Abstract
Fibroblast growth factor receptors (FGFRs) have been implicated in the malignant transformation and chemoresistance of epithelial ovarian cancer; however, the underlying molecular mechanisms are poorly understood. Increased sialyltransferase activity that enhances protein sialylation is an important post-translational process promoting cancer progression and malignancy. In the present study, α2,6-sialyltransferase (ST6Gal-I) overexpression or knockdown cell lines were developed, and FGFR1 was examined to understand the effect of sialylation on migration and drug resistance, and the underlying mechanisms. It was identified that cells with ST6Gal-I overexpression had increased cell viability and migratory ability upon serum deprivation. Moreover, ST6Gal-I overexpression cells had strong resistance to paclitaxel, as demonstrated by low growth inhibition rate and cell apoptosis level. A mechanistic study showed that ST6Gal-I overexpression induced high α2,6-sialylation of FGFR1 and increased the expression of phospho-ERK1/2 and phospho-focal adhesion kinase. Further study demonstrated that the FGFR1 inhibitor PD173047 reduced cell viability and induced apoptosis; however, ST6Gal-I overexpression decreased the anticancer effect of PD173047. In addition, ST6Gal-I overexpression attenuated the effect of Adriamycin on cancer cells. Collectively, these results suggested that FGFR1 sialylation plays an important role in cell migration and drug chemoresistance in ovarian cancer cells.
Keywords: ovarian cancer, ST6Gal-I, FGFR1, chemoresistance
Introduction
Fibroblast growth factor receptors (FGFRs), which belong to the receptor tyrosine kinase (RTK) family, are known to signal from the cell membrane as well as from endosomal compartments (1). There are four FGFRs: FGFR1, FGFR2, FGFR3 and FGFR4; these FGFs bind their receptors and >20 known ligands to these receptors, resulting in diverse effects in many different target cells (2). FGFR signaling plays an important role in cell proliferation, angiogenesis and many normal biological processes (3); however, FGFR signaling dysregulation has been implicated in aberrant pathologies associated with tumor growth, including ovarian, colon, breast, prostate, soft tissue sarcomas, melanoma and lung cancer (4–9).
Despite advances in treatment over the past decades, ovarian cancer has the highest mortality among gynecologic malignancies (10). Limited prognosis remains a key obstacle for the treatment of patients with advanced ovarian cancer (11). Upregulation of all four members of the FGFR family and other various fibroblast growth factors has been found in epithelial ovarian carcinoma tissue (10,12), suggesting that dysregulated FGFR signaling contributes to ovarian carcinogenesis and may represent a suitable therapeutic target (13). The FGFR4 GlyArg388 polymorphism has been shown to predict prolonged survival and platinum sensitivity in advanced ovarian cancer (14). FGFR1 and FGFR2 mutations have also been demonstrated to promote ovarian cancer progression and invasion (15,16). The mechanisms of FGFR1 in other cancer types have been studied; for example, the upregulation of FGFR1 in carcinoma cells is critical for prostate cancer progression and invasion (17). Furthermore, the FGFR1 pathway recruits macrophages to the mammary epithelium and promotes paracrine interactions between tumor cells and macrophages, thus inducing tumor growth (18,19). However, to the best of the authors' knowledge, not many studies on the role of FGFR1 in ovarian cancer exist, and how FGFR1 functions in ovarian cancer is unclear.
Genetic evidence and structure analysis indicated that the N-glycosylation of FGFR may constitute an important regulatory input (20). The disruption of N-glycosylation can cause the mutation of an asparagine residue in the extracellular domain of FGFR2 and FGFR3, and result in skeletal growth defects. Abnormal cellular glycosylation has been shown to play a key role in cancer progression and malignancy (21–23). Therefore, understanding the regulation of FGFR glycosylation may provide novel insight into cancer biology and result in developing possible therapeutic strategies. Glycosylation is regulated by various glycosyltransferases, such as fucosyl-, sialyl- and galactosyltransferases (24). The β galactoside α2,6-sialyltransferase, CMP-NeuAc: Galβ (1,4) GlcNAc: α2,6-sialyltransferase (ST6Gal-I) is a vital sialyltransferase that adds sialic acid residues to N-linked oligosaccharides (25). ST6Gal-I has been reported to induce adhesion and migration, and promote drug resistance in various cancer cells (26–29). However, the possible biological effect of ST6Gal-I on FGFR1 in ovarian cancer has not been clearly established.
In the present study, ST6Gal-I knockdown or overexpression OVCAR3 ovarian cell lines were prepared and characterized, to investigate the sialylation of FGFR1 and its effects on cancer cell proliferation and migration, and sensitivity to anticancer drugs. It was identified that ST6Gal-I overexpression induced high sialylation levels of FGFR1, and activated ERK and focal adhesion kinase (FAK) signaling in cells. ST6Gal-I overexpression decreased the effects of anticancer drugs, but ST6Gal-I knockdown resulted in the opposite effect. Collectively, these data suggested that FGFR1 sialylation affects FGFR1-mediated cell growth and chemotherapeutic drug sensitivity in human ovarian cancer cells. FGFR1 sialylation levels are hypothesized to be a reliable biomarker for anti-FGFR1 therapy.
Materials and methods
Cell culture and transfection
OVCAR3 ovarian cancer cells, purchased from The American Type Culture Collection, were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.) with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a 5% CO2-humidified atmosphere. Stable ST6Gal-I overexpression (oe-ST6Gal-I), knockdown small hairpin-ST6Gal-I (sh-ST6Gal-I) or empty vector cell lines were established, as previously described (30). In brief, pcDNA3.1(−)/ST6Gal-I, small hairpin (sh)-ST6Gal-I and empty vector plasmids (10 µg/ml) were purchased from Invitrogen; Thermo Fisher Scientific, Inc., and transfected into OVCAR3 ovarian cancer cells with Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.). A limiting dilution was applied to obtain subcell line clones after 24 h of transfection. Blasticidin S Hcl (1.5 µg/ml, Invitrogen; Thermo Fisher Scientific, Inc.) was used to select the sh-ST6Gal-I clone, and Geneticin® Selective Antibiotic G418 (350 µg/ml, Invitrogen; Thermo Fisher Scientific, Inc.) was utilized to select the ST6Gal-I overexpression clone. ST6Gal-I overexpression or knockdown cell lines were verified by reverse transcription (RT)-semi-quantitative (q)PCR and immunoblotting.
RT-qPCR
Total RNA was isolated from the cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), and cDNA was synthesized using a PrimeScript® RT Master Mix kit (Takara Bio) according to the manufacturer's protocol: 37°C for 60 min, 85°C for 5 min and hold at 4°C. RT-qPCR was performed on a Real-Time PCR Detection System (Bio-Rad Laboratories). PCR cycles were: Pretreatment at 95°C for 10 min, 93°C for 15 sec, 67°C for 45 sec (45 cycles), then 93°C for 15 sec, 67°C for 1 min, 95°C for 15 sec, 75°C for 10 min and hold at 4°C. The primer sequences used for the real-time PCR assays were as follows: Forward, 5′-CCTCTGGGATGCTTGGTATC-3′; and reverse, 5′-GTGCAGGCACTATCGAAGAA-3′ for ST6Gal-I; forward, 5′-AGCCTCAAGATCATCAGC-3′ and reverse, 5′-GAGTCCTTCCACGATACC-3′ for GAPDH (BGI, Inc.). The gene expression was determined using the 2−ΔΔCq method (31).
ST6Gal-I activity assay
Lectin staining was conducted to measure ST6Gal-I activity. Cells were stained with FITC-conjugated SNA lectin (EY Laboratories, Inc.), which is specific for 2–6 sialic acids. Cells were stained for 40 min at 4°C with SNA-FITC (1:200) and analyzed by fluorescence-activated cell sorting (FACS; Becton Dickinson). In addition, cells were stained with SNA-FITC (1:100) for 4 h at room temperature for an immunofluorescence assay using a Leica DM2500 LED microscope (Leica Microsystems GmbH) at 200× magnification.
Scratch wound healing assay
After the oe-ST6Gal-I and sh-ST6Gal-I subcell line clones were verified, a wound healing assay was used to assess cell migration (32). Cells (105 cells/well) were seeded in a 6-well plate, and the tip of a 200-µl micropipette was used to make a straight scratch on a confluent monolayer of cells to create a wound. The detached cells were rinsed with PBS twice, and serum-free DMEM was then added. The wound closure in the area was imaged in brightfield using a microscope (Olympus Corporation; magnification, ×100) after incubation for 0, 12 and 24 h. The wound closure areas were selected randomly and the width of the wound was quantified in ImageJ (v1.8, National Institutes of Health) to show the wound closure at each time point. The results of four independent experiments were imaged under a microscope and quantified.
Cell Counting Kit-8 (CCK-8) assay
To measure the proliferation of different transfected cloned cell lines, a CCK-8 detection kit (Dojindo Molecular Technologies, Inc.) was used, according to the manufacturer's protocol. In total, ~3,000 cells were seeded into a 96-well plate in quintuplicate for 6 h, and complete medium was then changed to DMEM with different concentrations (0, 10, 100, 1,000 and 10,000 nM) of paclitaxel, Adriamycin or PD173074 (Sigma-Aldrich; Merck KGaA) for 24 h or the cells were cultured without FBS for 0, 24, 48, 72, 96, 120 and 144 h at 37°C. Next, 10 µl CCK-8 reagent was added to each well, and the absorbance value was measured at 490 nm using a Multiskan Spectrum spectrophotometer (BioTek Instruments, Inc.).
Apoptosis analysis by flow cytometry
Cells were incubated with different concentrations of paclitaxel, Adriamycin or PD173074 (Sigma-Aldrich; Merck KGaA) for 24 h and collected for staining. After centrifugation at 200 × g at 4°C, the cell pellets were resuspended and placed in 100 µl Annexin V binding buffer containing 5 µl Annexin V-Phycoerythrin (PE) and 2 µl 7-aminoactinomycin D (7-AAD; BD Biosciences). The cells were incubated with PE-labeled Annexin V binding buffer in the dark at room temperature for 20 min. Staining controls were prepared and were single-stained or unstained. A positive apoptotic control was obtained by incubating cells for 15 h with 1 mM hydrogen peroxide (Sigma-Aldrich; Merck KGaA). The stained cell populations were analyzed using a FACSCalibur flow cytometer (BD Biosciences), and the cell cycle distributions were analyzed using FlowJo software v10 (FlowJo LLC).
Immunoprecipitation and immunoblotting
Cells were lysed using cell lysis buffer (PBS, 1% NP40, 1% sodium deoxycholate and 0.1% SDS, 100 µg/ml PMSF, 1 mmol/l sodium orthovanadate and 1 protease inhibitor tablet/10 ml), and the protein concentration was measured using a bicinchoninic acid assay kit (Beyotime Institute of Biotechnology). Equal amounts of denatured proteins (20 µg) were separated by SDS-PAGE on 10% gels and transferred onto PVDF membranes. Antibodies against ST6Gal-I (1:200, cat. no. AF5924, R&D Systems), FGFR1 (1:200, cat. no. ab156031, Abcam), phosphorylated (p)-FGFR1 (1:200, cat. no. ab59194, Abcam), ERK (1:200, cat. no. sc514302, Santa Cruz Biotechnology), p-ERK(1:200, cat. no. sc156521, Santa Cruz Biotechnology), FAK (1:200, cat. no. ab72140, Abcam), p-FAK (1:200, cat. no. ab4792, Abcam), cleaved caspase-3 (1:200, cat. no. ab49822, Abcam) and GAPDH (1:1,000, cat. no. sc32233, Santa Cruz Biotechnology, Inc.) were used as the primary antibodies. For SNA-FGFR1, 100 µl SNA-conjugated agarose (EY Laboratories, Inc.) was added to the lysed protein for 1 h, and the beads were collected. α2–6 sialylated proteins bound to SNA-agarose beads were precipitated by centrifugation (200 × g) at 4°C for 10 min and washed extensively with lysis buffer. Sialylated proteins were released from the complexes and then boiled in SDS-PAGE sample buffer and immunoblotted for FGFR1 (Santa Cruz Biotechnology, Inc.). The membranes were blocked with 5% non-fat milk at room temperature, and then incubated with a primary antibody and horseradish peroxidase-conjugated secondary antibody (Goat IgG Horseradish Peroxidase-conjugated Antibody, cat. no. HAF019, R&D Systems, Oakville, Canada; Mouse IgG Horseradish Peroxidase-conjugated Antibody, cat. no. sc516132, Santa Cruz Biotechnology, Inc.; Rabbit IgG Horseradish Peroxidase-conjugated Antibody, cat. no. HAF008, R&D Systems Oakville, Canada), and detected using an ECL kit (GE Healthcare) according to the manufacturer's protocol. The relative amount of protein was determined by densitometry using ImageJ software (version v1.8.0, National Institutes of Health).
Statistical analysis
All the experiments were repeated 3 times. The data are presented as the mean ± SD and analyzed by GraphPad Prism 6 (GraphPad Software Inc.). One-way ANOVA with the Least Significant Difference post hoc test was performed to determine statistical significance between groups. P<0.05 was considered to indicate a statistically significant difference.
Results
Establishment of oe-ST6Gal-I or sh-ST6Gal-I subcell clones of OVCAR3 human ovarian cancer cells
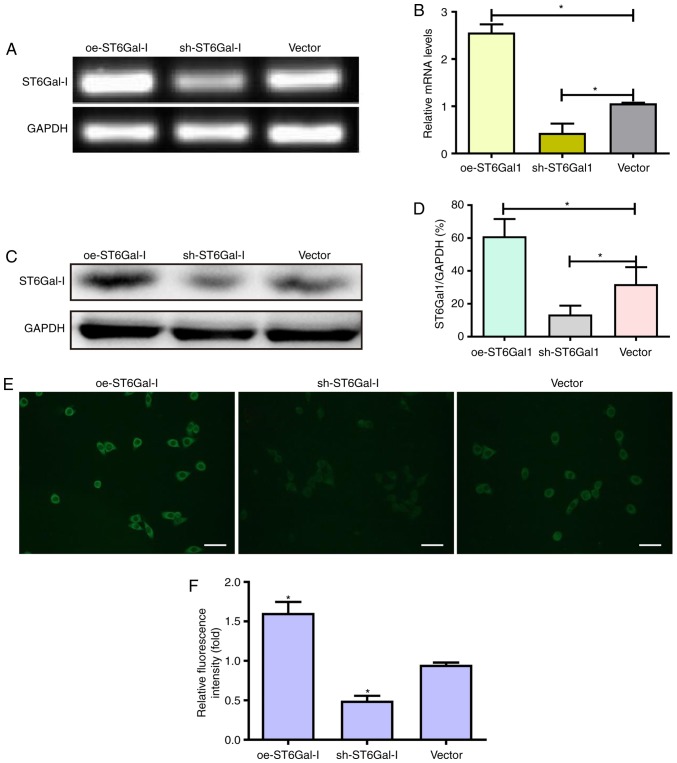
The majority of previous FGFR1 studies have focused on FGFR1 amplification and activating mutations (33–35), whereas regulation of FGFR1 activity via post-translational modifications, such as glycosylation, fucosylation and sialylation, has been studied considerably less. To the best of the authors' knowledge, there are no studies that have investigated the relevance of FGFR1 α2,6-sialylation in the poor prognosis and treatment of ovarian cancer. Therefore, the present study aimed to evaluate the effects of ST6Gal-I on FGFR1 and ovarian cancer progression. An overexpressing plasmid and shRNA vector of ST6Gal-I were constructed and transfected into OVCAR3 cells. After limiting dilution and persistent culture, stable subcell line clones were established. The expression of ST6Gal-I was further confirmed by PCR and western blotting assays. The endogenous ST6Gal-I gene and protein were stably overexpressed in the oe-ST6Gal-I clone, whereas the ST6Gal-I gene and protein were decreased in the sh-ST6Gal-I cell line compared with their expression in the empty vector cell line (Fig. 1A-D). These data indicated that ST6Gal-I was stably overexpressed or knocked down in OVCAR3 cells.
Figure 1.
Establishment of oe-ST6Gal-I and sh-ST6Gal-I subcell clones of OVCAR3 human ovarian cancer cells. (A and B) OVCAR3 cells were transfected with oe-ST6Gal-I, sh-ST6Gal-I and empty vector, and ST6Gal-I stable expression in clones was analyzed by reverse transcription-PCR. Protein levels of ST6Gal-I in oe-ST6Gal-I or sh-ST6Gal-I cells were assessed by (C) western blotting and (D) subsequent densitometry. *P<0.05. (E) α2,6-Linked sialic acid levels were detected with FITC-conjugated SNA lectin by fluorescence microscopy. Scale bar, 100 µm. (F) Relative fluorescence intensity of oe-ST6Gal-I or sh-ST6Gal-I cells. *P<0.05 vs. vector. oe, overexpression; ST6Gal-I, α2,6-sialyltransferase; sh, small hairpin.
To assess the effect of ST6Gal-I upregulation or downregulation on the FGFR1 receptor in tumor cells, FITC-conjugated SNA lectin was used to recognize α2,6-linked sialic acids by immunofluorescence microscopy. The immunofluorescence results showed that oe-ST6Gal-I cells expressed significantly higher levels of α2,6-linked sialic acids than vector cells; conversely, sh-ST6Gal-I cells expressed lower levels of α2,6-linked sialic acids than vector control cells (Fig. 1E and F).
Cells with high ST6Gal-I expression enhance tumor cell viability and migratory ability
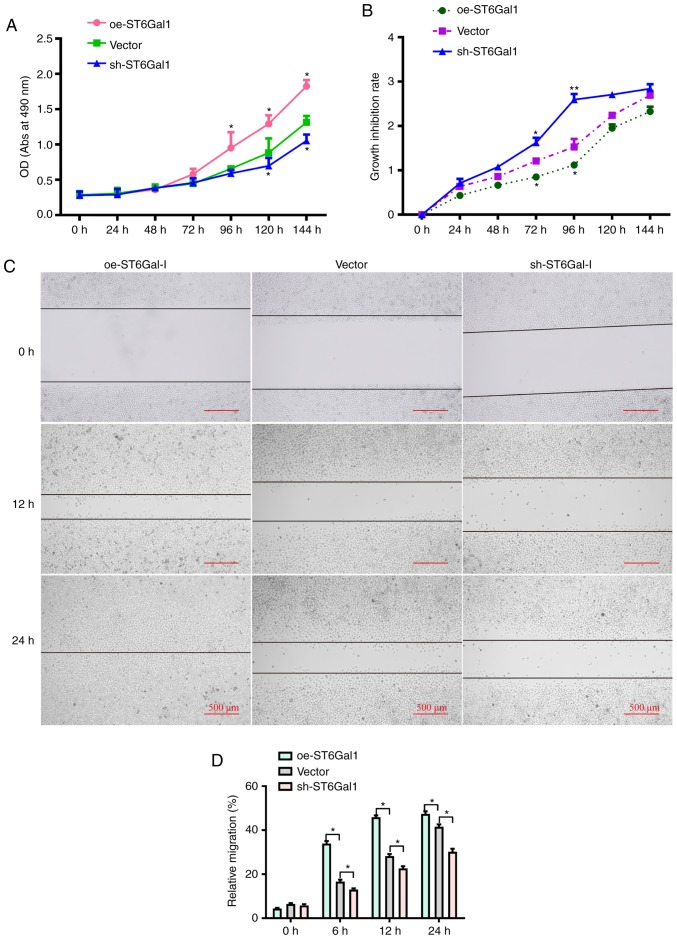
To test whether ST6Gal-I expression affected ovarian cancer cell viability, a CCK-8 assay was conducted. It was observed that the growth rate and cell viability were markedly higher in oe-ST6Gal-I cells than in sh-ST6Gal-I cells and vector cells incubated with complete culture medium (Fig. 2A). To determine the role of ST6Gal-I in protecting against serum withdrawal, oe-ST6Gal-I, sh-ST6Gal-I and vector cells were cultured under serum starvation conditions for 0, 24, 48, 72, 96, 120 or 144 h. As shown in Fig. 2B, the CCK-8 results indicated that the growth inhibition rate and cytotoxicity were lower in oe-ST6Gal-I cells than in sh-ST6Gal-I cells. Additionally, scratch-wound healing assays were conducted to detect cell migration. The results showed that compared with vector cells, the increased ST6Gal-I expression in oe-ST6Gal-I cells significantly promoted cell migration, and the decreased ST6Gal-I expression in sh-ST6Gal-I cells significantly attenuated cell migration at 6, 12 and 24 h (Fig. 2C and D). Collectively, these results suggested that ST6Gal-I overexpression promoted the proliferation and migration of ovarian cancer cells.
Figure 2.
Cells with high ST6Gal-I expression have enhanced viability and migratory ability. (A) Viability in oe-ST6Gal-I and sh-ST6Gal-I cells was measured by a CCK-8 assay. (B) Growth inhibition rates in serum-starved oe-ST6Gal-I and sh-ST6Gal-I cells were assessed by a CCK-8 assay. *P<0.05 vs. respective vector. (C) Scratch wound healing ability of oe-ST6Gal-I and sh-ST6Gal-I cells was determined by imaging cells under a microscope after incubation for 0, 12 and 24 h. (D) Quantitative analysis of the migratory ability of oe-ST6Gal-I and sh-ST6Gal-I cells. *P<0.05. ST6Gal-I; **P<0.01 vs. vector group/α2,6-sialyltransferase; oe, overexpression; sh, small hairpin; CCK-8, Cell Counting Kit-8; OD, optical density; Abs, absorbance.
Cells with high ST6Gal-I expression have reduced cell apoptosis and increased chemoresistance
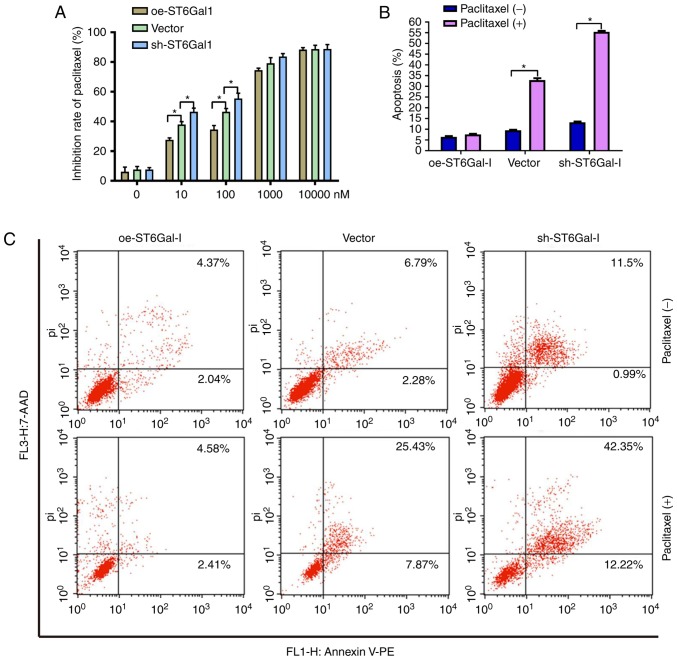
To test the effect of ST6Gal-I expression status on the anticancer efficacy of paclitaxel, a cell viability assay and FACS analysis were performed in ovarian cancer cells. The data showed that the growth inhibition of paclitaxel was dose-dependent in each group. The growth inhibition was higher in sh-ST6Gal-I stable clone cells than in oe-ST6Gal-I cells at drug concentrations of 10–1,000 nM, and most of the cells died at the high concentration of 10 µM (Fig. 3A). Furthermore, PE-labeled Annexin V and 7-AAD staining were analyzed by FACS to determine cell apoptosis. The Annexin V receptor is a phosphatidylserine, which is normally an asymmetric resident of the inner membrane. Only when its asymmetric distribution is lost can a population with increased Annexin V staining be detected. As shown in Fig. 3B and C, FACS analysis of oe-ST6Gal-I cells showed only low surface staining for Annexin V, but Annexin V staining was distinctly higher in sh-ST6Gal-I and vector cells after treatment with paclitaxel. A small population of oe-ST6Gal-I cells showed apoptosis (both early- and late-stage), whereas sh-ST6Gal-I cells had a large population undergoing apoptosis. Collectively, these results suggested that increased α2,6 sialylation in cancer cells reduced cell apoptosis and increased paclitaxel resistance.
Figure 3.
Cells with high ST6Gal-I expression have reduced apoptosis and increased chemoresistance. (A) Inhibition rates of oe-ST6Gal-I and sh-ST6Gal-I cells treated with different concentrations of paclitaxel were assessed by a Cell Counting Kit-8 assay. (B) Quantitative analysis of apoptotic populations of oe-ST6Gal-I and sh-ST6Gal-I cells treated with or without paclitaxel; each analysis was performed in triplicate. *P<0.05. (C) Representative apoptotic populations of oe-ST6Gal-I and sh-ST6Gal-I cells treated with or without paclitaxel were stained with FITC Annexin V and 7-ADD. Lower right quadrant, Annexin V positive; upper right quadrant, Annexin V and 7-AAD positive. ST6Gal-I, α2,6-sialyltransferase; oe, overexpression; sh, small hairpin; 7-AAD, 7-aminoactinomycin D; PE, phycoerythrin; pi, propidium iodide.
α2,6 sialylation of FGFR1 affects the ERK and FAK signaling pathways
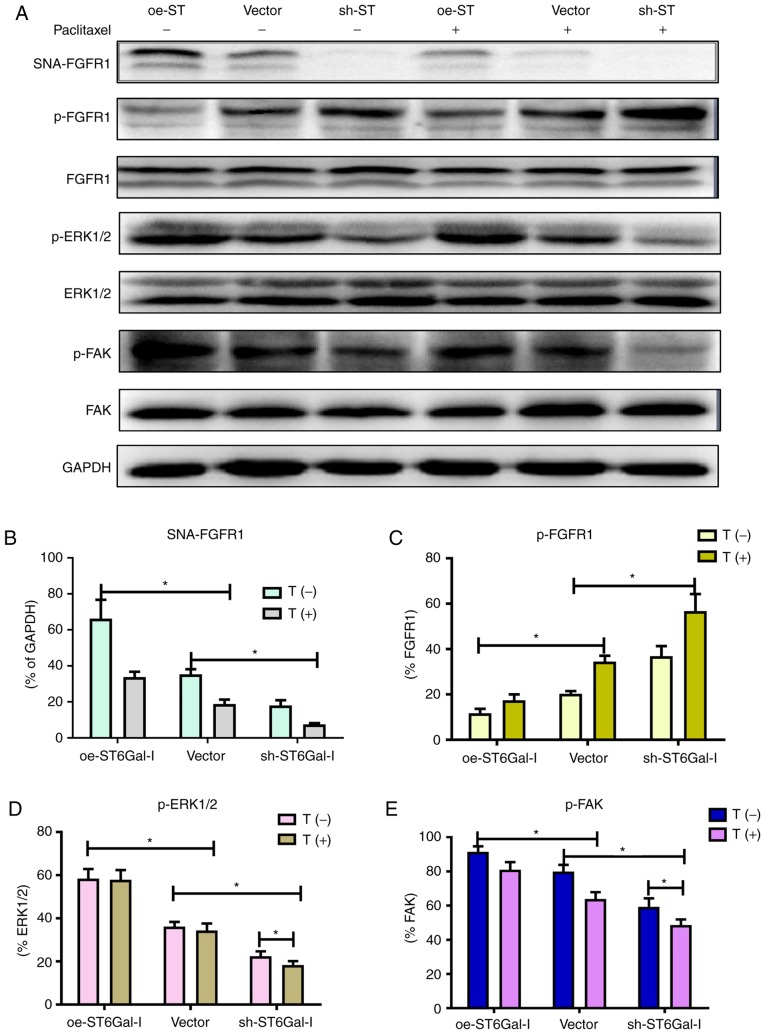
FGFR1 signaling was significantly correlated with tumorigenesis and metastasis in different types of cancer (10,17). A previous study demonstrated that FGFR1 phosphorylation can activate downstream ERK signaling cascades, which play a vital role in the proliferation and survival of cancer cells (36). Prior findings strongly suggested that FGFR1 and β3 integrin work in a complex, leading to the activation of FAK signaling to drive tumor metastasis. To investigate whether α2,6 sialylation of FGFR1 affected FGFR1 phosphorylation and the downstream ERK and FAK signaling pathways, a western blotting assay was used to detect the protein levels of FGFR1, ERK and FAK in oe-ST6Gal-I and sh-ST6Gal-I cells with or without paclitaxel treatment. As shown in Fig. 4A and B, α2,6-sialylated proteins in oe-ST6Gal-I and sh-ST6Gal-I cells bound by SNA-agarose were isolated by SDS-PAGE and immunoblotted for FGFR1. The SNA precipitation results demonstrated that SNA-FGFR1 expression was notably higher in oe-ST6Gal-I cells than in sh-ST6Gal-I cells. ST6Gal-I overexpression in hypersialylated FGFR1 cells, and paclitaxel treatment attenuated this effect. p-FGFR1 expression was notably lower in oe-ST6Gal-I cells than in sh-ST6Gal-I cells, and p-FGFR1 expression was enhanced after 10 µM paclitaxel treatment. The total FGFR1 expression was similar in each group (Fig. 4A and C). ERK1/2 and FAK phosphorylation levels were higher in oe-ST6Gal-I cells than in sh-ST6Gal-I cells, and paclitaxel decreased ERK and FAK phosphorylation levels in cancer cells, especially in sh-ST6Gal-I cells (Fig. 4A, D and E). The present results showed that ST6Gal-I overexpression in cancer cells increased the α2,6 sialylation of proteins and decreased the phosphorylation of FGFR1. Both α2,6 sialylation and phosphorylation of FGFR1 can activate downstream ERK and FAK signaling. Therefore, it was suggested that the decreased FGFR1 phosphorylation did not attenuate the effect of high α2,6 sialylation on downstream cascade activation.
Figure 4.
α2,6 sialylation of FGFR1 affects the ERK and FAK signaling pathways. (A) oe-ST6Gal-I and sh-ST6Gal-I cells were treated with or without paclitaxel for 24 h, and SNA-FGFR1, p-FGFR1, FGFR1, p-ERK1/2, ERK1/2, p-FAK, FAK and GAPDH protein levels were then measured by western blotting. Relative protein intensities of (B) SNA-FGFR1, (C) p-FGFR1, (D) p-ERK1/2 and (E) p-FAK were detected using ImageJ. *P<0.05. FGFR1, fibroblast growth factor receptor 1; FAK, focal adhesion kinase; ST6Gal-I/ST, α2,6-sialyltransferase; oe, overexpression; sh, small hairpin; p, phosphorylated; T, paclitaxel.
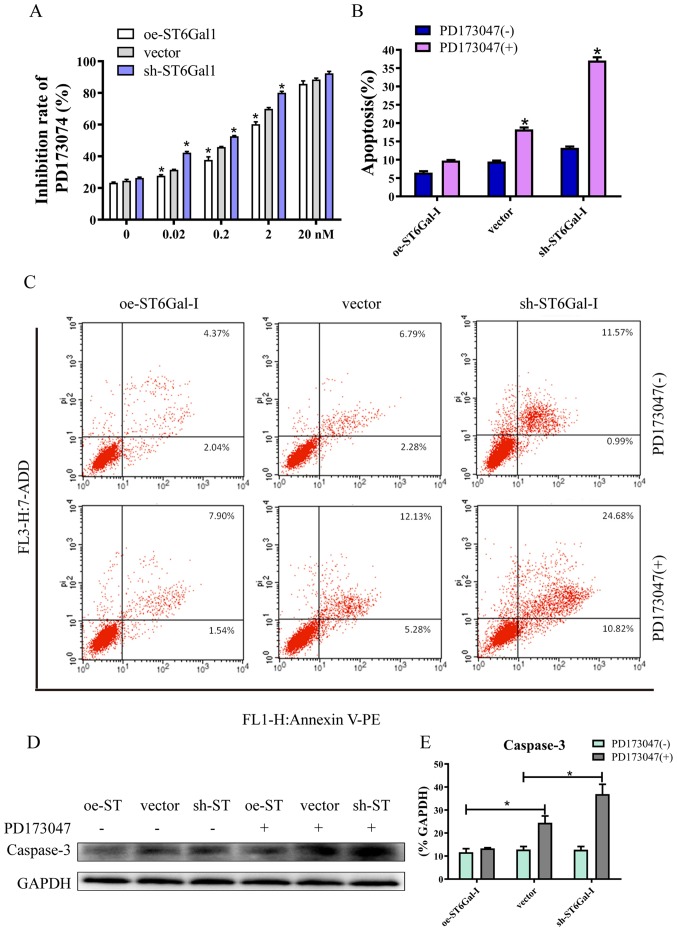
High ST6Gal-I expression attenuates FGFR1 inhibitor-induced cell apoptosis
Since aberrant FGFR activity has been implicated in various cancer types, several FGFR inhibitors are currently in the early phases of clinical development (37). PD173074 has reportedly shown both high affinity and selectivity for the FGFR family, and is being used as an FGFR inhibitor in the clinical settings and in experiments (38,39). The CCK-8 assay results showed that PD173074 inhibited cancer cell growth in a dose-dependent manner, and the inhibition rate was lower in oe-ST6Gal-I cells than in sh-ST6Gal-I cells. Moreover, there was no significant difference between oe-ST6Gal-I cells and sh-ST6Gal-I cells when the concentration of PD173074 reached 20 nM, and most of the cells died (Fig. 5A). The FACS results demonstrated that the number of apoptotic cells (including early and late apoptotic cells) was increased with PD173074 treatment in oe-ST6Gal-I cells. The number of apoptotic sh-ST6Gal-I cells was notably higher with PD173074 treatment than the oe-ST6Gal-I cells (Fig. 5B and C). In agreement with the flow cytometry results, the western blotting results indicated that the apoptotic marker caspase-3 was significantly increased in sh-ST6Gal-I cells treated with PD173074, whereas caspase-3 levels in oe-ST6Gal-I cells were slightly increased after PD173074 treatment compared with vector cells (Fig. 5D and E). Taken together, these data suggested that FGFR1 inhibitors can effectively induce cell death in vector cells and sh-ST6Gal-I cells; however, ST6Gal-I overexpression reduced this anticancer effect.
Figure 5.
High ST6Gal-I expression attenuates FGFR1 inhibitor-induced cell apoptosis. (A) PD173074 is an FGFR1 inhibitor, and the inhibition rate of PD173074 in oe-ST6Gal-I and sh-ST6Gal-I cells was analyzed by a Cell Counting Kit-8 assay. *P<0.05 vs. respective vector. (B) Quantitative analysis of apoptotic populations of oe-ST6Gal-I and sh-ST6Gal-I cells incubated with or without PD173074. *P<0.05 vs. respective PD173047 (−). (C) Representative apoptotic populations of oe-ST6Gal-I and sh-ST6Gal-I cells incubated with or without PD173074 were stained with FITC Annexin V and 7-ADD. (D) Caspase-3 levels were analyzed in oe-ST6Gal-I and sh-ST6Gal-I cells incubated with or without PD173074 by western blotting. (E) Quantitative analysis of caspase-3 protein intensity. *P<0.05. FGFR1, fibroblast growth factor receptor 1; ST6Gal-I, α2,6-sialyltransferase; oe, overexpression; sh, small hairpin; 7-ADD, 7-aminoactinomycin D; PE, phycoerythrin; pi, propidium iodide.
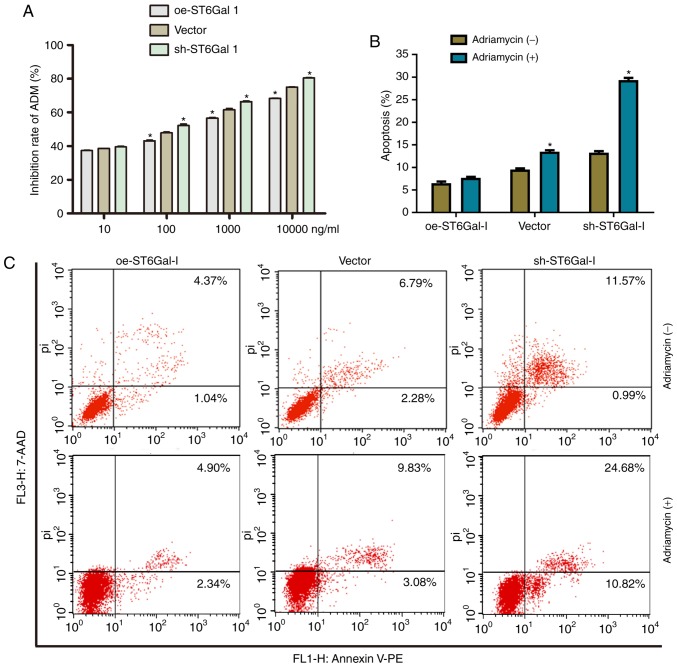
High ST6Gal-I expression protects cancer cells from Adriamycin
To further investigate the drug-resistant effects of ST6Gal-I on another chemotherapy drug, the inhibition rate of cells and cell apoptosis after treatment with Adriamycin were assessed. Similar to the paclitaxel results, the growth inhibition of Adriamycin was dose-dependent in each group. The growth inhibition was higher in sh-ST6Gal-I stable clone cells than in oe-ST6Gal-I cells at drug concentrations of 10–10000 nM (Fig. 6A). Furthermore, the FACS results showed that only a small population of oe-ST6Gal-I cells showed apoptosis (7.24%), whereas sh-ST6Gal-I cells had a large population undergoing apoptosis (29.34%) with Adriamycin treatment (Fig. 6B and C). In conclusion, ST6Gal-I overexpression enhances the chemoresistance of ovarian cancer cells; conversely, ST6Gal-I knockdown decreases chemoresistance.
Figure 6.
High ST6Gal-I expression protected cancer cells from Adriamycin. (A) Inhibition rate of Adriamycin in oe-ST6Gal-I or sh-ST6Gal-I cells was analyzed by a Cell Counting Kit-8 assay. *P<0.05 vs. respective vector. (B) Quantitative analysis of apoptotic populations of oe-ST6Gal-I and sh-ST6Gal-I cells incubated with or without Adriamycin. *P<0.05 vs. respective Adriamycin (−). (C) Representative apoptotic populations of oe-ST6Gal-I and sh-ST6Gal-I cells incubated with or without Adriamycin were stained with FITC Annexin V and 7-ADD. ST6Gal-I, α2,6-sialyltransferase; oe, overexpression; sh, small hairpin; 7-AAD, 7-aminoactinomycin D; PE, phycoerythrin; pi, propidium iodide.
Discussion
Ovarian cancer is characterized by a lack of early symptoms or screening methods, which often lead to late diagnosis in advanced stages and a high mortality rate (40). ST6Gal-I has been demonstrated to confer radiation resistance in colon cancer cell lines (41). However, the functional contribution of ST6Gal-I to ovarian cancer has yet to be elucidated. Accumulating evidence suggests that ST6Gal-I is a major inhibitor of cell death pathways initiated by Fas, TNFR1 and galectins (27,42). RTKs, such as epidermal growth factor receptor and FGFR, are highly expressed or activated in ovarian cancer (10,43,44). Previous studies have demonstrated that the amplification and mutation of FGFR1 are associated with poor ovarian cancer prognosis and malignancy (45,46). In the present study, it was verified that ST6Gal-I overexpression leads to high sialylation of FGFR1 in ovarian cancer and the subcellular mechanism was specifically investigated; FGFR1 signaling regulates cancer cell behavior. The present results provide novel insight for the role of α2,6 sialylated FGFR1 in ovarian cancer drug resistance.
In the present study, it was observed that ST6Gal-I overexpression induced high FGFR1 sialylation, leading to high proliferation and a low growth inhibition rate under serum deprivation conditions. Moreover, compared with sh-ST6Gal-I cells, ST6Gal-I overexpression promoted cancer cell migration after scratch wounding. In addition, it was identified that ST6Gal-I overexpression attenuated paclitaxel-induced apoptosis. These findings are consistent with the hypothesis that ST6Gal-I activity might underlie the survival or drug resistance of cancer cell populations (29,47).
FGFR1 is a member of the FGFR family of RTKs; FGFR1 activation leads to downstream signaling via the ERK and FAK pathways, which are central to growth, survival migration and angiogenesis in many cancer types (48,49). ERK activation might be associated with the progression of a wide variety of neoplasias, as well as poor prognosis and chemotherapeutic resistance in cancer cells (50,51). Accumulating evidence supports that FAK also plays a vital role in tumor cell proliferation, survival and migration (52,53). In agreement with this evidence, it was observed that ST6Gal-I overexpression increased the α2,6-linked sialic acids of FGFR1 and enhanced ERK- and FAK-mediated cell signaling pathways. Although ST6Gal-I overexpression decreased FGFR1 phosphorylation, the high FGFR1 sialylation levels could activate the ERK- and FAK-mediated signaling pathways through other mechanisms. Therefore, further investigation is needed to thoroughly explore the underlying mechanism.
Multiple FGFR inhibitors are in development. Many of these are multi-targeted tyrosine kinase inhibitors against targeted receptors, including FGFR1 (54). In the present study, it was identified that the FGFR1 inhibitor PD173074 suppressed cancer cell proliferation and induced cell apoptosis; however, ST6Gal-I overexpression attenuated the effects of PD173074. Consistent with these results, ST6Gal-I overexpression also weakened the effect of Adriamycin on cancer cells. A hypothesis is that ST6Gal-I overexpression in cancer cells may protect cells from multiple apoptotic stimuli, thus promoting cell proliferation and migration.
In conclusion, the present data suggested that ST6Gal-I overexpression induces high levels of protein α2,6-sialylation and that FGFR1 is one of the targeted molecules. Sialylated FGFR1 activated the ERK and FAK pathways, thus promoting cell proliferation and migration. Overall, the present study provides new insight into how ST6Gal-I and FGFR1 signaling regulate cancer progression and drug resistance in ovarian cancer cells.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- FGFR1
fibroblast growth factor receptor 1
- ST6Gal-I
α2,6-sialyltransferase
- FAK
focal adhesion kinase
Funding
This study was supported by The National Natural Science Foundation of China (grant nos. 81071751 and 81703120), Guangdong Provincial Medical Research Foundation (grant no. A2016360) and Natural Science Foundation of Guangdong Province (grant no. 2017A030310365).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
LO and XH performed the experiments. SL and XW secured the funding for the present study. SL designed the present study, and made final corrections to the manuscript; XW provided ovarian cancer cell lines and analyzed the chemical resistant data. XH and ZD sourced and prepared the equipment and reagents for the experiments. NL and YS performed data collection. JL and LG analyzed and interpreted the data. LO wrote the manuscript. LO and LG revised the manuscript. LO responded to the comments and questions and revised the paper accordingly and LG edited the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 2.Cotton LM, O'Bryan MK, Hinton BT. Cellular signaling by fibroblast growth factors (FGFs) and their receptors (FGFRs) in male reproduction. Endocr Rev. 2008;29:193–216. doi: 10.1210/er.2007-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Touat M, Ileana E, Postel-Vinay S, André F, Soria JC. Targeting FGFR signaling in cancer. Clin Cancer Res. 2015;21:2684–2694. doi: 10.1158/1078-0432.CCR-14-2329. [DOI] [PubMed] [Google Scholar]
- 4.Manchado E, Weissmueller S, Morris JP, IV, Chen CC, Wullenkord R, Lujambio A, de Stanchina E, Poirier JT, Gainor JF, Corcoran RB, et al. A combinatorial strategy for treating KRAS-mutant lung cancer. Nature. 2016;534:647–651. doi: 10.1038/nature18600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turkington RC, Longley DB, Allen WL, Stevenson L, McLaughlin K, Dunne PD, Blayney JK, Salto-Tellez M, Van Schaeybroeck S, Johnston PG. Fibroblast growth factor receptor 4 (FGFR4): A targetable regulator of drug resistance in colorectal cancer. Cell Death Dis. 2014;5:e1046. doi: 10.1038/cddis.2014.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ye T, Wei X, Yin T, Xia Y, Li D, Shao B, Song X, He S, Luo M, Gao X, et al. Inhibition of FGFR signaling by PD173074 improves antitumor immunity and impairs breast cancer metastasis. Breast Cancer Res Treat. 2014;143:435–446. doi: 10.1007/s10549-013-2829-y. [DOI] [PubMed] [Google Scholar]
- 7.Wan X, Corn PG, Yang J, Palanisamy N, Starbuck MW, Efstathiou E, Li Ning Tapia EM, Zurita AJ, Aparicio A, Ravoori MK, et al. Prostate cancer cell-stromal cell crosstalk via FGFR1 mediates antitumor activity of dovitinib in bone metastases. Sci Transl Med. 2014;6:252ra122. doi: 10.1126/scitranslmed.3009332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schweiger N, Hauck M, Steinhoff H, Sampl S, Reifinger M, Walter I, Kreilmeier T, Marian B, Grusch M, Berger W, et al. Canine and human sarcomas exhibit predominant FGFR1 expression and impaired viability after inhibition of signaling. Mol Carcinog. 2015;54:841–852. doi: 10.1002/mc.22155. [DOI] [PubMed] [Google Scholar]
- 9.Garay T, Molnár E, Juhász É, László V, Barbai T, Dobos J, Schelch K, Pirker C, Grusch M, Berger W, et al. Sensitivity of melanoma cells to EGFR and FGFR activation but not inhibition is influenced by oncogenic BRAF and NRAS mutations. Pathol Oncol Res. 2015;21:957–968. doi: 10.1007/s12253-015-9916-9. [DOI] [PubMed] [Google Scholar]
- 10.Cole C, Lau S, Backen A, Clamp A, Rushton G, Dive C, Hodgkinson C, McVey R, Kitchener H, Jayson GC. Inhibition of FGFR2 and FGFR1 increases cisplatin sensitivity in ovarian cancer. Cancer Biol Ther. 2010;10:495–504. doi: 10.4161/cbt.10.5.12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meng QH, Xu E, Hildebrandt MA, Liang D, Lu K, Ye Y, Wagar EA, Wu X. Genetic variants in the fibroblast growth factor pathway as potential markers of ovarian cancer risk, therapeutic response, and clinical outcome. Clin Chem. 2014;60:222–232. doi: 10.1373/clinchem.2013.211490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Birrer MJ, Johnson ME, Hao K, Wong KK, Park DC, Bell A, Welch WR, Berkowitz RS, Mok SC. Whole genome oligonucleotide-based array comparative genomic hybridization analysis identified fibroblast growth factor 1 as a prognostic marker for advanced-stage serous ovarian adenocarcinomas. J Clin Oncol. 2007;25:2281–2287. doi: 10.1200/JCO.2006.09.0795. [DOI] [PubMed] [Google Scholar]
- 13.Ivan M, Matei D. Blockade of FGF signaling: Therapeutic promise for ovarian cancer. Cancer Biol Ther. 2010;10:505–508. doi: 10.4161/cbt.10.5.13023. [DOI] [PubMed] [Google Scholar]
- 14.Marmé F, Hielscher T, Hug S, Bondong S, Zeillinger R, Castillo-Tong DC, Sehouli J, Braicu I, Vergote I, Isabella C, et al. Fibroblast growth factor receptor 4 gene (FGFR4) 388Arg allele predicts prolonged survival and platinum sensitivity in advanced ovarian cancer. Int J Cancer. 2012;131:E586–E591. doi: 10.1002/ijc.27329. [DOI] [PubMed] [Google Scholar]
- 15.Zhao X, Zhou Y, Chen YU, Yu F. miR-494 inhibits ovarian cancer cell proliferation and promotes apoptosis by targeting FGFR2. Oncol Lett. 2016;11:4245–4251. doi: 10.3892/ol.2016.4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zecchini S, Bombardelli L, Decio A, Bianchi M, Mazzarol G, Sanguineti F, Aletti G, Maddaluno L, Berezin V, Bock E, et al. The adhesion molecule NCAM promotes ovarian cancer progression via FGFR signalling. EMBO Mol Med. 2011;3:480–494. doi: 10.1002/emmm.201100152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang F, Zhang Y, Ressler SJ, Ittmann MM, Ayala GE, Dang TD, Wang F, Rowley DR. FGFR1 is essential for prostate cancer progression and metastasis. Cancer Res. 2013;73:3716–3724. doi: 10.1158/0008-5472.CAN-12-3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Welm BE, Freeman KW, Chen M, Contreras A, Spencer DM, Rosen JM. Inducible dimerization of FGFR1: Development of a mouse model to analyze progressive transformation of the mammary gland. J Cell Biol. 2002;157:703–714. doi: 10.1083/jcb.200107119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bohrer LR, Schwertfeger KL. Macrophages promote fibroblast growth factor receptor-driven tumor cell migration and invasion in a CXCR2-dependent manner. Mol Cancer Res. 2012;10:1294–1305. doi: 10.1158/1541-7786.MCR-12-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duchesne L, Tissot B, Rudd TR, Dell A, Fernig DG. N-glycosylation of fibroblast growth factor receptor 1 regulates ligand and heparan sulfate co-receptor binding. J Biol Chem. 2006;281:27178–27189. doi: 10.1074/jbc.M601248200. [DOI] [PubMed] [Google Scholar]
- 21.Taniguchi N, Korekane H. Branched N-glycans and their implications for cell adhesion, signaling and clinical applications for cancer biomarkers and in therapeutics. BMB Rep. 2011;44:772–781. doi: 10.5483/BMBRep.2011.44.12.772. [DOI] [PubMed] [Google Scholar]
- 22.Dall'Olio F, Malagolini N, Trinchera M, Chiricolo M. Mechanisms of cancer-associated glycosylation changes. Front Biosci (Landmark Ed) 2012;17:670–699. doi: 10.2741/3951. [DOI] [PubMed] [Google Scholar]
- 23.Schultz MJ, Swindall AF, Bellis SL. Regulation of the metastatic cell phenotype by sialylated glycans. Cancer Metastasis Rev. 2012;31:501–518. doi: 10.1007/s10555-012-9359-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dall'Olio F, Malagolini N, Trinchera M, Chiricolo M. Sialosignaling: Sialyltransferases as engines of self-fueling loops in cancer progression. Biochim Biophys Acta. 2014;1840:2752–2764. doi: 10.1016/j.bbagen.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 25.Harduin-Lepers A, Vallejo-Ruiz V, Krzewinski-Recchi MA, Samyn-Petit B, Julien S, Delannoy P. The human sialyltransferase family. Biochimie. 2001;83:727–737. doi: 10.1016/S0300-9084(01)01301-3. [DOI] [PubMed] [Google Scholar]
- 26.Lee M, Park JJ, Lee YS. Adhesion of ST6Gal I-mediated human colon cancer cells to fibronectin contributes to cell survival by integrin beta1-mediated paxillin and AKT activation. Oncol Rep. 2010;23:757–761. [PubMed] [Google Scholar]
- 27.Swindall AF, Bellis SL. Sialylation of the Fas death receptor by ST6Gal-I provides protection against Fas-mediated apoptosis in colon carcinoma cells. J Biol Chem. 2011;286:22982–22990. doi: 10.1074/jbc.M110.211375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhuo Y, Chammas R, Bellis SL. Sialylation of beta1 integrins blocks cell adhesion to galectin-3 and protects cells against galectin-3-induced apoptosis. J Biol Chem. 2008;283:22177–22185. doi: 10.1074/jbc.M800015200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park JJ, Yi JY, Jin YB, Lee YJ, Lee JS, Lee YS, Ko YG, Lee M. Sialylation of epidermal growth factor receptor regulates receptor activity and chemosensitivity to gefitinib in colon cancer cells. Biochem Pharmacol. 2012;83:849–857. doi: 10.1016/j.bcp.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 30.Britain CM, Dorsett KA, Bellis SL. The glycosyltransferase ST6Gal-I protects tumor cells against serum growth factor withdrawal by enhancing survival signaling and proliferative potential. J Biol Chem. 2017;292:4663–4673. doi: 10.1074/jbc.M116.763862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Saporiti F, Piacentini L, Alfieri V, Bono E, Ferrari F, Chiesa M, Colombo GI. Melanocortin-1 receptor positively regulates human artery endothelial cell migration. Cell Physiol Biochem. 2019;52:1339–1360. doi: 10.33594/000000094. [DOI] [PubMed] [Google Scholar]
- 33.Park JS, Lee JS, Kim EY, Jung JY, Kim SK, Chang J, Kim DJ, Lee CY, Jung I, Kim JH. The frequency and impact of FGFR1 amplification on clinical outcomes in Korean patients with small cell lung cancer. Lung Cancer. 2015;88:325–331. doi: 10.1016/j.lungcan.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 34.Preusser M, Berghoff AS, Berger W, Ilhan-Mutlu A, Dinhof C, Widhalm G, Dieckmann K, Wöhrer A, Hackl M, von Deimling A, et al. High rate of FGFR1 amplifications in brain metastases of squamous and non-squamous lung cancer. Lung Cancer. 2014;83:83–89. doi: 10.1016/j.lungcan.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 35.Gessi M, Moneim YA, Hammes J, Goschzik T, Scholz M, Denkhaus D, Waha A, Pietsch T. FGFR1 mutations in Rosette-forming glioneuronal tumors of the fourth ventricle. J Neuropathol Exp Neurol. 2014;73:580–584. doi: 10.1097/NEN.0000000000000080. [DOI] [PubMed] [Google Scholar]
- 36.Wu J, Wei T, Tang Q, Weng B, Li W, Jiang X, Ding T, Li X, Liang G, Cai Y, Ji J. Discovery and anti-cancer evaluation of two novel non-ATP-competitive FGFR1 inhibitors in non-small-cell lung cancer. BMC Cancer. 2015;15:276. doi: 10.1186/s12885-015-1307-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turner N, Grose R. Fibroblast growth factor signalling: From development to cancer. Nat Rev Cancer. 2010;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 38.Mohammadi M, Froum S, Hamby JM, Schroeder MC, Panek RL, Lu GH, Eliseenkova AV, Green D, Schlessinger J, Hubbard SR. Crystal structure of an angiogenesis inhibitor bound to the FGF receptor tyrosine kinase domain. EMBO J. 1998;17:5896–5904. doi: 10.1093/emboj/17.20.5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen PT, Tsunematsu T, Yanagisawa S, Kudo Y, Miyauchi M, Kamata N, Takata T. The FGFR1 inhibitor PD173074 induces mesenchymal-epithelial transition through the transcription factor AP-1. Br J Cancer. 2013;109:2248–2258. doi: 10.1038/bjc.2013.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 41.Lee M, Lee HJ, Bae S, Lee YS. Protein sialylation by sialyltransferase involves radiation resistance. Mol Cancer Res. 2008;6:1316–1325. doi: 10.1158/1541-7786.MCR-07-2209. [DOI] [PubMed] [Google Scholar]
- 42.Zhuo Y, Bellis SL. Emerging role of alpha2,6-sialic acid as a negative regulator of galectin binding and function. J Biol Chem. 2011;286:5935–5941. doi: 10.1074/jbc.R110.191429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gui T, Shen K. The epidermal growth factor receptor as a therapeutic target in epithelial ovarian cancer. Cancer Epidemiol. 2012;36:490–496. doi: 10.1016/j.canep.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 44.McKie AB, Vaughan S, Zanini E, Okon IS, Louis L, de Sousa C, Greene MI, Wang Q, Agarwal R, Shaposhnikov D, et al. The OPCML tumor suppressor functions as a cell surface repressor-adaptor, negatively regulating receptor tyrosine kinases in epithelial ovarian cancer. Cancer Discov. 2012;2:156–171. doi: 10.1158/2159-8290.CD-11-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gorringe KL, Jacobs S, Thompson ER, Sridhar A, Qiu W, Choong DY, Campbell IG. High-resolution single nucleotide polymorphism array analysis of epithelial ovarian cancer reveals numerous microdeletions and amplifications. Clin Cancer Res. 2007;13:4731–4739. doi: 10.1158/1078-0432.CCR-07-0502. [DOI] [PubMed] [Google Scholar]
- 46.Whitworth MK, Backen AC, Clamp AR, Wilson G, McVey R, Friedl A, Rapraeger AC, David G, McGown A, Slade RJ, et al. Regulation of fibroblast growth factor-2 activity by human ovarian cancer tumor endothelium. Clin Cancer Res. 2005;11:4282–4288. doi: 10.1158/1078-0432.CCR-04-1386. [DOI] [PubMed] [Google Scholar]
- 47.Chen X, Wang L, Zhao Y, Yuan S, Wu Q, Zhu X, Niang B, Wang S, Zhang J. ST6Gal-I modulates docetaxel sensitivity in human hepatocarcinoma cells via the p38 MAPK/caspase pathway. Oncotarget. 2016;7:51955–51964. doi: 10.18632/oncotarget.10192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singleton KR, Hinz TK, Kleczko EK, Marek LA, Kwak J, Harp T, Kim J, Tan AC, Heasley LE. Kinome RNAi screens reveal synergistic targeting of MTOR and FGFR1 pathways for treatment of lung cancer and HNSCC. Cancer Res. 2015;75:4398–4406. doi: 10.1158/0008-5472.CAN-15-0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anderson HJ, Galileo DS. Small-molecule inhibitors of FGFR, integrins and FAK selectively decrease L1CAM-stimulated glioblastoma cell motility and proliferation. Cell Oncol (Dordr) 2016;39:229–242. doi: 10.1007/s13402-016-0267-7. [DOI] [PubMed] [Google Scholar]
- 50.Wales CT, Taylor FR, Higa AT, McAllister HA, Jacobs AT. ERK-dependent phosphorylation of HSF1 mediates chemotherapeutic resistance to benzimidazole carbamates in colorectal cancer cells. Anticancer Drugs. 2015;26:657–666. doi: 10.1097/CAD.0000000000000231. [DOI] [PubMed] [Google Scholar]
- 51.Xiao Z, Ding N, Xiao G, Wang S, Wu Y, Tang L. Reversal of multidrug resistance by gefitinib via RAF1/ERK pathway in pancreatic cancer cell line. Anat Rec (Hoboken) 2012;295:2122–2128. doi: 10.1002/ar.22552. [DOI] [PubMed] [Google Scholar]
- 52.Zhang J, Hochwald SN. The role of FAK in tumor metabolism and therapy. Pharmacol Ther. 2014;142:154–163. doi: 10.1016/j.pharmthera.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee BY, Timpson P, Horvath LG, Daly RJ. FAK signaling in human cancer as a target for therapeutics. Pharmacol Ther. 2015;146:132–149. doi: 10.1016/S1525-0016(16)33940-5. [DOI] [PubMed] [Google Scholar]
- 54.Katoh M. FGFR inhibitors: Effects on cancer cells, tumor microenvironment and whole-body homeostasis (Review) Int J Mol Med. 2016;38:3–15. doi: 10.3892/ijmm.2016.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.