Abstract
Food producing animal is a global challenge in terms of antimicrobial resistance spread. Extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae are relevant opportunistic pathogens that may spread in many ecological niches of the One Health approach as human, animal and environment due to intestinal selection of antimicrobial resistant commensals in food production animals. Cattle production is a relevant ecological niche for selection of commensal bacteria with antimicrobial resistance from microbiota. Enterobacteriaceae show importance in terms of circulation of resistant-bacteria and antimicrobial resistance genes via food chain creating a resistance reservoir, setting up a threat for colonization of humans and consequent health risk. ESBL-producing Enterobacteriaceae are a threat in terms of human health responsible for life threatening outbreaks and silent enteric colonization of community populations namely the elder population. Food associated colonization is a risk difficult to handle and control. In a time of globalization of food trading, population intestinal colonization is a mirror of food production and in that sense this work aims to make a picture of ESBL-producing Enterobacteriaceae in animal production for food over the world in order to make some light in this reality of selection of resistant threats in food producing animal.
Keywords: Microbiology, Food microbiology, Cattle, Antibiotic resistance, Antibiotic resistant bacteria
Microbiology; Food microbiology; Cattle; Antibiotic resistance; Antibiotic resistant bacteria
1. Introduction
Extended-spectrum beta-lactamases (ESBL) are enzymes responsible for the hydrolysis of oxyimino-beta-lactam antibiotics, which are important therapeutic agents for the treatment of serious human and animal infections. ESBL were first described in 1983 in Enterobacteriaceae (new taxonomy Enterobacterales) and since then, with the research of the scientific community, it has been observed that ESBL-producing Enterobacteriaceae (E-ESBL) are a real threat to human health, being responsible for 1700 deaths in the USA due to therapeutic failure in severe infections in 2013 (Adeolu et al., 2016; CDC, 2013; Knothe et al., 1983). However, E-ESBL are not only limited to hospital environment, they are also present as human intestinal commensals (Gonçalves et al., 2016; Karanika et al., 2016). The presence of E-ESBL in several ecological niches, as commensals in humans and animals and as environmental contaminants, is reported worldwide, however, in the last decades a niche that has raised great concern, for being able to function as a reservoir and vehicle of transmission and dissemination of E-ESBL is the production animals due to their direct connection with the food chain (Madec et al., 2017).
Cattle are one of the main sources of animal protein, becoming one of the most consumed meat around the world and milk, one of the main constituents of the human food chain (Alexandratos and Bruinsma, 2012). It is also one of the main sources of biological fertilizers, due to the high production of faecal mass of these animals (Smith and Williams, 2016). All this, highlights the importance of cattle production in the context of the food chain and the contaminated environment as reservoir and transmitting/disseminating vehicle of E-ESBL, thus configuring a threat to the world public health. This circulation of E-ESBL within our ecosystem creates a consensual concern of the scientific community and of the authority involved in the One health approach (Robinson et al., 2016).
The ESBL are enzymes that are classified in several types, being CTX-M, SHV and TEM the most prevalent around the world (Paterson and Bonomo, 2005). However, there are other ESBL such as OXA, PER, VEB, BES, GES, SFO, TLA, and IBC (Paterson and Bonomo, 2005). The CTX-M are enzymes with environmental origins which are currently the most widespread type of ESBL and are commonly associated with E-ESBL reports (Cantón et al., 2012). Variants such as CTX-M-15, responsible for infectious outbreaks around the world, are associated with a clone responsible for extraintestinal E. coli infections resistant to antibiotics, the ST131 (Price et al., 2013).
The objective of this study was to make an insight about the epidemiology of the spread of E-ESBL and the ESBL genes distribution in cattle around the world, in order to update the current scenario of E-ESBL dissemination through cattle production in all continents.
2. ESBL producing Enterobacteriaceae in cattle - a global view
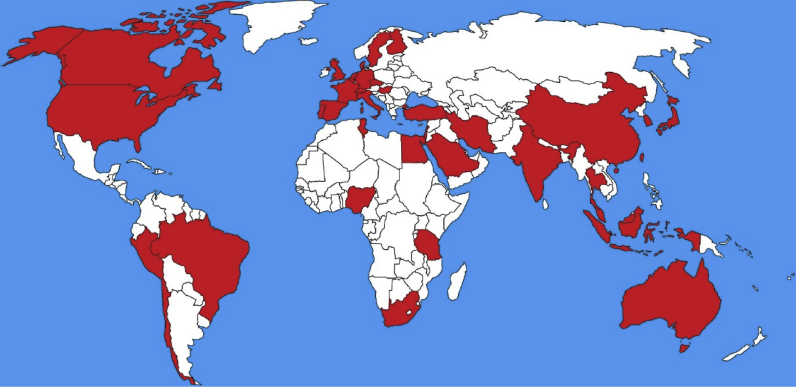
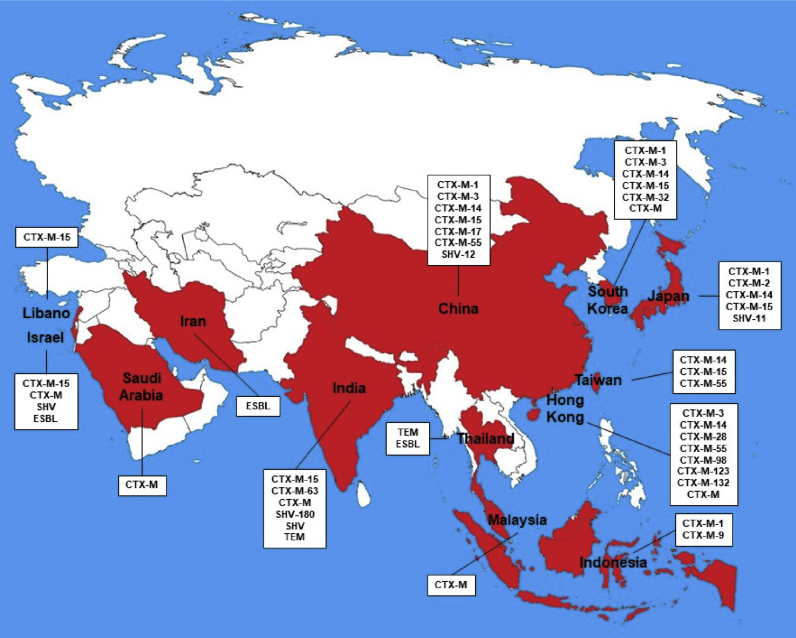
The first description of an E-ESBL in cattle was in Japan, where a CTX-M-2 E. coli producer was detected in cattle faeces from an important region close to the centre of the country (Shiraki et al., 2004). From the first description to the present, E-ESBL has already been described in cattle production in 39 countries, with more concentration in Europe (n = 16) and Asia (n = 13) as shown in Figure 1. The origins of E-ESBL are diverse, being isolated from healthy animals (faecal samples) or from veterinary clinical origin (mastitis, diarrheal processes, infections or with any other pathological picture). The countries with the highest reports on E-ESBL in cattle, are the United Kingdom (n = 14), Germany (n = 11), France (n = 9) and the United States (n = 9), the last one being the world's largest cattle producer, and the 3 Europeans, 4th, 3rd and 1st, respectively, in number of cattle in Europe (Eurostat, 2016; USDA et al., 2017). Within the 5 world largest cattle producers (United States, Brazil, the European Union, China and India) there have been reports of animals harbouring commensal or clinical E-ESBL. The Table 1 shows all the descriptions of E-ESBL in cattle around the World, including the source, species and ESBL gene.
Figure 1.
World map illustrating the countries with description of E-ESBL in cattle.
Table 1.
Relation of ESBL described in cattle by type of ESBL, described species, country of report, origin of the isolate and bibliographic reference of the description.
| Beta-lactamase | Enterobacteriaceae | Country | Source | Reference |
|---|---|---|---|---|
| CTX-M-1 | Escherichia coli | Europe | ||
| Germany | Faecal | (Wieler et al., 2011) | ||
| Mastitis | (Freitag et al., 2017; Michael et al., 2017) | |||
| Diarrheic | (Ewers et al., 2014) | |||
| Sick | (Michael et al., 2017) | |||
| Infection | (Brennan et al., 2016) | |||
| Denmark | Commensal | (Kjeldsen et al., 2015) | ||
| ND | (Garcia-Fernandez et al., 2011; Jakobsen et al., 2015) | |||
| Spain | Mastitis | (Briñas et al., 2005) | ||
| Finland | Faecal | (Päivärinta et al., 2016) | ||
| France | Faecal | (Haenni et al., 2014; Hartmann et al., 2012; Madec et al., 2008; Meunier et al., 2006) | ||
| Mastitis | (Dahmen et al., 2013) | |||
| Diarrheic | (Hartmann et al., 2012) | |||
| Sick | (Madec et al., 2008; Valat et al., 2016) | |||
| Infection | (Meunier et al., 2006) | |||
| Hungary | Infection | (Toth et al., 2013) | ||
| Portugal | Faecal | (Ramos et al., 2013) | ||
| Mayotte | ND | (Gay et al., 2018) | ||
| Netherlands | Faecal | (Ceccarelli et al., 2019; Heuvelink et al., 2019; Hordijk et al., 2013a, b, c) | ||
| United Kingdom | Faecal | (Velasova et al., 2019) | ||
| Infection | (Hunter et al., 2010) | |||
| ND | (Stokes, 2014) | |||
| Czech Republic | Faecal | (Dolejska et al., 2011b) | ||
| Sick | (Dolejska et al., 2013) | |||
| Réunion | ND | (Gay et al., 2018) | ||
| Slovakia | Faecal | (Kmeť and Bujňáková, 2018) | ||
| Sweden | Faecal | (Duse et al., 2015) | ||
| Switzerland | Faecal | (Endimiani et al., 2012; Geser et al., 2012a; Zurfluh et al., 2015) | ||
| Turkey | Faecal | (AslantaŞ et al., 2017; Pehlivanoglu et al., 2016) | ||
| North America | ||||
| Canada | ND | (Cormier et al., 2019) | ||
| USA | Faecal | (Mir et al., 2016; Mollenkopf et al., 2012; Wittum et al., 2010) | ||
| Asia | ||||
| China | Mastitis | (Ali et al., 2016, 2017) | ||
| South Korea | Mastitis | (Tark et al., 2017) | ||
| Indonesia | Faecal | (Sudarwanto et al., 2016) | ||
| Japan | Mastitis | (Ohnishi et al., 2013b) | ||
| Klebsiella pneumoniae | Europe | |||
| Italy | Mastitis | (Locatelli et al., 2010) | ||
| Klebsiella ozaenae | Italy | Faecal | (Stefani et al., 2014) | |
| Salmonella enterica | Germany | ND | (Rodríguez et al., 2009) | |
| CTX-M-1/61 | E. coli | Europe | ||
| Germany | Faecal | (Dahms et al., 2015) | ||
| CTX-M-2 | E. coli | South America | ||
| Brazil | Faecal | (Palmeira et al., 2018) | ||
| Europe | ||||
| Germany | Mastitis | (Eisenberger et al., 2017; Freitag et al., 2017; Michael et al., 2017) | ||
| Sick | (Michael et al., 2017) | |||
| Netherlands | Faecal | (Ceccarelli et al., 2019; Heuvelink et al., 2019) | ||
| North America | ||||
| Canada | Faecal | (Cormier et al., 2016) | ||
| Asia | ||||
| Japan | Faecal | (Shiraki et al., 2004) | ||
| Diarrheic | (Ohnishi et al., 2013a) | |||
| Infection | (Asai et al., 2011) | |||
| K. pneumoniae | Japan | Mastitis | (Ohnishi et al., 2013a, b; Saishu et al., 2014) | |
| Klebsiella oxytoca | Japan | Mastitis | (Ohnishi et al., 2013b) | |
| Citrobacter freundii | Japan | Mastitis | (Ohnishi et al., 2013a) | |
| Citrobacter koseri | Japan | Mastitis | (Ohnishi et al., 2013b) | |
| Enterobacter cloacae | Japan | Mastitis | (Ohnishi et al., 2013a) | |
| Enterobacter aerogenes | Japan | Mastitis | (Ohnishi et al., 2013b) | |
| CTX-M-2/97 | E. coli | Europe | ||
| Netherlands | Faecal | (Hordijk et al., 2013a, b, c) | ||
| CTX-M-3 | E. coli | Europe | ||
| Germany | Mastitis | (Michael et al., 2017) | ||
| France | Faecal | (Haenni et al., 2014) | ||
| Netherlands | Faecal | (Ceccarelli et al., 2019; Hordijk et al., 2013b) | ||
| United Kingdom | Infection | (Hunter et al., 2010) | ||
| Turkey | Faecal | (AslantaŞ et al., 2017; Pehlivanoglu et al., 2016) | ||
| Asia | ||||
| China | Faecal | (Zheng et al., 2019) | ||
| Mastitis | (Ali et al., 2016, 2017) | |||
| South Korea | Mastitis | (Tark et al., 2017) | ||
| Hong Kong | Faecal | (Ho et al., 2013) | ||
| CTX-M-8 | E. coli | Europe | ||
| Netherlands | Faecal | (Ceccarelli et al., 2019) | ||
| CTX-M-9 | E. coli | Europe | ||
| Netherlands | Faecal | (Ceccarelli et al., 2019) | ||
| North America | ||||
| USA | Faecal | (Poole et al., 2017) | ||
| Asia | ||||
| Indonesia | Faecal | (Sudarwanto et al., 2016) | ||
| Africa | ||||
| Egypt | Faecal | (Braun et al., 2016) | ||
| Oceania | ||||
| Australia | Sick | (Abraham et al., 2015) | ||
| S. enteric | Australia | ND | (Sparham et al., 2017) | |
| CTX-M-14 | E. coli | Europe | ||
| Germany | Mastitis | (Eisenberger et al., 2017; Freitag et al., 2017; Michael et al., 2017) | ||
| Sick | (Michael et al., 2017) | |||
| Belgium | Faecal | (Pardon et al., 2017) | ||
| Infection | (Pardon et al., 2017) | |||
| France | Faecal | (Haenni et al., 2014; Madec et al., 2008) | ||
| Mastitis | (Dahmen et al., 2013) | |||
| Sick | (Madec et al., 2008) | |||
| Wales | Sick | (Tyrrell et al., 2016) | ||
| Netherlands | Faecal | (Ceccarelli et al., 2019; Heuvelink et al., 2019; Hordijk et al., 2013a, b, c) | ||
| United Kingdom | Faecal | (Cottell et al., 2011; Horton et al., 2011; Randall et al., 2014; Snow et al., 2011) | ||
| Infection | (Hunter et al., 2010) | |||
| ND | (Stokes, 2014; Stokes et al., 2013) | |||
| Switzerland | Faecal | (Geser et al., 2012a; Zurfluh et al., 2015) | ||
| Mastitis | (Geser et al., 2012a) | |||
| Asia | ||||
| China | Faecal | (Zheng et al., 2012, 2019) | ||
| Mastitis | (Ali et al., 2016, 2017) | |||
| South Korea | Faecal | (Rayamajhi et al., 2011; Tamang et al., 2013a) | ||
| Sick | (Lim et al., 2009) | |||
| Hong Kong | Faecal | (Ho et al., 2011, 2013) | ||
| Japan | Mastitis | (Ohnishi et al., 2013b) | ||
| Diarrheic | (Ohnishi et al., 2013a) | |||
| Taiwan | Mastitis | (Su et al., 2016) | ||
| North America | ||||
| Canada | ND | (Cormier et al., 2019) | ||
| USA | Faecal | (Mollenkopf et al., 2012) | ||
| Oceania | ||||
| Australia | Sick | (Abraham et al., 2015) | ||
| K. pneumoniae | Europe | |||
| France | Mastitis | (Dahmen et al., 2013) | ||
| Asia | ||||
| Japan | Mastitis | (Ohnishi et al., 2013b) | ||
| CTX-M-15 | E. coli | South America | ||
| Brazil | Faecal | (Sartori et al., 2017) | ||
| Europe | ||||
| Germany | Faecal | (Fischer et al., 2014; Wieler et al., 2011) | ||
| Mastitis | (Eisenberger et al., 2017; Freitag et al., 2017; Michael et al., 2017) | |||
| Sick | (Michael et al., 2017) | |||
| France | Faecal | (Haenni et al., 2014) | ||
| Sick | (Madec et al., 2008) | |||
| Infection | (Madec et al., 2012; Meunier et al., 2006) | |||
| Madagascar | ND | (Gay et al., 2018) | ||
| Mayotte | ND | (Gay et al., 2018) | ||
| Netherlands | Faecal | (Ceccarelli et al., 2019; Heuvelink et al., 2019; Hordijk et al., 2013a, b, c) | ||
| United Kingdom | Faecal | (Horton et al., 2011; Randall et al., 2014; Watson et al., 2012) | ||
| Mastitis | (Timofte et al., 2014) | |||
| Infection | (Hunter et al., 2010) | |||
| Sweden | Faecal | (Duse et al., 2015) | ||
| Switzerland | Faecal | (Endimiani et al., 2012; Geser et al., 2012a; Zurfluh et al., 2015) | ||
| Turkey | Faecal | (AslantaŞ et al., 2017; Pehlivanoglu et al., 2016) | ||
| Asia | ||||
| China | Faecal | (Zheng et al., 2019) | ||
| Mastitis | (Ali et al., 2016, 2017) | |||
| Israel | ND | (Lifshitz et al., 2018) | ||
| South Korea | Faecal | (Tamang et al., 2013a) | ||
| Mastitis | (Tark et al., 2017) | |||
| Japan | Faecal | (Usui et al., 2013) | ||
| Mastitis | (Ohnishi et al., 2013b) | |||
| Diarrheic | (Ohnishi et al., 2013a) | |||
| Lebanon | Faecal | (Diab et al., 2016) | ||
| Taiwan | Mastitis | (Su et al., 2016) | ||
| North America | ||||
| Canada | ND | (Cormier et al., 2019) | ||
| Faecal | (Cormier et al., 2016) | |||
| USA | Faecal | (Mir et al., 2016; Mollenkopf et al., 2012) | ||
| Africa | ||||
| Egypt | Faecal | (Braun et al., 2016) | ||
| Tanzania | Faecal | (Seni et al., 2016) | ||
| Tunisia | Faecal | (Grami et al., 2014) | ||
| Mastitis | (Saidani et al., 2018) | |||
| K. pneumoniae | Europe | |||
| France | Faecal | (Haenni et al., 2014) | ||
| Asia | ||||
| India | Mastitis | (Koovapra et al., 2016) | ||
| K. ozaenae | Europe | |||
| Italy | Faecal | (Stefani et al., 2014) | ||
| K. oxytoca | Asia | |||
| Egypt | Mastitis | (Ahmed and Shimamoto, 2011) | ||
| CTX-M-15/28 | E. coli | Europe | ||
| United Kingdom | Faecal | (Snow et al., 2011) | ||
| CTX-M-17 | E. coli | Asia | ||
| China | Faecal | (Zheng et al., 2019) | ||
| CTX-M-17/18 | E. coli | Europe | ||
| United Kingdom | Faecal | (Liebana et al., 2006) | ||
| CTX-M-20 | E. coli | Europe | ||
| United Kingdom | Infection | (Hunter et al., 2010) | ||
| CTX-M-22 | E. coli | Europe | ||
| Netherlands | Faecal | (Heuvelink et al., 2019) | ||
| CTX-M-24 | E. coli | North America | ||
| Canada | Faecal | (Cormier et al., 2016) | ||
| CTX-M-27 | E. coli | Europe | ||
| Netherlands | Faecal | (Ceccarelli et al., 2019) | ||
| North America | ||||
| USA | Faecal | (Tadesse et al., 2018) | ||
| ND | (Afema et al., 2018) | |||
| Canada | Faecal | (Cormier et al., 2016) | ||
| ND | (Cormier et al., 2019) | |||
| CTX-M-28 | E. coli | Asia | ||
| Hong Kong | Faecal | (Ho et al., 2011) | ||
| CTX-M-32 | E. coli | Europe | ||
| France | Faecal | (Haenni et al., 2014) | ||
| Germany | Mastitis | (Eisenberger et al., 2017) | ||
| Mayotte | ND | (Gay et al., 2018) | ||
| Netherlands | Faecal | (Ceccarelli et al., 2019; Heuvelink et al., 2019; Hordijk et al., 2013a, b, c) | ||
| Portugal | Faecal | (Ramos et al., 2013) | ||
| United Kingdom | Infection | (Hunter et al., 2010) | ||
| North America | ||||
| Canada | Faecal | (Cormier et al., 2016) | ||
| ND | (Cormier et al., 2019) | |||
| USA | Faecal | (Cottell et al., 2013; Poole et al., 2017) | ||
| Asia | ||||
| South Korea | Faecal | (Tamang et al., 2013a) | ||
| CTX-M-55 | E. coli | Europe | ||
| France | Sick | (Haenni et al., 2018; Lupo et al., 2018) | ||
| Netherlands | Faecal | (Ceccarelli et al., 2019; Heuvelink et al., 2019) | ||
| Spain | Faecal | (Hernández et al., 2017) | ||
| North America | ||||
| Canada | Faecal | (Cormier et al., 2016) | ||
| ND | (Cormier et al., 2019) | |||
| Asia | ||||
| China | Faecal | (Zheng et al., 2012, 2019) | ||
| Mastitis | (Ali et al., 2016, 2017) | |||
| Hong Kong | Faecal | (Ho et al., 2011, 2013) | ||
| Taiwan | Mastitis | (Su et al., 2016) | ||
| CTX-M-57 | E. coli | Europe | ||
| France | Faecal | (Haenni et al., 2014) | ||
| CTX-M-61 | E. coli | North America | ||
| Canada | Faecal | (Cormier et al., 2016) | ||
| CTX-M-63 | K. pneumoniae | Asia | ||
| India | Mastitis | (Koovapra et al., 2016) | ||
| CTX-M-65 | E. coli | Europe | ||
| Netherlands | Faecal | (Ceccarelli et al., 2019; Heuvelink et al., 2019) | ||
| North America | ||||
| Canada | Faecal | (Cormier et al., 2016) | ||
| ND | (Cormier et al., 2019) | |||
| S. enterica | USA | ND | (Tate et al., 2017) | |
| CTX-M-79 | E. coli | Europe | ||
| Netherlands | Faecal | (Hordijk et al., 2013c) | ||
| North America | ||||
| USA | Faecal | (Wittum et al., 2010) | ||
| CTX-M-98 | E. coli | Asia | ||
| Hong Kong | Faecal | (Ho et al., 2011) | ||
| CTX-M-115 | E. coli | North America | ||
| Canada | Faecal | (Cormier et al., 2016) | ||
| CTX-M-117 | E. coli | Europe | ||
| Switzerland | Faecal | (Hächler et al., 2013; Zurfluh et al., 2015) | ||
| CTX-M-123 | E. coli | Asia | ||
| Hong Kong | Faecal | (Ho et al., 2015) | ||
| CTX-M-132 | E. coli | Asia | ||
| Hong Kong | Faecal | (Ho et al., 2015) | ||
| CTX-M-172 | E. coli | North America | ||
| Canada | Faecal | (Cormier et al., 2016) | ||
| CTX-M – without variant description | E. coli | Europe | ||
| Germany | Faecal | (Wu et al., 2013) | ||
| France | Diarrheic | (Valat et al., 2012) | ||
| Netherlands | Faecal | (Wu et al., 2013) | ||
| United Kingdom | Faecal | (Wu et al., 2013) | ||
| Switzerland | Faecal | (Geser et al., 2011) | ||
| North America | ||||
| Canada | Faecal | (Awosile et al., 2018) | ||
| USA | Faecal | (Davis et al., 2015) | ||
| Asia | ||||
| China | Mastitis | (Yang et al., 2018) | ||
| Saudi Arabia | ND | (Hassan et al., 2015) | ||
| South Korea | Faecal | (Tamang et al., 2013b) | ||
| Hong Kong | Faecal | (Duan et al., 2006) | ||
| India | Faecal | (Borah et al., 2014) | ||
| Mastitis | (Bandyopadhyay et al., 2015; Ghatak et al., 2013; Kar et al., 2015) | |||
| Israel | Faecal | (Adler et al., 2015) | ||
| Malaysia | Faecal | (Kamaruzzaman, 2015) | ||
| Africa | ||||
| South Africa | Faecal | (Iweriebor et al., 2015) | ||
| Nigeria | Faecal | (Olowe et al., 2015) | ||
| K. pneumoniae | South America | |||
| Brazil | Mastitis | (Nóbrega et al., 2013) | ||
| Asia | ||||
| India | Mastitis | (Das et al., 2017) | ||
| Salmonella sp | North America | |||
| USA | Sick | (Frye and Fedorka-Cray, 2007) | ||
| SHV-2 | E. coli | North America | ||
| Canada | Faecal | (Cormier et al., 2016) | ||
| SHV-5 | E. coli | Europe | ||
| Turkey | Faecal | (Kucukbasmaci et al., 2008) | ||
| C. freundii | Turkey | Faecal | (Kucukbasmaci et al., 2008) | |
| C. brakii | Turkey | Faecal | (Kucukbasmaci et al., 2008) | |
| SHV-11 | E. coli | Asia | ||
| Japan | Diarrheic | (Ohnishi et al., 2013a) | ||
| K. pneumoniae | Japan | Diarrheic | (Ohnishi et al., 2013a) | |
| SHV-12 | E. coli | Europe | ||
| Germany | Sick | (Michael et al., 2017) | ||
| France | Faecal | (Haenni et al., 2014; Madec et al., 2008) | ||
| Netherlands | Faecal | (Ceccarelli et al., 2019; Hordijk et al., 2013c) | ||
| Turkey | Faecal | (Kucukbasmaci et al., 2008; Pehlivanoglu et al., 2016) | ||
| Asia | ||||
| China | Mastitis | (Ali et al., 2016, 2017) | ||
| K. pneumoniae | Europe | |||
| France | Faecal | (Haenni et al., 2014) | ||
| United Kingdom | Mastitis | (Timofte et al., 2014) | ||
| Africa | ||||
| Egypt | Mastitis | (Ahmed and Shimamoto, 2011) | ||
| K. oxytoca | Egypt | Mastitis | (Ahmed and Shimamoto, 2011) | |
| E. cloacae | Egypt | Mastitis | (Ahmed and Shimamoto, 2011) | |
| SHV-28 | Serratia marcescens | Africa | ||
| Egypt | Mastitis | (Ahmed and Shimamoto, 2011) | ||
| SHV-180 | K. pneumoniae | Asia | ||
| India | Mastitis | (Koovapra et al., 2016) | ||
| SHV – without variant description | E. coli | Europe | ||
| Netherlands | Faecal | (Wu et al., 2013) | ||
| Asia | ||||
| India | Faecal | (Borah et al., 2014) | ||
| Mastitis | (Kar et al., 2015) | |||
| Israel | Faecal | (Adler et al., 2015) | ||
| K. pneumoniae | South America | |||
| Brazil | Mastitis | (Nóbrega et al., 2013) | ||
| Salmonella spp. | North America | |||
| USA | Sick | (Frye and Fedorka-Cray, 2007) | ||
| TEM-20 | E. coli | Europe | ||
| Netherlands | Faecal | (Hordijk et al., 2013c) | ||
| TEM-24 | K. ozaenae | Europe | ||
| Italy | Faecal | (Stefani et al., 2014) | ||
| TEM-52 | E. coli | Europe | ||
| Germany | Faecal | (Wieler et al., 2011) | ||
| Sick | (Michael et al., 2017) | |||
| France | Diarrheic | (Haenni et al., 2012) | ||
| Netherlands | Faecal | (Ceccarelli et al., 2019; Heuvelink et al., 2019; Hordijk et al., 2013a, b, c) | ||
| TEM-71 | E. coli | Europe | ||
| France | Faecal | (Hartmann et al., 2012) | ||
| Diarrheic | (Hartmann et al., 2012) | |||
| TEM-126 | E. coli | Europe | ||
| France | Sick | (Madec et al., 2008) | ||
| TEM-186 | E. coli | Europe | ||
| Switzerland | Faecal | (Geser et al., 2012a) | ||
| TEM-190 | E. coli | Europe | ||
| Netherlands | Faecal | (Ceccarelli et al., 2019) | ||
| TEM – without variant description | E. coli | North America | ||
| USA | Faecal | (Donaldson et al., 2006; Mir et al., 2016) | ||
| Asia | ||||
| India | Faecal | (Borah et al., 2014) | ||
| Thailand | Mastitis | (Hinthong et al., 2017) | ||
| Africa | ||||
| Egypt | Faecal | (Braun et al., 2016) | ||
| OXA-10 | E. coli | Europe | ||
| Turkey | Faecal | (Kucukbasmaci et al., 2008) | ||
| C. freundii | Turkey | Faecal | (Kucukbasmaci et al., 2008) | |
| C. brakii | Turkey | Faecal | (Kucukbasmaci et al., 2008) | |
| OXA-30 | K. oxytoca | Africa | ||
| Egypt | Mastitis | (Ahmed and Shimamoto, 2011) | ||
| E. cloacae | Egypt | Mastitis | (Ahmed and Shimamoto, 2011) | |
| ESBL producers - without beta-lactamase description (ND-ESBL | E. coli | South America | ||
| Brazil | Mastitis | (Santos, 2006) | ||
| Chile | Mastitis | (Gonzalez, 2006) | ||
| Peru | Faecal | (Mendoza, 2017) | ||
| Europe | ||||
| Germany | Faecal | (Friese et al., 2013) | ||
| Spain | Faecal | (Briñas et al., 2005) | ||
| País de Gales | ND | (Teale et al., 2005) | ||
| Netherlands | Faecal | (Gonggrijp et al., 2016) | ||
| Switzerland | Faecal | (Reist et al., 2013) | ||
| North America | ||||
| Canada | Faecal | (Lussier, 2010) | ||
| Asia | ||||
| Iran | Faecal | (Barzan et al., 2017) | ||
| Thailand | Mastitis | (Hinthong et al., 2017) | ||
| Africa | ||||
| Nigeria | Faecal | (Ogefere et al., 2017) | ||
| Tanzania | Faecal | (Mkala and Azizi, 2017) | ||
| K. pneumoniae | Asia | |||
| Israel | Faecal | (Adler et al., 2015) | ||
| Klebsiella spp. | Africa | |||
| Nigeria | Faecal | (Ogefere et al., 2017) | ||
| Salmonella spp. | Nigeria | Faecal | (Ogefere et al., 2017) | |
| C. youngae | Europe | |||
| Switzerland | Faecal | (Reist et al., 2013) | ||
| E. cloacae | South America | |||
| Peru | Faecal | (Mendoza, 2017) | ||
| Europe | ||||
| Switzerland | Faecal | (Reist et al., 2013) | ||
| Proteus mirabilis | Africa | |||
| Nigeria | Faecal | (Ogefere et al., 2017) | ||
| Proteus vulgaris | Nigeria | Faecal | (Ogefere et al., 2017) | |
| Providencia spp. | Nigeria | Faecal | (Ogefere et al., 2017) | |
| Shigella spp. | Nigeria | Faecal | (Ogefere et al., 2017) | |
Source: ND – Not described isolate source.
The most frequent ESBL types in E-ESBL in cattle, as expected, were the ones of CTX-M-1 group with higher prevalence for CTX-M-1, CTX-M-14 and CTX-M-15. CTX-M-1 was reported in 20 countries, most frequently in Europe (n = 14), being found in Germany, Denmark, Spain, Finland, France, Hungary, Portugal, Netherlands, United Kingdom, Czech Republic, Slovakia, Sweden, Switzerland and Turkey. CTX-M-1 was first described in human E-ESBL in 1989 in Germany, and it has also been reported in other European countries such as Spain, France, Italy and United Kingdom as well as in Asia and North America (Cantσn and Coque, 2006; Moosavian and Ahmadkhosravy, 2016; Wang et al., 2013).
CTX-M-15 and CTX-M-14 are the most important CTX-M enzymes due to their large diffusion and relation to outbreaks and severe extraintestinal infections (Cantón et al., 2012; Matsumura et al., 2015; Price et al., 2013). CTX-M-14 was described in E-ESBL in cattle in 13 countries, mainly in Europe (Germany, Belgium, France, Netherlands, United Kingdom, and Switzerland) and in Asia (China, South Korea, Hong Kong, Japan, and Taiwan), as well as the United States and Oceania. CTX-M-14 was first described in 2002 in E-ESBL from a hospital in China (Chanawong et al., 2002; Ma et al., 2002). E-ESBL producers isolated from human of CTX-M-14 type are described in Europe, Asia, North and South America, Africa and Oceania, many times related to pandemic clones such as E. coli ST131 responsible for outbreaks in the last years (Cantón et al., 2008; Chen et al., 2014; Giedraitienė et al., 2017; Peirano et al., 2010, 2011; Pitout et al., 2005; Shin et al., 2011; Silva and Lincopan, 2012; Zong et al., 2008).
CTX-M-15 was first described in 2001 in E-ESBL isolate in a hospital in New Delhi, India, and today is the most widespread ESBL in the various niches and the most important of all, due to its high relation to important, for human health, E-ESBL clones (Cantón et al., 2012; Clermont et al., 2008; Karim et al., 2001; Kim et al., 2017; Price et al., 2013; Woodford et al., 2004). E-ESBL producing CTX-M-15 in cattle were described in 21 countries around the world, present in most of Europe, being reported in Germany, France, Italy, Netherlands, United Kingdom, Sweden, Switzerland and Turkey. In Asia they were described in China, South Korea, India, Israel, Japan, Lebanon and Taiwan and also reported in North and South America (Brazil, Canada and United States) and Africa (Egypt, Tanzania and Tunisia).
CTX-M-15 has been reported in all continents (Europe, North America, South America, Asia, Africa, Oceania and Antarctica with reports in all major ecological niches (humans, animals, and environment), these E-ESBL producers of CTX-M-15 are an excellent example of the public health threat that involves circulation of resistant Enterobacteriaceae and resistance genes among the different ecological niches that is currently evidenced under the prism of the “One Health” approach (Chen et al., 2014; Dia et al., 2016; Fam et al., 2011; Hasan et al., 2016; Hernández et al., 2012; Liao et al., 2017; Poirel et al., 2013; Ruiz et al., 2011; Sidjabat et al., 2010).
The virulent and multi-resistant CTX-M-15-producing E. coli O25b-ST131 clone is certainly one of the most well adapted circulant clones among E-ESBL, which is responsible for outbreaks and deaths around the world and is not related only to infectious processes, but is also reported in human intestinal colonization (elderly, adults and children) and animals (terrestrial and aquatic) and environmental contamination (Badran et al., 2016; Brahmi et al., 2015; Dolejska et al., 2011a; Ewers et al., 2010; Gonçalves et al., 2016; Namaei et al., 2017; Naseer et al., 2007; Olesen et al., 2013; Oteo et al., 2009; Owens et al., 2011; Zhong et al., 2015).
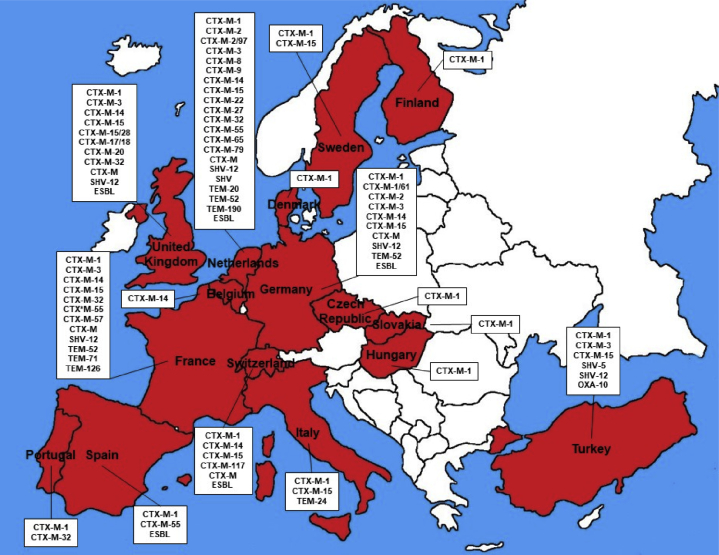
3. Europe
Europe is the continent with more number of countries (n = 16) with description of E-ESBL in cattle, Figure 2 shows all countries. Belgium, Czech Republic, Denmark, Finland, France, Germany, Hungary, Italy, Netherlands, Portugal, Slovakia, Spain, Sweden, Switzerland, Turkey and United Kingdom, presented at least one report of E-ESBL in cattle (AslantaŞ et al., 2017; Briñas et al., 2005; Dolejska et al., 2011b; Duse et al., 2015; Hordijk et al., 2013c; Hunter et al., 2010; Kjeldsen et al., 2015; Kmeť and Bujňáková, 2018; Michael et al., 2017; Päivärinta et al., 2016; Pardon et al., 2017; Ramos et al., 2013; Stefani et al., 2014; Toth et al., 2013; Valat et al., 2016; Zurfluh et al., 2015). CTX-M-1 was described in 14 of the 16 countries with E-ESBL in cattle, CTX-M-15 present in 7, CTX-M-14 in 6 and CTX-M-3 and SHV-12 in 5 (AslantaŞ et al., 2017; Briñas et al., 2005; Dolejska et al., 2011b; Duse et al., 2015; Gay et al., 2018; Haenni et al., 2014; Hordijk et al., 2013b; Hordijk et al., 2013c; Hunter et al., 2010; Kjeldsen et al., 2015; Kmeť and Bujňáková, 2018; Michael et al., 2017; Päivärinta et al., 2016; Pardon et al., 2017; Pehlivanoglu et al., 2016; Ramos et al., 2013; Timofte et al., 2014; Toth et al., 2013; Valat et al., 2016; Zurfluh et al., 2015).
Figure 2.
Illustrative map of Europe showing countries with description of E-ESBL in cattle and beta-lactamases type.
Germany presented the description of CTX-M-1, CTX-M-1/61, CTX-M-2, CTX-M-3, CTX-M-14, CTX-M-15, CTX-M without the variant description (CTX-M), SHV-12, TEM-52 and E-ESBL without the description or detection of beta-lactamase (ND-ESBL) described in E. coli and Salmonella enterica from faecal and clinical samples (Dahms et al., 2015; Friese et al., 2013; Michael et al., 2017; Wieler et al., 2011; Wu et al., 2013). CTX-M-1, CTX-M-3, CTX-M-14, CTX-M-15 and SHV-12 have already been reported in E-ESBL in humans in Germany associated with infections (Gerhold et al., 2016; Mshana et al., 2009; Schmitt et al., 2007).
The Netherlands, German neighbours, also presented high diversity and number of reports of E-ESBL in cattle. There were identified CTX-M-1, CTX-M-2, CTX-M-2/97, CTX-M-3, CTX-M-8, CTX-M-9, CTX-M-14, CTX-M-15, CTX-M-22, CTX-M-27, CTX-M-32, CTX-M-55, CTX-M-65, CTX-M-79, CTX-M, SHV-12, SHV without description of variant (SHV), TEM-20, TEM-52, TEM-190 and ND-ESBL, all of them described in faecal E. coli. CTX-M-1, CTX-M-2, CTX-M-14, CTX-M-15, SHV-12 and TEM-52 have already been described in E-ESBL of human (faecal and hospital) origin in the Netherlands (Naiemi et al., 2006; Overdevest et al., 2011).
The biggest European cattle producer, France, including 2 French departments in Africa (Mayotte and Réunion) has reported E-ESBL in cattle with CTX-M-1, CTX-M-3, CTX-M-14, CTX-M-15, CTX-M-32, CTX-M-55, CTX-M-57, CTX-M, SHV-12, TEM-52, TEM-71 and TEM-126 reported in E. coli and K. pneumoniae of faecal and clinical origin. All of the described beta-lactamases (CTX-M-1, CTX-M-3, CTX-M-14, CTX-M-15, CTX-M-32, CTX-M-57, SHV-12, TEM-52, TEM-71 and TEM-126) in cattle have been reported in E-ESBL from humans of hospital origin in France (De Champs et al., 2004; Robin et al., 2017).
The United Kingdom was the European country with the highest number of reported CTX-M variants. The CTX-M-1, CTX-M-3, CTX-M-14, CTX-M-15, CTX-M-15/28, CTX-M-17/18, CTX -M-20, CTX-M-32, CTX-M, SHV-12 and ND-ESBL were reported in E-ESBL in cattle. E. coli and K. pneumoniae were the species that harbour the genes and these are of faecal and clinical origin. CTX-M-3, CTX-M-14, CTX-M-15, CTX-M-17/18 and SHV-12 have already been described in humans in E-ESBLs (Batchelor et al., 2005; Doumith et al., 2012).
Switzerland presented E-ESBL in cattle with CTX-M-1, CTX-M-14, CTX-M-15, CTX-M-117, CTX-M, TEM-186 and ND-ESBL (Geser et al., 2011, 2012a; Reist et al., 2013; Zurfluh et al., 2015). They were reported in 3 species: E. coli, C. youngae and E. cloacae. All reports were of faecal origin, with the exception of 1 case of mastitis (CTX-M-14) (Geser et al., 2012a). E-ESBL of human origin have been reported to harbour CTX-M-1, CTX-M-14 and CTX-M-15 in Switzerland (Geser et al., 2012b).
CTX-M-1, CTX-M-15 and TEM-24 were described in E-ESBL in cattle in Italy. Reported in K. pneumoniae (mastitis) and K. ozaenae (faecal carriage). The three types of ESBL described in cattle, CTX-M-1, CTX-M-15 and TEM-24, have also been described in human clinical isolates (Mugnaioli et al., 2006; Perilli et al., 2011).
Turkey is the second largest cattle producer in Europe and presented E-ESBL in cattle with CTX-M-1, CTX-M-3, CTX-M-15, SHV-5, SHV-12 and OXA-10 in E. coli, C. freundii and C. brakii of faecal origin (AslantaŞ et al., 2017; Kucukbasmaci et al., 2008; Pehlivanoglu et al., 2016). CTX-M-1, CTX-M-3, CTX-M-15, SHV-5 and SHV-12 were reported in human clinical isolates in Turkish hospitals (Gur et al., 2008; Tasli and Bahar, 2005).
The Nordic countries have few reports and diversity of beta-lactamases, with E-ESBL in cattle harbouring CTX-M-1 in Denmark and Finland and CTX-M-1 and CTX-M-15 in Sweden all in E. coli and of intestinal origin (Duse et al., 2015; Kjeldsen et al., 2015; Päivärinta et al., 2016). When analysed the description in E-ESBL of human origin, CTX-M-1 has been described in Denmark, Finland and Sweden and CTX-M-15 in Sweden (Brolund et al., 2014; Forssten et al., 2010; Jakobsen et al., 2015).
In the Iberian Peninsula, Spain and Portugal also presented E-ESBL in cattle, with descriptions on Spain of CTX-M-1, CTX-M-55 and ND-ESBL and in Portugal of CTX-M-1 and CTX-M-32 (Briñas et al., 2005; Hernández et al., 2017; Ramos et al., 2013). The descriptions in both countries were in E. coli, but in Spain they were of faecal and clinical origin and in Portugal only faecal. CTX-M-1 has already been described in E- ESBL in human in Spain and CTX-M-1 and CTX-M-32 in Portugal (Fernandes et al., 2014; Novais et al., 2007).
CTX-M-1 was described in Hungary, Czech Republic and Slovakia and CTX-M-14 in Belgium in E-ESBL in cattle. All in E. coli and with faecal and clinical origins (Dolejska et al., 2013; Kmeť and Bujňáková, 2018; Pardon et al., 2017; Toth et al., 2013). In E-ESBL of human origin, CTX-M-1 has already been reported in Hungary, Czech Republic and CTX-M-14 in Belgium (Dolejska et al., 2013; Ebrahimi, 2016; Rodriguez-Villalobos et al., 2011).
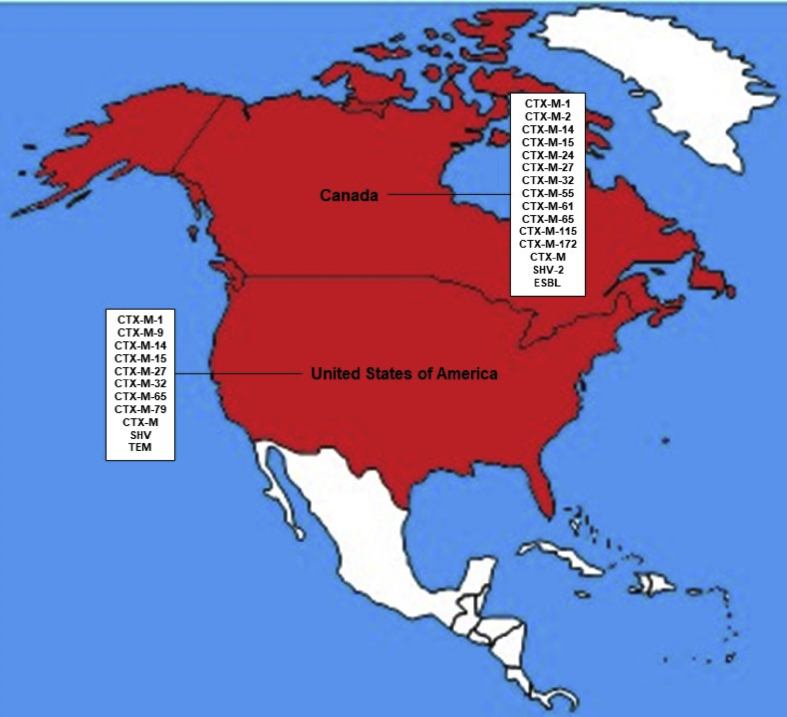
4. North America
In North America only 2 countries registered E-ESBL description in cattle, Canada and the United States, the last one being the world's largest cattle producer (USDA et al., 2017). CTX-M-15, CTX-M-32 and CTX-M-65 were the only beta-lactamases described in both countries, the Figure 3 shows the complete described ESBL from cattle in the North America (Cormier et al., 2016; Mir et al., 2016; Poole et al., 2017; Tate et al., 2017).
Figure 3.
Illustrative map of North America with the countries with description of E-ESBL in cattle and the diversity of beta-lactamases presented.
In the United States there were described in E-ESBL in cattle the CTX-M-1, CTX-M-9, CTX-M-14, CTX-M-15, CTX-M-27, CTX-M-32, CTX-M-65, CTX-M-79, CTX-M, SHV and TEM. E-ESBL of clinical and faecal origin in E. coli, Salmonella enteric and Salmonella spp (Afema et al., 2018; Davis et al., 2015; Frye and Fedorka-Cray, 2007; Mir et al., 2016; Mollenkopf et al., 2012; Poole et al., 2017; Tate et al., 2017; Wittum et al., 2010). CTX-M- 1, CTX-M-14, CTX-M-15 and CTX-M-65 were reported in E-ESBL isolates from humans (Chen et al., 2014; Li et al., 2015; Tate et al., 2017; Wang et al., 2013).
CTX-M-1, CTX-M-2, CTX-M-14, CTX-M-15, CTX-M-24, CTX-M-27, CTX-M-32, CTX-M-55, CTX-M-61, CTX-M-65, CTX-M-115, CTX-M-172, CTX-M, SHV-2 and ND-ESBL were described in E-ESBL in cattle in Canada, which was the country where the largest variety of CTX-M's types were described (Awosile et al., 2018; Cormier et al., 2016; Lussier, 2010). All E-ESBLs were E. coli of faecal origin. In E-ESBL of human origin in Canada were detected similarly to cattle the CTX-M-2, CTX-M-15, CTX-M-24, CTX-M-27, CTX-M-55, CTX-M-65 and SHV-2 (Denisuik et al., 2013, 2015; Peirano et al., 2010; Pitout et al., 2008).
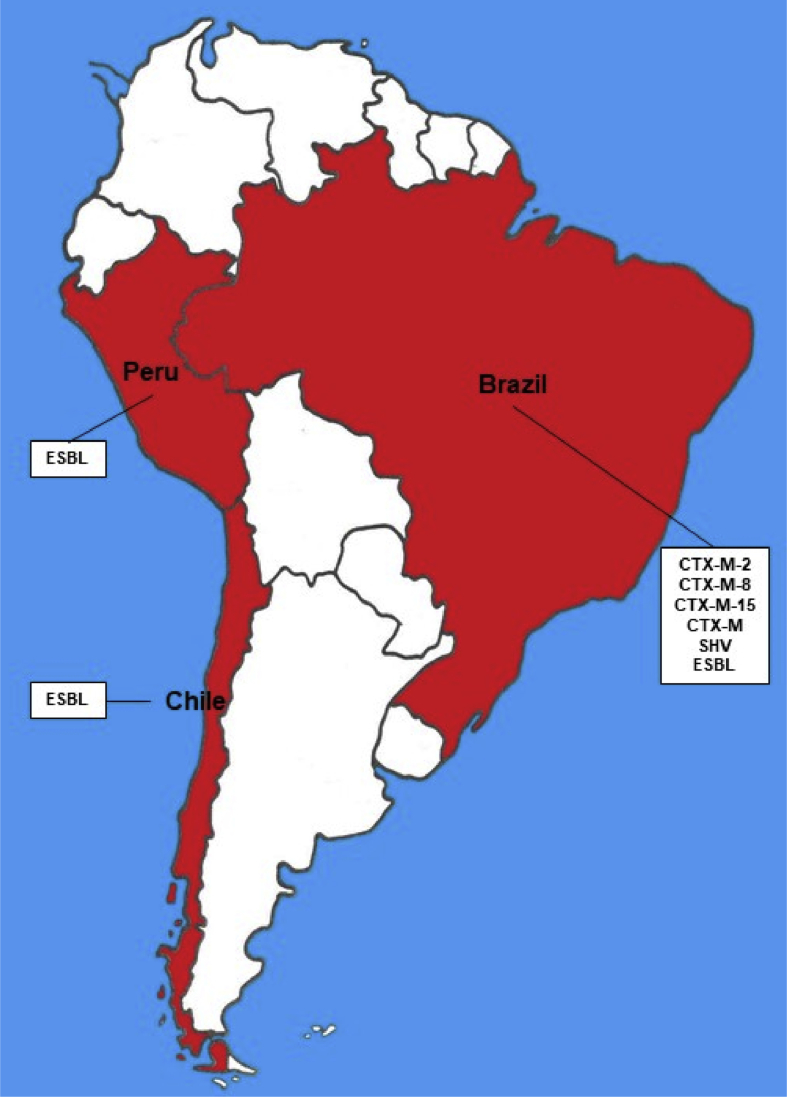
5. South America
In South America E-ESBL is still poorly described, being few and incomplete, from the molecular point of view, the reports of E-ESBL in cattle in the countries of this continent. Only Brazil, Chile and Peru present reports of E-ESBL in cattle. Figure 4 shows what E-ESBL has already been described in these countries.
Figure 4.
Map of South America illustrating the countries with description of E-ESBL in cattle and the diversity of beta-lactamases presented.
Brazil is the world's second largest cattle producer and the world's second largest exporter of cattle (USDA et al., 2017). The presence of E-ESBL in cattle with CTX-M-2, CTX-M-15, CTX-M, SHV and ND-ESBL has already been described in the country (Nóbrega et al., 2013; Palmeira et al., 2018; Santos, 2006; Sartori et al., 2017). Originated from mastitis or with faecal origin in E. coli and K. pneumoniae. There are several reports in human clinical E-ESBL of CTX-M-2, CTX-M-15, CTX-M and SHV in Brazil (Sampaio and Gales, 2016).
Chile and Peru describe the presence of E-ESBL, but not described the enzymes responsible for ESBL phenotype. These reports were in E. coli in mastitis in Chile and in E. coli and E. cloacae of faecal origin in Peru (Gonzalez, 2006; Mendoza, 2017). Both countries have reports of E-ESBL in humans (Colquechagua Aliaga et al., 2015; Hernandez et al., 2013).
6. Asia
The second continent with the highest number of countries reporting E-ESBL in cattle is Asia, which shows description of E-ESBL in 13 countries (China, Hong Kong, India, Indonesia, Iran, Israel, Japan, Lebanon, Malaysia, Saudi Arabia, South Korea, Thailand and Taiwan), highlighting China and India which are the fourth and fifth largest cattle producers in the world (Adler et al., 2015; Ali et al., 2017; Barzan et al., 2017; Diab et al., 2016; Hassan et al., 2015; Hinthong et al., 2017; Ho et al., 2015; Kamaruzzaman, 2015; Koovapra et al., 2016; Ohnishi et al., 2013a; Su et al., 2016; Sudarwanto et al., 2016; Tark et al., 2017). They have different roles in the import and export scenario, as India is the world's largest exporter of cattle and China the largest importer, since its internal production is not sufficient for internal consumption (USDA et al., 2017). Figure 5 shows the countries and the diversity of beta-lactamases found in each of them.
Figure 5.
Asia illustrative map of countries with description of E-ESBL in cattle and the diversity of beta-lactamases presented.
China presents a description of E-ESBL in cattle harbouring CTX-M-1, CTX-M-3, CTX-M-14, CTX-M-15, CTX-M-17, CTX-M-28, CTX-M-55, CTX-M-65, CTX-M-88, CTX-M-98, CTX-M-102, CTX-M-103, CTX-M-123, CTX-M and SHV-12. All descriptions were in E. coli and faecal origin and mastitis (Ali et al., 2017; Yang et al., 2018; Zheng et al., 2019). CTX-M-3, CTX-M-14, CTX-M-15, CTX-M-55 and SHV-12 were described isolated from E-ESBL of human origin in China (Hu et al., 2013; Tian et al., 2012).
In India CTX-M-15, CTX-M-63, CTX-M, SHV-180, SHV and TEM in E-ESBL from cattle have already been described. Faecal origin and mastitis, being detected in E. coli and K. pneumoniae (Borah et al., 2014; Das et al., 2017; Koovapra et al., 2016). The presence of CTX-M-15 and SHV in E-ESBL in India has been reported in humans (Hawkey, 2008).
Japan and South Korea, the 3rd and 4th largest world importers of cattle, presented E-ESBL in cattle, respectively, with CTX-M-1, CTX-M-2, CTX-M-14, CTX-M-15 and SHV-11 and CTX-M-1, CTX-M-3, CTX-M-14, CTX-M-15, CTX-M-32, and CTX-M. These were detected in faecal E-ESBL and clinical in the animals and in Japan in E. coli, K. pneumonia, K. oxytoca, C. freundii, C. koseri, E. cloacae and E. aerogenes and in South Korea only in E. coli (Ohnishi et al., 2013a, b; Shiraki et al., 2004; Tamang et al., 2013a, b; Tark et al., 2017). In Japan CTX-M-14, CTX-M-15 and SHV-11 have been described in E-ESBL of human origin (Kuroda et al., 2012; Saito et al., 2014). The CTX-M-3, CTX-M-14, CTX-M-15 and CTX-M-32 beta-lactamases have already been described in E-ESBL of human origin in South Korea (Lee et al., 2009).
Hong Kong and Taiwan presented an E-ESBL profile in cattle only with reports of beta-lactamases of the CTX-M type. Hong Kong with CTX-M-3, CTX-M-14, CTX-M-28, CTX-M-55, CTX-M-98, CTX-M-123, CTX-M-132 and CTX-M (Duan et al., 2006; Ho et al., 2011, 2013, 2015). Taiwan already has CTX-M-14, CTX-M-15 and CTX-M-55 (Su et al., 2016). All in E. coli of faecal origin and mastitis. In Hong Kong the E-ESBL description of human origin has been reported well for CTX-M-14 and in Taiwan for CTX-M-14 and CTX-M-15 (Yan et al., 2006; Yeung, 2011).
Indonesia, Malaysia, and Thailand reported E-ESBL in cattle, respectively, for CTX-M-1 and CTX-M-9; CTX-M; and TEM and ND-ESBL. All in E. coli of origin in mastitis or faecal (Hinthong et al., 2017; Kamaruzzaman, 2015; Sudarwanto et al., 2016). In humans, reports of E-ESBL have been described for CTX-M-1 in Indonesia, CTX-M in Malaysia and ND-ESBL in Thailand (Bagus Wasito et al., 2017; Ho et al., 2012; Kiratisin et al., 2008).
In the Middle East region there is a description of E-ESBL in cattle in Saudi Arabia (CTX-M), Iran (ND-ESBL), Israel (CTX-M-15, CTX-M, SHV and ND-ESBL) and in Lebanon (CTX-M-15). They were identified in E. coli and K. pneumoniae in faecal samples (Adler et al., 2015; Barzan et al., 2017; Diab et al., 2016; Hassan et al., 2015; Lifshitz et al., 2018). All E-ESBL profiles in cattle described above in these countries are also found described in humans (Bazzaz et al., 2009; Chmelnitsky et al., 2005; Hassan and Abdalhamid, 2014; Moubareck et al., 2005).
7. Africa
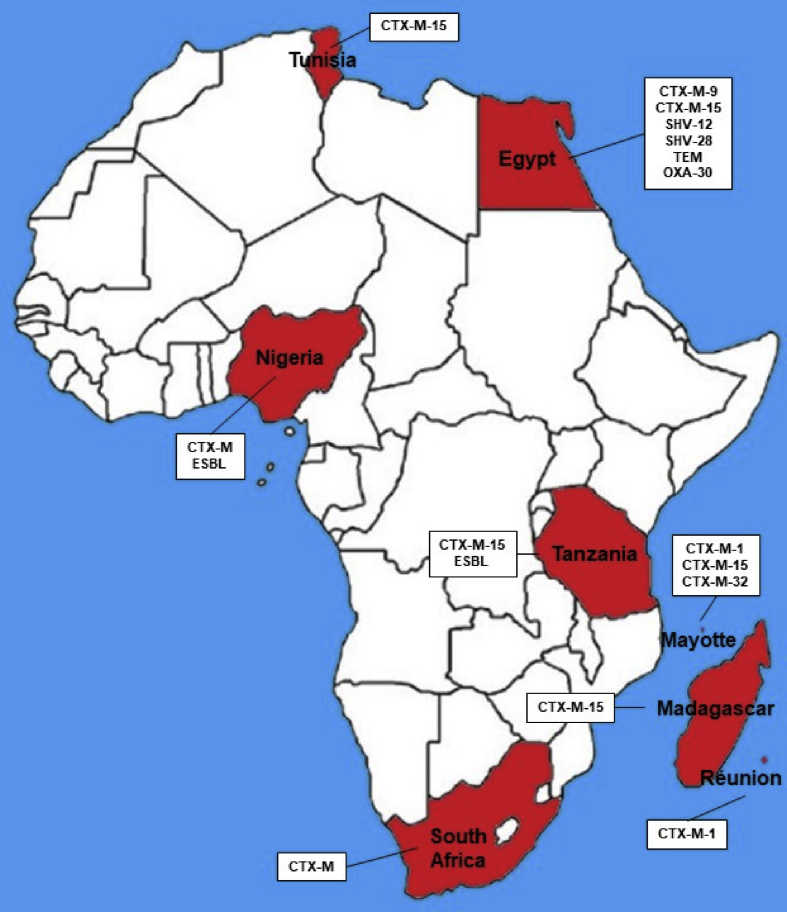
In Africa, only 6 countries presented reports of E-ESBL in cattle. They were Egypt, Madagascar, Nigeria, South Africa, Tanzania and Tunisia (Braun et al., 2016; Gay et al., 2018; Iweriebor et al., 2015; Mkala and Azizi, 2017; Olowe et al., 2015; Saidani et al., 2018; Seni et al., 2016). There are not numerous nor descriptive reports on this continent. They are also not very prominent countries within the world economic cattle cycle, highlighting only Egypt which is the 8th biggest importer of cattle in the world (USDA et al., 2017). Figure 6 shows the countries with E-ESBL reported in cattle and what beta-lactamase type has been described in them.
Figure 6.
Map of Africa illustrating the countries with description of E-ESBL in cattle and the diversity of beta-lactamases presented.
In Egypt E-ESBL was described in cattle harbouring CTX-M-9, CTX-M-15, SHV-12, SHV-28, TEM and OXA-30. Described in E. coli, K. pneumoniae, K. oxytoca, E. cloacae and S. marcescens of faecal origin and mastitis (Ahmed and Shimamoto, 2011; Braun et al., 2016). There are reports in human E-ESBL also with CTX-M-9, CTX-M-15 and SHV-12 (Fam et al., 2011; Hamdy Mohammed et al., 2016; Newire et al., 2013).
South Africa (CTX-M), Madagascar (CTX-M-15), Nigeria (CTX-M and ND-ESBL), Tanzania (CTX-M-15) and Tunisia (CTX-M-15) also presented reports of E-ESBL in cattle. The reports were E. coli, Klebsiella sp, Salmonella sp, P. mirabilis, P. vulgaris, Providencia sp e Shigella spp (Gay et al., 2018; Iweriebor et al., 2015; Ogefere et al., 2017; Olowe et al., 2015; Saidani et al., 2018; Seni et al., 2016). In all countries the origin was faecal (except Tunisia, with mastitis also). All types of descriptions presented in these 5 countries for E-ESBL in cattle also present reports in humans (Abbassi et al., 2008; Iroha et al., 2012; Manyahi et al., 2017; Ouedraogo et al., 2016).
8. Oceania
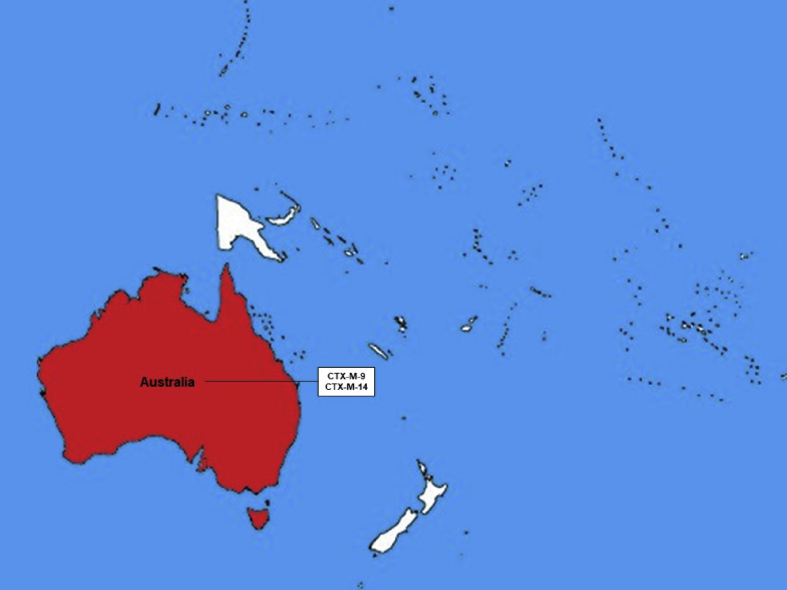
In Oceania as one might imagine, due to its size and number of countries, there exist few reports of E-ESBL in cattle production. The only country with reports is Australia, which is the 6th largest world producer country and the 3rd biggest exporter (USDA et al., 2017). Figure 7 shows the countries with E-ESBL description in cattle and what ESBL type has been described in them.
Figure 7.
Map of Oceania illustrating countries with description of E-ESBL in cattle and the diversity of beta-lactamases presented.
Australia presented a description of CTX-M-9 and CTX-M-14 in E-ESBL in cattle. The description was carried out in clinical isolated and in E. coli and in S. enterica (Abraham et al., 2015; Sparham et al., 2017). Both variants, CTX-M-9 and CTX-M-14, have already been described in human clinical isolates in Australia (Zong et al., 2008).
9. Conclusion
E-ESBL are a threat to human health and are now scattered around the world in intestinal colonization and clinical processes of cattle in Europe, the Americas, Asia, Africa and Oceania. These are described in 6 of the 7 major world cattle producers and certainly these E-ESBLs are contributing to the circulation of these and the ESBL genes through the ecosystems. A circulation that does not only concern the internal level of each country, since the circulation trade of cattle and their derivatives between countries is increasing, with E-ESBL being found in the animals of the world 5 largest meat exporters.
Further studies in the various areas of each country, as well as in other countries without data, are necessary for a better understanding of the presence and circulation of these E- ESBL through cattle and the food chain to assist in the implementation of measures to help in the surveillance and control of the E-ESBL dissemination and propagation.
Declarations
Author contribution statement
Josman Dantas Palmeira, Helena Maria Neto Ferreira: Analyzed and interpreted the data; Wrote the paper.
Funding statement
This work was supported by a scholarship from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- Hächler H., Kotsakis S.D., Tzouvelekis L.S., Geser N., Lehner A., Miriagou V., Stephan R. Characterisation of CTX-M-117, a Pro174Gln variant of CTX-M-15 extended-spectrum β-lactamase, from a bovine Escherichia coli isolate. Int. J. Antimicrob. Agents. 2013;41:94–95. doi: 10.1016/j.ijantimicag.2012.09.011. [DOI] [PubMed] [Google Scholar]
- Jakobsen L., Bortolaia V., Bielak E., Moodley A., Olsen S.S., Hansen D.S., Frimodt-Møller N., Guardabassi L., Hasman H. Limited similarity between plasmids encoding CTX-M-1 β-lactamase in Escherichia coli from humans, pigs, cattle, organic poultry layers and horses in Denmark. J. Glob. Antimicrob. Resist. 2015;3:132–136. doi: 10.1016/j.jgar.2015.03.009. [DOI] [PubMed] [Google Scholar]
- Lee S.-G., Jeong S.H., Lee H., Kim C.K., Lee Y., Koh E., Chong Y., Lee K. Spread of CTX-M–type extended-spectrum β-lactamases among bloodstream isolates of Escherichia coli and Klebsiella pneumoniae from a Korean hospital. Diagn. Microbiol. Infect. Dis. 2009;63:76–80. doi: 10.1016/j.diagmicrobio.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Abbassi M.S., Torres C., Achour W., Vinue L., Saenz Y., Costa D., Bouchami O., Ben Hassen A. Genetic characterisation of CTX-M-15-producing Klebsiella pneumoniae and Escherichia coli strains isolated from stem cell transplant patients in Tunisia. Int. J. Antimicrob. Agents. 2008;32:308–314. doi: 10.1016/j.ijantimicag.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Abraham S., Jordan D., Wong H.S., Johnson J.R., Toleman M.A., Wakeham D.L., Gordon D.M., Turnidge J.D., Mollinger J.L., Gibson J.S., Trott D.J. First detection of extended-spectrum cephalosporin- and fluoroquinolone-resistant Escherichia coli in Australian food-producing animals. J. Glob. Antimicrob. Resist. 2015;3:273–277. doi: 10.1016/j.jgar.2015.08.002. [DOI] [PubMed] [Google Scholar]
- Adeolu M., Alnajar S., Naushad S., R S.G. Genome-based phylogeny and taxonomy of the 'Enterobacteriales': proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2016;66:5575–5599. doi: 10.1099/ijsem.0.001485. [DOI] [PubMed] [Google Scholar]
- Adler A., Sturlesi N.a., Fallach N., Zilberman-Barzilai D., Hussein O., Blum S.E., Klement E., Schwaber M.J., Carmeli Y. Prevalence, risk factors, and transmission dynamics of extended-spectrum-β-lactamase-producing enterobacteriaceae: a national survey of cattle farms in Israel in 2013. J. Clin. Microbiol. 2015;53:3515–3521. doi: 10.1128/JCM.01915-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afema J.A., Ahmed S., Besser T.E., Jones L.P., Sischo W.M., Davis M.A. Molecular epidemiology of dairy cattle-associated Escherichia coli carrying blaCTX-M genes in Washington state. Appl. Environ. Microbiol. 2018;84 doi: 10.1128/AEM.02430-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed A.M., Shimamoto T. Molecular characterization of antimicrobial resistance in Gram-negative bacteria isolated from bovine mastitis in Egypt. Microbiol. Immunol. 2011;55:318–327. doi: 10.1111/j.1348-0421.2011.00323.x. [DOI] [PubMed] [Google Scholar]
- Alexandratos N., Bruinsma J. Global Perspective Studies Team. Food and Agriculture Organization of the United Nations; 2012. World agriculture towards 2030/2050 - the 2012 revision. [Google Scholar]
- Ali T., ur Rahman S., Zhang L., Shahid M., Zhang S., Liu G., Gao J., Han B. ESBL-producing Escherichia coli from cows suffering mastitis in China contain clinical class 1 integrons with CTX-M linked to ISCR1. Front. Microbiol. 2016;7 doi: 10.3389/fmicb.2016.01931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali T., Rahman S.U., Zhang L., Shahid M., Han D., Gao J., Zhang S., Ruegg P.L., Saddique U., Han B. Characteristics and genetic diversity of multi-drug resistant extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli isolated from bovine mastitis. Oncotarget. 2017;8:90144–90163. doi: 10.18632/oncotarget.21496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T., Masani K., Sato C., Hiki M., Usui M., Baba K., Ozawa M., Harada K., Aoki H., Sawada T. Phylogenetic groups and cephalosporin resistance genes of Escherichia coli from diseased food-producing animals in Japan. Acta Vet. Scand. 2011;53:52. doi: 10.1186/1751-0147-53-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AslantaŞ Ö., ElmacioĞLu S., Yilmaz E.Ş. Prevalence and characterization of ESBL- and AmpC-producing Escherichia coli from cattle. Kafkas Universitesi Veteriner Fakultesi Dergisi. 2017;23:63–67. [Google Scholar]
- Awosile B., McClure J., Sanchez J., Rodriguez-Lecompte J.C., Keefe G., Heider L.C. Salmonella enterica and extended-spectrum cephalosporin-resistant Escherichia coli recovered from Holstein dairy calves from 8 farms in New Brunswick, Canada. J. Dairy Sci. 2018;101:3271–3284. doi: 10.3168/jds.2017-13277. [DOI] [PubMed] [Google Scholar]
- Badran E.F., Qamer Din R.A., Shehabi A.A. Low intestinal colonization of Escherichia coli clone ST131 producing CTX-M-15 in Jordanian infants. J. Med. Microbiol. 2016;65:137–141. doi: 10.1099/jmm.0.000210. [DOI] [PubMed] [Google Scholar]
- Bagus Wasito E., Shigemura K., Osawa K., Fardah A., Kanaida A., Raharjo D., Kuntaman K., Hadi U., Harijono S., Marto Sudarmo S., Nakamura T., Shibayama K., Fujisawa M., Shirakawa T. Antibiotic susceptibilities and genetic characteristics of extended-spectrum beta-lactamase-producing Escherichia coli isolates from stools of pediatric diarrhea patients in Surabaya, Indonesia. Jpn. J. Infect. Dis. 2017;70:378–382. doi: 10.7883/yoken.JJID.2016.234. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S., Samanta I., Bhattacharyya D., Nanda P.K., Kar D., Chowdhury J., Dandapat P., Das A.K., Batul N., Mondal B., Dutta T.K., Das G., Das B.C., Naskar S., Bandyopadhyay U.K., Das S.C., Bandyopadhyay S. Co-infection of methicillin-resistant Staphylococcus epidermidis, methicillin-resistant Staphylococcus aureus and extended spectrum β-lactamase producing Escherichia coli in bovine mastitis – three cases reported from India. Vet. Q. 2015;35:56–61. doi: 10.1080/01652176.2014.984365. [DOI] [PubMed] [Google Scholar]
- Barzan M., Gharibi D., Ghorbanpoor M., Haji Hajikolaei M., Pourmehdi-Boroujeni M. Phylogenetic grouping and phenotypic detection of extended-spectrum β-lactamases among Escherichia coli from calves and dairy cows in Khuzestan, Iran. Int. J. Enteric. Pathog. 2017;5:24–29. [Google Scholar]
- Batchelor M., Hopkins K., Threlfall E.J., Clifton-Hadley F.A., Stallwood A.D., Davies R.H., Liebana E. bla(CTX-M) genes in clinical Salmonella isolates recovered from humans in England and Wales from 1992 to 2003. Antimicrob. Agents Chemother. 2005;49:1319–1322. doi: 10.1128/AAC.49.4.1319-1322.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzaz B.S., Naderinasab M., Mohamadpoor A.H., Farshadzadeh Z., Ahmadi S., Yousefi F. The prevalence of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae among clinical isolates from a general hospital in Iran. Acta Microbiol. Immunol. Hung. 2009;56:89–99. doi: 10.1556/AMicr.56.2009.1.7. [DOI] [PubMed] [Google Scholar]
- Borah V.V., Bora P., Roy M., Saikia K.K. High prevalence of antibiotic resistance in Escherichia coli isolated from fecal sample of cows and assessment of antibacterial efficacy of indigenous medicinal plants from Assam, India. Austin J. Biotechnol. Bioeng. 2014;1:6. [Google Scholar]
- Brahmi S., Dunyach-Remy C., Touati A., Lavigne J.P. CTX-M-15-producing Escherichia coli and the pandemic clone O25b-ST131 isolated from wild fish in Mediterranean Sea. Clin. Microbiol. infect. : Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2015;21:e18–20. doi: 10.1016/j.cmi.2014.09.019. [DOI] [PubMed] [Google Scholar]
- Braun S.D., Ahmed M.F.E., El-Adawy H., Hotzel H., Engelmann I., Weiß D., Monecke S., Ehricht R. Surveillance of extended-spectrum beta-lactamase-producing Escherichia coli in dairy cattle farms in the nile Delta, Egypt. Front. Microbiol. 2016;7:1020. doi: 10.3389/fmicb.2016.01020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan E., Martins M., McCusker M.P., Wang J., Alves B.M., Hurley D., El Garch F., Woehrlé F., Miossec C., McGrath L., Srikumar S., Wall P., Fanning S. Multidrug-resistant Escherichia coli in bovine animals, Europe. Emerg. Infect. Dis. 2016;22:1650–1652. doi: 10.3201/eid2209.160140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briñas L., Moreno M.A., Teshager T., Sáenz Y., Porrero M.C., Domínguez L., Torres C. Monitoring and characterization of extended-spectrum β-lactamases in Escherichia coli strains from healthy and sick animals in Spain in 2003. Antimicrob. Agents Chemother. 2005;49:1262–1264. doi: 10.1128/AAC.49.3.1262-1264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brolund A., Edquist P.J., Mäkitalo B., Olsson-Liljequist B., Söderblom T., Wisell K.T., Giske C.G. Epidemiology of extended-spectrum β-lactamase-producing Escherichia coli in Sweden 2007–2011. Clin. Microbiol. Infect. 2014;20:O344–O352. doi: 10.1111/1469-0691.12413. [DOI] [PubMed] [Google Scholar]
- Cantón R., Novais A., Valverde A., Machado E., Peixe L., Baquero F., Coque T.M. Prevalence and spread of extended-spectrum β-lactamase-producing enterobacteriaceae in Europe. Clin. Microbiol. Infect. 2008;14:144–153. doi: 10.1111/j.1469-0691.2007.01850.x. [DOI] [PubMed] [Google Scholar]
- Cantón R., González-Alba J.M., Galán J.C. CTX-M enzymes: origin and diffusion. Front. Microbiol. 2012;3:110. doi: 10.3389/fmicb.2012.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantσn R., Coque T.M. The CTX-M β-lactamase pandemic. Curr. Opin. Microbiol. 2006;9:466–475. doi: 10.1016/j.mib.2006.08.011. [DOI] [PubMed] [Google Scholar]
- CDC, C.f.D.C.a.P. U. S. Departament of Health and Human Services; 2013. Antibiotic Resistance Threats in United States in 2013. [Google Scholar]
- Ceccarelli D., Kant A., van Essen-Zandbergen A., Dierikx C., Hordijk J., Wit B., Mevius D.J., Veldman K.T. Diversity of plasmids and genes encoding resistance to extended spectrum cephalosporins in commensal Escherichia coli from Dutch livestock in 2007–2017. Front. Microbiol. 2019;10 doi: 10.3389/fmicb.2019.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanawong A., M'Zali F.H., Heritage J., Xiong J.-H., Hawkey P.M. Three cefotaximases, CTX-M-9, CTX-M-13, and CTX-M-14, among enterobacteriaceae in the People's Republic of China. Antimicrob. Agents Chemother. 2002;46:630–637. doi: 10.1128/AAC.46.3.630-637.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.F., Freeman J.T., Nicholson B., Keiger A., Lancaster S., Joyce M., Woods C.W., Cook E., Adcock L., Louis S., Cromer A.L., Sexton D.J., Anderson D.J. Widespread dissemination of CTX-M-15 genotype extended-spectrum-β-lactamase-producing enterobacteriaceae among patients presenting to community hospitals in the southeastern United States. Antimicrob. Agents Chemother. 2014;58:1200–1202. doi: 10.1128/AAC.01099-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmelnitsky I., Carmeli Y., Leavitt A., Schwaber M.J., Navon-Venezia S. CTX-M-2 and a new CTX-M-39 enzyme are the major extended-spectrum beta-lactamases in multiple Escherichia coli clones isolated in Tel Aviv, Israel. Antimicrob. Agents Chemother. 2005;49:4745–4750. doi: 10.1128/AAC.49.11.4745-4750.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clermont O., Lavollay M., Vimont S., Deschamps C., Forestier C., Branger C., Denamur E., Arlet G. The CTX-M-15-producing Escherichia coli diffusing clone belongs to a highly virulent B2 phylogenetic subgroup. J. Antimicrob. Chemother. 2008;61:1024–1028. doi: 10.1093/jac/dkn084. [DOI] [PubMed] [Google Scholar]
- Colquechagua Aliaga F., Sevilla Andrade C., Gonzales Escalante E. [Extended-spectrum beta-lactamase (esbl)-producing enterobacteriaceae in fecal samples at the National Institute of Child Health, Peru] Rev. Peru. Med. Exp. Salud Pública. 2015;32:26–32. [PubMed] [Google Scholar]
- Cormier A.C., Chalmers G., McAllister T.A., Cook S., Zaheer R., Scott H.M., Booker C., Read R., Boerlin P. Extended-spectrum-Cephalosporin resistance genes in Escherichia coli from beef cattle. Antimicrob. Agents Chemother. 2016;60:1162–1163. doi: 10.1128/AAC.02516-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier A., Zhang P.L.C., Chalmers G., Weese J.S., Deckert A., Mulvey M., McAllister T., Boerlin P. Diversity of CTX-M-positive Escherichia coli recovered from animals in Canada. Vet. Microbiol. 2019;231:71–75. doi: 10.1016/j.vetmic.2019.02.031. [DOI] [PubMed] [Google Scholar]
- Cottell J.L., Webber M.A., Coldham N.G., Taylor D.L., Cerdeño-Tárraga A.M., Hauser H., Thomson N.R., Woodward M.J., Piddock L.J.V. Complete sequence and molecular epidemiology of IncK epidemic plasmid encoding bla(CTX-M-14) Emerg. Infect. Dis. 2011;17:645–652. doi: 10.3201/eid1704.101009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottell J.L., Kanwar N., Castillo-Courtade L., Chalmers G., Scott H.M., Norby B., Loneragan G.H., Boerlin P. blaCTX-M-32 on an IncN plasmid in Escherichia coli from beef cattle in the United States. Antimicrob. Agents Chemother. 2013;57:1096–1097. doi: 10.1128/AAC.01750-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmen S., Métayer V., Gay E., Madec J.-Y., Haenni M. Characterization of extended-spectrum beta-lactamase (ESBL)-carrying plasmids and clones of Enterobacteriaceae causing cattle mastitis in France. Vet. Microbiol. 2013;162:793–799. doi: 10.1016/j.vetmic.2012.10.015. [DOI] [PubMed] [Google Scholar]
- Dahms C., Hübner N.-O., Kossow A., Mellmann A., Dittmann K., Kramer A. Occurrence of ESBL-producing Escherichia coli in livestock and farm workers in Mecklenburg-Western Pomerania, Germany. PLoS One. 2015;10 doi: 10.1371/journal.pone.0143326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A., Guha C., Biswas U., Jana P.S., Chatterjee A., Samanta I. Detection of emerging antibiotic resistance in bacteria isolated from subclinical mastitis in cattle in West Bengal. Vet. World. 2017;10:517–520. doi: 10.14202/vetworld.2017.517-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M.A., Sischo W.M., Jones L.P., Moore D.A., Ahmed S., Short D.M., Besser T.E. Recent emergence of Escherichia coli with cephalosporin resistance conferred by blaCTX-M on Washington state dairy farms. Appl. Environ. Microbiol. 2015;81:4403–4410. doi: 10.1128/AEM.00463-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Champs C., Chanal C., Sirot D., Baraduc R., Romaszko J.P., Bonnet R., Plaidy A., Boyer M., Carroy E., Gbadamassi M.C., Laluque S., Oules O., Poupart M.C., Villemain M., Sirot J. Frequency and diversity of Class A extended-spectrum beta-lactamases in hospitals of the Auvergne, France: a 2 year prospective study. J. Antimicrob. Chemother. 2004;54:634–639. doi: 10.1093/jac/dkh395. [DOI] [PubMed] [Google Scholar]
- Denisuik A.J., Lagacé-Wiens P.R.S., Pitout J.D., Mulvey M.R., Simner P.J., Tailor F., Karlowsky J.A., Hoban D.J., Adam H.J., Zhanel G.G., Zhanel G.G., Hoban D.J., Adam H.J., Karlowsky J.A., Baxter M.R., Nichol K.A., Lagacé-Wiens P.R.S., Walkty A. Molecular epidemiology of extended-spectrum β-lactamase-, AmpC β-lactamase- and carbapenemase-producing Escherichia coli and Klebsiella pneumoniae isolated from Canadian hospitals over a 5 year period: CANWARD 2007–11. J. Antimicrob. Chemother. 2013;68:i57–i65. doi: 10.1093/jac/dkt027. [DOI] [PubMed] [Google Scholar]
- Denisuik A.J., Adam H.J., Lagacé-Wiens P., Simner P.J., Mulvey M.R., Baxter M., Gilmour M., Karlowsky J.A., Hoban D.J., Zhanel G.G. ICAAC/ICC 2015. 2015. Rates of extended-spectrum β-lactamase-producing Escherichia coli quadruple in Canadian hospitals over an 8-year period: CANWARD 2007-2014. San Diego, CA, USA. [Google Scholar]
- Dia M.L., Ngom B., Diagne R., Ka R., Lo S., Cisse M.F., Arlet G., Sow A.I. Molecular detection of CTX-M-15-type β-lactamases in Escherichia coli strains from Senegal. New Microbes New Infections. 2016;9:45–46. doi: 10.1016/j.nmni.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diab M., Hamze M., Madec J.-Y., Haenni M. High prevalence of non-ST131 CTX-M-15-producing Escherichia coli in healthy cattle in Lebanon. Microb. Drug Resist. 2016;23:261–266. doi: 10.1089/mdr.2016.0019. [DOI] [PubMed] [Google Scholar]
- Dolejska M., Frolkova P., Florek M., Jamborova I., Purgertova M., Kutilova I., Cizek A., Guenther S., Literak I. CTX-M-15-producing Escherichia coli clone B2-O25b-ST131 and Klebsiella spp. isolates in municipal wastewater treatment plant effluents. J. Antimicrob. Chemother. 2011;66:2784–2790. doi: 10.1093/jac/dkr363. [DOI] [PubMed] [Google Scholar]
- Dolejska M., Jurcickova Z., Literak I., Pokludova L., Bures J., Hera A., Kohoutova L., Smola J., Cizek A. IncN plasmids carrying blaCTX-M-1 in Escherichia coli isolates on a dairy farm. Vet. Microbiol. 2011;149:513–516. doi: 10.1016/j.vetmic.2010.11.032. [DOI] [PubMed] [Google Scholar]
- Dolejska M., Villa L., Hasman H., Hansen L., Carattoli A. Characterization of IncN plasmids carrying blaCTX-M-1 and qnr genes in Escherichia coli and Salmonella from animals, the environment and humans. J. Antimicrob. Chemother. 2013;68:333–339. doi: 10.1093/jac/dks387. [DOI] [PubMed] [Google Scholar]
- Donaldson S.C., Straley B.A., Hegde N.V., Sawant A.A., DebRoy C., Jayarao B.M. Molecular epidemiology of ceftiofur-resistant Escherichia coli isolates from dairy calves. Appl. Environ. Microbiol. 2006;72:3940–3948. doi: 10.1128/AEM.02770-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doumith M., Dhanji H., Ellington M.J., Hawkey P., Woodford N. Characterization of plasmids encoding extended-spectrum β-lactamases and their addiction systems circulating among Escherichia coli clinical isolates in the UK. J. Antimicrob. Chemother. 2012;67:878–885. doi: 10.1093/jac/dkr553. [DOI] [PubMed] [Google Scholar]
- Duan R.S., Sit T.H., Wong S.S., Wong R.C., Chow K.H., Mak G.C., Yam W.C., Ng L.T., Yuen K.Y., Ho P.L. Escherichia coli producing CTX-M beta-lactamases in food animals in Hong Kong. Microb. Drug Resist. 2006;12:145–148. doi: 10.1089/mdr.2006.12.145. [DOI] [PubMed] [Google Scholar]
- Duse A., Waller K.P., Emanuelson U., Unnerstad H.E., Persson Y., Bengtsson B. Risk factors for antimicrobial resistance in fecal Escherichia coli from preweaned dairy calves. J. Dairy Sci. 2015;98:500–516. doi: 10.3168/jds.2014-8432. [DOI] [PubMed] [Google Scholar]
- Ebrahimi F. University of Debrecen; 2016. Epidemiology of Faecal Carriage of Extended-Spectrum Beta-Lactamases in Healthy Individuals and in Different Patient Populations. PhD. [Google Scholar]
- Eisenberger D., Carl A., Balsliemke J., Kampf P., Nickel S., Schulze G., Valenza G. Molecular characterization of extended-spectrum beta-lactamase-producing Escherichia coli isolates from milk samples of dairy cows with mastitis in Bavaria, Germany. Microb. Drug Resist. 2017 doi: 10.1089/mdr.2017.0182. [DOI] [PubMed] [Google Scholar]
- Endimiani A., Rossano A., Kunz D., Overesch G., Perreten V. First countrywide survey of third-generation cephalosporin-resistant Escherichia coli from broilers, swine, and cattle in Switzerland. Diagn. Microbiol. Infect. Dis. 2012;73:31–38. doi: 10.1016/j.diagmicrobio.2012.01.004. [DOI] [PubMed] [Google Scholar]
- Eurostat . 2016. Livestock Population, 2015.http://ec.europa.eu/eurostat/statistics-explained/index.php/File:Livestock_population,_2015_(million_head)_T1.png#file [Google Scholar]
- Ewers C., Grobbel M., Stamm I., Kopp P.A., Diehl I., Semmler T., Fruth A., Beutlich J., Guerra B., Wieler L.H., Guenther S. Emergence of human pandemic O25:H4-ST131 CTX-M-15 extended-spectrum-β-lactamase-producing Escherichia coli among companion animals. J. Antimicrob. Chemother. 2010;65:651–660. doi: 10.1093/jac/dkq004. [DOI] [PubMed] [Google Scholar]
- Ewers C., Stamm I., Stolle I., Guenther S., Kopp P.A., Fruth A., Wieler L.H., Scheufen S., Bauerfeind R., Bethe A., Prenger-Berninghoff E. Detection of Shiga toxin- and extended-spectrum beta-lactamase-producing Escherichia coli O145:NM and Ont:NM from calves with diarrhoea. J. Antimicrob. Chemother. 2014;69:2005–2007. doi: 10.1093/jac/dku042. [DOI] [PubMed] [Google Scholar]
- Fam N., Leflon-Guibout V., Fouad S., Aboul-Fadl L., Marcon E., Desouky D., El-Defrawy I., Abou-Aitta A., Klena J., Nicolas-Chanoine M.H. CTX-M-15-producing Escherichia coli clinical isolates in Cairo (Egypt), including isolates of clonal complex ST10 and clones ST131, ST73, and ST405 in both community and hospital settings. Microb. Drug Resist. 2011;17:67–73. doi: 10.1089/mdr.2010.0063. [DOI] [PubMed] [Google Scholar]
- Fernandes R., Amador P., Oliveira C., Prudêncio C. Molecular characterization of ESBL-producing enterobacteriaceae in northern Portugal. Sci. World J. 2014;2014:782897. doi: 10.1155/2014/782897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer J., Rodríguez I., Baumann B., Guiral E., Beutin L., Schroeter A., Kaesbohrer A., Pfeifer Y., Helmuth R., Guerra B. blaCTX-M-15-carrying Escherichia coli and Salmonella isolates from livestock and food in Germany. J. Antimicrob. Chemother. 2014;69:2951–2958. doi: 10.1093/jac/dku270. [DOI] [PubMed] [Google Scholar]
- Forssten S.D., Kolho E., Lauhio A., Lehtola L., Mero S., Oksaharju A., Jalava J., Tarkka E., Vaara M., Vuopio-Varkila J. Emergence of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae during the years 2000 and 2004 in Helsinki, Finland. Clin. Microbiol. Infect. 2010;16:1158–1161. doi: 10.1111/j.1469-0691.2010.03068.x. [DOI] [PubMed] [Google Scholar]
- Freitag C., Michael G.B., Kadlec K., Hassel M., Schwarz S. Detection of plasmid-borne extended-spectrum β-lactamase (ESBL) genes in Escherichia coli isolates from bovine mastitis. Vet. Microbiol. 2017;200:151–156. doi: 10.1016/j.vetmic.2016.08.010. [DOI] [PubMed] [Google Scholar]
- Friese A., Schulz J., Laube H., von Salviati C., Hartung J., Roesler U. Faecal occurrence and emissions of livestock-associated methicillin-resistant Staphylococcus aureus (laMRSA) and ESbl/AmpC-producing E. coli from animal farms in Germany. Berl. Münchener Tierärztliche Wochenschr. 2013;126:175–180. [PubMed] [Google Scholar]
- Frye J.G., Fedorka-Cray P.J. Prevalence, distribution and characterisation of ceftiofur resistance in Salmonella enterica isolated from animals in the USA from 1999 to 2003. Int. J. Antimicrob. Agents. 2007;30:134–142. doi: 10.1016/j.ijantimicag.2007.03.013. [DOI] [PubMed] [Google Scholar]
- Garcia-Fernandez A., Villa L., Moodley A., Hasman H., Miriagou V., Guardabassi L., Carattoli A. Multilocus sequence typing of IncN plasmids. J. Antimicrob. Chemother. 2011;66:1987–1991. doi: 10.1093/jac/dkr225. [DOI] [PubMed] [Google Scholar]
- Gay N., Leclaire A., Laval M., Miltgen G., Jego M., Stephane R., Jaubert J., Belmonte O., Cardinale E. Risk factors of extended-spectrum beta-lactamase producing enterobacteriaceae occurrence in farms in Reunion, Madagascar and Mayotte Islands, 2016-2017. Vet. Sci. 2018;5 doi: 10.3390/vetsci5010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhold G., Schulze M.H., Gross U., Bohne W. Multilocus sequence typing and CTX-M characterization of ESBL-producing E. coli: a prospective single-centre study in Lower Saxony, Germany. Epidemiol. Infect. 2016;144:3300–3304. doi: 10.1017/S0950268816001412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geser N., Stephan R., Kuhnert P., Zbinden R., Kaeppeli U., Cernela N., Haechler H. Fecal carriage of extended-spectrum beta-lactamase-producing Enterobacteriaceae in swine and cattle at slaughter in Switzerland. J. Food Prot. 2011;74:446–449. doi: 10.4315/0362-028X.JFP-10-372. [DOI] [PubMed] [Google Scholar]
- Geser N., Stephan R., Hächler H. Occurrence and characteristics of extended-spectrum β-lactamase (ESBL) producing Enterobacteriaceae in food producing animals, minced meat and raw milk. BMC Vet. Res. 2012;8:21. doi: 10.1186/1746-6148-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geser N., Stephan R., Korczak B.M., Beutin L., Hächler H. Molecular identification of extended-spectrum-β-lactamase genes from enterobacteriaceae isolated from healthy human carriers in Switzerland. Antimicrob. Agents Chemother. 2012;56:1609–1612. doi: 10.1128/AAC.05539-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghatak S., Singha A., Sen A., Guha C., Ahuja A., Bhattacharjee U., Das S., Pradhan N.R., Puro K., Jana C., Dey T.K., Prashantkumar K.L., Das A., Shakuntala I., Biswas U., Jana P.S. Detection of New Delhi metallo-beta-lactamase and extended-spectrum beta-lactamase genes in Escherichia coli isolated from mastitic milk samples. Transbound. Emerg. Dis. 2013;60:385–389. doi: 10.1111/tbed.12119. [DOI] [PubMed] [Google Scholar]
- Giedraitienė A., Vitkauskienė A., Pavilonis A., Patamsytė V., Genel N., Decre D., Arlet G. Prevalence of O25b-ST131 clone among Escherichia coli strains producing CTX-M-15, CTX-M-14 and CTX-M-92 β-lactamases. Infect. Dis. 2017;49:106–112. doi: 10.1080/23744235.2016.1221531. [DOI] [PubMed] [Google Scholar]
- Gonggrijp M.A., Santman-Berends I.M.G.A., Heuvelink A.E., Buter G.J., van Schaik G., Hage J.J., Lam T.J.G.M. Prevalence and risk factors for extended-spectrum β-lactamase- and AmpC-producing Escherichia coli in dairy farms. J. Dairy Sci. 2016;99:9001–9013. doi: 10.3168/jds.2016-11134. [DOI] [PubMed] [Google Scholar]
- Gonzalez C.M.A. Universidad de Chile; 2006. Susceptibilidad microbiana: Un test rapido para el analisis de resistencia bacteriana en cepas aisladas de mastitis clinicasBachelor. [Google Scholar]
- Gonçalves D., Cecílio P., Ferreira H. Nursing homes and long-term care facilities: reservoirs of CTX-M-15-producing Escherichia coli O25b-ST131 in Portugal. J. Glob. Antimicrob. Resist. 2016;7:69–71. doi: 10.1016/j.jgar.2016.08.001. [DOI] [PubMed] [Google Scholar]
- Grami R., Dahmen S., Mansour W., Mehri W., Haenni M., Aouni M., Madec J.Y. blaCTX-M-15-carrying F2:A-:B- plasmid in Escherichia coli from cattle milk in Tunisia. Microb. Drug Resist. 2014;20:344–349. doi: 10.1089/mdr.2013.0160. [DOI] [PubMed] [Google Scholar]
- Gur D., Gulay Z., Akan O.A., Aktas Z., Kayacan C.B., Cakici O., Erac B., Gultekin M., Ogunc D., Soyletir G., Unal N., Uysal S. [Resistance to newer beta-lactams and related ESBL types in gram-negative nosocomial isolates in Turkish hospitals: results of the multicentre HITIT study] Mikrobiyol. Bul. 2008;42:537–544. [PubMed] [Google Scholar]
- Haenni M., Saras E., Métayer V., Doublet B., Cloeckaert A., Madec J.-Y. Spread of the blaTEM-52 gene is mainly ensured by IncI1/ST36 plasmids in Escherichia coli isolated from cattle in France. J. Antimicrob. Chemother. 2012;67:2774–2776. doi: 10.1093/jac/dks282. [DOI] [PubMed] [Google Scholar]
- Haenni M., Châtre P., Métayer V., Bour M., Signol E., Madec J.-Y., Gay E. Comparative prevalence and characterization of ESBL-producing Enterobacteriaceae in dominant versus subdominant enteric flora in veal calves at slaughterhouse, France. Vet. Microbiol. 2014;171:321–327. doi: 10.1016/j.vetmic.2014.02.023. [DOI] [PubMed] [Google Scholar]
- Haenni M., Beyrouthy R., Lupo A., Châtre P., Madec J.-Y., Bonnet R. Epidemic spread of Escherichia coli ST744 isolates carrying mcr-3 and blaCTX-M-55 in cattle in France. J. Antimicrob. Chemother. 2018;73:533–536. doi: 10.1093/jac/dkx418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdy Mohammed E.s., Elsadek Fakhr A., Mohammed El sayed H., Al Johery S.a.E., Abdel Ghani Hassanein W. Spread of TEM, VIM, SHV, and CTX-M β-lactamases in imipenem-resistant gram-negative bacilli isolated from Egyptian hospitals. Int. J. Microbiol. 2016;2016:8382605. doi: 10.1155/2016/8382605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann A., Amoureux L., Locatelli A., Depret G., Jolivet C., Gueneau E., Neuwirth C. Occurrence of CTX-M producing Escherichia coli in soils, cattle, and farm environment in France (Burgundy region) Front. Microbiol. 2012;3 doi: 10.3389/fmicb.2012.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan B., Laurell K., Rakib M.M., Ahlstedt E., Hernandez J., Caceres M., Järhult J.D. Fecal carriage of extended-spectrum β-lactamases in healthy humans, poultry, and wild birds in León, Nicaragua—a shared pool of blaCTX-M genes and possible interspecies clonal spread of extended-spectrum β-lactamases-producing Escherichia coli. Microb. Drug Resist. 2016;22:682–687. doi: 10.1089/mdr.2015.0323. [DOI] [PubMed] [Google Scholar]
- Hassan H., Abdalhamid B. Molecular characterization of extended-spectrum beta-lactamase producing Enterobacteriaceae in a Saudi Arabian tertiary hospital. J. Infect. Dev. Ctries. 2014;8:282–288. doi: 10.3855/jidc.3809. [DOI] [PubMed] [Google Scholar]
- Hassan S., Gherbawy Y., Altalhi A.D. 2015. Genetic Heterogeneity of CTX-M Type Extended-Spectrum β-lactamase Producing Escherichia coli Strains from Diverse Sources in Saudi Arabia. [Google Scholar]
- Hawkey P.M. Prevalence and clonality of extended-spectrum β-lactamases in Asia. Clin. Microbiol. Infect. 2008;14:159–165. doi: 10.1111/j.1469-0691.2007.01855.x. [DOI] [PubMed] [Google Scholar]
- Hernández J., Stedt J., Bonnedahl J., Molin Y., Drobni M., Calisto-Ulloa N., Gomez-Fuentes C., Astorga-España M.S., González-Acuña D., Waldenström J., Blomqvist M., Olsen B. Human-associated extended-spectrum β-lactamase in the Antarctic. Appl. Environ. Microbiol. 2012;78:2056–2058. doi: 10.1128/AEM.07320-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez J., Johansson A., Stedt J., Bengtsson S., Porczak A., Granholm S., González-Acuña D., Olsen B., Bonnedahl J., Drobni M. Characterization and comparison of extended-spectrum β-lactamase (ESBL) resistance genotypes and population structure of Escherichia coli isolated from Franklin's Gulls (Leucophaeus pipixcan) and humans in Chile. PLoS One. 2013;8 doi: 10.1371/journal.pone.0076150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández M., Iglesias M.R., Rodríguez-Lázaro D., Gallardo A., Quijada N., Miguela-Villoldo P., Campos M.J., Píriz S., López-Orozco G., de Frutos C., Sáez J.L., Ugarte-Ruiz M., Domínguez L., Quesada A. Co-occurrence of colistin-resistance genes mcr-1 and mcr-3 among multidrug-resistant Escherichia coli isolated from cattle, Spain, September 2015. Euro Surveill. 2017;22:30586. doi: 10.2807/1560-7917.ES.2017.22.31.30586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuvelink A.E., Gonggrijp M.A., Buter R.G.J., ter Bogt-Kappert C.C., van Schaik G., Velthuis A.G.J., Lam T.J.G.M. Prevalence of extended-spectrum and AmpC β-lactamase-producing Escherichia coli in Dutch dairy herds. Vet. Microbiol. 2019;232:58–64. doi: 10.1016/j.vetmic.2019.04.005. [DOI] [PubMed] [Google Scholar]
- Hinthong W., Pumipuntu N., Santajit S., Kulpeanprasit S., Buranasinsup S., Sookrung N., Chaicumpa W., Aiumurai P., Indrawattana N. Detection and drug resistance profile of Escherichia coli from subclinical mastitis cows and water supply in dairy farms in Saraburi Province, Thailand. PeerJ. 2017;5 doi: 10.7717/peerj.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho P.L., Chow K.H., Lai E.L., Lo W.U., Yeung M.K., Chan J., Chan P.Y., Yuen K.Y. Extensive dissemination of CTX-M-producing Escherichia coli with multidrug resistance to ‘critically important’ antibiotics among food animals in Hong Kong, 2008–10. J. Antimicrob. Chemother. 2011;66:765–768. doi: 10.1093/jac/dkq539. [DOI] [PubMed] [Google Scholar]
- Ho W.S., Balan G., Puthucheary S., Kong B.H., Lim K.T., Tan L.K., Koh X.P., Yeo C.C., Thong K.L. Prevalence and characterization of multidrug-resistant and extended-spectrum beta-lactamase-producing Escherichia coli from pediatric wards of a Malaysian hospital. Microb. Drug Resist. 2012;18:408–416. doi: 10.1089/mdr.2011.0222. [DOI] [PubMed] [Google Scholar]
- Ho P.L., Chan J., Lo W.U., Law P.Y., Li Z., Lai E.L., Chow K.H. Dissemination of plasmid-mediated fosfomycin resistance fosA3 among multidrug-resistant Escherichia coli from livestock and other animals. J. Appl. Microbiol. 2013;114:695–702. doi: 10.1111/jam.12099. [DOI] [PubMed] [Google Scholar]
- Ho P.-L., Liu M.C.-J., Lo W.-U., Lai E.L.-Y., Lau T.C.-K., Law O.-K., Chow K.-H. Prevalence and characterization of hybrid blaCTX-M among Escherichia coli isolates from livestock and other animals. Diagn. Microbiol. Infect. Dis. 2015;82:148–153. doi: 10.1016/j.diagmicrobio.2015.02.010. [DOI] [PubMed] [Google Scholar]
- Hordijk J., Mevius D.J., Kant A., Bos M.E.H., Graveland H., Bosman A.B., Hartskeerl C.M., Heederik D.J.J., Wagenaar J.A. Within-farm dynamics of ESBL/AmpC-producing Escherichia coli in veal calves: a longitudinal approach. J. Antimicrob. Chemother. 2013;68:2468–2476. doi: 10.1093/jac/dkt219. [DOI] [PubMed] [Google Scholar]
- Hordijk J., Wagenaar J.A., Kant A., van Essen-Zandbergen A., Dierikx C., Veldman K., Wit B., Mevius D. Cross-sectional study on prevalence and molecular characteristics of plasmid mediated ESBL/AmpC-Producing Escherichia coli isolated from veal calves at slaughter. PLoS One. 2013;8 doi: 10.1371/journal.pone.0065681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hordijk J., Wagenaar J.A., van de Giessen A., Dierikx C., van Essen-Zandbergen A., Veldman K., Kant A., Mevius D. Increasing prevalence and diversity of ESBL/AmpC-type beta-lactamase genes in Escherichia coli isolated from veal calves from 1997 to 2010. J. Antimicrob. Chemother. 2013;68:1970–1973. doi: 10.1093/jac/dkt132. [DOI] [PubMed] [Google Scholar]
- Horton R.A., Randall L.P., Snary E.L., Cockrem H., Lotz S., Wearing H., Duncan D., Rabie A., McLaren I., Watson E., La Ragione R.M., Coldham N.G. Fecal carriage and shedding density of CTX-M extended-spectrum β-lactamase-producing Escherichia coli in cattle, chickens, and pigs: implications for environmental contamination and food production. Appl. Environ. Microbiol. 2011;77:3715–3719. doi: 10.1128/AEM.02831-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y.-Y., Cai J.-C., Zhou H.-W., Chi D., Zhang X.-F., Chen W.-L., Zhang R., Chen G.-X. Molecular typing of CTX-M-producing Escherichia coli isolates from environmental water, swine feces, specimens from healthy humans, and human patients. Appl. Environ. Microbiol. 2013;79:5988–5996. doi: 10.1128/AEM.01740-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter P.A., Dawson S., French G.L., Goossens H., Hawkey P.M., Kuijper E.J., Nathwani D., Taylor D.J., Teale C.J., Warren R.E., Wilcox M.H., Woodford N., Wulf M.W., Piddock L.J.V. Antimicrobial-resistant pathogens in animals and man: prescribing, practices and policies. J. Antimicrob. Chemother. 2010;65:i3–i17. doi: 10.1093/jac/dkp433. [DOI] [PubMed] [Google Scholar]
- Iroha I.R., Esimone C.O., Neumann S., Marlinghaus L., Korte M., Szabados F., Gatermann S., Kaase M. First description of Escherichia coli producing CTX-M-15- extended spectrum beta lactamase (ESBL) in out-patients from south eastern Nigeria. Ann. Clin. Microbiol. Antimicrob. 2012;11:19. doi: 10.1186/1476-0711-11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iweriebor B.C., Iwu C.J., Obi L.C., Nwodo U.U., Okoh A.I. Multiple antibiotic resistances among Shiga toxin producing Escherichia coli O157 in feces of dairy cattle farms in Eastern Cape of South Africa. BMC Microbiol. 2015;15:213. doi: 10.1186/s12866-015-0553-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamaruzzaman E.A. Universiti Putra; Malaysia: 2015. Occurrence of Extended Spectrum Beta-Lactamase Producing escherichia Coli in Dairy Cattle, Farm Environment and Milk. Master. [Google Scholar]
- Kar D., Bandyopadhyay S., Bhattacharyya D., Samanta I., Mahanti A., Nanda P.K., Mondal B., Dandapat P., Das A.K., Dutta T.K., Bandyopadhyay S., Singh R.K. Molecular and phylogenetic characterization of multidrug resistant extended spectrum beta-lactamase producing Escherichia coli isolated from poultry and cattle in Odisha, India. Infect. Genet. Evol. 2015;29:82–90. doi: 10.1016/j.meegid.2014.11.003. [DOI] [PubMed] [Google Scholar]
- Karanika S., Karantanos T., Arvanitis M., Grigoras C., Mylonakis E. Fecal colonization with extended-spectrum beta-lactamase–producing enterobacteriaceae and risk factors among healthy individuals: a systematic review and metaanalysis. Clin. Infect. Dis. 2016;63:310–318. doi: 10.1093/cid/ciw283. [DOI] [PubMed] [Google Scholar]
- Karim A., Poirel L., Nagarajan S., Nordmann P. Plasmid-mediated extended-spectrum β-lactamase (CTX-M-3 like) from India and gene association with insertion sequence ISEcp1. FEMS (Fed. Eur. Microbiol. Soc.) Microbiol. Lett. 2001;201:237–241. doi: 10.1111/j.1574-6968.2001.tb10762.x. [DOI] [PubMed] [Google Scholar]
- Kim J.S., Park J., Shin E., Kim S., Oh S.S., Yang H.J., Kim D.W., Oh K.H., Kim Y., Kim M., Kwon M.J., Na K., Lee J., Cho E.H., Kang B.H., Kwak H.S., Seong W.K., Kim J. Outbreak of CTX-M-15-producing enterotoxigenic Escherichia coli O159:H20 in the Republic of Korea in 2016. Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.00339-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiratisin P., Apisarnthanarak A., Laesripa C., Saifon P. Molecular characterization and epidemiology of extended-spectrum- β-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolates causing health care-associated infection in Thailand, where the CTX-M family is endemic. Antimicrob. Agents Chemother. 2008;52:2818–2824. doi: 10.1128/AAC.00171-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjeldsen T.S.B., Overgaard M., Nielsen S.S., Bortolaia V., Jelsbak L., Sommer M., Guardabassi L., Olsen J.E. CTX-M-1 β-lactamase expression in Escherichia coli is dependent on cefotaxime concentration, growth phase and gene location. J. Antimicrob. Chemother. 2015;70:62–70. doi: 10.1093/jac/dku332. [DOI] [PubMed] [Google Scholar]
- Kmeť V., Bujňáková D. Antimicrobial resistance Escherichia coli isolated from calves. J. Microbiol. Biotechnol. Food Sci. 2018;7:412–415. [Google Scholar]
- Knothe H., Shah P., Krcmery V., Antal M., Mitsuhashi S. Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection. 1983;11:315–317. doi: 10.1007/BF01641355. [DOI] [PubMed] [Google Scholar]
- Koovapra S., Bandyopadhyay S., Das G., Bhattacharyya D., Banerjee J., Mahanti A., Samanta I., Nanda P.K., Kumar A., Mukherjee R., Dimri U., Singh R.K. Molecular signature of extended spectrum β-lactamase producing Klebsiella pneumoniae isolated from bovine milk in eastern and north-eastern India. Infect. Genet. Evol. 2016;44:395–402. doi: 10.1016/j.meegid.2016.07.032. [DOI] [PubMed] [Google Scholar]
- Kucukbasmaci O., Ciftcioglu G., Midilli K., Issa G. Detection of extended spectrum β-Lactamase producing Enterobacteriaceae from food animals in Turkey. Rev. Méd. Vét. 2008;159:586–592. [Google Scholar]
- Kuroda H., Yano H., Hirakata Y., Arai K., Endo S., Kanamori H., Yamamoto H., Ichimura S., Ogawa M., Shimojima M., Komatsu M., Jonai T., Itagaki S., Nonomiya Y., Suwabe A., Kaku M. Molecular characteristics of extended-spectrum β-lactamase-producing Escherichia coli in Japan: emergence of CTX-M-15-producing E. coli ST131. Diagn. Microbiol. Infect. Dis. 2012;74:201–203. doi: 10.1016/j.diagmicrobio.2012.06.011. [DOI] [PubMed] [Google Scholar]
- Li J.-J., Spychala C.N., Hu F., Sheng J.-F., Doi Y. Complete nucleotide sequences of blaCTX-M-harboring IncF plasmids from community-associated Escherichia coli strains in the United States. Antimicrob. Agents Chemother. 2015;59:3002–3007. doi: 10.1128/AAC.04772-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao K., Chen Y., Wang M., Guo P., Yang Q., Ni Y., Yu Y., Hu B., Sun Z., Huang W., Wang Y., Wu A., Feng X., Luo Y., Hu Z., Chu Y., Chen S., Cao B., Su J., Gui B., Duan Q., Zhang S., Shao H., Kong H., Xu Y. Molecular characteristics of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae causing intra-abdominal infections from 9 tertiary hospitals in China. Diagn. Microbiol. Infect. Dis. 2017;87:45–48. doi: 10.1016/j.diagmicrobio.2016.10.007. [DOI] [PubMed] [Google Scholar]
- Liebana E., Batchelor M., Hopkins K.L., Clifton-Hadley F.A., Teale C.J., Foster A., Barker L., Threlfall E.J., Davies R.H. Longitudinal farm study of extended-spectrum β-lactamase-mediated resistance. J. Clin. Microbiol. 2006;44:1630–1634. doi: 10.1128/JCM.44.5.1630-1634.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifshitz Z., Sturlesi N., Parizade M., Blum S.E., Gordon M., Taran D., Adler A. Distinctiveness and similarities between extended-spectrum beta-lactamase-producing Escherichia coli isolated from cattle and the community in Israel. Microb. Drug Resist. 2018 doi: 10.1089/mdr.2017.0407. [DOI] [PubMed] [Google Scholar]
- Lim S.-K., Lee H.-S., Nam H.-M., Jung S.-C., Bae Y.-c. CTX-M-Type β-lactamase in Escherichia coli isolated from sick animals in Korea. Microb. Drug Resist. 2009;15:139–142. doi: 10.1089/mdr.2009.0901. [DOI] [PubMed] [Google Scholar]
- Locatelli C., Scaccabarozzi L., Pisoni G., Moroni P. CTX-M1 ESBL-producing Klebsiella pneumoniae subsp. pneumoniae isolated from cases of bovine mastitis. J. Clin. Microbiol. 2010;48:3822–3823. doi: 10.1128/JCM.00941-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupo A., Saras E., Madec J.-Y., Haenni M. Emergence of blaCTX-M-55 associated with fosA, rmtB and mcr gene variants in Escherichia coli from various animal species in France. J. Antimicrob. Chemother. 2018:dkx489. doi: 10.1093/jac/dkx489. [DOI] [PubMed] [Google Scholar]
- Lussier P. University of Lethbridge; 2010. Characterization of Putative Extended-Spectrum β-lactamases (ESBL) Producing Escherichia coli Isolated from Feedlot Cattle in Southern Alberta. Master. [Google Scholar]
- Ma L., Ishii Y., Chang F.-Y., Yamaguchi K., Ho M., Siu L.K. CTX-M-14, a plasmid-mediated CTX-M type extended-spectrum β-lactamase isolated from Escherichia coli. Antimicrob. Agents Chemother. 2002;46:1985–1988. doi: 10.1128/AAC.46.6.1985-1988.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madec J.-Y., Lazizzera C., Châtre P., Meunier D., Martin S., Lepage G., Ménard M.-F., Lebreton P., Rambaud T. Prevalence of fecal carriage of acquired expanded-spectrum cephalosporin resistance in enterobacteriaceae strains from cattle in France. J. Clin. Microbiol. 2008;46:1566–1567. doi: 10.1128/JCM.02299-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madec J.-Y., Poirel L., Saras E., Gourguechon A., Girlich D., Nordmann P., Haenni M. Non-ST131 Escherichia coli from cattle harbouring human-like blaCTX-M-15-carrying plasmids. J. Antimicrob. Chemother. 2012;67:578–581. doi: 10.1093/jac/dkr542. [DOI] [PubMed] [Google Scholar]
- Madec J.Y., Haenni M., Nordmann P., Poirel L. Extended-spectrum beta-lactamase/AmpC- and carbapenemase-producing Enterobacteriaceae in animals: a threat for humans? Clin. Microbiol. infect. : Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2017 doi: 10.1016/j.cmi.2017.01.013. [DOI] [PubMed] [Google Scholar]
- Manyahi J., Moyo S.J., Tellevik M.G., Ndugulile F., Urassa W., Blomberg B., Langeland N. Detection of CTX-M-15 beta-lactamases in Enterobacteriaceae causing hospital- and community-acquired urinary tract infections as early as 2004, in Dar es Salaam, Tanzania. BMC Infect. Dis. 2017;17:282. doi: 10.1186/s12879-017-2395-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura Y., Johnson J.R., Yamamoto M., Nagao M., Tanaka M., Takakura S., Ichiyama S. CTX-M-27- and CTX-M-14-producing, ciprofloxacin-resistant Escherichia coli of the H30 subclonal group within ST131 drive a Japanese regional ESBL epidemic. J. Antimicrob. Chemother. 2015;70:1639–1649. doi: 10.1093/jac/dkv017. [DOI] [PubMed] [Google Scholar]
- Mendoza M.V. Universidad Peruana Cayetano Heredia; 2017. Identificación de enterobacterias resistentes a antibióticos en el vampiro común (Desmodus rotundus) y en animales de traspatio en el departamento de Lima, PerúBachelors. [Google Scholar]
- Meunier D., Jouy E., Lazizzera C., Kobisch M., Madec J.-Y. CTX-M-1- and CTX-M-15-type β-lactamases in clinical Escherichia coli isolates recovered from food-producing animals in France. Int. J. Antimicrob. Agents. 2006;28:402–407. doi: 10.1016/j.ijantimicag.2006.08.016. [DOI] [PubMed] [Google Scholar]
- Michael G.B., Kaspar H., Siqueira A.K., de Freitas Costa E., Corbellini L.G., Kadlec K., Schwarz S. Extended-spectrum β-lactamase (ESBL)-producing Escherichia coli isolates collected from diseased food-producing animals in the GERM-Vet monitoring program 2008–2014. Vet. Microbiol. 2017;200:142–150. doi: 10.1016/j.vetmic.2016.08.023. [DOI] [PubMed] [Google Scholar]
- Mir R.A., Weppelmann T.A., Johnson J.A., Archer D., Morris J.G., Jr., Jeong K.C. Identification and characterization of cefotaxime resistant bacteria in beef cattle. PLoS One. 2016;11 doi: 10.1371/journal.pone.0163279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mkala R.S., Azizi K. 2017. Prevalence and Antimicrobial Resistance Patterns of Extended Spectrum Beta Lactamase Producing Enterohemorrhagic Escherichia coli Strain O157:H7 from Cattle and Humans in Moshi, Northern Tanzania. [Google Scholar]
- Mollenkopf D.F., Weeman M.F., Daniels J.B., Abley M.J., Mathews J.L., Gebreyes W.A., Wittum T.E. Variable within- and between-herd diversity of CTX-M cephalosporinase-bearing Escherichia coli isolates from dairy cattle. Appl. Environ. Microbiol. 2012;78:4552–4560. doi: 10.1128/AEM.00373-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moosavian M., Ahmadkhosravy N. Survey of CTX-M gene frequency in extended-spectrum beta-lactamase-producing enterobacteriaceae isolates using the combination disk and PCR methods in Ahvaz, Iran. Jundishapur J. Microbiol. 2016;9 doi: 10.5812/jjm.40423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moubareck C., Daoud Z., Hakime N.I., Hamze M., Mangeney N., Matta H., Mokhbat J.E., Rohban R., Sarkis D.K., Doucet-Populaire F. Countrywide spread of community- and hospital-acquired extended-spectrum beta-lactamase (CTX-M-15)-producing Enterobacteriaceae in Lebanon. J. Clin. Microbiol. 2005;43:3309–3313. doi: 10.1128/JCM.43.7.3309-3313.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mshana S.E., Imirzalioglu C., Hossain H., Hain T., Domann E., Chakraborty T. Conjugative IncFI plasmids carrying CTX-M-15 among Escherichia coli ESBL producing isolates at a University hospital in Germany. BMC Infect. Dis. 2009;9:97. doi: 10.1186/1471-2334-9-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugnaioli C., Luzzaro F., De Luca F., Brigante G., Perilli M., Amicosante G., Stefani S., Toniolo A., Rossolini G.M. CTX-M-Type extended-spectrum β-lactamases in Italy: molecular epidemiology of an emerging countrywide problem. Antimicrob. Agents Chemother. 2006;50:2700–2706. doi: 10.1128/AAC.00068-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naiemi N., Bart A., de Jong M.D., Vandenbroucke-Grauls C.M., Rietra P.J.G.M., Debets-Ossenkopp Y.J., Wever P.C., Spanjaard L., Bos A.J., Duim B. Widely distributed and predominant CTX-M extended-spectrum β-lactamases in Amsterdam, The Netherlands. J. Clin. Microbiol. 2006;44:3012–3014. doi: 10.1128/JCM.01112-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namaei M.H., Yousefi M., Ziaee M., Salehabadi A., Ghannadkafi M., Amini E., Askari P. First report of prevalence of CTX-M-15-producing Escherichia coli O25b/ST131 from Iran. Microb. Drug Resist. 2017;23:879–884. doi: 10.1089/mdr.2016.0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naseer U., NatÅS O.B., Haldorsen B.C., Bue B., Grundt H., Walsh T.R., Sundsfjord A. Nosocomial outbreak of CTX-M-15-producing E. coli in Norway. APMIS. 2007;115:120–126. doi: 10.1111/j.1600-0463.2007.apm_547.x. [DOI] [PubMed] [Google Scholar]
- Newire E.A., Ahmed S.F., House B., Valiente E., Pimentel G. Detection of new SHV-12, SHV-5 and SHV-2a variants of extended spectrum beta-lactamase in Klebsiella pneumoniae in Egypt. Ann. Clin. Microbiol. Antimicrob. 2013;12:16. doi: 10.1186/1476-0711-12-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nóbrega D.B., Guiduce M.V.S., Guimarães F.F., Riboli D.F., Cunha M.L.R.S., Langoni H.l., Pantoja J.C.F., Lucheis S.B. Molecular epidemiology and extended-spectrum 2-lactamases production of Klebsiella pneumoniae isolated from three dairy herds. Pesqui. Vet. Bras. 2013;33:855–859. [Google Scholar]
- Novais Â., Cantón R., Moreira R., Peixe L., Baquero F., Coque T.M. Emergence and dissemination of enterobacteriaceae isolates producing CTX-M-1-like enzymes in Spain are associated with IncFII (CTX-M-15) and broad-host-range (CTX-M-1, -3, and -32) plasmids. Antimicrob. Agents Chemother. 2007;51:796–799. doi: 10.1128/AAC.01070-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogefere H.O., Agbe S.O., Ibadin E.E. Detection of extended spectrum beta-lactamases among gram negative bacilli recovered from cattle feces in Benin city, Nigeria. Not. Sci. Biol. 2017;9:177. [Google Scholar]
- Ohnishi M., Okatani A.T., Esaki H., Harada K., Sawada T., Murakami M., Marumo K., Kato Y., Sato R., Shimura K., Hatanaka N., Takahashi T. Herd prevalence of Enterobacteriaceae producing CTX-M-type and CMY-2 β-lactamases among Japanese dairy farms. J. Appl. Microbiol. 2013;115:282–289. doi: 10.1111/jam.12211. [DOI] [PubMed] [Google Scholar]
- Ohnishi M., Okatani A.T., Harada K., Sawada T., Marumo K., Murakami M., Sato R., Esaki H., Shimura K., Kato H., Uchida N., Takahashi T. Genetic characteristics of CTX-M-type extended-spectrum-β-lactamase (ESBL)-Producing enterobacteriaceae involved in mastitis cases on Japanese dairy farms, 2007 to 2011. J. Clin. Microbiol. 2013;51:3117–3122. doi: 10.1128/JCM.00920-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesen B., Hansen D.S., Nilsson F., Frimodt-Møller J., Leihof R.F., Struve C., Scheutz F., Johnston B., Krogfelt K.A., Johnson J.R. Prevalence and characteristics of the epidemic multiresistant Escherichia coli ST131 clonal group among extended-spectrum beta-lactamase-producing E. coli isolates in Copenhagen, Denmark. J. Clin. Microbiol. 2013;51:1779–1785. doi: 10.1128/JCM.00346-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olowe O.A., Adewumi O., Odewale G., Ojurongbe O., Adefioye O.J. Phenotypic and molecular characterisation of extended-spectrum beta-lactamase producing Escherichia coli obtained from animal fecal samples in Ado Ekiti, Nigeria. J. Environ. Public Health. 2015;2015:7. doi: 10.1155/2015/497980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oteo J., Orden B., Bautista V., Cuevas O., Arroyo M., Martinez-Ruiz R., Perez-Vazquez M., Alcaraz M., Garcia-Cobos S., Campos J. CTX-M-15-producing urinary Escherichia coli O25b-ST131-phylogroup B2 has acquired resistance to fosfomycin. J. Antimicrob. Chemother. 2009;64:712–717. doi: 10.1093/jac/dkp288. [DOI] [PubMed] [Google Scholar]
- Ouedraogo A.-S., Sanou M., Kissou A., Sanou S., Solaré H., Kaboré F., Poda A., Aberkane S., Bouzinbi N., Sano I., Nacro B., Sangaré L., Carrière C., Decré D., Ouégraogo R., Jean-Pierre H., Godreuil S. High prevalence of extended-spectrum ß-lactamase producing enterobacteriaceae among clinical isolates in Burkina Faso. BMC Infect. Dis. 2016;16:326. doi: 10.1186/s12879-016-1655-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overdevest I., Willemsen I., Rijnsburger M., Eustace A., Xu L., Hawkey P., Heck M., Savelkoul P., Vandenbroucke-Grauls C., van der Zwaluw K., Huijsdens X., Kluytmans J. Extended-spectrum β-lactamase genes of Escherichia coli in chicken meat and humans, The Netherlands. Emerg. Infect. Dis. 2011;17:1216–1222. doi: 10.3201/eid1707.110209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens R.C., Johnson J.R., Stogsdill P., Yarmus L., Lolans K., Quinn J. Community transmission in the United States of a CTX-M-15-producing sequence type ST131 Escherichia coli strain resulting in death. J. Clin. Microbiol. 2011;49:3406–3408. doi: 10.1128/JCM.00993-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Päivärinta M., Pohjola L., Fredriksson-Ahomaa M., Heikinheimo A. Low occurrence of extended-spectrum β-lactamase-Producing Escherichia coli in Finnish food-producing animals. Zoonoses and Public Hlth. 2016;63:624–631. doi: 10.1111/zph.12277. [DOI] [PubMed] [Google Scholar]
- Palmeira J.D., Ferreira H., Madec J.-Y., Haenni M. Draft genome of an ST443 mcr-1- and blaCTX-M-2-carrying Escherichia coli from cattle in Brazil. J. Glob. Antimicrob. Resist. 2018 doi: 10.1016/j.jgar.2018.05.010. [DOI] [PubMed] [Google Scholar]
- Pardon B., Smet A., Butaye P., Argudín M.A., Valgaeren B., Catry B., Haesebrouck F., Deprez P. Nosocomial intravascular catheter infections with extended-spectrum beta-lactamase-producing Escherichia coli in calves after strain introduction from a commercial herd. Transbound. Emerg. Dis. 2017;64:130–136. doi: 10.1111/tbed.12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson D.L., Bonomo R.A. Extended-spectrum β-lactamases: a clinical update. Clin. Microbiol. Rev. 2005;18:657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pehlivanoglu F., Turutoglu H., Ozturk D., Yardimci H. Acta Veterinaria. 2016. Molecular characterization of ESBL-producing Escherichia coli isolated from healthy cattle and sheep; p. 520. [Google Scholar]
- Peirano G., Richardson D., Nigrin J., McGeer A., Loo V., Toye B., Alfa M., Pienaar C., Kibsey P., Pitout J.D.D. High prevalence of ST131 isolates producing CTX-M-15 and CTX-M-14 among extended-spectrum-β-lactamase-producing Escherichia coli isolates from Canada. Antimicrob. Agents Chemother. 2010;54:1327–1330. doi: 10.1128/AAC.01338-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirano G., van Greune C.H., Pitout J.D. Characteristics of infections caused by extended-spectrum beta-lactamase-producing Escherichia coli from community hospitals in South Africa. Diagn. Microbiol. Infect. Dis. 2011;69:449–453. doi: 10.1016/j.diagmicrobio.2010.11.011. [DOI] [PubMed] [Google Scholar]
- Perilli M., Segatore B., Mugnaioli C., Celenza G., Rossolini G.M., Stefani S., Luzzaro F., Pini B., Amicosante G. Persistence of TEM-52/TEM-92 and SHV-12 extended-spectrum beta-lactamases in clinical isolates of Enterobacteriaceae in Italy. Microb. Drug Resist. 2011;17:521–524. doi: 10.1089/mdr.2011.0059. [DOI] [PubMed] [Google Scholar]
- Pitout J.D.D., Gregson D.B., Church D.L., Elsayed S., Laupland K.B. Community-wide outbreaks of clonally related CTX-M-14 β-lactamase-producing Escherichia coli strains in the calgary health region. J. Clin. Microbiol. 2005;43:2844–2849. doi: 10.1128/JCM.43.6.2844-2849.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitout J.D.D., Le P., Church D.L., Gregson D.B., Laupland K.B. Antimicrobial susceptibility of well-characterised multiresistant CTX-M-producing Escherichia coli: failure of automated systems to detect resistance to piperacillin/tazobactam. Int. J. Antimicrob. Agents. 2008;32:333–338. doi: 10.1016/j.ijantimicag.2008.04.023. [DOI] [PubMed] [Google Scholar]
- Poirel L., Nordmann P., Ducroz S., Boulouis H.-J., Arné P., Millemann Y. Extended-spectrum β-lactamase CTX-M-15-producing Klebsiella pneumoniae of sequence type ST274 in companion animals. Antimicrob. Agents Chemother. 2013;57:2372–2375. doi: 10.1128/AAC.02622-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole T.L., Callaway T.R., Norman K.N., Scott H.M., Loneragan G.H., Ison S.A., Beier R.C., Harhay D.M., Norby B., Nisbet D.J. Transferability of antimicrobial resistance from multidrug-resistant Escherichia coli isolated from cattle in the United States to Escherichia coli and Salmonella Newport recipients. J. Glob. Antimicrob. Resist. 2017 doi: 10.1016/j.jgar.2017.08.001. [DOI] [PubMed] [Google Scholar]
- Price L.B., Johnson J.R., Aziz M., Clabots C., Johnston B., Tchesnokova V., Nordstrom L., Billig M., Chattopadhyay S., Stegger M., Andersen P.S., Pearson T., Riddell K., Rogers P., Scholes D., Kahl B., Keim P., Sokurenko E.V. The epidemic of extended-spectrum-β-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-Rx. mBio. 2013;4 doi: 10.1128/mBio.00377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos S., Igrejas G., Silva N., Jones-Dias D., Capelo-Martinez J.-L., Caniça M., Poeta P. First report of CTX-M producing Escherichia coli, including the new ST2526, isolated from beef cattle and sheep in Portugal. Food Control. 2013;31:208–210. [Google Scholar]
- Randall L., Heinrich K., Horton R., Brunton L., Sharman M., Bailey-Horne V., Sharma M., McLaren I., Coldham N., Teale C., Jones J. Detection of antibiotic residues and association of cefquinome residues with the occurrence of Extended-Spectrum β-Lactamase (ESBL)-producing bacteria in waste milk samples from dairy farms in England and Wales in 2011. Res. Vet. Sci. 2014;96:15–24. doi: 10.1016/j.rvsc.2013.10.009. [DOI] [PubMed] [Google Scholar]
- Rayamajhi N., Cha S.B., Shin S.W., Jung B.Y., Lim S.-K., Yoo H.S. Plasmid typing and resistance profiling of Escherichia fergusonii and other enterobacteriaceae isolates from south Korean farm animals. Appl. Environ. Microbiol. 2011;77:3163–3166. doi: 10.1128/AEM.02188-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reist M., Geser N., Hächler H., Schärrer S., Stephan R. ESBL-producing enterobacteriaceae: occurrence, risk factors for fecal carriage and strain traits in the Swiss slaughter cattle population younger than 2 Years sampled at Abattoir level. PLoS One. 2013;8 doi: 10.1371/journal.pone.0071725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin F., Beyrouthy R., Bonacorsi S., Aissa N., Bret L., Brieu N., Cattoir V., Chapuis A., Chardon H., Degand N., Doucet-Populaire F., Dubois V., Fortineau N., Grillon A., Lanotte P., Leyssene D., Patry I., Podglajen I., Recule C., Ros A., Colomb-Cotinat M., Ponties V., Ploy M.C., Bonnet R. Inventory of extended-spectrum-β-lactamase-producing enterobacteriaceae in France as assessed by a multicenter study. Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.01911-16. e01911-01916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson T.P., Bu D.P., Carrique-Mas J., Fèvre E.M., Gilbert M., Grace D., Hay S.I., Jiwakanon J., Kakkar M., Kariuki S., Laxminarayan R., Lubroth J., Magnusson U., Thi Ngoc P., Van Boeckel T.P., Woolhouse M.E.J. Antibiotic resistance is the quintessential One Health issue. Trans. R. Soc. Trop. Med. Hyg. 2016;110:377–380. doi: 10.1093/trstmh/trw048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez I., Barownick W., Helmuth R., Mendoza M.C., Rodicio M.R., Schroeter A., Guerra B. Extended-spectrum β-lactamases and AmpC β-lactamases in ceftiofur-resistant Salmonella enterica isolates from food and livestock obtained in Germany during 2003–07. J. Antimicrob. Chemother. 2009;64:301–309. doi: 10.1093/jac/dkp195. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Villalobos H., Bogaerts P., Berhin C., Bauraing C., Deplano A., Montesinos I., de Mendonça R., Jans B., Glupczynski Y. Trends in production of extended-spectrum β-lactamases among Enterobacteriaceae of clinical interest: results of a nationwide survey in Belgian hospitals. J. Antimicrob. Chemother. 2011;66:37–47. doi: 10.1093/jac/dkq388. [DOI] [PubMed] [Google Scholar]
- Ruiz S.J., Montealegre M.C., Ruiz-Garbajosa P., Correa A., Briceño D.F., Martinez E., Rosso F., Muñoz M., Quinn J.P., Cantón R., Villegas M.V. First characterization of CTX-M-15-producing Escherichia coli ST131 and ST405 clones causing community-onset infections in South America. J. Clin. Microbiol. 2011;49:1993–1996. doi: 10.1128/JCM.00045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saidani M., Messadi L., Soudani A., Daaloul-Jedidi M., Chatre P., Ben Chehida F., Mamlouk A., Mahjoub W., Madec J.Y., Haenni M. Epidemiology, antimicrobial resistance, and extended-spectrum beta-lactamase-producing enterobacteriaceae in clinical bovine mastitis in Tunisia. Microb. Drug Resist. 2018 doi: 10.1089/mdr.2018.0049. [DOI] [PubMed] [Google Scholar]
- Saishu N., Ozaki H., Murase T. CTX-M-Type extended-spectrum β-lactamase-producing Klebsiella pneumoniae isolated from cases of bovine mastitis in Japan. J. Vet. Med. Sci. 2014;76:1153–1156. doi: 10.1292/jvms.13-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito R., Takahashi R., Sawabe E., Koyano S., Takahashi Y., Shima M., Ushizawa H., Fujie T., Tosaka N., Kato Y., Moriya K., Tohda S., Tojo N., Koike R., Kubota T. First report of KPC-2 carbapenemase-producing Klebsiella pneumoniae in Japan. Antimicrob. Agents Chemother. 2014;58:2961–2963. doi: 10.1128/AAC.02072-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampaio J.L.M., Gales A.C. Antimicrobial resistance in Enterobacteriaceae in Brazil: focus on 2-lactams and polymyxins. Braz. J. Microbiol. 2016;47:31–37. doi: 10.1016/j.bjm.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos C.D.M. Federal University of Uberlândia; 2006. Staphylococcus sp e enterobactérias isoladas de mastite recorrente em oito rebanhos da região de Uberlândia-MG : perfil de suscetibilidade aos antimicrobianos. Master. [Google Scholar]
- Sartori L., Fernandes M.R., Ienne S., de Souza T.A., Gregory L., Cerdeira L., Lincopan N. Draft genome sequences of two fluoroquinolone-resistant CTX-M-15-producing Escherichia coli ST90 (ST23 complex) isolated from a calf and a dairy cow in South America. J. Glob. Antimicrob. Resist. 2017;11:145–147. doi: 10.1016/j.jgar.2017.10.009. [DOI] [PubMed] [Google Scholar]
- Schmitt J., Jacobs E., Schmidt H. Molecular characterization of extended-spectrum beta-lactamases in Enterobacteriaceae from patients of two hospitals in Saxony, Germany. J. Med. Microbiol. 2007;56:241–249. doi: 10.1099/jmm.0.46670-0. [DOI] [PubMed] [Google Scholar]
- Seni J., Falgenhauer L., Simeo N., Mirambo M.M., Imirzalioglu C., Matee M., Rweyemamu M., Chakraborty T., Mshana S.E. Multiple ESBL-producing Escherichia coli sequence types carrying quinolone and aminoglycoside resistance genes circulating in companion and domestic farm animals in Mwanza, Tanzania, harbor commonly occurring plasmids. Front. Microbiol. 2016;7:142. doi: 10.3389/fmicb.2016.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Kim D.H., Ko K.S. Comparison of CTX-M-14- and CTX-M-15-producing Escherichia coli and Klebsiella pneumoniae isolates from patients with bacteremia. J. Infect. 2011;63:39–47. doi: 10.1016/j.jinf.2011.05.003. [DOI] [PubMed] [Google Scholar]
- Shiraki Y., Shibata N., Doi Y., Arakawa Y. Escherichia coli Producing CTX-M-2 β-Lactamase in Cattle, Japan. Emerg. Infect. Dis. J. 2004;10:69. doi: 10.3201/eid1001.030219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidjabat H.E., Derrington P., Nimmo G.R., Paterson D.L. Escherichia coli ST131 producing CTX-M-15 in Australia. J. Antimicrob. Chemother. 2010;65:1301–1303. doi: 10.1093/jac/dkq098. [DOI] [PubMed] [Google Scholar]
- Silva K.C.d., Lincopan N. Epidemiologia das betalactamases de espectro estendido no Brasil: impacto clínico e implicações para o agronegócio. J. Bras. Patol. Med. Lab. 2012;48:91–99. [Google Scholar]
- Smith K.A., Williams A.G. Production and management of cattle manure in the UK and implications for land application practice. Soil Use Manag. 2016;32:73–82. [Google Scholar]
- Snow L.C., Wearing H., Stephenson B., Teale C.J., Coldham N.G. Investigation of the presence of ESBL-producing Escherichia coli in the North Wales and West Midlands areas of the UK in 2007 to 2008 using scanning surveillance. Vet. Rec. 2011;169:656. doi: 10.1136/vr.100037. [DOI] [PubMed] [Google Scholar]
- Sparham S.J., Kwong J.C., Valcanis M., Easton M., Trott D.J., Seemann T., Stinear T.P., Howden B.P. Emergence of multidrug resistance in locally-acquired human infections with Salmonella Typhimurium in Australia owing to a new clade harbouring blaCTX-M-9. Int. J. Antimicrob. Agents. 2017;50:101–105. doi: 10.1016/j.ijantimicag.2017.02.014. [DOI] [PubMed] [Google Scholar]
- Stefani S., Giovanelli I., Anacarso I., Condo C., Messi P., de Niederhausern S., Bondi M., Iseppi R., Sabia C. Prevalence and characterization of extended-spectrum beta-lactamase-producing Enterobacteriaceae in food-producing animals in Northern Italy. New Microbiol. 2014;37:551–555. [PubMed] [Google Scholar]
- Stokes M.O. Kingston University; 2014. Comparative Genomics of blaCTX-M Plasmids from Veterinary and Human ' Escherichia coli ' and Methods for Their Identification and Differentiation. PhD. [Google Scholar]
- Stokes M.O., AbuOun M., Umur S., Wu G., Partridge S.R., Mevius D.J., Coldham N.G., Fielder M.D. Complete sequence of pSAM7, an IncX4 plasmid carrying a novel blaCTX-M-14b transposition unit isolated from Escherichia coli and Enterobacter cloacae from cattle. Antimicrob. Agents Chemother. 2013;57:4590–4594. doi: 10.1128/AAC.01157-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., Yu C.-Y., Tsai Y., Wang S.-H., Lee C., Chu C. Fluoroquinolone-resistant and extended-spectrum β-lactamase-producing Escherichia coli from the milk of cows with clinical mastitis in Southern Taiwan. J. Microbiol. Immunol. Infect. 2016;49:892–901. doi: 10.1016/j.jmii.2014.10.003. [DOI] [PubMed] [Google Scholar]
- Sudarwanto M.B., Lukman D.W., Latif H., Pisestyani H., Sukmawinata E., Akineden Ö., Usleber E. CTX-M producing Escherichia coli isolated from cattle feces in Bogor slaughterhouse, Indonesia. Asian Pac. J. Trop. Biomed. 2016;6:605–608. [Google Scholar]
- Tadesse D.A., Li C., Mukherjee S., Hsu C.-H., Bodeis J.S., Gaines S.A., Kabera C., Loneragan G.H., Torrence M., Harhay D.M., McDermott P.F., Zhao S. Whole-Genome sequence analysis of CTX-M containing Escherichia coli isolates from retail meats and cattle in the United States. Microb. Drug Resist. 2018;24:939–948. doi: 10.1089/mdr.2018.0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamang M.D., Nam H.-M., Gurung M., Jang G.-C., Kim S.-R., Jung S.-C., Park Y.H., Lim S.-K. Molecular characterization of CTX-M β-lactamase and associated addiction systems in Escherichia coli circulating among cattle, farm workers, and the farm environment. Appl. Environ. Microbiol. 2013;79:3898–3905. doi: 10.1128/AEM.00522-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamang M.D., Nam H.M., Kim S.R., Chae M.H., Jang G.C., Jung S.C., Lim S.K. Prevalence and molecular characterization of CTX-M beta-lactamase-producing Escherichia coli isolated from healthy swine and cattle. Foodb. Pathog. Dis. 2013;10:13–20. doi: 10.1089/fpd.2012.1245. [DOI] [PubMed] [Google Scholar]
- Tark D.-S., Moon D.C., Kang H.Y., Kim S.-R., Nam H.-M., Lee H.-S., Jung S.-C., Lim S.-K. Antimicrobial susceptibility and characterization of extended-spectrum β-lactamases in Escherichia coli isolated from bovine mastitic milk in South Korea from 2012 to 2015. J. Dairy Sci. 2017;100:3463–3469. doi: 10.3168/jds.2016-12276. [DOI] [PubMed] [Google Scholar]
- Tasli H., Bahar I.H. Molecular characterization of TEM- and SHV-derived extended-spectrum beta-lactamases in hospital-based Enterobacteriaceae in Turkey. Jpn. J. Infect. Dis. 2005;58:162–167. [PubMed] [Google Scholar]
- Tate H., Folster J.P., Hsu C.H., Chen J., Hoffmann M., Li C., Morales C., Tyson G.H., Mukherjee S., Brown A.C., Green A., Wilson W., Dessai U., Abbott J., Joseph L., Haro J., Ayers S., McDermott P.F., Zhao S. Comparative analysis of extended-spectrum-beta-lactamase CTX-M-65-producing Salmonella enterica serovar infantis isolates from humans, food animals, and retail chickens in the United States. Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.00488-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teale C.J., Barker L., Foster A.P., Liebana E., Batchelor M., Livermore D.M., Threlfall E.J. Extended-spectrum beta-lactamase detected in E coli recovered from calves in Wales. Vet. Rec. 2005;156:186–187. [PubMed] [Google Scholar]
- Tian G.-B., Wang H.-N., Zhang A.-Y., Zhang Y., Fan W.-Q., Xu C.-W., Zeng B., Guan Z.-B., Zou L.-K. Detection of clinically important β-lactamases in commensal Escherichia coli of human and swine origin in western China. J. Med. Microbiol. 2012;61:233–238. doi: 10.1099/jmm.0.036806-0. [DOI] [PubMed] [Google Scholar]
- Timofte D., Maciuca I.E., Evans N.J., Williams H., Wattret A., Fick J.C., Williams N.J. Detection and molecular characterization of Escherichia coli CTX-M-15 and Klebsiella pneumoniae SHV-12 β-lactamases from bovine mastitis isolates in the United Kingdom. Antimicrob. Agents Chemother. 2014;58:789–794. doi: 10.1128/AAC.00752-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth A., Juhasz-Kaszanyitzky E., Mag T., Hajbel-Vekony G., Paszti J., Damjanova I. Characterization of extended-spectrum beta-lactamase (ESBL) producing Escherichia coli strains isolated from animal and human clinical samples in Hungary in 2006-2007. Acta Microbiol. Immunol. Hung. 2013;60:175–185. doi: 10.1556/AMicr.60.2013.2.8. [DOI] [PubMed] [Google Scholar]
- Tyrrell J.M., Wootton M., Toleman M.A., Howe R.A., Woodward M., Walsh T.R. Genetic & virulence profiling of ESBL-positive E. coli from nosocomial & veterinary sources. Vet. Microbiol. 2016;186:37–43. doi: 10.1016/j.vetmic.2016.02.007. [DOI] [PubMed] [Google Scholar]
- USDA, United, State, Departament, Agriculture, o. 2017. Livestock and Poultry: World Markets and Trade. [Google Scholar]
- Usui M., Iwasa T., Fukuda A., Sato T., Okubo T., Tamura Y. The role of flies in spreading the extended-spectrum beta-lactamase gene from cattle. Microb. Drug Resist. 2013;19:415–420. doi: 10.1089/mdr.2012.0251. [DOI] [PubMed] [Google Scholar]
- Valat C., Auvray F., Forest K., Métayer V., Gay E., Peytavin de Garam C., Madec J.-Y., Haenni M. Phylogenetic grouping and virulence potential of extended-spectrum-β-lactamase-producing Escherichia coli strains in cattle. Appl. Environ. Microbiol. 2012;78:4677–4682. doi: 10.1128/AEM.00351-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valat C., Goldstone R.J., Hirchaud E., Haenni M., Smith D.G.E., Madec J.-Y. Draft genome sequences of enterohemorrhagic Escherichia coli encoding extended-spectrum beta-lactamases. Genome Announc. 2016;4 doi: 10.1128/genomeA.01633-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasova M., Smith R.P., Lemma F., Horton R.A., Duggett N.A., Evans J., Tongue S.C., Anjum M.F., Randall L.P. Detection of extended-spectrum β-lactam, AmpC and carbapenem resistance in Enterobacteriaceae in beef cattle in Great Britain in 2015. J. Appl. Microbiol. 2019;126:1081–1095. doi: 10.1111/jam.14211. [DOI] [PubMed] [Google Scholar]
- Wang G., Huang T., Surendraiah P.K.M., Wang K., Komal R., Zhuge J., Chern C.-R., Kryszuk A.A., King C., Wormser G.P. CTX-M β-Lactamase–producing Klebsiella pneumoniae in Suburban New York city, New York, USA. Emerg. Infect. Dis. 2013;19:1803–1810. doi: 10.3201/eid1911.121470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson E., Jeckel S., Snow L., Stubbs R., Teale C., Wearing H., Horton R., Toszeghy M., Tearne O., Ellis-Iversen J., Coldham N. Epidemiology of extended spectrum beta-lactamase E. coli (CTX-M-15) on a commercial dairy farm. Vet. Microbiol. 2012;154:339–346. doi: 10.1016/j.vetmic.2011.07.020. [DOI] [PubMed] [Google Scholar]
- Wieler L.H., Semmler T., Eichhorn I., Antao E.M., Kinnemann B., Geue L., Karch H., Guenther S., Bethe A. No evidence of the Shiga toxin-producing E. coli O104:H4 outbreak strain or enteroaggregative E. coli (EAEC) found in cattle faeces in northern Germany, the hotspot of the 2011 HUS outbreak area. Gut Pathog. 2011;3:17. doi: 10.1186/1757-4749-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittum T.E., Mollenkopf D.F., Daniels J.B., Parkinson A.E., Mathews J.L., Fry P.R., Abley M.J., Gebreyes W.A. CTX-M-type extended-spectrum beta-lactamases present in Escherichia coli from the feces of cattle in Ohio, United States. Foodb. Pathog. Dis. 2010;7:1575–1579. doi: 10.1089/fpd.2010.0615. [DOI] [PubMed] [Google Scholar]
- Woodford N., Ward M.E., Kaufmann M.E., Turton J., Fagan E.J., James D., Johnson A.P., Pike R., Warner M., Cheasty T., Pearson A., Harry S., Leach J.B., Loughrey A., Lowes J.A., Warren R.E., Livermore D.M. Community and hospital spread of Escherichia coli producing CTX-M extended-spectrum beta-lactamases in the UK. J. Antimicrob. Chemother. 2004;54:735–743. doi: 10.1093/jac/dkh424. [DOI] [PubMed] [Google Scholar]
- Wu G., Day M.J., Mafura M.T., Nunez-Garcia J., Fenner J.J., Sharma M., van Essen-Zandbergen A., Rodríguez I., Dierikx C., Kadlec K., Schink A.-K., Wain J., Helmuth R., Guerra B., Schwarz S., Threlfall J., Woodward M.J., Woodford N., Coldham N., Mevius D. Comparative analysis of ESBL-positive Escherichia coli isolates from animals and humans from the UK, The Netherlands and Germany. PLoS One. 2013;8 doi: 10.1371/journal.pone.0075392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J.-J., Hsueh P.-R., Lu J.-J., Chang F.-Y., Shyr J.-M., Wan J.-H., Liu Y.-C., Chuang Y.-C., Yang Y.-C., Tsao S.-M., Wu H.-H., Wang L.-S., Lin T.-P., Wu H.-M., Chen H.-M., Wu J.-J. Extended-spectrum β-lactamases and plasmid-mediated AmpC enzymes among clinical isolates of Escherichia coli and Klebsiella pneumoniae from seven medical centers in Taiwan. Antimicrob. Agents Chemother. 2006;50:1861–1864. doi: 10.1128/AAC.50.5.1861-1864.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Zhang S., Shang X., Wang L., Li H., Wang X. Characteristics of quinolone-resistant Escherichia coli isolated from bovine mastitis in China. J. Dairy Sci. 2018 doi: 10.3168/jds.2017-14156. [DOI] [PubMed] [Google Scholar]
- Yeung M.-k. University of Hong Kong; 2011. Epidemiology of CTX-M Type Extended-Spectrum Beta-Lactamaseproducing escherichia Coli Among Blood Culture Isolates in Hong Kong. Master. [Google Scholar]
- Zheng H., Zeng Z., Chen S., Liu Y., Yao Q., Deng Y., Chen X., Lv L., Zhuo C., Chen Z., Liu J.-H. Prevalence and characterisation of CTX-M β-lactamases amongst Escherichia coli isolates from healthy food animals in China. Int. J. Antimicrob. Agents. 2012;39:305–310. doi: 10.1016/j.ijantimicag.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Zheng B., Feng C., Xu H., Yu X., Guo L., Jiang X., Song X. Detection and characterization of ESBL-producing Escherichia coli expressing mcr-1 from dairy cows in China. J. Antimicrob. Chemother. 2019;74:321–325. doi: 10.1093/jac/dky446. [DOI] [PubMed] [Google Scholar]
- Zhong Y.-M., Liu W.-E., Liang X.-H., Li Y.-M., Jian Z.-J., Hawkey P.M. Emergence and spread of O16-ST131 and O25b-ST131 clones among faecal CTX-M-producing Escherichia coli in healthy individuals in Hunan Province, China. J. Antimicrob. Chemother. 2015;70:2223–2227. doi: 10.1093/jac/dkv114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong Z., Partridge S.R., Thomas L., Iredell J.R. Dominance of bla(CTX-M) within an Australian extended-spectrum β-lactamase gene pool. Antimicrob. Agents Chemother. 2008;52:4198–4202. doi: 10.1128/AAC.00107-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurfluh K., Cernela N., Stephan R. Quinolone resistance mechanisms among extended-spectrum beta-lacta- mase (ESBL)-producing Escherichia coli isolated from farm animals in Switzerland. Schweizer Archiv fur Tierheilkunde. 2015;157:59–62. doi: 10.17236/sat00006. [DOI] [PubMed] [Google Scholar]