Abstract
Precise quantification of the energetics and interactions that stabilize membrane proteins in a lipid bilayer is a long-sought goal. Toward this end, atomic force microscopy has been used to unfold individual membrane proteins embedded in their native lipid bilayer, typically by retracting the cantilever at a constant velocity. Recently, unfolding intermediates separated by as few as two amino acids were detected using focused-ion-beam-modified ultrashort cantilevers. However, unambiguously discriminating between such closely spaced states remains challenging, in part because any individual unfolding trajectory only occupies a subset of the total number of intermediates. Moreover, structural assignment of these intermediates via worm-like-chain analysis is hindered by brief dwell times compounded with thermal and instrumental noise. To overcome these issues, we moved the cantilever in a sawtooth pattern of 6–12 nm, offset by 0.25–1 nm per cycle, generating a “zigzag” force ramp of alternating positive and negative loading rates. We applied this protocol to the model membrane protein bacteriorhodopsin (bR). In contrast to conventional studies that extract bR’s photoactive retinal along with the first transmembrane helix, we unfolded bR in the presence of its retinal. To do so, we introduced a previously developed enzymatic-cleavage site between helices E and F and pulled from the top of the E helix using a site-specific, covalent attachment. The resulting zigzag unfolding trajectories occupied 40% more states per trajectory and occupied those states for longer times than traditional constant-velocity records. In total, we identified 31 intermediates during the unfolding of five helices of EF-cleaved bR. These included a previously reported, mechanically robust intermediate located between helices C and B that, with our enhanced resolution, is now shown to be two distinct states separated by three amino acids. Interestingly, another intermediate directly interacted with the retinal, an interaction confirmed by removing the retinal.
Significance
For almost two decades, atomic force microscopy has been a valuable tool to probe the energetics and stabilizing interactions of membrane proteins by mechanically unfolding individual proteins from lipid bilayers. Recent advances in spatiotemporal resolution of atomic force microscopy have enabled resolving tens of metastable unfolding intermediates, with some separated by as few as two amino acids. This rich unfolding pathway creates the challenge of reliably localizing stabilizing interactions to individual amino acids. Here, we developed an improved data acquisition protocol that solves this problem. Applied to the model membrane protein bacteriorhodopsin, we used this protocol to localize an interaction to a residue contacting the photoactive retinal and validated this assignment by removing the retinal.
Introduction
Membrane proteins perform key biological functions, including signaling and transport. Recent advances in cryo-electron tomography have accelerated the structural characterization of membrane proteins (1, 2, 3). Yet, there remains a longstanding need to better quantify the forces and interactions that stabilize folded membrane proteins (4, 5, 6). One valuable approach uses atomic force microscopy (AFM) to unfold individual membrane proteins embedded in lipid bilayers (Fig. 1 A; (7,8)). Such unfolding assays avoid the thermodynamically ambiguous denatured state present in chemical denaturation experiments (9) and the confounding structural effects of detergent solubilization (10). Mechanical unfolding proceeds via a series of abrupt drops in force, corresponding to transitions between partially unfolded, structurally stable states (Fig. 1 B, black). Failure to discriminate between closely spaced, transiently occupied states leads to multiple complications. It obscures the underlying free-energy landscape (11), leads to anomalous composite states that exhibit corrupted kinetic properties (12), and incorrectly assigns stabilizing interactions. Hence, precise discrimination between such intermediate states requires an AFM-based assay that provides high force precision and temporal resolution.
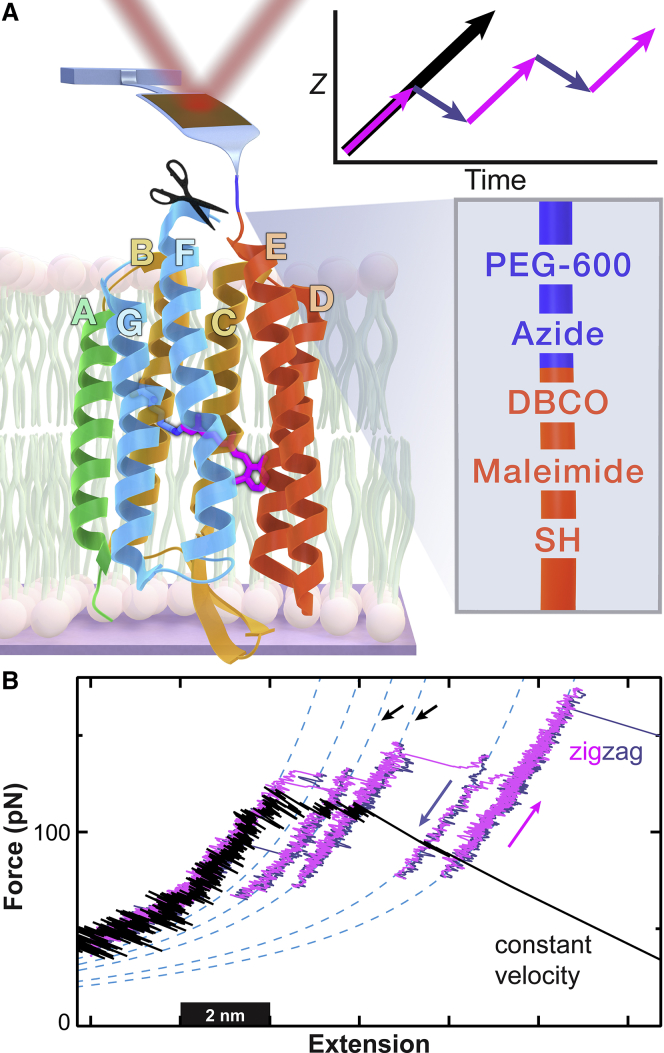
Figure 1.
Unfolding bacteriorhodopsin (bR) via a zigzag protocol enhances state detection. (A) An illustration shows a modified AFM cantilever unfolding bR. To unfold bR starting from helix E, a cysteine was introduced within an expanded EF loop containing a protease cleavage site (scissors). This pulling geometry allowed helices E–A to be extracted while maintaining the GF helix pair and the associated photoactive retinal in the lipid bilayer. (Lower inset) A scheme depicts site-specific attachment of DBCO-labeled bR to an azide-functionalized, PEG-coated AFM tip. DBCO is a copper-free click chemistry reagent. (Upper inset) A position-versus-time sketch shows the position of the base of the cantilever in the traditional constant velocity assay (black arrow) and the zigzag protocol (magenta and blue arrows). (B) Force-extension curves show the unfolding of the ED helix pair via constant-velocity retraction (black) and the zigzag force ramp consisting of alternating segments of constant-velocity retraction and approach (magenta and blue, respectively). Segments of the force-extension curves were well described by a WLC model (cyan) and corresponded to the stretching of the unfolded portion of bR while the rest of the bR remained folded in the lipid bilayer. The two states indicated by black arrows are present in both curves but in the zigzag protocol were sampled over a sevenfold larger force range. The zigzag protocol also led to more states being occupied. Data were smoothed to 1 kHz.
Focused-ion-beam-modified cantilevers provide both enhanced force precision and temporal resolution (13, 14, 15). Modified ultrashort (L = 9 μm) cantilevers, for example, provide a 10-fold higher force precision and a 100-fold higher temporal resolution (14) compared with traditional AFM-based studies of membrane proteins (8). When these cantilevers were applied to pulling on the model membrane protein bacteriorhodopsin (bR) from its C-terminal G helix, three “major states” corresponding to pulling on the top of a transmembrane helix (i.e., E, C, and A helices) were well resolved (11). Moreover, 19 previously undetected intermediates were resolved, including states separated by as few as two amino acids (aa) and states occupied for as little as 8 μs. Here, we define an “intermediate” as a state observed during the partial unfolding of transmembrane helix pairs or of the terminal helix.
Nonetheless, it remains challenging to unambiguously distinguish and structurally assign this multitude of closely spaced states, especially because any individual unfolding trajectory only populates a subset of the total number of intermediates. To explain more fully, we review the canonical AFM assay for unfolding bR as it has typically been implemented (7,8). Such an assay is initiated by pressing the tip of an AFM cantilever into bR embedded in its native lipid bilayer (i.e., purple membrane) at high force (∼1 nN) to promote nonspecific attachment. The cantilever is then retracted away from the surface at constant velocity while measuring force via cantilever deflection. The resulting force-extension curves show mechanically stable major states followed by a set of sparsely populated intermediates, as seen in constant-velocity data we collected using site-specific attachment and a lower tip-sample contact force (100 pN) (Fig. 1 B, black). Segments of force-extension curves that are well described by a worm-like-chain (WLC) model represent individual structural states (Fig. 1 B, cyan curves). Fitting WLC curves to such segments yields the change in contour length (ΔLo) between states, typically computed from the preceding major state. This ΔLo is then converted to the number of amino acids unfolded based on the elastic properties of the taut unfolded polypeptide chain and the known structure of bR (7,8).
Biophysical conclusions are enhanced by assigning structural states with high precision, ideally 1 aa. For instance, an imprecision of 2 aa precludes determining which side of an α-helical turn is involved in a stabilizing interaction. High-precision determination of ΔLo is challenging to achieve in unfolding trajectories that only briefly occupy a certain state. This effect is particularly pronounced for the partial helix unwinding intermediates, which rupture at lower forces than the major states at the top of each helix (8,16). As a result, only a fraction of unfolding trajectories populate these nonobligate unfolding intermediates. Moreover, only a small portion of the arc length of the WLC curve corresponding to such states is sampled experimentally because of brief state occupancy (Fig. 1 B, black). Because of this limited sampling, discrimination between states separated by a few amino acids is hampered by thermal and instrumental noise.
The precision of structural assignment increases when individual unfolding trajectories occupy adjacent states and when state occupancy times are extended. Occupancy of adjacent states increases confidence that the states are truly distinct and are not erroneous manifestations of variable pulling geometry between separate trajectories or of instrumental noise. Longer occupancy times better define the unique WLC curve associated with a particular state, allowing for a more precise determination of ΔLo.
Here, we improved upon conventional constant-velocity experiments by implementing a nonmonotonic “zigzag” motion of the cantilever with alternating positive and negative loading rate (∂F/∂t). Whereas sawtooth-like motions of the cantilever have been used previously to promote refolding of globular protein domains (17) and of pairs of transmembrane helices (18), the scale of the cantilever motion in this study was finer, and the pattern was chosen to promote enhanced detection of bR-unfolding intermediates (Fig. 1 A, upper inset). The resulting individual zigzag records exhibited longer state lifetimes and occupied more distinct states (Fig. 1 B, magenta-blue zigzag versus black constant-velocity curves). In a particularly illustrative example, two intermediates separated by 3 aa showed 95-fold longer lifetimes and spanned a sevenfold larger force range when using the zigzag pulling protocol in comparison to a conventional constant-velocity unfolding record (Fig. 1 B, black arrows).
To demonstrate the utility of this zigzag protocol, we revisited the unfolding of bR by pulling on the cytoplasmic side of the E helix after cleaving the loop between the E and F helices (7). This pulling geometry was first implemented in the pioneering bR-unfolding experiments of Oesterhelt et al. to help establish that the mechanical unfolding of membrane proteins occurs via pairwise extraction of transmembrane helices (7). In this work, this configuration allowed us to unfold helices E through A in the presence of the GF helix pair and bR’s photoactive retinal. This experiment was motivated by a recent AFM study that demonstrated that the retinal attached to helix G at Lys216 led to the most mechanically stable state in bR’s unfolding pathway and that removal of the retinal decreased the unfolding force by ∼2-fold (19). To maintain tip-sample attachment at high forces over the extended duration of the zigzag protocol (2–5 s vs. ∼0.2 s in standard constant-velocity assays), we covalently attached the bR to a modified ultrashort AFM cantilever using a recently developed attachment scheme involving a comparatively gentle tip-sample contact force (100 pN) (Fig. 1 A, lower inset) (19). The integrated application of this mechanically robust attachment with the zigzag protocol allowed us to identify 31 intermediates in the unfolding of EF-cleaved bR, compared to the single intermediate previously reported (7). This dramatic increase is similar to that seen in recent work on unfolding bR from its C-terminus when using a modified ultrashort cantilever (11). The large number of unfolding intermediates seen in both cases illustrates the need for enhanced state determination and assignment.
Materials and Methods
Preparation of cleaved bR sample for site-specific stretching
We prepared cleaved bR in its native purple membrane following a previously published method (20). Briefly, we replaced the E-F loop of native bR with the longer loop found in bovine rhodopsin. This longer loop facilitated cleavage by proteinase V8, which cut the E-F loop at two glutamic-acid residues. Importantly, prior work demonstrated that this loop substitution preserved the assembly of bR into the native trigonal lattice of purple membrane and the correct incorporation of retinal (20,21). To achieve site-specific attachment of EF-cleaved bR to azide-functionalized AFM tips via copper-free click chemistry (19,22), we introduced a cysteine (Q167C) into the expanded loop adjacent to one of the cleavage sites. We functionalized an estimated 1% of the bR with a maleimide-dibenzocyclooctyl (DBCO) cross-linker (Click Chemistry Tools, Scottsdale, AZ). Such sparse labeling avoided stretching multiple bR molecules in parallel given the dense two-dimensional packing of bR in purple membrane. Full details of the purification and labeling, including buffer conditions at different steps, are provided in the Supporting Materials and Methods.
For the experiments in which the retinal was removed, chemical hydrolysis of the EF-cleaved sample was performed as in Yu et al. (19) by treatment with 200 mM hydroxylamine in the presence of light from a mercury arc lamp, passed through a 495-nm long-pass filter, for 3 h. Removal of the retinal was confirmed by the disappearance of the peak in the sample absorption spectrum at 570 nm and the increase of the peak at 370 nm, both compared to a reference peak at 280 nm.
Functionalization of micromachined cantilevers
To stretch bR with a mechanically robust tip-sample attachment, we made azide-functionalized, PEG-coated cantilevers using a recently developed protocol (22). First, ultrashort cantilevers (L = 9 μm (Olympus BioLever Fast, Tokyo, Japan)) were modified with a focused-ion beam (14,23). In particular, we used a Warhammer geometry (15) for better force stability at a slight reduction in time resolution (2 μs) (19). After modification, the cantilevers had a stiffness of 16–42 pN/nm and a quality factor (Q) in water near 0.5 when positioned ∼100 nm over the substrate. Cantilever stiffness was determined in air using the thermal method (24,25) before functionalization. The mechanical properties of a representative cantilever used are shown in Fig. S1.
The cantilevers were prepared for silanization by placing them in an ultraviolet-ozone system (Novascan, Boone, IA) for 30 min. We then incubated the cantilevers in a silane-PEG-azide (Nanocs, Boston, MA) toluene solution (0.15 mg/mL) at 60°C for 3 h. Two different, short lengths of PEG were used (MW = 600 and 3400 Da). After the 3-h incubation, the cantilevers were sequentially rinsed in toluene, isopropanol, and deionized water. Functionalized cantilevers were stored in phosphate-buffered saline buffer (10 mM phosphate buffer (pH 7.4), 137 mM NaCl, and 3 mM KCl) at 4°C. To reuse cantilevers, we removed the bR and other surface-bound molecules in an oxygen plasma cleaner (O2 flow at 100 sccm, 300 W for 180 s, PlasmaSTAR; AXIC, Santa Clara, CA) to generate a bare silicon-nitride surface. Reuse of modified ultrashort cantilevers was limited by handling and/or bending during plasma cleaning, with typically two to three refunctionalizations per cantilever. We obtained an average of four traces that met our final selection criteria per cantilever per refunctionalization in this study.
Single-molecule AFM assay
The single-molecule assay followed the methods of Yu et al. in attaching the AFM tip to the bR via a site-specific, covalent bond (19). To prepare the sample, we first diluted labeled bR to ∼20 ng/μL in absorption buffer (10 mM TrisHCl (pH 7.8), 300 mM KCl), sonicated for 3 min under a pulsing protocol (2 s on at 100% intensity, 2 s off (Vibra-cell VCX; Sonics, Newton, CT)) and then deposited 50 μL onto freshly cleaved mica for 1 h. The mica was next rinsed with 1 mL imaging buffer by pipetting up and down (10 mM TrisHCl (pH 7.8), 150 mM KCl) and then loaded into the AFM (Cypher ES; Asylum Research, Santa Barbara, CA), which has a temperature-regulated, closed-fluidic sample chamber. To measure deflection of the modified ultrashort cantilever, we used a custom-built small-spot-size illumination module (14) that yielded a 2.8-μm circular spot. Patches of bR-containing purple membrane were located on the mica surface by tapping-mode imaging.
To initiate individual measurements, the azide-functionalized AFM tip was gently pressed into bR patches at ∼100 pN for 3 s and then retracted, either using the zigzag force ramp or conventional constant-velocity retraction (v = 300 nm/s). The zigzag force ramp was implemented by programming a time-varying set point for the feedback loop regulating the vertical position (Z) of the base of the cantilever. We used two Z(t) patterns: one a simple rising sawtooth and the other with a speed that varied between different portions of the cycle. The parameters of each pattern are shown in Fig. S2. Cantilever deflection was digitized at 50 kHz and 5 MHz simultaneously. For subsequent analysis, 5-MHz data were smoothed using a 501-point second-order Savitzky-Golay filter (26) to yield a 100-μs time resolution. For presentation, all the data were smoothed to 1 kHz.
The resulting records of force as a function of Z showed a significant optical-interference artifact similar to prior AFM experiments using ultrashort cantilevers (11,27). During data acquisition, we subtracted a coarse fourth-order polynomial fit of this artifact in real time to achieve reasonably accurate surface contact forces. In the final analysis, we more carefully removed this artifact using a previously described method (11) (albeit in this analysis, we used a smoothing spline rather than a phenomenological analytical function).
Identification and structural assignment of unfolding intermediates
Force-extension curves that exhibited three major states—corresponding to pulling on the top of the E, C, and A helices—and minimal tip-sample adhesion (<50 pN) were selected for further analysis. To identify unfolding intermediates, we used the general strategy developed by Yu et al. (11) but modified it to account for the effect of variable PEG linker length (19). The intermediate-identification process consisted of three main steps: finding the contour lengths of the intermediates in each force-extension curve, combining these results to obtain the consensus set of contour lengths across all records, and assigning those contour length values to amino acid residues in the bR structure. Details of each step are given in the Supporting Materials and Methods.
Results and Discussion
As a practical validation of the zigzag methodology, we revisited the unfolding of EF-cleaved bR. The resulting data clearly showed longer dwells in individual states and more states occupied per single-molecule record when compared to traditional constant-velocity pulling. Additionally, the study provided new, to our knowledge, insight into the EF-cleaved bR system in its own right.
Zigzag force spectroscopy of bR increased state dwell times and occupancy
In general, the sparse population of unfolding intermediates in standard constant-velocity assays arises because intermediates after the major states at the top of each helix pair (or of the terminal helix) become less stable as the unfolding pathway proceeds (8,16). Hence, intermediate N + 1 typically unfolds at a lower average force than intermediate N. The average lifetime of state N + 1 at a particular force (F) is thus also reduced, causing many states to remain unresolved in any given unfolding trajectory. A small reduction in F dramatically reduces the rate of unfolding and thereby leads to longer state occupancy because of the approximately exponential dependence of unfolding rate on F (28). Hence, if the loading rate is negative when the system enters state N + 1 or becomes negative shortly thereafter, the reduced force extends the state occupancy until the force is raised again. Furthermore, reduced F also promotes local refolding (Fig. 2 B, inset; Fig. S3). Refolding behavior is especially prevalent in a 14-aa region at the top of helix E (Fig. S3) and in a 21-aa region spanning helix C. Refolding transitions increase confidence in identifying closely spaced states and, more generally, provide information about the underlying free-energy landscape (29,30).
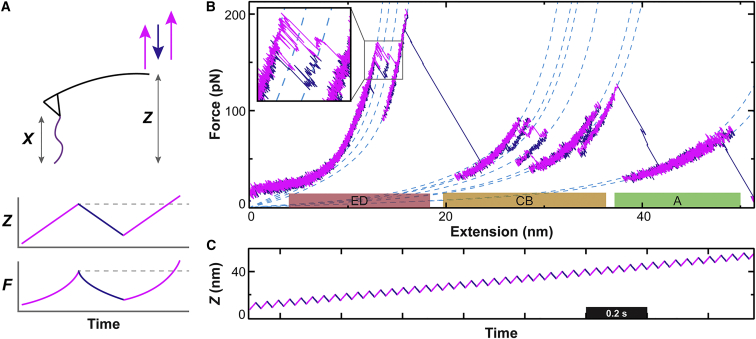
Figure 2.
Zigzag force ramp. (A) In the zigzag protocol, the position of the cantilever base (Z) repeatedly cycles back and forth vertically during a net slow retraction from the surface (magenta, upward motion; blue, downward motion). This protocol results in cyclically increasing and decreasing force (F) as a function of time. (B) A representative force-extension curve shows the application of the zigzag protocol to EF-cleaved bR. Colored bars indicate the helix pair or terminal helix being unfolded. (Inset) Local refolding occurred between three states, each separated by 4 aa or one helical turn. (C) A plot of Z(t) shows the pattern used to obtain these data.
Our approach built on earlier studies that have used periodic cantilever trajectories to promote refolding of globular protein domains (17) and pairs of transmembrane helices (18). The earliest membrane protein study to do so (18) used a preprogrammed cantilever motion with a large amplitude to induce refolding of entire helix pairs. Here, we used a finer-scale motion of 6–10 nm at ∼10 Hz. Similar finer-scale motion, but at much higher frequencies (3 kHz), has also been previously used to study the viscoelasticity of bR (31). Although this prior study identified three additional intermediates using this method, it lacked the force precision and temporal resolution—facilitated by our use of micromachined cantilevers—necessary to resolve a multitude of closely spaced, transiently occupied states (11).
Force-extension curves of EF-cleaved bR acquired using the zigzag protocol showed longer state occupancy and more intermediates (Figs. 1 B and 2 B) as compared to a set of reference constant-velocity trajectories. More quantitatively, the average total dwell time in unfolding intermediates—excluding major states at the tops of helices—was 21-fold longer in zigzag records than for constant-velocity pulling (1.5 ± 0.1 s (mean ± SEM; N = 39) vs. 0.068 ± 0.003 s (N = 10), respectively). Longer occupancy in an individual state leads to a longer arc length of the WLC curve associated with a particular state being sampled, which allows ΔLo to be fitted with greater precision. For example, the statistical uncertainty in ΔLo between the highlighted states in Fig. 1 B decreases from 0.09 nm in the constant-velocity record to 0.01 nm in the zigzag record, using a 95% confidence interval. Additionally, the average number of states per record increased 40% from 10.7 ± 0.7 (mean ± SEM) for the constant-velocity records to 15.2 ± 0.5 for the zigzag protocol. This simultaneous increase in the dwell time per state and the number of states detected per trace increases the confidence in distinguishing and assigning the resulting multitude of closely spaced states.
Note that the improvements in the zigzag method are not merely a consequence of the longer data acquisition time. For example, Fig. S4 shows a constant-velocity trace acquired at 27 nm/s that leads to having the same total acquisition time as one of our zigzag protocols (protocol #1, Fig. S2). Even at such a slow pulling speed, the constant-velocity protocol disfavored resolving closely spaced states in which the second unfolding force was comparable to or lower than the first (e.g., states and ).
EF-cleaved bR unfolds through 31 intermediates
Oesterhelt et al. developed the EF-cleaved bR assay in their pioneering AFM study of bR (7). They pulled from the top of the E helix so that the GF helix pair and the retinal remained in place during unfolding. They identified two major states associated with pulling on the top of helices C and A. However, tip-sample surface adhesion at low extensions prevented analysis of the first pair of helices to be extracted (i.e., the ED helix pair), a ubiquitous problem in AFM force spectroscopy when using nonspecific tip-sample attachment. In addition to the two major states, Oesterhelt et al. identified a single and particularly stable intermediate in the loop between helices B and C. Interestingly, this intermediate was not observed in their contemporaneous unfolding of wild-type bR from the C-terminus of helix G, indicating that it is stabilized by interactions with the GF helix pair and/or the retinal.
We revisited the unfolding EF-cleaved bR with improved precision by integrating a series of advancements. First, we characterized EF-cleaved bR using the zigzag force ramp. Second, we covalently attached the bR to the cantilever. This achieved a mechanically robust tip-sample attachment while also suppressing tip-sample adhesion because of the tip’s PEG coating and the ∼10-fold lower contact forces needed to promote attachment (19). Covalently bound bR does limit the total number of attachments in an experiment; we averaged four usable traces per cantilever in this study. Cantilevers may be reused after plasma cleaning. We note that, in practice, traditional assays relying on nonspecific attachment are also limited in the number of molecules that can be probed per cantilever. In this study, the lower number of attachments per cantilever was offset by reduced tip-sample adhesion, allowing interpretation of the initial unfolding of the EF helix pair, and mechanically robust tip-bR attachment, enabling the multisecond-long zigzag protocol used here. Finally, we used focused-ion-beam-modified ultrashort cantilevers that provided a 100-fold improvement in time resolution and a 10-fold improvement in force precision (14). Unlike the original EF-cleaved bR study, this set of advancements yielded interpretable force-extension curves starting from the top of helix E, the first helix unfolded. Hence, similar to recent work using site-specific coupling to pull from the top of helix G (19), we characterized bR starting from its fully folded state when all of its tertiary interactions were intact, including those with the retinal cofactor.
By applying these metrological improvements, we identified 31 intermediates in the unfolding of EF-cleaved bR in addition to the expected three major states corresponding to pulling on the tops of the E, C, and A helices (Fig. 3). These intermediates were broadly distributed, with 12 occurring in the ED helix pair (Fig. 3 A), 15 in the CB helix pair (Fig. 3 B), and 4 in helix A (Fig. 3 C). The one intermediate reported in the prior EF-cleaved bR measurements (7) was among the intermediates detected in the CB helix pair. Interestingly, 3 aa ahead of this state, a second mechanically stable intermediate was also detected, suggesting that the originally reported state combined these two states into a single composite state.
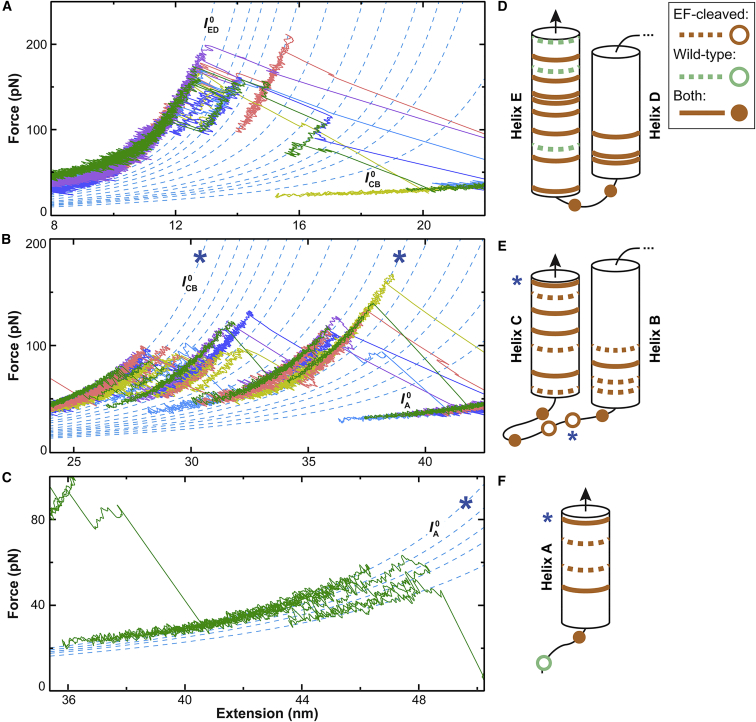
Figure 3.
Zigzag force spectroscopy revealed 31 intermediates in the unfolding of EF-cleaved bR. (A–C) Representative force-extension curves show the unfolding of (A) the ED helix pair, (B) the CB helix pair, and (C) helix A. Major and intermediate states correspond to force-extension segments lying along particular WLC curves (dashed blue lines). All states identified in our analysis are shown. For clarity, we do not show representative traces occupying all states and only show one trajectory in (C). The previously reported two major states and single intermediate are denoted with blue asterisks (7). (D–F) Schematic depictions of state locations within (D) the ED helix pair, (E) the CB helix pair, and (F) helix A are given. States within transmembrane helices are depicted as bands and those within loops as circles. The states depicted are further distinguished by whether they were found in both this study of EF-cleaved bR and the previous high-resolution unfolding of wild-type bR (11) (solid brown line/solid circle), in EF-cleaved bR only (dashed/open brown), or in wild-type bR only (dashed/open green).
Each identified intermediate was assigned to a specific amino acid residue in bR. In doing so, we made the standard assumptions in membrane protein single-molecule force spectroscopy analysis: that residues that have not yet unfolded remain in their native locations and that the unfolded protein retains no secondary structure (8). These assumptions are reasonable given the high rupture forces seen for many bR transitions—typically >50 pN—when compared to the unfolding of α-helical globular proteins (32), the densely packed nature of the purple membrane, and the favorable tertiary interactions in bR (33). We schematically depict the assigned locations of the intermediates in Fig. 3, D–F and list the specific residue assignments in Table S1. Because most of the detected intermediates have an uncertainty in ΔL0 of 0.1–0.3 nm, these amino acid assignments are expected to be accurate to within one or two residues.
Comparing the number and location of intermediates with prior results
Two prior studies provide an important basis for comparing our results. The first study is the original EF-cleaved bR assay of Oesterhelt et al. (7). That work, like this one, retained the GF helix pair and retinal in the lipid bilayer during the unfolding of the E–A helices. The prior work reported a single intermediate that, interestingly, was within the B-C loop (Fig. 3, B and E, asterisks). Oesterhelt et al. attributed the presence of this intermediate to an all-or-none unfolding of helix B arising from its proximity to helix G. Our higher-resolution data, however, cast doubt on this explanation. First, we found that helix B unfolds via five intermediates. Moreover, as discussed above, our enhanced resolution shows that the previously detected intermediate corresponds to two closely spaced intermediates assigned to Val69 and Leu66 (denoted and ).
In the second study for comparison by Yu et al. (11), bR was unfolded from the C-terminus of helix G using modified ultrashort cantilevers. As with most traditional studies of bR by AFM that used nonspecific adhesion to attach the bR to the tip (8), this study did not quantify the unfolding of the GF helix pair but only the unfolding of helices E–A. Notably, the enhanced resolution provided by using modified ultrashort cantilevers revealed 14 intermediates in the ED helix pair, 8 in the CB helix pair, and 4 in helix A. Most of these intermediates (83%) were also seen in our study of EF-cleaved bR (Fig. 3, D–F, solid bars/circles).
Interestingly, the noncommon intermediates divided into two classes. In the ED helix pair, the traditional pulling geometry starting with the G helix by Yu et al. showed three additional intermediates despite our study using a shorter, less compliant linker and the zigzag force ramp (Fig. 3 D, green dashed bars). This result suggests that the additional stabilization provided by the GF helix pair and the retinal in the EF-cleaved construct led to higher rupture forces and the suppression of subsequent intermediates. Indeed, we found that the average constant-velocity rupture force of coincident states occurring in the ED helix pair of both EF-cleaved and wild-type bR was 76% larger in the EF-cleaved sample (Fig. S5). In contrast, the EF-cleaved bR and zigzag protocol yielded six additional states in the CB helix pair and two in helix A (Fig. 3, E and F, dashed brown) and no difference in rupture forces was seen for coincident states in helix A (Fig. S5).
Verifying a retinal-stabilized intermediate in helix E
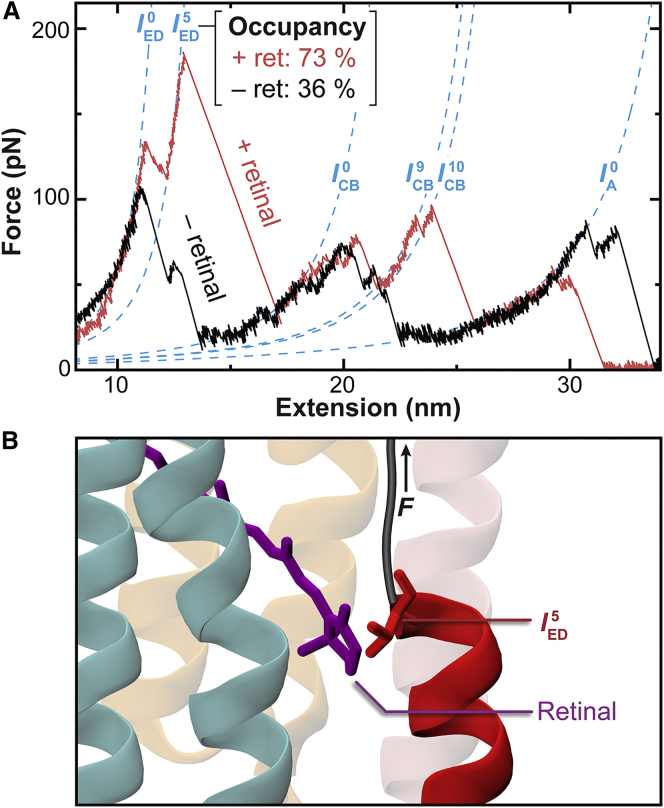
Inspection of the EF-cleaved bR intermediates in helix E revealed that one intermediate () occurred in a notably larger fraction of unfolding trajectories (61% of all records) than intermediates before and after it (Fig. 4 A, red; Table S1). Contour length analysis assigned this intermediate to Thr142. Structurally, Thr142 lines the retinal pocket (Fig. 4 B; (34,35)). Hence, we hypothesized that the enhanced stabilization of arose from its interaction with the retinal.
Figure 4.
Localization of a retinal-stabilized intermediate verified by removing the retinal. (A) Constant-velocity force-extension curves of EF-cleaved bR with (red) and without (black) the retinal are shown. The percentage of unfolding trajectories occupying decreased in the absence of retinal. (B) Detailed view of the bR crystal structure (Protein Data Bank, PDB: 1C3W (35)) shows the proximity of Thr142 to the retinal. State corresponds to unfolding bR up to this residue, as represented by the taut polyprotein (black). Helix color coding is the same as in Fig. 1.
We tested this hypothesis by removing the retinal from the EF-cleaved sample and performing constant-velocity unfolding measurements (v = 300 nm/s). The occupancy of state decreased from 73% (N = 8 of 11) with the retinal intact to 36% in the absence of retinal (N = 10 of 28). This decrease was statistically significant (p = 0.04) when using the conservative two-tailed test (36). In contrast, the percentage occupancy of the immediately preceding state showed no significant change (45.5% with retinal vs. 46.4% without; p = 0.96). Hence, contour length analysis localized a stabilizing interaction to a particular amino acid, and this interaction was verified by removing the retinal without significantly altering a neighboring intermediate.
In contrast to the retinal-stabilized intermediate , we find no obvious native-state structural rationalization for the strongly stabilized intermediates and in the B-C loop. These intermediates are absent in both the original bR pulling from the C-terminus of helix G (7) and in the more recent high-precision study using modified ultrashort cantilevers (11). We speculate that these intermediates could arise from non-native hydrogen bonding between the unfolded β-sheet in the B-C loop and the unsatisfied hydrogen bonding sites of the remaining transmembrane helices (35). A more complete explanation awaits future experiments and would likely benefit from complementary steered molecular dynamic simulations (37).
Conclusions
To fully leverage recent developments in AFM-based single-molecule force spectroscopy, we combined advances in cantilevers and site-specific anchoring with improvements in data acquisition protocols. The zigzag force ramp improved upon the conventional constant-velocity assay by sampling unfolding intermediates over both a longer span of time and a broader domain of the force-extension curve and by promoting the occupancy of more states per single-molecule unfolding trajectory. Contour length analysis localized a robust unfolding intermediate to a particular amino acid and led to the hypothesis that this intermediate was retinal stabilized. We verified this hypothesis by removing retinal. Looking forward, this work lays the foundation for future applications of the zigzag protocol to characterize the energetics of point mutants. Such studies will benefit from merging the near-equilibrium fluctuations seen here with recent developments in force spectroscopy theory that increase the information extracted from trajectories showing near-equilibrium folding (30).
Author Contributions
D.R.J. and T.T.P. designed research. L.U. prepared protein samples. D.R.J. performed research and analyzed data. D.R.J. and T.T.P. wrote the manuscript.
Acknowledgments
We thank James Bowie for providing cell lines and expression plasmids for making bR mutants and Matthew Siewny and Hao Yu for helpful discussions.
This work was supported by the National Science Foundation (MCB-1716033), a National Research Council Research Associateship (D.R.J.), and National Institutue of Standards and Technology (NIST). Mention of commercial products is for information only; it does not imply NIST’s recommendation or endorsement. T.T.P. is a staff member of the NIST Quantum Physics Division.
Editor: Keir Neuman.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.12.003.
Supporting Citations
References (38, 39, 40, 41, 42, 43, 44, 45) appear in the Supporting Material.
Supporting Material
References
- 1.Fernandez-Leiro R., Scheres S.H. Unravelling biological macromolecules with cryo-electron microscopy. Nature. 2016;537:339–346. doi: 10.1038/nature19948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park E., Campbell E.B., MacKinnon R. Structure of a CLC chloride ion channel by cryo-electron microscopy. Nature. 2017;541:500–505. doi: 10.1038/nature20812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liang Y.L., Khoshouei M., Sexton P.M. Phase-plate cryo-EM structure of a class B GPCR-G-protein complex. Nature. 2017;546:118–123. doi: 10.1038/nature22327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.White S.H., Wimley W.C. Membrane protein folding and stability: physical principles. Annu. Rev. Biophys. Biomol. Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 5.Bowie J.U. Solving the membrane protein folding problem. Nature. 2005;438:581–589. doi: 10.1038/nature04395. [DOI] [PubMed] [Google Scholar]
- 6.Cymer F., von Heijne G., White S.H. Mechanisms of integral membrane protein insertion and folding. J. Mol. Biol. 2015;427:999–1022. doi: 10.1016/j.jmb.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oesterhelt F., Oesterhelt D., Müller D.J. Unfolding pathways of individual bacteriorhodopsins. Science. 2000;288:143–146. doi: 10.1126/science.288.5463.143. [DOI] [PubMed] [Google Scholar]
- 8.Bippes C.A., Muller D.J. High-resolution atomic force microscopy and spectroscopy of native membrane proteins. Rep. Prog. Phys. 2011;74:086601. [Google Scholar]
- 9.Krishnamani V., Hegde B.G., Lanyi J.K. Secondary and tertiary structure of bacteriorhodopsin in the SDS denatured state. Biochemistry. 2012;51:1051–1060. doi: 10.1021/bi201769z. [DOI] [PubMed] [Google Scholar]
- 10.Chang Y.C., Bowie J.U. Measuring membrane protein stability under native conditions. Proc. Natl. Acad. Sci. USA. 2014;111:219–224. doi: 10.1073/pnas.1318576111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu H., Siewny M.G., Perkins T.T. Hidden dynamics in the unfolding of individual bacteriorhodopsin proteins. Science. 2017;355:945–950. doi: 10.1126/science.aah7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stigler J., Ziegler F., Rief M. The complex folding network of single calmodulin molecules. Science. 2011;334:512–516. doi: 10.1126/science.1207598. [DOI] [PubMed] [Google Scholar]
- 13.Bull M.S., Sullan R.M., Perkins T.T. Improved single molecule force spectroscopy using micromachined cantilevers. ACS Nano. 2014;8:4984–4995. doi: 10.1021/nn5010588. [DOI] [PubMed] [Google Scholar]
- 14.Edwards D.T., Faulk J.K., Perkins T.T. Optimizing 1-μs-resolution single-molecule force spectroscopy on a commercial atomic force microscope. Nano Lett. 2015;15:7091–7098. doi: 10.1021/acs.nanolett.5b03166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edwards D.T., Faulk J.K., Perkins T.T. Force spectroscopy with 9-μs resolution and sub-pN stability by tailoring AFM cantilever geometry. Biophys. J. 2017;113:2595–2600. doi: 10.1016/j.bpj.2017.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Müller D.J., Kessler M., Gaub H. Stability of bacteriorhodopsin alpha-helices and loops analyzed by single-molecule force spectroscopy. Biophys. J. 2002;83:3578–3588. doi: 10.1016/S0006-3495(02)75358-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rief M., Gautel M., Gaub H.E. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 18.Kessler M., Gottschalk K.E., Gaub H.E. Bacteriorhodopsin folds into the membrane against an external force. J. Mol. Biol. 2006;357:644–654. doi: 10.1016/j.jmb.2005.12.065. [DOI] [PubMed] [Google Scholar]
- 19.Yu H., Heenan P.R., Perkins T.T. Quantifying the initial unfolding of bacteriorhodopsin reveals retinal stabilization. Angew. Chem. Int. Ed. 2019;58:1710–1713. doi: 10.1002/anie.201812072. [DOI] [PubMed] [Google Scholar]
- 20.Heymann J.B., Pfeiffer M., Müller D.J. Conformations of the rhodopsin third cytoplasmic loop grafted onto bacteriorhodopsin. Structure. 2000;8:643–653. doi: 10.1016/s0969-2126(00)00151-9. [DOI] [PubMed] [Google Scholar]
- 21.Geiser A.H., Sievert M.K., Ruoho A.E. Bacteriorhodopsin chimeras containing the third cytoplasmic loop of bovine rhodopsin activate transducin for GTP/GDP exchange. Protein Sci. 2006;15:1679–1690. doi: 10.1110/ps.062192306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walder R., LeBlanc M.A., Perkins T.T. Rapid characterization of a mechanically labile α-helical protein enabled by efficient site-specific bioconjugation. J. Am. Chem. Soc. 2017;139:9867–9875. doi: 10.1021/jacs.7b02958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faulk J.K., Edwards D.T., Perkins T.T. Improved force spectroscopy using focused-ion-beam-modified cantilevers. Methods Enzymol. 2017;582:321–351. doi: 10.1016/bs.mie.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 24.Hutter J.L., Bechhoefer J. Calibration of atomic-force microscope tips. Rev. Sci. Instrum. 1993;64:1868–1873. [Google Scholar]
- 25.Burnham N.A., Chen X., Tendler S.J.B. Comparison of calibration methods for atomic-force microscopy cantilevers. Nanotechnology. 2003;14:1–6. [Google Scholar]
- 26.Savitzky A., Golay M.J.E. Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem. 1964;36:1627–1639. [Google Scholar]
- 27.Rico F., Gonzalez L., Scheuring S. High-speed force spectroscopy unfolds titin at the velocity of molecular dynamics simulations. Science. 2013;342:741–743. doi: 10.1126/science.1239764. [DOI] [PubMed] [Google Scholar]
- 28.Bell G.I. Models for the specific adhesion of cells to cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 29.Hummer G., Szabo A. Free energy profiles from single-molecule pulling experiments. Proc. Natl. Acad. Sci. USA. 2010;107:21441–21446. doi: 10.1073/pnas.1015661107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y., Dudko O.K. A transformation for the mechanical fingerprints of complex biomolecular interactions. Proc. Natl. Acad. Sci. USA. 2013;110:16432–16437. doi: 10.1073/pnas.1309101110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Janovjak H., Müller D.J., Humphris A.D. Molecular force modulation spectroscopy revealing the dynamic response of single bacteriorhodopsins. Biophys. J. 2005;88:1423–1431. doi: 10.1529/biophysj.104.052746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffmann T., Dougan L. Single molecule force spectroscopy using polyproteins. Chem. Soc. Rev. 2012;41:4781–4796. doi: 10.1039/c2cs35033e. [DOI] [PubMed] [Google Scholar]
- 33.Popot J.L., Gerchman S.E., Engelman D.M. Refolding of bacteriorhodopsin in lipid bilayers. A thermodynamically controlled two-stage process. J. Mol. Biol. 1987;198:655–676. doi: 10.1016/0022-2836(87)90208-7. [DOI] [PubMed] [Google Scholar]
- 34.Henderson R., Baldwin J.M., Downing K.H. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J. Mol. Biol. 1990;213:899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- 35.Luecke H., Schobert B., Lanyi J.K. Structure of bacteriorhodopsin at 1.55 A resolution. J. Mol. Biol. 1999;291:899–911. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- 36.Campbell I. Chi-squared and Fisher-Irwin tests of two-by-two tables with small sample recommendations. Stat. Med. 2007;26:3661–3675. doi: 10.1002/sim.2832. [DOI] [PubMed] [Google Scholar]
- 37.Kappel C., Grubmüller H. Velocity-dependent mechanical unfolding of bacteriorhodopsin is governed by a dynamic interaction network. Biophys. J. 2011;100:1109–1119. doi: 10.1016/j.bpj.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DasSarma S., RajBhandary U.L., Khorana H.G. High-frequency spontaneous mutation in the bacterio-opsin gene in Halobacterium halobium is mediated by transposable elements. Proc. Natl. Acad. Sci. USA. 1983;80:2201–2205. doi: 10.1073/pnas.80.8.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oesterhelt D., Stoeckenius W. Isolation of the cell membrane of Halobacterium halobium and its fractionation into red and purple membrane. Methods Enzymol. 1974;31:667–678. doi: 10.1016/0076-6879(74)31072-5. [DOI] [PubMed] [Google Scholar]
- 40.Bustamante C., Marko J.F., Smith S. Entropic elasticity of λ-phage DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 41.Marko J.F., Siggia E.D. Stretching DNA. Macromolecules. 1995;28:8759–8770. [Google Scholar]
- 42.Bouchiat C., Wang M.D., Croquette V. Estimating the persistence length of a worm-like chain molecule from force-extension measurements. Biophys. J. 1999;76:409–413. doi: 10.1016/s0006-3495(99)77207-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sapra K.T., Besir H., Muller D.J. Characterizing molecular interactions in different bacteriorhodopsin assemblies by single-molecule force spectroscopy. J. Mol. Biol. 2006;355:640–650. doi: 10.1016/j.jmb.2005.10.080. [DOI] [PubMed] [Google Scholar]
- 44.Petrosyan R., Bippes C.A., Müller D.J. Single-molecule force spectroscopy of membrane proteins from membranes freely spanning across nanoscopic pores. Nano Lett. 2015;15:3624–3633. doi: 10.1021/acs.nanolett.5b01223. [DOI] [PubMed] [Google Scholar]
- 45.Sullivan D.B., Allan D.W., Walls E.L., editors. Characterization of Clocks and Oscillators. U.S. Government Printing Office; Washington, DC: 1990. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.