Abstract
Receptor interacting protein kinase 4 (RIPK4) is a serine/threonine kinase that plays an important role in the regulation of cell proliferation, invasion and metastasis in several malignancies; however, its clinical significance and biological function in osteosarcoma (OS) remains unknown. In the present study, the RIPK4 expression level was significantly upregulated in OS tissues and cell lines. High RIPK4 expression was positively associated with larger sized tumors, advanced Enneking stage and poor prognosis in patients with OS. Furthermore, the results revealed that RIPK4 knockdown in the OS cell lines MG-63 and U2OS reduced cell migration and invasion via the inhibition of epithelial-mesenchymal transition (EMT) process, whereby E-cadherin expression was increased and N-cadherin and vimentin expression decreased. Mechanistically, RIPK4 knockdown inhibited EMT by inactivating the Wnt/β-catenin signaling pathway. These findings suggest that RIPK4 may be a novel potential therapeutic target for the treatment of metastases in patients with OS.
Keywords: receptor interacting protein kinase 4, epithelial-mesenchymal transition, osteosarcoma, Wnt/β-catenin
Introduction
Osteosarcoma (OS) is the most common primary malignant bone tumor found in children and adolescents, which mostly occurs in the metaphyseal part of tubular bones such as the distal femur, proximal humerus and proximal tibia (1,2). A previous study revealed that the introduction of neoadjuvant chemotherapy and advanced surgical techniques have significantly increased the overall survival rate from 20 to 70% (3). However, the overall survival rate of patients with lung metastases is only 10–30% (4). Further studies are necessary to understand the underlying mechanisms of OS metastasis and thereby identify novel therapeutic targets.
Tumor metastasis is a complex, multistage process. An increasing body of evidence has suggested that EMT plays an important role in tumor progression and metastasis (5–7). EMT is a physiological process whereby epithelial cells lose their epithelial characteristics and convert to a mesenchymal phenotype (8). At the molecular level, E-cadherin expression is repressed, while mesenchymal marker expression (including N-cadherin and vimentin) is increased (9). Many signaling pathways (TGF-β, Notch, Wnt/β-catenin, Hedgehog and Hippo) induce and modulate the EMT process (10–14). However, complex molecular networks that influence EMT in OS have yet to be clarified.
Receptor interacting protein kinase 4 (RIPK4), a member of the RIP kinase family, is comprised of a homologous N-terminal serine/threonine kinase domain, an intermediate domain and a C-terminal region containing ankyrin repeat domain 11 (15,16). Early RIPK4 studies focused mainly on keratinocyte proliferation and differentiation (17). Recent studies have demonstrated that RIPK4 was overexpressed in various types of human cancers, including cervical, skin, colorectal and ovarian cancers (18,19). Furthermore, RIPK4 overexpression has been associated with progression and poor prognosis. However, RIPK4 acted as a tumor suppressor in other types of cancers, such as tongue cancer and hepatocellular carcinoma (20,21). The biological function of RIPK4 in OS is largely unknown. The present study detected the expression levels of RIPK4 in OS tissues and cell lines. Furthermore, its relationship with clinicopathological parameters and prognosis in OS patients were analyzed, and its effects on EMT and the underlying molecular mechanisms were investigated.
Materials and methods
Tissue specimens
A total of 36 OS tissues and 15 normal bone tissues were obtained from The Second Hospital of Lanzhou University (Gansu, China) from January 2010 to December 2015, and the clinicopathological parameters of the 36 OS patients were recorded (Table I). The 36 OS patients were followed up by telephone, outpatient service and letters from the date of operation. The deadline was May 2016. The follow-up period was 5–72 months. The median follow-up time was 27 months. Furthermore, 4 fresh specimens of OS and normal bone tissue were collected and stored at −80°C for western blotting. None of the patients were undergoing chemotherapy or radiotherapy treatment. The present study was approved by the Medical Ethics Committee of Lanzhou University and informed consent forms were signed by the patients.
Table I.
Relationship between expression of RIPK4 and clinicopathologic parameters, RIPK4, receptor interacting protein kinase 4.
| RIPK4 expression level | |||||
|---|---|---|---|---|---|
| Clinicopathological parameters | Total n | High | Low | χ2 | P-value |
| Sex | |||||
| Male | 20 | 13 | 7 | 0.024 | 0.877 |
| Female | 16 | 10 | 6 | ||
| Age/year | |||||
| >14 | 31 | 19 | 12 | 0.653 | 0.419 |
| ≤14 | 5 | 4 | 1 | ||
| Tumor size, cm | |||||
| >8 | 17 | 14 | 3 | 4.760 | 0.029a |
| ≤8 | 19 | 9 | 10 | ||
| Tumor site | |||||
| Femur | 17 | 9 | 8 | 1.798 | 0.407 |
| Tibia and Fibula | 9 | 7 | 2 | ||
| Other | 10 | 7 | 3 | ||
| Histological type | |||||
| Osteoblastic | 21 | 12 | 9 | 3.739 | 0.291 |
| Chondroblastic | 6 | 3 | 3 | ||
| Fibroblastic | 5 | 4 | 1 | ||
| Mixed | 4 | 4 | 0 | ||
| Enneking stage | |||||
| I/IIA | 9 | 2 | 7 | 9.030 | 0.003b |
| IB/III | 27 | 21 | 6 | ||
| Metastasis | |||||
| Yes | 13 | 10 | 3 | 1.498 | 0.221 |
| No | 23 | 13 | 10 | ||
P<0.05
P<0.01.
Cell culture and transfection
The human OS cell lines MG-63 and U2OS, and the normal osteoblastic cell line hFOB 1.19 were obtained from the Chinese Academy of Medical Sciences. MG-63 cells were cultured in Minimum Essential Medium, U2OS in McCoy's 5A medium, hFOB 1.19 in Dulbecco's modified Eagle's medium/F-12 medium (all from Hyclone; GE Healthcare Life Sciences), and supplemented with 10% fetal bovine serum (FBS; PAN-Biotech GmbH) and 100 U/ml penicillin/streptomycin, at 37°C in a humidified atmosphere containing 5% CO2. For RIPK4 expression silencing, the 6 µl RIPK4 small interfering (si)-RNA and 6 µl negative control siRNA products (Santa Cruz Biotechnology, Inc.) were transfected into 80% confluence MG-63 and U2OS cells using a siRNA reagent system (Santa Cruz Biotechnology, Inc.) according to the manufacturer's specifications. RIPK4-siRNA sense sequences: 5′-CACACGCAGUAUGAAGAUA-3′; antisense sequences: 5′-UAUCUUCAUACUGCGUGUG-3′; NC-siRNA sense sequences: 5′-UUGUGGCCUGUUAGCUUCA-3′; antisense sequences: 5′-UGAAGCUAACAGGCCACAA-3′. Following incubation for 48 h, RIPK4 expression levels were detected by western blot analysis.
Western blot analysis
Tissues and cells were lysed with RIPA buffer (Beyotime Institute of Biotechnology) on ice containing a protease inhibitor supplemented with 1 mmol/l phenylmethylsulfonyl fluoride. Protein concentration was detected by a BCA protein assay kit (Beyotime Institute of Biotechnology). Then equal amounts of protein samples (40 µg) were separated by 8 or 10% SDS-PAGE and transferred onto PVDF membranes. The membranes were blocked with 5% non-fat dry milk for 2 h at room temperature, and subsequently incubated overnight at 4°C with primary antibodies, namely: Anti-RIPK4 (1:200, cat no. ab124649), anti-E-cadherin (1:500, cat no. ab152102), anti-N-cadherin (1:1,000, cat no. ab76011), anti-vimentin (1:500, cat no. ab8978), anti-β-catenin (1:500, cat no. ab16051; all Abcam), anti-β-actin (1:5,000, cat no. 1978, Sigma-Aldrich; Merck KGaA) and anti-Lamin B1 (1:5,000, cat no. bsm-33442M, BIOSS). After washing three times with Tris-buffered saline and 0.05% Tween (pH 7.4), the membranes were incubated with the horseradish peroxidase (HRP)-conjugated anti-rabbit or anti-mouse immunoglobulin (Ig)-G secondary antibodies (1:5,000, catalog no. A0208; Beyotime Institute of Biotechnology) for 2 h at room temperature. The protein bands were then detected using an ECL system (EMD Millipore) and imaged using a ImageQuant TL A600 imaging system (GE Healthcare Biosciences). The density of each band was analyzed with Pro-Plus 6.0 Software (Media Cybernetics, Inc.).
Cell proliferation assay
The cell proliferation assay was evaluated with a Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.). At 48 h after the transfection incubation period, cells were seeded in a 96-well plate at 3,000 cells/well with 100 µl culture medium. At 0, 24, 48 and 72 h, 10 µl CCK-8 reagent solution was added to each well and incubated for 4 h at 37°C. Absorbance was measured using a microplate reader set at a 450 nm wavelength.
Migration and invasion assay
For the migration and invasion assays, 24-well plates containing 8.0 µm pore size Transwell chambers (Corning Incorporated) were used according to the manufacturer's specifications. For the migration assay, ~1×105 cells/well (48 h post transfection incubation period) were seeded in the upper chambers with 100 µl of serum-free medium. Thereafter, 500 µl medium supplemented with 10% FBS as a chemoattractant was added to the lower chambers of the wells. After 24 h of incubation at 37°C, non-migrating cells on the upper surface of the membrane were removed with cotton swabs, while migrating cells on the lower surface of the membrane were fixed with 4% paraformaldehyde for 30 min, stained with 0.1% crystal violet for 10 min at 37°C and photographed under an inverted microscope (magnification, ×200). The invasion assay was performed in a similar manner, except 100 µl of 1:10 serum-free medium-diluted Matrigel (BD Biosciences) was also added to each well before the cells were seeded in the membrane.
Wound healing assay
Following 48 h of transfection, the cells were trypsinized by 0.25% trypsin, and 1×105 cells seeded into 6-well plates. When cell confluency reached 95%, a sterile 200 µl pipette tip was used to make a scratch. After washing with PBS, cells were then incubated in serum-free medium for 24 h at 37°C. The width of the scratch was measured under an inverted microscope at 0 and 24 h, respectively.
Immunofluorescence staining
Cells (1×105) were cultured on glass coverslips in a 35 mm diameter dish. After 24 h, the cells were fixed with 4% paraformaldehyde for 30 min at 37°C, permeabilized with 0.1% Triton X-100 in PBS for 15 min at 37°C, blocked with 5% bovine serum albumin (BSA, Beyotime Institute of Biotechnology) in PBS for 1 h, and incubated with primary antibodies against E-cadherin (1:100, cat. no. ab152102), N-cadherin (1:100, cat. no. ab76011), vimentin (1:100, cat. no. ab8978) and β-catenin (1:100, cat. no. ab16051; all from Abcam) at 4°C overnight. The cells were then washed three times with PBS and incubated with fluorescein isothiocyanate-conjugated goat anti-mouse or anti-rabbit secondary antibodies (1:5,000, cat. nos. A0412 and A0423; Beyotime Institute of Biotechnology) for 2 h at 37°C. Subsequently the nuclei were counterstained with 4′,6-diamidino-2-phenylindole for 10 min at 37°C, and images were captured using a fluorescence microscope (Olympus Corporation).
Immunohistochemical staining
Tissue samples were fixed with 10% formalin for 30 min at 37°C, embedded in paraffin, and cut into 4 µm sections. The sections were dewaxed in xylene and rehydrated in gradients of high ethanol concentration to distilled water. Endogenous peroxidase activity was quenched by 3% H2O2 for 15 min at 37°C. The sections were heated for 30 min at 95°C in 10 nM, pH 6.0 sodium citrate buffer for antigen retrieval, blocked with 5% BSA (Beyotime Institute of Biotechnology) at 37°C for 1 h and incubated overnight at 4°C with primary antibody against RIPK4 (1:200, cat. no. ab124649, Abcam). After rinsing with PBS, the sections were incubated 1 h at room temperature with HRP-conjugated goat anti-mouse IgG antibody (1:5,000, cat. no. ZB-2301, Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd.). Then the sections were stained at 37°C for 3 min using 3,3′-diaminobenzidine, counterstained for 1 min with Gill's hematoxylin and observed under a light microscope (magnification, ×200). The stained results were scored independently by two pathologists. The percentage of positive staining cells were scored as follows: 0, <5%; 1, 5–25%; 2, 26–50%; 3, 51–75%; and 4, >75%. Staining intensity was graded as follows: 0, no staining; 1, light brown; 2, brown; and 3, dark brown. The final immunoreactive score was calculated by multiplying the percentage of positive staining cells with the staining intensity. Scores of 0–3 and 4–12 were designated as low and high expression, respectively.
Statistical analysis
All statistical analyses was performed using SPSS version 19.0 software (IBM Corp.). All data were presented as mean ± standard deviation. Continuous variables were analyzed by a two-tailed Student's t-test or one-way analysis of variance followed by Tukey's post hoc test. The association between RIPK4 expression levels and the clinicopathological parameters of the patients was assessed using the Chi-squared test. Overall survival was evaluated using the Kaplan-Meier method and a log-rank test. P<0.05 was considered to indicate a statistically significant difference. All experiments were independently repeated at least three times.
Results
RIPK4 expression is upregulated in human OS tissues and is associated with poor prognosis
To investigate RIPK4 expression in OS, 36 OS tissues and 15 normal bone tissues were analyzed via immunohistochemistry. The results revealed that high RIPK4 expression was detected in 63.9% (23/36) of OS tissues, which was significantly higher than that observed in normal bone tissues (0/15; Fig. 1A). Additionally, western blotting was conducted to analyze the protein levels of RIPK4 in 4 fresh OS and normal bone tissues. Consistent with the immunochemistry results, RIPK4 protein levels were upregulated in OS tissues when compared with those in normal bone tissues (Fig. 1B). The present study also analyzed the relationship between RIPK4 expression and clinicopathological parameters in OS. High RIPK4 expression was significantly associated with larger tumor sizes and advanced Enneking stage. There were no significant associations between high RIPK4 expression and other clinicopathological parameters, including sex, age, tumor site, histological type and metastasis (Table I).
Figure 1.
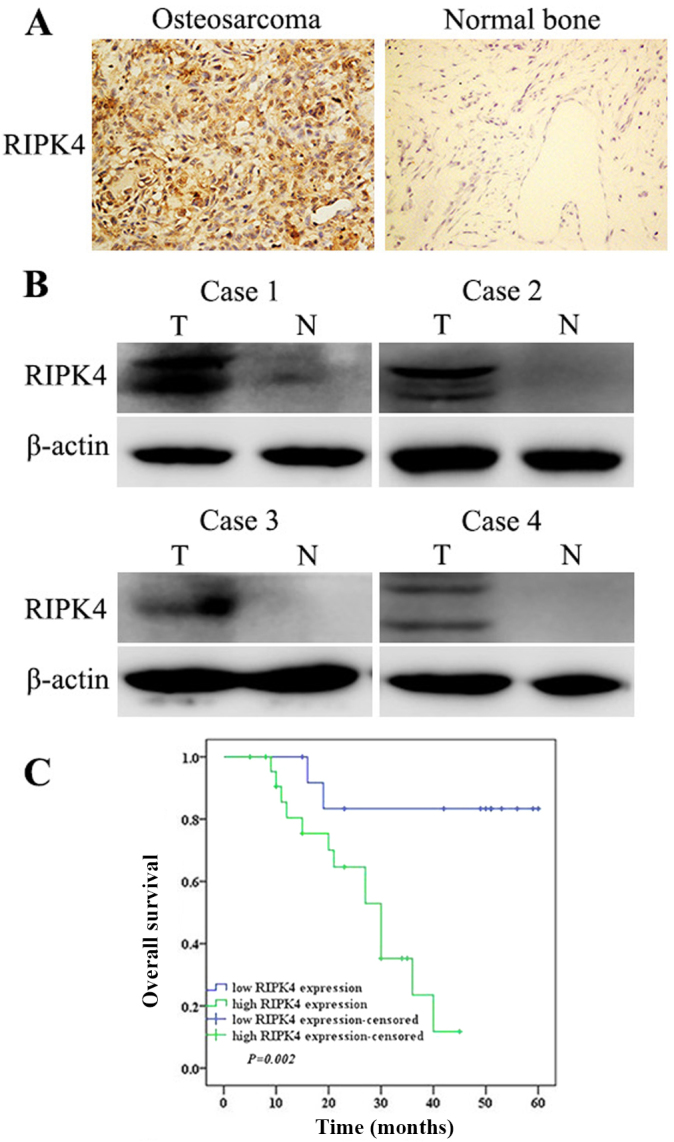
RIPK4 expression is upregulated in OS and is associated with poor prognosis. (A) Immunohistochemical staining of RIPK4 in OS and normal bone tissues (magnification, ×200). (B) Western blot analysis of RIPK4 protein expression in 4 fresh specimens of OS and normal bone tissue. β-actin was used as the loading control. The top bands are specific for RIPK4, and the bottom bands shown are non-specific. (C) Kaplan-Meier survival analysis of OS patients with high and low RIPK4 expression. RIPK4, receptor interacting protein kinase 4; OS, osteosarcoma; T, tumor; N, normal.
All OS patients had follow-up consultations from their surgery date until they succumbed to the disease or the last follow-up of the present study (range, 5–72 months; median follow-up, 27 months). Kaplan-Meier survival analysis was used to evaluate the association between RIPK4 expression and the prognosis survival of patients with OS. Patients with high RIPK4 expression had a significantly shorter survival time (median survival time, 21 months) as opposed to those with low RIPK4 expression (median survival time, 42 months; Fig. 1C). Taken together, these results suggest that RIPK4 is associated with OS progression and prognosis.
Silencing of RIPK4 inhibits cell proliferation, migration and invasion in OS cells
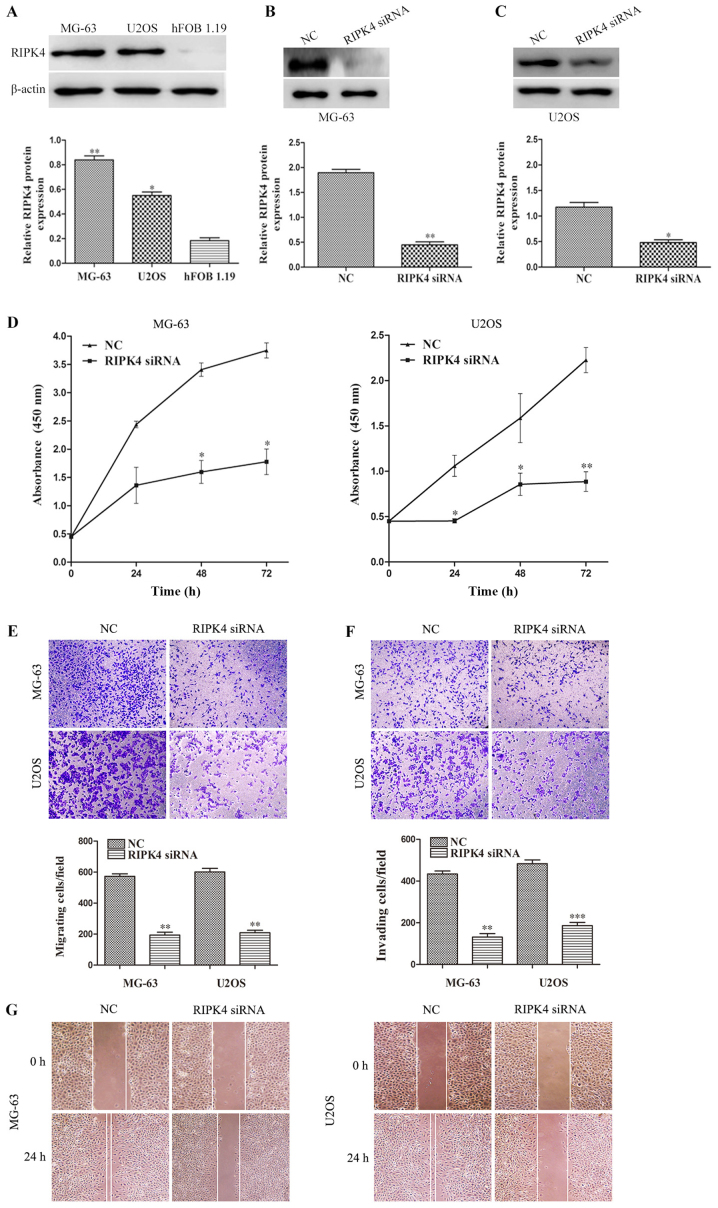
In the present study, RIPK4 expression was examined in two human OS cell lines (MG-63 and U2OS) and a normal human osteoblastic cell line (hFOB 1.19). Compared with hFOB 1.19 cells, RIPK4 expression was upregulated in OS MG-63 and U2OS cells (Fig. 2A). To further investigate the role of RIPK4 in OS, the specific siRNA targeting the RIPK4 gene (RIPK4-siRNA) was transfected by liposome into MG-63 and U-2OS cells. The silencing efficiency was analyzed by western blotting. The results demonstrated that RIPK4 expression was significantly decreased after transfection with RIPK4-siRNA, indicating that the stable RIPK4 gene knockdown MG-63 and U-2OS cell lines were successfully established (Fig. 2B and C). Subsequently, the effect of RIPK4 knockdown on cell proliferation, migration and invasion was examined. The CCK-8 assay revealed that the proliferation of MG-63 at 48 and 72 h, and U2OS cells at 24, 48 and 72 h, was significantly suppressed after RIPK4 knockdown (Fig. 2D). The wound healing and migration assays indicated that RIPK4 knockdown markedly inhibited cell migration in MG-63 and U2OS cells (Fig. 2E and G). The invasion assay results showed that RIPK4 knockdown markedly reduced the invasion potential of MG-63 and U2OS cells (Fig. 2F). These results suggest that RIPK4 silencing inhibits the proliferation, migration and invasion abilities of OS cells.
Figure 2.
RIPK4 silencing inhibits cell proliferation, migration and invasion in MG-63 and U2OS. (A) Western blot analysis of RIPK4 expression in two human OS cell lines (MG-63 and U2OS) and a human normal osteoblastic cell line (hFOB 1.19). *P<0.05 and **P<0.01 vs. hFOB 1.19. (B) MG-63 and (C) U2OS cells were transfected with RIPK4 siRNA and NC siRNA. The expression levels of RIPK4 were assessed by western blotting after transfection. β-actin was used as the loading control. (D) Cell proliferation of MG-63 and U2OS after silencing RIPK4 was measured by Cell Counting Kit-8 assay. (E) Cell migration and (F) invasion of MG-63 and U2OS cells after RIPK4 silencing were measured by Transwell migration and invasion assays (magnification, ×200). (G) A wound healing assay was conducted to evaluate the migration of MG-63 and U2OS cells after RIPK4 silencing (magnification, ×200). Data were presented as the mean ± standard deviation of 3 independent experiments. *P<0.05, **P<0.01 and ***P<0.001 vs. NC. RIPK4, receptor interacting protein kinase 4; OS, osteosarcoma; NC, negative control; siRNA, small interfering RNA.
Silencing of RIPK4 suppresses EMT in OS cells
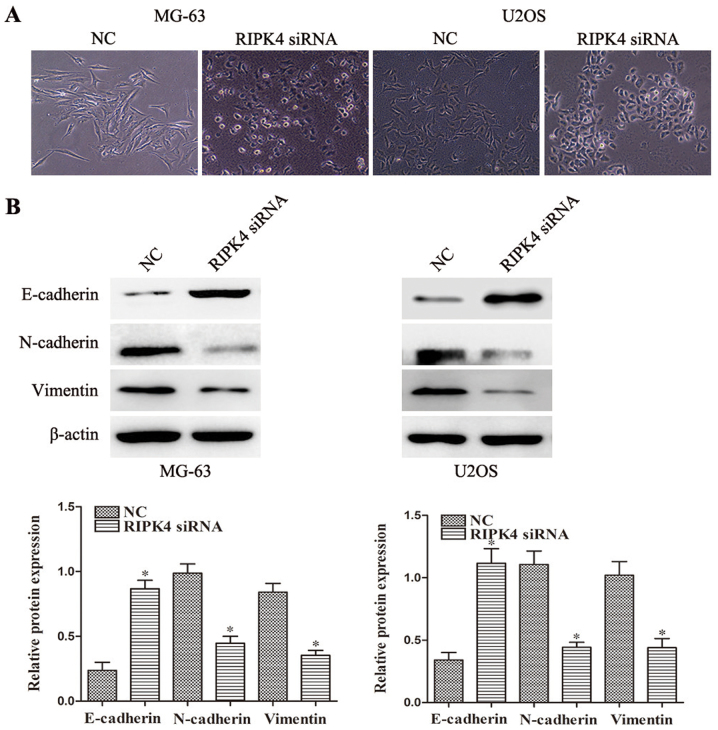
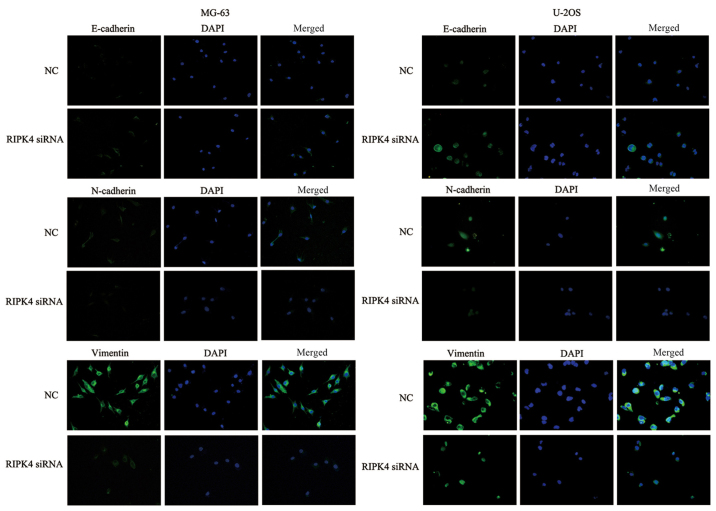
EMT is an essential process in tumor migration and invasion (5), therefore the present study further investigated whether RIPK4 silencing inhibits EMT in OS cells. RIPK4 knockdown led to morphological changes in the MG-63 and U2OS cells at 24 h, as characterized by a cobblestone-shaped appearance (Fig. 3A). The expression levels of the epithelial marker E-cadherin and the mesenchymal markers, N-cadherin and vimentin, were examined by western blotting and immunofluorescence staining. Significantly increased E-cadherin expression levels and decreased N-cadherin and vimentin expression levels were observed after RIPK4 silencing in MG-63 cells. Similar results were observed in U2OS cells (Fig. 3B). The immunofluorescence staining also supported these findings (Fig. 4). These results indicate that RIPK4 knockdown suppresses the EMT process in OS cells.
Figure 3.
Silencing of RIPK4 suppresses EMT in OS cells. (A) The morphological changes of OS MG-63 and U2OS cells induced by RIPK4 silencing that are associated with EMT were observed under an inverted optical microscope (magnification, ×200). (B) The epithelial maker E-cadherin and mesenchymal markers N-cadherin and vimentin in MG-63 and U2OS cells were assessed by western blotting. β-actin was used as the loading control. Data were presented as the mean ± standard deviation of 3 independent experiments. *P<0.05 vs. NC. RIPK4, receptor interacting protein kinase 4; OS, osteosarcoma; EMT, epithelial-mesenchymal transition; NC, negative control; siRNA, small interfering RNA.
Figure 4.
Immunofluorescence assay for E-cadherin, N-cadherin and vimentin expression in various cell groups. The targeted proteins were stained green and the nuclei were stained with DAPI (blue; magnification, ×200). DAPI, 4′,6-diamidino-2-phenylindole; RIPK4, receptor interacting protein kinase 4; NC, negative control; siRNA, small interfering RNA.
Silencing of RIPK4 suppresses EMT by inactivating the Wnt/β-catenin signaling pathway
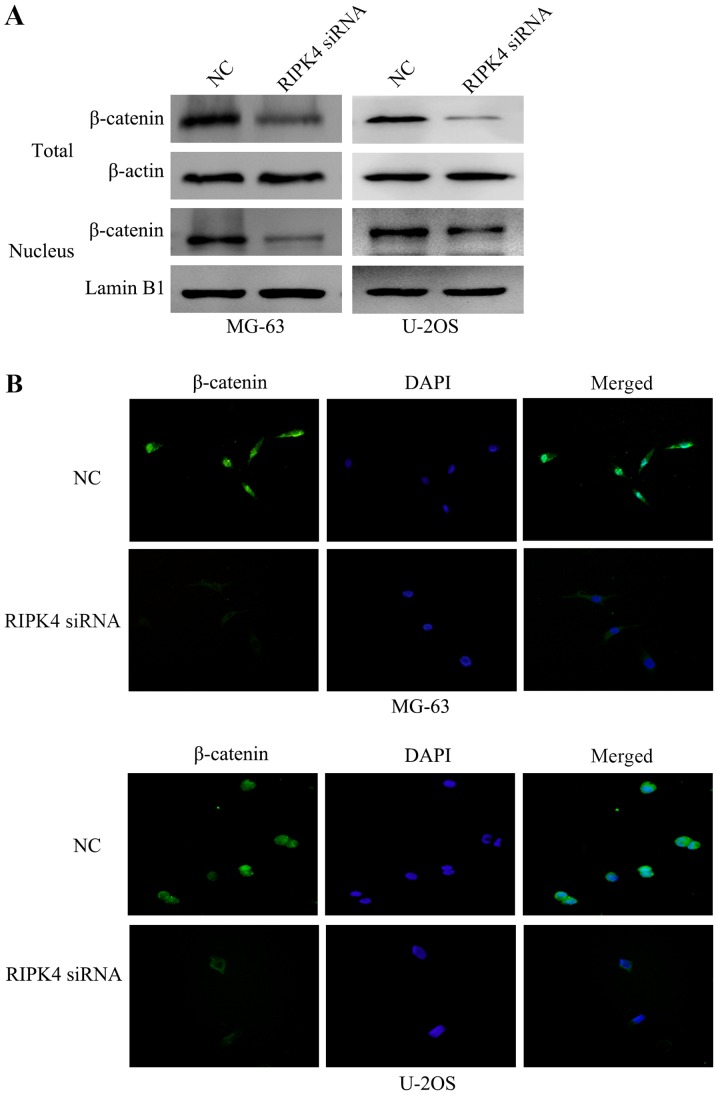
A previous study has reported that the Wnt/β-catenin signaling pathway regulates cell EMT (12). To investigate the underlying molecular mechanism of the EMT suppression induced by RIPK4 silencing, the present study examined the expression and localization of the key molecule of the Wnt/β-catenin signaling pathway, β-catenin, by western blotting. The results revealed that RIPK4 silencing inhibited the protein expression of total and nuclear β-catenin (Fig. 5A). In addition, immunofluorescence analysis confirmed that the levels of β-catenin were reduced following RIPK4 knockdown, in both the cytoplasm and the cell nucleus (Fig. 5B). These results suggest that RIPK4 silencing inhibits the EMT of OS cells by negatively regulating the Wnt/β-catenin signaling pathway.
Figure 5.
Silencing of RIPK4 suppresses the inactivation of the Wnt/β-catenin signaling pathway. (A) Western blot analysis revealed that RIPK4 silencing decreased total and nuclear β-catenin expression in MG-63 and U2OS cells. β-actin was used as the loading control for total expression. Lamin B1 was used as a loading control in the nucleus. (B) Immunofluorescent staining revealed that RIPK4 silencing reduced the levels of β-catenin. The targeted proteins were stained green and the nuclei were stained with DAPI (blue; magnification, ×200). RIPK4, receptor interacting protein kinase 4; DAPI, 4′,6-diamidino-2-phenylindole; NC, negative control; siRNA, small interfering RNA.
Discussion
Similar to other types of tumors, OS has a high propensity for metastasis. It has been reported that ~20% of OS patients with metastases were detected at the time of diagnosis, while 80% of patients would develop metastases at a later stage (22,23). Metastasis is the leading cause of death in patients with OS (24). Understanding the underlying mechanism of OS metastasis is an essential prognostic factor.
Recent studies have demonstrated that RIPK4 is important in the development and progression of many types of malignant tumors. Liu et al (18) reported that RIPK4 expression was significantly upregulated in cervical squamous cell carcinoma and this increased expression was associated with invasive and metastatic characteristics, including International Federation of Gynecology and Obstetrics stage, tumor size and distant metastasis. By contrast, other studies have shown that RIPK4 expression was significantly downregulated and acted as a tumor suppressor (20). Therefore, RIPK4 may trigger different carcinogenic mechanisms in different types of tumors. Little is known about the function of RIPK4 in OS. In the present study, RIPK4 expression was significantly upregulated in OS tissues and cell lines when compared with normal bone tissues and osteoblastic cell lines. In addition, RIPK4 overexpression was closely associated with a larger tumor size, advanced Enneking stage and poor prognosis of OS patients. These results indicated that RIPK4 might play a role the progression of OS.
EMT is a multistage process in which epithelial cells lose polarity and acquire migratory and invasive properties; it is considered to be a critical factor in invasion and metastasis (25). An increasing body of evidence has indicated that many sarcomas can undergo EMT-related processes, which may be associated with an aggressive clinical behavior. These processes may be particularly applicable to certain sarcoma subtypes, such as carcinosarcomas exhibiting a dual phenotype with mesenchymal and epithelial tumor characteristics (26). A previous study reported a depletion in RIPK4 expression using siRNA could inhibit a cervical cancer cell (18). In the present study, the results revealed that RIPK4 knockdown by siRNA suppressed tumor cell migration and invasion in OS. Furthermore, the mechanisms of the RIPK4-mediated suppression of migration and invasion were investigated. The results showed that RIPK4 knockdown significantly increased the expression of the epithelial marker E-cadherin and decreased the expression of the mesenchymal markers, N-cadherin and vimentin; it also induced morphological changes in OS cell lines, from spindle-shaped fibroblast to cobblestone-shaped epithelial-like morphology. These data suggested that the silencing of RIPK4 might prevent tumor cell migration and invasion by interfering with the EMT process in OS.
Further evidence has suggested the Wnt/β-catenin signaling pathway is involved in embryogenesis and tumor development (27,28). The aberrant activation of the Wnt/β-catenin pathway signaling could promote EMT progression in tumor cells, including OS cells (29). Huang et al (19) demonstrated that ectopic RIPK4 expression could induce cytosolic β-catenin accumulation and upregulate canonical Wnt target genes including Cyclin D1, lymphoid enhancer binding factor 1, Jun protooncogene AP-1 transcription factor subunit, Myc and transcription factor 7 in A2780 and COV434 ovarian cancer cells, thereby implying that RIPK4 regulates the Wnt/β-catenin signaling pathway. However, the association between RIPK4 and Wnt/β-catenin signaling in OS is still unclear. In the present study, the results showed that a significant decrease in total and nuclear β-catenin levels was induced by endogenous RIPK4 knockdown in OS cells. The translocation of β-catenin is an important molecular event in tumor formation (30). Consistent with these findings, immunofluorescence analysis confirmed that the levels of β-catenin were reduced following RIPK-4 knockdown, in both the cytoplasm and the cell nucleus. All of the above results suggest that RIPK4 knockdown could suppress cell EMT by deactivating the Wnt/β-catenin signaling pathway.
In conclusion, the present study revealed that RIPK4 was significantly upregulated in OS tissues and cell lines, and its high expression was associated with larger tumor sizes, advanced Enneking stage and poor prognosis. Furthermore, RIPK4 knockdown inhibited cell migration and invasion by interfering with the EMT process, which was mediated by the inactivation the Wnt/β-catenin signaling pathway. This may provide a novel therapeutic target for preventing OS cell metastasis. However, the precise regulatory mechanisms need to be investigated further.
Acknowledgements
Not applicable.
Funding
The present study was supported by Basic Research Innovative Group Project of Gansu Province (grant no. 1308RJIA004).
Availability of data and materials
The analyzed datasets used and/or analyzed during the study are available from the corresponding author on reasonable request.
Authors' contributions
ZY and PD conceived and designed the experiments. ZY, YP and RG performed the experiments. YC, XR and WL analyzed the data. WL, ZY, YP, RG and PD wrote the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Written informed consent was obtained from all of the patients and the research protocols were approved by the Ethics Committee of Second hospital of Lanzhou University (Ganzu, China).
Patient consent for publication
All patients within the present study provided consent for the publication of their data.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Moore DD, Luu HH. Osteosarcoma. Cancer Treat Res. 2014;162:65–92. doi: 10.1007/978-3-319-07323-1_4. [DOI] [PubMed] [Google Scholar]
- 2.Arndt CA, Rose PS, Folpe AL, Laack NN. Common musculoskeletal tumors of childhood and adolescence. Mayo Clin Proc. 2012;87:475–487. doi: 10.1016/j.mayocp.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daw NC, Billups CA, Rodriguez-Galindo C, McCarville MB, Rao BN, Cain AM, Jenkins JJ, Neel MD, Meyer WH. Metastatic osteosarcoma. Cancer. 2006;106:403–412. doi: 10.1002/cncr.21626. [DOI] [PubMed] [Google Scholar]
- 4.Farfalli GL, Albergo JI, Lobos PA, Smith DE, Streitenberger PD, Pallotta Rodríguez MG, Aponte-Tinao LA. Osteosarcoma lung metastases. Survival after chemotherapy and surgery. Medicina (B Aires) (Article in Spanish) 2015;75:87–90. [PubMed] [Google Scholar]
- 5.Diepenbruck M, Christofori G. Epithelial-mesenchymal transition (EMT) and metastasis: Yes, no, maybe. Curr Opin Cell Biol. 2016;43:7–13. doi: 10.1016/j.ceb.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 6.Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, Wu CC, LeBleu VS, Kalluri R. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527:525–530. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mittal V. Epithelial mesenchymal transition in aggressive lung cancers. Adv Exp Med Biol. 2016;890:37–56. doi: 10.1007/978-3-319-24932-2_3. [DOI] [PubMed] [Google Scholar]
- 8.Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 9.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moustakas A, Heldin CH. Mechanisms of TGFβ-induced epithelial-mesenchymal transition. J Clin Med. 2016;5(pii):E63. doi: 10.3390/jcm5070063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, Ma J, Qian X, Wu Q, Xia J, Miele L, Sarkar FH, Wang Z. Regulation of EMT by Notch signaling pathway in tumor progression. Curr Cancer Drug Targets. 2013;13:957–962. doi: 10.2174/15680096113136660101. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez DM, Medici D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci Signal. 2014;7:re8. doi: 10.1126/scisignal.2005189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu X, Yun F, Shi L, Li ZH, Luo NR, Jia YF. Roles of signaling pathways in the epithelial-mesenchymal transition in cancer. Asian Pac J Cancer Prev. 2015;16:6201–6206. doi: 10.7314/APJCP.2015.16.15.6201. [DOI] [PubMed] [Google Scholar]
- 14.van Rensburg HJ, Yang X. The roles of the Hippo pathway in cancer metastasis. Cell Signal. 2016;28:1761–1772. doi: 10.1016/j.cellsig.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 15.Meylan E, Tschopp J. The RIP kinases: Crucial integrators of cellular stress. Trends Biochem Sci. 2005;30:151–159. doi: 10.1016/j.tibs.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 16.Meylan E, Martinon F, Thome M, Gschwendt M, Tschopp J. RIP4 (DIK/PKK), a novel member of the RIP kinase family, activates NF-kappa B and is processed during apoptosis. EMBO Rep. 2002;3:1201–1208. doi: 10.1093/embo-reports/kvf236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adams S, Munz B. RIP4 is a target of multiple signal transduction pathways in keratinocytes: Implications for epidermal differentiation and cutaneous wound repair. Exp Cell Res. 2010;316:126–137. doi: 10.1016/j.yexcr.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 18.Liu DQ, Li FF, Zhang JB, Zhou TJ, Xue WQ, Zheng XH, Chen YB, Liao XY, Zhang L, Zhang SD, et al. Increased RIPK4 expression is associated with progression and poor prognosis in cervical squamous cell carcinoma patients. Sci Rep. 2015;5:11955. doi: 10.1038/srep11955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang X, McGann JC, Liu BY, Hannoush RN, Lill JR, Pham V, Newton K, Kakunda M, Liu J, Yu C, et al. Phosphorylation of dishevelled by protein kinase ripk4 regulates wnt signaling. Science. 2013;339:1441–1445. doi: 10.1126/science.1232253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, Zhu W, Zhou Y, Xu W, Wang H. RIPK4 is downregulated in poorly differentiated tongue cancer and is associated with migration/invasion and cisplatin-induced apoptosis. Int J Biol Markers. 2014;29:e150–159. doi: 10.5301/jbm.5000073. [DOI] [PubMed] [Google Scholar]
- 21.Heim D, Cornils K, Schulze K, Fehse B, Lohse AW, Brümmendorf TH, Wege H. Retroviral insertional mutagenesis in telomerase-immortalized hepatocytes identifies RIPK4 as novel tumor suppressor in human hepatocarcinogenesis. Oncogene. 2015;34:364–372. doi: 10.1038/onc.2013.551. [DOI] [PubMed] [Google Scholar]
- 22.Bhattasali O, Vo AT, Roth M, Geller D, Randall RL, Gorlick R, Gill J. Variability in the reported management of pulmonary metastases in osteosarcoma. Cancer Med. 2015;4:523–531. doi: 10.1002/cam4.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayden JB, Hoang BH. Osteosarcoma: Basic science and clinical implications. Orthop Clin North Am. 2006;37:1–7. doi: 10.1016/j.ocl.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 24.Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 25.Kotiyal S, Bhattacharya S. Events of molecular changes in epithelial-mesenchymal transition. Crit Rev Eukaryot Gene Expr. 2016;26:163–171. doi: 10.1615/CritRevEukaryotGeneExpr.2016016307. [DOI] [PubMed] [Google Scholar]
- 26.Sannino G, Marchetto A, Kirchner T, Grünewald TGP. Epithelial-to-mesenchymal and mesenchymal-to-epithelial transition in mesenchymal tumors: A paradox in sarcomas. Cancer Res. 2017;77:4556–4561. doi: 10.1158/0008-5472.CAN-17-0032. [DOI] [PubMed] [Google Scholar]
- 27.Caronia-Brown G, Anderegg A, Awatramani R. Expression and functional analysis of the Wnt/beta-catenin induced mir-135a-2 locus in embryonic forebrain development. Neural Dev. 2016;11:9. doi: 10.1186/s13064-016-0065-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene. 2017;36:1461–1473. doi: 10.1038/onc.2016.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang G, Yuan J, Li K. EMT transcription factors: Implication in osteosarcoma. Med Oncol. 2013;30:697. doi: 10.1007/s12032-013-0697-2. [DOI] [PubMed] [Google Scholar]
- 30.Prakash S, Swaminathan U. β-catenin in health: A review. J Oral Maxillofac Pathol. 2015;19:230–238. doi: 10.4103/0973-029X.164537. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed datasets used and/or analyzed during the study are available from the corresponding author on reasonable request.