Abstract
Average effects of alleles can show considerable differences between populations. The magnitude of these differences can be measured by the additive genetic correlation between populations (). This can be lower than one due to the presence of non-additive genetic effects together with differences in allele frequencies between populations. However, the relationship between the nature of non-additive effects, differences in allele frequencies, and the value of remains unclear, and was therefore the focus of this study. We simulated genotype data of two populations that have diverged under drift only, or under drift and selection, and we simulated traits where the genetic model and magnitude of non-additive effects were varied. Results showed that larger differences in allele frequencies and larger non-additive effects resulted in lower values of . In addition, we found that with epistasis, decreases with an increase of the number of interactions per locus. For both dominance and epistasis, we found that, when non-additive effects became extremely large, had a lower bound that was determined by the type of inter-allelic interaction, and the difference in allele frequencies between populations. Given that dominance variance is usually small, our results show that it is unlikely that true values lower than 0.80 are due to dominance effects alone. With realistic levels of epistasis, dropped as low as 0.45. These results may contribute to the understanding of differences in genetic expression of complex traits between populations, and may help in explaining the inefficiency of genomic trait prediction across populations.
Keywords: dominance, epistasis, genomic prediction, average effects, population divergence
Populations can differ considerably in the average effects of loci (i.e., , the difference between average effects of the two alleles, Falconer and Mackay (1996)). For a given genotype (i.e., individual), differences in between two populations lead to differences in the additive genetic values of that genotype, as expressed in both populations. The magnitude of these differences can be measured by the additive genetic correlation between populations (), defined as the correlation between the additive genetic values of a genotype expressed in population 1 and population 2. In reality, a single genotype cannot belong to two populations at the same time. This means that a trait expressed in two populations can be seen as a pair of traits that cannot be measured on the same individual, analogous to e.g., age at sexual maturity in males and females (Falconer and Mackay 1996). Although no phenotypic correlation exists between such pairs of traits, they can nevertheless be genetically correlated.
The can be lower than one due to genotype by environment interaction (GxE) (Falconer 1952), or due to non-additive genetic effects (GxG-interaction) together with differences in allele frequencies between populations (Fisher 1918). Knowledge of this correlation contributes to the understanding of the genetic architectures of polygenic traits (de Candia et al. 2013; Brown et al. 2016). Such understanding may lead to improved knowledge of genetics and can facilitate accurate prediction of traits, such as disease risk in humans and yield traits in crops (Forsberg et al. 2017). Furthermore, understanding the genetic mechanisms that determine may help in explaining the inefficiency of trait prediction across populations (Wientjes et al. 2015).
Following Falconer (1952), we can interpret a metric trait expressed in two populations as two different, genetically correlated traits. The additive genetic value of individual for the trait expressed in the population that belongs to (say, population 1) is
where is a column vector of additive genotypes (measured as allele counts, minus the mean allele count in the population) of individual at quantitative trait loci (QTL), and is a column vector of average effects at those QTL in population 1. The additive genetic value of individual for another population (say, population 2) is
where is a column vector of average effects in population 2. Conceptually, this can be thought of as the additive genetic value for an individual in population 2 that has the same genotype as individual . Here we define the additive genetic correlation between population 1 and population 2 () as the correlation between both additive genetic values for the individuals in population 1,
| (1) |
In other words, the is defined for individuals coming from population 1, which may be different from the defined for individuals coming from population 2 (See Discussion).
Equation (1) illustrates that the value of depends on the differences in average effects between populations. With non-additive effects, average effects depend on the allele frequencies in the population, and, therefore, larger differences in allele frequencies between populations are expected to result in lower values of .
Note that is the correlation between the additive genetic values, not the genotypic values (i.e., additive plus non-additive genetic values). In the absence of GxE-interaction, the genotypic correlation between both populations is equal to one irrespective of the presence of GxG-interactions, because the genotypic value of a genotype (i.e., individual) is the same in both populations. The additive genetic correlation may, however, be smaller than one because the partitioning of genotypic values into additive genetic values, dominance deviations and epistatic deviations depends on the allele frequencies (Fisher 1918; Cockerham 1954; Kempthorne 1954).
A deeper understanding of the relationship between non-additive genetic effects, allele frequencies and may help geneticists to predict the value of based on the importance of dominance and epistasis in the expression of the trait, and the genetic distance between populations. Wei et al. (1991) studied the impact of dominance on the additive genetic correlation between a purebred and crossbred population, known as . Using a two-locus model, they showed that indeed depends on both the magnitude of the dominance effect (), and on the difference in allele frequencies between the populations. We are not aware of any theoretical studies that investigated the relationship between the importance of dominance and between two purebred populations.
With epistasis, is also expected to depend on the magnitude of epistatic effects and on the difference in allele frequencies between populations. Epistasis in the functional (i.e., biological) sense means that the genotypic values of individuals depend on interactions between alleles or genotypes at different loci (Bateson and Mendel 1909), and there is substantial evidence for the existence of functional epistasis across species (Carlborg et al. 2003; Le Rouzic et al. 2008; Pettersson et al. 2011; Mackay 2015). Epistasis in the statistical sense is measured as the deviation of multi-locus genotypic values from the sum of the marginal effects (i.e., average and dominance effects) of the individual loci (Fisher 1918; Cockerham 1954). Although functional epistatic interactions do not necessarily lead to substantial statistical epistasis (Cheverud and Routman 1995; Hill et al. 2008; Mäki-Tanila and Hill 2014), epistasis can contribute significantly to the additive genetic variance because average effects of individual loci may capture a substantial part of the functional epistasis (Hill et al. 2008; Mäki-Tanila and Hill 2014; Monnahan and Kelly 2015). Furthermore, epistatic variance may be ‘converted’ into additive genetic variance due to genetic drift or due to selection (Cheverud and Routman 1996; Hill 2017). Thus, epistatic interactions modify average effects of individual loci when allele frequencies change, and may therefore play an important role in the value of and its change over time.
In summary, the between populations is affected by non-additive effects in combination with differences in allele frequencies between populations. For populations in the same environment (i.e., in the absence of GxE), is equal to 1 in the absence of non-additive effects or in the absence of allele frequency differences. So far, the relationship between the nature and magnitude of non-additive effects, differences in allele frequencies, and the value of remains unclear. Our objective was therefore to investigate the impact of non-additive effects on for populations that have diverged either under drift only, or under both drift and selection.
Methods
We aimed to investigate the relationship between non-additive effects and the additive genetic correlation between populations () with small effective size, as observed in livestock. For this purpose, we simulated genotypes of quantitative trait loci (QTL) for two populations that have diverged for a number of generations under either pure drift, or under drift and selection. The populations were assumed to be kept in the same environment, so there was no GxE. We simulated traits following several scenarios that differed in the type (i.e., genetic model) and the magnitude of non-additive effects (Table 1).
Table 1. Overview of scenarios with the genetic architecture of the trait, and their parameters for distributions of sampled dominance coefficients and epistatic coefficients.
| Parameters for distributions of non-additive effects | ||||
|---|---|---|---|---|
| Configuration | Small | Intermediate | Large | |
| D | Dominance | |||
| EAA | A*A | |||
| EDD | D*D | |||
| EC | Complementary | |||
| EM | Multiplicative | |||
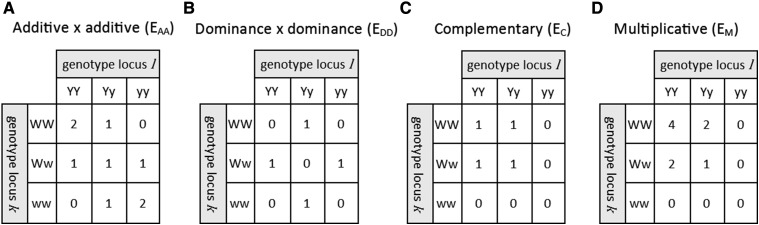
We considered six genetic models; a basic model with additive effects only (A), which served as a basis for comparison, and five alternative models with non-additive effects: one with only dominance effects (D), and four with only epistatic effects. With epistasis, we simulated interactions between pairs of loci that followed one of the configurations presented in Figure 1. We chose these genetic models so that there were scenarios with only dominance variance (D), scenarios with only additive by additive epistatic variance (EAA and EM), and scenarios with all types of non-additive variance (EC and EDD). For each genetic model, we considered three magnitudes of non-additive effects, labeled as small, intermediate, and large.
Figure 1.
Epistatic contrasts for four biological epistatic configurations.
Simulation
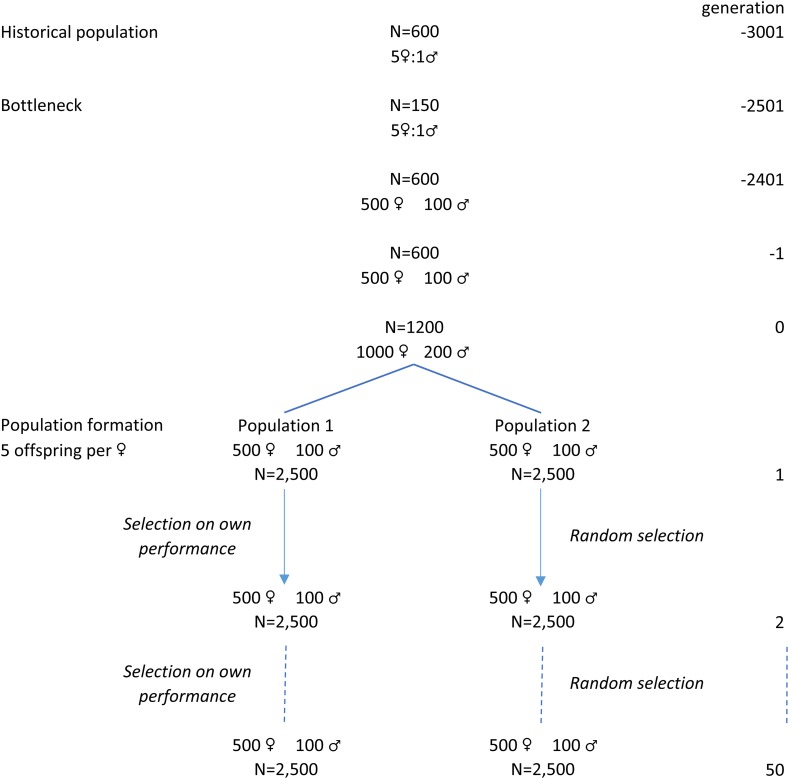
We simulated genotypes of two livestock populations (1 and 2) that diverged for 50 generations (Figure 2). For divergence, we considered two situations: one where the populations diverged due to drift only, and one where the populations diverged also due to selection in population 1 and drift in population 2.
Figure 2.
Overview of the simulated population structure.
Populations:
We simulated a historical population with QMSim (Sargolzaei and Schenkel 2009) by randomly mating 100 males and 500 females starting in generation -3001. From generation -3000 to generation -2501, we simulated a bottleneck by gradually decreasing population size to 150 (25 males and 125 females) to create initial linkage disequilibrium (LD), and population size was gradually increased again to 600 during the next 100 generations. The population size remained constant from generation -2400 until -1 to allow for the development of mutation-drift equilibrium. To provide a sufficient number of individuals for the development of populations 1 and 2, we doubled the number of individuals in the last historical generation (generation 0) to 200 males and 1,000 females. This simulation resulted in an average effective population size () of ∼285 generation 0, calculated as the harmonic mean of in each preceding generation, where is the number of males and is the number of females that become parents in a generation (Falconer and Mackay 1996).
After simulating the historical population, we simulated two current populations (1 and 2). We randomly sampled 100 males and 500 females from the last historical generation to become founders of population 1. The remaining 100 males and 500 females were the founders of population 2. We will refer to the generation of founders as generation 0. Within each population, simulation continued for 50 generations by randomly mating 100 selected males with 500 selected females. Each mating resulted in 5 offspring, resulting in a total of 2,500 offspring (exactly 1,250 males and 1,250 females) in each generation. Generations were non-overlapping, meaning that in each generation, the parents were selected from the previous generation only. In the drift scenario, animals in both populations were randomly selected to become parents of the next generation. Effective population size () in the drift scenario was ∼285 in the two populations. In the selection-drift scenario, animals in population 1 were selected based on their own phenotype (mass selection), while in population 2, selection was random. In this scenario, effective population size () in population 1 was ∼250, which was calculated as , where is the inbreeding rate estimated from the pedigree (Falconer and Mackay 1996). We simulated selection only in population 1 to reduce computation time.
Genome:
The simulated genome consisted of 10 chromosomes of 1 Morgan that each had 200 randomly positioned bi-allelic loci. In the first historical generation (generation -3001), we randomly sampled the allele frequencies of loci from a uniform distribution. Mutation rate was 2.5*10−5 during the historical generations. In generation 0, the distribution of allele frequencies had evolved to a U-shape, and we randomly selected 500 segregating loci to become QTL, which resulted in low linkage disequilibrium between QTL. There was no mutation from generation 0 to 50, because the QMSim software does not allow for mutation after the last historical generation.
Functional genetic effects:
Additive effects () of all 500 QTL were sampled from . We assumed that the size of the dominance and epistatic effects were proportional to the additive effects of the QTL involved in the interaction (Wellmann and Bennewitz 2011). We therefore sampled dominance coefficients () for all QTL from , from which dominance effects () were computed as . Similarly, we sampled epistatic coefficients () for all pairwise epistatic interactions from , from which functional epistatic effects () were computed as where and denote the QTL involved in the interaction. Each QTL had an epistatic interaction with 5 randomly sampled other QTL, resulting in a total of 1250 pairwise interactions.
For both dominance and epistasis, we considered 3 magnitudes of effects: small, intermediate, and large. For all magnitudes, the mean dominance coefficient () was 0.2, and the mean epistatic coefficient () was 0.0. The magnitude of dominance and epistatic effects were controlled by changing the standard deviation of dominance coefficients () or epistatic coefficients (). For dominance, was 0.3 with small effects, 0.7 with intermediate effects, and 1.5 with large effects (Table 1). The mean and standard deviation of small dominance coefficients were chosen based on empirical results of Bennewitz and Meuwissen (2010) and Sun and Mumm (2016). For epistasis, was scaled such that the total functional epistatic variance was comparable to the total functional dominance variance in the scenario with the same magnitude. To this end, was computed as , where is the number of epistatic interactions per QTL, and and are the squared mean and variance of dominance effects in the scenario with the corresponding magnitude. For example, with small epistatic effects, was computed as (Table 1).
From functional dominance and epistatic effects to statistically orthogonal effects:
We simulated dominance and epistasis by introducing functional dominance and epistatic effects that are independent of allele and genotype frequencies. Our interest, however, is in statistical average, dominance and epistatic effects of QTL, which do depend on genotype frequencies (Fisher 1918; Cheverud and Routman 1995). We will describe the general procedure to obtain these statistical effects, for a general situation where there can be dominance, epistasis, or both. Note, however, that our scenarios had either dominance or epistasis, but never both. After obtaining statistical effects, we describe how we computed the additive genetic value, genotypic value and phenotype for each individual. Although genotypic values themselves are independent of genotype frequencies, the partitioning of these genotypic values into additive, dominance, and epistatic components does depend on genotype frequencies. Additive genetic values of individuals in population 1 were needed to compute , and genotypic values and phenotypes were needed because selection in population 1 was based on own performance. In the following, we describe the procedure to obtain the average effects and dominance effects in population 1 (). The procedure to obtain these effects in population 2 () follows naturally by replacing the genotype and allele frequencies of population 1 with the frequencies in population 2.
The procedure starts by applying the natural and orthogonal interactions (NOIA) model (Álvarez-Castro and Carlborg 2007) for each epistatic interaction between two QTL. First, functional epistatic values for the 9 possible two-locus genotypes at QTL and were collected in a vector , where is a scalar representing the functional epistatic effect between QTL and , and is a 9 × 1 vector of epistatic contrasts for the 9 two-locus genotypes, ordered as (WWYY, WwYY, wwYY, WWYy, ..., wwyy). The simulated epistatic contrasts in followed one of four configurations: additive x additive (EAA), dominance x dominance (EDD), complementary (EC), or multiplicative (EM) (Figure 1). The contrasts in were centered and scaled to a standard deviation of one, so that the contrasts were comparable between configurations. We then used genotype frequencies of QTL and to partition the functional epistatic values in into 9 statistical genetic effects (Álvarez-Castro and Carlborg 2007; Vitezica et al. 2017)
where is a 9x9 diagonal matrix with each of the nine genotype frequencies in the same order as in . Matrix , where denotes the Kronecker product, and and are constructed as
| (1) |
where columns relate to orthogonal contrasts for the mean (), average effect (), and dominance effect () of QTL X and where , , and are the genotype frequencies of QTL X. The resulting vector of statistical genetic effects is
| (2) |
where and are the terms that contribute to average effects of QTL and . The other terms in contribute to dominance effects () of individual QTL and to epistatic effects of interacting QTL ().
We repeated this procedure of partitioning functional epistatic effects into statistical genetic effects for all pairwise interactions between QTL. Each QTL was involved in 5 epistatic interactions and therefore has 5 terms that contribute to its average effect. Following this reasoning, the average effect of QTL in population 1 with epistasis is
where is the frequency of the counted allele of QTL in population 1, is the set of loci that QTL interacts with, and . Note the difference between “additive effect” () and “average effect” (); the additive effect is half the difference in genotypic value between both opposing homozygotes, whereas the average effect () is the (statistical) marginal effect of the QTL. Throughout this manuscript, we will use the term “functional additive effect” to refer to , and “average effect” (i.e., statistical substitution effect) to refer to .
In our simulations, we needed to compute phenotypes of selection candidates in each generation, for which we needed the statistical dominance effect () of each QTL as well. The dominance effect of QTL in population 1 with epistasis is
Additive genetic values and phenotypes:
We computed additive genetic values () of selection candidates in population 1 for the trait expressed in both population 1 and 2. Their genotypic values () and phenotypes were only computed for the trait expressed in population 1. The additive genetic value of individual for the trait expressed in population 1 (2) were computed as (), and genotypic values for the trait expressed in population 1 were computed as
where is a column vector of additive genotype indicators for individual , and is column a vector of dominance genotype indicators for individual . These indicators were coded following the NOIA parameterization as denoted in the rows of and (Equation 1) for genotypes , , and , respectively. Phenotypes with a broad sense heritability of 0.5 were computed as , where , and was equal to the variance of genotypic values ().
Computing parameters of interest
The parameters of interest were (1) the genetic correlation between the trait in population 1 and the trait in population 2 (), and (2) the average absolute difference in allele frequencies between populations (). For each generation, we computed as the Pearson correlation between the additive genetic values of individuals in population 1 for the trait expressed in the two populations (equation (1)). Effectively, this is a weighted correlation between and , where the weights depend on the allele frequencies in population 1. Hence, the computed as the correlation of additive genetic values of individuals in population 2 may give different results because the genotypes sampled from population 2 result in different weights than those sampled from population 1 (see Discussion). For each generation, we computed as . We chose this parameter as a measure for population divergence, because we expect that there is a linear relationship between and . These parameters were computed for generation 1 to 5, and for every 5th generation after generation 5, to limit computation time.
Replicates
We ran the simulation with drift 50 times, resulting in 50 sets of genotypes (i.e., replicates). For each of those replicates, we computed and for each of the scenarios (i.e., genetic model and magnitude). We ran the simulations with both selection and drift for each scenario separately, because the selection of parents in population 1 depended on the genetic model. To limit computation time, we used 20 replicates for each scenario with selection.
Data availability
The data used in this study can be reproduced with the files and seeds in the following GitHub repository: https://git.wageningenur.nl/duenk002/rg-and-non-additive-effects. Supplemental material available at figshare: https://doi.org/10.25387/g3.10252856.
Results
First, for each scenario with selection, we show the change in mean genotypic value () and the change of additive genetic variance (VA) in population 1 across generations, to illustrate how population 1 evolved over time. Second, we report realized fractions of additive, dominance and epistatic variance in generation 1 and 50. Third, for scenarios with small non-additive effects, we show the effects of the genetic model and of applying selection on the additive genetic correlation () and the difference in allele frequency () between populations. Fourth, for each genetic model with selection, we investigate the impact of the magnitude of non-additive effects and the number of generations since divergence. Finally, we investigate the relationship between and across genetic models and within genetic models. All results presented refer to generation 50 and to scenarios with small non-additive effects, unless otherwise stated.
Mean genotypic value and variance components
With all scenarios, the mean genotypic value expressed in genetic standard deviations () in population 1 increased due to selection (Figure S 1). With all genetic models, the increase in was smaller when the magnitude of non-additive effects was larger. This result was expected, because the marginal effects of alleles may change over time in the presence of non-additive effects, reducing the effectiveness of selection. The increase in was largest with model A, and it was smallest with model EDD and large non-additive effects. There were only small differences in between models D, EAA, EC, and EM.
The additive genetic variance in population 1 (VA) decreased due to selection with all scenarios (Table 2 and Figure S 2). With genetic model A, EAA, EC and EM, about 95–98% of VA was lost after 50 generations of selection, whereas with D and EDD, 88–95% of VA was lost. A change in magnitude of non-additive effects did not substantially affect the decrease in VA, except with genetic models D and EDD, where more additive genetic variance was preserved with larger non-additive effects. In the drift scenario, the average loss of VA was about 7% for all scenarios (results not shown).
Table 2. Fractions of additive (VA), dominance (VD), and epistatic (VI) variances with respect to the total genetic variance in generation 1 and generation 50 with selection. Reported values are averages of 20 replicates.
| Generation 1 | Generation 50 | ||||||
|---|---|---|---|---|---|---|---|
| Scenario | Effect size | VA | VD | VI | VA | VD | VI |
| D | Small | 0.961 | 0.039 | 0.000 | 0.924 | 0.076 | 0.000 |
| Intermediate | 0.871 | 0.129 | 0.000 | 0.511 | 0.489 | 0.000 | |
| Large | 0.702 | 0.298 | 0.000 | 0.198 | 0.802 | 0.000 | |
| EAA | Small | 0.992 | 0.000 | 0.008 | 0.997 | 0.000 | 0.003 |
| Intermediate | 0.976 | 0.000 | 0.024 | 0.988 | 0.000 | 0.012 | |
| Large | 0.952 | 0.000 | 0.048 | 0.969 | 0.000 | 0.031 | |
| EDD | Small | 0.910 | 0.064 | 0.026 | 0.703 | 0.289 | 0.008 |
| Intermediate | 0.751 | 0.173 | 0.076 | 0.358 | 0.602 | 0.040 | |
| Large | 0.528 | 0.333 | 0.139 | 0.146 | 0.752 | 0.101 | |
| EC | Small | 0.985 | 0.012 | 0.003 | 0.947 | 0.051 | 0.001 |
| Intermediate | 0.947 | 0.044 | 0.009 | 0.737 | 0.250 | 0.013 | |
| Large | 0.871 | 0.105 | 0.024 | 0.471 | 0.511 | 0.017 | |
| EM | Small | 0.998 | 0.000 | 0.002 | 0.999 | 0.000 | 0.001 |
| Intermediate | 0.993 | 0.000 | 0.007 | 0.995 | 0.000 | 0.005 | |
| Large | 0.983 | 0.000 | 0.017 | 0.989 | 0.000 | 0.011 | |
In generation 1, scenarios that had only additive genetic (VA) and epistatic variance (VI), VA accounted for the largest, and VI for the smallest fraction of the total genetic variation (Table 2). The largest fraction of VI was realized with genetic model EAA (max. 0.048), followed by EDD (max. 0.033), EC (max. 0.024) and EM (max. 0.017). The largest fraction of dominance variance (VD) was realized with model EDD (max. 0.364), followed by D (max. 0.298) and EC (max. 0.105). With genetic models D, EDD and EC, the fraction VD increased and VA decreased across generations, especially with intermediate or large effects (Table 2, generation 50). The fraction VI remained relatively constant across generations with all scenarios.
Effect of genetic model and of selection on
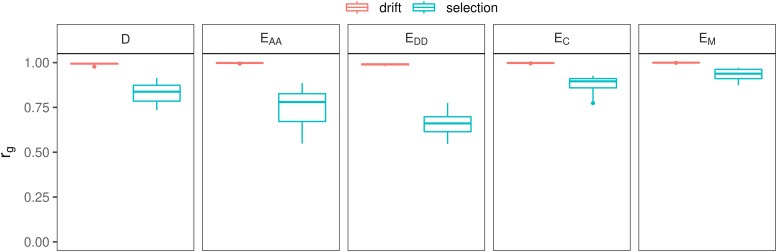
For all genetic models and small non-additive effects, was lower with selection than with drift only (Figure 3). With drift only, was between 0.99 and 1 for all genetic models. After 50 generations of selection, average was lowest with genetic model EDD (0.65), followed by EAA (0.75), D (0.83), EC (0.83) and finally EM (0.94). There was a tendency that scenarios with the largest non-additive variance in generation 1 had the smallest in generation 50 (Figure S 3). Note that the was always equal to 1 with the additive model (A) (results not shown).
Figure 3.
Effect of genetic model on with small non-additive effects, under drift only, or under drift and selection.
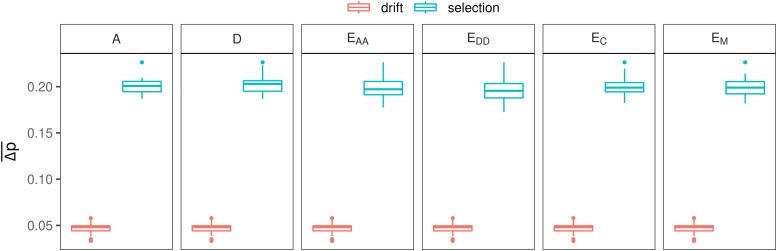
As expected, was larger with selection than with drift, and was the same across all genetic models with drift (0.05; Figure 4). With selection, with non-additive models was very similar (around 0.20) to the value with an additive model.
Figure 4.
Effect of genetic model on the difference in allele frequencies between populations, under drift only, or under drift and selection.
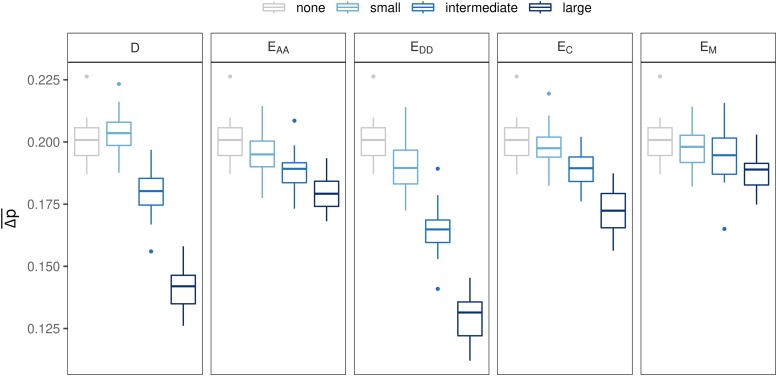
Effect of the magnitude of non-additive effects
For all genetic models and with selection, decreased with increasing magnitude of non-additive effects (Figure 5). With genetic model D, dropped about 31% from small to intermediate, and about 27% from intermediate to large dominance effects. With all epistatic models, the drop in with increasing magnitude was smaller (16–23%) than with D.
Figure 5.
Effect of magnitude of non-additive effects on for all genetic models, where population 1 was selected and population 2 was not selected.
For all genetic models with selection, the average absolute difference in allele frequency between lines () decreased with increasing magnitude of non-additive effects, especially with D and EDD (Figure 6). With model D, was 0.18 with intermediate dominance effects, and 0.141 with large effects. With EDD, was 0.162 with intermediate epistatic effects, and 0.130 with large effects. With the other epistatic models (EAA, EC and EM), the effect of an increase in magnitude on was much smaller (∼0.19 with intermediate and ∼0.18 with large effects).
Figure 6.
Effect of magnitude of non-additive effects on the difference in allele frequencies between populations under selection. In each subplot, the additive scenario (A) was included for reference (i.e., magnitude “none”).
Effect of number of generations since divergence
With all scenarios, decreased with the number of generations since divergence, and the rate of decrease was relatively small during the first five generations (), especially when the non-additive effects were small () (Figure 7). After the first five generations, the rate of decrease in differed across genetic models. There was a considerable difference between genetic models, the EM model showed the smallest decline of over time, and the EDD model showed the largest decline. With large non-additive effects, models EM and EAA tended to show an accelerated decrease in across generations, whereas models D, EC and EDD tended to show a decelerated decrease in (Figure 7).
Figure 7.
Effect of number of generations since divergence on for all genetic models with small (left) or large (right) non-additive effects.
With all scenarios, the average absolute difference in allele frequency between lines () increased with the number of generations since divergence (Figure 8). In contrast to the result of the genetic correlation with small non-additive effects (Figure 7), was remarkably similar between the genetic models (Figure 8). With large effects, models D and EDD showed a smaller than models EM, EAA, and EC.
Figure 8.
Effect of number of generations since divergence on the difference in allele frequencies between populations, for all genetic models and small (left) or large (right) non-additive effects. Dotted line represents additive trait.
In summary, for each genetic model, was smallest with selection, large non-additive effects and many generations since divergence. Overall, the smallest realized value of after 50 generations of divergence was achieved with genetic model D or EDD ( for both).
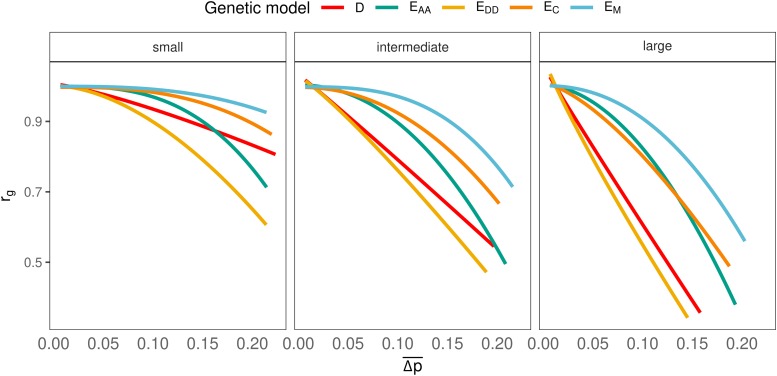
Relationship Between and
For all genetic models, there was a clear negative relationship between and (Figure 9), and the relationship was strongest for genetic models showing the strongest decline of with time (Figure 7). This result suggests that differences between genetic models in the decline of over time originate from different impacts of on , and not from differences in per se. For example, with small non-additive effects and after 50 generations of divergence, the value of was different between genetic models, whereas the realized was very similar (Figure 9). In other words, is a function of and of genetic architecture.
Figure 9.
Relationship between and the difference in allele frequencies between populations for all scenarios.
Discussion
Our objective was to investigate the relationship between non-additive effects, differences in allele frequencies between populations (), and the genetic correlation between populations (. We simulated genotype data of two populations that have diverged for a number of generations under drift only, or drift and selection, and we simulated traits where the genetic model and magnitude of non-additive effects were varied.
We computed as the correlation between additive genetic values of individuals in population 1, for the trait expressed in population 1 and 2. Effectively, this is a weighted correlation between average effects in population 1 () and 2 (), where the weights depend on the sample of genotypes that were used to compute the additive genetic values. This suggests that different values of could have been obtained when using the additive genetic values of individuals in population 2, because of differences in genotype frequencies between populations. We chose, however, to focus on population 1 because we were also interested in the change of allele frequencies over time due to selection. This approach leads to values of that indicate whether information from an unselected population (population 2) can be used to predict additive genetic values in a selected population (population 1).
Realized variance components
Because little is known about the quantity and magnitude of dominance and epistatic effects in reality, we considered a range of functional non-additive effect sizes and epistatic configurations. Realized proportions in our simulations (Table 2) did not always match with those observed in real data. For example, with large dominance effects, the fraction of dominance variance was 30%, which is uncommon in real data (Ertl et al. 2014; Lopes et al. 2016; Moghaddar and van der Werf 2017; Joshi et al. 2018). Similarly, scenario EDD also resulted in more dominance variance than expected in real populations, especially with large epistatic effects (33%). Empirical studies on livestock (Bennewitz and Meuwissen 2010) and crops (Sun and Mumm 2016) found that approximately 0.3% of loci show overdominance, which is comparable to our scenario with small dominance effects (0.5% overdominance). Furthermore, the scenario with small dominance effects resulted in a small proportion of dominance variance, and might therefore be most realistic for actual populations.
In contrast to our realized proportions of dominance variance, proportions of epistatic variance were lower (max. 5%) than estimates from an empirical study on litter size in pigs (about 26%) (Vitezica et al. 2018), though the standard error of that estimate was large (about 22%). Further evidence of statistical epistatic effects is scarce, probably because methods used for the detection of statistical epistasis are frequently underpowered (Wei et al. 2014). Furthermore, it has been suggested that incomplete LD between genomic markers and QTL may create the illusion of epistasis, making inference about the importance of epistasis from genome-wide regression studies difficult (Wei et al. 2014; Zan et al. 2018; de los Campos et al. 2019). In contrast to the lack of evidence of statistical epistasis, there is substantial evidence that physiological epistasis is abundant in several classes of organisms (Carlborg et al. 2003; Le Rouzic et al. 2008; Pettersson et al. 2011; Mackay 2015). Nevertheless, large epistatic effects between pairs of loci are believed to be unlikely (Wei et al. 2014), and the contribution of epistatic variance to the total genetic variance is expected to be small (Hill et al. 2008).
In summary, among the scenario’s we studied here, scenarios D and EDD with small effects, and scenarios EAA, EC and EM are probably most realistic, because these scenarios always resulted in little dominance (max. 7%) and epistatic (max. 5%) variance.
Effect of genetic model on
For the dominance model (D), we observed that decreased with increasing size of dominance effects and with increasing difference of allele frequencies between populations. In some cases, the can be negative due to dominance alone, as shown for a two-locus model (Wei et al. 1991). Such low values of were, however, only obtained with scenarios where both loci showed substantial overdominance, and where the difference in allele frequencies between the two populations was at least 0.3 for one of the loci. In our study, we considered many loci and the distributions of dominance effects was based on empirical results (Bennewitz and Meuwissen 2010; Sun and Mumm 2016). These distributions resulted in only a fraction of loci showing overdominance (i.e., 0.5% for small effects, 16% for intermediate effects, and 51% for large effects). Furthermore, our simulations resulted in U-shaped distributions of allele frequencies in the last generation of the historical population, which agrees with expectations based on neutral theory (Kimura and Crow 1964; Goddard 2001). After the two populations separated, allele frequency differences between populations were a result of drift and/or selection. We therefore believe that our simulations represent a more realistic model of quantitative traits and population divergence than those in Wei et al. (1991). In conclusion, given that dominance variance is usually small and overdominance does not occur frequently, our results show that it is unlikely that true values lower than 0.80 are due to dominance effects alone.
In another simulation study, where the fraction of loci showing overdominance was 12%, realized was 0.78 (Esfandyari et al. 2015). Although the fraction of loci showing overdominance in that study was comparable to our scenario with intermediate dominance effects, our realized in that scenario was much lower (0.57). This difference is likely due to the smaller number of generations that populations diverged in the study of Esfandyari et al. (2015).
With epistasis, decreased with increasing size of epistatic effects and with increasing difference of allele frequencies between populations, and the value of depended on the nature of the epistatic interaction (i.e., configuration). In addition, there was a tendency for configurations that resulted in large initial non-additive variance to result in smaller values of (Figure S 3). Even though large epistatic effects are unlikely and epistatic variance is expected to be small, could be as low as 0.45 for supposedly realistic epistatic scenarios.
To our knowledge, the relationship between the nature of epistasis and has not been studied before. The mechanism behind differences in between epistatic models can be illustrated with an example of two interacting loci. Suppose that both loci have an additive effect () of 1, an epistatic coefficient () of 0.5, and the allele frequency at locus 1 () is the same in both populations (here we use 0.10). Then, we study the effect of allele frequency difference between populations at locus 2 () on the difference in average effects between populations () for locus 1 and 2. Results show that EAA and EM interactions only affect the of the locus with fixed (locus 1), whereas EDD and EC interactions affect the at both loci (Figure S 4). Note that this result was the same with different values for , , or . This shows that, in general, EAA and EM interactions create a dependency of at a locus on the allele frequency of all loci it interacts with, whereas EDD and EC interactions also create a dependency of on the allele frequency of the locus itself. These mechanisms may contribute to the differences in between genetic models, because the interplay between differences in allele frequencies and depends on the genetic model.
Effect of magnitude of non-additive effects on
As expected, an increase in magnitude of dominance effects resulted in a lower , which is in line with results from Wei et al. (1991). Similarly, an increase in magnitude of epistatic effects also resulted in a lower . An important question is whether this decrease of due to an increase in magnitude continues until the theoretical limit of is reached. Additional analyses revealed that appears to asymptote with increasing magnitude of non-additive effects. In these analyses, we repeated our original simulations of genetic models D and EAA, using non-additive effects that were multiplied by 100 for all magnitudes. Results from those simulations showed that the difference in between “small”, “intermediate”, or “large” effects had indeed disappeared (Figure S 5), and that the lower bound of realized values for was ∼0.25 with scenario D and ∼0.36 with scenario EAA.
To show the mechanism behind this result, we again consider a two-locus model where, like before, both loci have an additive effect () of 1, the allele frequency of locus 1 () is 0.10 in both populations and . We studied the effect of the magnitude of the epistatic effect () on the absolute difference in average effects between populations, relative to the absolute value of in population 1 (). We observed that for all epistatic models, especially for larger values of , both and increase roughly linearly with , and that therefore stops increasing with large values of (Figure S 6). Note that the same mechanism was observed with dominance when was the same in both populations and . Hence, a change in magnitude equally affects the variance of ’s in the two populations, and the covariance between them. As a result, is unaffected by a change in size of non-additive effects when non-additive effects are already large. In conclusion, when non-additive effects are very large, no longer depends on the magnitude of non-additive effects relative to the magnitude of functional additive effects. At that point, there is a lower bound of that is determined by the nature of the non-additive effects (i.e., type of inter-allelic interaction) and by the difference in allele frequencies between populations.
Number of epistatic interactions
In the epistatic scenarios, we assumed that each locus interacted with 5 other loci. Because little is known about the number of interactions per locus () in reality, we tested whether our results were sensitive to a change in . For that purpose, we repeated all simulations of epistatic scenarios with . Note that the total functional epistatic variance with was the same as with , because the epistatic coefficients were scaled with , so that the product is constant. This analyses resulted in values of that were very similar to those of our original simulations (results not shown), suggesting that, in our simulations, the value of depends on the level of total functional epistatic variance, which scales similarly with or .
Effect of selection on
Non-additive effects and selection create a complex interplay between average effects, the difference in allele frequencies between populations () over time, and their effects on . For a trait with small dominance effects under selection, we observed that was almost the same as for an additive trait (Figure 8). We expected, however, that directional dominance would reduce , because the average effect at a locus can become smaller or even switch sign when the frequency of the favorable dominant allele increases (Falconer and Mackay 1996). This change in average effects would affect the change in allele frequencies over time due to selection in population 1, because the selection pressure at loci may change. A reduction in with small dominance effects was not observed, probably because only a small fraction of loci showed full- or over-dominance. Indeed, with large dominance coefficients (so that the fraction of loci showing over-dominance was much larger compared to with small dominance coefficients) was smaller (Figure 6). In real data, however, we do not expect a large fraction of loci that show full- or over-dominance (Wellmann and Bennewitz 2011). It is therefore unlikely that dominance significantly affects the change in allele frequencies over time due to selection, compared to a purely additive trait.
For a trait with epistatic effects under selection, we observed that was a bit smaller than that for a trait with only additive effects (Figure 8). Similar to the models with only dominance effects, this reduction in was expected because the average effect at a locus can become smaller or switch sign over time in the presence of epistasis. How epistasis affects the change in allele frequencies due to selection depends on the directionality of the epistatic interaction effect. Theory suggests that, compared to pure additivity, positive interactions (i.e., in the same direction as the additive effects) will promote the selection of favorable alleles, whereas negative interactions (i.e., in the opposite direction from the additive effects) will suppress the selection of favorable alleles (Carter et al. 2005; Hansen 2013; Paixão and Barton 2016). We chose to simulate both positive and negative interactions with equal probabilities, because empirical studies suggest that epistatic interactions are not biased in being either positive or negative (i.e., they are non-directional) (Mackay 2014). Our results showed that, for a trait with intermediate epistatic effects, the net effect of having both positive and negative interactions was a decrease in fixation rate of favorable alleles (i.e., with a positive ), and an increase in fixation rate of unfavorable alleles (i.e., with a negative ) compared to an additive trait (Figure S 7). Similar results were found by Esfandyari et al. (2017). In conclusion, epistatic effects may affect through two related mechanisms. First, with an epistatic model and when selection takes place in one of the populations, the difference in allele frequencies between populations may be smaller compared to an additive model. This reduction occurs because negative interactions decrease the fixation rates of favorable alleles, and increase those of unfavorable alleles. Second, for given allele frequency differences, the value of depends on the nature of the epistatic interaction.
Loss of additive genetic variance
Selection experiments in Drosophila, maize, and Escherichia coli have shown that additive genetic variation (VA) can be maintained for at least 100 generations (Hill 2016). Some researchers suggested that this preservation of VA may be due to the conversion of non-additive genetic variance to additive genetic variance (Cheverud and Routman 1996; Hallander and Waldmann 2007; Hill 2017). Simulation studies, however, have failed to show a preservation of VA due to this conversion (Carter et al. 2005; Esfandyari et al. 2017). Similarly, our simulations showed little conversion of non-additive genetic variance to VA with genetic models EAA and EM, and no conversion with other genetic models (Table 2). As a result, almost all additive genetic variance was lost after 50 generations (Figure S 2).
The large loss of additive genetic variance in our simulations may be due to two reasons. First, there was little epistatic variance in generation one that could be ‘converted’ to VA in subsequent generations (Hill et al. 2008; Mäki-Tanila and Hill 2014). This was largely because the allele frequency distribution was strongly U-shaped in generation one. Second, mutational variance was zero because there were no mutations simulated after the historical generation. Even though these mechanisms may explain some of the loss of in our simulations, the issue still remains that, to date, simulations have failed to convincingly reproduce the conservation of observed in reality (Johnson and Barton 2005; Walsh and Lynch 2018).
Practical relevance
In our simulations, there was selection in only one of the populations, while the other population was unselected. In reality, populations may have been divergently selected (e.g., Friesian Holstein vs. Angus cattle), resulting in larger differences in allele frequencies than simulated here. Hence, between divergently selected populations may be smaller than observed in our simulations.
In this study, we assumed that there were no genotype x environment interactions (GxE), so that values smaller than one were only due to non-additive effects. In reality, both non-additive effects and GxE may contribute to values being smaller than one. The relative importance of non-additive effects and GxE can be inferred from the difference between estimated , from a design where the populations were tested in different environments, and from a design where one of the populations was tested in the environment of the other population. This approach is similar to what was proposed by Wientjes and Calus (2017) to dissect the components of the genetic correlation between purebred and crossbred performance. However, to our knowledge, there are no studies that have used this approach to disentangle the effects of non-additive effects and GxE on . This study shows that, even without GxE, the can be substantially smaller than one, and sometimes even close to zero.
Estimated genetic correlations between two populations () may differ across traits (e.g., Lund et al. 2011; Karoui et al. 2012; Porto-Neto et al. 2015). For example, in dairy cattle, of fertility traits tended to be lower than those of fat yield and milk production (Karoui et al. 2012). The results from the present study suggest that such differences in may indicate differences in the underlying genetic model between traits (i.e., in the importance of non-additive effects). Although this may be the case, differences in between traits can arise through other mechanisms as well. First, often include a component due to GxE interactions. Such GxE interactions may be more important for some traits than for others, resulting in differences in between traits. Second, different traits are influenced by (at least partly) different QTL, and some traits may have been under stronger selection than others. As a result, the differences in allele frequencies at QTL between populations may vary across traits. These mechanisms may result in differences in between traits, even when the underlying genetic models of those traits are similar. It is therefore questionable whether inferences can be made about differences in genetic model among traits, based on differences in .
The results in this study may be relevant for the prediction of additive genetic values across populations using genomic information. In this strategy, termed across-population genomic prediction, average effects at markers are estimated in one population, and used to compute additive genetic values in another population (de Roos et al. 2009; Hayes et al. 2009). It has been suggested that the inefficiency of across-population genomic prediction is partly due to differences in linkage disequilibrium between markers and QTL. This insight has inspired the use of whole-genome sequence (WGS) data, because in WGS data, genotypes of the QTL themselves are included (Iheshiulor et al. 2016; Raymond et al. 2018a; Raymond et al. 2018b). The results of the current study suggest, however, that even when QTL genotypes are known and their average effects are accurately estimated in one population, across-population genomic prediction may be inefficient, because can differ considerably from one, even when genetic variance is mostly additive. This view is supported by the results of Raymond et al. (2018b), who reported that although the estimated from putative QTL was higher than the estimate from regular marker data, it was still lower than one.
Similar to across populations, genomic prediction from current to future generations may be inefficient because of changes in allele frequencies, and the subsequent changes in average effects at QTL. In other words, two different generations can be considered as two populations that have a genetic correlation between them that may be lower than unity. The results of this study may therefore partly explain the need for frequent retraining of genomic prediction models to achieve constant accuracy across generations (Sonesson and Meuwissen 2009; Wolc et al. 2011). We expect, however, that the change in allele frequency at a single QTL is relatively small across a few (4-5) generations, especially for traits that are highly polygenic. As a result, may be relatively high across a few generations. Nevertheless, the relative contribution of non-additive effects to the decline of genomic prediction accuracy across generations is currently unknown, and would be an interesting topic for future research.
Conclusion
Our findings show that the genetic correlation between populations ( is partly determined by the difference in allele frequencies between populations and the magnitude of non-additive effects. Large differences in allele frequencies and large non-additive effects resulted in low values of . For both dominance and epistasis, when non-additive effects become extremely large, has a lower bound that is determined by the nature of non-additive effects, and the difference in allele frequencies between populations. In addition, we found that with epistasis, depends on the level of total functional epistatic variance, which is a function of epistatic effect size and the number of interactions per locus. Given that dominance variance is usually small and there is not much overdominance, we expect that it is unlikely that values of below 0.8 are due to dominance alone. With supposedly realistic epistasis, could be as low as 0.45. These results may contribute to the understanding of differences in genetic expression of complex traits between populations, and may help in explaining the inefficiency of genomic prediction across populations.
Acknowledgments
The authors thank the Netherlands Organization of Scientific Research (NWO) and the Breed4Food consortium partners Cobb Europe, CRV, Hendrix Genetics, and Topigs Norsvin for their financial support. We thank the Wageningen Institute of Animal Science for a scholarship that allowed PD to visit the University of New England in Armidale, Australia. The use of the HPC cluster has been made possible by CAT-AgroFood (Shared Research Facilities Wageningen UR). The authors also thank Mehdi Sargolzaei for providing an improved version of QMSim.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25387/g3.10252856.
Communicating editor: J.-L. Jannink
Literature cited
- Álvarez-Castro J. M., and Carlborg Ö., 2007. A Unified Model for Functional and Statistical Epistasis and Its Application in Quantitative Trait Loci Analysis. Genetics 176: 1151–1167. 10.1534/genetics.106.067348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateson W., and Mendel G., 1909. Mendel’s principles of heredity. Cambridge University Press, Cambridge, UK: 10.5962/bhl.title.44575 [DOI] [Google Scholar]
- Bennewitz J., and Meuwissen T. H. E., 2010. The distribution of QTL additive and dominance effects in porcine F2 crosses. J. Anim. Breed. Genet. 127: 171–179. 10.1111/j.1439-0388.2009.00847.x [DOI] [PubMed] [Google Scholar]
- Brown B. C., Ye C. J., Price A. L., and Zaitlen N., 2016. Transethnic Genetic-Correlation Estimates from Summary Statistics. Am. J. Hum. Genet. 99: 76–88. 10.1016/j.ajhg.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlborg O., Kerje S., Schutz K., Jacobsson L., Jensen P. et al. , 2003. A global search reveals epistatic interaction between QTL for early growth in the chicken. Genome Res. 13: 413–421. 10.1101/gr.528003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter A. J., Hermisson J., and Hansen T. F., 2005. The role of epistatic gene interactions in the response to selection and the evolution of evolvability. Theor. Popul. Biol. 68: 179–196. 10.1016/j.tpb.2005.05.002 [DOI] [PubMed] [Google Scholar]
- Cheverud J. M., and Routman E. J., 1995. Epistasis and its contribution to genetic variance components. Genetics 139: 1455–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverud J. M., and Routman E. J., 1996. Epistasis as a source of increased additive genetic variance at population bottlenecks. Evolution 50: 1042–1051. 10.1111/j.1558-5646.1996.tb02345.x [DOI] [PubMed] [Google Scholar]
- Cockerham C. C., 1954. An extension of the concept of partitioning hereditary variance for analysis of covariances among relatives when epistasis is present. Genetics 39: 859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Candia T. R., Lee S. H., Yang J., Browning B. L., Gejman P. V. et al. , 2013. Additive genetic variation in schizophrenia risk is shared by populations of African and European descent. Am. J. Hum. Genet. 93: 463–470. 10.1016/j.ajhg.2013.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Campos G., Sorensen D. A., and Toro M. A., 2019. Imperfect Linkage Disequilibrium Generates Phantom Epistasis (&Perils of Big Data). G3 (Bethesda). 9: 1429–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Roos A. P., Hayes B. J., and Goddard M. E., 2009. Reliability of genomic predictions across multiple populations. Genetics 183: 1545–1553. 10.1534/genetics.109.104935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertl J., Legarra A., Vitezica Z. G., Varona L., Edel C. et al. , 2014. Genomic analysis of dominance effects on milk production and conformation traits in Fleckvieh cattle. Genet. Sel. Evol. 46: 40 10.1186/1297-9686-46-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esfandyari H., Henryon M., Berg P., Thomasen J. R., Bijma P. et al. , 2017. Response to Selection in Finite Locus Models with Nonadditive Effects. J. Hered. 108: 318–327. [DOI] [PubMed] [Google Scholar]
- Esfandyari H., Sorensen A. C., and Bijma P., 2015. A crossbred reference population can improve the response to genomic selection for crossbred performance. Genet. Sel. Evol. 47: 76 10.1186/s12711-015-0155-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer D., and Mackay T., 1996. Introduction to quantitative genetics, Pearson Education Limited, Harlow. [Google Scholar]
- Falconer D. S., 1952. The Problem of Environment and Selection. Am. Nat. 86: 293–298. 10.1086/281736 [DOI] [Google Scholar]
- Fisher R. A., 1918. The Correlation between Relatives on the Supposition of Mendelian Inheritance. Trans. R. Soc. Edinb. 52: 399–433. 10.1017/S0080456800012163 [DOI] [Google Scholar]
- Forsberg S. K. G., Bloom J. S., Sadhu M. J., Kruglyak L., and Carlborg Ö., 2017. Accounting for genetic interactions improves modeling of individual quantitative trait phenotypes in yeast. Nat. Genet. 49: 497–503. 10.1038/ng.3800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard M. E., 2001. The validity of genetic models underlying quantitative traits. Livest. Prod. Sci. 72: 117–127. 10.1016/S0301-6226(01)00272-X [DOI] [Google Scholar]
- Hallander J., and Waldmann P., 2007. The effect of non-additive genetic interactions on selection in multi-locus genetic models. Heredity 98: 349–359. 10.1038/sj.hdy.6800946 [DOI] [PubMed] [Google Scholar]
- Hansen T. F., 2013. Why epistasis is important for selection and adaptation. Evolution 67: 3501–3511. 10.1111/evo.12214 [DOI] [PubMed] [Google Scholar]
- Hayes B. J., Bowman P. J., Chamberlain A. C., Verbyla K., and Goddard M. E., 2009. Accuracy of genomic breeding values in multi-breed dairy cattle populations. Genet. Sel. Evol. 41: 51 10.1186/1297-9686-41-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W. G., 2016. Is Continued Genetic Improvement of Livestock Sustainable? Genetics 202: 877–881. 10.1534/genetics.115.186650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W. G., 2017. “Conversion” of epistatic into additive genetic variance in finite populations and possible impact on long-term selection response. J. Anim. Breed. Genet. 134: 196–201. 10.1111/jbg.12270 [DOI] [PubMed] [Google Scholar]
- Hill W. G., Goddard M. E., and Visscher P. M., 2008. Data and theory point to mainly additive genetic variance for complex traits. PLoS Genet. 4: e1000008 10.1371/journal.pgen.1000008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iheshiulor O. O. M., Woolliams J. A., Yu X., Wellmann R., and Meuwissen T. H. E., 2016. Within- and across-breed genomic prediction using whole-genome sequence and single nucleotide polymorphism panels. Genet. Sel. Evol. 48: 15 10.1186/s12711-016-0193-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T., and Barton N., 2005. Theoretical models of selection and mutation on quantitative traits. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360: 1411–1425. 10.1098/rstb.2005.1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi R., Woolliams J. A., Meuwissen T., and Gjoen H. M., 2018. Maternal, dominance and additive genetic effects in Nile tilapia; influence on growth, fillet yield and body size traits. Heredity 120: 452–462. 10.1038/s41437-017-0046-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karoui S., Carabaño M. J., Díaz C., and Legarra A., 2012. Joint genomic evaluation of French dairy cattle breeds using multiple-trait models. Genet. Sel. Evol. 44: 39 10.1186/1297-9686-44-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempthorne O., 1954. The Correlation between Relatives in a Random Mating Population. Proc R Soc Lond Ser B Biol sci 143: 103–113. [PubMed] [Google Scholar]
- Kimura M., and Crow J. F., 1964. The number of alleles that can be maintained in a finite population. Genetics 49: 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Rouzic A., Alvarez-Castro J. M., and Carlborg O., 2008. Dissection of the genetic architecture of body weight in chicken reveals the impact of epistasis on domestication traits. Genetics 179: 1591–1599. 10.1534/genetics.108.089300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M. S., Bastiaansen J. W. M., Janss L., Knol E. F., and Bovenhuis H., 2016. Genomic prediction of growth in pigs based on a model including additive and dominance effects. J. Anim. Breed. Genet. 133: 180–186. 10.1111/jbg.12195 [DOI] [PubMed] [Google Scholar]
- Lund M. S., de Roos A. P. W., de Vries A. G., Druet T., Ducrocq V. et al. , 2011. A common reference population from four European Holstein populations increases reliability of genomic predictions. Genet. Sel. Evol. 43: 43 10.1186/1297-9686-43-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F. C., 2014. Epistasis and quantitative traits: using model organisms to study gene–gene interactions. Nat. Rev. Genet. 15: 22–33. 10.1038/nrg3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F. C., 2015. Epistasis for Quantitative Traits in Drosophila. Epistasis: Methods and Protocols 1253: 47–70. 10.1007/978-1-4939-2155-3_4 [DOI] [PubMed] [Google Scholar]
- Mäki-Tanila A., and Hill W. G., 2014. Influence of gene interaction on complex trait variation with multilocus models. Genetics 198: 355–367. 10.1534/genetics.114.165282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghaddar N., and van der Werf J. H. J., 2017. Genomic estimation of additive and dominance effects and impact of accounting for dominance on accuracy of genomic evaluation in sheep populations. J. Anim. Breed. Genet. 00: 1–10. [DOI] [PubMed] [Google Scholar]
- Monnahan P. J., and Kelly J. K., 2015. Epistasis Is a Major Determinant of the Additive Genetic Variance in Mimulus guttatus. PLoS Genet. 11: e1005201 10.1371/journal.pgen.1005201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paixão T., and Barton N. H., 2016. The effect of gene interactions on the long-term response to selection. Proc. Natl. Acad. Sci. USA 113: 4422–4427. 10.1073/pnas.1518830113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson M., Besnier F., Siegel P. B., and Carlborg O., 2011. Replication and explorations of high-order epistasis using a large advanced intercross line pedigree. PLoS Genet. 7: e1002180 10.1371/journal.pgen.1002180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porto-Neto L., Barendse W., Henshall J., McWilliam S., Lehnert S. et al. , 2015. Genomic correlation: harnessing the benefit of combining two unrelated populations for genomic selection. Genet. Sel. Evol. 47: 84 10.1186/s12711-015-0162-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond B., Bouwman A. C., Schrooten C., Houwing-Duistermaat J., and Veerkamp R. F., 2018a Utility of whole-genome sequence data for across-breed genomic prediction. Genet. Sel. Evol. 50: 27 10.1186/s12711-018-0396-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond B., Bouwman A. C., Wientjes Y. C. J., Schrooten C., Houwing-Duistermaat J. et al. , 2018b Genomic prediction for numerically small breeds, using models with pre-selected and differentially weighted markers. Genet. Sel. Evol. 50: 49 10.1186/s12711-018-0419-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargolzaei M., and Schenkel F. S., 2009. QMSim: a large-scale genome simulator for livestock. Bioinformatics 25: 680–681. 10.1093/bioinformatics/btp045 [DOI] [PubMed] [Google Scholar]
- Sonesson A. K., and Meuwissen T. H., 2009. Testing strategies for genomic selection in aquaculture breeding programs. Genet. Sel. Evol. 41: 37 10.1186/1297-9686-41-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., and Mumm R. H., 2016. Method to represent the distribution of QTL additive and dominance effects associated with quantitative traits in computer simulation. BMC Bioinformatics 17: 73 10.1186/s12859-016-0906-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitezica Z. G., Legarra A., Toro M. A., and Varona L., 2017. Orthogonal Estimates of Variances for Additive, Dominance and Epistatic Effects in Populations. Genetics 206: 1297–1307. 10.1534/genetics.116.199406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitezica Z. G., Reverter A., Herring W., and Legarra A., 2018. Dominance and epistatic genetic variances for litter size in pigs using genomic models. Genet. Sel. Evol. 50: 71 10.1186/s12711-018-0437-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh B., and Lynch M., 2018. Maintenance of Quantitative Genetic Variation in Evolution and Selection of Quantitative Traits, Oxford University Press, Oxford. [Google Scholar]
- Wei M., van der Steen H. A. M., van der Werf J. H. J., and Brascamp E. W., 1991. Relationship between purebred and crossbred parameters. J. Anim. Breed. Genet. 108: 253–261. 10.1111/j.1439-0388.1991.tb00183.x [DOI] [Google Scholar]
- Wei W.-H., Hemani G., and Haley C. S., 2014. Detecting epistasis in human complex traits. Nat. Rev. Genet. 15: 722–733. 10.1038/nrg3747 [DOI] [PubMed] [Google Scholar]
- Wellmann R., and Bennewitz J., 2011. The contribution of dominance to the understanding of quantitative genetic variation. Genet. Res. 93: 139–154. 10.1017/S0016672310000649 [DOI] [PubMed] [Google Scholar]
- Wientjes Y. C. J., and Calus M. P. L., 2017. BOARD INVITED REVIEW: The purebred-crossbred correlation in pigs: A review of theory, estimates, and implications. J. Anim. Sci. 95: 3467–3478. [DOI] [PubMed] [Google Scholar]
- Wientjes Y. C. J., Veerkamp R. F., Bijma P., Bovenhuis H., Schrooten C. et al. , 2015. Empirical and deterministic accuracies of across-population genomic prediction. Genet. Sel. Evol. 47: 5 10.1186/s12711-014-0086-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolc A., Arango J., Settar P., Fulton J. E., O’Sullivan N. P. et al. , 2011. Persistence of accuracy of genomic estimated breeding values over generations in layer chickens. Genet. Sel. Evol. 43: 23 10.1186/1297-9686-43-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan Y., Forsberg S.K.G., and Carlborg Ö., 2018. On the Relationship Between High-Order Linkage Disequilibrium and Epistasis. G3 (Bethesda). 8: 2817–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used in this study can be reproduced with the files and seeds in the following GitHub repository: https://git.wageningenur.nl/duenk002/rg-and-non-additive-effects. Supplemental material available at figshare: https://doi.org/10.25387/g3.10252856.