Abstract
Cancer cells use aerobic glycolysis to sustain their proliferation. Long non-coding RNA brain cytoplasmic RNA 1 (BCYRN1) has been reported to act as an oncogene in non-small-cell lung cancer (NSCLC). The present study investigated the role of BCYRN1 in NSCLC glycolysis. BCYRN1 expression was detected in NSCLC cells and tissues using reverse transcription-quantitative PCR. The effect of BCYRN1 on aerobic glycolysis was examined by measuring NSCLC cell glucose catabolism and lactate synthesis. The relationships between BCYRN1 and microRNA (miR)-149, and between miR-149 and pyruvate kinase M1/2 (PKM2) were measured using a dual-luciferase reporter assay. Cell proliferation and invasion were analyzed by the Cell Counting kit-8 assay and the Matrigel invasion assay, respectively. High BCYRN1 expression was observed in NSCLC tissues and cells compared with the corresponding controls. BCYRN1 induced glycolysis and upregulated the expression levels of PKM2 in NSCLC cells. In addition, BCYRN1 regulated miR-149 expression levels, and miR-149 inhibitor rescued the effects of si-BCYRN1 on glucose consumption and lactate production. miR-149 knockdown significantly enhanced the expression of PKM2. Furthermore, PKM2 inhibition significantly reversed the effects of miR-149 inhibitor on glucose catabolism and lactate synthesis. Furthermore, PKM2 was involved in NSCLC cell proliferation and invasion, and BCYRN1 knockdown and miR-149 overexpression inhibited both processes. The present study suggested that BCYRN1 was involved in cell glycolysis, proliferation and invasion during NSCLC via regulating miR-149 and PKM2.
Keywords: non-small-cell lung cancer, brain cytoplasmic RNA 1, glycolysis, microRNA-149, pyruvate kinase M1/2
Introduction
Lung cancer has become the most common malignant tumor worldwide, contributing to the highest mortality and morbidity rate compared with all cancer types (1). Non-small-cell lung cancer (NSCLC) accounts for >80% of all lung cancer cases (1). Patients with NSCLC typically have a poor prognosis, which is primarily due to distant metastasis and local recurrence (2). Several patients with NSCLC are diagnosed at the late stages with local infiltration and distant metastases; at which point, surgery is not an effective treatment option. Therefore, identifying novel targets to aid with the early diagnosis of NSCLC is required for improving the efficacy of clinical treatment.
The Warburg effect is a phenomenon that cancer cells rely on to sustain their proliferation by increasing the uptake of glucose and the production of lactate. This phenomenon is a unique energy metabolism present in cancer cells (3). Increasing evidence suggests that enzymes involved in this process might be potential targets for cancer therapy (4). Pyruvate kinase (PK) is a key rate-limiting enzyme involved in tumor glycolysis, which catalyzes the conversion of phosphoenolpyruvic acid and ADP into pyruvate acid and ATP (5). In humans, four isoforms of PK have been identified, including red blood cell PK, liver-type PK, and PK muscle isozyme M1 and M2 (PKM1 and PKM2, respectively) (6). PKM2 has been reported to be upregulated in tumor cells and cancer stem cells, playing a role in glycolysis and tumor malignancy (7). Furthermore, a number of previous studies have indicated that glycolysis is closely related to cell proliferation and metastasis in NSCLC (8–11).
Long non-coding RNA (lncRNA) brain cytoplasmic RNA 1 (BCYRN1) is associated with the progress of numerous diseases (12). In patients with Alzheimer's disease, neurons display high levels of BCYRN1 expression, which regulates cell viability and apoptosis by targeting β-site amyloid precursor protein-cleaving enzyme 1 (13). BCYRN1 also acts as either an oncogene or a tumor suppressor in numerous types of human cancer, including cervical, colorectal, breast and ovarian cancer, as well as esophageal squamous cell carcinoma (14). During NSCLC, BCYRN1 enhances cell migration and invasion by upregulating matrix metalloproteinases (12). BCYRN1 has also been identified as an oncogene in NSCLC (12); however, the mechanisms of action of BCYRN1 are not completely understood.
The aim of the present study was to investigate the role of BCYRN1 in NSCLC glycolysis. BCYRN1 was highly expressed in NSCLC tissues and cells compared with the corresponding controls. The function of BCYRN1 on the Warburg effect in NSCLC cell lines was examined. Mechanistically, the results suggested that BCYRN1 induced cell glycolysis and tumor progression via the microRNA (miRNA/miR)-149/PKM2 signaling pathway.
Materials and methods
Clinical samples
Written informed consent was obtained from patients with NSCLC (male patients, 12 males; female patients, 8; mean age, 37.6 years; age range, 25–48 years) admitted to The Fourth Affiliated Hospital of Harbin Medical University (Harbin, China) between March 2017 and December 2018. Subsequently, 20 primary NSCLC and paired normal tissues were collected, snap-frozen and stored at −80°C. The matched ‘normal tissue’ was obtained from a 5 cm distance from the tumor margin, which were confirmed by a pathologist to not contain tumor cells. The inclusion criteria were as follows: i) Patients diagnosed with lung adenocarcinoma or lung squamous carcinoma by histopathological examination; ii) tumors identified as TNM stage I or IIIa; and iii) patients were subjected to radical operation, and had not received systemic chemotherapy or radiotherapy prior to surgery (15). Patients with autoimmune disease or incomplete case data were excluded. The present study was approved by the Clinical Research Ethics Committee of The Fourth Affiliated Hospital of Harbin Medical University.
Cell lines and transfection
Human NSCLC cell lines (A549, H460 and H1299) and the normal human bronchial epithelial cell line 16HBE were obtained from the American Type Culture Collection and were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2.
miR-149 mimic, pre-negative control (NC), miR-149 inhibitor, NC, small interfering (si)RNA targeting BCYRN1 (si-BCYRN1) or targeting PKM2 (si-PKM2), and si-NC were synthesized by Guangzhou RiboBio Co., Ltd. The corresponding sequences were as follows: miR-149 mimic, 5′-UCUGGCUCCGUGUCUUCACUCCC-3′; pre-NC, 5′-UUCUCCGAACGUGUCACGU-3′; miR-149 inhibitor, 5′-GGGAGUGAAGACACGGAGCCAGA-3′; NC, 5′-CAGUACUUUUGUGUAGUACAA-3′; si-BCYRN1, 5′-CUCCAGAAAAAGGAAAAAAAAAA-3′; si-PKM2, 5′-GGAAAGAACAUCAAGAUUAUC-3′; si-NC, 5′-UUCUCCGAACGUGUCACGU-3′. Plasmid pcDNA-BCYRN1 or pcDNA-PKM2 were constructed by Shanghai GenePharma Co., Ltd. for the overexpression of BCYRN1 or PKM2 in cells. A459 or H1299 cells (2×105) were transfected with 50 nM plasmid or 50 nM RNAs using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. After a 48-h incubation at 37°C, cells were harvested for subsequent experiments. An empty plasmid was used as the negative control.
Cell Counting kit-8 (CCK-8) assay
Transfected A549 cells (1×104 cells/well) were seeded in 96-well plates. After a 48-h culture, CCK-8 solution (Dojindo Molecular Technologies, Inc.) (10 µl) was added into each well and incubated for 4 h at 37°C. The absorbance was measured at a wavelength of 450 nm by a microplate reader to analyze cell proliferation. The cell survival rate=[(Aexperiment group-Ablank)/(Acontrol group-Ablank)] ×100%
Matrigel invasion assay
Transfected A549 cells (2.5×103 cells/well) in serum-free DMEM were seeded into the upper chamber of Transwell inserts pre-coated with Matrigel. DMEM supplemented with 10% FBS was plated in the lower chamber. After incubation for 24 h at 37°C, the invasive cells were stained with 0.3% crystal violet at room temperature for 20 min. Subsequently, the number of invading cells in the lower chamber were counted using an inverted fluorescence microscope IX71 (magnification, ×100).
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from the cells or tissues using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. After quantification using a spectrophotometer, total RNA was reverse transcribed into cDNA using the PrimeScript RT Reagent kit (Takara Bio, Inc.), according to the manufacturer's protocol. The conditions for RT were as follows: 70°C for 3 min, 42°C for 60 min and 70°C for 15 min. Subsequently, qPCR was performed using the SYBR Green kit (Takara Bio, Inc.) on an ABI 7900 system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The primer pairs that were used for qPCR are presented in Table I. The following thermocycling conditions were used for qPCR: Initial denaturation at 95°C for 30 sec, followed by 40 cycles at 95°C for 5 sec and at 60°C for 45 sec. Relative expression levels were quantified using the 2−ΔΔCq method (16) and GAPDH or U6 was used as the internal reference gene for mRNA and miRNA, respectively.
Table I.
Primer sequences used for reverse transcription--quantitative PCR.
| Gene | Primer sequences (5′-3′) |
|---|---|
| BCYRN1 | F: GCCTGTAATCCCAGCTCTCA |
| R: GGGTTGTTGCTTTGAGGGAA | |
| microRNA-149 | F: AGCGCGUCUGGCUCCGUGUCUUC |
| R: ATCCAGTGCAGGGTCCGAGG | |
| PKM2 | F: GCACACCGTATTCAGCTCTG |
| R: TCCAGGAATGTGTCAGCCAT | |
| GAPDH | F: ACCACAGTCCATGCCATCAC |
| R: TCCACCACCCTGTTGCTGTA | |
| U6 | F: GCTTCGGCAGCACATATACTAAAAT |
| R: TACTGTGCGTTTAAGCACTTCGC |
BCYRN1, brain cytoplasmic RNA 1; F, forward; PKM2, pyruvate kinase M1/2; R, reverse.
Western blotting
Total protein was extracted from A549 and H1299 cells using the IP cell lysis buffer (Pierce; Thermo Fisher Scientific, Inc.). Bicinchoninic acid protein assay kit (Takara Biotechnology Co., Ltd.) was used to quantify protein concentration. Proteins (30 µg/lane) were separated by SDS-PAGE on 10% gels. Subsequently, the separated proteins were transferred to PVDF membranes (EMD Millipore) and blocked in 5% milk at 37°C for 2 h. The membranes were incubated overnight at 4°C with the primary antibodies targeted against: PKM2 (1:1,000; cat. no. ab137852; Abcam) and β-actin (1:2,000; cat. no. ab8227; Abcam). Following the primary antibody incubation, the membranes were incubated with a horseradish peroxidase conjugated secondary antibody (1:2,000; cat. no. ab7090; Abcam) for 2 h at room temperature. Protein bands were visualized using the enhanced chemiluminescence reagent (EMD Millipore).
Glucose consumption and lactate production assay
Following transfection, the culture media and the cells were collected separately. A Glucose assay kit (Sigma-Aldrich; cat. no. GAHK20; Merck KGaA) was used to measure the glucose levels in the culture media according to the manufacturer's protocol. Lactate levels in the culture media were determined using a Lactic Acid assay kit (Biovision, Inc.; cat. no. K606-100), according to the manufacturer's protocol.
Dual-luciferase reporter assay
The potential miRNAs that bind to BCYRN1 were predicted using DIANA (diana.imis.athena-innovation.gr/DianaTools/index.php) and the potential binding site between miR-149 and PKM2 was predicted using Microrna (http://www.microrna.org/microrna/home.do). To investigate the interaction between BCYRN1 and miR-149, and between miR-149 and PKM2, the wild-type (WT) or mutant (MUT) BCYRN1 sequences and 3′-untranslated regions (3′-UTRs) of wild-type (WT) or mutant (MUT) PKM2, were synthesized by Guangzhou RiboBio Co., Ltd. H1299 cells (2×105) were co-transfected with the firefly luciferase reporter plasmid pmirGLO (Promega Corporation) and 50 nM miR-149 mimic or pre-NC using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. After a 48-h incubation at room temperature, the luciferase activity was detected using a Dual-Luciferase Reporter assay system (Promega Corporation), according to the manufacturer's protocol. Firefly luciferase activity was normalized to Renilla luciferase activity.
Statistical analysis
Statistical analyses were performed using SPSS software (version 23; IBM Corp.). Data are presented as the mean ± SD from three replicates. A paired Student's t-test was used to assess statistical differences for the BCYRN1 express level between tumor and adjacent healthy tissues. An unpaired Student's t-test or one-way ANOVA followed by Tukey's post hoc test was used to assess statistical differences between two groups or multiple groups, respectively. P<0.05 was considered to indicate a statistically significant difference.
Results
BCYRN1 induces glycolysis and increases the expression levels of PKM2 in NSCLC cells
The expression of BCYRN1 in NSCLC tissues was detected by RT-qPCR and the results suggested that BCYRN1 expression was significantly higher in tumor tissues compared with adjacent normal tissues (Fig. 1A). Additionally, BCYRN1 was significantly upregulated in a number of NSCLC cell lines, including A549, H460 and H1299 cells, compared with 16HBE cells (Fig. 1B). The expression of BCYRN1 was significantly reduced by 57% in A549 cells and increased by 3.85-fold in H1299 cells following transfection with si-BCYRN1 or pcDNA-BCYRN1, respectively, compared with the corresponding control (Fig. 1C). Glucose consumption and lactate production assays were performed to examine the effects of BCYRN1 on glycolysis. BCYRN1 knockdown significantly decreased glucose consumption by 60% and lactate production by 49% in A549 cells, and BCYRN1 overexpression significantly increased glucose consumption and lactate production in H1299 cells, compared with the corresponding controls (Fig. 1D and E). The mRNA and protein expression levels of PKM2 were downregulated in A549 cells following BCYRN1 knockdown, and conversely, BCYRN1 overexpression upregulated the expression levels of PKM2 in H1299 cells, compared with the corresponding controls (Fig. 1F and G).
Figure 1.
BCYRN1 induces glycolysis and increases the expression levels of PKM2 in NSCLC cells. Expression of BCYRN1 in NSCLC (A) tissues and (B) cell lines. (C) Expression of BCYRN1 in NSCLC cells transfected with si-BCYRN1 or pcDNA-BCYRN1. (D) Glucose consumption and (E) lactate production in NSCLC cells transfected with si-BCYRN1 or pcDNA-BCYRN1. PKM2 (F) mRNA and (G) protein expression levels in NSCLC cells transfected with si-BCYRN1 or pcDNA-BCYRN1. *P<0.05 vs. the corresponding control. BCYRN1, brain cytoplasmic RNA 1; NC, negative control; NSCLC, non-small-cell lung cancer; si, small interfering RNA; PKM2, pyruvate kinase M1/2.
BCYRN1 regulates miR-149 expression levels during glycolysis
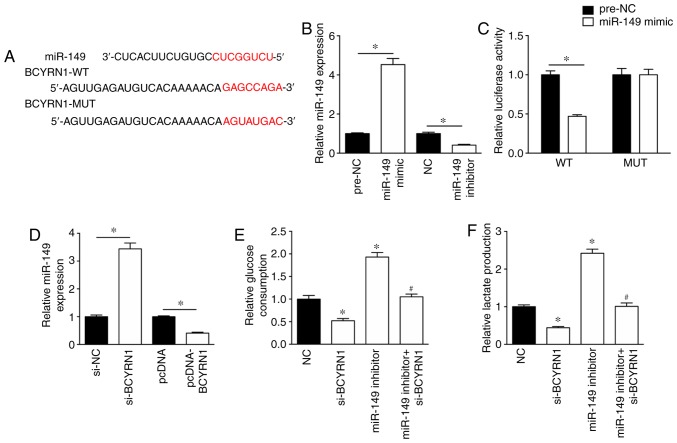
To investigate the mechanism underlying the role of BCYRN1 during glycolysis, the potential miRNAs that bind to BCYRN1 were predicted using DIANA (diana.imis.athena-innovation.gr/DianaTools/index.php). In silico results suggested that the sequence of BCYRN1 contained a possible miR-149 binding site (Fig. 2A). The miR-149 mimic significantly enhanced the expression levels of miR-149 in A549 cells by 4.53-fold, whereas the expression levels of miR-149 were significantly reduced by 41% in A549 cells transfected with miR-149 inhibitor, compared with the corresponding controls (Fig. 2B). In addition, the miR-149 mimic inhibited the luciferase activity of the BCYRN1-WT luciferase plasmid, but had no effect on the luciferase activity of the BCYRN1-MUT luciferase plasmid in A549 cells (Fig. 2C). RT-qPCR was performed to assess the effect of BCYRN1 on miR-149 expression. The results suggested that miR-149 expression levels were significantly increased by 3.44-fold in A549 cells transfected with si-BCYRN1 compared with cells transfected with si-NC (Fig. 2D). Additionally, miR-149 expression levels were significantly downregulated in A549 cells following BCYRN1 overexpression compared with cells transfected with the empty pcDNA vector (Fig. 2D). The results of the glucose consumption and lactate production assays suggested that the miR-149 inhibitor rescued si-BCYRN1-induced inhibition of glucose consumption and lactate production, compared with the si-BCYRN1 (Fig. 2E and F).
Figure 2.
miR-149 is involved in BCYRN1-regulated glycolysis. (A) Predicted binding site between BCYRN1 and miR-149. (B) miR-149 expression levels following transfection with miR-149 mimic or miR-149 inhibitor. (C) Luciferase assay suggested a relationship between BCYRN1 and miR-149 expression. (D) miR-149 expression levels were regulated by BCYRN1. miR-149 knockdown rescued the si-BCYRN1-induced decrease in (E) glucose consumption and (F) lactate production compared with the si-BCYRN1 group. *P<0.05 vs. si-NC or NC; #P<0.05 vs. si-BCYRN1. BCYRN1, brain cytoplasmic RNA 1; miR, microRNA; NC, negative control; si, small interfering RNA.
miR-149 regulates the expression of PKM2
The potential mechanism underlying the effects of BCYRN1 and miR-149 in A549 cells was further investigated. miR-149 was predicted to target the 3′-UTR of PKM2 (Fig. 3A). Notably, miR-149 overexpression significantly inhibited the luciferase activity of the PKM2-WT plasmid, but had no effect on the luciferase activity of the PKM2-MUT plasmid (Fig. 3A). miR-149 knockdown significantly upregulated the expression levels of PKM2 and rescued the decreased expression levels of PKM2 induced by si-BCYRN1 (Fig. 3B and C). To further explore the role of PKM2 in BCYRN1/miR-149 function, si-PKM2 was used to inhibit the expression of PKM2 and pcDNA-PKM2 was used to overexpress PKM2 in A549 cells (Fig. 3D). PKM2 knockdown significantly reversed the effects of the miR-149 inhibitor on glucose consumption and lactate production (Fig. 3E and F).
Figure 3.
miR-149 regulates the expression of PKM2. (A) Binding site between miR-149 and 3′-untranslated region of PKM2 was predicted and a luciferase assay was performed. miR-149 knockdown rescued the si-BCYRN1-induced decrease in PKM2 (B) mRNA and (C) protein expression levels. (D) si-PKM2 decreased PKM2 expression levels and pcDNA-PKM2 increased PKM2 expression levels. si-PKM2 reversed the miR-149 inhibitor-induced increase in (E) glucose consumption and (F) lactate production. *P<0.05 vs. si-NC or NC; #P<0.05 vs. si-BCYRN1; &P<0.05 vs. si-BCYRN1 + miR-149 inhibitor. miR, microRNA; NC, negative control; PKM2, pyruvate kinase M1/2; si, small interfering RNA.
PKM2 is associated with the function of BCYRN1 in A549 cells
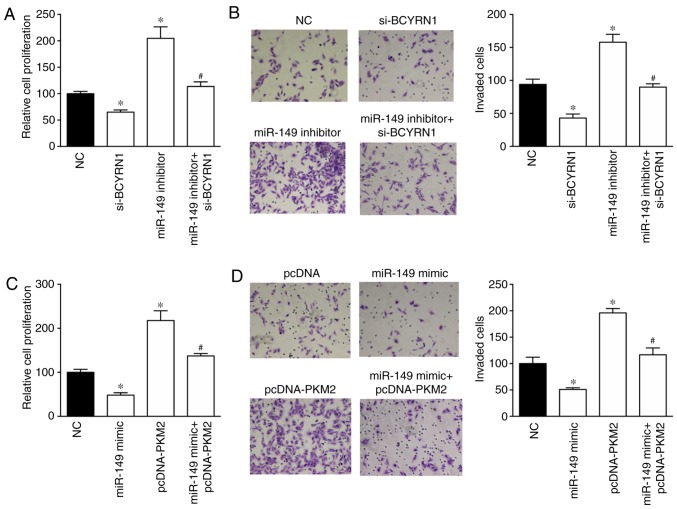
The effects of the BCYRN1/miR-149/PKM2 regulatory pathway on NSCLC cell proliferation and invasion were investigated using the CCK-8 and invasion assays. Cell proliferation and invasion were inhibited by BCYRN1 knockdown and enhanced by the miR-149 inhibitor in A549 cells (Fig. 4A and B). Notably, the miR-149 inhibitor rescued the si-BCYRN1-induced inhibition of cell proliferation and invasion. Furthermore, PKM2 overexpression induced a 2.17- and 1.96-fold increase in proliferation and invasion in A549 cells compared with NC group, respectively. In addition, PKM2 overexpression reversed the miR-149 mimic-induced inhibition of cell proliferation and invasion (Fig. 4C and D).
Figure 4.
PKM2 is involved in the biological effects of BCYRN1 in NSCLC cells. miR-149 knockdown rescued the si-BCYRN1-induced decrease in cell (A) proliferation and (B) invasion. Magnification, ×100. pcDNA-PKM2 reversed the miR-149 mimic-induced decrease in cell (C) proliferation and (D) invasion. Magnification, ×100. *P<0.05 vs. NC; #P<0.05 vs. si-BCYRN1 or miR-149 mimic. BCYRN1, brain cytoplasmic RNA 1; miR, microRNA; NC, negative control; NSCLC, non-small-cell lung cancer; PKM2, pyruvate kinase M1/2; si, small interfering RNA.
Discussion
Aerobic glycolysis, as one of the ‘hallmarks of cancer’, converts glucose into pyruvate, which is then further oxidized into lactate (17). The resulting lactate contributes to destruction of the extracellular matrix and provides a metastatic advantage for cancer cells, leading to rapid proliferation and resistance to apoptosis (18). Aerobic glycolysis during cancer has attracted increasing attention in recent years, as it has been suggested that targeting glycolysis might be a promising treatment for cancer (19–21). Emerging data suggests that glycolysis could be regulated by certain lncRNAs in cancer. For example, Li et al (22) reported that urothelial cancer-associated 1 was involved in the proliferation and glycolysis of bladder cancer cells via the induction of hexokinase 2 (HK2) expression. c-Myc-activated lncRNA macrophage migration inhibitory factor inhibited aerobic glycolysis and tumorigenesis in cancer cells by downregulating c-Myc and upregulating F-box and WD repeat domain containing 7 expression (23). Furthermore, Pvt1 oncogene has been reported to promote osteosarcoma cell glucose metabolism, cell proliferation and motility via the miR-497/HK2 signaling pathway (24).
In the present study, increased glucose consumption and lactate production was observed in NSCLC cells overexpressing BCYRN1. Additionally, higher expression levels of BCYRN1 were observed in NSCLC tissues and cell lines compared with normal control tissues and cell lines. Consistent with these results, Hu and Lu (12) also reported that BCYRN1 acted as an oncogene in NSCLC as evidenced by the high cell proliferation and metastasis induced by BCYRN1. BCYRN1 has been reported to function as an oncogene in colorectal and cervical cancer, as well as in gastric carcinoma (25–27). Mechanistically, BCYRN1 may regulate gene expression by targeting miRNAs. For example, BCYRN1 targeted miR-138 or miR-150 to alter the proliferation and metastasis of cervical cancer cells or airway smooth muscle cells, respectively (25,28).
To further investigate the molecular mechanisms underlying the effects of BCYRN1 on NSCLC, bioinformatics analysis was performed to identify the potential targets of BCYRN1 and suggested that miR-149 might be a target of BCYRN1. The results of the luciferase activity assay further suggested that BCYRN1 interacted with miR-149 and regulated the expression of miR-149 in NSCLC cells. In addition, the miR-149 inhibitor rescued the si-BCYRN1-induced effects on glucose consumption, lactate production and cell proliferation and invasion. miR-149 has been identified as a tumor suppressor in several types of cancer, including gastric cancer, and renal cell and colonic carcinoma (29–31). Zhao et al (32) reported that miR-149 decreased human NSCLC growth and metastasis by inhibiting the forkhead box M1/cyclin D1/matrix metallopeptidase 2 axis. Furthermore, in cervical cancer cells, miR-149 regulated proliferation and apoptosis via regulating GIT ArfGAP1 (33).
The coding gene downstream of the BCYRN1/miR-149 axis was further investigated in NSCLC cells. Finally, PKM2 was identified as a potential target of miR-149 and was upregulated by the miR-149 inhibitor. PKM2, as the critical isoenzyme of PK, serves a role in maintaining the metabolic program of cancer cells (34). Inhibition of PKM2 expression was previously reported to decrease glucose consumption and lactate production (35), and elevated expression of PKM2 has been reported in colon and lung cancer (36,37). Previous studies have suggested that PKM2 contributes to cancer metabolism by promoting hypoxia-inducible factor-1 expression or coordinating with c-Myc (38,39). In the present study, PKM2 knockdown significantly reversed the miR-149 inhibitor-induced effects on glucose consumption and lactate production. PKM2 overexpression also reversed the miR-149 mimic-induced reduction of cell proliferation and invasion.
Collectively, the present study suggested that BCYRN1 participated in glycolysis during NSCLC via regulating the miR-149/PKM2 axis, and accelerating cell proliferation and invasion. The results suggested that the BCYRN1/miR-149/PKM2 signaling pathway was related to regulation of the Warburg effect and therefore might be a potential therapeutic target for NSCLC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
NL, CW and JZ designed the study. NL performed the experiments and wrote the paper. CW performed the statistical analyses. FS, TW and HAC performed the cell experiments.
Ethics approval and consent to participate
The present study was approved by the Clinical Research Ethics Committee of the Fourth Affiliated Hospital of Harbin Medical University. All patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S, Shafi S, Johnson DH, Mitter R, Rosenthal R, et al. Tracking the evolution of non-small-cell lung cancer. N Engl J Med. 2017;376:2109–2121. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 2.Detterbeck FC, Boffa DJ, Kim AW, Tanoue LT. The eighth edition lung cancer stage classification. Chest. 2017;151:193–203. doi: 10.1016/j.chest.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 3.Schwartz L, Supuran CT, Alfarouk KO. The Warburg effect and the hallmarks of cancer. Anticancer Agents Med Chem. 2017;17:164–170. doi: 10.2174/1871520616666161031143301. [DOI] [PubMed] [Google Scholar]
- 4.Ngo DC, Ververis K, Tortorella SM, Karagiannis TC. Introduction to the molecular basis of cancer metabolism and the Warburg effect. Mol Biol Rep. 2015;42:819–823. doi: 10.1007/s11033-015-3857-y. [DOI] [PubMed] [Google Scholar]
- 5.Allen AE, Locasale JW. Glucose metabolism in cancer: The saga of pyruvate kinase continues. Cancer Cell. 2018;33:337–339. doi: 10.1016/j.ccell.2018.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Israelsen WJ, Vander Heiden MG. Pyruvate kinase: Function, regulation and role in cancer. Semin Cell Dev Biol. 2015;43:43–51. doi: 10.1016/j.semcdb.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu J, Chen M, Gao S, Yuan J, Zhu Z, Zou X. LY294002 inhibits the Warburg effect in gastric cancer cells by downregulating pyruvate kinase M2. Oncol Lett. 2018;15:4358–4364. doi: 10.3892/ol.2018.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J, Cheng D, Zhu M, Yu H, Pan Z, Liu L, Geng Q, Pan H, Yan M, Yao M. OTUB2 stabilizes U2AF2 to promote the Warburg effect and tumorigenesis via the AKT/mTOR signaling pathway in non-small cell lung cancer. Theranostics. 2019;9:179–195. doi: 10.7150/thno.29545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu QF, Liu Y, Fan Y, Hua SN, Qu HY, Dong SW, Li RL, Zhao MY, Zhen Y, Yu XL, et al. Alpha-enolase promotes cell glycolysis, growth, migration, and invasion in non-small cell lung cancer through FAK-mediated PI3K/AKT pathway. J Hematol Oncol. 2015;8:22. doi: 10.1186/s13045-015-0117-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao W, Li W, Dai W, Huang N, Qiu J. LINK-A promotes cell proliferation through the regulation of aerobic glycolysis in non-small-cell lung cancer. Onco Targets Ther. 2018;11:6071–6080. doi: 10.2147/OTT.S171216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen H, Zhang M, Zhang W, Li Y, Zhu J, Zhang X, Zhao L, Zhu S, Chen B. Downregulation of BarH-like homeobox 2 promotes cell proliferation, migration and aerobic glycolysis through Wnt/β-catenin signaling, and predicts a poor prognosis in non-small cell lung carcinoma. Thorac Cancer. 2018;9:390–399. doi: 10.1111/1759-7714.12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu T, Lu YR. BCYRN1, a c-MYC-activated long non-coding RNA, regulates cell metastasis of non-small-cell lung cancer. Cancer Cell Int. 2015;15:36. doi: 10.1186/s12935-015-0183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, Zheng L, Jiang A, Mo Y, Gong Q. Identification of the biological affection of long noncoding RNA BC200 in Alzheimer's disease. Neuroreport. 2018;29:1061–1067. doi: 10.1097/WNR.0000000000001057. [DOI] [PubMed] [Google Scholar]
- 14.Booy EP, McRae EK, Koul A, Lin F, McKenna SA. The long non-coding RNA BC200 (BCYRN1) is critical for cancer cell survival and proliferation. Mol Cancer. 2017;16:109. doi: 10.1186/s12943-017-0679-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edge SB, Compton CC. The American Joint Committee on Cancer: The 7th Edition of the AJCC Cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Lim SO, Li CW, Xia W, Lee HH, Chang SS, Shen J, Hsu JL, Raftery D, Djukovic D, Gu H, et al. EGFR signaling enhances aerobic glycolysis in triple-negative breast cancer cells to promote tumor growth and immune escape. Cancer Res. 2016;76:1284–1296. doi: 10.1158/0008-5472.CAN-15-2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kong XZ, Hu SS, Sun Z, Zuo LH, Kang J, Zhu ZF, Tian X, Zhang XJ. Regulation of aerobic glycolysis by long non-coding RNAs in cancer. Biochem Biophys Res Commun. 2016;479:28–32. doi: 10.1016/j.bbrc.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 19.Goncalves MD, Cantley LC. A glycolysis outsider steps into the cancer spotlight. Cell Metab. 2018;28:3–4. doi: 10.1016/j.cmet.2018.06.017. [DOI] [PubMed] [Google Scholar]
- 20.Chen R, Zhu S, Fan XG, Wang H, Lotze MT, Zeh HJ, III, Billiar TR, Kang R, Tang D. High mobility group protein B1 controls liver cancer initiation through yes-associated protein-dependent aerobic glycolysis. Hepatology. 2018;67:1823–1841. doi: 10.1002/hep.29663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Lu JH, Wu QN, Jin Y, Wang DS, Chen YX, Liu J, Luo XJ, Meng Q, Pu HY, et al. LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Mol Cancer. 2019;18:174. doi: 10.1186/s12943-019-1105-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li T, Sun X, Jiang X. UCA1 involved in the metformin-regulated bladder cancer cell proliferation and glycolysis. Tumour Biol. 2017;39:1010428317710823. doi: 10.1177/1010428317710823. [DOI] [PubMed] [Google Scholar]
- 23.Zhang P, Cao L, Fan P, Mei Y, Wu M. LncRNA-MIF, a c-Myc-activated long non-coding RNA, suppresses glycolysis by promoting Fbxw7-mediated c-Myc degradation. EMBO Rep. 2016;17:1204–1220. doi: 10.15252/embr.201642067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song J, Wu X, Liu F, Li M, Sun Y, Wang Y, Wang C, Zhu K, Jia X, Wang B, Ma X. Long non-coding RNA PVT1 promotes glycolysis and tumor progression by regulating miR-497/HK2 axis in osteosarcoma. Biochem Biophys Res Commun. 2017;490:217–224. doi: 10.1016/j.bbrc.2017.06.024. [DOI] [PubMed] [Google Scholar]
- 25.Peng J, Hou F, Feng J, Xu SX, Men XY. Long non-coding RNA BCYRN1 promotes the proliferation and metastasis of cervical cancer via targeting microRNA-138 in vitro and in vivo. Oncol Lett. 2018;15:5809–5818. doi: 10.3892/ol.2018.8015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu L, Lu L, Zhou D, Liu Z. Long noncoding RNA BCYRN1 promotes the proliferation of colorectal cancer cells via Up-regulating NPR3 expression. Cell Physiol Biochem. 2018;48:2337–2349. doi: 10.1159/000492649. [DOI] [PubMed] [Google Scholar]
- 27.Ren H, Yang X, Yang Y, Zhang X, Zhao R, Wei R, Zhang X, Zhang Y. Upregulation of LncRNA BCYRN1 promotes tumor progression and enhances EpCAM expression in gastric carcinoma. Oncotarget. 2017;9:4851–4861. doi: 10.18632/oncotarget.23585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang XY, Tang XY, Ma LJ, Guo YL, Li XS, Zhao LM, Tian CJ, Cheng DJ, Chen ZC, Zhang LX. Schisandrin B down-regulated lncRNA BCYRN1 expression of airway smooth muscle cells by improving miR-150 expression to inhibit the proliferation and migration of ASMC in asthmatic rats. Cell Prolif. 2017;50:e12382. doi: 10.1111/cpr.12382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin L, Li Y, Liu J, Yang S, Gui Y, Mao X, Nie G, Lai Y. Tumor suppressor miR-149-5p is associated with cellular migration, proliferation and apoptosis in renal cell carcinoma. Mol Med Rep. 2016;13:5386–5392. doi: 10.3892/mmr.2016.5205. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y, Zheng X, Zhang Z, Zhou J, Zhao G, Yang J, Xia L, Wang R, Cai X, Hu H, et al. MicroRNA-149 inhibits proliferation and cell cycle progression through the targeting of ZBTB2 in human gastric cancer. PLoS One. 2012;7:e41693. doi: 10.1371/journal.pone.0041693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang F, Ma YL, Zhang P, Shen TY, Shi CZ, Yang YZ, Moyer MP, Zhang HZ, Chen HQ, Liang Y, Qin HL. SP1 mediates the link between methylation of the tumour suppressor miR-149 and outcome in colorectal cancer. J Pathol. 2013;229:12–24. doi: 10.1002/path.4078. [DOI] [PubMed] [Google Scholar]
- 32.Zhao L, Liu L, Dong Z, Xiong J. miR-149 suppresses human non-small cell lung cancer growth and metastasis by inhibiting the FOXM1/cyclin D1/MMP2 axis. Oncol Rep. 2017;38:3522–3530. doi: 10.3892/or.2017.6047. [DOI] [PubMed] [Google Scholar]
- 33.Qian B, Zhao L, Wang X, Xu J, Teng F, Gao L, Shen R. RETRACTED: miR-149 regulates the proliferation and apoptosis of cervical cancer cells by targeting GIT1. Biomed Pharmacother. 2018;105:1106–1116. doi: 10.1016/j.biopha.2018.06.075. [DOI] [PubMed] [Google Scholar]
- 34.Dayton TL, Jacks T, Vander Heiden MG. PKM2, cancer metabolism, and the road ahead. EMBO Rep. 2016;17:1721–1730. doi: 10.15252/embr.201643300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong G, Mao Q, Xia W, Xu Y, Wang J, Xu L, Jiang F. PKM2 and cancer: The function of PKM2 beyond glycolysis. Oncol Lett. 2016;11:1980–1986. doi: 10.3892/ol.2016.4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Papadaki C, Sfakianaki M, Lagoudaki E, Giagkas G, Ioannidis G, Trypaki M, Tsakalaki E, Voutsina A, Koutsopoulos A, Mavroudis D, et al. PKM2 as a biomarker for chemosensitivity to front-line platinum-based chemotherapy in patients with metastatic non-small-cell lung cancer. Br J Cancer. 2014;111:1757–1764. doi: 10.1038/bjc.2014.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He J, Xie G, Tong J, Peng Y, Huang H, Li J, Wang N, Liang H. Overexpression of microRNA-122 re-sensitizes 5-FU-resistant colon cancer cells to 5-FU through the inhibition of PKM2 in vitro and in vivo. Cell Biochem Biophys. 2014;70:1343–1350. doi: 10.1007/s12013-014-0062-x. [DOI] [PubMed] [Google Scholar]
- 38.Wang HJ, Hsieh YJ, Cheng WC, Lin CP, Lin YS, Yang SF, Chen CC, Izumiya Y, Yu JS, Kung HJ, Wang WC. JMJD5 regulates PKM2 nuclear translocation and reprograms HIF-1α-mediated glucose metabolism. Proc Natl Acad Sci USA. 2014;111:279–284. doi: 10.1073/pnas.1311249111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luan W, Wang Y, Chen X, Shi Y, Wang J, Zhang J, Qian J, Li R, Tao T, Wei W, et al. PKM2 promotes glucose metabolism and cell growth in gliomas through a mechanism involving a let-7a/c-Myc/hnRNPA1 feedback loop. Oncotarget. 2015;6:13006–13018. doi: 10.18632/oncotarget.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.