to investigate the molecular mechanisms of salt tolerance (Qi et al., 2010; Sun et al., 2013; Zhang L. et al., 2017; Zheng Y. et al., 2017). Two CDKs have been identified in A. thaliana: AtCDC2a regulates the G1/S and G2/M transitions, whereas AtCDC2b is mainly expressed in S and G2 phases in meristem tissues. Upon NaCl treatment, the transcription of AtCDC2a and CYCA2;1 was decreased in the vascular cylinder of the root, which correlated with reduced lateral root formation. In root tips, AtCDC2a, CYCA2;1, and CYCB1;1 were downregulated along with reduced root growth (S et al., 2004; West et al., 2004). West et al. (2004) demonstrated that, in Arabidopsis thaliana roots, mild salt stress leads to loss of CDK activity and reduced promoter activity of CYCB1;2. Severe salt stress transiently decreases the expression levels of the cyclins CYCA2;1 and CYCB;1. In addition, Zhao L. et al. (2014) revealed that heat and cold treatments both induced a pronounced cell accumulation in G2/M transition and NaCl treatment results in an extensive inhibition in both S and G2/M phase by analyzing DNA content in maize (Zea MAYS l). Furthermore, the perturbed cell cycle progression is attributed to the dynamic histone acetylation change which participated in the control of CDKs and cyclins transcription. Thus, high salinity affects cell cycle regulation via control the cell cycle regulators. However, molecular players which perceive Na+ ions and confer salt stress to regulated cyclins and CDKs are still need to be unraveled.
Interestingly, there exists a group of salt-tolerant plants which grow in highly saline soil or water called halophyte (Zhao S. et al., 2010; Zhao et al., 2011; Zhang T. et al., 2015). An example of a halophyte is Suaeda salsa (Chen et al., 2010; Cheng et al., 2014; Guo et al., 2015; Guo et al., 2018), which produces dimorphic seeds on the same plant, brown seeds but not black seeds are able to germinate under conditions of high salinity owing to the presence of a seed coat (Wang et al., 2015; Song et al., 2016; Xu et al., 2016; Song et al., 2017). In this species, ion transport signaling contributes to salt tolerance through the action of various genes including HIGH-AFFINITY POTASSIUM TRANSPORTER (SsHKT)1 (Shao et al., 2014), SODIUM/HYDROGEN EXCHANGER (SsNHX)1 (Liu et al., 2018), and ASCORBATE PEROXIDASE (SsAPX) (Song and Wang, 2015). Besides, S. salsa treated with 200 mM NaCl generated healthier seeds with higher starch, soluble sugar, protein, and lipid contents than those treated with other concentrations of NaCl (Sui et al., 2010; Yang et al., 2010; Zhang et al., 2010; Li X. et al., 2012; Sun et al., 2015; Zhou et al., 2016). Another salt-tolerant plant is Limonium bicolor, a recretohalophyte with salt glands and a bladder that enable the secretion of excess salt into the environment (Yuan et al., 2013; Yuan et al., 2014; Yuan et al., 2015a; Yuan et al., 2015b; Yuan et al., 2016a; Yuan et al., 2016b). Recent studies show that the rate of secretion is determined by K+ and Ca2+ concentration (Ding et al., 2010; Feng et al., 2014; Feng et al., 2015; Leng et al., 2018). In Thellungiella salsuginea, microRNAs and unsaturated fatty acids enhance resistance to high salinity (Zhang et al., 2013; Sui and Han, 2014). It’s meaningful to detect whether the genes or microRNAs or unsaturated fatty acids mentioned above help to sustain the normal expression of cyclins and the activities of CDKs to some extent.
In conclusion, halophytes have evolved functional adaptations, ensuring their survival in saline environment. Analysis the difference between halophytes and glycophytes in transcriptomics and proteomics contributes to discover genes which is responsible for salt tolerance. Moreover, although most crop plants are glycophytes and are unable to tolerate high levels of salt stress, the abilities of glycophytes to withstand salt stress differ. This natural variation can be utilized to identify genetic components underlying salinity tolerance in glycophytes. Furthermore, screening proteins interacting with cyclins or CDKs under normal or stressed conditions are conductive to find molecules which confer salt stress signal to manipulated cell cycle regulators. Crops can be genetically engineered to resist high salinity through the expression of salt-responsive genes from halophytes; this has been demonstrated by the generation of salt-tolerant varieties of tomato, wheat, soybean, tobacco, peanut, cotton, and A. thaliana (Kong et al., 2011; Deng et al., 2016; Yang Z. et al., 2017).
Temperature Stress
Plants are continually exposed to changes in the ambient temperature. Both chilling and hot can negatively affect plant growth and development. Temperature stress triggers Ca2+ fluxes, kinase cascades, and accumulation of abscisic acid. These factors activate cell cycle checkpoints that delay entry into mitosis (Kadota et al., 2004; Kadota et al., 2005). In plants, more and more evidence has emerged to reveal the molecular mechanisms that regulate CDK functions in adapt to temperature stress. Under mild heat stress, proliferating cells are transiently arrested at G1/S or G2/M phase in BY2 cells, depending on the stage at which the stress was applied (Jang et al., 2005). The arrested cell cycle partially due to the reduced transcription of CYCA and CYCB and, as a consequence, decreased CDK activity. On the other hand, high temperatures induce a signaling cascade that leads to programmed cell death (Larkindale and Knight, 2002; Vacca et al., 2004).
Rymen et al. (2007) demonstrated that the increased cell cycle time in meristem of leaves exposed to chilling was associated with the up-regulated expression of cell cycle inhibitors such as KPRS. Similarly, the EL2 gene from rice (Orysa;E2L) was induced to transcript by low temperature. Orysa;E2L is a plant specific inhibitor of CDK that inhibit CDKA1 activation during G1/S transition through direct or indirect binding to CYCD (Peres et al., 2007). In addition, cold stress stimulates the expression of the transcription factor OsMYB3R2, which increases the expression of several G2/M-specific genes such as OsCYCB1. Furthermore, OsCYCB1 overexpression increased resistance to cold stress (Ma et al., 2009). Moreover, many other cold-responsive genes have been identified in T. salsuginea and rice by RNA-sequencing (Zhou et al., 2014; Liu et al., 2016; Wang et al., 2017). The T. salsuginea gene FILAMENTOUS TEMPERATURE-SENSITIVE H8 was shown to alleviate cold-induced photoinhibition (Liu et al., 2016). In rice (Oryza sativa), LOW-TEMPERATURE RESPONSE PROTEIN KINASE 1 contributes to cold resistance by regulating cytoskeletal rearrangement. Meanwhile, phytochrome B diminishes cold tolerance by modulating the expression of DEHYDRATION-RESPONSIVE ELEMENT-BINDING 1 and unsaturated fatty acid content (Liu et al., 2012; Yang et al., 2013; Zhou et al., 2014; He et al., 2016). Ectopic expression of the above genes increases the ability of crops to resist cold temperatures, although the underlying mechanisms are unclear. It’s of interest to investigate whether the different stress responsive genes work in the same signal pathway.
Put together, plants adjust to high-temperature and low-temperature via regulating the expression of different cell cycle machinery components. The fact that plants have larger number of cyclins compared with other eukaryotes may indicate cyclins function in stress response or perception to some degree. Most studies have revealed that temperature halt cell cycle progression through decrease or increase the transcript levels of related genes, however, we should take posttranslational modification of CDKs into consideration. Despite all of this, additional studies are needed to clarify the molecular basis for cell cycle regulators in response to hot or cold conditions.
Drought Stress
Drought stress is one of the abiotic stresses that limit plant growth and crop yield. Drought induces physiological changes in plants that enable survival, such as stomatal closure, perturbation of cell growth, and inhibition of photosynthesis. These processes involve the activation or repression of transcription factors, protein kinases, and metabolism-related enzymes. Here, we focus on the effect of drought on cell cycle proliferation and cell expansion. The growth of an organ is roughly divided into two stages: the cell proliferation stage, in which the cell number increases; and the cell expansion stage, in which cells expand to their final size. In Arabidopsis and sunflowers, water deficit affects both cell division and cell expansion. The reduced cell number in sunflower leaves was owing to the arrested G1/S transition. Progression to S phase is mediated by the activity of cyclin-dependent kinase A (CDKA). We can speculate that drought damages CDKA activity more or less. In wheat seedlings subjected to mild water stress, leaf elongation rate and mitotic activity were reduced in mesophyll cells due to decreased activity of CDKA1, which is required for entry into mitosis. This was accompanied by downregulation of cyclins and the accumulation of cells in G1 or G2 phase (Schuppler et al., 1998). Similarly, in maize leaves, cell division and CDKA1 activity was decreased in response to a mild water deficit, resulting in the inhibition of endosperm development. However, the amount of CDKA1 protein remained unaffected suggesting that posttranslational regulation of CDKA1 was responsible (TL and BA, 2001). In another drought-tolerant Brachypodium leaves, cell expansion is affected while cell proliferation is not, which is opposite to Arabidopsis and maize. The molecules response to drought stress strongly dependent on the developmental stage. In the leaf’s proliferation zone, drought stress had no effect on the expression of genes related to cell division. By Affymetrix tiling array analysis, the researchers identified numerous drought-responsive genes which were mainly expressed in the mature leaf zone. In summary, our current understanding of stress-regulated growth is still very fragmentary, partly because studies combining detailed growth analysis and molecular characterization of growing tissues are relatively scarce. Molecular characterization of the stress responses of growing tissues has to be investigated by further step. Broadening our knowledge of the mechanisms underlying growth reduction under stress is an important prerequisite to further improve crop productivity.
Conclusion and Perspectives
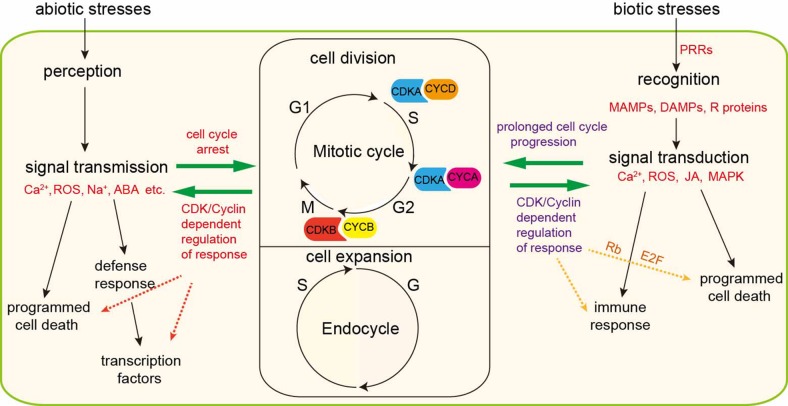
As sessile organisms, plants are frequently challenged as their environment changes during their lifetime. The ability of plants to perceive and adapt to these changes facilitates survival and production. Similarly, both biotic stress and abiotic stress perturb cell cycle progression and, at worst, lead to programmed cell death. Plants have evolved multiple mechanisms to defense against stresses. In brief, perception of biotic and abiotic stress signals activates signaling cascades that trigger ion fluxes, kinase cascades, reactive oxygen species production, and accumulation of hormones, such as abscisic acid (ABA) and jasmonic acid (JA). Theses signaling molecules suppress the activities of CDKs via controlling the expression level of cyclins or regulating the posttranslational modification of CDKs and, as a consequence, arrest the cell cycle or even exit cell cycle. This review highlights the role of cell cycle modulation in the adaption of plants to biotic and abiotic stresses and discusses the associated mechanisms proposed by recent studies ( Figure 2 ). Even though the existing studies indicate that CDKs and cyclins are important for the detection and adaptation to stress by plants, there still exist many outstanding questions regarding the relationship between plant defense responses and cell cycle kinetics. For instance, it is unclear whether cyclins are directly involved in sensing stresses and whether the cyclin molecules differ in sensing abiotic stress and biotic stress. Additionally, plant proteins that detect stress and transmit signals to delay cell cycle progression remain to be identified. Moreover, it’s unknown why elevating the transcription level of cyclins or manipulating CDK activities could, to some extent, increase the ability of plants to defense against stresses. Single cell RNA sequencing approach, high-throughput, genome-wide transcriptome and proteomic technologies will help us to identify new genes and proteins involved in stress response and then broad our knowledge about cell cycle regulation and response adaption. Fully understanding the molecular signal network which regulate stress response and cell growth is the prerequisite for researchers to elevate crop plants’ resistance to stresses and improve crop plants yield.
Figure 2.
Interactions between stress/response signaling cascades and cell cycle regulation. Plant growth depends on cell division and expansion. Upon biotic stress, cell surface pattern recognition receptors (PRRs) recognize conserved microbe-associated molecular patterns (MAMPs) or damage-associated molecular patterns (DAMPs) or resistant (R) proteins and then transduce primary signal to secondary signal molecules including Ca2+ flux, ROS, jasmonates (JA) and MAPK and eventually initiate immunity. Immune response induces prolonged cell cycle progression or programmed cell death. Overexpression of CDKs or cyclins could enhance immune responses. Besides, G1-S phase checkpoint proteins Rb and E2F engaged in immune-related programmed cell death. Similarly, under abiotic stress conditions, plant cells sense and percept the signals and transmit them to downstream signal molecules, such as Ca2+, Na+, ABA, and ROS. These signaling cascades halt cell cycle progression through inhibiting the transcription of CDK/cyclins-related genes. Manipulating the level of CDK or cyclins could change the defense response abilities. Severe abiotic stresses trigger programmed cell death.
Author Contributions
FQ and FZ wrote the manuscript and FQ critically revised the manuscript. All authors approved the final version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict interest.
References
- Achard P., Gusti A., Cheminant S., Alioua M., Dhondt S., Coppens F., et al. (2009). Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr. Biol. 19 (14), 1188–1193. 10.1016/j.cub.2009.05.059 [DOI] [PubMed] [Google Scholar]
- An J., Hou L., Li C., Wang C. X., Xia H., Zhao C. Z., et al. (2015). Cloning and expression analysis of four DELLA genes in peanut. Russ. J. Plant Physiol. 62 (1), 116–126. 10.1134/S1021443715010021 [DOI] [Google Scholar]
- Ascencio-Ibanez J. T., Sozzani R., Lee T. J., Chu T. M., Wolfinger R. D., Cella R., et al. (2008). Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 148 (1), 436–454. 10.1104/pp.108.121038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F. M. (2005). Are innate immune signaling pathways in plants and animals conserved? Nat. Immunol. 6 (10), 973–979. 10.1038/ni1253 [DOI] [PubMed] [Google Scholar]
- Bai B., Zhao J., Li Y. P., Zhang F., Zhou J. J., Chen F., et al. (2016). OsBBX14 delays heading date by repressing florigen gene expression under long and short-day conditions in rice. Plant Sci. 247, 25–34. 10.1016/j.plantsci.2016.02.017 [DOI] [PubMed] [Google Scholar]
- Bao Z., Hua J. (2015). Linking the cell cycle with innate immunity in Arabidopsis. Mol. Plant 8 (7), 980–982. 10.1016/j.molp.2015.03.013 [DOI] [PubMed] [Google Scholar]
- Bao Z., Yang H., Hua J. (2013). Perturbation of cell cycle regulation triggers plant immune response via activation of disease resistance genes. Proc. Natl. Acad. Sci. U.S.A. 110 (6), 2407–2412. 10.1073/pnas.1217024110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastas K. K. (2014). Importance of reactive oxygen species in plants-pathogens interactions. Selcuk J. Agric. Food Sci. 28 (1), 11–21. [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. 10.1146/annurev.arplant.57.032905.105346 [DOI] [PubMed] [Google Scholar]
- Cao S., Du X. H., Li L. H., Liu Y. D., Zhang L., Pan X., et al. (2017). Overexpression of Populus tomentosa cytosolic ascorbate peroxidase enhances abiotic stress tolerance in tobacco plants. Russ. J. Plant Physiol. 64 (2), 224–234. 10.1134/S1021443717020029 [DOI] [Google Scholar]
- Capron A., Okrész L., Genschik P. (2003). First glance at the plant APC/C, a highly conserved ubiquitin-protein ligase. Trends Plant Sci. 8 (2), 83–89. 10.1016/S1360-1385(02)00028-6 [DOI] [PubMed] [Google Scholar]
- Chandran D., Tai Y. C., Hather G., Dewdney J., Denoux C., Burgess D. G., et al. (2009). Temporal global expression data reveal known and novel salicylate-impacted processes and regulators mediating powdery mildew growth and reproduction on Arabidopsis. Plant Physiol. 149 (3), 1435–1451. 10.1104/pp.108.132985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran D., Rickert J., Huang Y., Steinwand M. A., Marr S. K. K., Wildermuth M. C. C. (2014). Atypical E2F transcriptional repressor DEL1 acts at the intersection of plant growth and immunity by controlling the hormone salicylic acid. Cell Host Microbe 15 (4), 506–513. 10.1016/j.chom.2014.03.007 [DOI] [PubMed] [Google Scholar]
- Chang J., Zhang E. L., Liu E. F., Sun W. W., Langdon P. G., Shulmeister J. (2018). A 2500-year climate and environmental record inferred from subfossil chironomids from Lugu Lake, southwestern China. Hydrobiologia 811 (1), 193–206. 10.1007/s10750-017-3488-5 [DOI] [Google Scholar]
- Chen M., Song J., Wang B. S. (2010). NaCl increases the activity of the plasma membrane H+-ATPase in C-3 halophyte Suaeda salsa callus. Acta Physiol. Plant. 32 (1), 27–36. 10.1007/s11738-009-0371-7 [DOI] [Google Scholar]
- Chen Q., Sun J. Q., Zhai Q. Z., Zhou W. K., Qi L. L., Xu L., et al. (2011). The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 23 (9), 3335–52. 10.1105/tpc.111.089870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Zhang W. H., Lv Z. W., Zhang S. L., Hidema J., Shi F. M., et al. (2013). Abscisic acid is involved in the response of Arabidopsis mutant sad2-1 to ultraviolet-B radiation by enhancing antioxidant enzymes. South Afr. J. Bot. 85, 79–86. 10.1016/j.sajb.2012.11.006 [DOI] [Google Scholar]
- Chen T. S., Yuan F., Song J., Wang B. S. (2016). Nitric oxide participates in waterlogging tolerance through enhanced adventitious root formation in the euhalophyte Suaeda salsa. Funct. Plant Biol. 43 (3), 244–253. 10.1071/fp15120 [DOI] [PubMed] [Google Scholar]
- Cheng S., Yang Z., Wang M. J., Song J., Sui N., Fan H. (2014). Salinity improves chilling resistance in Suaeda salsa. Acta Physiol. Plant. 36 (7), 1823–1830. 10.1007/s11738-014-1555-3 [DOI] [Google Scholar]
- Chisholm S. T., Coaker G., Day B., Staskawicz B. J. (2006). Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124 (4), 803–814. 10.1016/j.cell.2006.02.008 [DOI] [PubMed] [Google Scholar]
- Cui L. L., Yang G. W., Pan J., Zhang C. (2011). Tumor necrosis factor alpha knockout increases fertility of mice. Theriogenology 75 (5), 867–876. 10.1016/j.theriogenology.2010.10.029 [DOI] [PubMed] [Google Scholar]
- Cui F., Sui N., Duan G. Y., Liu Y. Y., Han Y., Liu S. S., et al. (2018). Identification of metabolites and transcripts involved in salt stress and recovery in peanut. Front. Plant Sci. 9. 10.3389/fpls.2018.00217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo J. C., Diaz-Trivino S., Cisneros N., Gutierrez C. (2006). The balance between cell division and endoreplication depends on E2FC-DPB, transcription factors regulated by the ubiquitin-SCFSKP2A pathway in Arabidopsis . Plant Cell 18 (9), 2224–2235. 10.1105/tpc.105.039651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y. Q., Feng Z. T., Yuan F., Guo J. R., Suo S. S., Wang B. S. (2015). Identification and functional analysis of the autofluorescent substance in Limonium bicolor salt glands. Plant Physiol. Biochemi. 97, 20–27. 10.1016/j.plaphy.2015.09.007 [DOI] [PubMed] [Google Scholar]
- Deng Y. Q., Bao J., Yuan F., Liang X., Feng Z. T., Wang B. S. (2016). Exogenous hydrogen sulfide alleviates salt stress in wheat seedlings by decreasing Na+ content. Plant Growth Regul. 79 (3), 391–399. 10.1007/s10725-015-0143-x [DOI] [Google Scholar]
- Ding F., Chen M., Sui N., Wang B. S. (2010). Ca2+ significantly enhanced development and salt-secretion rate of salt glands of Limonium bicolor under NaCl treatment. South Afr. J. Bot. 76 (1), 95–101. 10.1016/j.sajb.2009.09.001 [DOI] [Google Scholar]
- Feng Z. T., Sun Q. J., Deng Y. Q., Sun S. F., Zhang J. G., Wang B. S. (2014). Study on pathway and characteristics of ion secretion of salt glands of Limonium bicolor. Acta Physiol. Plant. 36 (10), 2729–2741. 10.1007/s11738-014-1644-3 [DOI] [Google Scholar]
- Feng Z. T., Deng Y. Q., Zhang S. C., Liang X., Yuan F., Hao J. L., et al. (2015). K+ accumulation in the cytoplasm and nucleus of the salt gland cells of Limonium bicolor accompanies increased rates of salt secretion under NaCl treatment using NanoSIMS. Plant Sci. 238, 286–296. 10.1016/j.plantsci.2015.06.021 [DOI] [PubMed] [Google Scholar]
- Gong W. P., Han R., Li H. S., Song J. M., Yan H. F., Li G. Y., et al. (2017). Agronomic traits and molecular marker identification of Wheat-Aegilops caudata addition lines. Front. Plant Sci. 8. 10.3389/fpls.2017.01743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F. D., Liu C. L., Xia H., Bi Y. P., Zhao C. Z., Zhao S. Z., et al. (2013). Induced Expression of AtLEC1 and AtLEC2 differentially promotes somatic embryogenesis in transgenic tobacco plants. Plos One 8 (8), e71714. 10.1371/journal.pone.0071714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J. R., Suo S. S., Wang B. S. (2015). Sodium chloride improves seed vigour of the euhalophyte Suaeda salsa. Seed Sci. Res. 25 (3), 335–344. 10.1017/S0960258515000239 [DOI] [Google Scholar]
- Guo J. R., Li Y. D., Han G. L., Song J., Wang B. S. (2018). NaCl markedly improved the reproductive capacity of the euhalophyte Suaeda salsa. Funct. Plant Biol. 45 (3), 350–361. 10.1071/FP17181 [DOI] [PubMed] [Google Scholar]
- Halder G., Johnson R. L. (2011). Hippo signaling: growth control and beyond. Development 138 (1), 9–22. 10.1242/dev.045500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C. Q., Liu Y. X., Wang H. M., Hou P. L., He H. B., Ding N. Z. (2016). New genetic mechanism, origin and population dynamic of bovine ephemeral fever virus. Vet. Microbiol. 182, 50–56. 10.1016/j.vetmic.2015.10.029 [DOI] [PubMed] [Google Scholar]
- He Y. A., Li Y. P., Cui L. X., Xie L. X., Zheng C. K., Zhou G. H., et al. (2016). Phytochrome B negatively affects cold tolerance by regulating OsDREB1 gene expression through phytochrome interacting factor-like protein OsPIL16 in Rice. Front. Plant Sci. 7 (1963). 10.3389/fpls.2016.01963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L., Chen L. J., Wang J. Y., Xu D. F., Dai L. X., Zhang H., et al. (2012). Construction of Stress responsive synthetic promoters and analysis of their activity in transgenic Arabidopsis thaliana. Plant Mol. Biol. Rep. 30 (6), 1496–1506. 10.1007/s00425-014-2135-x [DOI] [Google Scholar]
- Hou L., Liu W., Li Z., Huang C., Fang X. L., Wang Q., et al. (2014). Identification and expression analysis of genes responsive to drought stress in peanut. Russ. J. Plant Physiol. 61 (6), 842–852. 10.1134/s1021443714060089 [DOI] [Google Scholar]
- Hou P. L., Wang H. M., Zhao G. M., He C. Q., He H. B. (2017). Rapid detection of infectious bovine Rhinotracheitis virus using recombinase polymerase amplification assays. Bmc Vet. Res. 13 (1), 386. 10.1186/s12917-017-1284-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou P. L., Zhao G. M., He C. Q., Wang H. M., He H. B. (2018). Biopanning of polypeptides binding to bovine ephemeral fever virus G(1) protein from phage display peptide library. Bmc Vet. Res. 14 (1), 3. 10.1186/s12917-017-1315-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. H., Wang X. J., Zhang F., Huo X. B., Fu R. S., Liu J. J., et al. (2013). The Identification of a Bacterial strain BGI-1 Isolated From the Intestinal Flora of Blattella Germanica, and Its Anti-Entomopathogenic Fungi Activity. J. Econ. Entomol, 106 (1), 43–49. 10.1603/ec12120 [DOI] [PubMed] [Google Scholar]
- Jang S. J., Shin S. H., Yee S. T., Hwang B., Im K. H., Park K. Y. (2005). Effects of abiotic stresses on cell cycle progression in Tobaco BY-2 Cells. Mol. Cells 20 (1), 136–141. [PubMed] [Google Scholar]
- Kadota Y., Watanabe T., Fujii S., Higashi K., Sano T., Nagata T., et al. (2004). Crosstalk between elicitor-induced cell death and cell cycle regulation in tobacco BY-2 cells. Plant J. 40 (1), 131–142. 10.1111/j.1365-313X.2004.02197.x [DOI] [PubMed] [Google Scholar]
- Kadota Y., Watanabe T., Fujii S., Maeda Y., Ohno R., Higashi K., et al. (2005). Cell cycle dependence of elicitor-induced signal transduction in tobacco BY-2 cells. Plant Cell Physiol. 46 (1), 156–165. 10.1093/pcp/pci008 [DOI] [PubMed] [Google Scholar]
- Kim S. H., Son G. H., Bhattacharjee S., Kim H. J., Nam J. C., Nguyen P. D., et al. (2014). The Arabidopsis immune adaptor SRFR1 interacts with TCP transcription factors that redundantly contribute to effector-triggered immunity. Plant J. 78 (6), 978–989. 10.1111/tpj.12527 [DOI] [PubMed] [Google Scholar]
- Kong X. Q., Gao X. H., Sun W., An J., Zhao Y. X., Zhang H. (2011). Cloning and functional characterization of a cation-chloride cotransporter gene OsCCC1. Plant Mol. Biol. 75 (43363), 567–578. 10.1007/s11103-011-9744-6 [DOI] [PubMed] [Google Scholar]
- Kong L. A., Xie Y., Hu L., Si J. S., Wang Z. S. (2017). Excessive nitrogen application dampens antioxidant capacity and grain filling in wheat as revealed by metabolic and physiological analyses. Sci. Rep. 7 (743363). 10.1038/srep43363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkindale J., Knight M. R. (2002). Protection against heat stress-induced oxidative damage in Arabidopsis involves calcium, abscisic acid, ethylene, and salicylic acid. Plant Physiol. 128 (2), 682–695. 10.1104/pp.128.2.682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng B. Y., Yuan F., Dong X. X., Wang J., Wang B. S. (2018). Distribution pattern and salt excretion rate of salt glands in two recretohalophyte species of Limonium (Plumbaginaceae). South Afr. J. Bot. 115, 74–80. 10.1016/j.sajb.2018.01.002 [DOI] [Google Scholar]
- Li K., Pang C. H., Ding F., Sui N., Feng Z. T., Wang B. S. (2012). Overexpression of Suaeda salsa stroma ascorbate peroxidase in Arabidopsis chloroplasts enhances salt tolerance of plants. South Afr. J. Bot. 78, 235–245. 10.1016/j.sajb.2011.09.006 [DOI] [Google Scholar]
- Li X., Liu Y., Chen M., Song Y. P., Song J., Wang B. S., et al. (2012). Relationships between ion and chlorophyll accumulation in seeds and adaptation to saline environments in Suaeda salsa populations. Plant Biosyst. 146, 142–149. 10.1080/11263504.2012.727880 [DOI] [Google Scholar]
- Li Z. Y., Li B., Dong A. W. (2012). The Arabidopsis transcription factor AtTCP15 regulates endoreduplication by modulating expression of key cell-cycle genes. Mol. Plant 5 (1), 270–280. 10.1093/mp/ssr086 [DOI] [PubMed] [Google Scholar]
- Li H., Zhang F. M., Guo H. Y., Zhu Y. Y., Yuan J. D., Yang G. W., et al. (2013). Molecular characterization of hepcidin gene in common carp (Cyprinus carpio L.) and its expression pattern responding to bacterial challenge. Fish Shellfish Immunol. 35 (3), 1030–1038. 10.1016/j.fsi.2013.07.001 [DOI] [PubMed] [Google Scholar]
- Li H., Yang G. W., Ma F., Li T., Yang H. T., Rombout J., et al. (2017). Molecular characterization of a fish-specific toll-like receptor 22 (TLR22) gene from common carp (Cyprinus carpio L.): Evolutionary relationship and induced expression upon immune stimulants. Fish Shellfish Immunol. 63, 74–86. 10.1016/j.fsi.2017.02.009 [DOI] [PubMed] [Google Scholar]
- Li T., Li H., Peng S. Q., Zhang F. M., An L. G., Yang G. W. (2017). Molecular characterization and expression pattern of X box-binding protein-1 (XBP1) in common carp (Cyprinus carpio L.): Indications for a role of XBP1 in antibacterial and antiviral immunity. Fish Shellfish Immunol. 67, 667–674. 10.1016/j.fsi.2017.06.055 [DOI] [PubMed] [Google Scholar]
- Liang J. W., Tian F. L., Lan Z. R., Huang B., Zhuang W. Z. (2010). Selection characterization on overlapping reading frame of multiple-protein-encoding P gene in Newcastle disease virus. Vet. Microbiol. 144 (3-4), 257–263. 10.1016/j.vetmic.2009.12.029 [DOI] [PubMed] [Google Scholar]
- Liu X. Y., Ju Z. H., Wang L. L., Zhang Y., Huang J. M., Li Q. L., et al. (2011). Six novel single-nucleotide polymorphisms in SPAG11 gene and their association with sperm quality traits in Chinese Holstein bulls. Anim. Reprod. Sci. 129 (1-2), 14–21. 10.1016/j.anireprosci.2011.10.003 [DOI] [PubMed] [Google Scholar]
- Liu D. S., Hu J. F., Horvath D. P., Zhang X. J., Bian X. Y., Chang G. L., et al. (2012). Invasions and Impacts of Alligatorweed in the Upper Xiaoqing River Basin of Northern China. J. Aquat. Plant Manag. 50, 19–24. [Google Scholar]
- Liu W., Ji S. X., Fang X. L., Wang Q. G., Li Z., Yao F. Y., et al. (2013). Protein Kinase LTRPK1 Influences cold adaptation and microtubule stability in rice. J. Plant Growth Regul. 32 (3), 483–490. 10.1007/s00344-012-9314-4 [DOI] [Google Scholar]
- Liu X. X., Fu C., Yang W. W., Zhang Q., Fan H., Liu J. (2016). The involvement of TsFtsH8 in Thellungiella salsuginea tolerance to cold and high light stresses. Acta Physiol. Plant. 38 (3). 10.1007/s11738-016-2080-3 [DOI] [Google Scholar]
- Liu S. S., Wang W. Q., Li M., Wan S. B., Sui N. (2017). Antioxidants and unsaturated fatty acids are involved in salt tolerance in peanut. Acta Physiol. Plant. 39 (9). 207 10.1007/s11738-017-2501-y [DOI] [Google Scholar]
- Liu T. T., Liu S., Ma L., Li F. L., Zheng Z. D., Chai R. F., et al. (2017). Oogenesis, vitellogenin-mediated ovarian degeneration and immune response in the annual fish Nothobranchius guentheri. Fish Shellfish Immunol. 66, 86–92. 10.1016/j.fsi.2017.05.015 [DOI] [PubMed] [Google Scholar]
- Liu Q. Q., Liu R. R., Ma Y. C., Song J. (2018). Physiological and molecular evidence for Na+ and Cl- exclusion in the roots of two Suaeda salsa populations. Aquat. Bot. 146, 1–7. 10.1016/j.aquabot.2018.01.001 [DOI] [Google Scholar]
- Ma Q., Dai X., Xu Y., Guo J., Liu Y., Chen N., et al. (2009). Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiol. 150 (1), 244–256. 10.1104/pp.108.133454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Yang D. Y., Li X. D., Zhao S. Y., Sui N., Meng Q. W. (2015). Physiological changes in fruit ripening caused by overexpression of tomato SlAN2, an R2R3-MYB factor. Plant Physiol. Biochemi. 89, 24–30. 10.1016/j.plaphy.2015.02.005 [DOI] [PubMed] [Google Scholar]
- Peres A., Churchman M. L., Hariharan S., Himanen K., Verkest A., Vandepoele K., et al. (2007). Novel plant-specific cyclin-dependent kinase inhibitors induced by biotic and abiotic stresses. J. Biol. Chem. 282 (35), 25588–25596. 10.1074/jbc.M703326200 [DOI] [PubMed] [Google Scholar]
- Petutschnig E. K., Jones A. M. E., Serazetdinova L., Lipka U., Lipka V. (2010). The lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J. Biol. Chem. 285 (37), 28902–28911. 10.1074/jbc.M110.116657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y. C., Liu W. Q., Qiu L. Y., Zhang S. M., Ma L., Zhang H. (2010). Overexpression of glutathione S-transferase gene increases salt tolerance of arabidopsis. Russ. J. Plant Physiol. 57 (2), 233–240. 10.1134/s102144371002010x [DOI] [Google Scholar]
- Rymen B., Fiorani F., Kartal F., Vandepoele K., Inzé D., Beemster G. T. (2007). Cold nights impair leaf growth and cell cycle progression in maize through transcriptional changes of cell cycle genes. Plant Physiol. 143 (3), 1429–1438. 10.1104/pp.106.093948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuppler U., He. P. H., John. P. C., Munns. R. (1998). Effect of water stress on cell division and Cdc2-Like cell cycle Kinase activity in wheat leaves. Plant Physiol. 117 (2), 667–678. 10.1104/pp.117.2.667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan S. J., Qi C. C., Zhu Y. Y., Li H., An L. G., Yang G. W. (2016). Expression profile of carp IFN correlate with the up-regulation of interferon regulatory factor-1 (IRF-1) in vivo and in vitro: the pivotal molecules in antiviral defense. Fish Shellfish Immunol. 52, 94–102. 10.1016/j.fsi.2016.03.019 [DOI] [PubMed] [Google Scholar]
- Shan S. J., Liu D. Z., Liu R. R., Zhu Y. Y., Li T., Zhang F. M., et al. (2018). Non-mammalian Toll-like receptor 18 (Tlr18) recognizes bacterial pathogens in common carp (Cyprinus carpio L.): Indications for a role of participation in the NF-kappa B signaling pathway. Fish Shellfish Immunol. 72, 187–198. 10.1016/j.fsi.2017.09.081 [DOI] [PubMed] [Google Scholar]
- Shao Q., Han N., Ding T. L., Zhou F., Wang B. S. (2014). SsHKT1;1 is a potassium transporter of the C-3 halophyte Suaeda salsa that is involved in salt tolerance. Funct. Plant Biol. 41 (8), 790–802. 10.1071/FP13265 [DOI] [PubMed] [Google Scholar]
- Shen X. Y., Wang Z. L., Song X. F., Xu J. J., Jiang C. Y., Zhao Y. X., et al. (2014). Transcriptomic profiling revealed an important role of cell wall remodeling and ethylene signaling pathway during salt acclimation in Arabidopsis. Plant Mol. Biol. 86 (3), 303–317. 10.1007/s11103-014-0230-9 [DOI] [PubMed] [Google Scholar]
- Song J., Wang B. S. (2015). Using euhalophytes to understand salt tolerance and to develop saline agriculture: Suaeda salsa as a promising model. Ann. Bot. 115 (3), 541–553. 10.1093/aob/mcu194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Shi G. W., Gao B., Fan H., Wang B. S. (2011). Waterlogging and salinity effects on two Suaeda salsa populations. Physiol. Plant. 141 (4), 343–351. 10.1111/j.1399-3054.2011.01445.x [DOI] [PubMed] [Google Scholar]
- Song J., Zhou J. C., Zhao W. W., Xu H. L., Wang F. X., Xu Y. G., et al. (2016). Effects of salinity and nitrate on production and germination of dimorphic seeds applied both through the mother plant and exogenously during germination in Suaeda salsa. Plant Species Biol. 31 (1), 19–28. 10.1111/1442-1984.12071 [DOI] [Google Scholar]
- Song J., Shi W. W., Liu R. R., Xu Y. G., Sui N., Zhou J. C., et al. (2017). The role of the seed coat in adaptation of dimorphic seeds of the euhalophyte Suaeda salsa to salinity. Plant Species Biol. 32 (2), 107–114. 10.1111/1442-1984.12132 [DOI] [Google Scholar]
- Sui N., Han G. L. (2014). Salt-induced photoinhibition of PSII is alleviated in halophyte Thellungiella halophila by increases of unsaturated fatty acids in membrane lipids. Acta Physiol. Plant. 36 (4), 983–992. 10.1007/s11738-013-1477-5 [DOI] [Google Scholar]
- Sui N., Li M., Li K., Song J., Wang B. S. (2010). Increase in unsaturated fatty acids in membrane lipids of Suaeda salsa L. enhances protection of photosystem II under high salinity. Photosynthetica 48 (4), 623–629. 10.1007/s11099-010-0080-x [DOI] [Google Scholar]
- Sui N., Tian S. S., Wang W. Q., Wang M. J., Fan H. (2017). Overexpression of Glycerol-3-Phosphate Acyltransferase from Suaeda salsa Improves Salt Tolerance in Arabidopsis. Front. Plant Sci. 8 (1337). 10.3389/fpls.2017.01337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui N. (2015). Photoinhibition of Suaeda salsa to chilling stress is related to energy dissipation and water-water cycle. Photosynthetica 53 (2), 207–212. 10.1007/s11099-015-0080-y [DOI] [Google Scholar]
- Sun Y. L., Li F., Su N., Sun X. L., Zhao S. J., Meng Q. W. (2010). The increase in unsaturation of fatty acids of phosphatidylglycerol in thylakoid membrane enhanced salt tolerance in tomato. Photosynthetica 48 (3), 400–408. 10.1007/s11099-010-0052-1 [DOI] [Google Scholar]
- Sun G. J., Pan J., Liu K. C., Wang S. F., Wang X., Wang X. M. (2012). Molecular cloning and expression analysis of P-selectin glycoprotein ligand-1 from zebrafish (Danio rerio). Fish Physiol. Biochemi. 38 (2), 555–564. 10.1007/s10695-011-9535-7 [DOI] [PubMed] [Google Scholar]
- Sun Z. B., Qi X. Y., Wang Z. L., Li P. H., Wu C. X., Zhang H., et al. (2013). Overexpression of TsGOLS2, a galactinol synthase, in Arabidopsis thaliana enhances tolerance to high salinity and osmotic stresses. Plant Physiol. Biochemi. 69, 82–89. 10.1016/j.plaphy.2013.04.009 [DOI] [PubMed] [Google Scholar]
- Sun W., Li Y., Zhao Y. X., Zhang H. (2015). The TsnsLTP4, a nonspecific lipid transfer protein involved in wax deposition and stress tolerance. Plant Mol. Biol. Rep. 33 (4), 962–974. 10.1007/s11105-014-0798-x [DOI] [Google Scholar]
- Tang G. Y., Wei L. Q., Liu Z. J., Bi Y. P., Shan L. (2012). Ectopic expression of peanut acyl carrier protein in tobacco alters fatty acid composition in the leaf and resistance to cold stress. Biologia Plant. 56 (3), 493–501. 10.1007/s10535-012-0057-7 [DOI] [Google Scholar]
- Tang G. Y., Shao F. X., Xu P. L., Shan L., Liu Z. J. (2017). Overexpression of a peanut NAC gene, AhNAC4, confers enhanced drought tolerance in tobacco. Russ. J. Plant Physiol. 64 (4), 525–535. 10.1134/S1021443717040161 [DOI] [Google Scholar]
- Tian W., Zhang H. Y., Zhang J., Zhao L., Miao M. S., Huang H. (2017). Responses of Zooplankton community to environmental factors and phytoplankton biomass in lake Nansihu, China. Pak. J. Zool. 49 (2), 493–504. 10.17582/journal.pjz/2017.49.2.461.470 [DOI] [Google Scholar]
- TL S., BA F. (2001). Water deficit inhibits cell division and expression of transcripts involved in cell proliferation and endoreduplication in maize endosperm. J. Exp. Bot. 52 (360), 1401–1408. 10.1093/jexbot/52.360.1401 [DOI] [PubMed] [Google Scholar]
- Vacca R. A., de Pinto M. C., Valenti D., Passarella S., Marra E., De Gara L. (2004). Production of reactive oxygen species, alteration of cytosolic ascorbate peroxidase, and impairment of mitochondrial metabolism are early events in heat shock-induced programmed cell death in tobacco Bright-Yellow 2 cells. Plant Physiol. 134 (3), 1100–1112. 10.1104/pp.103.035956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbon E. H., Post J. A., Boonstra J. (2012). The influence of reactive oxygen species on cell cycle progression in mammalian cells. Gene 511 (1), 1–6. 10.1016/j.gene.2012.08.038 [DOI] [PubMed] [Google Scholar]
- Wang S. S., Wang F., Tan S. J., Wang M. X., Sui N., Zhang X. S. (2014). Transcript profiles of maize embryo sacs and preliminary identification of genes involved in the embryo sac-pollen tube interaction. Front. Plant Sci. 5 (702). 10.3389/fpls.2014.00702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. G., Huang J. M., Feng M. Y., Ju Z. H., Wang C. F., Yang G. W., et al. (2014). Regulatory mutations in the A2M gene are involved in the mastitis susceptibility in dairy cows. Anim. Genet. 45 (1), 28–37. 10.1111/age.12099 [DOI] [PubMed] [Google Scholar]
- Wang F. X., Xu Y. G., Wang S., Shi W. W., Liu R. R., Feng G., et al. (2015). Salinity affects production and salt tolerance of dimorphic seeds of Suaeda salsa. Plant Physiol. Biochemi. 95, 41–48. 10.1016/j.plaphy.2015.07.005 [DOI] [PubMed] [Google Scholar]
- Wang F. R., Zhang C. Y., Liu G. D., Chen Y., Zhang J. X., Qiao Q. H., et al. (2016). Phenotypic variation analysis and QTL mapping for cotton (Gossypium hirsutum L.) fiber quality grown in different cotton-producing regions. Euphytica 211 (2), 169–183. 10.1007/s10681-016-1728-9 [DOI] [Google Scholar]
- Wang J. S., Zhang Q., Cui F., Hou L., Zhao S. Z., Xia H., et al. (2017). Genome-Wide analysis of gene expression provides new insights into cold responses in Thellungiella salsuginea. Front. Plant Sci. 8 (713). 10.3389/fpls.2017.00713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng L., Tian Z. X., Feng X. Z., Li X., Xu S. L., Hu X. H., et al. (2011). Petal development in Lotus japonicus. J. Integr. Plant Biol. 53 (10), 770–782. 10.1111/j.1744-7909.2011.01072.x [DOI] [PubMed] [Google Scholar]
- West G., Inze D., Beemster G. T. (2004). Cell cycle modulation in the response of the primary root of Arabidopsis to salt stress. Plant Physiol. 135 (2), 1050–1058. 10.1104/pp.104.040022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing N., Ji L. Z., Song J., Ma J. C., Li S. G., Ren Z. M., et al. (2017). Cadmium stress assessment based on the electrocardiogram characteristics of zebra fish (Danio rerio): QRS complex could play an important role. Aquat. Toxicol. 191, 236–244. 10.1016/j.aquatox.2017.08.015 [DOI] [PubMed] [Google Scholar]
- Xu Y. G., Liu R. R., Sui N., Shi W. W., Wang L., Tian C. Y., et al. (2016). Changes in endogenous hormones and seed-coat phenolics during seed storage of two Suaeda salsa populations. Aust. J. Bot. 64 (4), 325–332. 10.1071/bt16014 [DOI] [Google Scholar]
- Yang M. F., Song J., Wang B. S. (2010). Organ-Specific responses of vacuolar H+-ATPase in the shoots and roots of C-3 Halophyte Suaeda salsa to NaCl. J. Integr. Plant Biol. 52 (3), 308–314. CNKISUNZWXB.0.2010-03-007 [DOI] [PubMed] [Google Scholar]
- Yang J. C., Li M., Xie X. Z., Han G. L., Sui N., Wang B. S. (2013). Deficiency of Phytochrome B alleviates chilling-induced photoinhibition in rice. Am. J. Bot. 100 (9), 1860–1870. 10.3732/ajb.1200574 [DOI] [PubMed] [Google Scholar]
- Yang H. T., Zou S. S., Zhai L. J., Wang Y., Zhang F. M., An L. G., et al. (2017). Pathogen invasion changes the intestinal microbiota composition and induces innate immune responses in the zebrafish intestine. Fish Shellfish Immunol. 71, 35–42. 10.1016/j.fsi.2017.09.075 [DOI] [PubMed] [Google Scholar]
- Yang Z., Wang Y., Wei X. C., Zhao X., Wang B. S., Sui N. (2017). Transcription Profiles of genes related to hormonal regulations under salt stress in sweet Sorghum. Plant Mol. Biol. Rep. 35 (6), 586–599. 10.1007/s11105-017-1047-x [DOI] [Google Scholar]
- Yoshida H., Lareau C. A., Ramirez R. N., Rose S. A., Maier B., Wroblewska A., et al. (2019). The cis-Regulatory Atlas of the mouse immune system. Cell 176 (4), 897–912 e820. 10.1016/j.cell.2018.12.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Wu J. Q., Zhang Y. Y., Guo L. H., Cong X. Y., Du Y. J., et al. (2012). Concurrent highly pathogenic porcine reproductive and respiratory syndrome virus infection accelerates Haemophilus parasuis infection in conventional pigs. Vet. Microbiol. 158 (3-4), 316–321. 10.1016/j.vetmic.2012.03.001 [DOI] [PubMed] [Google Scholar]
- Yuan F., Chen M., Leng B. Y., Wang B. S. (2013). An efficient autofluorescence method for screening Limonium bicolor mutants for abnormal salt gland density and salt secretion. South Afr. J. Bot. 88, 110–117. 10.1016/j.sajb.2013.06.007 [DOI] [Google Scholar]
- Yuan F., Chen M., Yang J. C., Leng B. Y., Wang B. S. (2014). A system for the transformation and regeneration of the recretohalophyte Limonium bicolor. Vitro Cell. Dev. Biology-Plant 50 (5), 610–617. 10.1007/s11627-014-9611-7 [DOI] [Google Scholar]
- Yuan F., Chen M., Yang J. C., Song J., Wang B. S. (2015. a). The optimal dosage of Co-60 gamma irradiation for obtaining salt gland mutants of exo-recreyohalophyte limonium bicolor (Bunge) O. Kuntze. Pak. J. Bot. 47 (1), 71–76. [Google Scholar]
- Yuan F., Lyu M. J. A., Leng B. Y., Zheng G. Y., Feng Z. T., Li P. H., et al. (2015. b). Comparative transcriptome analysis of developmental stages of the Limonium bicolor leaf generates insights into salt gland differentiation. Plant Cell Environ. 38 (8), 1637–1657. 10.1111/pce.12514 [DOI] [PubMed] [Google Scholar]
- Yuan F., Leng B. Y., Wang B. S. (2016. a). Progress in studying salt secretion from the salt glands in recretohalophytes: how do plants secrete salt? Front. Plant Sci. 7 (977). 10.3389/fpls.2016.00977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan F., Lyu M. J. A., Leng B. Y., Zhu X. G., Wang B. S. (2016. b). The transcriptome of NaCl-treated Limonium bicolor leaves reveals the genes controlling salt secretion of salt gland. Plant Mol. Biol. 91 (3), 241–256. 10.1007/s11103-016-0460-0 [DOI] [PubMed] [Google Scholar]
- Zebell S. G., Dong X. (2015). Cell-Cycle Regulators and Cell Death in Immunity. Cell Host Microbe 18 (4), 402–407. 10.1016/j.chom.2015.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. R., Song J., Wang H., Feng G. (2010). Effect of salinity on seed germination, ion content and photosynthesis of cotyledons in halophytes or xerophyte growing in Central Asia. J. Plant Ecol. 3 (4), 259–267. 10.1093/jpe/rtq005 [DOI] [Google Scholar]
- Zhang Q., Zhao C. Z., Li M., Sun W., Liu Y., Xia H., et al. (2013). Genome-wide identification of Thellungiella salsuginea microRNAs with putative roles in the salt stress response. Bmc Plant Biol. 13. 10.1186/1471-2229-13-180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Wang X. J., Huang Y. H., Zhao Z. G., Zhang S. S., Gong X. S., et al. (2014). Differential expression of hemolymph proteins between susceptible and insecticide-resistant Blattella germanica (Blattodea: Blattellidae). Environ. Entomol, 43 (4), 1117–1123. 10.1603/EN13351 [DOI] [PubMed] [Google Scholar]
- Zhang F. M., Liu D. Z., Wang L., Li T., Chang Q., An L. G., et al. (2015). Characterization of IgM-binding protein: A pIgR-like molecule expressed by intestinal epithelial cells in the common carp (Cyprinus carpio L.). Vet. Immunol. Immunopathol. 167 (1-2), 30–35. 10.1016/j.vetimm.2015.06.015 [DOI] [PubMed] [Google Scholar]
- Zhang T., Song J., Fan J. L., Feng G. (2015). Effects of saline-waterlogging and dryness/moist alternations on seed germination of halophyte and xerophyte. Plant Species Biol. 30 (3), 231–236. 10.1111/1442-1984.12056 [DOI] [Google Scholar]
- Zhang L. Y., Zhang X. J., Fan S. J. (2017). Meta-analysis of salt-related gene expression profiles identifies common signatures of salt stress responses in Arabidopsis. Plant Syst. Evol. 303 (6), 757–774. 10.1007/s00606-017-1407-x [DOI] [Google Scholar]
- Zhang Y. J., Kong X. Q., Dai J. L., Luo Z., Li Z. H., Lu H. Q., et al. (2017). Global gene expression in cotton (Gossypium hirsutum L.) leaves to waterlogging stress. Plos One 12 (9). e0185075. 10.1371/journal.pone.0185075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Wang Z., Bao Z., Yang L., Wu D., Shu X., et al. (2018). MOS1 functions closely with TCP transcription factors to modulate immunity and cell cycle in Arabidopsis. Plant J. 93 (1), 66–78. 10.1111/tpj.13757 [DOI] [PubMed] [Google Scholar]
- Zhao K. F., Song J., Fan H., Zhou S., Zhao M. (2010). Growth response to ionic and osmotic stress of NaCl in Salt-tolerant and Salt-sensitive Maize. J. Integr. Plant Biol. 52 (5), 468–475. 10.1111/j.1744-7909.2010.00947.x [DOI] [PubMed] [Google Scholar]
- Zhao S. Z., Sun H. Z., Chen M., Wang B. S. (2010). Light-regulated betacyanin accumulation in euhalophyte Suaeda salsa calli. Plant Cell Tissue Organ Cult. 102 (1), 99–107. 10.1007/s11240-010-9710-z [DOI] [Google Scholar]
- Zhao S. Z., Sun H. Z., Gao Y., Sui N., Wang B. S. (2011). Growth regulator-induced betacyanin accumulation and dopa-4,5-dioxygenase (DODA) gene expression in euhalophyte Suaeda salsa calli. Vitro Cell. Dev. Biology-Plant 47 (3), 391–398. 10.1007/s11627-011-9339-6 [DOI] [Google Scholar]
- Zhao L., Ding Q., Zeng J., Wang F. R., Zhang J., Fan S. J., et al. (2012). An improved CTAB-Ammonium Acetate method for total RNA isolation from cotton. Phytochem. Anal. 23 (6), 647–650. 10.1002/pca.2368 [DOI] [PubMed] [Google Scholar]
- Zhao L., Wang P., Hou H., Zhang H., Wang Y., Yan S., et al. (2014). Transcriptional regulation of cell cycle genes in response to abiotic stresses correlates with dynamic changes in histone modifications in maize. PLoS One. 9 (8), e106070. 10.1371/journal.pone.0106070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X. Y., Zheng Z. W., Shang Z. Y., Wang J., Cheng R. Q., Qian J. L. (2014). Climatic information recorded in stable carbon isotopes in tree rings of Cryptomeria fortunei, Tianmu Mountain, China. Dendrochronologia 32 (3), 256–265. 10.1016/j.dendro.2014.06.002 [DOI] [Google Scholar]
- Zhao B. T., Dai A. H., Wei H. C., Yang S. X., Wang B. S., Jiang N., et al. (2016). Arabidopsis KLU homologue GmCYP78A72 regulates seed size in soybean. Plant Mol. Biol. 90 (1-2), 33–47. 10.1007/s11103-015-0392-0 [DOI] [PubMed] [Google Scholar]
- Zhao S. S., Jiang Y. X., Zhao Y., Huang S. J., Yuan M., Zhao Y. X., et al. (2016). CASEIN KINASE1-LIKE PROTEIN2 regulates actin filament stability and stomatal Closure via Phosphorylation of Actin Depolymerizing Factor. Plant Cell 28 (6), 1422–1439. 10.1105/tpc.16.00078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S., Wu X., Zhang L., Xin C., Liu Y., Shi J., et al. (2017). The occurrence of porcine circovirus 3 without clinical infection signs in Shandong Province. Transbound. Emerg. Dis. 64 (5), 1337–1341. 10.1111/tbed.12667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Liao C. C., Zhao S. S., Wang C. W., Guo Y. (2017). The Glycosyltransferase QUA1 regulates Chloroplast-Associated calcium signaling during salt and drought stress in Arabidopsis. Plant Cell Physiol. 58 (2), 329–341. 10.1093/pcp/pcw192 [DOI] [PubMed] [Google Scholar]
- Zheng S., Shi J., Wu X., Peng Z., Xin C., Zhang L., et al. (2018). Presence of Torque teno sus virus 1 and 2 in porcine circovirus 3-positive pigs. Transbound. Emerg. Dis. 65 (2), 327–330. 10.1111/tbed.12792 [DOI] [PubMed] [Google Scholar]
- Zhou J. J., Liu Q. Q., Zhang F., Wang Y. Y., Zhang S. Y., Cheng H. M., et al. (2014). Overexpression of OsPIL15, a phytochromeinteracting factor- like protein gene, represses etiolated seedling growth in rice. J. Integr. Plant Biol. 56 (4), 373–387. 10.1111/jipb.12137 [DOI] [PubMed] [Google Scholar]
- Zhou J. C., Fu T. T., Sui N., Guo J. R., Feng G., Fan J. L., et al. (2016). The role of salinity in seed maturation of the euhalophyte Suaeda salsa. Plant Biosyst. 150 (1), 83–90. 10.1080/11263504.2014.976294 [DOI] [Google Scholar]
- Zhu Y. Y., Qi C. C., Shan S. J., Zhang F. M., Li H., An L. G., et al. (2016. a). Characterization of common carp (Cyprinus carpio L.) interferon regulatory factor 5 (IRF5) and its expression in response to viral and bacterial challenges. Bmc Vet. Res. 12 (1), 127 10.1186/s12917-016-0750-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y. Y., Xing W. X., Shan S. J., Zhang S. Q., Li Y. Q., Li T., et al. (2016. b). Characterization and immune response expression of the Rig-I-like receptor mda5 in common carp Cyprinus carpio. J. Fish Biol. 88 (6), 2188–2202. 10.1111/jfb.12981 [DOI] [PubMed] [Google Scholar]