Abstract
In-silico activity prediction was performed to predict new inhibitory activities of 2, 9-disubstituted 8-phenylthio/phenylsulfinyl-9h-purine derivatives as anti-proliferative agents using QSAR technique. The anti-proliferative agents were optimized using Density Functional Theory (DFT) method utilizing the B3LYP/6-31G* level of theory. Genetic Function Algorithm (GFA) was used to build the QSAR models. Out of the models built, the best one was selected and reported because of its fitness statistically with the following assessment parameters: R2trng = 0.919035, R2adj = 0.893733, Q2cv = 0.866475, R2test = 0.636217, and LOF = 0.215884. The selected model was further subjected to other assessment such as VIF, Y-scrambling test, applicability domain and found to be statistically significant. The binding mode of some selected 2, 9-disubstituted 8-phenylthio/phenylsulfinyl-9H-purine (ligands) in the active site of EGFR-tyrosine kinase (EGFR-TK) (receptor) was studied via Molecular docking. Molecule 22 was identified to have the highest binding energy (-10.4 kcal/mol) among the other selected ligands which it might be as a result of hydrogen interactions formed with MET793 (2.48599 Å, 2.04522 Å) & THR854 (3.76616 Å) amino acid residues and hydrophobic/other interactions with amino acid residues (LEU718, LEU844, MET766, VAL726, ALA743, LYS745 and MET790) in the active site of EGFR-tyrosine kinase (EGFR-TK). The drug-likeness of these selected anti-proliferative agents were predicted via the pharmacokinetics profile of the molecules utilizing SWISS ADME. The anti-proliferative agents were found to be orally safe by not having more than 1 violation of the Lipinski's rule of five. This research proposed a way for designing potent anti-proliferative agents against their target enzyme.
Keywords: Physical chemistry, Theoretical chemistry, NSCLC, DFT, B3LYP/6-31G*, EGFR, ADME
Physical chemistry; Theoretical chemistry, NSCLC; DFT; B3LYP/6-31G*; EGFR; ADME
1. Introduction
Receptor tyrosine kinase (RTK) is a Protein kinase which plays an essential role in signal transduction pathways [1]. Epidermal growth factor receptor (EGFR) is a member of tyrosine kinase (TK) which belongs to the ErbB family and regulates essential cellular roles, including reproduction, survival, movement, and differentiation. Overexpression, intensification, and modification of EGFR occur in a broad range of human cancers and are connected with tumour progression and decrease sensitivity to antitumor agents. Thus EGFR has been recognised as one of the principal anticancer targets [2].
The most common and deadly type of all cancers around the globe is lung cancer which accounts for 25% of the cancer deaths every year [3, 4]. Among the types of lung cancer with about 1.5 million patients and less than 20% survival rate is non-small cell lung cancer (NSCLC) [5]. Most lung cancer statistics include both small cell lung cancer (SCLC) and NSCLC. In general, about 10%–15% of all lung cancers are SCLC. According to the American Cancer Society's estimates for lung cancer in the United States for 2019, there were about 228,150 new cases of lung cancer in which about 116,440 cases were in men and 111,710 cases were in women. And the report on the death statistic was about 142,670. Out of the figure mentioned, about 76,650 were in men and 66,020 were in women [6].
To mitigate the problem of NSCLC, several medications were developed for several generations. The first generation (gefitinib and erlotinib) was designed to treat EGFRL858R mutations [7, 8]. The second generation was designed to treat EGFRT790M mutations examples were afatinib, dacomtinib and neratinib. The second-generation inhibitors share common structural properties of quinazoline pharmacophore and acrylamide structure [9]. While in the case of third generation, they were developed to treat EGFRT790M/L790M double mutations example AZD9291 [10]. The third-generation tyrosine kinase inhibitors (TKI), osimertinib and rociletinib of EGFR are highly active against T790M. Gradually, this third-generation tyrosine kinase inhibitors developed resistance rapidly. EGFR C797S mutation was reported to be a leading mechanism of resistance to the third-generation inhibitors. The C797S mutation appears to be an ideal target for overcoming the acquired resistance to the third-generation inhibitors. This summarizes the latest development on the discovery of a fourth-generation EGFR TKI (EAI045s) [11].
3D-QSAR modelling is a molecular modelling method which quantitatively correlates response variable (biological activities) and molecular descriptors (physicochemical properties) of a molecule [12]. Also, the 3D-QSAR technique of computer-aided drug design plays a significant role in predicting the biological activities of small molecules that have not been synthesised [13]. Furthermore, 3D-QSAR modelling reduces the number of compounds to be synthesized by helping in the selection of the most promising candidates and thereby reducing a lengthy time and cost in drug development. Many achievements of 3D-QSAR have attracted the medicinal chemists to investigate the relationships of structural properties with biological activity [14].
Virtual screening is a molecular modelling method used to analyzed large databases of compounds to identify potential hit candidates [15]. Molecular docking is another molecular modelling technique used to explore the interaction between 3D structures of a ligand and a receptor and how the ligand bind in the active site of the receptor tightly. Its also contribute to the virtual screening of a library of compounds at the pre-clinical stage of drug development [16].
The assessment of the absorption, distribution, metabolism and excretion (ADME) in drug development involved earlier in the discovery process, at a step when considered compounds are many due to restriction in the access to the physical samples [17]. Predicting ADME properties and drug-likeness of a small molecule (drug) in hit-to-lead and lead-optimization campaigns played an essential function in drug research and development [18]. This work aims at carrying out in-silico activity prediction on some anti-proliferative agents using QSAR technique, study the nature of interactions between the anti-proliferative agents and EGFR-tyrosine kinase (EGFR-TK) via docking and also to predict the ADME properties and drug-likeness of these anti-proliferative agents.
2. Computational method
2.1. Dataset collection
Thirty [30] 2, 9-disubstituted 8-phenylthio/phenylsulfinyl-9H-purine derivatives as anti-proliferative agents with their anti-proliferator inhibitory activities (IC50) in nM against human lung carcinoma cell line HCC827 were selected from the work of Hei et al [19]. The anti-proliferator inhibitory activities (IC50) of all the dataset were then converted to their corresponding negative logarithms (pIC50) using Eq. (1) [20]. Table 3 presents the molecular formula, pIC50, Predicted pIC50 and residuals and docking scores for all the data set and the standard drug (Gefitinib) used in this research.
| pIC50 = -log IC50 × 10−9 | (1) |
Table 3.
The Molecular formula, pIC50, Predicted pIC50, the residual values and binding energy for the studied molecules.
| S/No | Molecular formula | pIC50 | Predicted pIC50 | Residuals | Binding energy (kcal/mol) |
|---|---|---|---|---|---|
| 1 | C25H29N7S | 6.537752 | 6.675639 | -0.13789 | -9.3 |
| 2 | C25H27N7S | 6.516413 | 6.369865 | 0.146548 | -9.5 |
| 3 | C26H29N7S | 6.180522 | 6.347182 | -0.16666 | -9.4 |
| 4 | C27H31N7S | 7.29073 | 7.213299 | 0.077431 | -9.9 |
| 5 | C27H31N7OS | 7.37059 | 7.429651 | -0.05906 | -9.1 |
| 6 | C28H33N7OS | 7.319664 | 7.329735 | -0.01007 | -9.1 |
| 7 | C26H30N6O2S | 7.353596 | 7.636959 | -0.28336 | -9.6 |
| 8 | C27H33N7OS | 6.302074 | 6.453033 | -0.15096 | -9.7 |
| 9 | C29H35N7OS | 7.531653 | 7.149655 | 0.381998 | -9.6 |
| 10 | C28H32N6O2S | 6.854493 | 6.95115 | -0.09666 | -10.1 |
| 11y | C26H29FN6O2S | 7.289037 | 7.289037 | 0.065794 | -9.3 |
| 12 | C26H29FN6O2S | 7.320572 | 6.295373 | 0.079107 | -9.4 |
| 13 | C25H29N7OS | 6.668573 | 5.973654 | 0.3732 | -9.2 |
| 14 | C25H27N7OS | 5.788399 | 6.109828 | -0.18526 | -9.8 |
| 15 | C26H29N7OS | 6.076601 | 7.001534 | -0.03323 | -9.5 |
| 16 | C27H31N7OS | 7.189096 | 7.365658 | 0.187562 | -10.2 |
| 17y | C27H31N7O2S | 6.386581 | 6.386581 | 0.386269 | -10.3 |
| 18y | C28H33N7O2S | 6.571379 | 6.571379 | 0.3376 | -9.2 |
| 19 | C26H30N6O3S | 7.298432 | 7.298432 | -0.06723 | -9.4 |
| 20y | C27H33N7O2S | 5.876377 | 5.876377 | -0.2037 | -9.5 |
| 21 | C29H35N7O2S | 5.890219 | 5.890219 | -0.2963 | -9 |
| 22 | C28H32N6O3S | 6.069204 | 6.069204 | 0.003431 | -10.4 |
| 23 | C26H29FN6O3S | 6.445269 | 6.445269 | 0.057485 | -9.7 |
| 24y | C26H29FN6O3S | 6.257196 | 6.257196 | 0.066387 | -9.7 |
| 25 | C25H28N8S | 5.053911 | 5.053911 | 0.092906 | -9.1 |
| 26 | C27H32N8S | 6.286258 | 6.286258 | -0.14532 | -9.5 |
| 27y | C27H32N8OS | 6.301378 | 6.301378 | 0.300808 | -9.7 |
| 28 | C28H33N7OS | 6.97265 | 6.97265 | 0.232312 | -9.0 |
| 29y | C27H32N8OS | 5.628323 | 5.628323 | 0.590811 | -9.9 |
| 30y | C28H33N7O2S | 6.363412 | 6.363412 | -0.39023 | -9.0 |
| Gefitinib | C22H24ClFN4O3 | 33.3 | 5.505041 | -1.97251 | -8.0 |
y = Test set.
2.2. Structure generation and stable geometry calculations
In this research, after data collection, the next thing is the drawing of the 2D-structures of the dataset. The 2D-structures of the dataset was done using Chemdraw 12.0 software [21]. After 2D-structure generation of the studied molecules, the 2D structures were automatically converted to 3D by the Spartan 14 software prior to energy minimization. Energy minimization was carried out to reduce constraint in the structures before finding the most stable geometry of the studied molecules on potential energy surface using the same software. DFT at B3LYP/6-311G* level of theory was used in finding the most stable structures of all the studied molecules on global minima on the potential energy surface (PES) [22].
2.3. 1D, 2D and 3D descriptors generation, data pre-treatment and daataset splitting
The calculation of the independent variables (descriptors) was done using PaDEL descriptor tool kit. It calculates both 1D, 2D and 3D descriptors [23].
The dataset was pre-treated manually to eliminate redundant and constant descriptors. After pre-treating the data, Data division software was further used in dividing the data into a training set and test set utilizing Kennard-Stone algorithm [24]. The model building/training set was used for the generation of the models while the validation/test set was used for assessing the generated models [12].
2.4. Model development
The models were generated utilizing Genetic Function Approximation (GFA) method with the actual pIC50 as the response variable and the descriptors as independent variables. In the case of variable selection, GFA selects the most highly correlated descriptors to develop so many models which is one of the distinct characteristics of GFA.
2.5. Validation of the selected model
The most widely used assessment terms for QSAR models are the; Square correlation coefficient of the training and test set (R2training and R2 test), Adjusted R2 (R2 adj), and Cross-validation coefficient (Qcv2. The high value of these parameters appear to be necessary but not enough [25].
Because of this, the inter-correlation between descriptors can be detected using their variation inflation factors (VIF), to see whether these descriptors are highly correlated with one another or not. If the computed VIF values are up to 1 it means there is no inter-correlation between the descriptors; if it falls between 1–5, the model can be accepted; and if it is higher than 10, the model cannot be accepted. It can be calculated using the equation below:
| (2) |
where R2 is the correlation coefficient of the selected model [26].
The evaluation of significance and contribution of each descriptor to the selected model is performed using the value of mean effect of each descriptor. The mean effect is defined by equation below:
| (3) |
where MFj is the mean effect of a descriptor j in a model, βj is the coefficient of the descriptor J in that model and dij is the value of the descriptor in the data matrix for each molecule in the model building set, m is the number of descriptor that appear in the model and n is the number of molecules in the model building set [27].
To confirm the robustness of the reported model and that the model was not obtained by chance correlation, Y-Scrambling test was performed. The test was done by reshuffling the actual activities and keeping the descriptors unchanged to generate new QSAR models for several trials. The new built QSAR models were anticipated to give low Q2 and R2 value. The validation parameter for this test is cRp (cR2p >0.5) [28].
2.6. Applicability domain
A QSAR model is considered valid and void if it is subjected to the applicability domain (AD) and found that the model can make a reliable prediction of new activities of the training and test molecules. As such, the reported model was subjected to AD to find out whether there are influential or outliers molecules in the studied ones [29]. One of the methods used in assessing the AD is leverage approach and is given as hi:
| hi = xi (XT X)−K xiT (i = A,…, Z) | (4) |
where the training set matrix I is given by xi, n × k descriptor matrix of the training set is represented by X and XT is the transpose matrix X used in generating the model. The thresh-hold for the value of X is the warning threshold (h*) which is presented in the equation below:
| h*= 3(x+1)/q | (5) |
where the number of chemicals of the model building set is given by q, and the number of the descriptors in the model under evaluation is represented by x.
2.7. Molecular docking
A Dell Latitude E6520 computer system, with the following specification: Intel ® Core™ i7 Dual CPU, M330 @2.75 GHz 2.75GHz, 8GB of RAM was utilized to explore the nature of interactions between the active site of EGFR-tyrosine kinase (EGFR-TK) and some selected anti-proliferative agents (ligands) with the help of Pyrex virtual screening software, Chimera, PyMOL and Discovery studio.
Before the docking analysis, ligands were prepared from the optimized structures in 2.2 above saved in pdb file format using Spartan’14 [13]. The 3D structure of EGFR-tyrosine kinase (EGFR-TK) was downloaded from the protein data bank (with pdb ID: 4zau) [30] The enzyme was prepared with help of Discovery Studio Visualizer for the docking analysis. In the course of the preparation, hydrogen was added. Water molecule, heteroatoms and co-ligands were eliminated from the crystal structure saved in pdb file.
The docking of the ligands to the active site of EGFR-tyrosine kinase (EGFR-TK) was achieved with the help of Pyrex software using Autodock vina [12]. After successful docking protocol, re-formation of the complexes (ligand-receptor) for further investigation was also achieved utilizing Chimera software. Discovery studio visualizer and PyMOL were used to investigate the interactions of the complexes.
2.8. ADME properties and drug-likeness prediction
ADME properties and drug-likeness prediction of some selected anti-proliferative agents among the data set was carried out using SwissADME a free web tool used in evaluating ADME properties and drug-likeness of small molecules [17]. The Lipinski's rule of five is useful at pre-clinical stage of drug discovery which state that if any chemical violate more than 2 of these criteria (Molecular weight ˂ 500, Number of hydrogen bond donors ≤5, Number of hydrogen bond acceptors ≤10, Calculated Log p ≤ 5 and Polar surface area (PSA) ˂140 Å2), the chemical is said to be impermeable or badly absorbed Guangzhe et la., (2019) [31].
3. Result and discussion
3.1. 3D-QSAR modeling
The model reported was found to have passed the minimum requirement for the assessment of a reliable QSAR models with the following assessment parameters: R2 of 0.919035, R2adj of 0.893733, Qcv2 of 0.866475, R2test of 0.636217 and LOF of 0.215884 as reported by [32] (Table 1).
| pIC50 = - 12.417755021 (AATS7e) + 5.879592939 (AATS8e) - 0.433185723 (ATSC3e) - 22.018131847 (MATS7m) + 0.333566302 (VR3_D) + 46.983337086 |
Table 1.
General minimum required value for the assessment of QSAR model.
| Symbol | Name | Recommended Value | Reported Model |
|---|---|---|---|
| R2 | Co-efficient of determination | 0.6 | 0.919035 |
| Q2 | Cross-Validation Co-efficient | 0.5 | 0.866475 |
| R2- Q2 | Difference between R2 and Q2 | 0.3 | 0.05256 |
| N(ext, test set) | Minimum number of external test set | 5 | 8 |
| R2ext. | Co-efficient of determination of external and test set | 0.5 | 0.636217 |
The details of the descriptors in the reported model were presented in Table 2. The descriptors with negative coefficients (AATS7e, ATSC3e and MATS7m) highlighted the negative correlation of these descriptors to the anti-proliferator inhibitory activities of the anti-proliferative agents. The lesser in the number of these descriptors in the structures of anti-proliferative agents the more the action of anti-proliferative agents against EGFR-tyrosine kinase (EGFR-TK). Looking at the descriptors with positive co-efficient (AATS8e and VR3_D), it indicates positive correlation the descriptors to the anti-proliferator inhibitory activities of anti-proliferative agents that is the more you have these types of descriptors the more the anti-proliferator inhibitory activities of the anti-proliferative agents against EGFR-tyrosine kinase (EGFR-TK).
Table 2.
The symbols, descriptions and classes of descriptors for the selected model.
| S/no | Symbol | Description | Class |
|---|---|---|---|
| 1 | AATS7e | Average Broto-Moreau autocorrelation - lag 7/weighted by Sanderson electronegativities | 2D |
| 2 | AATS8e | Average Broto-Moreau autocorrelation - lag 8/weighted by Sanderson electronegativities | 2D |
| 3 | ATSC3e | Centered Broto-Moreau autocorrelation - lag 3/weighted by Sanderson electronegativities | 2D |
| 4 | MATS7m | Moran autocorrelation - lag 7/weighted by mass | 2D |
| 5 | VR3_D | Logarithmic Randic-like eigenvector-based index from Barysz matrix/weighted by atomic number | 3D |
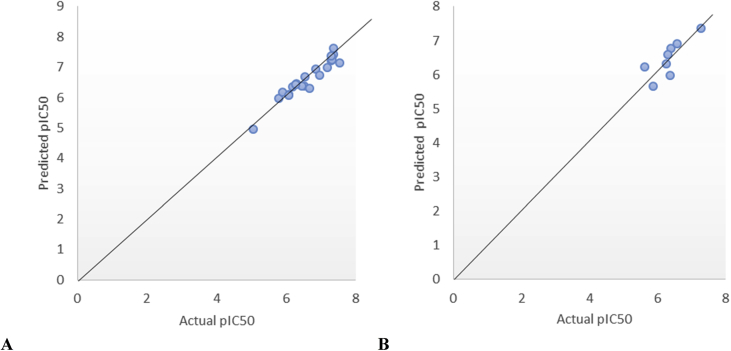
The XY (Scatter) plot of predicted activities of both the test and training sets against the Actual pIC50 was shown in Figure 1A & 1B. It can be seen from the two plots that the values were plotted around the straight line which shows the significant of the selected model. Also the R2 values from the plots agree with those of the training and test for the internal and external assessment.
Figure 1.
(A) XY (Scatter) Plot of the actual pIC50 against predicted pIC50 of training set (B) XY (Scatter) Plot of the actual pIC50 against predicted pIC50 of test set of the selected model.
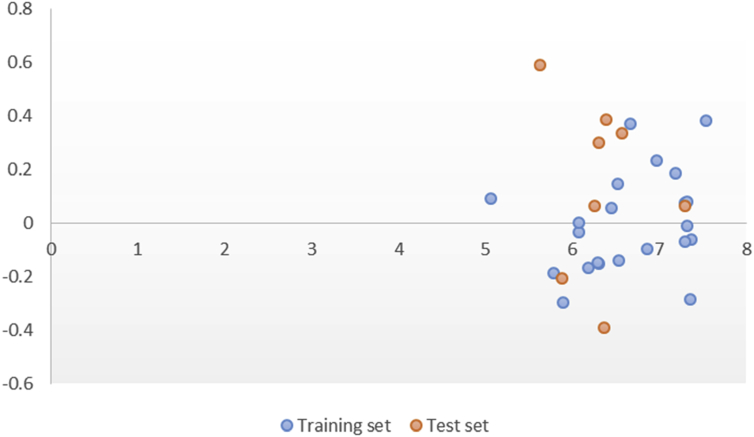
Also XY (Scatter) plot of Actual pIC50 against the residuals of both the training and test was also shown (Figure 2.). The irregular appearance of these residuals on either side of zero on the plot shows the non-existence of methodological error in the selected model.
Figure 2.
XY (Scatter) Plot of actual pIC50 against the residuals of both the test and training sets of the selected model.
The Molecular formula, pIC50, Predicted pIC50, the residual values and docking scores for all the studied molecules and the standard drug Gefitinib were presented in Table 3. The low residual values (The difference between the actual and the predicted activities is termed residual) noted in the table and other validation assessments verified the reliability of the reported model.
The Pearson's correlation matrix statistics of the descriptors in the reported model was carried out (Table 4) and the descriptors were found to have no correlation with one another. This shows the high quality pf the physicochemical parameters (descriptors) utilized in generating the reported model. To further confirm whether there is relationship or not between the descriptors in the reported model, The VIF values of these descriptors in the training set were computed and obtained to be less than 10 (Table 4) indicating the fitness of the reported model and the descriptors were independent of one another. The MF value (Table 4) gives the contribution of a descriptor in contrast to other descriptors in the best model. The signs point the various direction of either increase or decrease in the values of these descriptors which will improve the anti-proliferator inhibitory activities of the studies molecules.
Table 4.
MF, VIF and correlation between descriptors of the selected model.
| AATS7e | AATS8e | ATSC3e | MATS7m | VR3_D | VIF | ME | |
|---|---|---|---|---|---|---|---|
| AATS7e | 1 | 7.726502 | 2.33884 | ||||
| AATS8e | 0.585238 | 1 | 8.27239 | -1.09383 | |||
| ATSC3e | -0.07618 | -0.45661 | 1 | 1.457493 | -0.0027 | ||
| MATS7m | -0.39681 | 0.419105 | -0.28992 | 1 | 5.410414 | -0.05726 | |
| VR3_D | 0.621244 | 0.675433 | -0.30276 | 0.085265 | 1 | 2.190966 | -0.18505 |
Table 5 shows the result of Y-scrambling test for ten [10] differently generated random models. The newly randomly generated models were found to have low R2 and Q2 values. This has proven the obtainability of the reported model was actually not by chance and further confirm its robustness.
Table 5.
Y-scrambling test.
| Model | R | R2 | Q2 |
|---|---|---|---|
| Original | 0.842447 | 0.709717 | 0.504914 |
| Random 1 | 0.63572 | 0.40414 | 0.013407 |
| Random 2 | 0.577014 | 0.332946 | -0.05171 |
| Random 3 | 0.440502 | 0.194042 | -0.35335 |
| Random 4 | 0.252932 | 0.063975 | -0.53688 |
| Random 5 | 0.631791 | 0.39916 | -0.04994 |
| Random 6 | 0.25435 | 0.064694 | -0.46379 |
| Random 7 | 0.279886 | 0.078336 | -0.59263 |
| Random 8 | 0.307425 | 0.09451 | -0.59972 |
| Random 9 | 0.424458 | 0.180165 | -0.3578 |
| Random 10 | 0.627664 | 0.393962 | -0.14752 |
| Average r: | 0.443174 | ||
| Average r2: | 0.220593 | ||
| Average Q2: | -0.31399 | ||
| cRp2: | 0.603579 |
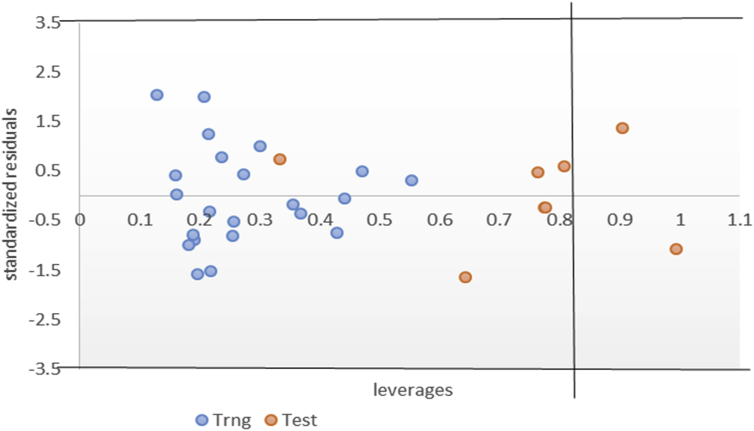
The Williams plot presented in Figure 3 identified two [2] influential compounds from which were all in the test set. It is very paramount to decipher that these molecules with leverage value greater than the threshold h*(h* = 0.82) are not put into consideration when designing new Anti-proliferative agents. These molecules might be structurally different from those used to generate the reported model and, thus may have different mechanism of action.
Figure 3.
Williams Plot of the selected model.
3.2. Molecular docking
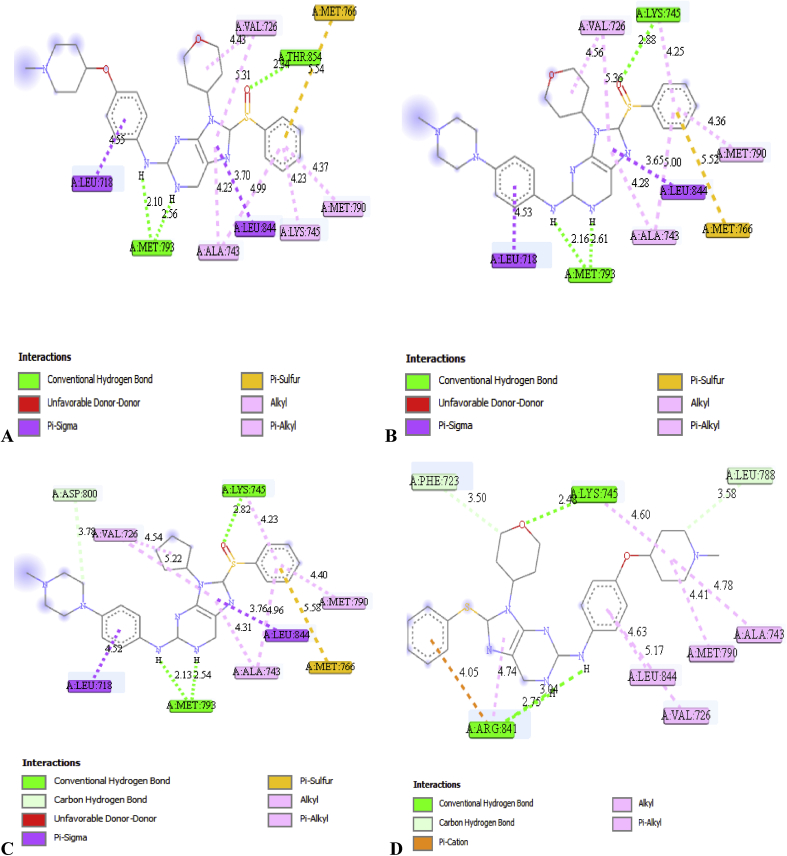
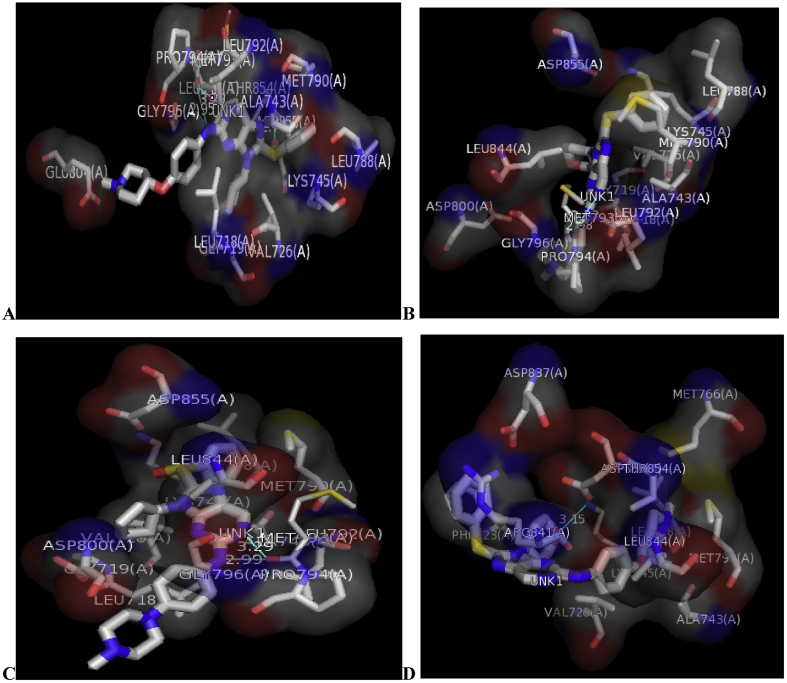
The binding mode of some selected 2, 9-disubstituted 8-phenylthio/phenylsulfinyl-9H-purine (ligands) in the active site of EGFR-tyrosine kinase (EGFR-TK) (receptor) was studied via Molecular docking (Table 6). From Table 6, molecule 22 was identified to have the highest binding energy (-10.4 kcal/mol) among the other selected ligands which is probably as a result of hydrogen interactions formed with MET793 (2.48599 Å, 2.04522 Å) & THR854 (3.76616 Å) amino acid residues in the active site of EGFR-tyrosine kinase (EGFR-TK). Besides hydrogen bond, it also formed hydrophobic and other interactions with amino acid residues (LEU718, LEU844, MET766, VAL726, ALA743, LYS745 and MET790) of EGFR-tyrosine kinase (EGFR-TK). On the other hand, molecule 17 with the binding energy of -10.3 kcal/mol formed similar interaction with molecule 22 in the active site of EGFR-tyrosine kinase (EGFR-TK). Looking at molecule 16 which is third in the ranking in terms of binding energy (-10.2 kcal/mol). It formed hydrogen bond with MET793, LYS745 and ASP800 amino acid residues in the active site of EGFR-tyrosine kinase (EGFR-TK) with bond distance of 2.53982 Å, 2.1281 Å, 2.816 Å and 3.7817 Å respectively. Not only hydrogen bond has it interacted with, but also interacted with LEU718, LEU844, MET766, VAL726, ALA743, LYS745 & MET790 in the active site of EGFR-tyrosine kinase (EGFR-TK) via hydrophobic. The last in the ranking (ligand 10) also interact very well in the active site of the EGFR-tyrosine kinase (EGFR-TK) with binding energy of -10.1 kcal/mol. Hydrogen bond between the ligand and these amino acids ARG841 (2.75436 Å, 3.04097 Å) LYS745 (2.48106 Å), LEU788 (3.58374 Å) & PHE723 (3.49783 Å) of the enzyme was observed. A part from the hydrogen bond it also interact with these amino acid residues ALA743, LYS745, MET790, ARG841, VAL726 & LEU844 in the active site of EGFR-tyrosine kinase (EGFR-TK) through hydrophobic. The 3D and 2D structures of complex 22, 17, 16 and 10 are shown in Figures 4 and 5. In order to validate the docking protocol and productivity, the co-crystalized ligand (AEE788) was also docked to the binding site of EGFR-tyrosine kinase (EGFR-TK) with the binding affinity of -9.3 kcal/mol and the RMSD value for both upper and lower bounds were measured (0.0) which confirmed the docking protocol and productivity.
Table 6.
The binding energy, Amino acid residues, Hydrogen bond (bond length Å) of some selected ligands.
| S/N | Binding energy (Kcal/mol) | Amino Acid Residues | Hydrogen Bond (bond length Å) |
|---|---|---|---|
| 22 | -10.4 | LEU718, LEU844, MET766, VAL726 ALA743, LYS745 & MET790 |
MET793(2.49, 2.05 & THR854(3.77) |
| 17 | -10.3 | LEU718, LEU844, MET766, VAL726, LA743 LYS745 & MET790 | MET793(2.61, 2.16) & LYS745(2.88) |
| 16 | -10.2 | LEU718, LEU844, MET766, VAL726, ALA743, LYS745 & MET790 | MET793 (2.54, 2.13) LYS745 (2.82) & ASP800 (3.78) |
| 10 | -10.1 | ALA743, LYS745, MET790, ARG841, VAL726 & LEU844 | ARG841(2.75, 3.04) LYS745(2.48), LEU788(3.58) & PHE723(3.50) |
Figure 4.
2D structures of (A) Complex 22, (B) Complex 17, (C) Complex 16 and (D) Complex 10 with bond distances using Discovery studio visualizer.
Figure 5.
3D structures of (A) Complex 22, (B) Complex 17, (C) Complex 16 and (D) Complex 10 using PyMOL.
3.3. ADME and drug-likeness properties
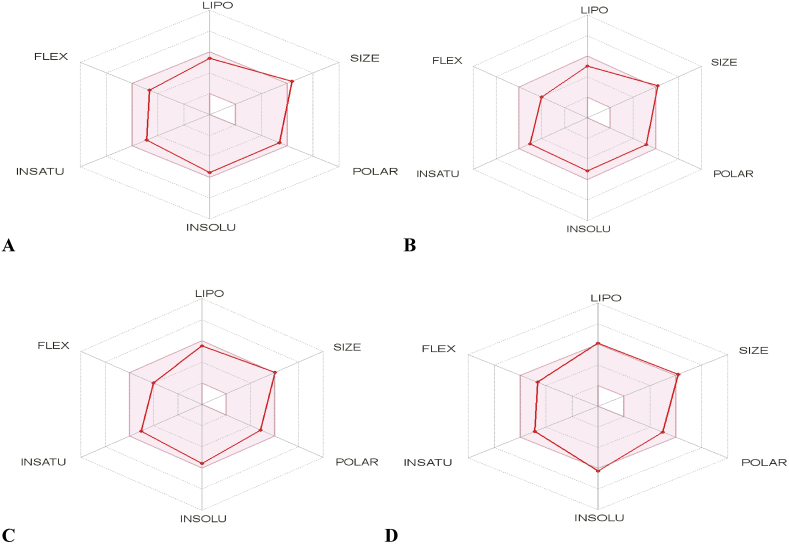
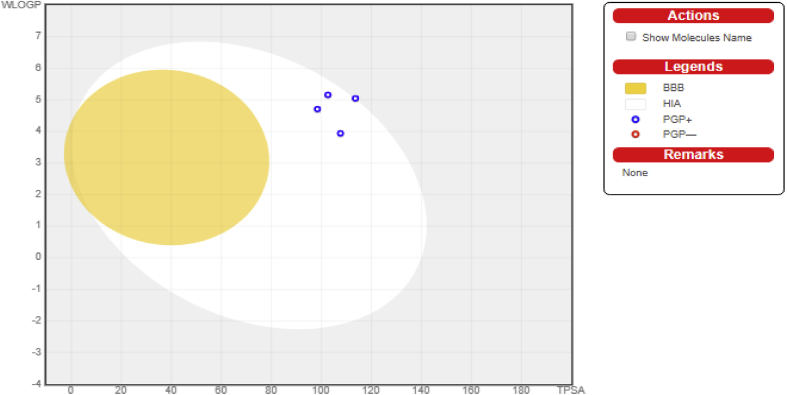
The ADME and drug-likeness results of these selected 2, 9-disubstituted 8-phenylthio/phenylsulfinyl-9H-purine are shown in Tables Table 7a, Table 7b. From the Table 7a it can be seen that none of the molecules violate more than 1 of the criteria stated by Lipinski's rule of five, it means there is a high tendency all of these molecules might be pharmacologically active. In a null shell these molecules are said have good absorption, low toxicity level, orally bioavailable and permeable. The Bioavailability Radar gives an overview of the drug-likeness of a molecule (Figure 6). The region painted pink indicates the range for each properties. The Boiled-egg plot between WLOGP and TPSA to predict gastrointestinal absorption and brain penetration of the selected molecules was shown in Figure 7. It can be seen from the plot that none of the molecules possess the BBB permeant but they are within the GI absorption region. The ADME and Drug-likeness properties for the remaining 26 compounds were presented in the Supplementary Table 1a and Supplementary Table 1b. The remaining 26 compounds have good absorption and low toxicity level. Also, they are orally bioavailable and permeable.
Table 7a.
ADME properties.
| S/N | MW | HB donor | HB acceptor | WLOGP | TPSA | Lipinski violations |
|---|---|---|---|---|---|---|
| 22 | 532.66 | 1 | 7 | 5.05 | 113.61 | 1 |
| 17 | 517.65 | 1 | 6 | 3.94 | 107.62 | 1 |
| 16 | 501.65 | 1 | 5 | 4.71 | 98.39 | 1 |
| 10 | 516.66 | 1 | 6 | 5.16 | 102.63 | 1 |
Table 7b.
Drug-likeness properties.
| S/N | GI absorption | BBB permeant | Pgp substrate | Bioavailability Score |
|---|---|---|---|---|
| 22 | High | No | Yes | 0.55 |
| 17 | High | No | Yes | 0.55 |
| 16 | High | No | Yes | 0.55 |
| 10 | High | No | Yes | 0.55 |
Figure 6.
The Bioavailability Radar of (A) Molecule 22 (B) Molecule 17 (C) Molecule 16 (D) Molecule 10 with the highest docking score.
Figure 7.
The Boiled-egg plot of the selected molecules.
4. Conclusion
GFA-MLR analysis was used to perform 3D-QSAR modeling to predict new inhibitory activities of 2, 9-disubstituted 8-phenylthio/phenylsulfinyl-9H-purine as anti-proliferative agents. DFT method at B3LYP/6-31G* level of theory was utilized to ascertained the equilibrium structures of these Anti-proliferative agents. The reported model was selected because of its fitness with the following assessment parameters: R2trng = 0.919035, R2adj = 0.893733, Q2cv = 0.866475, R2test = 0.636217, and LOF = 0.215884. The predict power, reliability and robustness of the reported model was confirmed by passing different validation techniques such as Applicability domain, VIF and Y-scrambling test.
The Molecular docking result of some selected 2, 9-disubstituted 8-phenylthio/phenylsulfinyl-9H-purine (ligands) in the active site of EGFR-tyrosine kinase (EGFR-TK) (receptor) shows that molecule 22 has the highest binding energy of -10.4 kcal/mol among co-ligands. The results of the 3D-QSAR modeling and molecular docking agrees with one another in which the molecule with the lowest residual values has the highest binding energy.
The ADME properties and drug-likeness of these selected ant-proliferative agents were predicted via pharmacokinetic profile of these molecules utilizing SWISS ADME. These molecules were found to be orally bioavailable by not violating more than the minimum recommended criteria set by the Lipinski's rule of five filters.
Declarations
Author contribution statement
A Uzairu: Conceived and designed the experiments.
Muhammad Tukur Ibrahim: Performed the experiments; Wrote the paper.
G. A. Shallangwa, S. Uba: Analyzed and interpreted the data.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgments
The authors sincerely acknowledge Ahmadu Bello University, Zaria for its technical support in the course of this research.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Zhang H., Wu W., Feng C., Liu Z., Bai E., Wang X., Lei M., Cheng H., Feng H., Shi J. Design, synthesis, SAR discussion, in vitro and in vivo evaluation of novel selective EGFR modulator to inhibit L858R/T790M double mutants. Eur. J. Med. Chem. 2017;135:12–23. doi: 10.1016/j.ejmech.2017.04.036. [DOI] [PubMed] [Google Scholar]
- 2.Hanan E.J., Baumgardner M., Bryan M.C., Chen Y., Eigenbrot C., Fan P., Gu X.-H., La H., Malek S., Purkey H.E. 4-Aminoindazolyl-dihydrofuro [3, 4-d] pyrimidines as non-covalent inhibitors of mutant epidermal growth factor receptor tyrosine kinase. Bioorg. Med. Chem. Lett. 2016;26(2):534–539. doi: 10.1016/j.bmcl.2015.11.078. [DOI] [PubMed] [Google Scholar]
- 3.Chico L.K., Van Eldik L.J., Watterson D.M. Targeting protein kinases in central nervous system disorders. Nat. Rev. Drug Discov. 2009;8(11):892. doi: 10.1038/nrd2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song Z., Ge Y., Wang C., Huang S., Shu X., Liu K., Zhou Y., Ma X. Challenges and perspectives on the development of small-molecule EGFR inhibitors against T790M-mediated resistance in non-small-cell lung cancer: miniperspective. J. Med. Chem. 2016;59(14):6580–6594. doi: 10.1021/acs.jmedchem.5b00840. [DOI] [PubMed] [Google Scholar]
- 5.Tiseo M., Bartolotti M., Gelsomino F., Bordi P. Emerging role of gefitinib in the treatment of non-small-cell lung cancer (NSCLC) Drug Des. Dev. Ther. 2010;4:81. doi: 10.2147/dddt.s6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.2019. https://www.cancer.org/content/cancer/en/cancer/lung-cancer/about/key-statistics.html
- 7.Maemondo M., Inoue A., Kobayashi K., Sugawara S., Oizumi S., Isobe H., Gemma A., Harada M., Yoshizawa H., Kinoshita I. Gefitinib or chemotherapy for non–small-cell lung cancer with mutated EGFR. N. Engl. J. Med. 2010;362(25):2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 8.Tsao M.-S., Sakurada A., Cutz J.-C., Zhu C.-Q., Kamel-Reid S., Squire J., Lorimer I., Zhang T., Liu N., Daneshmand M. Erlotinib in lung cancer—molecular and clinical predictors of outcome. N. Engl. J. Med. 2005;353(2):133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- 9.Solca F., Dahl G., Zoephel A., Bader G., Sanderson M., Klein C., Kraemer O., Himmelsbach F., Haaksma E., Adolf G.R. Target binding properties and cellular activity of afatinib (BIBW 2992), an irreversible ErbB family blocker. J. Pharmacol. Exp. Ther. 2012;343(2):342–350. doi: 10.1124/jpet.112.197756. [DOI] [PubMed] [Google Scholar]
- 10.Cross D.A., Ashton S.E., Ghiorghiu S., Eberlein C., Nebhan C.A., Spitzler P.J., Orme J.P., Finlay M.R.V., Ward R.A., Mellor M.J. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov. 2014;4(9):1046–1061. doi: 10.1158/2159-8290.CD-14-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang S., Song Y., Liu D. EAI045: the fourth-generation EGFR inhibitor overcoming T790M and C797S resistance. Cancer Lett. 2017;385:51–54. doi: 10.1016/j.canlet.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 12.Ibrahim M.T., Uzairu A., Shallangwa G.A., Ibrahim A. In-silico studies of some oxadiazoles derivatives as anti-diabetic compounds. J. King Saud Univ. Sci. 2018 [Google Scholar]
- 13.Abdulfatai U., Uba S., Umar B.A., Ibrahim M.T. Molecular design and docking analysis of the inhibitory activities of some α_substituted acetamido-N-benzylacetamide as anticonvulsant agents. SN Appl. Sci. 2019;1(5):499. [Google Scholar]
- 14.Verma J., Khedkar V.M., Coutinho E.C. 3D-QSAR in drug design-a review. Curr. Top. Med. Chem. 2010;10(1):95–115. doi: 10.2174/156802610790232260. [DOI] [PubMed] [Google Scholar]
- 15.Medina-Franco J.L., Yoo J., Dueñas-González A. Epigenetic Technological Applications. Elsevier; 2015. DNA methyltransferase inhibitors for cancer therapy; pp. 265–290. [Google Scholar]
- 16.Bello A., Uzairu A., Ibrahim M., Gatugel Y. Quantum modelling analysis of some potent indole derivatives on ns5b polymerase inhibitors. Sci. World J. 2019;14(1):32–37. [Google Scholar]
- 17.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferreira L.L., Andricopulo A.D. ADMET modeling approaches in drug discovery. Drug Discov. Today. 2019 doi: 10.1016/j.drudis.2019.03.015. [DOI] [PubMed] [Google Scholar]
- 19.Hei Y.-Y., Shen Y., Wang J., Zhang H., Zhao H.-Y., Xin M., Cao Y.-X., Li Y., Zhang S.-Q. Synthesis and evaluation of 2, 9-disubstituted 8-phenylthio/phenylsulfinyl-9H-purine as new EGFR inhibitors. Bioorg. Med. Chem. 2018;26(8):2173–2185. doi: 10.1016/j.bmc.2018.03.025. [DOI] [PubMed] [Google Scholar]
- 20.Abdullahia M., Shallangwaa G.A., Ibrahima M.T., Bello A.U., Arthura D.E., Uzairua A., Mamzaa P. 2018. QSAR Studies on Some C14-Urea Tetrandrine Compounds as Potent Anti-cancer Agents against Leukemia Cell Line (K562). JKS-S. [Google Scholar]
- 21.Ibrahim M.T., Uzairu A., Shallangwa G.A., Ibrahim A. Computational studies of some biscoumarin and biscoumarin thiourea derivatives as⍺-glucosidase inhibitors. J Eng Exact Sci. 2018;4(2):276–285. [Google Scholar]
- 22.Kohn W., Becke A.D., Parr R.G. Density functional theory of electronic structure. J. Phys. Chem. 1996;100(31):12974–12980. [Google Scholar]
- 23.Yap C.W. PaDEL-descriptor: an open source software to calculate molecular descriptors and fingerprints. J. Comput. Chem. 2011;32(7):1466–1474. doi: 10.1002/jcc.21707. [DOI] [PubMed] [Google Scholar]
- 24.Kennard R.W., Stone L.A. Computer aided design of experiments. Technometrics. 1969;11(1):137–148. [Google Scholar]
- 25.Tropsha A., Bajorath J.r. ACS Publications; 2015. Computational Methods for Drug Discovery and Design. [DOI] [PubMed] [Google Scholar]
- 26.Beheshti A., Pourbasheer E., Nekoei M., Vahdani S. QSAR modeling of antimalarial activity of urea derivatives using genetic algorithm–multiple linear regressions. J. Saudi Chem. Soc. 2016;20(3):282–290. [Google Scholar]
- 27.Adedirin O., Uzairu A., Shallangwa G.A., Abechi S.E. QSAR AND molecular docking based design of some n-benzylacetamide as γ-aminobutyrate-aminotransferase inhibitors. J. Eng. Exact Sci. 2018;4(1) 0065-0084. [Google Scholar]
- 28.Ibrahim M.T., Uzairu A., Shallangwa G.A., Uba S. QSAR modelling and docking analysis of some thiazole analogues as⍺-glucosidase inhibitors. J Eng Exact Sci. 2019;5(3):257–270. [Google Scholar]
- 29.Tropsha A., Gramatica P., Gombar V.K. The importance of being earnest: validation is the absolute essential for successful application and interpretation of QSPR models. Mol. Inform. 2003;22(1):69–77. [Google Scholar]
- 30.Yosaatmadja Y., Silva S., Dickson J.M., Patterson A.V., Smaill J.B., Flanagan J.U., McKeage M.J., Squire C.J. Binding mode of the breakthrough inhibitor AZD9291 to epidermal growth factor receptor revealed. J. Struct. Biol. 2015;192(3):539–544. doi: 10.1016/j.jsb.2015.10.018. [DOI] [PubMed] [Google Scholar]
- 31.Ismail S.Y., Uzairu A., Sagagi B., Sabiu M. In silico molecular docking and pharmacokinetic study of selected phytochemicals with estrogen and progesterone receptors as anticancer agent for breast cancer. 2018;5(3):1337–1350. [Google Scholar]
- 32.Veerasamy R., Rajak H., Jain A., Sivadasan S., Varghese C.P., Agrawal R.K. Validation of QSAR models-strategies and importance. Int J Drug Design Discov. 2011;3:511–519. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.