Abstract
The application of statistical methods to comparatively framed questions about the molecular dynamics (MD) of proteins can potentially enable investigations of biomolecular function beyond the current sequence and structural methods in bioinformatics. However, the chaotic behavior in single MD trajectories requires statistical inference that is derived from large ensembles of simulations representing the comparative functional states of a protein under investigation. Meaningful interpretation of such complex forms of big data poses serious challenges to users of MD. Here, we announce Detecting Relative Outlier Impacts from Molecular Dynamic Simulation (DROIDS) 3.0, a method and software package for comparative protein dynamics that includes maxDemon 1.0, a multimethod machine learning application that trains on large ensemble comparisons of concerted protein motions in opposing functional states generated by DROIDS and deploys learned classifications of these states onto newly generated MD simulations. Local canonical correlations in learning patterns generated from independent, yet identically prepared, MD validation runs are used to identify regions of functionally conserved protein dynamics. The subsequent impacts of genetic and/or drug class variants on conserved dynamics can also be analyzed by deploying the classifiers on variant MD simulations and quantifying how often these altered protein systems display opposing functional states. Here, we present several case studies of complex changes in functional protein dynamics caused by temperature, genetic mutation, and binding interactions with nucleic acids and small molecules. We demonstrate that our machine learning algorithm can properly identify regions of functionally conserved dynamics in ubiquitin and TATA-binding protein (TBP). We quantify the impact of genetic variation in TBP and drug class variation targeting the ATP-binding region of Hsp90 on conserved dynamics. We identify regions of conserved dynamics in Hsp90 that connect the ATP binding pocket to other functional regions. We also demonstrate that dynamic impacts of various Hsp90 inhibitors rank accordingly with how closely they mimic natural ATP binding.
Significance
We propose a statistical method and graphically interfaced software pipeline for comparing simulations of the complex motions of proteins (i.e., dynamics) in different functional states. We also provide both method and software to apply artificial intelligence (i.e., machine learning methods) that enable the computer to recognize complex functional differences in protein dynamics on new simulations and report them to the user. This method can identify conserved dynamics important for protein function and quantify how the motions of molecular variants differ from these important functional dynamic states. This method of analysis allows the impacts of different genetic backgrounds or drug classes to be examined within the context of functionally conserved motions of the specific protein system under investigation.
Introduction
The physicist Richard Feynman once said, “if we were to name the most powerful assumption of all … in an attempt to understand life, it is …that everything that living things do can be understood in terms of the jigglings and wigglings of atoms” (1). Restated with more precision, Feynman’s conjecture would imply that all biological function can ultimately be understood by analyzing rapid molecular motions in biomolecular structures as they alter or shift their functional state(s). Many decades later, these functional shifts in molecular dynamics (MD) are being illuminated by structural and computational biology. Examples of functionally altered dynamics include the destabilization of interresidue contacts during signal activation and disease (2, 3, 4, 5), the stabilization of interresidue contacts during protein folding and the formation of larger complexes (6, 7, 8), and the dynamic complexity of binding interactions of many proteins to a variety of small molecules (9,10). And although the functional role of rapid vibrations revealed by short-term MD simulations has been debated in the past, more recent empirical and computational studies have clearly demonstrated that differences in both rapid and directed vibrations can drive longer-term functional conformational change (11,12). From a broader perspective, if Feynman’s conjecture is true, then specific details of a given protein system’s biomolecular dynamics will represent a potentially large source of latent variability in our functional understanding of the genome, a problem largely ignored by those disciplines currently generating the vast amount of static forms of “omic”-type data (i.e., DNA sequence, transcript level, and protein structure) (13). However, in the last decade, simultaneous advances in the development of massively parallelized graphics hardware and more accurate biomolecular force fields have elevated our ability to computationally simulate MD long enough to capture ns-to-μs timescales for moderately sized proteins (14,15) and to accurately simulate some of their functionally relevant motions. And now, the application of proper statistical comparisons of ensembles of randomly spaced, short-time-framed MD simulation can potentially enable meaningful interpretations of comparative questions about protein dynamics (16). But because of the richly complex structure of data underlying the moving images generated by MD software, functional interpretation of modern MD simulations poses a serious challenge to current users. This is especially problematic with comparatively framed questions, in which large ensembles of many production runs need to be generated and subsequently analyzed statistically. A potential solution to this problem exists with the application of machine learning to the feature extraction and classification of the dynamic differences between ensembles of MD runs. These ensembles can be designed to represent pair-wise functional states of biomolecular systems (e.g., before/after environmental change, chemical mutation, or binding interaction). Therefore, the high-performance accelerated computation used to generate simulated protein motions for comparison can be effectively partnered with high-performance methods for optimally extracting and learning the underlying dynamic feature differences that define the different functional states of proteins. Although machine learning has recently been applied to individual MD studies for a variety of specific tasks (17, 18, 19), there is no current software platform for the general application of machine learning to general comparative problems in protein dynamics.
In 2018, we released Detecting Relative Outlier Impacts from Molecular Dynamic Simulation (DROIDS) v1.2 and v2.0, a GPU-accelerated software pipeline designed for calculating and visualizing statistical comparisons of protein dynamics drawn from large repeated ensembles of short dynamic simulations representing two protein states (16). This application allowed simple visual and statistical comparison of protein MD ensembles set up in any way the user wanted to define them. Here, we announce the release of DROIDS v3.0, which now offers multiple pipelines tailored for specific functional comparisons of systems made up of combinations of proteins, nucleic acids, and small ligand molecules. Comparisons can include different temperatures, different protein binding states (i.e., to DNA, drugs, toxins, or natural ligands), or divergent genetic/epigenetic mutant states. We also include a major new machine learning tool, maxDemon v1.0, a multimachine learning postprocessing application for DROIDS that trains on the data that represent the comparatively divergent functional dynamic states occurring in a functional binding interaction (i.e., bound versus unbound protein dynamics) and subsequently identifies these states when deployed upon new MD simulations. This allows for the determination of regions of functionally conserved dynamics in independent yet identically prepared validation runs, as well as the quantification of impacts upon dynamics in different genetic and drug class variants. Thus, much like James Clerk Maxwell’s mythical creature (20), maxDemon derives important information from the classification of all atom resolution observations of dynamic motion. The three primary features/aims of our expanded software are to 1) ease the generation of MD run ensembles for statistical comparison, 2) enable the local detection of functionally conserved protein dynamics, and 3) enable the assessment of the local dynamic impacts of both genetic and drug class variants within the conserved functional context of protein systems of interest. Because the machine learning model we employ is trained on MD data that represent two contrasting functional dynamic states of a protein, this metric of impact is highly context dependent with regard to how a given mutation or drug impacts a specific protein. Thus, it potentially gives considerably more functional relevance to the analysis of variants when compared with more-general database-derived metrics of mutational tolerance (e.g., SIFT, PolyPhen2, etc. (21,22)). In our online user manual and tutorial, we present many examples of methodological pipelines available in DROIDS 3.0 (with maxDemon 1.0) to address functional questions in comparative protein dynamics. In our Results and Discussion here, we present data on three case studies of functional protein dynamics that include feature extraction and classification of 1) functional and nonfunctional shifts in ubiquitin dynamics, 2) mutation-specific impacts on functional binding of TATA-binding protein (TBP) to DNA, and 3) comparison of binding dynamics of drug class variants that mimic ATP binding in Hsp90 to varying degrees.
Materials and Methods
Overview of comparative dynamics and visualization with DROIDS v3.0
Our DROIDS method/software leverages several important key concepts when making comparisons between MD runs. The method utilizes structural alignment to restrict comparison of dynamics between individual homologous amino acids. The method also restricts dynamic comparison to non-side-chain atoms in the protein backbone (C, N, O, and Cα). The method also employs statistical ensembles to make a robust comparison between protein dynamics in different functional states (16). Although this is computationally intensive, it is necessary because of the inherent chaotic nature and unpredictability of single protein trajectory projections. This logic likens individual MD runs to the many storm tracks repeatedly modeled by meteorologists to gain statistical confidence in a hurricane weather forecast, in which an ensemble of model runs all with slightly different initial conditions has far more predictive power than any single simulation. In DROIDS, the user can decide how large the MD ensembles need to be based upon the inherent stability of the protein under investigation. The dynamics is summarized by calculation of root mean-square fluctuations (rmsf) over constant time intervals represented by a constant number of image frames defined by the user (allowing rmsf values to be sampled repeatedly on an identical and comparable scale). The default number of frames (n) in the software for a given time slice is n = 50, representing 0.01 ns of simulation time (pulled at a rate of 0.0002 ns/frame). Users can adjust the number of frames for time slices at the command prompt. The rmsf value is thus
| (1) |
where v represents the set of XYZ atom coordinates for i backbone atoms (C, N, O, and Cα) for a given amino acid residue over j time points, and w represents the average coordinate structure for each MD production run for a given ensemble (using the “atomicfluct” function from cpptraj software (23)). Therefore, rmsf values as defined here represent MD at the resolution of a single amino acid backbone segment and the same resolution at which fine-scale protein-level molecular evolution operates via amino acid replacement, insertion, and deletion. Two ensembles of rmsf values (a query set and a reference set) are compared to calculate average delta rmsf (dRMSF). The user can choose to see the average angstrom difference between sets of values, or more preferably, the user can calculate the symmetric Kullback-Leibler (KL) divergence (24) (i.e., relative entropy) between the two empirical statistical distributions of rmsf. The KL divergence generally provides a richer, more informative view of dynamic differences with less loss of information than simple averaging. Thus, dRMSF comparing rmsf values for two ensembles of size m for a given amino acid is
| (2) |
or
| (3) |
The resulting dRMSF or “dFLUX” values are color mapped to either still structures or movie images of the dynamics according to either a “temperature” scale in which (+) dRMSF = amplified vibration is red and (−) dRMSF = dampened vibration is blue. A “stoplight” scale in which (+) dRMSF is green and (−) dRMSF is red is also available. On both scales, neutral values are shaded toward white.
Functional classification of new MD simulation with maxDemon v1.0
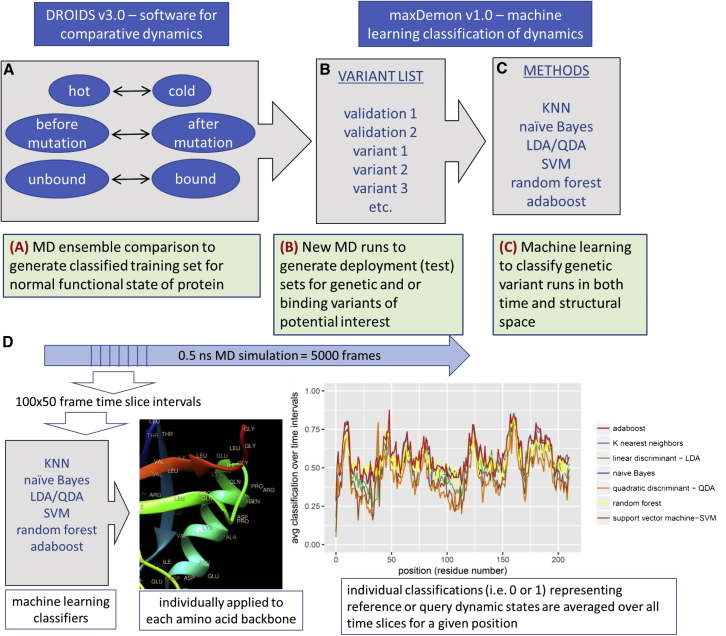
Although users can easily employ DROIDS 3.0 to examine ensemble differences between functional genetic or binding states, the application of this knowledge to new MD simulation is nearly impossible because of the inherent complexity of the moving protein behavior. Our new postprocessing software, maxDemon 1.0, uses machine learning to label or classify the differences learned by a previous DROIDS query/reference state comparison when subsequently applied to one or more new MD runs. The machine-learning-based detection of variant impacts on functional protein dynamics presented here is outlined schematically in Fig. 1. Similar to the statistics for comparative dynamics, the learning algorithms are also applied individually to each amino acid backbone’s ensemble of rmsf values. This allows for similar single-residue resolution in the results. Learners are also applied within the same user-defined time slices of rmsf, allowing for visualization of time resolution of the classification of functional dynamic behaviors as well. The learning performance is summarized by tallying the average classification over all time slices for each amino acid. Individual classifications are either 0 or 1; therefore, an average performance of 0.5 would indicate that the learners are not finding the functional states defined by and trained by the initial DROIDS comparative analysis. Local canonical correlations in the positional performance plots are then used in detecting sequence-encoded functionally conserved dynamics regions, as well as genetic and drug class variant impacts to these functional regions. This is described with more formality below.
Figure 1.
Figure360 Schematic overview of DROIDS 3.0 + maxDemon 1.0 software for machine-learning-based detection of variant impacts on functionally conserved protein dynamics. The pipeline starts with (A) generation of two large ensembles of molecular dynamic (MD) simulations that represent a functional comparison of protein states (e.g., mutation, binding, or environmental change). The rmsf of protein backbone atoms in these ensembles are comparatively analyzed/visualized (i.e., using DROIDS) and are also later used as preclassified training data sets for machine learning (i.e., using maxDemon). Note: the pictured DROIDS analysis of nucleosome shows overall dampening of rmsf in the histone core with maximal dampening where the histone tails cross the DNA helix. (B) New MD simulations are generated on two structures self-similar to the query state of training as well as a list of functional variants, and (C) up to seven machine learning methods are employed to classify the MD in the self-similar and variant runs according to the functional comparison defined by the initial training step. (D) The performance of learning is defined by average value of classification (i.e., 0 or 1) over 50 frame time slices for each amino acid position, and regions of functionally conserved dynamics are later identified by significant canonical correlations in this learning efficiency (i.e., Wilk’s lambda) in self-similar MD validation runs. The impacts of variants are later defined by relative entropy of genetic/drug class variant MD compared with the MD in the self-similar runs (data not shown). To see this figure in color, go online.
Machine learning training and validation
The feature vectors (X) for machine learning are collections of rmsf values (xi) that represent amino acid backbone atoms C, N, O, and Cα, which are labeled according to a query (q) and reference state (r) that are defined by the DROIDS MD comparison (i.e., where labels yi are q = 1 and r = 0).
| (4) |
The length of the vector (N) is defined by the length of the MD production run chosen by the user and the size of the ensemble of MD production runs taken. Thus, if the user chooses an ensemble of 200 MD production runs each at a time length of 0.5 ns (=2500 frames) and uses the default time interval of 50 frames to calculate any given interval of rmsf, then the resulting feature vector will contain 20,000 data values for training (i.e., 10,000 values each for q and r).
MaxDemon creates a “stacked model” or metamodel containing up to seven different machine learning classification algorithms, including K-nearest neighbors (KNN), naïve Bayes, linear discriminant analysis, quadratic discriminant analysis, random forest, adaptive boosting, and support vector machine (with kernel options including parameter tuned linear, polynomial, laplace, and radial basis functions). R packages employed here are KNN, MASS, kernlab, randomForest, and ada (25,26). We restricted machine learning to “shallow” learning methods because of the relatively small data sets created when resolving dynamics of protein systems to short slices of time over single amino acids and also because of the robustness of the R packages when applied sequentially over large amounts of total time and large amounts of structural space. Therefore, we do not yet support implementation of deep-learning neural networks. For methodologically robust results on small proteins, we generally recommend users select all seven available methods. As real features of dynamics should be detectable by any method of learning, the agreement of classification obtained by the creation of a stacked model utilizing different learning methods makes the learning less sensitive to methodological artifacts. Depending on system resources, users can choose to include or omit methods from four categories of learning (i.e., instance-based = K-nearest neighbors (KNN), probabilistic = naive Bayes (NB)/linear discriminant analysis (LDA)/quadratic discriminant analysis (QDA), black box = supportive vector machine (SVM), and ensemble learning = random forest/adaptive boosting). Users will want to use as many models as their system resources can handle; however, for faster processing, a minimum of three of the seven learning methods can be chosen. Currently, all algorithms except KNN are programmed to use all available CPU cores found on the system. SVM and adaptive boosting are sometimes the slower methods for larger protein systems and can be omitted first when more than 500 residues are present in the protein simulation. However, these methods are better at classifying complex differences in dynamic behavior and should be retained whenever possible.
After learners are trained on the query and reference ensembles, they are validated on two new MD runs that match the state of the reference MD runs during training. For example, when analyzing a binding interaction in which the reference ensemble of training runs are conducted in the unbound protein state, a new run will be conducted in the unbound state, and a line plot of the machine learning performance (i.e., precision, recall, and accuracy) will be generated for all positions on the protein. It would be expected that if comparative differences in dynamics observed in the training set have a genuine relation to function(s) defined during training, they will display repeated behavior in the new reference run and be identified by the stacked learning model, which generates local peaks in learning performance (i.e., accuracy) at functional regions (Fig. 1 D). Learner performance for a given machine learning method is defined as
| (5) |
where TP, TN, FP, and FN are true positive, true negative, false positive, and false negative classifications, respectively. The zero value terms arise because the validation is conducted on simulations representing just the reference state of the DROIDS comparison (where yi = 0). Therefore, accuracy and precision are algebraically collapsed to a single equivalent performance metric, whereas recall is always equal to 1.
Identifying regions of conserved dynamics
Functionally conserved dynamics are defined as repeated (i.e., self-similar) local sequence-dependent dynamics discovered after training machine learners on the functional state ensembles derived with DROIDS. Conserved dynamics are detected via significant canonical correlations in position-specific learning performance patterns after the deployment of learners on the two new MD simulation runs that were set up identically to the reference dynamic state defined by the MD ensemble training set. We expect that functionally conserved dynamics will be sequence encoded and therefore should display a repeated position-dependent signature in our learned pattern profiles whenever MD runs are set up identically to MD upon which learners were trained. Therefore, a significant local canonical correlation (i.e., Wilk’s lambda (27)) between learning performance profiles of self-similar MD runs can be used to detect local regions of conserved protein dynamics.
To detect functionally conserved dynamics after training, two additional MD runs matching the functional reference state are created. The learning performance of these runs are compared using a canonical correlation analysis conducted using all selected learners (i.e., the stacked model) across both space and time (i.e., fluctuations of backbone atoms of individual amino acids over subdivided time intervals). Any sequence-dependent or “functionally conserved” dynamics can be recognized through a significant canonical correlation in the profile of the overall learning performance along the amino acid positions for the two similar state runs. In effect, this metric defines dynamics that are functionally conserved by capturing a signal of significant self-similarity in dynamics that colocalizes to a specific part of the protein backbone.
| (6) |
Significantly conserved regions calculated within a user-defined sliding window (default value = 20 residues with cutoff of p < 0.01) are plotted upon the positional local correlational value profile (i.e., R value) and also mapped to the reference structure of the protein, colored in dark gray on a light background.
Variant impact assessment
By extension, mutational impacts of genetic or drug class variants on the functionally conserved dynamics can be quantified by their effects that range significantly beyond those observed in the self-similar validation runs that identify functionally conserved dynamic regions. Thus, when canonical correlations of variants differ significantly from the self-correlation observed in functionally conserved regions, we can plot the magnitude of impact defining how the variant’s dynamics differs from the routine self-similar dynamics of the normally functioning protein. The impacts of dissimilar states caused by altered amino acid sequence or different binding partners are assessed through their local effect on the same canonical correlation identifying conserved dynamics. We introduce a metric of relative entropy relating the canonical correlations in both the self-similar and altered variant state. In essence, this is a metric of the “impact” of a given genetic or drug class variant within the context of normal functioning dynamics. For example, when trained on a natural binding interaction (e.g., DROIDS analysis comparing a DNA binding protein in its bound and unbound states), novel MD simulations with a variety of amino acid replacements can be deployed to see whether the learners can still recognize the functional dynamics in the mutant forms. In this case, functionally tolerated mutations will result in functionally conserved dynamics that do not vary outside of ±3 standard deviation bounds of the self-similar validation runs, whereas functionally intolerant mutations will result in significant deviations from self-similarity of motion. An overall impact of a genetic and/or drug class binding variant on the conserved dynamic regions is calculated by
| (7) |
Comparative plots of local variant impacts outside of the 3 standard deviation bound determined by the validation run are generated within a user-defined sliding window. Thus, this variant impact metric is designed to identify variant regions with dynamics that potentially alter conserved dynamic features of the normally functioning protein system.
Three example applications (case studies)
To demonstrate the performance and utility of DROIDS 3.0 with maxDemon 1.0, we ran the following three comparative case studies using the Protein Data Bank (PDB) identifiers mentioned below. Bound and unbound files were created by deleting binding partners in UCSF Chimera (28) and resaving PDBs (e.g., 3t0z_bound.pdb, 3t0z_unbound, and 3t0z_ligand). Each MD run ensemble consisted of 200 production runs at 0.5 ns explicitly solvated in a 12-nm octahedral water box using TIP3P solvent model (29) with constant temperature under an Anderson thermostat (30) using particle mesh Ewald summation implemented on pmemd.cuda (15). The models were charge neutralized with both Na+ and Cl− ions. The heating and equilibration runs before production were 0.3 and 10 ns, respectively. Before heating, 2000 steps of energy minimization were also performed. All seven available machine learning classifiers were trained on the functional MD ensembles and deployed upon new 5-ns production runs for each variant analyzed. The force fields applied were ff14SB (31), DNA.OL15 (32), and GAFF2 (33), when appropriate.
Case study 1 (Figs. 2 and 3; PDB: 1ubq and 2oob)—to analyze self-stability, effect of temperature shift in ubiquitin, and functional binding of ubiquitin to ubiquitin ligase.
Figure 2.
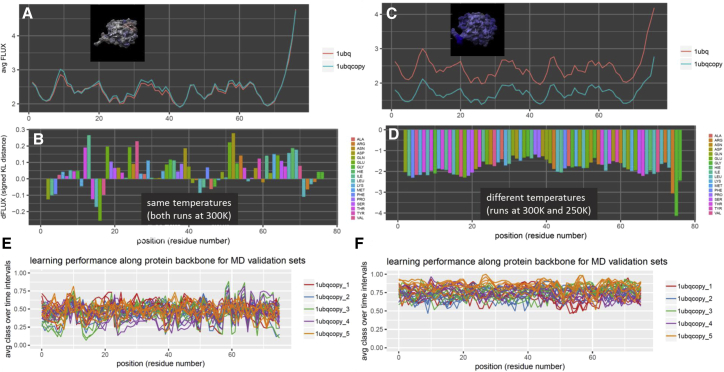
Analysis of environmental temperature change on nonfunctional ubiquitin dynamics (PDB: 1ubq). Shown are DROIDS image and analysis of random ubiquitin dynamics compared at the same (A, B, and E) and different (C, D, and F) temperatures. Note: inset images (A and C) show the KL divergence in dynamics (B and D), with blue color indicating damped atom fluctuation (decreased rmsf) at temperatures lowered by 50 K. Note that machine learning performance is much higher when a temperature difference is modeled (E and F); however, as expected, neither comparison offers the machine learners a sequence-dependent profile correlation by which to establish a signal of conserved dynamics. To see this figure in color, go online.
Figure 3.
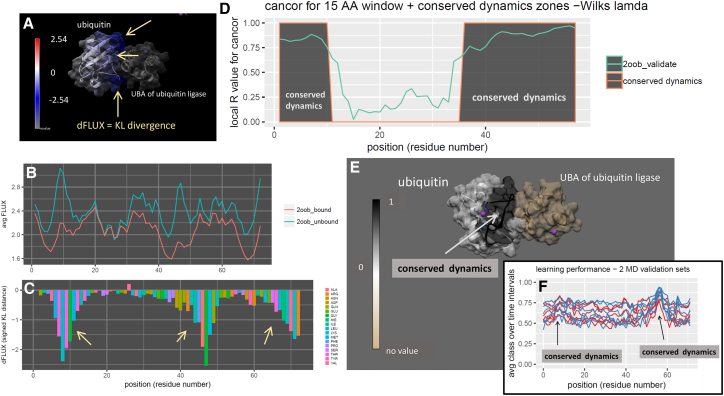
Analysis of mutational impact and tolerance on functional ubiquitin dynamics (PDB: 2oob). (A) Shown are DROIDS image and analysis of ubiquitin bound to the UBA of ubiquitin ligase. Note: blue color quantifies dampened atom fluctuation (rmsf) at the binding interface (i.e., negative dFLUX on same range scale in (C)) and also by the (B) respective rmsf profiles of bound and unbound training states and (C) the KL divergence or dFLUX profile colored by residue. Arrows indicate the most prominent dampening of rmsf near loops at Thr9, Ala46, and C-terminus. Regions of functionally conserved dynamics, determined via significant local canonical correlation, are shown in dark gray in both (D) traditional N- to C-terminal plot as well as (E) structural image. (F) Local learning performance of each machine learning method in self-similar testing runs is shown color coded by validation runs on ubiquitin bound to ubiquitin ligase (PDB: 2oob). Note the two prominent local regions of correlated (and sequence-dependent) learning performance used as indicators of functionally conserved dynamics. To see this figure in color, go online.
Case study 2 (Fig. 4; PDB: 1cdw) —to analyze functional binding of TBA to DNA and the impacts of several genetic variants.
Figure 4.
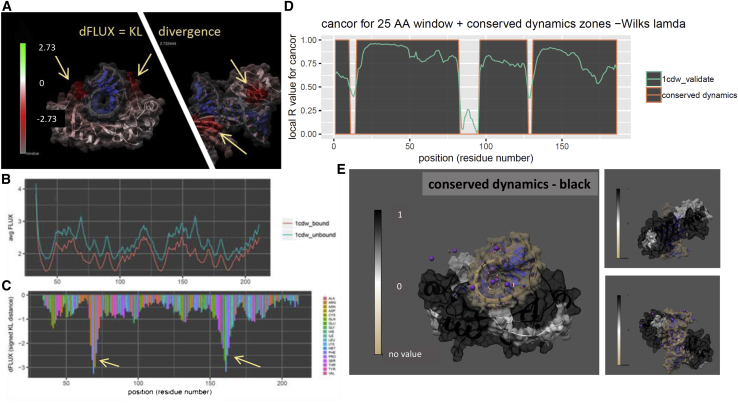
Analysis of mutational impact and tolerance on DNA binding in TBP (PDB: 1cdw). Shown are DROIDS image and analysis of TBP in DNA-bound and -unbound states showing (A) KL divergence colored TBP structure, (B) respective rmsf profiles, and (C) KL divergence (dFLUX) plot. Note: arrows indicate functional binding loops in the DNA minor groove, and red color indicates dampened rmsf. Shown is the maxDemon analysis (D and E) identifying conserved dynamics supporting both minor groove binding loops and connecting them through the central region of the β-sheet in the main body of TBP closest to the DNA. To see this figure in color, go online.
Case study 3 (Fig. 5; PDB: 3t0z plus six variants (Fig. S2)) —to analyze functional ATP binding in Hsp90 and subsequent impacts of six Hsp90-inhibitor drug variants.
Figure 5.
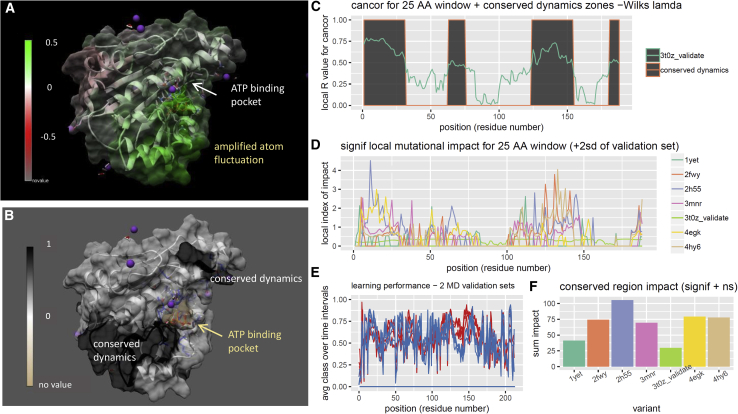
Analysis of drug class variant binding in the ATP-binding domain of Hsp90 (PDB: 1yet, 2fwy, 2h55, 3t0z (ATP), 3mnr, 4egk, 4hy6). Shown are DROIDS image and analysis of Hsp90 in ATP-bound and -unbound states showing (A) KL divergence colored Hsp90 structure. Note: arrows and green color indicate regions where rmsf is amplified in response to ATP binding. Shown is a maxDemon analysis identifying conserved dynamics connecting the ATP-binding pocket and region of amplified rmsf (B and C); mutational impacts of six drug class variants targeting the ATP-binding pocket of Hsp90 are plotted locally as line graphs and in total as a bar chart. Note how geldanamycin (1yet), which mimics the ATP in its binding contacts, has overall dynamic impact very similar to that of ATP (3t0z). See Fig. S3 for more details regarding drug class variant contacts to the ATP-binding pocket. To see this figure in color, go online.
Improvements and upgrades over previous versions
A complete list of improved features and upgrades can be found on page 2 of the DROIDS 3.0 user manual. General advice for implementation can also be found here. A 22-page, step-by-step, illustrated tutorial is also available (DROIDS + maxDemonTUTORIAL.pdf) on our website and GitHub repository below. At GitHub, please follow the link to “Releases” and download the latest release as a .tar.gz or .zip file. This software is distributed under open source GPL v3 license. See COPYING.txt in the GitHub repo for more details (https://github.com/gbabbitt/DROIDS-3.0-comparative-protein-dynamics; https://doi.org/10.5281/zenodo.3358976 concurrent with this publication; https://zenodo.org/record/3567555#.Xe4MJ5NKiiQ).
The software is also hosted at our laboratory’s website, https://people.rit.edu/gabsbi, along with a short video description; information regarding its utility, system requirements, and implementation; and a step-by-step tutorial for new users. Bugs can be reported to the to the corresponding author of this paper.
We also periodically post examples using DROIDS, video tutorials, and ongoing student projects here to https://www.youtube.com/channel/UCJTBqGq01pBCMDQikn566Kw.
Major dependencies are as follows: Amber 16/18, Ambertools (cpptraj), CUDA 9.0, UCSF Chimera, and R software. Note that there are more minor package dependencies from Debian, perl, python, and R, so please use the installer perl script DROIDS + AMBERinstaller.pl included in the download. Command prompts allow users to skip over the UCSF Chimera, Amber, Ambertools, CUDA, and base R installations if the system already has these in place. Building from a fresh Linux Mint install might work best for most users who are experienced with building Linux-based Amber systems. We are currently working on a Docker container and virtual machine environment for DROIDS to allow for high-performance installations on GPU-enabled servers and GPU-enabled cloud services.
Results and Discussion
To demonstrate the variety of comparative analyses that can be addressed with the new release of DROIDS 3.0 and maxDemon 1.0, we chose three different case studies of comparative protein dynamics. These included 1) an analysis of self-stability and temperature effects in single ubiquitin structure and a subsequent analysis of ubiquitin and ubiquitin ligase binding interaction, 2) a functional genetic variant analysis of DNA binding in TBA, and 3) a drug class variant analysis of compounds targeting the ATP-binding region of the Hsp90 heat shock protein.
Machine learning analysis of functional and nonfunctional dynamics of ubiquitin
We first simulated a null comparison as a “sanity check” by running a query and reference ubiquitin (34) MD at the same temperatures (both 300 K) and same solvent conditions (PDB: 1ubq). The DROIDS analysis (Fig. 2, A and B) showed identical atom fluctuation profiles along the backbone and a random dFLUX profile indicative of nonsignificant differences due to small random local thermal differences in the training sets. The machine learning classification plots on new MD runs vary randomly around 0.5, reflecting the fact that the learning algorithms effectively had no features to train on (Fig. 2 E). By contrast, a protein dynamic comparison run with a 50 K temperature difference (Fig. 2, C and D) shows a much higher machine learner performance upon deployment (i.e., 70–80% successful classification; Fig. 2 F). Because environmental temperature shifts are not expected to reflect evolutionarily conserved dynamics and are not position dependent in their effect, they subsequently do not result in significant canonical correlations in the learning profiles (data not shown). Representative time slices of the positional classifications in each of these experiments indicate that our machine learning is capable of extracting and identifying simple differences in dynamics due to temperature. Another interesting observation here was the slightly higher learning performance of the simpler machine learning methods QDA and LDA over others at all sites in the temperature-shifted example. We interpret this to be related to the fact that underlying rmsf distributions are probably Gaussian, a critical assumption of these two models, with unequal variances caused by steric hindrances on the backbone. This would predict that QDA might outperform other learners in this situation, and it appears that it does. We note that when more complex functional dynamics are concerned, the more sophisticated learning methods, such as support vector machine and adaboost, often perform slightly better than others. However, we also note that these performance differences are usually quite small and that all learning methods generally come to similar local conclusions about functional dynamics.
We now move on to examine machine learning performance regarding more functional binding dynamics in ubiquitin. To examine functional dynamics in ubiquitin, we conducted a DROIDS analysis comparing its two functional states, bound and unbound, to the ubiquitin-associated binding (UBA) domain of ubiquitin ligase (Fig. 3 A; (35)). This binding domain is highly conserved among the many other proteins that interact directly with ubiquitin. The binding interaction greatly reduces the atom fluctuation in ubiquitin at three characteristic positions involving two loop structures centered at Leu8 and Ala46 and a portion of β-sheet at the C-terminus (Fig. 3, B and C). These three regions also drive significant differences in dynamics across the whole protein. In novel self-similar MD validation runs on the bound state, we successfully detect significant canonical correlations, determined by Wilk’s lambda, and indicate functionally conserved dynamics in these three regions covering a broad expanse of the known conserved regions across the UBA domain (Fig. 3, D and E). These regions correspond to prominent local peaks in the machine learning performance after training upon the bound and unbound dynamics (Fig. 3 F).
Machine learning analysis of impacts of genetic variants on DNA binding interaction
TBP is a general transcription factor that binds DNA upstream in most highly regulated eukaryotic gene promoter regions (36). Although relatively small, it is a mechanically dynamic protein with a C-clamp-like structure that highly distorts the rigid DNA double helix by inserting four phenylalanine side chains between basepairs. It is thought that this bending allows TBP to be more rapidly released from the TATA element, as opposed to TATA-less promoters, subsequently allowing more highly controlled regulatory responses in TATA box genes (37). Because of its obvious symmetry and ability to impart large forces during binding, we thought that it would represent a good candidate for comparison of its dynamics during its binding interaction with DNA. We conducted a DROIDS analysis comparing human TBP (38) in its functionally bound and unbound states (Fig. 4, A and C). TBP exhibits a characteristically large signature of dampening of atom fluctuation throughout its entire structure, with most pronounced effects in two loop regions that interact with the minor groove of DNA (arrows in Fig. 4, A and C). Canonical correlations in new self-similar MD runs marking increased performance in classification were observed in these regions (Fig. 4 D) along with corresponding regions of conserved dynamics identified by significant Wilk’s lambda (Fig. 4 E). Conserved dynamics from these loop areas are connected through the chains in the β-sheet region of TBP spanning the DNA major groove contact. Mutational impacts of four variants affecting the binding loop most proximal to the C-terminal exhibited followed our expectation of increasing impact ordering from R192Q, R192K, R192polyD, and R192polyW (Fig. S1). The polyD and polyW mutations incorporated five sequential Asp or Trp residues centered at R192, both causing the loop region to become more rigid (causing increased negative dFLUX). We expected that the strong functional binding effect observed across nearly all residues in this system would make it relatively highly tolerant to single amino acid substitutions, even when located in the most functional binding loop. In accordance with our expectations, we found the most impactful multiple mutation (i.e., R192polyW) significantly affected the dynamics of nearly six times more local residues than the least impactful single substitution (i.e., R192Q).
Machine learning analysis of impacts of drug class variants targeting the ATP-binding region of Hsp90
In contrast to TBP binding to DNA, we wanted to use our method to examine a small-molecule binding interaction in a protein with potentially more complex impacts on MD. Hsp90 is a well-known chaperone protein that assists the folding of many proteins, thereby mitigating many environmental stresses in the cell. Hsp90 also capacitates the evolutionary process by allowing potential phenotypic variation exhibited under stress to be hidden from natural selection until needed in response to environmental change (39). Hsp90 contains a highly conserved N-terminal domain in which ATP binding and activation occur (i.e., the Bergerat fold). The binding of ATP physically changes motions in this region, creating a “lid” that is closed during ATP binding and open when conversion to ADP occurs. Dimerization of two N-terminal domains occurs as part of the ATPase cycle. Because of the role of Hsp90 in stress mitigation in most tumors, it is a common drug target for ATP inhibitors in many cancer therapies (40,41). The amino acid residues that interact with ATP in this region are well-known, and the inhibitor geldanamycin is known to mimic nearly all the local ATP contacts as well (42). Other more modern inhibitors interact with the ATP-binding pocket quite differently (41,43,44), so we hypothesized that this system would be a good candidate for comparative analysis of drug class variants with our software.
We conducted a DROIDS analysis comparing the dynamics of this ATP-binding N-terminal domain of Hsp90, a common drug target for inhibitors in many cancer therapies, in both its ATP-bound and -unbound states. The binding of ATP was discovered to significantly destabilize three colocalized α-helical regions of the protein adjacent to and extending from the ATP-binding site (Fig. 5 A), possibly functioning to unbalance solvent interactions on the structure and subsequently enhance the dimer formation between adjacent N-terminal domains. MaxDemon analysis confirmed the dynamics of this region to be highly conserved in new MD runs (Fig. 5, B and C). We also analyzed the impacts of the six drug class variants targeting the ATP site (42, 43, 44, 45, 46) but interacting differently with residues in this region (Fig. 5, D–F). The contacts in the ATP-binding site are listed in Figure360. An Author Presentation of Fig. 1, Document S2. Article plus Supporting Material A. Although the localized patterns of impacts of the drug variants were all quite similar to ATP (Fig. S2 B), the drug variants that most closely mimicked the contacts of ATP (i.e., geldanamycin, PDB: 1yet) had far less impact on conserved dynamics than variants that interacted very differently with the binding pocket (i.e., benzamide SNX1321 and inhibitor FJ1 (PDB: 3mnr and 4hy6). See Fig. 5 F. We feel that this finding not only demonstrates the potential utility of our method quite well but also suggests that although it is important to be able to target a druggable protein binding site (47), researchers should also consider how these various small molecules might alter, or fail to alter, the natural dynamics of the ligand binding in the target protein. For situations in which a drug might too closely mimic the dynamic effects of a natural activator like ATP, a hyperactivation response of phosphorylation signaling in pathways might occur (48, 49, 50). Alternatively, other situations may require drug targeting that does not alter the natural dynamic behavior too much, potentially activating proteolytic systems in the cell. Our software allows more detailed investigations of these potential dynamic impacts of drug class variants.
Conclusion
We provide a valid method and well-documented, user-friendly software pipeline for conducting statistically sound comparative studies of large ensembles of comparative protein dynamics. The method/software provides machine-learning-based quantification of effects on novel MD simulations that represent various functional variants of interest to the user. Although there are currently several other software packages allowing users to connect sequence-based evolutionary metrics to protein dynamics (51, 52, 53), our method/software is unique in that regions of functional conservation are identified by analyzing self-similar features of dynamics themselves rather than relying upon linking analysis of dynamics to traditional, static, sequence-based approaches to molecular evolutionary inference, which do not necessarily assume that a conserved function region has a strong dynamic component. By providing a systematic way of comparing protein dynamics at single-residue resolution, our method/software provides an important step beyond traditional sequence-based bioinformatics, allowing investigators another valuable method by which to gain a more biophysically grounded view of functional and evolutionary change (52, 53, 54). Another advantage to our method/software is that our functional impacts (i.e., mutational tolerance) are defined solely within the context of a protein dynamic system simulation. This provides a much deeper look into protein-specific function than current genomic and proteomic database methods of predicting mutational tolerance (21,22) currently allow. Although MD analysis is currently often limited by molecular system size and timescale, as off-the-shelf GPU technology for both IT servers and the PC gaming community continues to advance at a rapid pace over the next few years, our method/software may have future potential applications to the development of precision and personalized medicine. This is especially true when detailed understanding of the interactions between genetic and drug class variants are needed within the context of the dynamics of specific disease-related protein systems.
Author Contributions
G.A.B. and E.P.F. conceived the project and method. All authors contributed to the code base. G.A.B. and L.E.A. worked on β-testing and debugging.
Acknowledgments
We acknowledge the laboratory of Dr. Andre O. Hudson for β-testing our software during its early development. We also acknowledge Dr. Daniel Wysocki (Astrophysics program-CCRG at Rochester Institute of Technology) for very helpful suggestions regarding early methodological discussions. We also acknowledge Dr. Miranda Lynch and her colleagues at the Hauptman-Woodward Medical Research Institute for many helpful comments.
We acknowledge the Nvidia corporation for a hardware support grant.
Editor: Alan Grossfield.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.12.008.
Supporting Material
References
- 1.Feynman R.P., Leighton R.B., Sands M. First Edition. Pearson P T R; Reading, MA: 1970. The Feynman Lectures on Physics. [Google Scholar]
- 2.Gao M., Zhou H., Skolnick J. Insights into disease-associated mutations in the human proteome through protein structural analysis. Structure. 2015;23:1362–1369. doi: 10.1016/j.str.2015.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tokuriki N., Stricher F., Tawfik D.S. How protein stability and new functions trade off. PLoS Comput. Biol. 2008;4:e1000002. doi: 10.1371/journal.pcbi.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shoichet B.K., Baase W.A., Matthews B.W. A relationship between protein stability and protein function. Proc. Natl. Acad. Sci. USA. 1995;92:452–456. doi: 10.1073/pnas.92.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martelli P.L., Fariselli P., Casadio R. Large scale analysis of protein stability in OMIM disease related human protein variants. BMC Genomics. 2016;17(Suppl 2):397. doi: 10.1186/s12864-016-2726-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sathyapriya R., Duarte J.M., Lappe M. Defining an essence of structure determining residue contacts in proteins. PLoS Comput. Biol. 2009;5:e1000584. doi: 10.1371/journal.pcbi.1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saha R.P., Bahadur R.P., Chakrabarti P. Interresidue contacts in proteins and protein-protein interfaces and their use in characterizing the homodimeric interface. J. Proteome Res. 2005;4:1600–1609. doi: 10.1021/pr050118k. [DOI] [PubMed] [Google Scholar]
- 8.Faure G., Bornot A., de Brevern A.G. Protein contacts, inter-residue interactions and side-chain modelling. Biochimie. 2008;90:626–639. doi: 10.1016/j.biochi.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 9.Greives N., Zhou H.X. Both protein dynamics and ligand concentration can shift the binding mechanism between conformational selection and induced fit. Proc. Natl. Acad. Sci. USA. 2014;111:10197–10202. doi: 10.1073/pnas.1407545111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mobley D.L., Dill K.A. Binding of small-molecule ligands to proteins: “what you see” is not always “what you get”. Structure. 2009;17:489–498. doi: 10.1016/j.str.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henzler-Wildman K.A., Lei M., Kern D. A hierarchy of timescales in protein dynamics is linked to enzyme catalysis. Nature. 2007;450:913–916. doi: 10.1038/nature06407. [DOI] [PubMed] [Google Scholar]
- 12.Niessen K.A., Xu M., Markelz A.G. Moving in the right direction: protein vibrations steering function. Biophys. J. 2017;112:933–942. doi: 10.1016/j.bpj.2016.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Babbitt G.A., Coppola E.E., Hudson A.O. Can all heritable biology really be reduced to a single dimension? Gene. 2016;578:162–168. doi: 10.1016/j.gene.2015.12.043. [DOI] [PubMed] [Google Scholar]
- 14.Götz A.W., Williamson M.J., Walker R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized born. J. Chem. Theory Comput. 2012;8:1542–1555. doi: 10.1021/ct200909j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salomon-Ferrer R., Götz A.W., Walker R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J. Chem. Theory Comput. 2013;9:3878–3888. doi: 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- 16.Babbitt G.A., Mortensen J.S., Liao J.K. DROIDS 1.20: a GUI-based pipeline for GPU-accelerated comparative protein dynamics. Biophys. J. 2018;114:1009–1017. doi: 10.1016/j.bpj.2018.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Díaz Ó., Dalton J.A.R., Giraldo J. Artificial intelligence: a novel approach for drug discovery. Trends Pharmacol. Sci. 2019;40:550–551. doi: 10.1016/j.tips.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 18.Plante A., Shore D.M., Weinstein H. A machine learning approach for the discovery of ligand-specific functional mechanisms of GPCRs. Molecules. 2019;24:E2097. doi: 10.3390/molecules24112097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Terayama K., Iwata H., Tsuda K. Machine learning accelerates MD-based binding pose prediction between ligands and proteins. Bioinformatics. 2018;34:770–778. doi: 10.1093/bioinformatics/btx638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maxwell J.C. Ninth Reprint Edition. Dover Publications; Mineola, NY: 2001. Theory of Heat. [Google Scholar]
- 21.Adzhubei I., Jordan D.M., Sunyaev S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013;Chapter 7:Unit7.20. doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 23.Roe D.R., Cheatham T.E., III PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 24.Kullback S., Leibler R.A. On information and sufficiency. Ann. Math. Stat. 1951;22:79–86. [Google Scholar]
- 25.Breiman L. Random forests. Mach. Learn. 2001;45:5–32. [Google Scholar]
- 26.Venables W.N., Ripley B.D. New York; 2010. Modern Applied Statistics with S. Springer Publishing Company, Incorporated, [Google Scholar]
- 27.2007. Canonical Correlation Analysis. In: Applied Multivariate Statistical Analysis, W. Härdle and L. Simar, eds. (Springer Berlin Heidelberg), pp. 321–330.
- 28.Pettersen E.F., Goddard T.D., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 29.Mao Y., Zhang Y. Thermal conductivity, shear viscosity and specific heat of rigid water models. Chem. Phys. Lett. 2012;542:37–41. [Google Scholar]
- 30.Andersen H.C. Molecular dynamics simulations at constant pressure and/or temperature. J. Chem. Phys. 1980;72:2384–2393. [Google Scholar]
- 31.Maier J.A., Martinez C., Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dans P.D., Ivani I., Orozco M. How accurate are accurate force-fields for B-DNA? Nucleic Acids Res. 2017;45:4217–4230. doi: 10.1093/nar/gkw1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J., Wolf R.M., Case D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 34.Vijay-Kumar S., Bugg C.E., Cook W.J. Structure of ubiquitin refined at 1.8 A resolution. J. Mol. Biol. 1987;194:531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 35.Peschard P., Kozlov G., Gehring K. Structural basis for ubiquitin-mediated dimerization and activation of the ubiquitin protein ligase Cbl-b. Mol. Cell. 2007;27:474–485. doi: 10.1016/j.molcel.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 36.Kornberg R.D. The molecular basis of eukaryotic transcription. Proc. Natl. Acad. Sci. USA. 2007;104:12955–12961. doi: 10.1073/pnas.0704138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tora L., Timmers H.T. The TATA box regulates TATA-binding protein (TBP) dynamics in vivo. Trends Biochem. Sci. 2010;35:309–314. doi: 10.1016/j.tibs.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 38.Nikolov D.B., Chen H., Burley S.K. Crystal structure of a human TATA box-binding protein/TATA element complex. Proc. Natl. Acad. Sci. USA. 1996;93:4862–4867. doi: 10.1073/pnas.93.10.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rutherford S.L., Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- 40.Neckers L., Workman P. Hsp90 molecular chaperone inhibitors: are we there yet? Clin. Cancer Res. 2012;18:64–76. doi: 10.1158/1078-0432.CCR-11-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuno A., Lee M.J., Trepel J.B. Clinical evaluation and biomarker profiling of Hsp90 inhibitors. Methods Mol. Biol. 2018;1709:423–441. doi: 10.1007/978-1-4939-7477-1_29. [DOI] [PubMed] [Google Scholar]
- 42.Stebbins C.E., Russo A.A., Pavletich N.P. Crystal structure of an Hsp90-geldanamycin complex: targeting of a protein chaperone by an antitumor agent. Cell. 1997;89:239–250. doi: 10.1016/s0092-8674(00)80203-2. [DOI] [PubMed] [Google Scholar]
- 43.Immormino R.M., Kang Y., Gewirth D.T. Structural and quantum chemical studies of 8-aryl-sulfanyl adenine class Hsp90 inhibitors. J. Med. Chem. 2006;49:4953–4960. doi: 10.1021/jm060297x. [DOI] [PubMed] [Google Scholar]
- 44.Li J., Sun L., He J. Structure insights into mechanisms of ATP hydrolysis and the activation of human heat-shock protein 90. Acta Biochim. Biophys. Sin. (Shanghai) 2012;44:300–306. doi: 10.1093/abbs/gms001. [DOI] [PubMed] [Google Scholar]
- 45.Fadden P., Huang K.H., Hall S.E. Application of chemoproteomics to drug discovery: identification of a clinical candidate targeting hsp90. Chem. Biol. 2010;17:686–694. doi: 10.1016/j.chembiol.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 46.Austin C., Pettit S.N., Hughes D.E. Fragment screening using capillary electrophoresis (CEfrag) for hit identification of heat shock protein 90 ATPase inhibitors. J. Biomol. Screen. 2012;17:868–876. doi: 10.1177/1087057112445785. [DOI] [PubMed] [Google Scholar]
- 47.Vajda S., Beglov D., Whitty A. Cryptic binding sites on proteins: definition, detection, and druggability. Curr. Opin. Chem. Biol. 2018;44:1–8. doi: 10.1016/j.cbpa.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poulikakos P.I., Zhang C., Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hatzivassiliou G., Song K., Malek S. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464:431–435. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 50.Cichowski K., Jänne P.A. Drug discovery: inhibitors that activate. Nature. 2010;464:358–359. doi: 10.1038/464358a. [DOI] [PubMed] [Google Scholar]
- 51.Bakan A., Dutta A., Bahar I. Evol and ProDy for bridging protein sequence evolution and structural dynamics. Bioinformatics. 2014;30:2681–2683. doi: 10.1093/bioinformatics/btu336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Münz M., Lyngsø R., Biggin P.C. Dynamics based alignment of proteins: an alternative approach to quantify dynamic similarity. BMC Bioinformatics. 2010;11:188. doi: 10.1186/1471-2105-11-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nevin Gerek Z., Kumar S., Banu Ozkan S. Structural dynamics flexibility informs function and evolution at a proteome scale. Evol. Appl. 2013;6:423–433. doi: 10.1111/eva.12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fortin C.H., Schulze K.V., Babbitt G.A. TRX-LOGOS - a graphical tool to demonstrate DNA information content dependent upon backbone dynamics in addition to base sequence. Source Code Biol. Med. 2015;10:10. doi: 10.1186/s13029-015-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.