Abstract
Non-small cell lung cancer (NSCLC) is the most common type of lung cancer and the most common cause of mortality in patients with lung cancer. The efficacy of cisplatin-based chemotherapy in NSCLC is limited by drug resistance, therefore, the development of novel anticancer agents is required to overcome cisplatin resistance. The present study investigated the anticancer activity of gambogenic acid (GNA), derived from gamboge, in the cisplatin-resistant NSCLC cell line A549/Cis. GNA was revealed to have a potent inhibitory effect on cell growth in A549/Cis cells by blocking the cell cycle and inducing apoptosis. The investigation of the molecular mechanisms identified that GNA arrested the cell cycle at the G1 phase through the downregulation of cyclin Ds, cyclin dependent kinase (CDK)4 and CDK6, and the upregulation of p53 and p21. In addition, GNA induced apoptosis by increasing the activation of caspase 3 and caspase 7, in addition to the cleavage of poly(ADP-ribose) polymerase. The results of the present study supported the potential application of GNA in cisplatin-resistant NSCLC.
Keywords: gambogenic acid, A549/Cis cells, cell cycle arrest, apoptosis, RNA sequencing
Introduction
Lung cancer is the most commonly diagnosed cancer (11.6% of the total cases) and the leading cause of cancer-associated mortality (18.4% of the total cancer-associated mortality cases) globally (1). Non-small cell lung cancer (NSCLC) accounts for ~83% of lung cancer cases, and the majority of patients with advanced NSCLC are treated with chemotherapy (2). Platinum-based drugs, particularly cisplatin (Cis), are used in the treatment of numerous cancer types, including NSCLC (3). Cis is the most widely used drug in cancer therapy and the first Food and Drug Administration-approved platinum compound for lung cancer treatment (4,5). However, the use of Cis in lung cancer chemotherapy is limited by the resistance of cells to the drug (6–8). In order to enhance the therapeutic efficacy of currently available cytotoxic drugs and to identify novel anti-tumor drugs, it is important to identify alternative strategies to overcome Cis resistance and identify novel therapies.
Natural products are promising sources of anticancer agents and contribute substantially to cancer therapy (9). Gambogenic acid (GNA), a natural compound derived from gamboge, has long been used in traditional Chinese medicine (10). GNA exerts cytotoxicity in numerous human cancer lines (11–16). In addition, previous studies have demonstrated that GNA may also enhance chemosensitivity in certain cancer cell types (17,18). However, to the best of our knowledge, it is not known whether GNA serves a function in the treatment of Cis-resistant lung cancer. The chemical structure of GNA has an active bond (19), which may bind with the target protein, but the detailed molecular mechanism underlying its anticancer effects have not yet been fully elucidated. The present study investigated the anticancer effects and mechanisms of GNA on Cis-resistant NSCLC cells. The present results suggested that GNA efficiently inhibited the proliferation of Cis-resistant NSCLC cells in vitro, highlighting the potential application of GNA in NSCLC clinical studies.
Materials and methods
Cell culture
The human NSCLC cell line A549 and A549/Cis cells were obtained from The Cell Resource Center, Institute of Basic Medicine, Chinese Academy of Medical Sciences. The cells were cultured in DMEM (Corning Inc.), supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.). All cells were cultured at 37°C and 5% CO2. A549/Cis cells were cultured in the presence of 2 µM Cis (Selleck Chemicals), which were collected subsequent to two generations prior to the experiments being performed. These cell lines grew in monolayers and when the confluence reached 70–80%, they were passaged.
MTT assay
Briefly, A549 and A549/Cis cells (3×103 cells/well) were added to 96-well plates in 100 µl cell culture medium. The cells were treated with various concentrations of Cis (0, 2.5, 5, 10 or 20 µM) or GNA (0, 0.5, 1, 2, 4 or 6 µM; Fig. 1; purity >98%; Nanjing Dasf Biotechnology Co., Ltd.). GNA has poor aqueous solubility, and was dissolved in DMSO as the stock solution at a concentration of 20 mM, and then diluted to working concentration as required in complete culture medium immediately prior to use. The cells were then incubated at 37°C and 5% CO2 for 24, 48 or 72 h. Following the treatments, 20 µl/well MTT (Biosharp; 5 mg/ml) was added for further incubation at 37°C for 4 h. The supernatant was then discarded and the formazan product was dissolved in DMSO. The optical density was measured at 490 nm in a Synergy H1 Hybrid Multi-Mode Microplate Reader (BioTeke Coporation) and the experimental results were recorded.
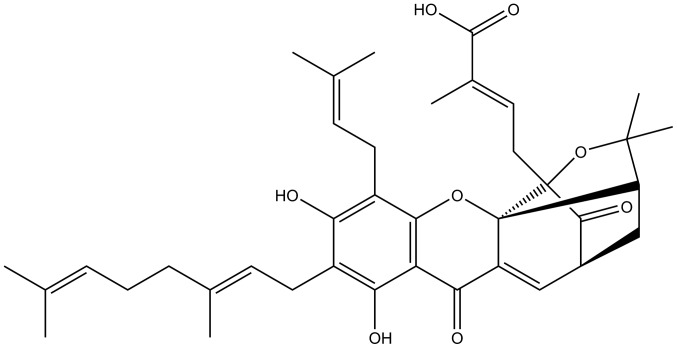
Figure 1.
Chemical structure of gambogenic acid.
Hoechst 33342 staining analysis
Briefly, A549/Cis cells (5×104 cells/well) were seeded into 12-well plates and cultured at 37°C for 24 h. Prior to GNA treatment, cells were observed by microscopy and recorded as the initial point. Then, the cells were treated with GNA (0, 2 or 4 µM) at 37°C for 24 and 48 h. At the end of the various treatments, the cells were stained with Hoechst 33342 (10 µg/ml; Beijing Solarbio Science & Technology Co., Ltd.) for 20 min at 37°C and changes in the cell morphology were observed by fluorescence microscopy (magnification, ×200; Leica Microsystems GmbH).
Analysis of cell cycle and apoptosis
The cell cycle and apoptosis were analyzed by flow cytometry. For the cell cycle analysis, cells were harvested and washed twice with cold phosphate buffered saline (PBS), then fixed in precooled 70% alcohol overnight at −20°C. Subsequent to washing in cold PBS 3 times, cells were incubated with RNase (Tiangen Biotech Co., Ltd.; 0.5 mg/ml) solution at 37°C for 1 h. Subsequently, propidium iodide (Beijing Solarbio Science & Technology Co., Ltd.; 10 µg/ml) was added to the cells and incubated at room temperature for 15 min. Then, the cell cycle analysis was performed using a BD FACSCelesta Flow Cytometer (BD Biosciences). The data were analyzed using Modifit LT 5.0 software (Verity Software House, Inc.). Cell apoptosis was analyzed using an Annexin V-APC/7-amino-actinomycin (7-AAD) Apoptosis Detection kit (BioLegend, Inc.), according to the manufacturer's protocol. The stained cells were analyzed using a BD FACSCelesta Flow Cytometer (BD Biosciences) and the data were analyzed using FlowJo 7.6.1 software (FlowJo LLC).
RNA sequencing (RNA-seq)
The total RNA of A549/Cis cells treated with 4 µM GNA for 24 h was extracted using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer's protocol. The RNA-seq was performed by LC-Bio Technologies (Hangzhou) Co., Ltd. using an Illumina X10 (LC Sciences) following the manufacturer's protocol. StringTie (20) was used to analyze the expression levels for mRNAs by calculating the fragments per kilobase of transcript per million mapped reads. The differentially expressed mRNAs and genes were selected with log2 (fold change)>1 or log2 (fold change)<-1 and with statistical significance (P-value <0.05) using the R package - Ballgown, as described previously (21). For the Gene Ontology enrichment (22,23) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (24–26) pathway analysis, the Database for Annotation, Visualization and Integrated Discovery web server (http://david.ncifcrf.gov/) was used (27,28).
RNA extraction and reverse transcription-quantitative PCR (RT-qPCR)
A549/Cis cells were harvested 24 h after 4 µM GNA treatment and the total RNA was extracted using the GeneJET RNA Purification kit (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. RT was conducted using the First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.). The following RT temperature protocol was used: 65°C for 5 min, 50°C for 50 min and 85°C for 5 min. qPCR was subsequently performed using the SYBR® Select Master Mix kit (Thermo Fisher Scientific, Inc.) and a QuantStudio 3 Real-Time PCR system (Thermo Fisher Scientific, Inc.). The thermocycling conditions were as follows: 95°C for 10 min, followed by 40 cycles at 95°C for 15 sec and 60°C for 1 min. The primer sequences used for the qPCR are listed in Table I. Quantification was performed using the comparative 2−∆∆Cq method (29). The Cq value for each sample was normalized to the value of GAPDH. The experiments were performed in triplicate.
Table I.
Primer pairs for reverse transcription-quantitative PCR.
| Gene | Primer sequence (5′→3′) |
|---|---|
| GAPDH | F: ACATCGCTCAGACACCATG |
| R: TGTAGTTGAGGTCAATGAAGGG | |
| GADD45A | F: GGAGAGCAGAAGACCGAAAG |
| R: AGGCACAACACCACGTTATC | |
| CCND3 | F: AACTGTGCATCTACACCGAC |
| R: GCCAGGAAATCATGTGCAATC | |
| CCNB1 | F: GGCTTTCTCTGATGTAATTCTTGC |
| R: GTATTTTGGTCTGACTGCTTGC | |
| CDC20 | F: GATGTAGAGGAAGCCAAGATCC |
| R: AAGGAATGTAACGGCAGGTC | |
| CDC25B | F: CCGAGAGCTGATTGGAGATTAC |
| R: CACGATGTTGCTGAACTTGC | |
| PCNA | F: GTCTCTTTGGTGCAGCTCA |
| R: ATCTTCGGCCCTTAGTGTAATG | |
| PLK1 | F: ACAGTTTCGAGGTGGATGTG |
| R: GGTTGATGTGCTTGGGAATAC | |
| MCM2 | F: ATTTCGTCCTGGGTCCTTTC |
| R: CGCTGGTAGTTCTGATAGATGG | |
| MCM3 | F: AGCGAAGTGAGGATGAATCAG |
| R: CTGTGTCACTGAAGTCATAGGG | |
| MCM7 | F: GATGCCACCTATACTTCTGCC |
| R: TCCTTTGACATCTCCATTAGCC | |
| SERPINE1 | F: GTGGACTTTTCAGAGGTGGAG |
| R: GAAGTAGAGGGCATTCACCAG | |
| THBS1 | F: CTCCCCTATGCTATCACAACG |
| R: AGGAACTGTGGCATTGGAG | |
| CASPASE7 | F: CCTCGATACAAGATCCCAGTG |
| R: GATTTCCAGGTCTTTTCCGTG | |
| TNFRSF10B | F: ACCACGACCAGAAACACAG |
| R: CATTCGATGTCACTCCAGGG |
GADD45A, growth arrest and DNA damage-inducible α; CCND3, cyclin D3; CCNB1, cyclin B1; CDC20, cell division cycle 20; CDC25B, cell division cycle 25B; PCNA, proliferating cell nuclear antigen; PLK1, polo like kinase 1; MCM, minichromosome maintenance complex component; SERPINE1, serpin family E member 1; THBS1, thrombospondin 1; TNFRSF10B, TNF receptor superfamily member 10b; F, forward; R, reverse.
Western blotting
Total protein was extracted from cells using RIPA lysis buffer (Beijing Solarbio Science & Technology Co., Ltd.) which contains 1% (V/V) phenylmethanesulfonyl fluoride (Beijing Solarbio Science & Technology Co., Ltd.). Nuclear proteins were prepared using a Nuclear and Cytoplasmic Protein Extraction kit (Wanleibio Co., Ltd.), according to the manufacturer's protocol. A BCA protein assay kit (Thermo Fisher Scientific, Inc.) was used to determine the protein concentration. Samples containing equal amounts of protein (30 µg) were loaded onto 10% gels (p53, nuclear p53, CDK6, Lamin B and GAPDH proteins) or 12% gels [p21, CDK4, growth arrest and DNA damage-inducible α (GADD45A), cyclin D1, cyclin D3, cyclin B1, caspase 3/7, cleaved-caspase 3/7, poly(ADP-ribose) polymerase (PARP), cleaved-PARP and GAPDH proteins], separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes (EMD Millipore). The membrane was blocked with TBS with Tween-20 (1:1,000; TBST) containing 1% BSA (Beijing Solarbio Science & Technology Co., Ltd.) for 1 h at room temperature and incubated overnight at 4°C with specific primary antibodies against p21 (1:1,000; cat. no. 2947; Cell Signaling Technology, Inc.), GADD45A (1:1,000; cat. no. 4632; Cell Signaling Technology, Inc.), cyclin D3 (1:2,000; cat. no. 2936; Cell Signaling Technology, Inc.), cyclin D1 (1:500; cat. no. WL01435a; Wanleibio Co., Ltd.), cyclin B1 (1:500; cat. no. WL01760; Wanleibio Co., Ltd.), CDK4 (1:500; cat. no. WL02274; Wanleibio Co., Ltd.), CDK6 (1:500; cat. no. WL01676; Wanleibio Co., Ltd.), caspase 3 (1:500; cat. no. WL01927; Wanleibio Co., Ltd.), cleaved-caspase 3 (1:500; cat. no. WL01992; Wanleibio Co., Ltd.), cleaved-caspase 7 (1:500; cat. no. WL02360; Wanleibio Co., Ltd.), PARP/cleaved-PARP (1:500; cat. no. WL01932; Wanleibio Co., Ltd.), lamin B (1:500; cat. no. WL01775; Wanleibio Co., Ltd.), caspase 7 (1:300; cat. no. sc-56063; Santa Cruz Biotechnology, Inc.), p53 (1:500; cat. no. bsm-33058M; BIOSS) and GAPDH (1:1,000; cat. no. bs-0755R; BIOSS). Subsequent to washing three times with TBST, the membranes were incubated with the following horseradish peroxidase (HRP)-conjugated secondary antibodies: Goat anti-mouse (1:3,000; cat. no. bs-0296G-HRP; BIOSS) and goat anti-rabbit (1:3,000; cat. no. bs-0295G-HRP; BIOSS) for 2 h at room temperature. Following washing in TBST three times, immunoblots were visualized using an enhanced chemiluminescent substrate kit (Beijing Solarbio Science & Technology Co., Ltd.) under an Amersham Imager 600 system (GE Healthcare Life Sciences). Expression levels were quantified using ImageJ version 1.52 software (National Institutes of Health).
Statistical analysis
All data are presented as the mean ± SEM, unless otherwise stated, and represent three independent experimental repeats. SPSS 20.0 software (IBM Corp.) and GraphPad Prism 5.0 software (GraphPad Software, Inc.) were used for statistical analysis. One-way ANOVAs were conducted to examine the differences among the multiple groups, followed by a Tukey's post-hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
GNA inhibits growth and induces the apoptosis of A549/Cis cells
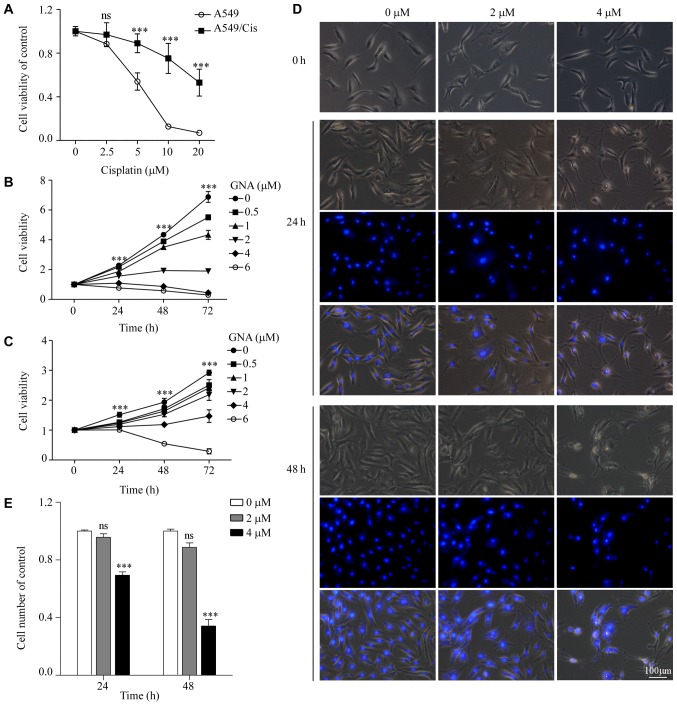
A549/Cis is a Cis-resistant cell line derived from the A549 lung cancer cell line. To confirm its chemoresistance to Cis, A549 cells were exposed to Cis in vitro to establish A549/Cis cells. The MTT assay demonstrated that A549/Cis cells were significantly more resistant to Cis compared with the parental cells (P<0.001; Fig. 2A). The cytotoxic effect of GNA on A549 and A549/Cis cells was determined. Cells were treated with increasing concentrations of GNA for 24, 48 and 72 h. Cell viability was measured using an MTT assay. As presented in Fig. 2B and C, GNA significantly decreased the viability of A549 and A549/Cis cells compared with the untreated group (P<0.001). GNA induced a high degree of cell death at a concentration of 6 µM only after 24 h. Accordingly, 2 and 4 µM GNA was used in the subsequent experiments. Hoechst 33342 staining further demonstrated the inhibitory effect of GNA in A549/Cis cells (Fig. 2C and D). Compared with the untreated cells, the cells treated with GNA had inhibited proliferation and exhibited morphological alterations. Furthermore, the nuclear condensation of GNA-treated cells was also observed.
Figure 2.
GNA inhibits the cell growth of A549 and A549/Cis cells. (A) MTT assay was used to confirm the cell viability of A549 and A549/Cis cell lines subsequent to treatment with various concentrations of Cis for 48 h. (B) Cell viability of A549 cells treated with a range of concentrations of GNA for 24, 48 and 72 h was measured by an MTT assay. (C) Cell viability of A549/Cis cells treated with a range of concentrations of GNA for 24, 48 and 72 h. (D) A549/Cis cells treated with specified concentrations of GNA were observed under an inverted fluorescent contrast phase microscope for the indicated time periods. Scale bar, 100 µm; magnification, ×200. (E) Quantification of cell counts. ***P<0.001 vs. control. GNA, gambogenic acid; Cis, cisplatin; ns, not significant.
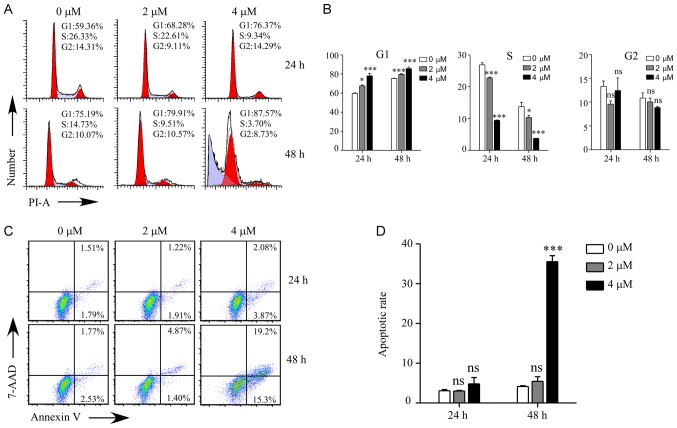
GNA induces cell cycle arrest and apoptosis in A549/Cis cells
To investigate the cellular process responsible for the inhibited proliferation by GNA treatment, the cell cycle and apoptosis were examined by flow cytometry in A549/Cis cells (Fig. 3). As presented in Fig. 3A and B, the cell cycle of A549/Cis cells was significantly arrested at the G1 phase following GNA treatment for 24 and 48 h compared with the untreated group (P<0.5). There was a significantly higher sub-G1 population in the cells treated with 4 µM GNA for 48 h compared with the untreated group (P<0.001). Cell cycle arrest may induce cell death, which was measured using flow cytometry. The annexin V/7-AAD double staining assay revealed that the apoptosis rate was significantly increased compared with the control group when A549/Cis cells were treated with 4 µM GNA for 48 h (P<0.001; Fig. 3C and D).
Figure 3.
Effects of GNA on cell cycle arrest and apoptosis in A549/Cis cells. (A) Ratio of the cell cycle phases of A549/Cis cells following GNA treatment for 24 and 48 h. (B) Cell cycle populations following GNA treatment were estimated. (C) Percentage of apoptotic A549/Cis cells subsequent to GNA treatment for 24 and 48 h. (D) Quantification of apoptosis. Data are presented as the mean ± standard deviation of triplicate measurements. *P<0.05 and ***P<0.001 vs. control. GNA, gambogenic acid; Cis, cisplatin; ns, not significant.
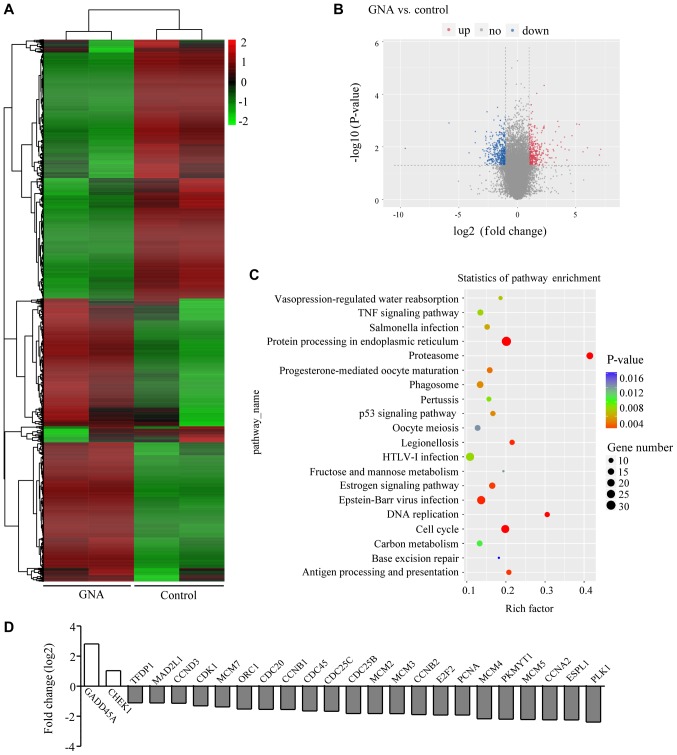
Differential gene expression and enrichment analysis in A549/Cis cells treated with GNA
To understand how GNA inhibits cell growth and promotes cell death in A549/Cis cells, an RNA-seq assay was performed using samples from the control and GNA-treated cells. Data analysis indicated that GNA treatment induced a global gene expression change (Fig. 4A). All genes whose threshold was restricted with a P-value <0.05, fold change ≥2 or ≤0.5 were identified as differentially expressed genes (DEGs). There were 353 upregulated DEGs and 425 downregulated DEGs in the cells treated with GNA; the DEGs are visualized in the volcano plot (Fig. 4B). To further investigate the function of DEGs, KEGG pathway analysis was performed. It was identified that the DEGs influenced by GNA treatment were mostly enriched in pathways involved in the cell cycle, DNA replication and p53 signaling pathways associated with tumor proliferation (Fig. 4C). In addition, the DEGs associated with the cell cycle were further enriched (Fig. 4D).
Figure 4.
Gene expression profiling of GNA-treated and untreated A549/Cis cells. (A) Heat map of the hierarchical clustering of significant differential genes (P-value <0.05) identified in A549/Cis cells treated with 4 µM GNA for 24 h. (B) Volcano plot presenting the significant differentially up- and downregulated genes in A549/Cis cells subsequent to GNA treatment compared with untreated cells. (C) Top 20 enrichment Kyoto Encylopedia of Genes and Genomes pathways of the DEGs. (D) Expression of DEGs associated with the cell cycle pathway. GNA, gambogenic acid; Cis, cisplatin; DEGs, differentially expressed genes.
Mechanisms of the anticancer effects of GNA in A549/Cis cells
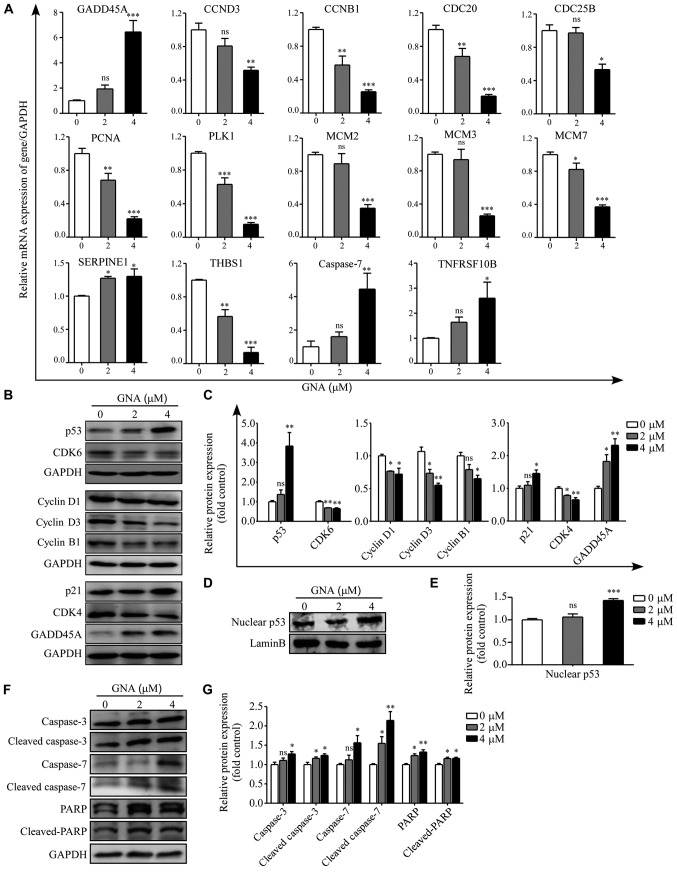
To validate the RNA-seq results, 14 DEGs were selected for RT-qPCR analysis and GAPDH was used to normalize the gene expression data. Among these genes, 4 were verified to be upregulated in GNA-treated A549/Cis cells whereas the other 10 were identified as downregulated. In total, 10 genes [GADD45A, cyclin D3 (CCND3), cyclin B1 (CCNB1), cell division cycle 20, cell division cycle 25B, proliferating cell nuclear antigen, polo like kinase 1, minichromosome maintenance complex component (MCM)2, MCM 3 and MCM 7] were involved in the cell cycle, 6 genes [GADD45A, CCND3, CCNB1, SERPINE1, thrombospondin 1 and TNF receptor superfamily member 10b (TNFRSF10B)] were associated with the p53 signaling pathway and two genes (CASPASE7 and TNFRSF10B) were associated with apoptosis. The results of RT-qPCR were consistent with the RNA-seq analysis (P<0.05; Fig. 5A). To further investigate the underlying mechanisms of GNA-induced cell cycle arrest and apoptosis, a western blotting assay was performed. The protein expression levels of the hub genes associated with the G1 phase cell cycle checkpoint and apoptosis are presented in Fig. 5B, D and F. The data revealed that the protein levels of cyclin D1, cyclin D3, CDK4 and CDK6 were significantly downregulated (P<0.05), while the expression of p21, GADD45A, p53 and nuclear p53 were significantly upregulated in A549/Cis cells following GNA treatment compared with the untreated group (P<0.05). Furthermore, the regulators of apoptosis were additionally detected subsequent to GNA treatment. GNA was able to significantly increase the protein levels of the precursor forms of caspase 3/7 and their active forms as well (P<0.05). The hallmark of apoptosis, PARP, which is associated with DNA repair, was revealed to be significantly upregulated (P<0.05), and its cleavage was significantly enhanced by GNA treatment in A549/Cis cells (P<0.05).
Figure 5.
Mechanisms underlying GNA-induced cell cycle arrest and apoptosis. (A) Relative expression of GADD45A, CCND3, CCNB1, CDC20, CDC25B, PCNA, PLK1, MCM2, MCM3, MCM7, THBS1, SERPINE1, CASPASE7, TNFRSF10B and GAPDH were identified in A549/Cis cells treated with different concentrations of GNA for 24 h by reverse transcription-quantitative PCR. (B) Western blot analysis and (C) quantitative results of p53, GADD45A, p21, cyclin D1, cyclin D3, CDK4, CDK6 and cyclin B1 in whole cell lysates of A549/Cis cells treated with different doses of GNA for 24 h. The levels of GAPDH were used as loading controls. (D) Western blot analysis and (E) quantitative results of p53 in the nucleus of A549/Cis cells treated with different doses of GNA for 24 h. The levels of lamin B were used as loading controls. (F) Representative blots and (G) quantitative results of the expression of caspase 3, caspase 7 and PARP subsequent to GNA treatments. *P<0.05, **P<0.01 and ***P<0.001 vs. untreated group. GNA, gambogenic acid; Cis, cisplatin; ns, not significant; GADD45A, growth arrest and DNA damage-inducible protein GADD45 α; CCND3, cyclin D3; CCNB1, cyclin B1; CDC20, cell division cycle 20; CDC25B, cell division cycle 25B; PCNA, proliferating cell nuclear antigen; PLK1, polo like kinase 1; MCM, minichromosome maintenance complex component; THBS1, thrombospondin 1; TNFRSF10B, TNF receptor superfamily member 10b; PARP, poly(ADP-ribose) polymerase.
Discussion
The present study investigated the anticancer activity of GNA in A549/Cis cells and investigated the underlying molecular mechanisms. It was demonstrated that GNA exhibited potent inhibitory activities in A549/Cis cells. Based on the present data, GNA induced G0/G1 phase cell cycle arrest mainly by regulating the expression of the cyclin D-CDK4/CDK6 complex, p21 and p53. The cell cycle arrest subsequently triggers apoptosis through the activation of caspases in A549/Cis cells.
GNA, an active ingredient isolated from the traditional Chinese medicine gamboge, was revealed to have anti-tumor activity in vitro and in vivo, and low toxicity to normal cells (11,13,14,30). In the present study, GNA exhibited anti-cancer effects on Cis-resistant NSCLC cells, indicating that it additionally has the potential to overcome drug resistance in cancer. However, despite these advantages, the clinical use of GNA was limited due to its poor aqueous solubility, short elimination half-life, low bio-availability and excessive irritation to blood vessels by intravenous administration (31). With the aim of overcoming these limitations, researchers have developed numerous nanocarrier drug delivery systems, including solid lipid nanoparticles, nanostructure lipid carriers (32), liquid crystal dispersions (33) and mixed polymeric micelles (34), which are able to enhance bio-availability and retain antitumor effects. These novel formulations of GNA may be developed further for its clinical application.
The cell cycle serves an important function in maintaining the dynamic balance between proliferation and cell death. Abnormal regulation of the cell cycle results in cancer development. Inhibition of the cell cycle is a known target for cancer therapy (35,36). Cell cycle progression is mainly regulated by cyclins, cyclin-dependent kinases and cyclin-dependent protein kinases inhibitors (CDKIs). The fluctuations in the cyclin levels during the cell cycle may activate CDK (37). The D-type cyclins (D1, D2 and D3) are the first cyclins sensing the mitogenic signals and activate CDK4 and CDK6 in the G1 phase. At the G1/S checkpoint, the cyclin D-CDK4/6 complex is crucial in regulating the G1/S phase transition. Phosphorylation of Rb by cyclin D-CDK4/6 and cyclin E-CDK2 releases E2F, allowing the expression of genes that encode products necessary for S-phase progression (38–40). The inhibition of the CDK4/6-cyclin D complex will cause cell cycle arrest (41). The present results were consistent with these observations. Previous studies also demonstrated that GNA may induce cell cycle arrest at the G1 phase in cancer cells (11,13). Consequently, the results of the present study on the regulatory proteins responsible for G1/S transition indicated that GNA arrested the cell cycle at the G1 phase by downregulating cyclin D-CDK4/6 complex proteins. The CDKI family member p21 (also known as p21WAF1/Cip1) serves an important function in binding to the cyclin-CDK complex and suppressing its catalytic activity, causing cell cycle arrest (42,43). Inactivation of the cyclin D-CDK4/6 complex by p21 may inhibit Rb phosphorylation and induce G1 phase cell cycle arrest (44). The present study observed that p21 was also altered by GNA. In addition, p21 is a direct transcriptional target of p53 and is required for p53-dependent cell cycle arrest (45). GADD45A is a protein that may inactivate cyclin B1 through a p53-dependent/independent pathway; upregulation of GADD45A may be regulated by p53. These results suggested that the elevated protein expression of p53 in response to GNA exposure may be involved in G1 phase arrest by increasing the p21 protein level.
Cell cycle inhibition would subsequently induce apoptosis in cancer therapy (46). In the present study, it was identified that the percentage of A549/Cis cells in the G1 phase increased following GNA treatment for 24 and 48 h. However, the apoptotic rate of A549/Cis cells had no statistical difference following 24 h GNA treatment and significant differences only occurred 48 h later following 4 µM GNA treatment. The present results demonstrated that apoptosis may be a result of cell cycle arrest, which supports the cell cycle inhibitory effect of GNA. Apoptosis is important as it serves a pivotal function in growth inhibition, and the agents that induce apoptosis in tumor cells may be useful for the treatment of malignancies (39). Apoptosis is primarily executed by a family of proteases known as the caspases (47). Caspase 3/7 are the executors of apoptosis, which are responsible for the definite cleavage of cellular components (48). Activation of caspase 3/7 may trigger PARP, which eventually results in apoptosis (49). In the present study, the activation of caspase 3/7, in addition to cleaved PARP, were observed following GNA treatment. In summary, it was hypothesized that GNA induced cell cycle arrest, which results in apoptosis via the activation of caspases in A549/Cis cells.
In conclusion, the present study demonstrated that GNA is a natural and effective compound that exerts anticancer effects against A549/Cis cells by inducing apoptosis via cell cycle arrest at the G1 phase. GNA induced G1 phase cell cycle arrest through the regulation of cyclin D1, cyclin D3, CDK4, CDK6, p21 and p53, which subsequently resulted in apoptosis via the activation of caspase 3/7 in A549/Cis cells. The present evidence supported the potential application of GNA as a promising drug in the treatment of Cis-resistant NSCLC.
Acknowledgements
Not applicable.
Funding
The present study was supported by The National Natural Science Foundation of China (grant nos. 81473569 and 81973735).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
CL and YW conceived and designed the study. DS and HN performed the experiments. DS wrote the manuscript. YW and DS analyzed the data. CL, YW, DS and HN reviewed and edited the manuscript. All authors read and approved the manuscript and agreed to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work is appropriately investigated and resolved.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, Stein KD, Alteri R, Jemal A. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66:271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 3.Arriagada R, Bergman B, Dunant A, Le Chevalier T, Pignon JP, Vansteenkiste J, International Adjuvant Lung Cancer Trial Collaborative Group Cisplatin-based adjuvant chemotherapy in patients with completely resected non-small-cell lung cancer. N Engl J Med. 2004;350:351–360. doi: 10.1056/NEJMoa031644. [DOI] [PubMed] [Google Scholar]
- 4.Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, Tsurutani J, Seto T, Satouchi M, Tada H, Hirashima T, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): An open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 5.Oliver TG, Mercer KL, Sayles LC, Burke JR, Mendus D, Lovejoy KS, Cheng MH, Subramanian A, Mu D, Powers S, et al. Chronic cisplatin treatment promotes enhanced damage repair and tumor progression in a mouse model of lung cancer. Genes Dev. 2010;24:837–852. doi: 10.1101/gad.1897010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stewart DJ. Mechanisms of resistance to cisplatin and carboplatin. Crit Rev Oncol Hematol. 2007;63:12–31. doi: 10.1016/j.critrevonc.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Maroun JA, Anthony LB, Blais N, Burkes R, Dowden SD, Dranitsaris G, Samson B, Shah A, Thirlwell MP, Vincent MD, Wong R. Prevention and management of chemotherapy-induced diarrhea in patients with colorectal cancer: A consensus statement by the Canadian working group on chemotherapy-induced diarrhea. Curr Oncol. 2007;14:13–20. doi: 10.3747/co.2007.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Gramont A, Figer A, Seymour M, Homerin M, Hmissi A, Cassidy J, Boni C, Cortes-Funes H, Cervantes A, Freyer G, et al. Leucovorin and fluorouracil with or without oxaliplatin as first-line treatment in advanced colorectal cancer. J Clin Oncol. 2000;18:2938–2947. doi: 10.1200/JCO.2000.18.16.2938. [DOI] [PubMed] [Google Scholar]
- 9.Luo F, Gu J, Chen L, Xu X. Systems pharmacology strategies for anticancer drug discovery based on natural products. Mol Biosyst. 2014;10:1912–1917. doi: 10.1039/c4mb00105b. [DOI] [PubMed] [Google Scholar]
- 10.Asano J, Chiba K, Tada M, Yoshii T. Cytotoxic xanthones from Garcinia hanburyi. Phytochemistry. 1996;41:815–820. doi: 10.1016/0031-9422(95)00682-6. [DOI] [PubMed] [Google Scholar]
- 11.Yan F, Wang M, Chen H, Su J, Wang X, Wang F, Xia L, Li Q. Gambogenic acid mediated apoptosis through the mitochondrial oxidative stress and inactivation of Akt signaling pathway in human nasopharyngeal carcinoma CNE-1 cells. Eur J Pharmacol. 2011;652:23–32. doi: 10.1016/j.ejphar.2010.11.018. [DOI] [PubMed] [Google Scholar]
- 12.Chen HB, Zhou LZ, Mei L, Shi XJ, Wang XS, Li QL, Huang L. Gambogenic acid-induced time- and dose-dependent growth inhibition and apoptosis involving Akt pathway inactivation in U251 glioblastoma cells. J Nat Med. 2012;66:62–69. doi: 10.1007/s11418-011-0553-7. [DOI] [PubMed] [Google Scholar]
- 13.Yu XJ, Han QB, Wen ZS, Ma L, Gao J, Zhou GB. Gambogenic acid induces G1 arrest via GSK3β-dependent cyclin D1 degradation and triggers autophagy in lung cancer cells. Cancer Lett. 2012;322:185–194. doi: 10.1016/j.canlet.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 14.Yan F, Wang M, Li J, Cheng H, Su J, Wang X, Wu H, Xia L, Li X, Chang HC, Li Q. Gambogenic acid induced mitochondrial-dependent apoptosis and referred to phospho-Erk1/2 and phospho-p38 MAPK in human hepatoma HepG2 cells. Environ Toxicol Pharmacol. 2012;33:181–190. doi: 10.1016/j.etap.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 15.Liu P, Wu X, Dai L, Ge Z, Gao C, Zhang H, Wang F, Zhang X, Chen B. Gambogenic acid exerts antitumor activity in hypoxic multiple myeloma cells by regulation of miR-21. J Cancer. 2017;8:3278–3286. doi: 10.7150/jca.19290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen F, Zhang XH, Hu XD, Zhang W, Lou ZC, Xie LH, Liu PD, Zhang HQ. Enhancement of radiotherapy by ceria nanoparticles modified with neogambogic acid in breast cancer cells. Int J Nanomedicine. 2015;10:4957–4969. doi: 10.2147/IJN.S82980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su J, Cheng H, Zhang D, Wang M, Xie C, Hu Y, Chang HC, Li Q. Synergistic effects of 5-fluorouracil and gambogenic acid on A549 cells: Activation of cell death caused by apoptotic and necroptotic mechanisms via the ROS-mitochondria pathway. Biol Pharm Bull. 2014;37:1259–1268. doi: 10.1248/bpb.b13-00972. [DOI] [PubMed] [Google Scholar]
- 18.He Y, Ding J, Lin Y, Li J, Shi Y, Wang J, Zhu Y, Wang K, Hu X. Gambogenic acid alters chemosensitivity of breast cancer cells to Adriamycin. BMC Complement Altern Med. 2015;15:181. doi: 10.1186/s12906-015-0710-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, Cao W, Zhang J, Yan M, Xu Q, Wu X, Wan L, Zhang Z, Zhang C, Qin X, et al. A covalently bound inhibitor triggers EZH2 degradation through CHIP-mediated ubiquitination. EMBO J. 2017;36:1243–1260. doi: 10.15252/embj.201694058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frazee AC, Pertea G, Jaffe AE, Langmead B, Salzberg SL, Leek JT. Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat Biotechnol. 2015;33:243–246. doi: 10.1038/nbt.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.The Gene Ontology Consortium: The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47(D1):D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanehisa M, Sato Y, Furumichi M, Morishima K, Tanabe M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019;47(D1):D590–D595. doi: 10.1093/nar/gky962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28:1947–1951. doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 28.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Huang T, Zhang H, Wang X, Xu L, Jia J, Zhu X. Gambogenic acid inhibits the proliferation of small-cell lung cancer cells by arresting the cell cycle and inducing apoptosis. Oncol Rep. 2019;41:1700–1706. doi: 10.3892/or.2018.6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang X, Chen YJ, Peng DY, Li QL, Wang XS, Wang DL, Chen WD. Solid lipid nanoparticles as delivery systems for gambogenic acid. Colloids Surf B Biointerfaces. 2013;102:391–397. doi: 10.1016/j.colsurfb.2012.08.058. [DOI] [PubMed] [Google Scholar]
- 32.Lin T, Huang X, Wang Y, Zhu T, Luo Q, Wang X, Zhou K, Cheng H, Peng D, Chen W. Long circulation nanostructured lipid carriers for gambogenic acid: Formulation design, characterization, and pharmacokinetic. Xenobiotica. 2017;47:793–799. doi: 10.1080/00498254.2016.1229084. [DOI] [PubMed] [Google Scholar]
- 33.Luo Q, Lin T, Zhang CY, Zhu T, Wang L, Ji Z, Jia B, Ge T, Peng D, Chen W. A novel glyceryl monoolein-bearing cubosomes for gambogenic acid: Preparation, cytotoxicity and intracellular uptake. Int J Pharm. 2015;493:30–39. doi: 10.1016/j.ijpharm.2015.07.036. [DOI] [PubMed] [Google Scholar]
- 34.Lin TY, Zhu TT, Xun Y, Tao YS, Yang YQ, Xie JL, Zhang XM, Chen SX, Ding BJ, Chen WD. A novel drug delivery system of mixed micelles based on poly(ethylene glycol)-poly(lactide) and poly(ethylene glycol)-poly(varepsilon-caprolactone) for gambogenic acid. Kaohsiung J Med Sci. 2019;35:757–764. doi: 10.1002/kjm2.12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sherr CJ, Roberts JM. CDK inhibitors: Positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 36.Goyeneche AA, Caron RW, Telleria CM. Mifepristone inhibits ovarian cancer cell growth in vitro and in vivo. Clin Cancer Res. 2007;13:3370–3379. doi: 10.1158/1078-0432.CCR-07-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu H, Wang Z, Jin S, Hao H, Zheng L, Zhou B, Zhang W, Lv H, Yuan Y. Dux4 induces cell cycle arrest at G1 phase through upregulation of p21 expression. Biochem Biophys Res Commun. 2014;446:235–240. doi: 10.1016/j.bbrc.2014.02.105. [DOI] [PubMed] [Google Scholar]
- 38.Bertoli C, Skotheim JM, de Bruin RA. Control of cell cycle transcription during G1 and S phases. Nat Rev Mol Cell Biol. 2013;14:518–528. doi: 10.1038/nrm3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giacinti C, Giordano A. RB and cell cycle progression. Oncogene. 2006;25:5220–5227. doi: 10.1038/sj.onc.1209615. [DOI] [PubMed] [Google Scholar]
- 40.Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer. 2011;11:558–572. doi: 10.1038/nrc3090. [DOI] [PubMed] [Google Scholar]
- 41.Rader J, Russell MR, Hart LS, Nakazawa MS, Belcastro LT, Martinez D, Li Y, Carpenter EL, Attiyeh EF, Diskin SJ, et al. Dual CDK4/CDK6 inhibition induces cell-cycle arrest and senescence in neuroblastoma. Clin Cancer Res. 2013;19:6173–6182. doi: 10.1158/1078-0432.CCR-13-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu S, Cetinkaya C, Munoz-Alonso MJ, von der Lehr N, Bahram F, Beuger V, Eilers M, Leon J, Larsson LG. Myc represses differentiation-induced p21CIP1 expression via Miz-1-dependent interaction with the p21 core promoter. Oncogene. 2003;22:351–360. doi: 10.1038/sj.onc.1206145. [DOI] [PubMed] [Google Scholar]
- 43.Lee EW, Lee MS, Camus S, Ghim J, Yang MR, Oh W, Ha NC, Lane DP, Song J. Differential regulation of p53 and p21 by MKRN1 E3 ligase controls cell cycle arrest and apoptosis. EMBO J. 2009;28:2100–2113. doi: 10.1038/emboj.2009.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tibbetts RS, Brumbaugh KM, Williams JM, Sarkaria JN, Cliby WA, Shieh SY, Taya Y, Prives C, Abraham RT. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev. 1999;13:152–157. doi: 10.1101/gad.13.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu G, Lozano G. p21 stability: Linking chaperones to a cell cycle checkpoint. Cancer Cell. 2005;7:113–114. doi: 10.1016/j.ccr.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 46.Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411:342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- 47.Li J, Yuan J. Caspases in apoptosis and beyond. Oncogene. 2008;27:6194–6206. doi: 10.1038/onc.2008.297. [DOI] [PubMed] [Google Scholar]
- 48.Thornberry NA, Lazebnik Y. Caspases: Enemies within. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 49.Saraste A, Pulkki K. Morphologic and biochemical hallmarks of apoptosis. Cardiovasc Res. 2000;45:528–537. doi: 10.1016/S0008-6363(99)00384-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.