Abstract
Circular RNAs (circRNAs) have been shown to be involved in the development of cancer. The aim of the present study was to investigate the role of circRNA SMARCA5 (cSMARCA5) in human cervical cancer. In the present study, cSMARCA5 expression was upregulated in cervical cancer tissues and cell lines. Furthermore, the proliferation rate of cells transduced with viral plasmids expressing small interfering RNA targeting cSMARCA5 was downregulated. Bioinformatics analysis predicted that microRNA (miR)-432 targeted cSMARCA5, and miR-432 was able to interact with epidermal growth factor receptor (EGFR) by binding to its 3′-untranslated region. The expression levels of EGFR, ERK1 and ERK2 were increased in cervical cancer tissues. Furthermore, correlation analysis revealed that cSMARCA5 levels were positively correlated with ERK1 and ERK2 levels. In conclusion, the present findings suggested that cSMARCA5 may play an important role in the progression of cervical cancer via the ERK signaling pathway by modulating miR-432.
Keywords: circular RNA SMARCA5, microRNA-432, epidermal growth factor receptor, cervical cancer
Introduction
Cervical cancer is one of the most common malignancies that occur in the cervical canal (1). It is estimated that ~528,000 cases of cervical cancer, with 266,000 mortalities, occur every year (2). Although not all of the causes of cervical cancer are known, human papillomavirus infections is considered to be the main risk factor (3). Currently, the standard tumor treatments for cancer include surgery, radiation and chemotherapy, alone or in combination (4). Although treatment strategies are improving, the 5-year survival rate for cervical cancer patients is <40% (5). Therefore, to further study cervical cancer development at the molecular level is necessary.
Circular RNAs (circRNAs) are single-stranded closed ring-like non-coding RNAs, with no 5′-terminal cap or a 3′-terminal poly-A tail (6). Due to their special stable structure, circRNAs cannot be degraded by RNA exonucleases (7). circRNAs have been shown to function as competitive endogenous RNAs and microRNA (miRNA) sponges (8,9). Recent studies have shown that altered circRNA levels play crucial roles in carcinogenesis (10,11). Circular RNA SMARCA5 (cSMARCA5; circBase ID: hsa_circ_0001445) is a novel circRNA derived from exons 15 and 16 of the SMARCA5 gene (12). A previous study reported that overexpression of cSMARCA5 could inhibit the proliferation and metastasis of hepatocellular carcinoma cells (13). However, to the best of our knowledge, the role of cSMARCA5 in cervical cancer has not been previously investigated.
miRNAs are a class of evolutionarily conserved, short non-coding RNA molecules of 18–25 nucleotides in length (14). miRNAs can modulate gene expression by binding to the 3′-untranslated regions (3′-UTRs) of target mRNAs, resulting in repression of protein translation or mRNA degradation (15,16). Previous studies have shown that miRNAs are involved in multiple cellular processes, including cell proliferation, cell cycle, apoptosis and cell differentiation (17). Abnormal expression of miRNAs has been identified in various types of malignancies (18–20). Several miRNAs have been demonstrated to be involved in cervical cancer initiation and progression (21–23). miRNA-432 (miR-432) dysregulation has been shown to be involved in the carcinogenesis of many cancers (24–26). However, a limited number of studies have reported the functional role of miR-432 in cervical cancer development.
The ERK pathway is involved in the regulation of a variety of growth and differentiation pathways through several phosphorylation cascades (27). Uncontrolled growth is a necessary step for the development of all cancers (28). In many cancer types, a defect in the mitogen-activated protein/ERK pathway is believed to contribute to uncontrolled proliferation (29). Upregulation of epidermal growth factor receptor (EGFR) is frequently detected in cervical cancer and is considered to be an independent predictor for the prognosis of cervical cancer (30).
The aim of the present study was to investigate the level of cSMARCA5 expression in human cervical cancer tissues and cell lines. In addition, the present study examined the function of cSMARCA and its underlying mechanism.
Materials and methods
Tissue samples
The present study was approved by the Medical Ethics Committee of Cangzhou Central Hospital. All patients (18–65 years) were informed of the study and signed written informed consent. Human cervical cancer tissues and adjacent normal tissues (n=56) were collected from patients who visited the Cangzhou Central Hospital from January 2016 to Decembr 2017. All the specimens were immediately snap-frozen and preserved in liquid nitrogen at −80°C until further use.
Cell culture and transfection
The human normal cervical epithelial cell line (Ect1/E6E7) and human cervical cancer cell lines HeLa, Ca-Ski, C-33A, and SiHa were purchased from VCANBIO Cell & Gene Engineering Corporation, Ltd. and cultured in DMEM with 10% FBS (both from Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2. Small interfering RNA (siRNA) targeting cSMARCA5 (si-cSMARCA5; 5′-AUUGGCGACUCAAUGGAUCAG-3′), miR-432 mimic (5′-CCUCGCGUUAUAACGUUAC-3′) and their corresponding negative controls (NCs; si-NC, 5′-UUCUCCGAACGUGUCA-3′; miR-NC, 5′-UUCUCCGAACGUGUCACGUAA-3′) were synthesized by Shanghai GenePharma, Co., Ltd. After culturing overnight at 37°C, HeLa and Ca-Ski cells (4×105 cells/well) were transfected with si-cSMARCA5, miR-432 mimic or their parental negative controls using Lipofectamine 2000 reagent (cat. no. 11668-019; Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The of siRNAs and miRNAs was 50 nM. Subsequent experiments were performed 24 h after transfection.
Reverse transcription-quantitative PCR (RT-qPCR)
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was added to the tissue and cell samples to extract total RNA. After that, total RNA was treated with RNase R (Invitrogen; Thermo Fisher Scientific, Inc.) to remove linear RNAs and enrich circRNAs. Total RNA was dissolved in RNase-free water, and the concentration was measured using a NanoDrop™ 2000 spectrophotometer. cDNA was synthesized using a TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) for miR-432 and a One-Step PrimerScript cDNA kit (Qiagen, Inc.) was used for cSMARCA5, EGFR, ERK1 and ERK2 using 50 ng total RNA with the temperature protocol of: 95°C for 30 sec and 60°C for 30 min. RT-qPCR was performed using SYBR Green PCR Master Mix kit (Thermo Fisher Scientific, Inc.). The thermocycling conditions were as follows: Initial denaturation at 95°C for 5 min; followed by 30 cycles of 95°C for 10 sec and annealing at 60°C for 45 sec; then a final extension for 10 min at 72°C. All primers were designed and synthesized by Shanghai GenePharma Co., Ltd. The primers were as follows: cSMARCA5 forward, 5′-GCTATCAAGCTCCATCCGCAT-3′ and reverse, 5′-TAAGACGAAGCACCGGA-3′; miR-432 forward, 5′-AACGAGACGACGACAGAC-3′ and reverse, 5′-CTTGGAGTAGGTCATTGGGT-3′; si-cSMARCA5 5′-CATGGTCCTCGAGGTTA-3′; si-NC 5′-UGGACAACAUGGGCUCU-3′; miR-432 mimic: 5′-AUCGAGACUACGUCUGAC-3′; miR-NC 5′-AGUGCAUGCGUACGAGCUGU-3′; EGFR forward, 5′-ATGGAATACCCTGGGTGT-3′ and reverse, 5′-GGACAAGCTGGTCAAGGT-3′; ERK1 forward, 5′-CCAGTTCCGAGAATAAGCGCA-3′ and reverse, 5′-CGTGTCGCCATGACACATGT-3′; ERK2 forward, 5′-TCATCCAACAGACAGACGTAGT-3′ and reverse, 5′-ACCAGAGCCATCAGACGA-3′; U6 forward, 5′-GCTCGCTTCGGCAGCACA-3′ and reverse, 5′-GAGGTATTCGCACCAGAGGA-3′; GAPDH forward, 5′-ACCACAGTCCATGCCATCCAC-3′ and reverse, 5′-TCCACCACCCTGTTGCTGTA-3′. GAPDH mRNA or U6 were used as endogenous reference genes. Gene expression was determined with the 2−ΔΔCq method (31).
MTT assay
Cells (3×103) were seeded in 96-well plates, incubated at 37°C for 24 h and stained with 0.5 mg/ml MTT at 37°C for 4 h. After removal of the supernatant, DMSO was added and thoroughly mixed for 15 min. The absorbance value in each well was measured at a wavelength of 490 nm.
Luciferase activity assay
Circinteractome (https://circinteractome.nia.nih.gov/) and TargetScan 7.2 (http://www.targetscan.org/vert_72/) were used to predict potential targets of miR-432, and binding sites between miR-432 and cSMARCA5/EGFR. The cSMARCA5 and EGFR 3′-UTR sequences were amplified and cloned into the pGL3 vector (Promega Corporation). For reporter assays, cells were cultured in 24-well plates at 37°C and cotransfected with wild type (WT)-cSMARCA5 or mutant (Mut)-cSMARCA5 (WT-EGFR or Mut EGFR) and 100 nM miR-432 mimics or miR-NC, using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). After 48 h, the luciferase activity was determined with a Dual-Luciferase Reporter System (Promega Corporation) according to the manufacturer's protocols. Firefly luciferase activity was normalized to Renilla luciferase activity using the pGL3 vector.
Cell invasion assay
For cell invasion assays, 1×105 HeLa and Ca-Ski cells were resuspended in 200 µl serum-free RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) and then plated into the upper chambers of Transwell inserts, which were coated with Matrigel. The lower chamber was filled with culture medium supplemented with 20% FBS (Gibco; Thermo Fisher Scientific, Inc.). After 24 h of incubation at 37°C, the cells on the bottom surface were fixed with 4% polyoxymethylene at room temperature for 30 min and stained with 0.1% crystal violet at room temperature for 20 min. Stained cells were counted and images were captured with an Olympus BX51 light microscope (magnification, ×200; Olympus Corporation).
Western blotting analysis
Proteins were extracted from cultured cells by RIPA buffer (Sigma-Aldrich; Merck KGaA) containing a mixture of protease inhibitors (100X; Beijing CoWin Biotech Co., Ltd.). Protein concentrations were quantified using a bicinchoninic acid assay (Beijing CoWin Biotech Co., Ltd.). Equal quantities of protein (30 µg/lane) were separated via 10% SDS-PAGE and then transferred to PVDF membranes. The membranes were blocked with 5% skimmed milk at room temperature for 1.5 h, followed by incubation with the following primary antibodies overnight at 4°C: EGFR (1:1,000; cat. no. ab32562; Abcam) and GAPDH (1:5,000; cat. no. ab185059; Abcam). Then, a horseradish peroxidase-conjugated goat anti-rabbit IgG secondary antibody (1:5,000; cat. no. sc-2054; Santa Cruz Biotechnology, Inc.) was incubated with the PVDF membranes for 1 h at room temperature. Protein signals were visualized using an Enhanced Chemiluminescence Plus reagent (GE Healthcare Life Sciences). Bands were quantified using Quantity One version 4.62 software (Bio-Rad Laboratories, Inc.).
Statistical analysis
Data are presented as the mean ± SEM. All statistical analyses were performed using SPSS software (version 19.0; IBM Corp) and GraphPad Prism software (version 5.0; GraphPad Software, Inc.). Data were analyzed using a Student's t-test for two-group comparisons, and one-way ANOVA with a Tukey's post-hoc test for multiple-group comparison. Spearman's correlation analysis was used to analyze the association between cSMARCA5 and ERK1 or ERK2 expression. P<0.05 was considered to indicate a statistically significant difference.
Results
cSMARCA5 expression is upregulated in cervical cancer tissues and cell lines
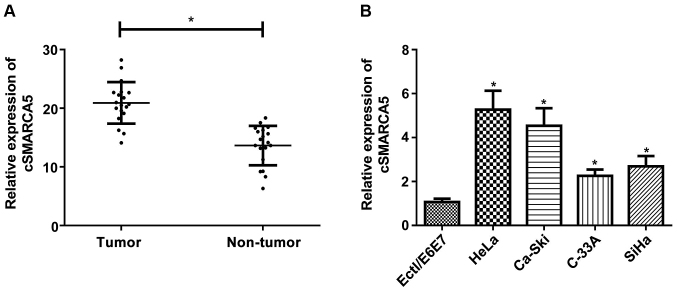
cSMARCA5 expression level in cervical cancer tissues was investigated using RT-qPCR. The expression level of cSMARCA5 was significantly increased in cervical cancer tissues (Fig. 1A) compared with non-tumor tissues. In addition, the present study examined the expression level of cSMARCA5 in four cervical cancer cell lines (HeLa, Ca-Ski, C-33A, and SiHa) and a human normal cervical epithelial cell line (Ect1/E6E7). A significant increase in cSMARCA5 expression level was found in the four cancer cell lines compared with Ect1/E6E7 cells (Fig. 1B).
Figure 1.
cSMARCA5 expression is upregulated in cervical cancer tissues and cell lines. (A) The expression level of cSMARCA5 was significantly upregulated in cervical cancer tissues compared with adjacent normal tissue samples. (B) The expression levels of cSMARCA5 were determined in the cervical cancer cell lines HeLa, C-33A, Ca-Ski and SiHa and a human normal cervical epithelial cell line Ect1/E6E7. *P<0.05 vs. the respective control. cSMARCA5, circular RNA SMARCA5.
cSMARCA5 silencing represses the proliferation and invasion of cervical cancer cells
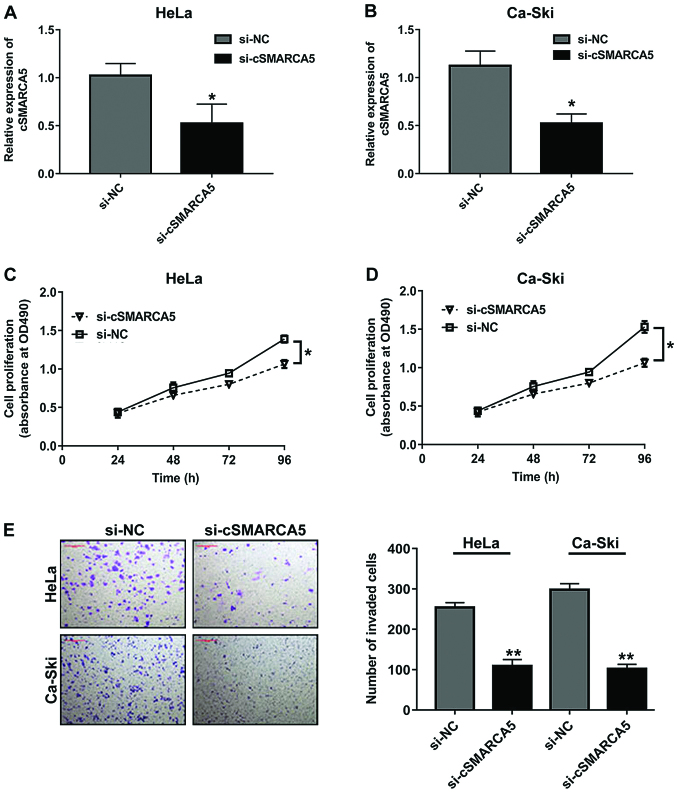
The oncogenic role of cSMARCA5 in cervical cancer cell lines was investigated, and it was observed that cSMARCA5 expression level was remarkably decreased in two cell lines (HeLa and Ca-Ski), which were transfected with si-cSMARCA5. The RT-qPCR results showed that cSMARCA5 expression level was significantly downregulated in the two cell lines transfected with si-cSMARCA5 (Fig. 2A and B). In addition, the MTT assay revealed that the proliferation rate of cells transduced with si-cSMARCA5 was significantly decreased at 96 h compared with cells transduced with si-NC in the two cell lines (Fig. 2C and D). Cell invasion assays revealed that si-cSMARCA5 suppressed the invasion of cervical cancer lines (Fig. 2E).
Figure 2.
cSMARCA5 silencing suppresses the proliferation and invasion of cervical cancer cells. (A) cSMARCA5 expression levels were significantly downregulated in HeLa cells transduced with si-cSMARCA5 compared with si-NC. (B) cSMARCA5 expression levels were significantly downregulated in Ca-Ski cells transduced with si-cSMARCA5 compared with si-NC. (C) The proliferation rate of cells transfected with si-cSMARCA5 was significantly decreased at 96 h compared with cells transfected with si-NC in HeLa cells. (D) The growth rate of cells transfected with si-cSMARCA5 was significantly decreased at 96 h compared with cells transfected with si-NC in Ca-Ski cells. (E) Cell invasion assays revealed that si-cSMARCA5 inhibited the invasive ability of HeLa cells and Ca-Ski cells. *P<0.05, **P<0.01 vs. respective control. Scale bar, 20 µm. cSMARCA5, circular RNA SMARCA5; si-NC, small interfering RNA-negative control; si-cSMARCA5, small interfering RNA-cSMARCA5.
miR-432 targets cSMARCA5
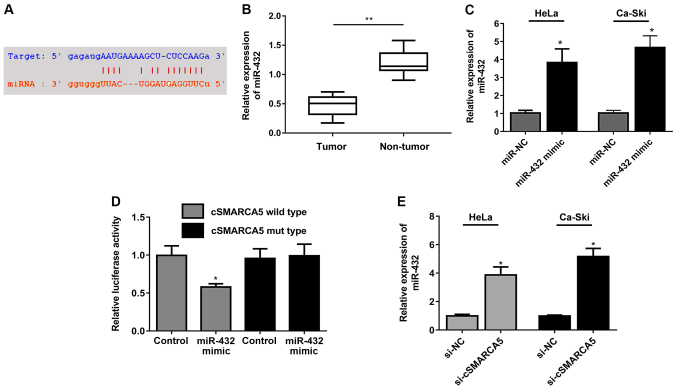
The binding sites between miR-432 and cSMARCA5 are presented in Fig. 3A. miR-432 expression level was significantly decreased in cervical cancer tissues (Fig. 3B). miR-432 expression was significantly increased in two cell lines (HeLa and Ca-Ski) following transfection of miR-432 mimic (Fig. 3C). Decreased luciferase activity was observed in cells transfected with miR-432 mimic and cSMARCA5 wild-type reporter, but not in cells transfected with the cSMARCA5 mutant reporter plasmid and miR-432 mimic (Fig. 3D). Moreover, miR-432 expression levels were significantly increased in cells transduced with si-cSMARCA5 compared with the control group (Fig. 3E).
Figure 3.
miR-432 targets cSMARCA5. (A) Binding sites between miR-432 and cSMARCA5. (B) miR-432 expression level was significantly decreased in cervical cancer tissues. (C) miR-432 expression level was remarkably increased in two cell lines (HeLa and Ca-Ski) that were transduced with miR-432 mimic. (D) Decreased luciferase activity was observed between cells transfected with miR-432 mimic and cSMARCA5 wild-type construct compared with cells transfected with NC-mimics. (E) miR-432 expression levels were significantly increased in cells transduced with si-cSMARCA5. *P<0.05, **P<0.01 vs. respective control. miR-432; microRNA-432; cSMARCA5, circular RNA SMARCA5; miR-NC, microRNA-negative control; si-NC, small interfering RNA-negative control; si-cSMARCA5, small interfering RNA-cSMARCA5.
miR-432 targets the 3′-UTR of EGFR
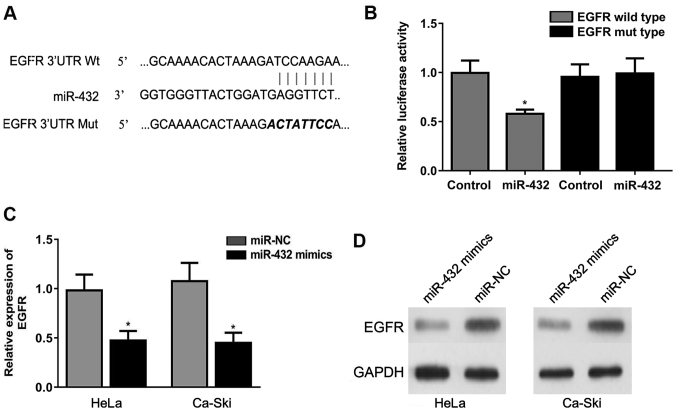
Bioinformatics analysis found that the 3′UTR of EGFR may interact with miR-432. The binding site between miR-432 and EGFR is shown in Fig. 4A. The miR-432 mimic significantly reduced the relative luciferase activity of the EGFR luciferase reporter in transfected cells (Fig. 4B), suggesting a direct interaction between miR-432 and EGFR. Moreover, the miR-432 mimic significantly downregulated EGFR expression in HeLa and Ca-Ski cells at the mRNA and protein levels (Fig. 4C and D). Collectively, the present results suggested that miR-432 inhibited the expression of EGFR by binding to the 3′-UTR of EGFR.
Figure 4.
miR-432 targets the 3′-UTR of EGFR. (A) The 3′ UTR of EGFR was found to be highly complementary to the sequence of miR-432. (B) Transfection of miR-432 mimics significantly reduced the relative luciferase activity in HeLa and Ca-Ski cells. miR-432 overexpression significantly downregulated EGFR expression in HeLa and Ca-Ski cells at the (C) mRNA and (D) protein levels. *P<0.05 vs. the respective control. miR-432; microRNA-432; miR-NC, microRNA-negative control; 3′-UTR, 3′-untranslated regions; EGFR, epidermal growth factor receptor.
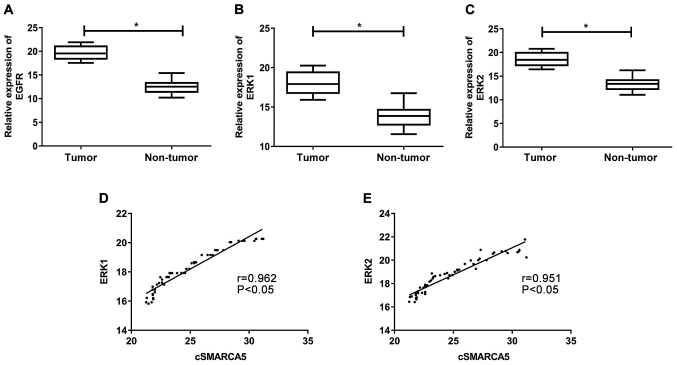
Expression levels of EGFR, ERK1 and ERK2 in cervical cancer tissues
EGFR expression in cervical cancer tissue samples was subsequently analyzed. EGFR levels were significantly increased in cervical cancer tissues compared with non-tumor tissues (Fig. 5A). The levels of ERK1 and ERK2 expression were significantly increased in cervical cancer tissues (Fig. 5B and C). Furthermore, correlation analysis revealed that cSMARCA5 levels were positively correlated with ERK1 (r=0.962, P<0.05; Fig. 5D) and ERK2 levels (r=0.951, P<0.05; Fig. 5E).
Figure 5.
Expression levels of EGFR, ERK1 and ERK2 in cervical cancer tissues. (A) Expression level of EGFR was significantly upregulated in cervical cancer tissues compared with adjacent normal tissue samples. Expression levels of (B) ERK1 and (C) ERK2 were significantly increased in cervical cancer tissues. Correlation analysis revealed that cSMARCA5 levels were positively correlated with (D) ERK1 and (E) ERK2 levels. *P<0.05. EGFR, epidermal growth factor receptor; cSMARCA5, circular RNA SMARCA5.
Discussion
In recent years, with the rapid development of bioinformatics technology, an increasing number of circRNAs have been reported to be involved in the development and progression of various malignant tumors (32). A previous study showed that hsa_circ_0005075 is involved in cell adhesion during hepatocellular carcinoma development (33). High levels of circular RNA CCDC66 promote colorectal cancer growth and metastasis by sponging miRNAs (34). In the present study, cSMARCA5 expression level was found to be upregulated in cervical cancer tissues. In addition, a significant increase in the cSMARCA5 level in the four cell lines was observed in the present study. The proliferation and invasion of tumor cells are important characteristics that affect the progression of tumors (35). The present results suggested that cSMARCA5 could promote cervical cancer cell proliferation and invasion. In addition, results suggested that cSMARCA5 may present tumor-promoting abilities in cervical cancer cells.
It is hypothesized that circRNAs modulate diverse biological processes, acting as miRNA ‘sponges’, as well as regulating transcription, protein binding and translation (36). In the present study, the miRNA ‘sponge’ theory was explored in relation to cSMARCA5 in cervical cancer. Using bioinformatics analysis, it was predicted that miR-432 was a target of cSMARCA5. The miR-432 expression level was increased following when cSMARCA5 knockdown. The present results suggested that cSMARCA5 may induce the progression of cervical cancer by targeting miR-432.
The human EGFR gene is localized on the 7th chromosome and encodes a glycoprotein composed of ~53 amino acids that is activated by binding to specific ligands, including EGF and transforming growth factor α (37). Biesterfeld et al (38) found that EGFR expression was upregulated in cervical carcinomas, suggesting that the expression of EGFR may correlate with the aggressive and proliferative phenotype of cervical carcinoma. In line with previous research, in the present study, upregulation of EGFR was detected in cervical cancer tissues. Using bioinformatics analysis, the present study showed that miR-432 directly targeted the 3′-UTR of EGFR, suggesting that the miR-432 mimic could directly downregulate EGFR expression.
As a highly conserved mitogen-activated protein kinase family member, ERK plays important roles in biological processes such as proliferation, differentiation and apoptosis (39). A previous study revealed that inhibition of the ERK pathway could exert antitumor effects (40). The present results showed that ERK1 and ERK2 expression levels increased significantly in cervical cancer tissues. Furthermore, the present study found a positive correlation between cSMARCA5 and ERK1/2, suggesting that cSMARCA5 could affect the progression of cervical cancer by upregulating the ERK1/2 signaling pathway. The present study suggested that cSMARCA5 promoted the progression of cervical cancer by modulating miR-432, and induced the proliferation and invasion of cervical cancer by upregulating the ERK1/2 signaling pathway.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
BQ, HY and LZ participated in data analysis and manuscript preparation. YL and QL performed the experiments and PH interpreted the data and drafted the manuscript.
Ethics approval and consent to participate
The present study was approved by The Medical Ethics Committee of Cangzhou Central Hospital. All patients were informed of the study and signed written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Pecorelli S. Revised FIGO staging for carcinoma of the vulva, cervix, and endometrium. Int J Gynaecol Obstet. 2009;105:103–104. doi: 10.1016/j.ijgo.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, Snijders PJ, Peto J, Meijer CJ, Muñoz N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 4.Yee GP, de Souza P, Khachigian LM. Current and potential treatments for cervical cancer. Curr Cancer Drug Targets. 2013;13:205–220. doi: 10.2174/1568009611313020009. [DOI] [PubMed] [Google Scholar]
- 5.Smith RA, Brooks D, Cokkinides V, Saslow D, Brawley OW. Cancer screening in the United States, 2013: A review of current American Cancer Society guidelines, current issues in cancer screening, and new guidance on cervical cancer screening and lung cancer screening. CA Cancer J Clin. 2013;63:88–105. doi: 10.3322/caac.21174. [DOI] [PubMed] [Google Scholar]
- 6.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 8.Salzman J. Circular RNA expression: Its potential regulation and function. Trends Genet. 2016;32:309–316. doi: 10.1016/j.tig.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 10.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan H, Li T, Jiang Y, Pan C, Ding Y, Huang Z, Yu H, Kong D. Overexpression of circular RNA ciRS-7 abrogates the tumor suppressive effect of miR-7 on gastric cancer via PTEN/PI3K/AKT signaling pathway. J Cell Biochem. 2018;119:440–446. doi: 10.1002/jcb.26201. [DOI] [PubMed] [Google Scholar]
- 12.Glazar P, Papavasileiou P, Rajewsky N. circBase: A database for circular RNAs. RNA. 2014;20:1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu J, Xu QG, Wang ZG, Yang Y, Zhang L, Ma JZ, Sun SH, Yang F, Zhou WP. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68:1214–1227. doi: 10.1016/j.jhep.2018.01.012. [DOI] [PubMed] [Google Scholar]
- 14.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 15.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 16.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 17.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lv D, Zhen Z, Huang D. MicroRNA-432 is downregulated in osteosarcoma and inhibits cell proliferation and invasion by directly targeting metastasis-associated in colon cancer-1. Exp Ther Med. 2019;17:919–926. doi: 10.3892/etm.2018.7029. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Wu K, Ma L, Zhu J. miR4835p promotes growth, invasion and selfrenewal of gastric cancer stem cells by Wnt/betacatenin signaling. Mol Med Rep. 2016;14:3421–3428. doi: 10.3892/mmr.2016.5603. [DOI] [PubMed] [Google Scholar]
- 20.Liu F, Cai Y, Rong X, Chen J, Zheng D, Chen L, Zhang J, Luo R, Zhao P, Ruan J. MiR-661 promotes tumor invasion and metastasis by directly inhibiting RB1 in non small cell lung cancer. Mol Cancer. 2017;16:122. doi: 10.1186/s12943-017-0698-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, Li F, Zhu L. Clinical significance and functions of microRNA-93/CDKN1A axis in human cervical cancer. Life Sci. 2018;209:242–248. doi: 10.1016/j.lfs.2018.08.021. [DOI] [PubMed] [Google Scholar]
- 22.Pedroza-Torres A, Campos-Parra AD, Millan-Catalan O, Loissell-Baltazar YA, Zamudio-Meza H, Cantú de León D, Montalvo-Esquivel G, Isla-Ortiz D, Herrera LA, Ángeles-Zaragoza Ó, et al. MicroRNA-125 modulates radioresistance through targeting p21 in cervical cancer. Oncol Rep. 2018;39:1532–1540. doi: 10.3892/or.2018.6219. [DOI] [PubMed] [Google Scholar]
- 23.Tan D, Zhou C, Han S, Hou X, Kang S, Zhang Y. MicroRNA-378 enhances migration and invasion in cervical cancer by directly targeting autophagy-related protein 12. Mol Med Rep. 2018;17:6319–6326. doi: 10.3892/mmr.2018.8645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang N, Chen WJ, Zhang JW, Xu C, Zeng XC, Zhang T, Li Y, Wang GY. Downregulation of miR-432 activates Wnt/β-catenin signaling and promotes human hepatocellular carcinoma proliferation. Oncotarget. 2015;6:7866–7879. doi: 10.18632/oncotarget.3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das E, Bhattacharyya NP. MicroRNA-432 contributes to dopamine cocktail and retinoic acid induced differentiation of human neuroblastoma cells by targeting NESTIN and RCOR1 genes. FEBS Lett. 2014;588:1706–1714. doi: 10.1016/j.febslet.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 26.Chen L, Kong G, Zhang C, Dong H, Yang C, Song G, Guo C, Wang L, Yu H. MicroRNA-432 functions as a tumor suppressor gene through targeting E2F3 and AXL in lung adenocarcinoma. Oncotarget. 2016;7:20041–20053. doi: 10.18632/oncotarget.7884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gollob JA, Wilhelm S, Carter C, Kelley SL. Role of Raf kinase in cancer: Therapeutic potential of targeting the Raf/MEK/ERK signal transduction pathway. Semin Oncol. 2006;33:392–406. doi: 10.1053/j.seminoncol.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 28.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 29.Leicht DT, Balan V, Kaplun A, Singh-Gupta V, Kaplun L, Dobson M, Tzivion G. Raf kinases: Function, regulation and role in human cancer. Biochim Biophys Acta. 2007;1773:1196–1212. doi: 10.1016/j.bbamcr.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kersemaekers AM, Fleuren GJ, Kenter GG, Van den Broek LJ, Uljee SM, Hermans J, Van de Vijver MJ. Oncogene alterations in carcinomas of the uterine cervix: Overexpression of the epidermal growth factor receptor is associated with poor prognosis. Clin Cancer Res. 1999;5:577–586. [PubMed] [Google Scholar]
- 31.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 32.Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, Wu YM, Dhanasekaran SM, Engelke CG, Cao X, et al. The landscape of circular RNA in cancer. Cell. 2019;176:869–881.e13. doi: 10.1016/j.cell.2018.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shang X, Li G, Liu H, Li T, Liu J, Zhao Q, Wang C. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular crcinoma development. Medicine (Baltimore) 2016;95:e3811. doi: 10.1097/MD.0000000000003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsiao KY, Lin YC, Gupta SK, Chang N, Yen L, Sun HS, Tsai SJ. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77:2339–2350. doi: 10.1158/0008-5472.CAN-16-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang PL, Liu B, Xia Y, Pan CF, Ma T, Chen YJ. Long non-coding RNA-low expression in tumor inhibits the invasion and metastasis of esophageal squamous cell carcinoma by regulating p53 expression. Mol Med Rep. 2016;13:3074–3082. doi: 10.3892/mmr.2016.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lasda E, Parker R. Circular RNAs: Diversity of form and function. RNA. 2014;20:1829–1842. doi: 10.1261/rna.047126.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gullick WJ, Marsden JJ, Whittle N, Ward B, Bobrow L, Waterfield MD. Expression of epidermal growth factor receptors on human cervical, ovarian, and vulval carcinomas. Cancer Res. 1986;46:285–292. [PubMed] [Google Scholar]
- 38.Biesterfeld S, Schuh S, Muys L, Rath W, Mittermayer C, Schroder W. Absence of epidermal growth factor receptor expression in squamous cell carcinoma of the uterine cervix is an indicator of limited tumor disease. Oncol Rep. 1999;6:205–209. doi: 10.3892/or.6.1.205. [DOI] [PubMed] [Google Scholar]
- 39.Yoshioka K. Scaffold proteins in mammalian MAP kinase cascades. J Biochem. 2004;135:657–661. doi: 10.1093/jb/mvh079. [DOI] [PubMed] [Google Scholar]
- 40.Kohno M, Pouyssegur J. Targeting the ERK signaling pathway in cancer therapy. Ann Med. 2006;38:200–211. doi: 10.1080/07853890600551037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.