Abstract
Of the different types of lung cancer, lung squamous cell cancer (LUSC) has the second highest rates of morbidity and mortality, which have been increasing in recent years. Epigenetic abnormalities may serve as potential biomarkers and diagnostic and/or therapeutic targets, which may help to monitor and improve the prognosis of patients with cancer. In the present study, data were obtained from The Cancer Genome Atlas database and survival and joint survival analyses were conducted using the R MethylMix package. Peptidase, mitochondrial processing a subunit pseudogene 1 (PMPCAP1), sosondowah ankyrin repeat domain family member C (SOWAHC) and zinc finger protein (ZNF) 454 were identified as independent prognosis-related hub methylation-driven genes (MDGs). Of these three genes, PMPCAP1 and SOWAHC, characterized by hypomethylation and high expression levels, were associated with poor prognosis in patients with LUSC, whilst ZNF454 was associated with an improved prognosis. In addition, pathway enrichment analysis suggested that PMPCAP1, SOWAHC and ZNF454 were primarily involved in gene expression or transcription pathways. Furthermore, 5, 1 and 10 key methylation sites of PMPCAP1, SOWAHC and ZNF454, respectively, were confirmed to be significantly relevant to gene expression, establishing a basis for further investigation into the mechanisms and more precise targets of these 3 genes. In conclusion, the MDGs PMPCAP1, SOWAHC and ZNF454 may be potential prognostic biomarkers of LUSC for guiding diagnosis and therapy options, as well as providing a theoretical basis for further investigation.
Keywords: lung squamous cell carcinoma, methylation-driven genes, The Cancer Genome Atlas, MethylMix, biomarker, prognosis
Introduction
The latest data released by the World Health Organization revealed that lung cancer has the highest global morbidity and mortality rates of all malignant tumors, and this trend is increasing yearly (1). Based on biological characteristics, treatment and prognosis, lung cancer is classified as non-small-cell lung cancer (NSCLC) or small-cell lung cancer (SCLC). NSCLC accounts for ~85% of all lung cancer cases, of which lung squamous cell cancer (LUSC) accounts for 20–30%, and has a five-year survival rate of <15% (2,3). With the recent rapid development of gene detection methods and targeted drugs, the overall survival time (OS) of patients with NSCLC has significantly improved (4). However, not all patients benefit from targeted therapy; in LUSC, the frequency of gene mutations sensitive to targeted drugs is relatively low, and this is accompanied by poor efficacy and the occurrence of drug resistance. In addition, the prognosis and OS of patients with early-stage lung cancer are markedly more favorable compared with those of patients at an advanced disease stage. Therefore, the identification of novel biomarkers and therapeutic targets is important for improving early diagnosis, treatment strategies and prognostic detection in patients with LUSC.
At present, the pathogenesis and progressive mechanisms of LUSC remain unclear, though the two most important mechanisms of tumorigenesis are gene mutations and epigenetic alterations (5,6). Relatively few gene mutation sites exist, particularly for early-stage patients; owing to severe fragmentization of tumor gene fragments in the blood, gene mutations are not suitable for the monitoring and diagnosis of early-stage cancer, and epigenetic changes provide a more suitable target (7,8). DNA methylation is easily detectable, and therefore the most studied, epigenetic modification, mediating the occurrence and development of cancer by regulating gene expression (8–10). It has also been indicated that DNA methylation may occur prior to gene mutation, deeming it more suitable for the early detection of cancer. Studies examining methylation and tumors have recently attracted increased attention, including a series of studies concerning targeted epigenetic therapy approaches for acute myeloid leukemia (11). Even in solid tumors, methylated or epigenetic signatures have become an area of increasing interest, in such malignancies as breast cancer (12), esophageal carcinoma (13,14), epithelial ovarian (15) and liver cancer (16). These studies indicated that the methylation of some specific genes may affect gene expression, and is closely associated with the diagnosis and prognosis of some types of cancer. Therefore, the identification of abnormal gene methylation signatures may provide a basis for the early diagnosis, prognosis and targeted therapy for patients with tumors.
In the context of the era of big data, bioinformatics analysis serves an important role in the comprehensive research of carcinomas, utilizing high-throughput databases such as The Cancer Genome Atlas (TCGA). The present study utilized data from TCGA, in which the genetic information profiles and corresponding clinical data of multiple cancer types can be effectively extracted for analysis, bridging the gap between molecular biology research and clinical application (17,18). Methylation-driven genes (MDGs) were the primary focus of the present study, defined as genes of differentially methylated states and significant predictive transcriptional function; as such, MDGs were identified by dissecting and integrating the correlation between methylation state and the level of gene expression. Previous studies have confirmed that MDGs are more comprehensive and representative tumor biomarkers compared with differentially methylated genes (DMGs) (14). In the present study, methylation, gene expression and patient survival information were extracted from TCGA, and the MethylMix algorithm and survival analysis were used to identify hub MDGs and their prognostic signatures, with a view to providing a basis for individualized precision treatment in patients with LUSC.
Materials and methods
Data acquisition and preprocessing
Firstly, the DNA methylation and gene expression quantification data of patients with LUSC were downloaded from TCGA (https://www.cancer.gov/tcga/), along with the corresponding clinical information, which included details of prognostic or survival analysis. According to the TCGA data number, the data were divided into two groups, LUSC samples and normal samples. In these data, the normal samples were tissues adjacent to the tumor. The Illumina Human Methylation 450 k platform was used to transform and normalize the initial DNA methylation data, which were expressed as β-values (range, 0–1) corresponding with low to high methylation states (19). Gene expression quantification data were obtained in RNA-Seq format.
Screening of differentially expressed genes (DEGs), DMGs and MDGs in LUSC
The R edge package (http://bioconductor.org/packages/edgeR/) was used to identify and analyze DEGs by comparing gene expression quantification data between normal and cancerous specimens, with a fold change (FC)=5 and adjusted P-value (padj)=0.01 as the threshold. The limma package (http://bioinf.wehi.edu.au/limma/) was used to compare the methylation states of normal and cancerous specimens, and DMGs were screened out using a false discovery rate of 0.01 and log2FC=1.
Next, MDGs were identified using the R MethylMix package (http://bioconductor.org/packages/3.9/bioc/html/MethylMix.html). MethylMix is a new algorithm developed by Gevaert et al (20,21), which uses univariate b mixture modeling to determine the methylation state of genes in cancer samples, and the Wilcoxon rank test to categorize these into hyper- and hypomethyled groups compared with the methylation state in normal tissues. It may also be used to determine the correlation between DNA methylation state and gene expression level, the absolute value of correlation coefficient (|Cor|) representing the degree of correlation. In the present study, the screening conditions of MDGs with significant inverse correlation were set as padj<0.05, log2FC=0 and Cor<-0.5.
Pathway analysis of MDGs in LUSC
To further investigate the dominating functional pathways of MDGs in LUSC, ConsensusPathDB (http://cpdb.molgen.mpg.de/) and Cytoscape.js was utilized to analyze and visualize the genetic interaction of high-throughput expression data (22,23). ConsensusPathDB is currently the most comprehensive database of functional interaction networks, integrating the functional aspects of genes, proteins, complexes and metabolites. Cytoscape.js is a graph library written in JavaScript that is used as visualization software for graph analysis (24). P<0.05 was set as the cut-off for minimum overlap criterion.
Survival and joint survival analysis of MDGs in LUSC
It is important to note that not all MDGs are significantly associated with cancer prognosis. In order to improve the understanding of the association between MDGs and patient survival, the MDGs independently associated with prognosis, classified as hub MDGs, were identified. Firstly, using the R package survival, Kaplan-Meier survival analysis and the log-rank test were conducted to determine the association between the methylation state of MDGs and the survival of patients with LUSC. P<0.05 was considered to indicate a statistically significant correlation.
Owing to the complexity of tumor tissue regulation by the combination of multiple factors, it was necessary and important to conduct joint survival analysis between the degree of methylation, the corresponding levels of MDG expression and patient survival. A joint survival curve was generated using the survival package, with P<0.05 as the cutoff value. It should be noted that the MDGs screened out using MethylMix were all characterized by a significant inverse correlation (Cor <-0.5). Therefore, of the joint survival analysis, there were only two cases to determine survival: Hypermethylation and low expression; and hypomethylation and high expression.
Finally, using the first two steps the common genes were identified. PMPCAP1, SOWAHC, ZNF454 AND LINC00668 were statistically significant in the survival analysis, while PMPCAP1, SOWAHC, ZNF454 and ADH7 were statistically significant in the joint survival analysis. So, PMPCAP1, SOWAHC and ZNF454 were taken as the hub MDGs.
Correlation analysis between methylation sites and the expression of hub MDGs
Finally, to further examine the internal mechanisms and more precise targets of the 3 identified hub MDGs, data corresponding to the initial methylation sites of these genes were downloaded. The present study focused on the correlation between the methylation of abnormal methylation sites and the corresponding gene expression of hub MDGs. Both P<0.05 and |Cor|>0.5 used as the cut-offs for the identification of key methylation sites.
Results
Identification of MDGs in LUSC
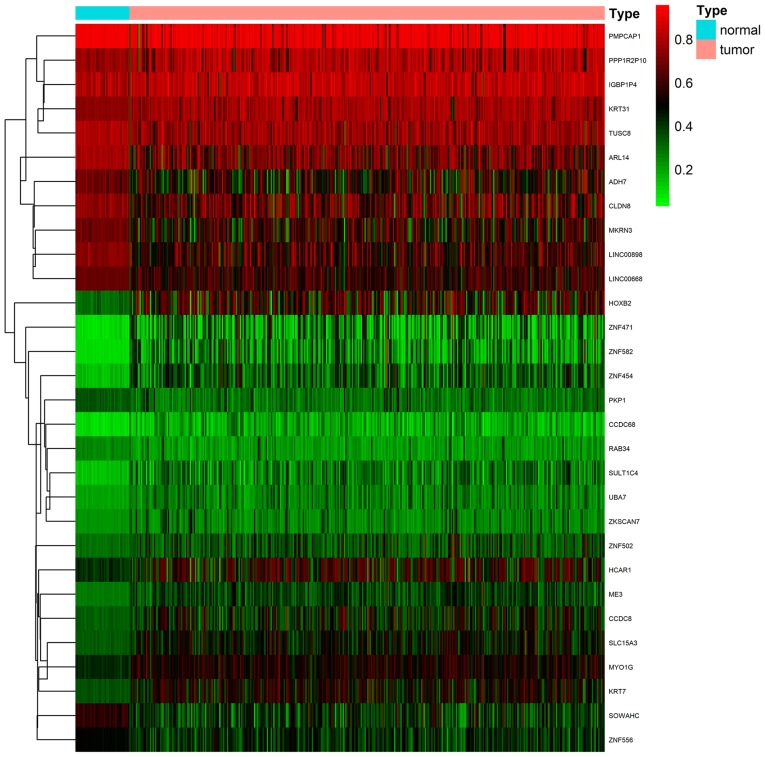
Firstly, a total of 370 LUSC samples and 42 normal samples (from 372 cases) with DNA methylation data were downloaded from TCGA database; 502 LUSC samples and 49 normal samples (from 501 cases) with gene expression quantification data were also downloaded. Additionally, 366 of the patients with LUSC also possessed clinical data for prognostic and survival analysis. Secondly, the R edge and limma packages were used to compare data between cancerous and normal samples, respectively, and to screen out 994 DEGs and 356 DMGs. Finally, the MethylMix algorithm (padj<0.05, log2FC=0 and Cor <-0.5) was used to identify 30 MDGs with strong inverse correlation between DNA methylation state and gene expression (Fig. 1; Table I). The methylation models are presented in Figs. 2 and S1, and the correlation plots are demonstrated in Figs. 3 and S2.
Figure 1.
Heat map of 30 abnormal MDGs in LUSC. The blue and pink colors represent normal and tumor samples, respectively. The scale from green to red represents a trend from low to high methylation state, respectively, with a β-value ranging from 0–1. MDG, methylation-driven gene; LUSC, lung squamous cell cancer.
Table I.
Output results of 30 MDGs using the MethylMix algorithm.
| Gene | Normal meana | Tumor meana | logFC | P-value | padj | Cor | P-value of Cor |
|---|---|---|---|---|---|---|---|
| ZNF582 | 0.102781 | 0.275603 | 1.423022 | 5.87×10−22 | 1.48×10−19 | −0.564313 | 1.69×10−32 |
| ME3 | 0.277294 | 0.382910 | 0.465587 | 7.11×10−22 | 1.79×10−19 | −0.553976 | 3.80×10−31 |
| ZNF454 | 0.157826 | 0.346334 | 1.133824 | 1.64×10−21 | 4.12×10−19 | −0.519207 | 6.28×10−27 |
| SLC15A3 | 0.333165 | 0.467888 | 0.489927 | 1.74×10−20 | 4.39×10−18 | −0.556532 | 1.78×10−31 |
| MYO1G | 0.426675 | 0.533590 | 0.322595 | 3.87×10−20 | 9.76×10−18 | −0.551080 | 8.92×10−31 |
| SOWAHC | 0.569583 | 0.405071 | −0.491730 | 1.12×10−19 | 2.82×10−17 | −0.522119 | 2.91×10−27 |
| CCDC68 | 0.103194 | 0.199258 | 0.949279 | 3.02×10−19 | 7.61×10−17 | −0.503257 | 3.77×10−25 |
| SULT1C4 | 0.152713 | 0.285577 | 0.903061 | 9.50×10−19 | 2.39×10−16 | −0.500977 | 6.65×10−25 |
| KRT7 | 0.350295 | 0.504885 | 0.527384 | 1.37×10−17 | 3.44×10−15 | −0.574797 | 6.43×10−34 |
| UBA7 | 0.202113 | 0.276516 | 0.452201 | 5.21×10−16 | 1.31×10−13 | −0.540067 | 2.12×10−29 |
| HOXB2 | 0.305355 | 0.536591 | 0.813338 | 3.38×10−14 | 8.52×10−12 | −0.611253 | 2.80×10−29 |
| LINC00898 | 0.734546 | 0.587677 | −0.321829 | 1.89×10−13 | 4.76×10−11 | −0.557094 | 1.50×10−31 |
| LINC00668 | 0.676192 | 0.576143 | −0.231005 | 2.25×10−13 | 5.66×10−11 | −0.584201 | 3.09×10−35 |
| ARL14 | 0.790435 | 0.661123 | −0.257730 | 3.39×10−12 | 8.53×10−10 | −0.505203 | 2.31×10−25 |
| MKRN3 | 0.691598 | 0.542513 | −0.350275 | 4.42×10−12 | 1.11×10−9 | −0.704902 | 7.51×10−57 |
| CCDC8 | 0.327849 | 0.448283 | 0.451379 | 5.46×10−12 | 1.38×10−9 | −0.579013 | 1.67×10−34 |
| ZNF471 | 0.101118 | 0.267232 | 1.402048 | 5.90×10−12 | 1.49×10−9 | −0.551843 | 7.13×10−31 |
| PKP1 | 0.341899 | 0.298902 | −0.193899 | 6.88×10−12 | 1.73×10−9 | −0.536918 | 5.14×10−29 |
| ADH7 | 0.680263 | 0.529756 | −0.360765 | 6.46×10−11 | 1.63×10−8 | −0.572790 | 1.21×10−33 |
| ZNF556 | 0.487939 | 0.414088 | −0.236764 | 2.67×10−10 | 6.74×10−8 | −0.638623 | 8.72×10−44 |
| KRT31 | 0.742604 | 0.772890 | 0.057671 | 6.79×10−10 | 1.71×10−7 | −0.542682 | 1.01×10−29 |
| RAB34 | 0.253476 | 0.227666 | −0.154930 | 5.53×10−9 | 1.39×10−6 | −0.523058 | 2.26×10−27 |
| CLDN8 | 0.746796 | 0.626198 | −0.254095 | 8.60×10−8 | 2.17×10−5 | −0.513212 | 3.00×10−26 |
| ZNF502 | 0.294033 | 0.369115 | 0.328093 | 3.42×10−7 | 8.61×10−5 | −0.668828 | 2.54×10−49 |
| PPP1R2P10 | 0.764206 | 0.790535 | 0.048868 | 7.83×10−7 | 1.97×10−4 | −0.568402 | 4.79×10−33 |
| HCAR1 | 0.408911 | 0.525195 | 0.361067 | 1.54×10−6 | 3.89×10−4 | −0.632664 | 9.13×10−43 |
| TUSC8 | 0.805850 | 0.753565 | −0.096780 | 2.95×10−6 | 7.43×10−4 | −0.515211 | 1.79×10−26 |
| PMPCAP1 | 0.925864 | 0.889418 | −0.057939 | 5.71×10−6 | 1.44×10−3 | −0.628458 | 4.65×10−42 |
| ZKSCAN7 | 0.229582 | 0.273455 | 0.252291 | 8.69×10−6 | 2.19×10−3 | −0.515312 | 1.74×10−26 |
| IGBP1P4 | 0.800055 | 0.826529 | 0.046965 | 1.59×10−4 | 4.00×10−2 | −0.556656 | 1.72×10−31 |
Normal mean and tumor mean values represent the mean value of the quantified methylation state data of each gene in normal and tumor specimens, respectively; FC, fold change; padj, adjusted P-value; Cor, correlation coefficient. The Cor value represents the degree of correlation between DNA methylation and gene expression of genes. The 30 MDGs were identified with the screening conditions of padj<0.05, log2FC=0 and Cor<-0.5.
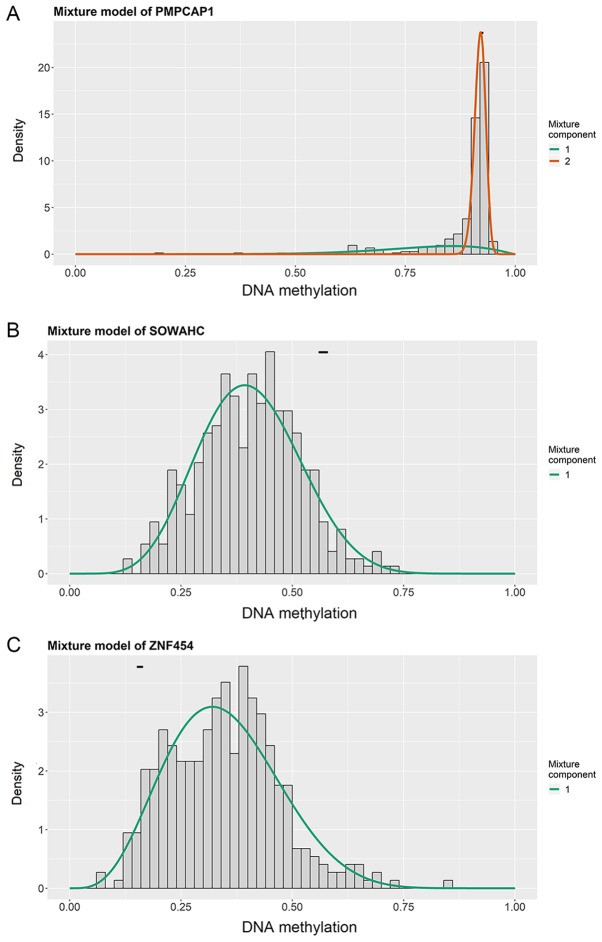
Figure 2.
Mixture models of PMPCAP1, SOWAHC and ZNF454 genes in LUSC. Mixture models of (A) PMPCAP1, (B) SOWAHC and (C) ZNF454. The density of tumor samples with different methylation states (range, 0–1) is represented by the histogram and curves, while the methylation state in the normal samples is represented by the horizontal short black bar. PMPCAP1, peptidase, mitochondrial processing a subunit pseudogene 1; SOWAHC, sosondowah ankyrin repeat domain family member C; ZNF454, zinc finger protein 454; LUSC, lung squamous cell cancer.
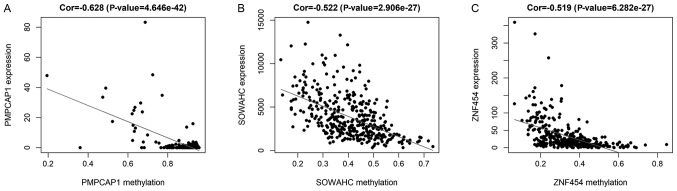
Figure 3.
Correlation between DNA methylation and gene expression for 3 hub MDGs in LUSC. Correlation between the methylation and expression of (A) PMPCAP1, (B) SOWAHC and (C) ZNF454. MDG, methylation-driven gene; LUSC, lung squamous cell cancer; PMPCAP1, peptidase, mitochondrial processing a subunit pseudogene 1; SOWAHC, sosondowah ankyrin repeat domain family member C; ZNF454, zinc finger protein 454; Cor, correlation coefficient.
Pathway analysis of MDGs in LUSC
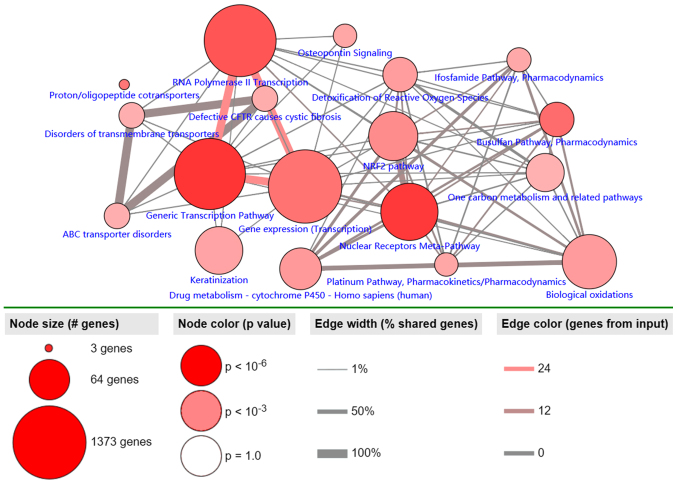
Pathway analysis of the 30 identified MDGs was conducted using the ConsensusPathDB database (Fig. 4). The results revealed 3 primary pathways: ‘generic transcription’, ‘RNA polymerase II transcription’ and ‘gene expression (transcription)’. The largest numbers of genes were associated with these 3 pathways, and ~100% of all shared genes, and to the most genes from input (P<0.001).
Figure 4.
Significantly enriched pathways of MDGs in LUSC. The node size corresponds to the number of genes, and the node color represents the P-value. This figure only includes the pathways with P<0.001. The edge width represents the percentage of shared genes, and the edge color represents the genes from input. MDG, methylation-driven gene; LUSC, lung squamous cell cancer.
Recognition of hub MDGs in LUSC
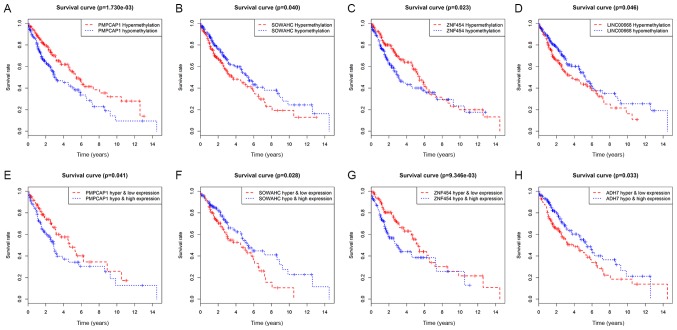
Initially, survival analysis between hyper- and hypomethylated MDGs revealed 4 genes with statistical importance: Peptidase, mitochondrial processing a subunit pseudogene 1 (PMPCAP; P=0.00173), sosondowah ankyrin repeat domain family member C (SOWAHC; P=0.04), zinc finger protein (ZNF) 454 (P=0.023) and LINC00668 (P=0.046; Figs. 5A-D and S3). Joint survival analysis was then conducted between the degree of methylation and the corresponding gene expression level of MDGs and survival, and PMPCAP1 (P=0.041), SOWAHC (P=0.028), ZNF454 (P=0.00935) and ADH7 (P=0.033) were identified as statistically significant (Figs. 5E-H and S4). By taking the common genes of the first two steps, 3 hub MDGs (PMPCAP1, SOWAHC and ZNF454) were deemed to be independently associated with prognosis in LUSC. Of these 3 hub MDGs, PMPCAP1 and SOWAHC, characterized by hypomethylation and high expression levels, were associated with poor prognosis in patients with LUSC, whilst ZNF454, characterized by hypermethylation and low expression level, was associated with an improved prognosis.
Figure 5.
Survival analysis curves of MDGs with statistical significance in LUSC. Kaplan-Meier survival curves, where the x-axis represents the overall survival time and the y-axis represents the survival rate. (A-D) Survival analysis comparing overall survival and the methylation state of (A) PMPCAP1, (B) SOWAHC, (C) ZNF454 and (D) LINC00668, respectively. (E-H) Joint survival analysis comparing overall survival between hypermethylation/low expression and hypomethylation/high expression of (E) PMPCAP1, (F) SOWAHC, (G) ZNF454 and (H) ADH7, respectively. P<0.05. MDGs, methylation-driven genes; LUSC, lung squamous cell cancer; PMPCAP1, peptidase, mitochondrial processing a subunit pseudogene 1; SOWAHC, sosondowah ankyrin repeat domain family member C; ZNF454, zinc finger protein 454.
Key methylation sites of hub MDGs in LUSC
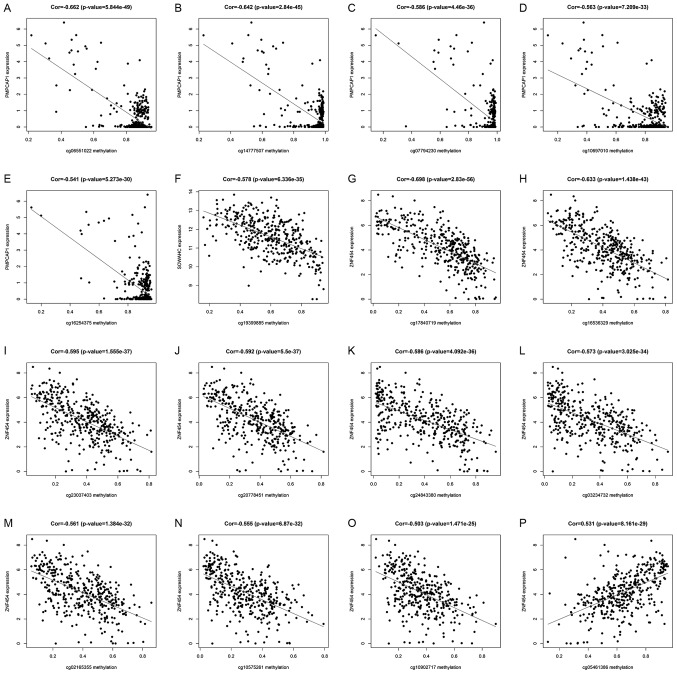
Using the associated R packages, key methylation sites statistically relevant to the expression of hub MDGs in LUSC were identified. The results of correlation analysis revealed 5 key methylation sites of the PMPCAP1 gene (cg06551022, cg14777507, cg07794230, cg10697010 and cg16254375), 1 key methylation site of the SOWAHC gene (cg19399885) and 10 key methylation sites of the ZNF454 gene (cg17840719, cg16536329, cg23037403, cg20778451, cg24843380, cg03234732, cg02165355, cg10575261, cg10902717 and cg05461386; Fig. 6 and Table II; P<0.001).
Figure 6.
Correlation between key methylation sites and expression of 3 hub MDGs. The x-axes of these graphs represent the site-specific methylation state, and the y-axis represents the corresponding gene expression. (A-E) Correlation graphs of the 5 key methylation sites in PMPCAP1: (A) cg06551022; (B) cg14777507; (C) cg07794230; (D) cg10697010; and (E) cg16254375. (F) Correlation diagram of the key methylation site in SOWAHC, cg19399885. (G-P) Correlation diagrams of the 10 key methylation sites in ZNF454: (G) cg17840719; (H) cg16536329; (I) cg23037403; (J) cg20778451; (K) cg24843380; (L) cg03234732; (M) cg02165355; (N) cg10575261; (O) cg10902717; and (P) cg05461386. MDG, methylation-driven gene; PMPCAP1, peptidase, mitochondrial processing a subunit pseudogene 1; SOWAHC, sosondowah ankyrin repeat domain family member C; ZNF454, zinc finger protein 454.
Table II.
Correlation between key methylation sites and expression of PMPCAP1, SOWAHC and ZNF454.
| Gene | Methylation site | Cor | P-value |
|---|---|---|---|
| PMPCAP1 | cg06551022 | −0.662 | 5.84×10−49 |
| cg14777507 | −0.642 | 2.84×10−45 | |
| cg07794230 | −0.586 | 4.46×10−36 | |
| cg10697010 | −0.563 | 7.21×10−33 | |
| cg16254375 | −0.541 | 5.27×10−30 | |
| SOWAHC | cg19399885 | −0.578 | 6.34×10−35 |
| ZNF454 | cg17840719 | −0.698 | 2.83×10−56 |
| cg16536329 | −0.633 | 1.44×10−43 | |
| cg23037403 | −0.595 | 1.56×10−37 | |
| cg20778451 | −0.592 | 5.50×10−37 | |
| cg24843380 | −0.586 | 4.09×10−36 | |
| cg03234732 | −0.573 | 3.03×10−34 | |
| cg02165355 | −0.561 | 1.38×10−32 | |
| cg10575261 | −0.555 | 6.87×10−32 | |
| cg10902717 | −0.503 | 1.47×10−25 | |
| cg05461386 | 0.531 | 8.16×10−29 |
Only the key methylation sites of PMPCAP1, SOWAHC and ZNF454 with both P<0.05 and |Cor|>0.5 are listed. Cor, correlation coefficient; |Cor|, the absolute value of correlation coefficient. PMPCAP1, peptidase, mitochondrial processing a subunit pseudogene 1; SOWAHC, sosondowah ankyrin repeat domain family member C; ZNF454: Zinc finger protein 454.
Discussion
The morbidity and mortality rates of LUSC are the second highest of all the pathological types of lung carcinoma, with poor prognosis depending on the biological characteristics of the specific subtype. Furthermore, >70% of patients with LUSC present with late-stage disease at the diagnosis, for which treatment options are limited, and clinical outcomes are far poorer compared with those in patients with early-stage disease (25). In order to decrease the mortality rate of LUSC, novel approaches for early diagnosis and treatment are required, as well as the identification of novel predictive biomarkers and therapeutic targets.
Compared with the rapid development of precise gene-targeted therapy in lung adenocarcinoma, there are a limited number of effective and distinctive targets to improve the prognosis of patients with LUSC. In addition to studies into gene mutations, the association between epigenetic changes (particularly DNA methylation) and LUSC has also attracted great attention. Epigenetic studies have revealed that genome-scale epigenetic modifications, including DNA methylation, histone modification and microRNA interference, are involved in the pathogenic mechanisms of malignancy (26). Due to its stability and ease of detection, accumulating evidence has demonstrated that aberrant gene methylation may serve as an effective, non-invasive diagnostic biomarker and therapeutic in carcinoma (27,28). Reports have indicated that the abnormal methylation of certain genes, such as ZNF671 (29), ADAMTS1 (30) and CD36 (31), may alter their functions, including the regulation of the cell cycle and signal transduction pathways, as well as transcriptional inhibition. Kiyozumi et al (32) demonstrated that indoleamine 2,3-dioygenase 1 promoter hypomethylation is associated with poor prognosis in esophageal cancer. Therefore, the accurate detection of methylated genes is likely to improve the clinical management of LUSC.
Studies have previously identified DMGs in LUSC (33–35); however, not all genes can be transcriptionally expressed. Therefore, as DMGs are not able to precisely demonstrate the relevance between genetic methylation and oncogenesis, MDGs are considered to be more representative (14,36). In the present study, high-throughput bioinformatics tools were used to identify and analyze MDGs associated with the prognosis of LUSC. Data extracted from TCGA were analyzed using packages from R, including edge, limma and MethylMix, and 30 LUSC-associated MDGs were derived. To improve the understanding of the functional pathways involving these MDGs, significant pathways were visualized using the ConsensusPathDB and Cytoscape.js library in the present study. The results identified 3 primary pathways: ‘generic transcription’; ‘RNA polymerase II transcription’; and ‘gene expression (transcription)’, which were affected by MDG interactions at a functional level. In other words, differential methylation of specific MDGs is able affect their expression and transcription. Furthermore, considering that not all MDGs are significantly associated with cancer prognosis, Kaplan-Meier survival and joint survival analyses were conducted using the R survival package, yielding 5 candidate prognosis-associated MDGs: PMPCAP1; SOWAHC; ZNF454; LINC00668; and ADH7 (P<0.05). The common genes showing significance in survival and joint survival analyses were chosen as the 3 hub MDGs (PMPCAP1, SOWAHC and ZNF454), which were identified to function as potential independent prognosis-associated markers for LUSC. The hub MDGs were determined by analyzing the association between hyper- or hypomethylation and survival, but also by integrating the degree of methylation and the expression of MDGs with survival.
PMPCAP1, a pseudogene of PMPCA1, is located on chromosome 4q22.1. To the best of our knowledge, the function of PMPCAP1 has not been investigated thus far. However, a number of studies have suggested that the functions of certain pseudogenes differ from those of normal homologous genes, but that the expression of associated non-coding (nc)RNAs plays an important regulatory role in the development of certain diseases (37–39). For example, PTENP expression may generate ncRNAs that competitively inhibit the function of PTEN, a known tumor-suppressor gene, and therefore inhibit cancer cell proliferation (40). Pseudogenes may also affect oncogenesis through epigenetic changes. The present study indicated that PMPCAP1 was hypomethylated in LUSC compared with normal tissues, which was associated with high expression levels, and ultimately, poor prognosis (P=0.041). Therefore, it may be speculated that the hypomethylation of PMPCAP1 in cancer tissue (and the subsequent increase in RNA expression) is an indicator of poor clinical outcome in patients with LUSC, though further investigation is required to confirm this hypothesis.
SOWAHC, also known as ankyrin repeat domain (ANKRD) 57, is a protein-coding gene. The principle biological function of the ANKRD family is to mediate interactions between proteins (41). Takahashi et al (42) identified that ANKRD1 was overexpressed in EGFR-TKIs-resistant NSCLC with EGFR mutation, and that by inhibiting ANKRD1 expression, resistant cells were re-sensitized to afatinib and osimertinib. Lei et al (43) also demonstrated that ANKRD1 regulated apoptosis in ovarian cancer cells and functioned as a potential target to increase sensitivity to chemotherapy in ovarian cancer. In addition, the prognostic value of SOWAHC has been confirmed in bladder cancer (41); however, its value in LUSC has not been elucidated thus far, to the best of our knowledge. In the present study, the results of the correlation analysis indicated that the methylation of SOWAHC was negatively associated with its expression, commonly presenting as hypomethylation and high expression. Joint survival analysis revealed a significant association between the combined methylation and expression data and survival (P=0.028), suggesting that hypomethylation and high expression levels denoted improved prognosis in LUSC. Therefore, SOWAHC may be a potential biomarker of LUSC.
ZNF454 is a protein-coding gene, which expresses a protein measuring 522 amino acids. The ZNF454 protein is primarily involved in functional pathways associated with gene expression and transcription, namely ‘DNA binding’, ‘DNA-binding transcription factor activity by RNA polymerase II-specific’, ‘nucleic acid binding’ and ‘metal ion binding’. To the best of our knowledge, no published studies of ZNF454 were available until now, while a number of other members of the ZNF family have been investigated. Previous studies have suggested that, as the largest family of transcription factors in humans, ZNFs serve numerous important roles, and were recently confirmed as potential tumor suppressors (44). For example, through ZNF545 promoter methylation-associated deactivation, ZNF545 inhibited tumor proliferation in colorectal cancer via the PI3K/AKT and MAPK/ERK signaling pathways (45). In the present study, ZNF454 was generally hypermethylated and expressed to a low degree in LUSC, and was associated with favorable prognoses. Therefore, ZNF454 may be a potential tumor-suppressor gene that functions as a transcriptional regulator, with potential use as a prognostic indicator.
Previous studies have demonstrated that site-specific methylation, such as that at the promoter or enhancer, particularly of CpG sites, may notably affect gene expression (46,47). In the present study, the specific methylation sites of 3 hub MDGs that were associated with gene expression were identified. The results indicated that the expression of PMPCAP1 is negatively associated with the methylation of 5 sites (cg06551022, cg14777507, cg07794230, cg10697010 and cg16254375). A single methylation site was associated with SOWAHC expression (cg19399885) and the expression of ZNF454 was associated with 10 methylation sites, including 9 negatively related sites (cg17840719, cg16536329, cg23037403, cg20778451, cg24843380, cg03234732, cg02165355, cg10575261 and cg10902717) and 1 positive site (cg05461386). Further studies on the effects of these methylation sites on gene expression are required, which may assist in identifying more precise diagnostic and therapeutic targets to improve the prognosis of patients with LUSC.
There were certain limitations to the present study: Firstly, due to the lack data from other databases, the results were not externally validated, which may have partially decreased reliability. Secondly, limited financial support prevented further mechanistic studies with lung cancer cell lines or human tissue samples, which is a potential future research prospect.
In conclusion, using the MethylMix algorithm, the present study identified 3 hub MDGs (PMPCAP1, SOWAHC and ZNF454) with independent prognostic values in LUSC. In patients with LUSC, PMPCAP1 and SOWAHC were hypomethylated and highly expressed, which was determined to be an indication of poor prognosis. By contrast, ZNF454 was hypermethylated and expressed to a low degree, which was associated with improved prognosis. In addition, specific sites of aberrant methylation were investigated to identify more precise targets for clinical application. Although the results require further experimental validation, the present study provides diagnostic, therapeutic and prognostic value for patients with LUSC, and may guide future clinical applications to some extent.
Supplementary Material
Acknowledgements
The results published here are in whole based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Glossary
Abbreviations
- NSCLC
non-small-cell lung cancer
- LUSC
lung squamous cell cancer
- TCGA
The Cancer Genome Atlas
- MDG
methylation-driven gene
- DEG
differentially expressed gene
- DMG
differentially methylated gene
- FC
fold change
- padj
adjusted P-value
- Cor
correlation coefficient
- |Cor|
absolute correlation coefficient value
- PMPCAP1
peptidase, mitochondrial processing a subunit pseudogene 1
- ZNF
zinc finger protein
- SOWAHC
sosondowah ankyrin repeat domain family member C
- ANKRD
ankyrin repeat domain
Funding
The present study was supported by the Key Technology Research and Development Program of Shandong Province (grant no. GG201710060039).
Availability of data and materials
The authors declare that the initial data are available from the TCGA database, and that all data generated and analyzed during the present study are included in this published article.
Authors' contributions
QZhu, JW and JL conceived and designed the study. JW and QZha performed the data analysis and generated the figures. FW and LF contributed analysis tools. QZhu drafted the manuscript. BS and CX also performed data analysis and critically revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Lim SL, Jia Z, Lu Y, Zhang H, Ng CT, Bay BH, Shen HM, Ong CN. Metabolic signatures of four major histological types of lung cancer cells. Metabolomics. 2018;14:118. doi: 10.1007/s11306-018-1417-x. [DOI] [PubMed] [Google Scholar]
- 3.Choi M, Kadara H, Zhang J, Parra ER, Rodriguez-Canales J, Gaffney SG, Zhao Z, Behrens C, Fujimoto J, Chow C, et al. Mutation profiles in early-stage lung squamous cell carcinoma with clinical follow-up and correlation with markers of immune function. Ann Oncol. 2017;28:83–89. doi: 10.1093/annonc/mdw437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong J, Li B, Lin D, Zhou Q, Huang D. Advances in targeted therapy and immunotherapy for non-small cell lung cancer based on accurate molecular typing. Front Pharmacol. 2019;10:230. doi: 10.3389/fphar.2019.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chakravarthi BV, Nepal S, Varambally S. Genomic and epigenomic alterations in cancer. Am J Pathol. 2016;186:1724–1735. doi: 10.1016/j.ajpath.2016.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mehta A, Dobersch S, Romero-Olmedo AJ, Barreto G. Epigenetics in lung cancer diagnosis and therapy. Cancer Metastasis Rev. 2015;34:229–241. doi: 10.1007/s10555-015-9563-3. [DOI] [PubMed] [Google Scholar]
- 7.Pfister SX, Ashworth A. Marked for death: Targeting epigenetic changes in cancer. Nat Rev Drug Discov. 2017;16:241–263. doi: 10.1038/nrd.2016.256. [DOI] [PubMed] [Google Scholar]
- 8.Baylin SB, Jones PA. Epigenetic determinants of cancer. Cold Spring Harb Perspect Biol. 2016;8:a019505. doi: 10.1101/cshperspect.a019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dawson MA, Kouzarides T. Cancer epigenetics: From mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 10.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 11.Wouters BJ, Delwel R. Epigenetics and approaches to targeted epigenetic therapy in acute myeloid leukemia. Blood. 2016;127:42–52. doi: 10.1182/blood-2015-07-604512. [DOI] [PubMed] [Google Scholar]
- 12.Győrffy B, Bottai G, Fleischer T, Munkácsy G, Budczies J, Paladini L, Børresen-Dale AL, Kristensen VN, Santarpia L. Aberrant DNA methylation impacts gene expression and prognosis in breast cancer subtypes. Int J Cancer. 2016;138:87–97. doi: 10.1002/ijc.29684. [DOI] [PubMed] [Google Scholar]
- 13.Lin DC, Wang MR, Koeffler HP. Genomic and epigenomic aberrations in esophageal squamous cell carcinoma and implications for patients. Gastroenterology. 2018;154:374–389. doi: 10.1053/j.gastro.2017.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu T, Chen D, Wang Y, Sun X, Li S, Miao S, Wo Y, Dong Y, Leng X, Du W, Jiao W. Identification of DNA methylation-driven genes in esophageal squamous cell carcinoma: A study based on the cancer genome atlas. Cancer Cell Int. 2019;19:52. doi: 10.1186/s12935-019-0770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gloss BS, Samimi G. Epigenetic biomarkers in epithelial ovarian cancer. Cancer Lett. 2014;342:257–263. doi: 10.1016/j.canlet.2011.12.036. [DOI] [PubMed] [Google Scholar]
- 16.Villanueva A, Portela A, Sayols S, Battiston C, Hoshida Y, Méndez-González J, Imbeaud S, Letouzé E, Hernandez-Gea V, Cornella H, et al. DNA methylation-based prognosis and epidrivers in hepatocellular carcinoma. Hepatology. 2015;61:1945–1956. doi: 10.1002/hep.27732. [DOI] [PubMed] [Google Scholar]
- 17.Tomczak K, Czerwińska P, Wiznerowicz M. The cancer genome atlas (TCGA): An immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19:A68–A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.No authors listed, corp-author. The TCGA Legacy. Cell. 2018;173:281–282. doi: 10.1016/j.cell.2018.03.049. [DOI] [PubMed] [Google Scholar]
- 19.Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, Delano D, Zhang L, Schroth GP, Gunderson KL, et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98:288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Gevaert O. MethylMix: An R package for identifying DNA methylation-driven genes. Bioinformatics. 2015;31:1839–1841. doi: 10.1093/bioinformatics/btv020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gevaert O, Tibshirani R, Plevritis SK. Pancancer analysis of DNA methylation-driven genes using MethylMix. Genome Biol. 2015;16:17. doi: 10.1186/s13059-014-0579-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamburov A, Stelzl U, Lehrach H, Herwig R. The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res. 2013;41:D793–D800. doi: 10.1093/nar/gks1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamburov A, Pentchev K, Galicka H, Wierling C, Lehrach H, Herwig R. ConsensusPathDB: Toward a more complete picture of cell biology. Nucleic Acids Res. 2011;39:D712–D717. doi: 10.1093/nar/gkq1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franz M, Lopes CT, Huck G, Dong Y, Sumer O, Bader GD. Cytoscape.js: A graph theory library for visualisation and analysis. Bioinformatics. 2016;32:309–311. doi: 10.1093/bioinformatics/btv557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blandin Knight S, Crosbie PA, Balata H, Chudziak J, Hussell T, Dive C. Progress and prospects of early detection in lung cancer. Open Biol. 2017;7 doi: 10.1098/rsob.170070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nebbioso A, Tambaro FP, Dell'Aversana C, Altucci L. Cancer epigenetics: Moving forward. PLoS Genet. 2018;14:e1007362. doi: 10.1371/journal.pgen.1007362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu J, Yao X. Use of DNA methylation for cancer detection: Promises and challenges. Int J Biochem Cell Biol. 2009;41:147–154. doi: 10.1016/j.biocel.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 28.Leygo C, Williams M, Jin HC, Chan MWY, Chu WK, Grusch M, Cheng YY. DNA methylation as a noninvasive epigenetic biomarker for the detection of cancer. Dis Markers. 2017;2017:3726595. doi: 10.1155/2017/3726595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mase S, Shinjo K, Totani H, Katsushima K, Arakawa A, Takahashi S, Lai HC, Lin RI, Chan MWY, Sugiura-Ogasawara M, Kondo Y. ZNF671 DNA methylation as a molecular predictor for the early recurrence of serous ovarian cancer. Cancer Sci. 2019;110:1105–1116. doi: 10.1111/cas.13936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eissa MAL, Lerner L, Abdelfatah E, Shankar N, Canner JK, Hasan NM, Yaghoobi V, Huang B, Kerner Z, Takaesu F, et al. Promoter methylation of ADAMTS1 and BNC1 as potential biomarkers for early detection of pancreatic cancer in blood. Clin Epigenetics. 2019;11:59. doi: 10.1186/s13148-019-0650-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun Q, Zhang W, Wang L, Guo F, Song D, Zhang Q, Zhang D, Fan Y, Wang J. Hypermethylated CD36 gene affected the progression of lung cancer. Gene. 2018;678:395–406. doi: 10.1016/j.gene.2018.06.101. [DOI] [PubMed] [Google Scholar]
- 32.Kiyozumi Y, Baba Y, Okadome K, Yagi T, Ogata Y, Eto K, Hiyoshi Y, Ishimoto T, Iwatsuki M, Iwagami S, et al. Indoleamine 2,3-dioxygenase 1 promoter hypomethylation is associated with a poor prognosis in patients with esophageal cancer. Cancer Sci. 2019;110:1863–1871. doi: 10.1111/cas.14028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carvalho RH, Hou J, Haberle V, Aerts J, Grosveld F, Lenhard B, Philipsen S. Genomewide DNA methylation analysis identifies novel methylated genes in non-small-cell lung carcinomas. J Thorac Oncol. 2013;8:562–573. doi: 10.1097/JTO.0b013e3182863ed2. [DOI] [PubMed] [Google Scholar]
- 34.Gao C, Zhuang J, Zhou C, Ma K, Zhao M, Liu C, Liu L, Li H, Feng F, Sun C. Prognostic value of aberrantly expressed methylation gene profiles in lung squamous cell carcinoma: A study based on The Cancer Genome Atlas. J Cell Physiol. 2019;234:6519–6528. doi: 10.1002/jcp.27389. [DOI] [PubMed] [Google Scholar]
- 35.Shi YX, Wang Y, Li X, Zhang W, Zhou HH, Yin JY, Liu ZQ. Genome-wide DNA methylation profiling reveals novel epigenetic signatures in squamous cell lung cancer. BMC Genomics. 2017;18:901. doi: 10.1186/s12864-017-4223-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao C, Zhuang J, Li H, Liu C, Zhou C, Liu L, Sun C. Exploration of methylation-driven genes for monitoring and prognosis of patients with lung adenocarcinoma. Cancer Cell Int. 2018;18:194. doi: 10.1186/s12935-018-0691-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao-Jie L, Ai-Mei G, Li-Juan J, Jiang X. Pseudogene in cancer: Real functions and promising signature. J Med Genet. 2015;52:17–24. doi: 10.1136/jmedgenet-2014-102785. [DOI] [PubMed] [Google Scholar]
- 38.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grandér D, Johnsson P. Pseudogene-expressed RNAs: Emerging roles in gene regulation and disease. Curr Top Microbiol Immunol. 2016;394:111–126. doi: 10.1007/82_2015_442. [DOI] [PubMed] [Google Scholar]
- 40.Johnsson P, Ackley A, Vidarsdottir L, Lui WO, Corcoran M, Grandér D, Morris KV. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat Struct Mol Biol. 2013;20:440–446. doi: 10.1038/nsmb.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Z, Liu A, Xiong Q, Xue Y, Liu F, Zeng S, Zhang Z, Li Y, Sun Y, Xu C. Prognostic value of differentially methylated gene profiles in bladder cancer. J Cell Physiol. 2019;234:18763–18772. doi: 10.1002/jcp.28515. [DOI] [PubMed] [Google Scholar]
- 42.Takahashi A, Seike M, Chiba M, Takahashi S, Nakamichi S, Matsumoto M, Takeuchi S, Minegishi Y, Noro R, Kunugi S, et al. Ankyrin repeat domain 1 overexpression is associated with common resistance to afatinib and osimertinib in EGFR-mutant lung cancer. Sci Rep. 2018;8:14896. doi: 10.1038/s41598-018-33190-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lei Y, Henderson BR, Emmanuel C, Harnett PR, DeFazio A. Inhibition of ANKRD1 sensitizes human ovarian cancer cells to endoplasmic reticulum stress-induced apoptosis. Oncogene. 2015;34:485–495. doi: 10.1038/onc.2013.566. [DOI] [PubMed] [Google Scholar]
- 44.Jen J, Wang YC. Zinc finger proteins in cancer progression. J Biomed Sci. 2016;23:53. doi: 10.1186/s12929-016-0269-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiang S, Xiang T, Xiao Q, Li Y, Shao B, Luo T. Zinc-finger protein 545 is inactivated due to promoter methylation and functions as a tumor suppressor through the Wnt/β-catenin, PI3K/AKT and MAPK/ERK signaling pathways in colorectal cancer. Int J Oncol. 2017;51:801–811. doi: 10.3892/ijo.2017.4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weigel C, Chaisaingmongkol J, Assenov Y, Kuhmann C, Winkler V, Santi I, Bogatyrova O, Kaucher S, Bermejo JL, Leung SY, et al. DNA methylation at an enhancer of the three prime repair exonuclease 2 gene (TREX2) is linked to gene expression and survival in laryngeal cancer. Clin Epigenetics. 2019;11:67. doi: 10.1186/s13148-019-0666-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pogribny IP, Pogribna M, Christman JK, James SJ. Single-site methylation within the p53 promoter region reduces gene expression in a reporter gene construct: Possible in vivo relevance during tumorigenesis. Cancer Res. 2000;60:588–594. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the initial data are available from the TCGA database, and that all data generated and analyzed during the present study are included in this published article.