Abstract
The present study investigated the effects and molecular mechanism of the second mitochondria-derived activator of caspase (SMAC) mimetic birinapant on the proliferation and apoptotic rate of liver cancer cells. Western blotting and reverse transcription-quantitative PCR were used to detect the protein and mRNA expression levels of cellular inhibitor of apoptosis 1 (cIAP1) and tumor necrosis factor receptor-associated factor 3 (TRAF3) in the liver cancer cell lines Huh7, H22 and HepG2, and the hepatocyte line AML12. Annexin V-FITC and Transwell assays were used to assess the effect of birinapant pretreatment on the apoptotic rate and invasive ability of liver cancer cells. Lentivirus-mediated silencing of TRAF3 was performed in liver cancer cells. Western blotting was used to detect the lentivirus silencing efficiency. A subcutaneous hepatocellular carcinoma model was established in nude mice and 15 days after tumor induction the subcutaneous tumors were measured in each group. Immunohistochemistry assays were used to detect the protein expression levels of proliferating cell nuclear antigen and caspase-3. The results suggested that the expression levels of cIAP1 and TRAF3 were lower in Huh7, H22 and HepG2 cells compared with AML12 cells. Pretreatment with birinapant promoted apoptosis and inhibited invasion of liver cancer cells by activating the cIAP1/TRAF3 axis. Birinapant also promoted apoptosis and inhibited the growth of subcutaneous hepatocellular carcinoma tumors in nude mice. The present results suggested that the SMAC mimetic birinapant may promote apoptosis, and inhibit the proliferation and invasion of liver cancer cells. The molecular mechanism responsible for the effects of birinapant may be related to activation of the cIAP1/TRAF3 signaling pathway by birinapant in liver cancer cells.
Keywords: birinapant, HCC, cIAP1, TRAF3, apoptosis
Introduction
Hepatocellular carcinoma (HCC) is the third leading cause of cancer-related mortality worldwide (1). While diverse treatments exist for HCC, the clinical prognosis of patients with HCC remains poor (2). Therefore, it is necessary to investigate the molecular mechanism underlying HCC. Second mitochondria-derived activator of caspase (SMAC) is a protein located in the mitochondria that can regulate apoptosis (3). SMAC can be released from mitochondria into the cytoplasm where it binds to cellular inhibitor of apoptosis 1 (cIAP1) and inhibits the anti-apoptotic function of cIAP1, thus promoting apoptosis (4,5). As a result of previous in-depth studies showing the structure and function of SMAC protein, the application of SMAC mimetics in the treatment of various diseases, especially tumors, has become a novel research hotspot (6–9). SMAC mimetics have been used for the treatment of cancer, including breast cancer, HCC and lung cancer (6,7). Birinapant is a SMAC mimetic that can promote apoptosis of tumor cells by binding to cIAP1 and inducing cIAP1 degradation (8,9). The caspase-8 inhibitor emricasan can combine with birinapant to induce necroptosis and treat acute myeloid leukemia (10). Additionally, birinapant can act as a nanomicellar carrier of paclitaxel delivery for cancer therapy (11); and it also has anti-inflammatory effects (12). Birinapant attenuates lipopolysaccharide (LPS)-induced liver injury by inhibiting tumor necrosis factor receptor-associated factor 3 (TRAF3) degradation in Kupffer cells (12). However, whether birinapant affects HCC growth and the possible underlying molecular mechanism of this effect are still unknown. In the present study, birinapant and liver cancer cell lines Huh7, H22 and HepG2 were used to study the effects and molecular mechanisms of birinapant on the proliferation and apoptotic rate of liver cancer cells. It was found that birinapant could promote apoptosis, as well as inhibiting the proliferation and invasion of liver cancer cells. The molecular mechanism may be related to the activation of the cIAP1/TRAF3 signaling pathway by birinapant in liver cancer cells.
Materials and methods
Cell culture and treatment
The liver cancer cell lines Huh7, HepG2 and H22, and the hepatocyte line AML12 were obtained from the Chinese Academy of Science. All cells were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS (PAN-Biotech GmbH) with 1% penicillin and 1% streptomycin. Cells were maintained at 37°C in a humidified atmosphere containing 5% CO2. Huh7, HepG2 and H22 cells were pretreated in a cell incubator with 300 nmol/ml birinapant (MedChemExpress) at 37°C for 24 h prior to subsequent experimentation.
Lentivirus-mediated knockdown of TRAF3 in Huh7 and H22 cells
The lentivirus was obtained from Shanghai GeneChem Co., Ltd. (shRNA-TRAF3 was inserted into these lentiviruses by Shanghai GeneChem Co., Ltd). The lentivirus-mediated knockdown of TRAF3 in Huh7 and H22 cells was performed as previously described (13). A total of 4 µl of lentivirus titer (1×108 TU/ml) with shRNA was added to 1×106 Huh7 and H22 cells. Lentivirus without shRNA served as the negative control, and the blank control group was Huh7 and H22 cells without the lentivirus vector, these cells were continuously cultured in a cell incubator at 37°C for 72 h. At 72 h later, green fluorescence observed under an inverted fluorescent microscope (×200) and western blotting were used to determine the efficiency of lentivirus-mediated knockdown.
Establishment of a subcutaneous tumor model in BALB/c nude mice
Male, 7-week-old BALB/c nude mice (~13 g) were obtained from the Animal Experimental Center of Chongqing Medical University. All mice were fed with common feed and sterile water ad libitum, and housed in 22°C with 50% humidity, with 12 h light/dark cycles in a cage of 5 mice. In total, 18 mice were divided into three groups (Control group; shRNA-TRAF3 + Birinapant group; and Birinapant group) with six mice in each group. Subcutaneous tumor models were prepared by subcutaneously injecting 2×107 H22 cells into the left armpit of each mouse. In the birinapant group, nude mice were injected intraperitoneally with 30 mg/kg body weight birinapant the day after the subcutaneous injection, and then again 1 week later. In the Control group, mice were injected with an equivalent volume of saline at the same time points as birinipant injection. The subcutaneous tumors were observed every day, and after 15 days the subcutaneous tumor tissues were removed for subsequent analyses.
Western blotting
After the cells were harvested, the cellular protein was extracted using a protein extraction kit (Nanjing KeyGen Biotech Co., Ltd.). Protein concentration was detected by BCA protein assay kit (Beyotime Institute of Biotechnology) and a microplate reader (Synergy™ HT; BioTek). Total cellular protein was mixed with loading buffer and boiled in water for 10 min. In total, 50 µg protein lysate was separated by SDS-PAGE with 10% gels and transferred to PVDF membranes (EMD Millipore), which were blocked with 5% skim milk for 1 h at room temperature. The membranes were then incubated overnight at 4°C with antibodies against cIAP1 (1:1,000; cat. no. ab189193; Abcam), TRAF3 (1:1,000; cat. no. ab36988; Abcam) and β-actin (1:1,000; cat. no. 4970; Cell Signaling Technology, Inc.). The membranes were further incubated with the corresponding horseradish peroxidase (HRP)-conjugated secondary antibody (1:10,000; cat. no. 7074; Cell Signaling Technology, Inc.) at 37°C for 1 h. Bands were analyzed and scanned using Quantity One software (version 5.2.1; Bio-Rad Laboratories, Inc.) after incubation with enhanced chemiluminescence reagent (EMD Millipore) at room temperature for 10 sec.
RNA isolation and reverse transcription-quantitative (RT-q)PCR
Total RNA was isolated from Huh7, H22, HepG2 and AML12 cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Then, the RNA was reverse transcribed to cDNA with the PrimeScript™ RT Reagent kit (Takara Biotechnology Co., Ltd.) at 37°C for 15 min, 85°C for 5 sec and 4°C hold. RT-qPCR was performed using SYBR-Green (Takara Biotechnology, Co., Ltd.) and an ABI PRISM 7900 Sequence Detection system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling conditions were as follows: 95°C for 30 sec for one cycle; 95°C for 5 sec and 60°C for 30 sec for 39 cycles; 65°C for 5 sec and 95°C for 15 sec for one cycle. The gene expression level of each target gene was normalized to that of GAPDH for each sample. The primers used were as follows: cIAP1, forward 5′-GATACGAATGAAAGGCCAAG-3′, reverse 5′-TCTCCAGGTCCAAAATGAAT-3′; TRAF3, forward 5′-CTCCCATCAAGAAGCCACC-3′, reverse 5′-TGTAGGGCCTCTTTGAGGTAA-3′; and GAPDH, forward 5′-CACCCACTCCTCCACCTTTG-3′ and reverse 5′-CCACCACCCTGTTGCTGTAG-3′. The relative gene expression was analyzed using the 2−∆∆Cq method (14).
Immunohistochemical staining
Immunohistochemical staining was performed as previously described (15). Subcutaneous tumor tissues (n=18) were fixed in 4% formalin at room temperature for 48 h and embedded in paraffin. Then, sections (4 µm) were cut and mounted on glass slides. The slides were dewaxed in 100% xylol for 30 min and rehydrated in 100, 95 and 85% graded alcohol solutions. Subsequently, antigen retrieval was performed in 0.01 M sodium citrate solution at 98°C for 15 min. Endogenous peroxidase activity was blocked with 5% normal goat serum (OriGene Technologies, Inc.) at 37°C for 1 h. The subcutaneous tumor tissues were incubated with a proliferating cell nuclear antigen (PCNA) antibody (1:100; cat. no. 13110; Cell Signaling Technology, Inc.) and caspase-3 antibody (1:100; cat. no. ab4051; Abcam) at 4°C overnight. Then, the slides were washed three times with PBS and incubated with an anti-rabbit lgG HRP-conjugated secondary antibody (1:10,000; cat. no. 7074; Cell Signaling Technology, Inc.) for 1 h at room temperature. The slides were then washed three times with PBS and 50 µl of DAB solution (Beyotime Institute of Biotechnology) was added for 3–5 min, followed by another wash. Then, the slides were stained with hematoxylin at room temperature for 5 min, followed by another wash. Next, 1% hydrochloric acid alcohol differentiation was used for 2 sec in order to stain the tissue blue, and then the sections were dehydrated and sealed with neutral gum after clearing. After drying, images of the slides were captured using an upright light microscope (×400). A total of 5 different fields of view were taken from each sample using magnification, ×400, and the number of brown cells in each field of view was counted. Statistical analyses were performed using SPSS 17.0 software (SPSS, Inc.).
Cell apoptosis and invasion assays
Apoptosis assays were performed using an Annexin V-FITC/PI Cell Apoptosis kit (Nanjing KeyGen Biotech, Co., Ltd.). In total, 100 µl suspension containing 5×105 cells was incubated with 5 µl Annexin V and 1 µl PI in the dark at room temperature for 15 min. The apoptotic rate (early + late apoptotic) was determined by flow cytometry (BD FACSCalibur; BD Biosciences) and the software used for analysis was FlowJo™ v10.4.0 (FlowJo LLC).
Cell invasion assays were performed as described in previous studies (8,9). In total, 1×105 Huh7 or 1×105 HepG2 cells were resuspended in FBS-free culture and was added to the upper Transwell chamber containing Matrigel (BD Biosciences), and 900 µl of culture medium with 10% FBS was added into the lower chamber. The cells were incubated at 37°C for 24 h and then fixed with 4% paraformaldehyde at room temperature for 10 min. The cells were removed from the upper surface of the filter membrane, and the invading cells were stained with a 0.1% crystal violet solution at room temperature for 10–15 min. Images were captured and the cells were counted using a light microscope (×200).
Statistical analysis
Data are presented as the mean ± SD of ≥3 independent experiments. Statistical analyses were performed using SPSS 17.0 software (SPSS, Inc.). Student's t-test was used for the comparison of parameters between two groups. One-way ANOVA and Tukey's test were used to compare the parameters among >2 groups. P<0.05 was considered to indicate a statistically significant difference.
Results
cIAP1 and TRAF3 are expressed at low levels in Huh7, H22 and HepG2 cells
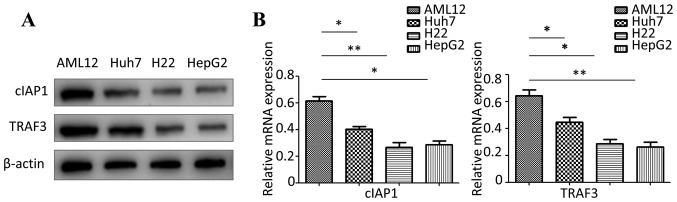
Western blotting and RT-qPCR were used to detect the protein and mRNA expression levels of cIAP1 and TRAF3 in Huh7, H22, HepG2 and AML12 cells. The present results suggested that the protein and mRNA expression levels of cIAP1 and TRAF3 were significantly lower in Huh7, H22 and HepG2 cells compared with in AML12 cells (Fig. 1).
Figure 1.
Expression levels of cIAP1 and TRAF3 in liver cancer cell lines. (A) Western blot analysis was used to detect the protein expression levels of cIAP1 and TRAF3 in AML12, Huh7, H22 and HepG2 cells. (B) Reverse transcription-quantitative PCR was used to detect the mRNA expression levels of cIAP1 and TRAF3 in AML12, Huh7, H22 and HepG2 cells. *P<0.05, **P<0.01. cIAP1, cellular inhibitor of apoptosis 1; TRAF3, tumor necrosis factor receptor-associated factor 3.
SMAC mimetic birinapant promotes apoptosis and inhibits invasion of liver cancer cells
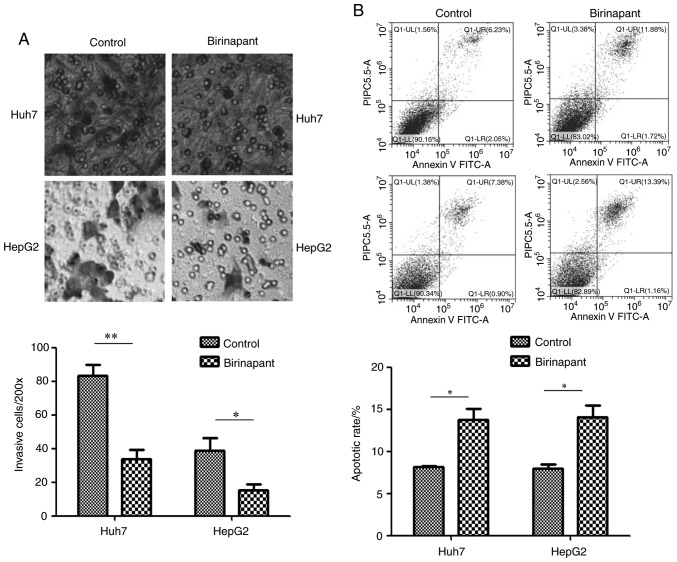
The experimental groups used in the present study included Huh7 and HepG2 cells treated with 300 nmol/ml birinapant for 24 h, and control cells that did not receive any treatment. Flow cytometry was used to detect cell apoptosis in the different groups. Transwell assays were used to detect cell invasion. The present results suggested that pretreatment with birinapant promoted apoptosis and inhibited the invasive ability of Huh7 and HepG2 cells (Fig. 2). H22 cells were not assessed for cell invasion as they do not adhere to well walls.
Figure 2.
Effect of birinapant on apoptosis and invasion in liver cancer cells. (A) Transwell assays were used to assess the invasive ability of Huh7 and HepG2 cells (magnification, ×200); ×200 represents the number of cells invaded under ×200 visual field. (B) Annexin V-FITC apoptosis assays were used to detect the apoptotic rate of Huh7 and HepG2 cells. Huh7: Control vs. birinapant, 8.28 vs. 13.60%; HepG2, Control vs. birinapant, 8.28 vs. 14.55%. *P<0.05, **P<0.01.
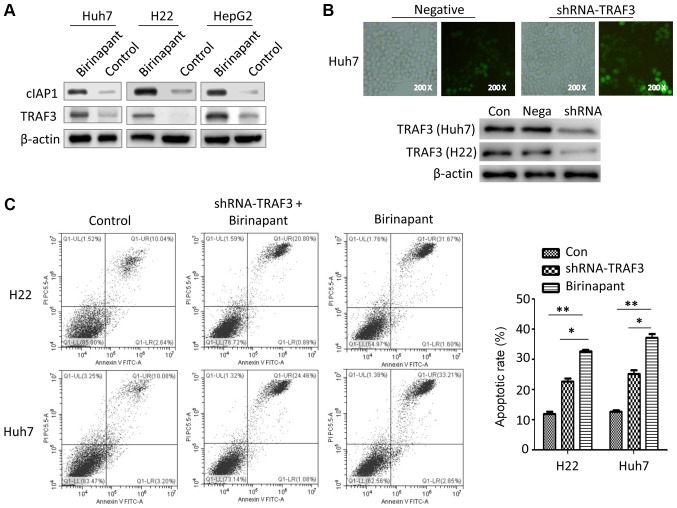
Silencing TRAF3 inhibits birinapant-mediated apoptosis in liver cancer cells
cIAP1 is the target protein of birinapant, a SMAC mimetic that regulates the development of several diseases, such as inflammatory and metabolic diseases, by modulating the cIAP1/TRAF signaling pathway (6,9,12). However, whether birinapant-induced apoptosis in liver cancer cells is related to the cIAP1/TRAF axis is unknown. The present results suggested that pretreatment with birinapant increased the protein expression levels of cIAP1 and TRAF3 in Huh7, H22 and HepG2 cells (Fig. 3A). Therefore, the present results suggested that birinapant may promote apoptosis in liver cancer cells by activating the cIAP1/TRAF3 signaling pathway. To investigate the role of TRAF3 in birinapant-mediated apoptosis in liver cancer cells, the present study knocked down TRAF3 in liver cancer cells using a lentivirus (Fig. 3B). There were three different experimental groups: Control (Con) group, the shRNA-TRAF3 + birinapant (shRNA) group and the birinapant group. The present results suggested that silencing TRAF3 expression inhibited birinapant-mediated apoptosis in H22 and Huh7 cells (Fig. 3C).
Figure 3.
Role of TRAF3 in birinapant-mediated apoptosis of liver cancer cells. (A) Western blotting was used to assess the protein expression levels of cIAP1 and TRAF3 in Huh7, H22 and HepG2 cells. (B) Green fluorescence (magnification, ×200) and western blotting were used to investigate the efficiency of lentivirus-mediated knockdown of TRAF3. (C) Annexin V-FITC apoptosis assays were used to assess the apoptotic rate in H22 and Huh7 cells. H22, Control vs. shRNA-TRAF3 + birinapant vs. birinapant, 12.68 vs. 21.69 vs. 32.27%; Huh7, Control vs. shRNA-TRAF3 + birinapant vs. birinapant, 13.28 vs. 25.54 vs. 36.06%. *P<0.05, **P<0.01. cIAP1, cellular inhibitor of apoptosis 1; Con, control; shRNA, short hairpin RNA; Nega, negative control; TRAF3, tumor necrosis factor receptor-associated factor 3.
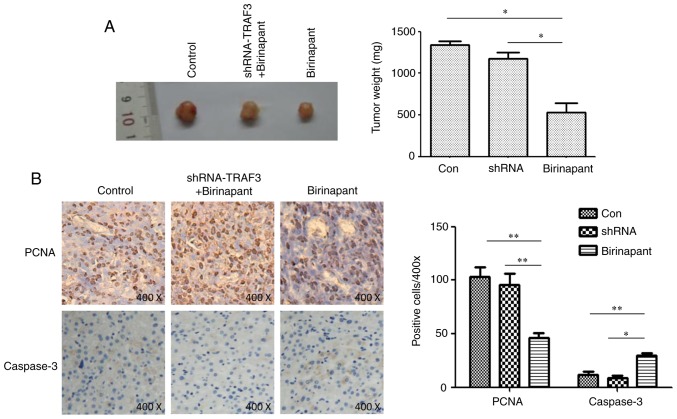
SMAC mimetic birinapant inhibits HCC growth by activating the cIAP1/TRAF3 signaling pathway
PCNA reflects the proliferative ability of tumor cells and caspase is a marker of cell apoptosis (16,17). In the present study, mice were subcutaneously injected with normal or shRNA-TRAF-infected H22 cells, and treated with saline or birinapant, and tumor growth was monitored for 15 days. Immunohistochemistry assays were used to detect the protein expression levels of PCNA and caspase-3 in the subcutaneous tumors. Birinapant inhibited the growth of subcutaneous tumors, and silencing TRAF3 reduced the inhibitory effect of birinapant on subcutaneous tumor growth (Fig. 4A). The present results suggested that birinapant inhibited the expression of PCNA and increased the expression of caspase-3, whereas silencing TRAF3 reversed these effects (Fig. 4B). Collectively, the present results suggested that the SMAC mimetic birinapant inhibited the growth of HCC and promoted apoptosis in liver cancer cells, possibly by activating the cIAP1/TRAF3 signaling pathway.
Figure 4.
Role and molecular mechanism of birinapant in hepatocellular carcinoma cell proliferation. (A) Subcutaneous tumors in each experimental group. (B) Immunohistochemistry was used to assess the protein expression levels of PCNA and caspase-3 in subcutaneous tumors; magnification, ×400. *P<0.05, **P<0.01. cIAP1, cellular inhibitor of apoptosis 1; Con, control; shRNA, short hairpin RNA; Nega, negative control; PCNA, proliferating cell nuclear antigen; TRAF3, tumor necrosis factor receptor-associated factor 3.
Discussion
HCC is a serious threat to human health due to a lack of effective treatments (1,2). Inducing apoptosis in tumor cells is currently an important strategy for inhibiting the malignant biological features of cancer (18,19). SMAC was discovered by Verhagen and Vaux (20) to localize in mitochondria and regulate cell apoptosis. SMAC may promote the apoptosis of tumor cells in several cancer types, including gastric cancer, ovarian cancer and non-Hodgkin lymphoma (21–23). This mechanism may be closely related to the fact that upon stimulation by certain factors, such as interferon and antitumor drugs, SMAC can be released from the mitochondria into the cytoplasm to bind cIAPs and inhibit the anti-apoptotic activity of cIAPs, thus promoting cell apoptosis and further inhibiting tumor growth (22,23). In addition, SMAC serves an important role in regulating immunity and inflammation. A previous in vitro study demonstrated that SMAC inhibits the LPS-mediated release of inflammatory cytokines from RAW264.7 macrophages by inhibiting the LPS-mediated degradation of TRAF3 and activation of the MAPK signaling pathway (24).
TRAF3 is expressed by numerous types of cell, including immune cells such as macrophages, B lymphocytes and T lymphocytes, and serves important roles in regulating the immune system (25). TRAF3 functions mainly via ubiquitination (Ub), including K48-linked Ub and K63-linked Ub (26). K48 polyubiquitination of TRAF3 induces TRAF3 degradation, which limits retinoic acid-inducible gene 1-induced type I interferon production in immune cells (24). TRAF3 is also subject to post-translational modification with K63-linked polyubiquitin chains, which is markedly different from K48-linked polyubiquitination (27). K63 polyubiquitination of TRAF3 does not induce degradation, but mediates PI3K activation in immune cells (28).
The structure and function of the SMAC protein, and the application of SMAC mimetics for the treatment of various tumors, has become a focus in research. SMAC mimetics have been used for the treatment of several types of cancer, such as breast cancer, prostate cancer and lung cancer (6,7). Birinapant, a typical SMAC mimetic, can inhibit the proliferation of head and neck cancer, myeloma and pancreatic cancer cells (29,30). However, whether birinapant affects the growth of HCC and its associated molecular mechanism are still unknown.
To the best of our knowledge, the present study was the first to suggest that cIAP1 and TRAF3 were expressed at low levels in liver cancer cells, and that the SMAC mimetic birinapant promoted apoptosis and inhibited invasion in liver cancer cells. In addition, the present results suggested that silencing TRAF3 inhibited birinapant-mediated apoptosis in liver cancer cells and that birinapant inhibited HCC growth in vivo. Therefore, the SMAC mimetic birinapant may promote apoptosis, and inhibit the proliferation and invasion of liver cancer cells. The present results suggested that the molecular mechanism may be related to activation of the cIAP1/TRAF3 signaling pathway by birinapant in liver cancer cells.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- SMAC
second mitochondria-derived activator of caspase
- HCC
hepatocellular carcinoma
- TRAF3
tumor necrosis factor receptor-associated factor 3
- cIAP1
cellular inhibitor of apoptosis 1
Funding
The present study was supported by The Enshi State Science and Technology Program Project (grant no. 2017-14).
Availability of data and materials
All data generated or analyzed during the present study are included in this published article.
Authors' contributions
JD and DQ performed most of the experiments and drafted the manuscript. YZ, QL and YL performed some experiments and collected the data. JD and JL designed the study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by The Research Ethics Committee of The Central Hospital of Enshi Tujia and Miao Autonomous Prefecture.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bruix J, Gores GJ, Mazzaferro V. Hepatocellular carcinoma: Clinical frontiers and perspectives. Gut. 2014;63:844–855. doi: 10.1136/gutjnl-2013-306627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hartke J, Johnson M, Ghabril M. The diagnosis and treatment of hepatocellular carcinoma. Semin Diagn Pathol. 2017;34:153–159. doi: 10.1053/j.semdp.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 3.Boddu P, Carter BZ, Verstovsek S, Pemmaraju N. SMAC mimetics as potential cancer therapeutics in myeloid malignancies. Br J Haematol. 2019;185:219–231. doi: 10.1111/bjh.15829. [DOI] [PubMed] [Google Scholar]
- 4.Cossu F, Milani M, Mastrangelo E, Lecis D. Targeting the BIR domains of inhibitor of apoptosis (IAP) proteins in cancer treatment. Comput Struct Biotechnol J. 2019;17:142–150. doi: 10.1016/j.csbj.2019.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pedersen J, LaCasse EC, Seidelin JB, Coskun M, Nielsen OH. Inhibitors of apoptosis (IAPs) regulate intestinal immunity and inflammatory bowel disease (IBD) inflammation. Trends Mol Med. 2014;20:652–665. doi: 10.1016/j.molmed.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 6.Chromik J, Safferthal C, Serve H, Fulda S. Smac mimetic primes apoptosis-resistant acute myeloid leukaemia cells for cytarabine-induced cell death by triggering necroptosis. Cancer Lett. 2014;344:101–109. doi: 10.1016/j.canlet.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 7.Nikkhoo A, Rostami N, Hojjat-Farsangi M, Azizi G, Yousefi B, Ghalamfarsa G, Jadidi-Niaragh F. Smac mimetics as novel promising modulators of apoptosis in the treatment of breast cancer. J Cell Biochem. 2019;120:9300–9314. doi: 10.1002/jcb.28205. [DOI] [PubMed] [Google Scholar]
- 8.Abhari BA, McCarthy N, Le Berre M, Kilcoyne M, Joshi L, Agostinis P, Fulda S. Smac mimetic suppresses tunicamycin-induced apoptosis via resolution of ER stress. Cell Death Dis. 2019;10:155. doi: 10.1038/s41419-019-1381-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerges S, Rohde K, Fulda S. Cotreatment with Smac mimetics and demethylating agents induces both apoptotic and necroptotic cell death pathways in acute lymphoblastic leukemia cells. Cancer Lett. 2016;375:127–132. doi: 10.1016/j.canlet.2016.02.040. [DOI] [PubMed] [Google Scholar]
- 10.Brumatti G, Ma C, Lalaoui N, Nguyen NY, Navarro M, Tanzer MC, Richmond J, Ghisi M, Salmon JM, Silke N, et al. The caspase-8 inhibitor emricasan combines with the SMAC mimetic birinapant to induce necroptosis and treat acute myeloid leukemia. Sci Transl Med. 2016;8:339ra69. doi: 10.1126/scitranslmed.aad3099. [DOI] [PubMed] [Google Scholar]
- 11.Shu X, Zhu Z, Cao D, Zheng L, Wang F, Pei H, Wen J, Yang J, Li D, Bai P, et al. PEG-derivatized birinapant as a nanomicellar carrier of paclitaxel delivery for cancer therapy. Colloids Surf B Biointerfaces. 2019;182:110356. doi: 10.1016/j.colsurfb.2019.110356. [DOI] [PubMed] [Google Scholar]
- 12.Liu H, Liao R, He K, Zhu X, Li P, Gong J. The SMAC mimetic birinapant attenuates lipopolysaccharide-induced liver injury by inhibiting the tumor necrosis factor receptor-associated factor 3 degradation in Kupffer cells. Immunol Lett. 2017;85:79–83. doi: 10.1016/j.imlet.2017.02.016. [DOI] [PubMed] [Google Scholar]
- 13.Ho PC, Tsui YC, Feng X, Greaves DR, Wei LN. NF-κB-mediated degradation of the coactivator RIP140 regulates inflammatory responses and contributes to endotoxin tolerance. Nat Immunol. 2012;13:379–386. doi: 10.1038/ni.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Liu Z, Mar KB, Hanners NW, Perelman SS, Kanchwala M, Xing C, Schoggins JW, Alto NM. A NIK-SIX signalling axis controls inflammation by targeted silencing of non-canonical NF-κB. Nature. 2019;68:249–253. doi: 10.1038/s41586-019-1041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mi L, Zhou Y, Wu D, Tao Q, Wang X, Zhu H, Gao X, Wang J, Ling R, Deng J, et al. ACSS2/AMPK/PCNA pathway-driven proliferation and chemoresistance of esophageal squamous carcinoma cells under nutrient stress. Mol Med Rep. 2019;20:5286–5296. doi: 10.3892/mmr.2019.10735. [DOI] [PubMed] [Google Scholar]
- 17.Sato Y, Yoshino H, Kazama Y, Kashiwakura I. Involvement of caspase-8 in apoptosis enhancement by cotreatment with retinoic acid-inducible gene-I-like receptor agonist and ionizing radiation in human non-small cell lung cancer. Mol Med Rep. 2018;18:5286–5294. doi: 10.3892/mmr.2018.9536. [DOI] [PubMed] [Google Scholar]
- 18.Razmkhah M, Abtahi S, Ghaderi A. Mesenchymal stem cells, immune cells and tumor cells crosstalk: A sinister triangle in the tumor microenvironment. Curr Stem Cell Res Ther. 2019;14:43–51. doi: 10.2174/1574888X13666180816114809. [DOI] [PubMed] [Google Scholar]
- 19.Jin Y, Meng X, Qiu Z, Su Y, Yu P, Qu P. Anti-tumor and anti-metastatic roles of cordycepin, one bioactive compound of Cordyceps militaris. Saudi J Biol Sci. 2018;25:991–995. doi: 10.1016/j.sjbs.2018.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verhagen AM, Vaux DL. Cell death regulation by the mammalian IAP antagonist Diablo/Smac. Apoptosis. 2002;7:163–166. doi: 10.1023/A:1014318615955. [DOI] [PubMed] [Google Scholar]
- 21.Soung YH, Park WS, Nam SW, Lee JY, Yoo NJ, Lee SH. SMAC/DIABLO mutation is uncommon in gastric and colorectal carcinomas. Pathology. 2016;38:85–87. doi: 10.1080/00313020500468848. [DOI] [PubMed] [Google Scholar]
- 22.Cekay MJ, Roesler S, Frank T, Knuth AK, Eckhardt I, Fulda S. Smac mimetics and type II interferon synergistically induce necroptosis in various cancer cell lines. Cancer Lett. 2017;410:228–237. doi: 10.1016/j.canlet.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 23.Bhatti IA, Abhari BA, Fulda S. Identification of a synergistic combination of Smac mimetic and Bortezomib to trigger cell death in B-cell non-Hodgkin lymphoma cells. Cancer Lett. 2017;405:63–72. doi: 10.1016/j.canlet.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 24.Tseng PH, Matsuzawa A, Zhang W, Mino T, Vignali DA, Karin M. Different modes of ubiquitination of the adaptor TRAF3 selectively activate the expression of type I interferons and proinflammatory cytokines. Nat Immunol. 2010;11:70–75. doi: 10.1038/ni.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin WW, Hostager BS, Bishop GA. TRAF3, ubiquitination, and B-lymphocyte regulation. Immunol Rev. 2015;266:46–55. doi: 10.1111/imr.12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wen J, Bai H, Chen N, Zhang W, Zhu X, Li P, Gong J. USP25 promotes endotoxin tolerance via suppressing K48-linked ubiquitination and degradation of TRAF3 in Kupffer cells. Mol Immunol. 2019;106:53–62. doi: 10.1016/j.molimm.2018.12.017. [DOI] [PubMed] [Google Scholar]
- 27.Guan K, Wei C, Zheng Z, Song T, Wu F, Zhang Y, Cao Y, Ma S, Chen W, Xu Q, et al. MAVS promotes inflammasome activation by targeting ASC for K63-linked ubiquitination via the E3 ligase TRAF3. J Immunol. 2015;194:4880–4890. doi: 10.4049/jimmunol.1402851. [DOI] [PubMed] [Google Scholar]
- 28.Muro I, Fang G, Gardella KA, Mahajan IM, Wright CW. The TRAF3 adaptor protein drives proliferation of anaplastic large cell lymphoma cells by regulating multiple signaling pathways. Cell Cycle. 2014;13:1918–1927. doi: 10.4161/cc.28895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eytan DF, Snow GE, Carlson S, Derakhshan A, Saleh A, Schiltz S, Cheng H, Mohan S, Cornelius S, Coupar J, et al. SMAC Mimetic birinapant plus radiation eradicates human head and neck cancers with genomic amplifications of cell death genes FADD and BIRC2. Cancer Res. 2016;76:5442–5454. doi: 10.1158/0008-5472.CAN-15-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu X, Shen X, Qu J, Straubinger RM, Jusko WJ. Multi-scale network model supported by proteomics for analysis of combined gemcitabine and birinapant effects in pancreatic cancer cells. CPT Pharmacometrics Syst Pharmacol. 2018;7:549–561. doi: 10.1002/psp4.12320. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during the present study are included in this published article.