Abstract
Liquid-liquid phase separation (LLPS) of proteins and nucleic acids has emerged as an important phenomenon in membraneless intracellular organization. We demonstrate that the linker histone H1 condenses into liquid-like droplets in the nuclei of HeLa cells. The droplets, observed during the interphase of the cell cycle, are colocalized with DNA-dense regions indicative of heterochromatin. In vitro, H1 readily undergoes LLPS with both DNA and nucleosomes of varying lengths but does not phase separate in the absence of DNA. The nucleosome core particle maintains its structural integrity inside the droplets, as demonstrated by FRET. Unexpectedly, H2A also forms droplets in the presence of DNA and nucleosomes in vitro, whereas the other core histones precipitate. The phase diagram of H1 with nucleosomes is invariant to the nucleosome length at physiological salt concentration, indicating that H1 is capable of partitioning large segments of DNA into liquid-like droplets. Of the proteins tested (H1, core histones, and the heterochromatin protein HP1α), this property is unique to H1. In addition, free nucleotides promote droplet formation of H1 nucleosome in a nucleotide-dependent manner, with droplet formation being most favorable with ATP. Although LLPS of HP1α is known to contribute to the organization of heterochromatin, our results indicate that H1 also plays a role. Based on our study, we propose that H1 and DNA act as scaffolds for phase-separated heterochromatin domains.
Significance
Histone proteins package cellular DNA into actively transcribed euchromatin domains as well as suppressed heterochromatin domains. Through in-cell and in vitro studies, we find that histones also contribute to heterochromatin formation through reversible liquid-liquid phase separation with DNA. The mesoscopic liquid droplets, rich in linker histone H1 and chromatin, likely govern the access of transcriptional factors and RNA to heterochromatin domains through charge balance, multicomponent interactions, and fluxional levels of small molecules such as ATP. In addition, the phase separation of the core histone H2A prompts new questions on the link between liquid-liquid phase-separation-mediated chromatin organization and epigenetic regulation.
Introduction
Studies over the past several years have established that many cellular proteins, typically containing intrinsically disordered regions (IDRs) rich in charged residues, are able to phase separate into distinct liquid-like droplets in both the presence and absence of nucleic acids (1, 2, 3, 4). Such liquid-liquid phase separation (LLPS) is driven by intermolecular interactions, including electrostatic, hydrophobic, π-stacking, and hydrogen-bonding interactions (5,6). It is thought that cells leverage this phenomenon for intracellular organization without a confining phospholipid membrane (2,4). Examples include P granules (1) and stress granules (7) in the cytoplasm, as well as the nucleoli (8) and Cajal bodies (9) in the nucleoplasm. Recent reports (10,11) have suggested that the separation of silenced heterochromatin from actively transcribed euchromatin is driven, at least in part, by LLPS. However, whether histones, which are key proteins for the structure and dynamics of chromatin, contribute to LLPS in cells remains unknown.
Histone proteins package genomic DNA as chromatin in the small volume of the cell nucleus and control access to the DNA by transcriptional factors (12, 13, 14, 15). The fundamental structural unit of chromatin is the nucleosome core particle (NCP), which consists of double-stranded DNA (dsDNA, ∼147 basepairs (bps)) wrapped around a histone octamer core consisting of dimers of H2A, H2B, H3, and H4 (16). The core histones have IDRs that constitute ∼50% of the protein. Linker histone H1, which binds to linker DNA at the DNA entry and exit site of NCPs, plays a key role in the higher-order structuring of NCPs (14). However, whether the higher-order structures formed in cells are in the 30-nm fiber architecture (17) or in a more disordered form is unclear (18). Furthermore, a large portion of H1 remains highly dynamic and mobile in the cell nuclei (19, 20, 21, 22), suggesting that H1 facilitates chromatin remodeling and dynamic organization (23).
It is becoming increasingly clear that H1 also plays an important role in epigenetic regulation (14,15,24,25). H1, known to bind many nuclear and cytosolic proteins (26,27), is found in high concentrations near suppressed genes (14,28, 29, 30), can bind directly to heterochromatin protein 1α (HP1α) (26,31, 32, 33), and recruits proteins responsible for histone modifications (14). H1 has a higher net positive charge and more structural disorder compared to core histones, with the longest IDR located at the C-terminus (Figs. S1, A and B and S2), and has been shown to readily undergo multicomponent LLPS with DNA in vitro (34,35). Shakya et al. demonstrated that a mixture of histones (the four core histones and the linker histone) undergoes LLPS with dsDNA oligomers, and that LLPS of H1 and DNA is more favorable (34). Turner et al. showed that the disordered C-terminal region of H1 remains dynamic upon complexation with 20- and 36-bp DNA (35). We hypothesized that H1 contributes to LLPS of heterochromatin because of its highly charged and disordered structure, direct interactions with HP1 (26,31, 32, 33), and elevated concentrations in heterochromatin (14,28, 29, 30). In HeLa cells expressing eGFP-tagged H1, we demonstrate that H1 condenses into puncta that are colocalized with HP1α and dense DNA of heterochromatin domains and that the puncta exhibit liquid-like behavior. Moreover, through in vitro studies using DNA and nucleosomes of varying lengths, we show that nucleosomes readily form droplets in presence of H1 in a manner invariant to the nucleosome length at physiological salt concentration. Inside the droplets, the nucleosomes maintain their structural integrity, as demonstrated by ensemble FRET measurements. Interestingly, we find that core histone H2A also forms droplets with DNA and nucleosomes in vitro, though to a lesser extent compared to H1. This is unexpected for a core histone because the other core histones (H2B, H3, and H4) precipitate under identical conditions. Furthermore, droplet formation of H1 and H2A with nucleosomes is promoted by the presence of free nucleotides in a nucleotide- and protein-dependent manner. This study suggests yet another role of histones in chromatin organization: LLPS-mediated formation of heterochromatin domains.
Materials and Methods
Materials
Nucleotide triphosphates (NTPs) (sodium salt), Tris-EDTA buffer (10 mM Tris-HCl, 1 mM EDTA (pH 7.4)), and sodium chloride were obtained from Sigma-Aldrich (St. Louis, MO). Fluorescently labeled NTPs (R6G-ATP, R110-GTP, R110-UTP, R110-CTP) were purchased from Perkin Elmer (Boston, MA). Calf thymus histone H1 was obtained from Sigma-Aldrich. Calf thymus H1 sequence contains 194 residues, has a net (+)53 charge, and is 77% disordered (see Supporting Materials and Methods). Human H2A, H2B, H3, H4, and HP1α were obtained from EpiCypher (Durham, NC) and EpiGex (Illkirch-Graffenstaden, France). The sequences of the proteins, including the human H1.2 used in in-cell experiments, are provided in the Supporting Materials and Methods. dsDNA (150-, 500-, and 1000-bp fragments of random sequences, Fig. S20 A) and AlexaFluor488 were obtained from Thermo Fisher Scientific (Waltham, MA). Nucleosomes (mononucleosomes (147 bp), polynucleosomes (trimers and heptamers), and FRET-pair-labeled mononucleosomes) were obtained from EpiCypher. The mononucleosomes were assembled from recombinant proteins with the 601 sequence DNA (147 bp) (Fig. S20 B). The polynucleosomes were polydisperse fragmented nucleosomes purified from cell extracts containing mostly trimers or heptamers. The concentrations of polynucleosome solutions were determined assuming the molecular weight of trimers (∼600 bp) or heptamers (∼1400 bp) (Fig. S20 B).
All samples for in vitro LLPS experiments were prepared in eight-well 1.0 borosilicate chambered glass slides (Nunc Lab Tek; Thermo Fisher Scientific). The glass slides were cleaned with RNase Zap (Ambion; Thermo Fisher Scientific) and nuclease-free water and passivated with 3.5% bovine serum albumin (Sigma-Aldrich) for 15 min at room temperature.
Cell culture
A HeLa cell line expressing eGFP-labeled H1.2 (labeled at the N-terminus) was purchased from EpiGex. Human H1.2 is the most common isoform in HeLa cells. Its sequence contains 215 residues, a net (+)54 charge, and is 77% disordered (see Supporting Materials and Methods). Cells were cultured in Dulbecco’s modified Eagle’s medium media (Sigma-Aldrich) with 10% fetal bovine serum, 2 mM glutamine, and 1% PenStrep at 37°C and under 5% CO2. Cells were grown in T-75 tissue culture flask (Thermo Fisher Scientific) until the cell culture density reached 90% confluency.
Live-cell imaging
For live-cell imaging, cell density of 2 × 104 cells/mL were plated on a confocal imaging dish (SPL Life Sciences, Naechon-Myeon, Republic of Korea) and incubated for two days at 37°C and under 5% CO2 before imaging. Three-dimensional live-cell imaging was carried out using an unmodified VT-iSIM microscope (VisiTech International, Sunderland, UK) equipped with an sCMOS camera (Zyla 5.5; Andor Technology, Oxford Instruments, Belfast, Northern Ireland, UK) and an incubator (Live Cell Instruments, Seoul, Republic of Korea). H1-eGFP imaging was carried out using 488 nm excitation and a 485–555 nm emission filter. For each experimental condition, measurements were carried out in triplicate. Cell culture conditions were maintained for live-cell imaging.
HP1α-antibody-AlexaFluor647 immunostaining
Cells were permeabilized using 0.2% Triton X-100 for 15 min and then blocked in 2% bovine serum albumin for 60 min. Cells were then incubated with 200 μM anti-HP1α-AlexaFluor647 (EPR5777; Abcam, Life Technologies, Cambridge, UK) and incubated overnight at 4°C. Cells were imaged on a Nikon A1R Confocal Microscope (Nikon, Tokyo, Japan) with simultaneous excitations at 487 nm with emission filter centered at 525 nm (GFP excitation) and 638 nm excitation with emission filter centered at 670 nm (AlexaFluor647 excitation).
DNA staining
DNA staining of HeLa cell nuclei was done by adding 5 μM (in final culture volume) DRAQ5 (Thermo Fisher Scientific) directly to the culture medium and incubating for 30 min at room temperature in the dark. Imaging was carried out using 647 nm excitation and a 652–732 nm emission filter.
ATP depletion
Cells were incubated with 10 mM sodium azide and 50 mM 2-deoxy glucose in Dulbecco’s modified Eagle’s medium for 30 min at 37°C to deplete cellular ATP and imaged under the same conditions as described in the live-cell imaging section.
LLPS of H1, H2A, H2B, H3, H4, and HP1α with 150-bp DNA
5.1 μM stock of 150-bp DNA was prepared in Tris-EDTA buffer with 150 mM NaCl. Stock solutions of histones were prepared at different concentrations (68.0 μM H1, 212.5 μM H2A, 200.0 μM H2B, 180.4 μM H3, and 212.5 μM H4). 1 μL of histone stock was added to 150-bp dsDNA solution such that the final N/P was ∼1. The N/P ratio was obtained assuming overall charges of 53 (H1), 17 (H2A), 18 (H2B), 20 (H3), and 18 (H4) per mole of histone. HP1α droplets were formed using stock solutions (21.4 μM) such that the final N/P was ∼0.9 (assuming an overall charge of −4). The protein-DNA mixtures were then loaded into surface-passivated borosilicate chambered glass slides, sealed to prevent evaporation, and imaged with a bright-field microscope. Each mixture was prepared in triplicate.
For experiments of H2B/H3/H4-DNA mixtures at different N/P ratios, dsDNA solution was prepared in Tris-EDTA buffer such that the final DNA concentration was 40 nM. Protein solutions were prepared in the same buffer, with concentrations tuned such that the final concentration ratio resulted in N/P ∼0.5, N/P ∼1, and N/P ∼2. The final salt concentration for all N/P ratios was 150 mM NaCl.
Salt-jump experiments, to test the reversibility of condensates, were performed by increasing the total NaCl concentration from 150 to 400 mM. Images were acquired 15 min after the addition of salt.
Phase diagrams of H1, H2A, and HP1α in the presence of DNA or polynucleosomes
For H1 and H2A, droplets were prepared at N/P ∼1. DNA solutions at final concentrations of 40.0 nM (150 bp), 12.0 nM (500 bp), and 6.0 nM (1000 bp) were prepared in Tris-EDTA buffer with NaCl concentrations of 150, 300, 400, and 500 mM. Polynucleosome solutions at final concentrations of 102.8 nM (147-bp monomer), 20.4 nM (600-bp trimer), and 8.8 nM (1400-bp heptamer) were prepared in Tris-EDTA buffer with NaCl concentrations of 150, 300, 400, and 500 mM. For H1-based droplets, H1 stock was added to the sample, resulting in a final concentration of 0.3 μM. For H2A-based droplets, H2A stock was added to the sample, resulting in a final concentration of 1.1 μM.
For HP1α, droplets were prepared at N/P ∼0.9. DNA solutions at final concentrations of 1.2 nM (150 bp), 0.4 nM (500 bp), and 0.2 nM (1000 bp) were prepared in Tris-EDTA buffer with NaCl concentrations of 150, 300, and 400 mM. Polynucleosome solutions at final concentrations of 1.8 nM (147-bp monomer), 0.6 nM (600-bp trimer), and 0.3 nM (1400-bp heptamer) were prepared in Tris-EDTA buffer with NaCl concentrations of 150, 300, and 400 mM. HP1α stock was added to the sample resulting in a final concentration of 0.1 μM. The mixtures were then loaded into surface-passivated borosilicate chambered glass slides, sealed to prevent evaporation, and imaged with bright-field microscopy. Each mixture was prepared in triplicate. For each experimental condition, 10 images per sample were analyzed to obtain the number of droplets, the size of the droplets, and the droplet surface coverage in the frame.
For experiments of H1/H2A-polynucleosome mixtures at different N/P ratios, nucleosome solutions were prepared such that the final concentration of nucleosomes were 102.8 nM (mononucleosomes), 20.4 nM (trimers), and 8.8 nM (heptamers) for all N/P ratios. Different amounts of H1/H2A were added to each nucleosome solution so as to reach final ratios of N/P ∼0.6, N/P ∼1, and N/P ∼1.75. The solutions were all prepared in Tris-EDTA buffer and 150 mM final NaCl concentration.
Role of free nucleotides in LLPS
To maintain the same overall total charge in the solution (N/P ∼1), the concentration of polynucleosome (1400-bp heptamer) was adjusted such that half of the total negative charge was accounted for by the added NTP. Polynucleosome (4.4 nM final concentration) and NTP (1.5 μM final concentration) in Tris-EDTA buffer with 150 mM NaCl were first mixed. Then, H1 stock solution (0.3 μM final concentration) was added to it such that the final N/P was ∼1. For the control, the same concentrations of polynucleosome (4.4 nM final concentration) and H1 (0.3 μM final concentration) but without ATP were mixed (Tris-EDTA buffer, 150 mM NaCl, final N/P ∼2.4). For confocal imaging, fluorescently labeled NTPs (R6G-ATP, R110-GTP, R110-UTP, R110-CTP) were added to the unlabeled NTP solution (5.0 nM final concentration).
Bright-field imaging
Bright-field microscopy was carried out on a Leica DMI6000 B microscope (Leica Microsystems, Wetzlar, Germany) equipped with a Grasshopper3 camera (Point Gray, Richmond, BC, Canada). Imaging was carried out with a 100× oil objective (1.2 NA). Final images were obtained after droplet merging had subsided (typically after ∼2 h at room temperature). The contrast and brightness were adjusted using ImageJ software (36).
Droplet analysis
In HeLa cells, puncta were analyzed using Particle Analysis feature in ImageJ software (36) with a lower size limit of diameter = 0.25 μm and upper size limit of diameter = 2 μm. For droplets formed in vitro, volumes were obtained assuming spherical shape. Images were recorded at the same z-section (distance from glass surface) for all samples. Surface coverage was obtained by taking the ratio of the total area of the camera field of view to the total area of condensates in the field of view.
Confocal and FRET
Fluorescently labeled mononucleosome (H2A-T120C-Cy5 and 5′-Cy3-DNA) was first mixed with unlabeled mononucleosomes (0.01 nM final fluorophore concentration). H1 or H2A was added to the mixture (as described above, 150 mM final NaCl concentration) to form droplets.
Confocal microscopy and FRET experiments on the droplets were carried out on an unmodified commercial Leica SP8× microscope (Leica Microsystems) equipped with pulsed laser excitation (NKT Photonics, Birkerød, Denmark), single-photon avalanche diodes (SPADs) (Micron Photon Devices, Bolzano, Italy), and external time-correlated single photon counting (TCSPC) detection (PicoQuant, Berlin, Germany). For confocal imaging of Cy3, excitation was carried out at 533 nm. Emission from Cy3 and Cy5 were filtered at 555–595 nm (Cy3 emission) and 721–749 nm (Cy5 emission) and detected on two detectors.
For ensemble FRET measurements, excitation of Cy3 was carried out at 533 nm. Emission was separated with a dichroic filter (620 nm), passed through emission filters of 555–595 nm (Cy3 emission) and 721–749 nm (Cy5 emission), and detected on two SPADs equipped with TCSPC. Spectral overlap was calibrated using dilute solutions (∼10.0 nM) of free Cy3 and Cy5 in solution. H1-mononucleosome and H2A-mononucleosome droplets were prepared in triplicate. Ensemble FRET measurements were performed three times on each sample. The error bars represent the standard deviation of a total of nine measurements.
For confocal imaging of fluorescently labeled NTPs, excitation was carried out at either 561 nm (R6G-ATP) or 514 nm (R110-GTP, R110-UTP, R110-CTP), with detection at either 570–620 or 525–575 nm, respectively. Images were collected over a 10 × 10 μm area.
Protein labeling for fluorescence correlation spectroscopy and fluorescence recovery after photobleaching experiments
An AlexaFluor488 protein labeling kit (Invitrogen, Thermo Fisher Scientific) was used for labeling of H1, H2A, and HP1α. A protein solution (∼20–70 μM) was added to a vial of reactive dye, and the reaction mixture was stirred for 1 h. Labeled protein was purified using purification resins and centrifuged to collect the purified protein in a collection tube. The degree of protein labeling was determined using absorption spectroscopy at 280 nm and 494 nm.
Fluorescence correlation spectroscopy
AlexaFluor488 (7.5 nM final concentration) was first mixed with DNA or nucleosome solutions. H1 was then added to the mixture (as described above, 150 mM final NaCl concentration) to form droplets.
Fluorescence correlation spectroscopy (FCS) of AlexaFluor488-labeled proteins (H1, H2A, and HP1α) in droplets or irregular condensates was also performed. For these experiments, condensates were prepared similarly to the method described above but with fluorescently labeled protein added (final concentration of 5 nM). Partitioning coefficients of the protein component into condensates for linear DNA (150 bp) and mononucleosome were estimated from the G0 value of the fluorescence intensity autocorrelation function. The G0 value is related to the concentration of the fluorescent label as c = (G0 × N × V)−1, where c is the concentration of fluorescent label, G0 is the value of the intensity autocorrelation function at τ = 0, N is Avogadro’s number, and V is the volume of the excitation spot.
The same Leica instrument used for confocal and FRET experiments was also used for FCS. Excitation at 488 nm was provided from a pulsed white-light laser passed through a 488-nm notch filter. The output power was reduced to reach 10% of the master power (70%). The fluorescence signal was passed through a pinhole (set to 1 Airy unit) and emission filtered at 500–550 nm for detection. The signal was detected on an SPAD with a TCSPC unit. SymPhoTime software was utilized to obtain the FCS autocorrelation curves and diffusion parameters inside the droplets.
Fluorescence recovery after photobleaching
Protein-DNA/nucleosome droplets were prepared (as described above) with added AlexaFluor488-labeled proteins (H1, H2A, HP1α) at a 5 nM final concentration. Fluorescence recovery after photobleaching (FRAP) experiments were performed on the same Leica instrument as used for confocal, FRET, and FCS experiments. Excitation at 488 nm was provided from a pulsed white-light laser passed through a 488 nm notch filter. The output power was reduced to reach 10% of the master power (70%). Bleaching was carried out by focusing 100% of the master power (70%) on a designated bleach point for 200 ms.
Sodium dodecyl sulfate-polyacrylamide-gel electrophoresis
10 μM histone protein sample was mixed with lysis buffer (Laemmli buffer), boiled for 10 min (100°C in water bath), and then centrifuged for 1 min at 5000 rpm. The samples were loaded into each well and allowed to run on electrophoresis at 120 V for 2–3 h. 0.75 mm thick gel (5% stacking gel and 15% resolving gel) was prepared in a vertical electrophoresis cell (Bio-Rad, Hercules, CA) according to the manufacturer’s instructions. The gel was treated with Coomassie blue G-250 (Sigma-Aldrich) for 2 h to stain the proteins and then placed in destaining buffer (10% acetic acid, 50% methanol, and 40% H2O) overnight before imaging.
DNA gel electrophoresis
1% agarose gels were prepared using agarose dissolved in 1×-TBE buffer by heating the solution in a microwave for 2–3 min. 6 μL of the sample (50–100 ng) was loaded into each well and run for 30 min at 100 V. DNA samples were mixed with the 6× loading dye and loaded into the well. Nucleosomes were first mixed with the 6× loading dye and heated at 75°C for 20 min to denature the protein, then cooled to room temperature, centrifuged, and loaded into the wells before imaging.
Results
LLPS of H1-chromatin in HeLa nuclei
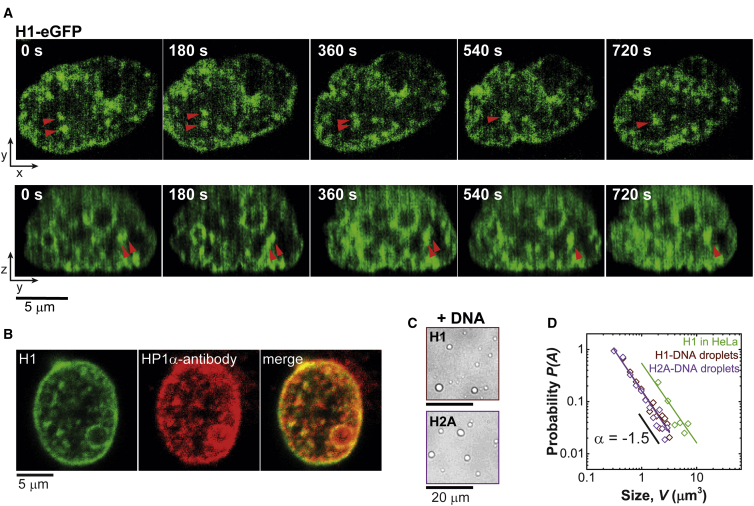
The concentration of H1 throughout the nucleus is heterogeneous (25), with high concentrations typically found near suppressed genes (14,28, 29, 30), which we hypothesized to play a role in LLPS of chromatin. In HeLa cells expressing eGFP-tagged H1, we monitored chromatin dynamics using confocal fluorescence microscopy. During the interphase of the cell cycle, H1 condenses into heterogeneously distributed puncta in HeLa nuclei (Fig. 1 A; Fig. S3). A typical cell nucleus contains ∼10 puncta, on average. Two-color confocal microscopy of H1-eGFP and HP1α-antibody-AlexaFluor647 confirm that the puncta incorporate heterochromatin, as indicated by colocalization of H1 and HP1α, a marker of heterochromatin (Fig. 1 B). The H1 puncta also colocalize with dense DNA (Fig. S4). Using confocal video microscopy, the dynamics of the puncta were observed over periods of 12–24 h. We find that in relatively rare events, puncta that come into contact with one another coalesce (Fig. 1 A; Fig. S3). Furthermore, the size distribution of the puncta follows a power law with an exponent of −1.5 (Fig. 1 D). Taken together, these observations indicate that the puncta are liquid-like droplets rich in H1 and chromatin (10,37). However, most of the puncta are not perfectly circular, unlike droplets with high surface tension and low viscosity, implying that the underlying chromatin structure provides a high energy barrier for relaxation to a circular shape. This is likely due to the local rigidity of chromatin partitioned in the puncta, which can lead to nonspherical condensates (34,38). Interestingly, under ATP-depleted conditions, the number of H1 puncta per cell nuclei was significantly reduced (Fig. S5), suggesting that ATP promotes the formation of H1-based condensates (38,39).
Figure 1.
H1 contributes to multicomponent LLPS in cells. (A) Time-series confocal microscopy images, sectioned in the x-y (top) and the z-y (bottom) planes, are given, showing the coalescence of distinct H1-eGFP puncta in HeLa nuclei. (B) Two-color confocal microscopy images of H1-eGFP (left) and immunostained anti-HP1α-AlexaFluor647 (middle) in the nucleus of a HeLa cell are shown. The merged image (right) shows colocalization of H1 with HP1α (marker for heterochromatin). (C) H1 (0.3 μM, top) and H2A (1.1 μM, bottom) undergo multicomponent LLPS in vitro in presence of 150-bp DNA (0.04 μM) at physiological salt concentration (N/P ∼1). (D) The size distribution of HeLa H1 puncta, as well as H1-DNA and H2A-DNA condensates, follows a power law with an exponent of −1.5. To see this figure in color, go online.
LLPS of histones with DNA in vitro
The linker histone H1, as well as the core histones, has significant intrinsic disorder and a large net positive charge (Figs. S1, A and B and S2). We therefore tested their ability to individually form liquid-like droplets in vitro. Unlike several multivalent and IDR-rich proteins that have been shown to undergo single-component LLPS in vitro (40), H1 alone does not form droplets (Fig. S1 D). We attribute this to the large overall positive charge that must be neutralized to form droplets. In the presence of 150-bp DNA, at an overall protein amino acid (positively charged)/DNA phosphate (negatively charged) charge ratio (N/P) ∼1, droplets are readily formed (Fig. 1 C; Fig. S1), consistent with previous observations with short DNA oligomers (34,35). Rapid diffusion of a small-molecule probe (AlexaFluor488) in H1-DNA and H1-nucleosome droplets is observed, as expected for a liquid-like environment. The fastest diffusion was observed for condensates formed with long DNA/nucleosome (Fig. S6). This is likely due to condensates formed from longer polymers having a larger mesh size and more free volume, thus allowing faster diffusion of small molecules (41). The droplet size distribution also follows the expected power law (Fig. 1 D). The associative LLPS of H1 with DNA under charge-balanced ratios suggests that both H1 and DNA act as scaffold components (42).
Unexpectedly, we also observe multicomponent LLPS of the core histone H2A in the presence of 150-bp DNA (droplet size follows the expected power-law distribution; Fig. 1, C and D). Droplet formation of H2A with DNA cannot be explained simply on the basis of overall positive charge and degree of disorder because the other core histones (H2B, H3, and H4), which have similar overall charge and disorder (Fig. S1, A and B), precipitate under identical conditions (Fig. S1 C). LLPS of H2B, H3, and H4 with 150-bp DNA was also tested at different charge ratios because the distribution of charged residues in structured and unstructured regions of the protein could shift the LLPS phase boundary away from charge-neutral conditions. However, even at different charge ratios (N/P ∼0.5, N/P ∼1, N/P ∼2), aggregates with DNA were formed (Fig. S7), suggesting other aspects of the protein structure besides overall charge and disorder (for instance, sequence and charge patterning), should also be considered (43, 44, 45). These aggregates were irreversible, as upon addition of salt, the condensates did not dissolve. The disordered regions of histone H2A contain a higher number of amino acids capable of hydrogen bonding compared to the other core histones (Fig. S2). Furthermore, specific interactions of the C-terminal tail (∼15 amino acids long) of H2A with DNA potentially contributes to the LLPS because it is known to make several contacts with nucleosomal and linker DNA (46, 47, 48, 49, 50). The C-terminal tail of H2A is also known to directly interact with H1 (49,51, 52, 53). Moreover, the C-terminal tail of H2A is subjected to post-translational modifications (54, 55, 56, 57) that could provide a mechanism for regulating LLPS of chromatin in cells.
LLPS of histones with nucleosomes and stability of the NCP
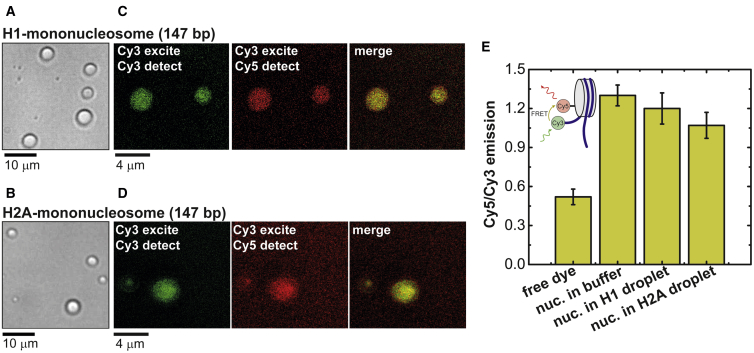
Because H1 and H2A readily form droplets with free DNA, we tested whether similar behavior is observed with nucleosomes in which the overall charge of the DNA is reduced because of binding of core histones, and, if so, whether the NCP remains structured inside the droplets. Droplet formation is observed for both H1 and H2A upon mixing with mononucleosomes (Fig. 2, A and B). Fluorescence imaging of FRET-labeled mononucleosomes (5′ Cy3-DNA, Cy5-H2A) reveal that nucleosomes partition into both H1- and H2A-based droplets (Fig. 2, C and D). Furthermore, ensemble FRET measurements indicate that the NCP remains structured inside the droplets (Fig. 2 E). It is interesting to consider these results within the context of experiments on Ddx4 protein droplets, which were shown to melt the duplex structure of short DNA oligomers (58). In the latter case, dsDNA partitioned into single-component Ddx4 droplets were found to denature, suggesting that under certain conditions, the environment within the condensates destabilize biological structures. In our experiments, we measure distances between a strand of the nucleosomal dsDNA and a core histone to report on the nucleosome stability. However, in general, the differences between the stability biological structures in condensates could be attributed to 1) enhanced stability of the duplex in the nucleosome structure or 2) the multicomponent nature of histone-DNA/nucleosome LLPS, which requires incorporation of DNA/nucleosome into the droplets to achieve charge balance. Single-component droplets, on the other hand, are charge balanced in the absence of DNA and could therefore destabilize structures with high charge densities. Additionally, specific interactions between H1/H2A and DNA (49,51, 52, 53) could contribute to the observed stability of nucleosomes in the droplets.
Figure 2.
The NCP remains structured when partitioned into H1-mononucleosome and H2A-mononucleosome droplets. Bright-field microscopy images of (A) H1-mononucleosome and (B) H2A-mononucleosome droplets in vitro at physiological salt concentration (N/P ∼1) are shown. (C and D) Confocal microscopy images of Cy3-Cy5-labeled mononucleosomes (Cy3-DNA, Cy5-H2A) showing the partitioning of the nucleosomes into the droplets are given. (E) Cy5/Cy3 emission ratio after Cy3 excitation shows FRET emission for labeled mononucleosome in buffer solution, H1-nucleosome droplets, and H2A-nucleosome droplets, indicating that the NCP remains intact within the droplets. Error bars represent the standard deviation of nine measurements. To see this figure in color, go online.
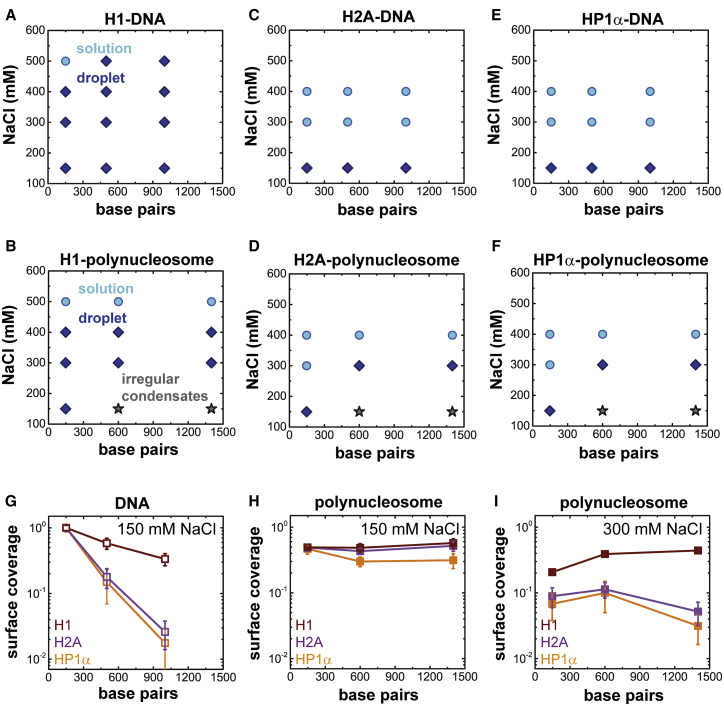
Phase diagram of H1/H2A/HP1α with DNA and polynucleosomes
To determine the ability of H1 and H2A to incorporate large DNA/nucleosomes into the liquid-like phase, we measured phase diagrams by varying DNA/polynucleosome length and salt concentration at a fixed charge ratio (N/P ∼1), keeping the protein concentration constant (Figs. 3 and 4; Figs. S8 and S9). For comparison, we also tested HP1α, which has recently been suggested to contribute to the LLPS of heterochromatin (11). HP1α has an overall negative charge (−4) and consists of a net charge-neutral disordered hinge region (Fig. S1 A; (59)). Note that because HP1α has an overall negative charge, it was not possible to reach charge-balanced conditions. Therefore, the experiments were performed at N/P ∼0.9.
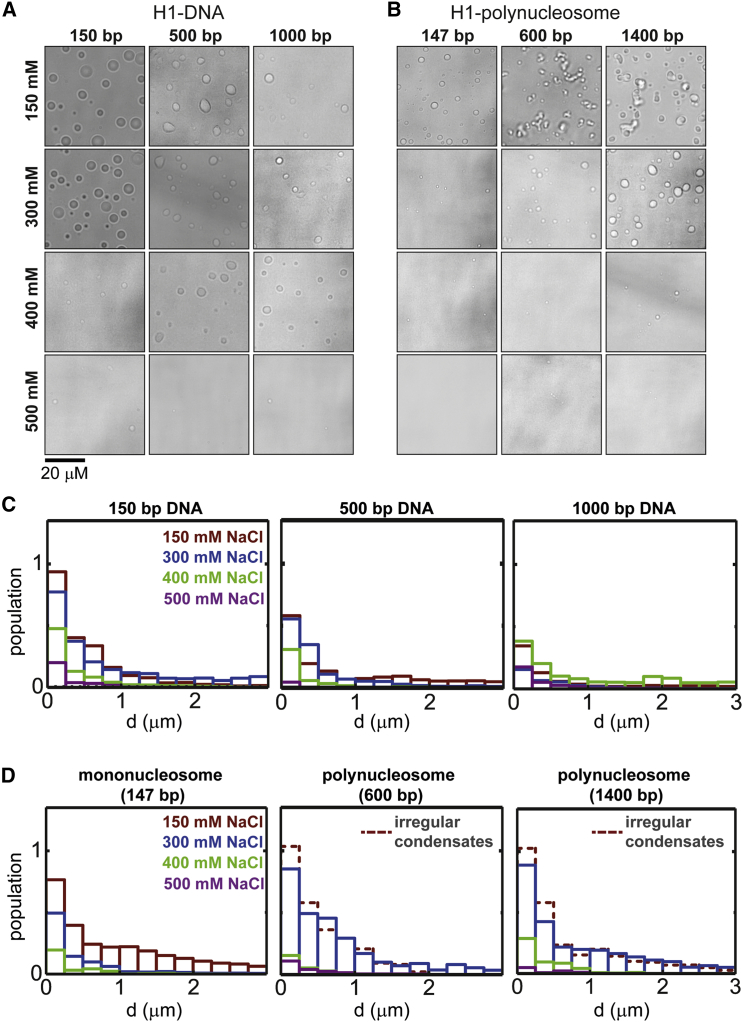
Figure 3.
LLPS of H1 is invariant to DNA or polynucleosome length. A phase diagram and droplet size analysis for H1-DNA and H1-polynucleosome are shown. Bright-field microscopy images of (A) H1-DNA and (B) H1-polynucleosome mixtures with varying salt concentrations and DNA or polynucleosome length (N/P ∼1) are given. Images were acquired ∼2 h after mixing. (C and D) Droplet population statistics versus droplet size (diameter, d) for each experimental condition is shown, obtained from the analysis of 10 images per experiment. To see this figure in color, go online.
Figure 4.
Phase diagrams for H1, H2A, and HP1α in the presence of DNA or polynucleosomes. A phase diagram as a function of salt concentration and DNA or polynucleosome length (expressed as number of DNA bps) (N/P ∼1) is shown for (A) H1-DNA and (B) H1-polynucleosome mixtures. (C and D) Corresponding phase diagrams for H2A-DNA and H2A-polynucleosome mixtures and (E and F) HP1α-DNA and HP1α-polynucleosome mixtures are shown. (G) Droplet surface coverage at [NaCl] = 150 mM is shown, quantified by analyzing bright-field microscopy images for H1, H2A, and HP1α droplets formed with DNA as a function of DNA length. LLPS decreases with increasing DNA length for all proteins, though to a lesser extent for H1. Droplet surface coverage at (H) [NaCl] = 150 mM and (I) [NaCl] = 300 mM for H1, H2A, and HP1α droplets formed with polynucleosomes as a function of polynucleosome length is shown. For H1, LLPS with polynucleosome is length invariant at 150 mM salt and scales favorably with length at 300 mM salt (at which spherical droplets are observed; Fig. S10) compared to H2A and HP1α. Error bars shown in G-I represent the standard deviation of analysis of ten images. To see this figure in color, go online.
LLPS was quantified by analyzing the surface coverage of droplets for each experimental condition (Fig. 3, C and D; Figs. S8, C and D and S9, C and D). H1-DNA mixtures form droplets across a wide range of salt concentrations (150–500 mM NaCl), with the most pronounced phase separation observed at 150 mM salt for all DNA lengths (150, 500, and 1000 bp linear DNA, Figs. 3 A and 4 A). H1-DNA LLPS is diminished with increasing DNA length (Fig. 4 G). A similar trend is observed for both H2A-DNA (Fig. S8, A and C) and HP1α -DNA mixtures (Fig. S9, A and C). However, LLPS is the most pronounced for H1, compared to H2A and HP1α, for all DNA lengths (Fig. 4 G).
H1-polynucleosome mixtures also undergo phase separation across a wide range of salt concentrations (150–400 mM NaCl) (Figs. 3 B and 4 B). However, at 150 mM salt, only the H1-mononucleosome (147 bp) mixture forms spherical droplets, whereas H1-trimer (600 bp) and H1-heptamer (1400 bp) mixtures form irregular condensates. These irregular condensates are, however, reversible, as determined by the salt-jump experiments discussed in the following section. Above 300 mM salt, spherical droplets are observed for all nucleosome lengths. Interestingly, unlike for H1-polynucleosome (Fig. 3, B and D), both H2A-polynucleosome (Fig. S8, B and D) and HP1α-polynucleosome (Fig. S9, B and D) mixtures show a decrease in formation of spherical droplets with increasing nucleosome length.
H1 shows a broad phase diagram for both H1-DNA and H1-polynucleosome mixtures, whereas in the case of H2A and HP1α, the phase diagrams are narrow (Fig. 4, A–F). Furthermore, for H1, LLPS with polynucleosome is length invariant at 150 mM salt and scales favorably with polynucleosme length at 300 mM salt (at which spherical droplets are observed) compared to H2A and HP1α (Fig. 4, G–I; Fig. S10). This indicates that H1 is capable of incorporating large segments of DNA/nucleosome into distinct droplets through LLPS. We suspect that this is due to the high positive-charge density of H1 (compared to H2A and HP1α) neutralizing the negative charge of the linker DNA, the high negative charge density of which is not screened by core histones.
LLPS of H1 and H2A with nucleosomes was also measured at different charge ratios (at 150 mM salt) to test whether the formation of irregular condensates observed for H1 and H2A with polynucleosomes was due to the protein structure rendering some charged residues inaccessible, therefore leading to an effective charge imbalance. For H1-mononucleosome, phase separation is not observed at an overall negative charge ratio (N/P ∼0.6), whereas at charge-neutral (N/P ∼1) and overall positive (N/P ∼1.75) charge ratios, spherical droplets are observed (Fig. S11). Therefore, overall charge-neutral or excess positive charge is required for droplet formation. For H1-polynucleosome LLPS (600 and 1400 bp), irregular condensates were formed at all charge ratios tested (Fig. S11). Similar general behavior is observed for H2A-nucleosome LLPS (Fig. S12).
Characterization of H1/H2A/HP1α-nucleosome condensates
At physiological salt concentration, LLPS of H1/H2A/HP1α with mononucleosomes forms spherical droplets, whereas LLPS with polynucleosomes forms irregularly shaped condensates. To characterize the reversibility of the condensates, we added salt to preformed condensates at 150 mM NaCl such that the final salt concentration increased to 400 mM. For all proteins studied, spherical condensates formed with mononucleosomes were found to be reversible upon an increase in salt concentration (Figs. S11–S13). This is expected for liquid-like droplets, in which cohesive protein-DNA interactions are weak and reversible (60). The irregular condensates formed with polynucleosomes are also found to be reversible upon an increase in salt concentration (Figs. S11–S13). To characterize whether these reversible irregular condensates are dynamically arrested (as expected when interactions are sufficiently strong such that the structure cannot relax into a spherical droplet typical of liquids, despite being reversible), we performed fluorescence recovery after photobleaching FRAP experiments as described below.
FRAP experiments were performed on AlexaFluor488-labeled H1, H2A, and HP1α partitioned into protein-nucleosome condensates at 150 mM NaCl. In H1-mononucleosome condensates (spherical droplets), FRAP experiments show clear photobleaching and fluorescence recovery (τ1 = 1 s, τ2 = 30 s) (Fig. S14, A and B), indicating that H1 is highly mobile within these condensates. In contrast, within H1-polynucleosome condensates (irregular condensates), no fluorescence recovery is observed after photobleaching (Fig. S14, A and B), indicating that H1 is dynamically arrested. In H2A- and HP1α-mononucleosome condensates, fluorescence recovery after photobleaching is observed, though on slower timescales relative to H1-mononucleosome condensates (H2A: τ1 = 2 s, τ2 = 500 s; HP1α: τ1 = 5 s, τ2 = 1000 s), whereas no fluorescence recovery is observed for H2A- and HP1α-polynucleosome condensates (Fig. S14, C–F).
FCS was used to measure the amount of partitioning of H1 and H2A in the condensates. In all cases, the slow diffusive dynamics of the protein are measured (Fig. S15), consistent with FRAP experiments. From the G0 values, H1 concentration is estimated to be higher in H1-mononucleosome droplets (2.4 mM) compared to H1-150-bp DNA droplets (1.4 mM). Similarly, H2A concentrations are estimated to be 0.8 mM in H2A-mononucleosome droplets and 0.4 mM in H2A-150-bp DNA (150-bp) droplets. For each protein, partitioning into protein-mononucleosome droplets is observed to be stronger. Furthermore, H1 is found to partition significantly stronger into droplets compared to H2A.
Free-nucleotide-facilitated droplet formation
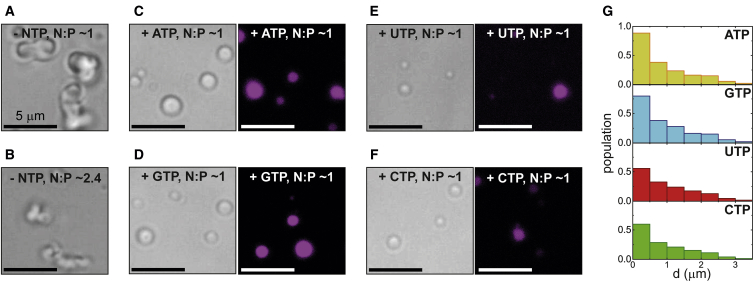
The unique phase diagram of H1 compared to H2A and HP1α suggests that H1 plays an important role in driving LLPS with chromatin. Although the biological role of LLPS in chromatin is not clearly understood, it is thought that LLPS of nuclear proteins contribute to heterochromatin formation as a mechanism of gene silencing (10,11). It is also probable that LLPS helps to regulate the local protein environment around silenced DNA because it has been shown that phase-separated droplets can act as selective biomolecular filters (58). In our experiments at physiological salt concentration, we find that only mononucleosomes form spherical droplets with H1, whereas polynucleosomes form irregular condensates (Fig. 5 A). Because, in HeLa cells, H1 clearly forms liquid-like puncta with chromatin (Fig. 1; Fig. S3), the question arises as to what additional factors promote LLPS of large polynucleosomes. Upon reducing the concentration of polynucleosomes, irregular condensates are still formed (Fig. 5 B), indicating that the absolute concentration of polynuclesomes is not the determining factor for droplet formation. Given the high concentration of ATP in the nucleus (61,62), we hypothesized that the presence of free nucleotides could promote LLPS of H1 and polynucleosomes. It has been previously demonstrated that ATP acts as a biological hydrotrope that promotes droplet formation or dissolution by suppressing aggregation (39). Furthermore, ATP was shown to promote LLPS in poly-L-lysine-dsDNA mixtures that otherwise precipitate because of the rigidity and high charge density of dsDNA (38). In presence of ATP, at N/P ∼1, H1-polynucleosomes form spherical droplets at physiological salt concentration (Fig. 5 C).
Figure 5.
H1-polynucleosomes form liquid-like droplets at physiological salt concentration in the presence of free NTPs. H1-polynucleosomes (1400 bp) form coarse condensates in absence of ATP at physiological salt concentration at (A) N/P ∼1 and (B) N/P ∼2.4 (reduced polynucleosome concentration). (C–F) Representative bright-field and confocal images of fluorescently labeled NTPs (C ATP; D GTP; E UTP; F CTP; all at a concentration of 1.5 μm) show the presence of free nucleotides promotes H1-polynucleosome (1400 bp) LLPS into spherical, liquid-like droplets at physiological salt concentration (N/P ∼1). (G) Droplet population statistics versus droplet size (diameter, d) is given for each NTP studied, showing higher propensity for droplet formation for ATP and GTP compared to UTP or CTP. To see this figure in color, go online.
We performed FRAP and FCS experiments on AlexaFluor488-labeled H1 and H2A in H1/H2A-heptanucleosome-ATP droplets. We observe that the protein component (H1 and H2A) is mobile in presence of ATP, as evidenced by rapid fluorescence recovery after photobleaching (H1-heptanucleosome-ATP τ1 = < 1 s, τ2 = 18 s; H2A-heptanucleosome-ATP τ1 = 6 s, τ2 = 45 s) (Fig. S16). This result is consistent with condensates having a spherical shape in presence of free nucleotides. In addition, FCS experiments on H1/H2A-heptanucleosome droplets (Fig. S17) show no decay in the intensity autocorrelation function, G(τ), over a timescale of 1 s in the absence of ATP. However, in the presence of ATP, the protein component is mobile, as evidenced by the complete decay of G(τ) within 1 s.
Interestingly, LLPS is promoted by other free nucleic acids (GTP, UTP, and CTP) as well, in a nucleotide-dependent manner (Fig. 5, D–F). Droplet population statistics versus droplet diameter demonstrate that the amount of LLPS is comparable for both ATP and GTP, whereas UTP and CTP show less phase separation (Fig. 5 G). In the case of H2A-polynucleosome condensates, free ATP was found to promote LLPS into spherical droplets at physiological salt concentration, whereas irregular condensates formed in presence of other nucleotides (Fig. S18). These results are consistent with in-cell experiments that show decreased puncta formation under ATP-depleted conditions (Fig. S5). These observations suggest that free nucleotides can modulate LLPS of H1/H2A and chromatin through fluxional concentration levels in cells in both nucleotide- and protein-dependent manners. It is not yet known how free ATP affects LLPS of chromatin because most of the studies are in the context of ATP hydrolysis. It has been speculated that the presence of ATP helps to keep proteins that are otherwise prone to aggregation soluble in the cells (39). It is possible that free ATP is also involved in maintaining the liquid-like environment of chromatin, in particular in heterochromatin, where the nucleosomal DNA is more tightly packed.
Discussion
The merging dynamics and the power-law (−1.5) size distribution of H1-rich puncta observed in the nuclei of HeLa cells during the interphase of the cell cycle indicate that the puncta are liquid-like droplets. Colocalization of the puncta with DNA-dense regions of the chromatin and HP1α indicate that these are heterochromatin domains (Fig. 1 B; Fig. S4). This study provides direct evidence that H1 contributes to LLPS of heterochromatin domains in cells. The multicomponent phase separation process likely involves participation from numerous nuclear proteins. This is consistent with recent observations of HP1α undergoing LLPS with chromatin (10,11) because H1 is also known to interact with HP1α (26,31, 32, 33), with the interactions modulated by post-translational modifications. For example, H1K85 acetylation is known to promote recruitment of HP1α and chromatin condensation (63), whereas H1 phosphorylation decreases the interaction with HP1α (64). Thus, it is possible that cells utilize epigenetically controlled co-phase separation of HP1α and H1 to form liquid-like heterochromatin domains. Within this context, it is important to note that acetylation (65) and phosphorylation (66) of intrinsically disordered proteins (IDPs) have been shown to modulate LLPS.
Given that the core histones also have a large net positive charge and possess IDRs, we tested the ability of all four core histones to form liquid-like droplets with DNA in vitro. Surprisingly, only H2A undergoes multicomponent LLPS with DNA, forming droplets, whereas the remaining core histones (H2B, H3, and H4) precipitate (Fig. 1, C and D; Fig. S1 C). The unique behavior of H2A, among all core histones, indicates that other factors must be considered besides overall charge and disorder of the proteins. For example, the C-terminal tail of H2A has a higher percentage of residues capable of hydrogen bonding (Fig. S2) compared to other core histones. H2A also forms droplets with nucleosomes, though the LLPS is strongly dependent on nucleosome length (Fig. 4). We attribute the LLPS of H2A to its C-terminal tail, which is known to form several contacts with the nucleosomal and linker DNA (16,46, 47, 48, 49, 50), hence allowing specific interactions with DNA, which could promote LLPS. Cross-linking experiments have shown that H2A can directly interact with H1 (51,52), the interaction being attributed to the last 15 amino acids of H2A C-terminal tail (49). The positioning of the C-terminal tail of H2A near the globular domain of H1 (48,49) likely predisposes protein-protein interactions that could facilitate phase separation in cell nuclei. Although studies have not demonstrated the presence of free core histones in live cells, it is well understood that the nucleosome is a dynamic structure that allows transient access of the nucleosomal DNA and histones by extranucleosomal proteins (15,67). Moreover, exchange of the canonical H2A with its variants mediated by remodeling proteins is also known (13), although most of the studies are in the context of H2A-H2B dimers. It is also possible that very low levels of free H2A in tandem with other phase-separating proteins could collectively contribute to LLPS, as has been observed in in vitro experiments with a natural mixture of histones and DNA oligomers (34).
Based on the LLPS of H1 with nucleosomes of various lengths, we rationalize that H1 plays a role in LLPS of chromatin in cells. However, we expect the interaction of H1 with other proteins such as HP1α and H2A to be important, in particular for regulation of LLPS associated with post-translational modifications. For example, ubiquitylation of lysine residues in the C-terminal tail of H2A (a post-translational modification that has been linked to transcription repression (57,68)) has been shown to enhance binding of H1 to nucleosomes in vitro (69). Consistently, deubiquitination of H2A has been shown to enhance phosphorylation and dissociation of H1 (70). The implication of heterochromatin formation by LLPS is that access to heterochromatin is likely regulated by rules that govern partitioning of small molecules and nuclear proteins into chromatin-rich droplets. As such, properties such as charge density, protein exchange, site-specific interactions, and fluxional levels of small molecules such as ATP are likely to be important.
Author Contributions
A.S. conceived and designed the project. A.S., S.P., and J.T.K. carried out in vitro experiments. N.R. carried out in-cell experiments. All authors contributed to data analysis, interpretation, and writing of the manuscript.
Acknowledgments
We thank Prof. François Amblard and Mrs. Azar Shahmoradi for helpful discussions on cell-based experiments.
The work was supported by the Korean Institute for Basic Science, project code IBS-R020-D1.
Editor: Tom Misteli.
Footnotes
Anisha Shakya and Seonyoung Park contributed equally to this work.
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.12.022.
Contributor Information
Anisha Shakya, Email: shanisha@unist.ac.kr.
John T. King, Email: jtking@unist.ac.kr.
Supporting Material
References
- 1.Brangwynne C.P., Eckmann C.R., Hyman A.A. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- 2.Hyman A.A., Weber C.A., Jülicher F. Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- 3.Zhang H., Elbaum-Garfinkle S., Gladfelter A.S. RNA controls polyQ protein phase transitions. Mol. Cell. 2015;60:220–230. doi: 10.1016/j.molcel.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitrea D.M., Kriwacki R.W. Phase separation in biology; functional organization of a higher order. Cell Commun. Signal. 2016;14:1–20. doi: 10.1186/s12964-015-0125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pak C.W., Kosno M., Rosen M.K. Sequence determinants of intracellular phase separation by complex coacervation of a disordered protein. Mol. Cell. 2016;63:72–85. doi: 10.1016/j.molcel.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang J., Choi J.M., Hyman A.A. A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell. 2018;174:688–699.e16. doi: 10.1016/j.cell.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Molliex A., Temirov J., Taylor J.P. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feric M., Vaidya N., Brangwynne C.P. Coexisting liquid phases underlie nucleolar subcompartments. Cell. 2016;165:1686–1697. doi: 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Machyna M., Heyn P., Neugebauer K.M. Cajal bodies: where form meets function. Wiley Interdiscip. Rev. RNA. 2013;4:17–34. doi: 10.1002/wrna.1139. [DOI] [PubMed] [Google Scholar]
- 10.Strom A.R., Emelyanov A.V., Karpen G.H. Phase separation drives heterochromatin domain formation. Nature. 2017;547:241–245. doi: 10.1038/nature22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larson A.G., Elnatan D., Narlikar G.J. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature. 2017;547:236–240. doi: 10.1038/nature22822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Misteli T. Concepts in nuclear architecture. Bioessays. 2005;27:477–487. doi: 10.1002/bies.20226. [DOI] [PubMed] [Google Scholar]
- 13.Venkatesh S., Workman J.L. Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 2015;16:178–189. doi: 10.1038/nrm3941. [DOI] [PubMed] [Google Scholar]
- 14.Fyodorov D.V., Zhou B.R., Bai Y. Emerging roles of linker histones in regulating chromatin structure and function. Nat. Rev. Mol. Cell Biol. 2018;19:192–206. doi: 10.1038/nrm.2017.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klemm S.L., Shipony Z., Greenleaf W.J. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 2019;20:207–220. doi: 10.1038/s41576-018-0089-8. [DOI] [PubMed] [Google Scholar]
- 16.Luger K., Mäder A.W., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 17.Tremethick D.J. Higher-order structures of chromatin: the elusive 30 nm fiber. Cell. 2007;128:651–654. doi: 10.1016/j.cell.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 18.Ou H.D., Phan S., O’Shea C.C. ChromEMT: Visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science. 2017;357 doi: 10.1126/science.aag0025. eaag0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lever M.A., Th’ng J.P., Hendzel M.J. Rapid exchange of histone H1.1 on chromatin in living human cells. Nature. 2000;408:873–876. doi: 10.1038/35048603. [DOI] [PubMed] [Google Scholar]
- 20.Misteli T., Gunjan A., Brown D.T. Dynamic binding of histone H1 to chromatin in living cells. Nature. 2000;408:877–881. doi: 10.1038/35048610. [DOI] [PubMed] [Google Scholar]
- 21.Contreras A., Hale T.K., Herrera R.E. The dynamic mobility of histone H1 is regulated by cyclin/CDK phosphorylation. Mol. Cell. Biol. 2003;23:8626–8636. doi: 10.1128/MCB.23.23.8626-8636.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernas T., Brutkowski W., Dobrucki J. Spatial heterogeneity of dynamics of H1 linker histone. Eur. Biophys. J. 2014;43:287–300. doi: 10.1007/s00249-014-0962-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Izzo A., Schneider R. The role of linker histone H1 modifications in the regulation of gene expression and chromatin dynamics. Biochim. Biophys. Acta. 2016;1859:486–495. doi: 10.1016/j.bbagrm.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 24.Wolffe A.P. Histone H1. Int. J. Biochem. Cell Biol. 1997;29:1463–1466. doi: 10.1016/s1357-2725(97)00026-5. [DOI] [PubMed] [Google Scholar]
- 25.Fan Y., Nikitina T., Skoultchi A.I. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 26.McBryant S.J., Lu X., Hansen J.C. Multifunctionality of the linker histones: an emerging role for protein-protein interactions. Cell Res. 2010;20:519–528. doi: 10.1038/cr.2010.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andreyeva E.N., Bernardo T.J., Fyodorov D.V. Regulatory functions and chromatin loading dynamics of linker histone H1 during endoreplication in Drosophila. Genes Dev. 2017;31:603–616. doi: 10.1101/gad.295717.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Izzo A., Kamieniarz-Gdula K., Schneider R. The genomic landscape of the somatic linker histone subtypes H1.1 to H1.5 in human cells. Cell Rep. 2013;3:2142–2154. doi: 10.1016/j.celrep.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 29.Nalabothula N., McVicker G., Fondufe-Mittendorf Y.N. The chromatin architectural proteins HMGD1 and H1 bind reciprocally and have opposite effects on chromatin structure and gene regulation. BMC Genomics. 2014;15:92–106. doi: 10.1186/1471-2164-15-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Millán-Ariño L., Islam A.B., Jordan A. Mapping of six somatic linker histone H1 variants in human breast cancer cells uncovers specific features of H1.2. Nucleic Acids Res. 2014;42:4474–4493. doi: 10.1093/nar/gku079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nielsen A.L., Oulad-Abdelghani M., Losson R. Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol. Cell. 2001;7:729–739. doi: 10.1016/s1097-2765(01)00218-0. [DOI] [PubMed] [Google Scholar]
- 32.Daujat S., Zeissler U., Schneider R. HP1 binds specifically to Lys26-methylated histone H1.4, whereas simultaneous Ser27 phosphorylation blocks HP1 binding. J. Biol. Chem. 2005;280:38090–38095. doi: 10.1074/jbc.C500229200. [DOI] [PubMed] [Google Scholar]
- 33.Iwasaki Y.W., Murano K., Saito K. Piwi modulates chromatin accessibility by regulating multiple factors including histone H1 to repress transposons. Mol. Cell. 2016;63:408–419. doi: 10.1016/j.molcel.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 34.Shakya A., King J.T. Non-Fickian molecular transport in protein-DNA droplets. ACS Macro Lett. 2018;7:1220–1225. doi: 10.1021/acsmacrolett.8b00565. [DOI] [PubMed] [Google Scholar]
- 35.Turner A.L., Watson M., Stott K. Highly disordered histone H1-DNA model complexes and their condensates. Proc. Natl. Acad. Sci. USA. 2018;115:11964–11969. doi: 10.1073/pnas.1805943115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brangwynne C.P., Mitchison T.J., Hyman A.A. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. USA. 2011;108:4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shakya A., King J.T. DNA local-flexibility-dependent assembly of phase-separated liquid droplets. Biophys. J. 2018;115:1840–1847. doi: 10.1016/j.bpj.2018.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patel A., Malinovska L., Hyman A.A. ATP as a biological hydrotrope. Science. 2017;356:753–756. doi: 10.1126/science.aaf6846. [DOI] [PubMed] [Google Scholar]
- 40.Zhou H.X., Nguemaha V., Qin S. Why do disordered and structured proteins behave differently in phase separation? Trends Biochem. Sci. 2018;43:499–516. doi: 10.1016/j.tibs.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doi M., Edwards S.F. Clarendon Press; Oxford, UK: 1988. The Theory of Polymer Dynamics. [Google Scholar]
- 42.Banani S.F., Rice A.M., Rosen M.K. Compositional control of phase-separated cellular bodies. Cell. 2016;166:651–663. doi: 10.1016/j.cell.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin Y.H., Forman-Kay J.D., Chan H.S. Sequence-specific polyampholyte phase separation in membraneless organelles. Phys. Rev. Lett. 2016;117:178101. doi: 10.1103/PhysRevLett.117.178101. [DOI] [PubMed] [Google Scholar]
- 44.Chang L.W., Lytle T.K., Perry S.L. Sequence and entropy-based control of complex coacervates. Nat. Commun. 2017;8:1273. doi: 10.1038/s41467-017-01249-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rumyantsev A.M., Jackson N.E., de Pablo J.J. Controlling complex coacervation via random polyelectrolyte sequences. ACS Macro Lett. 2019;8:1296–1302. doi: 10.1021/acsmacrolett.9b00494. [DOI] [PubMed] [Google Scholar]
- 46.Lindsey G.G., Orgeig S., Maeder D.L. Extended C-terminal tail of wheat histone H2A interacts with DNA of the “linker” region. J. Mol. Biol. 1991;218:805–813. doi: 10.1016/0022-2836(91)90268-b. [DOI] [PubMed] [Google Scholar]
- 47.Usachenko S.I., Bavykin S.G., Bradbury E.M. Rearrangement of the histone H2A C-terminal domain in the nucleosome. Proc. Natl. Acad. Sci. USA. 1994;91:6845–6849. doi: 10.1073/pnas.91.15.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee K.M., Hayes J.J. Linker DNA and H1-dependent reorganization of histone-DNA interactions within the nucleosome. Biochemistry. 1998;37:8622–8628. doi: 10.1021/bi980499y. [DOI] [PubMed] [Google Scholar]
- 49.Vogler C., Huber C., Schneider R. Histone H2A C-terminus regulates chromatin dynamics, remodeling, and histone H1 binding. PLoS Genet. 2010;6:e1001234. doi: 10.1371/journal.pgen.1001234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Z., Kono H. Distinct roles of histone H3 and H2A tails in nucleosome stability. Sci. Rep. 2016;6:31437. doi: 10.1038/srep31437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bonner W.M., Stedman J.D. Histone 1 is proximal to histone 2A and to A24. Proc. Natl. Acad. Sci. USA. 1979;76:2190–2194. doi: 10.1073/pnas.76.5.2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boulikas T., Wiseman J.M., Garrard W.T. Points of contact between histone H1 and the histone octamer. Proc. Natl. Acad. Sci. USA. 1980;77:127–131. doi: 10.1073/pnas.77.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou B.R., Feng H., Bai Y. Structural insights into the histone H1-nucleosome complex. Proc. Natl. Acad. Sci. USA. 2013;110:19390–19395. doi: 10.1073/pnas.1314905110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Downs J.A., Lowndes N.F., Jackson S.P. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature. 2000;408:1001–1004. doi: 10.1038/35050000. [DOI] [PubMed] [Google Scholar]
- 55.Downs J.A., Nussenzweig M.C., Nussenzweig A. Chromatin dynamics and the preservation of genetic information. Nature. 2007;447:951–958. doi: 10.1038/nature05980. [DOI] [PubMed] [Google Scholar]
- 56.Corujo D., Buschbeck M. Post-translational modifications of H2A histone variants and their role in cancer. Cancers (Basel) 2018;10:1–25. doi: 10.3390/cancers10030059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rape M. Ubiquitylation at the crossroads of development and disease. Nat. Rev. Mol. Cell Biol. 2018;19:59–70. doi: 10.1038/nrm.2017.83. [DOI] [PubMed] [Google Scholar]
- 58.Nott T.J., Craggs T.D., Baldwin A.J. Membraneless organelles can melt nucleic acid duplexes and act as biomolecular filters. Nat. Chem. 2016;8:569–575. doi: 10.1038/nchem.2519. [DOI] [PubMed] [Google Scholar]
- 59.Jacobs S.A., Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 60.Mészáros B., Erdős G., Pancsa R. PhaSePro: the database of proteins driving liquid-liquid phase separation. Nucleic Acids Res. 2019:gkz848. doi: 10.1093/nar/gkz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Traut T.W. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 62.Imamura H., Nhat K.P., Noji H. Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proc. Natl. Acad. Sci. USA. 2009;106:15651–15656. doi: 10.1073/pnas.0904764106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Y., Li Z., Zhu W.G. Histone H1 acetylation at lysine 85 regulates chromatin condensation and genome stability upon DNA damage. Nucleic Acids Res. 2018;46:7716–7730. doi: 10.1093/nar/gky568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hale T.K., Contreras A., Herrera R.E. Phosphorylation of the linker histone H1 by CDK regulates its binding to HP1alpha. Mol. Cell. 2006;22:693–699. doi: 10.1016/j.molcel.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 65.Saito M., Hess D., Matthias P. Acetylation of intrinsically disordered regions regulates phase separation. Nat. Chem. Biol. 2019;15:51–61. doi: 10.1038/s41589-018-0180-7. [DOI] [PubMed] [Google Scholar]
- 66.Monahan Z., Ryan V.H., Fawzi N.L. Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 2017;36:2951–2967. doi: 10.15252/embj.201696394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li G., Levitus M., Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nat. Struct. Mol. Biol. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- 68.Endoh M., Endo T.A., Koseki H. Histone H2A mono-ubiquitination is a crucial step to mediate PRC1-dependent repression of developmental genes to maintain ES cell identity. PLoS Genet. 2012;8:e1002774. doi: 10.1371/journal.pgen.1002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jason L.J., Finn R.M., Ausió J. Histone H2A ubiquitination does not preclude histone H1 binding, but it facilitates its association with the nucleosome. J. Biol. Chem. 2005;280:4975–4982. doi: 10.1074/jbc.M410203200. [DOI] [PubMed] [Google Scholar]
- 70.Zhu P., Zhou W., Rosenfeld M.G. A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol. Cell. 2007;27:609–621. doi: 10.1016/j.molcel.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.