Abstract
Zinc is essential for almost all living organisms, since it serves as a crucial cofactor for transcription factors and enzymes. However, it is toxic to cell growth when present in excess. The present work aims to investigate the toxicity mechanisms induced by zinc stress in yeast cells. To this end, 108 yeast single-gene deletion mutants were identified sensitive to 6 mM ZnCl2 through a genome-wide screen. These genes were predominantly related to the biological processes of vacuolar acidification and transport, polyphosphate metabolic process, cytosolic transport, the process utilizing autophagic mechanism. A result from the measurement of intracellular zinc content showed that 64 mutants accumulated higher intracellular zinc under zinc stress than the wild-type cells. We further measured the intracellular ROS (reactive oxygen species) levels of 108 zinc-sensitive mutants treated with 3 mM ZnCl2. We showed that the intracellular ROS levels in 51 mutants were increased by high zinc stress, suggesting their possible involvement in regulating ROS homeostasis in response to high zinc. The results also revealed that excess zinc could generate oxidative damage and then activate the expression of several antioxidant defenses genes. Taken together, the data obtained indicated that excess zinc toxicity might be mainly due to the high intracellular zinc levels and ROS levels induced by zinc stress in yeast cells. Our current findings would provide a basis to understand the molecular mechanisms of zinc toxicity in yeast cells.
Keywords: Saccharomyces cerevisiae, zinc toxicity, genetic screening, genomics, Reactive oxygen species (ROS)
The transition metal ion, zinc, is an essential cofactor for transcription factors and enzymes in all eukaryotic cells as well as an essential nutrient for life in all living organisms (Hambidge and Krebs 2007). However, it is toxic to cell growth when present in excess by generating reactive hydroxyl radicals and disturbing the cellular redox potential (Singh et al. 2017). In addition, excess zinc competes for the binding sites in functional proteins for other metals (King et al. 2000). Although Zinc can function as a member of antioxidant properties, it generates reactive oxygen species when yeast cells were exposed to high zinc levels (Pagani et al. 2007; Powell 2000; Hao and Maret 2005). Moreover, metal toxicities are often attributed mainly to the capacity to induce the unfolded protein response (UPR), the oxidative stress, DNA damage or even cell death (Nargund et al. 2008; Muthukumar and Nachiappan 2010; Liu et al. 2018). High intracellular ROS levels induced by zinc or other metals and stresses can trigger several biological molecules, such as DNA damage, lipid peroxidation and depletion of protein sulphydryl (Howlett and Avery 1997; Chrestensen et al. 2000; Serero et al. 2008). Therefore, the intracellular zinc levels must be tightly regulated to maintain zinc homeostasis in an optimal level regardless of its supply. As a model organism, the budding yeast Saccharomyces cerevisiae is used to study the basic mechanisms of many cellular processes, including zinc transport and zinc homeostasis (Wu et al. 2008).
In budding yeast, zinc homeostasis is tightly sustained via various transporters. Yeast cells assimilate the extracellular zinc through the high and low-affinity transport at the plasma membrane (Eide 2009). Cells uptake the extracellular zinc efficiently via a high-affinity zinc transporter Zrt1, and two low-affinity zinc transports, Zrt2 and Fet4, which are all regulated by the transcriptional factor Zap1 (Waters and Eide 2002; Zhao and Eide 1996a, 1996b). Inside the cell, two vacuolar zinc transporters Zrt3 and Zrc1 are responsible for transporting zinc out or into the vacuolar, respectively, and the heteromeric complex formed by Msc2 and Zrg17 transports the cytoplasm zinc to the endoplasmic reticulum when it is in excess. Interestingly, the three transporters Zrt3, Zrc1 and Zrg17 are also regulated by Zap1 in response to zinc level (MacDiarmid et al. 2000; Miyabe et al. 2000; Wu et al. 2011). The Fet4 transporter involved in uptake of iron and copper and the high-affinity phosphate transporter Pho84 can also uptake zinc (Waters and Eide 2002; Bun-Ya et al. 1991).
Zap1 was the first identified fungal zinc-responsive transcription factor from S. cerevisiae (Zhao and Eide 1997). Zap1 regulates the expression of about 80 genes by binding to their ZREs (Zinc Responsive Elements) in the promoter regions, including the genes required for zinc homeostasis or survival for a long period of zinc starvation (Zhao and Eide 1997; De Nicola et al. 2007; North et al. 2012; Zhao et al. 1998). At the transcriptional level, Zap1 autoregulates its coding gene ZAP1 by binding to a ZRE within its own promoter (Zhao et al. 1998). Zap1 contains seven C2H2-type zinc fingers. Five of these zinc fingers are needed for DNA binding, while the other two are involved in zinc sensing to regulate AD2 (Wilson and Bird 2016). For the post-translational/ regulation of Zap1, zinc regulates the activities of the Zap1 DNA binding domain, AD1 and AD2 independently (Bird et al. 2003; Herbig et al. 2005; Frey et al. 2011; Zhao et al. 1998). Hence, the activity of Zap1 is strongly enhanced in response to zinc limitation, inducing the expression of ZRT1 encoding a high-affinity zinc transporter while inhibiting the expression of ZRT2 encoding a low-affinity zinc transporter (Eide 2003; Wilson and Bird 2016). When espoused to high extracellular zinc concentrations, Zrt1 is removed from the plasma membrane rapidly by substrate-induced endocytosis (Schothorst et al. 2017; Gitan and Eide 2000; Gitan et al. 1998). The expression level of ZRT1 is induced over 100-fold in zinc-limited cells by Zap1, while ZRT2 is induced under mild zinc limitation but then inhibited by Zap1 in severe zinc limitation conditions (Zhao and Eide 1997; Bird et al. 2004).
Despite these discoveries, it is still largely unknown about the basic mechanisms by which proteins or pathways regulate the cytosolic zinc homeostasis or zinc toxicity. Using yeast as a model system, it is easy to know how a single-gene mutant regulates zinc homeostasis in response to high zinc (Bleackley et al. 2011). The present work aims to investigate the toxicity mechanisms induced by zinc stress in yeast cells. We first screened the zinc-sensitive mutants from a collection of S. cerevisiae deletion mutants. Specifically, the impact of high intracellular zinc concentration and ROS production were both addressed in this work. Also, we analyzed the expression of genes involved in antioxidant defenses in response to excess zinc.
Materials and Methods
Yeast strains and growth conditions
Diploid strains used in this work derived from the BY4743 genetic background. Yeast cells were grown at 30° in YPD medium (1% yeast extract, 2% peptone, 2% glucose). ZnCl2 was purchased from Sangon Biotech (Shanghai, China), and dihydroethidium was purchased from Sigma (Beijing, China).
Genome-wide screen for zinc-sensitive mutations
A collection of S. cerevisiae deletion mutants of 4,757 non-essential genes in BY4743 background were purchased from Thermo Fisher SCIENTIFIC (http://clones.thermofisher.com/cloneinfo.php?clone=yeast, catalog number: 95400.BY4743) and frozen at -80° in 96-well microtitre plates in liquid YPD medium containing 15% glycerol. The primary screen of zinc-sensitive mutations was first performed by transferring the deletion mutant library to fresh liquid YPD medium (pH ∼5.5) and cultured at 30° in new 96-well microtitre plates. Then, 20 μL of each mutant was transferred to 180 μL fresh liquid YPD medium with or without 3 mΜ ZnCl2 and incubated at 30° for 12 h. Mutants with a relative OD600 reduced by more than 30% in liquid YPD medium supplemented with ZnCl2 relative to wild-type cells but not in liquid YPD medium were considered zinc-sensitive. Mutants that appeared sensitive were confirmed by the serial dilution assay method in solid YPD plates, YPD plates with 6 mΜ ZnCl2, YPD plates with 6 mΜ ZnSO4 and YPD plates with 12 mΜ NaCl as described previously (Zhao et al. 2013). Namely, the individual deletion mutants grown overnight in liquid YPD at 30° and serially diluted by 10 times with ddH2O. Each dilution of 2.5 µL was spotted onto the above indicated plates and incubated at 30° for 2-3 days.
Determination of the intracellular zinc concentrations
To determinate of the total intracellular zinc concentrations in the 108 zinc-sensitive mutants, cells were first cultured in YPD media to middle log phase and then were treated with 3 mM ZnCl2 for 2 hr. Next, cells were collected and prepared for measuring the intracellular zinc concentration using an atomic absorption spectrometer in flame emission mode as described previously (Zhao et al. 2013). Three individual colonies for each mutant were measured, where the wild-type BY4743 cells served as a control.
Detection of the intracellular ROS accumulation
The intracellular ROS level was monitored with dihydroethidium (DHE) (Büttner et al. 2007). Before zinc treatment, cells were cultured in YPD media to the middle log phase and split into two aliquots. Subsequently, yeast cells were exposed to YPD supplemented with or without 3 mM ZnCl2 or 8 mM ZnCl2 for 2 hr or 5 hr, respectively. The relative fluorescence units (RFU) were measured in a Synergy H4 fluorescence reader (BioTek) at a fluorescence excitation of 485nm and an emission of 535nm. The RFU was corrected and normalized by the density of OD600 of each corresponding mutant. Three individual colonies for each mutant were measured, where wild-type BY4743 cells served as a control.
RNA extraction and quantitative real-time PCR (qPCR) assay
To extract the total RNA, yeast cells were first grown in YPD to a density of OD600 = 0.6∼0.8 and were spilt into aliquots Then cells were cultured in YPD supplemented with or without 3 mM ZnCl2 for an additional 1 hr. Cultures were then collected and the total RNA was extracted by the hot phenol method. The first-strand cDNA of each sample was synthesized via the Primer Script RT reagent kit (Cwbiotech, China) following the manufacturer’s instructions. Quantitative PCR (qPCR) method was used to detect the relative expression levels of TRR1, TRX2, SOD1, GSH1, CTT1, GPX2 with primer pairs listed in Table S1, respectively. The PGK1 gene was used as an internal control. qPCR reactions were performed in a Thermo Scientific CFX96 instrument using SYBR Premix Ex Taq (Cwbiotech, China). The results were abstained through the –ΔΔCt method (Livak and Schmittgen 2001). Each reaction was carried out in triplicate.
Meta-analysis and protein-protein interaction (PPI) network construction for the identified genes
Gene Ontology (GO) enrichment analyses of the zinc-sensitive genes were performed using the powerful web-based Metascape tool (http://metascape.org/gp/index.html#/main/step1). P-value < 0.01 was set as the cutoff criterion, and significance was ranked by enrichment score (− log 10 (P-value)). A web-based search tool, STRING (http://string-db.org), was selected to explore the potential protein-protein interactions of the screened zinc-sensitive genes. A reliability threshold of a combined score of >0.4 was considered as significant interaction pair and then protein-protein interaction (PPI) network was constructed and visualized by Cytoscape software (version 3.6.0, http://www.cytoscape.org/) (Shannon et al. 2003).
Data availability
Strains are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at figshare: https://doi.org/10.25387/g3.11365043.
Results
Identification of genes involved in zinc tolerance
To identify the genes involved in zinc tolerance, we screened the yeast diploid nonessential gene deletion library. The results indicated that a set of mutants for 108 genes showed reduced growth in exposing to 6 mM ZnCl2 (Table 1). The zinc sensitivity of 48 mutants has been reported (http://www.yeastgenome.org) (Table 1, underlined; Table S2), while the zinc sensitivity of the other 60 mutants is reported for the first time. The functional classes of these 108 are related to metabolism (11), cell cycle and DNA processing (18), transcription (15), Protein fate (synthesis, folding, modification, destination) (17), cellular transport, transport facilities and transport routes (32), as well as unclassified proteins (15) (Table 1).
Table 1. List of 108 genes whose deletion mutants are sensitive to 6 mM ZnCl2.
| Systemic name | Standard name | Systemic name | Standard name | Systemic name | Standard name | Systemic name | Standard name |
|---|---|---|---|---|---|---|---|
| Metabolism (11) | |||||||
| YDR127Wa | ARO1 | YHR106W | TRR2 | YNL129W | NRK1 | YPR060C | ARO7 |
| YGL234W | ADE5,7 | YJL216C | IMA5 | YNL220W | ADE12 | YDL133Wb | SRF1 |
| YHR004C | NEM1 | YLR425W | TUS1 | YPL268W | PLC1 | ||
| Cell cycle and DNA processing (18) | |||||||
| YAL015C | NTG1 | YDL047W | SIT4 | YJL092W | SRS2 | YNL271C | BNI1 |
| YAL020C | ATS1 | YDL101C | DUN1 | YJL208C | NUC1 | YPL031C | PHO85 |
| YAL047C | SPC72 | YDR137W | RGP1 | YJR043C | POL32 | YHR113W | APE4 |
| YCR077C | PAT1 | YER116C | SLX8 | YLL002W | RTT109 | ||
| YCR086W | CSM1 | YHR061C | GIC1 | YNL059C | ARP5 | ||
| Transcription (15) | |||||||
| YCR081W | SRB8 | YFL001W | DEG1 | YHR178W | STB5 | YLR182W | SWI6 |
| YDL160C | DHH1 | YFL049W | SWP82 | YIL154C | IMP2’ | YNR052C | POP2 |
| YDR028C | REG1 | YGL071W | AFT1 | YJL124C | LSM1 | YPL254W | HFI1 |
| YDR310C | SUM1 | YGR092W | DBF2 | YKL139W | CTK1 | ||
| Protein fate (synthesis, folding, modification, destination) (17) | |||||||
| YBR131W | CCZ1 | YDR283C | GCN2 | YJL004C | SYS1 | YNL069C | RPL16B |
| YCL001W | RER1 | YFL016C | MDJ1 | YJL204C | RCY1 | YPR133W-A | TOM5 |
| YCL045C | EMC1 | YGL124C | MON1 | YKL006W | RPL14A | ||
| YCR079W | PTC6 | YGL195W | GCN1 | YLL010C | PSR1 | ||
| YDL045W-A | MRP10 | YHR076W | PTC7 | YMR264W | CUE1 | ||
| Cellular transport, transport facilities and transport routes (32) | |||||||
| YBR127C | VMA2 | YFL004W | VTC2 | YJL012C | VTC4 | YLR261C | VPS63 |
| YCL038C | ATG22 | YGL212W | VAM7 | YJL024C | APS3 | YLR262C | YPT6 |
| YCR037C | PHO87 | YGL095C | VPS45 | YJL154C | VPS35 | YLR268W | SEC22 |
| YDL100C | GET3 | YGR105W | VMA21 | YJL198W | PHO90 | YLR396C | VPS33 |
| YDR089W | VTC5 | YHL031C | GOS1 | YLR242C | ARV1 | YMR243C | ZRC1 |
| YDR186C | SND1 | YHR026W | VMA16 | YKL080W | VMA5 | YNL323W | LEM3 |
| YDR276C | PMP3 | YHR094C | HXT1 | YKL119C | VPH2 | YOR270C | VPH1 |
| YDR456W | NHX1 | YHR108W | GGA2 | YKR020W | VPS51 | YPL045W | VPS16 |
| Unclassified proteins (15) | |||||||
| YCL007C | YHR112C | YKL044W | MMO1 | YNL204C | SPS18 | ||
| YDL041W | YHR151C | MTC6 | YLR149C | YPL261C | |||
| YDR203W | YJL211C | YLR232W | YPR123C | ||||
| YDR417C | YJL218W | YMR265C | |||||
Gene names were listed alphabetically according to their systemic names.
The mutations for 48 genes that were reported sensitive to zinc previously were underlined.
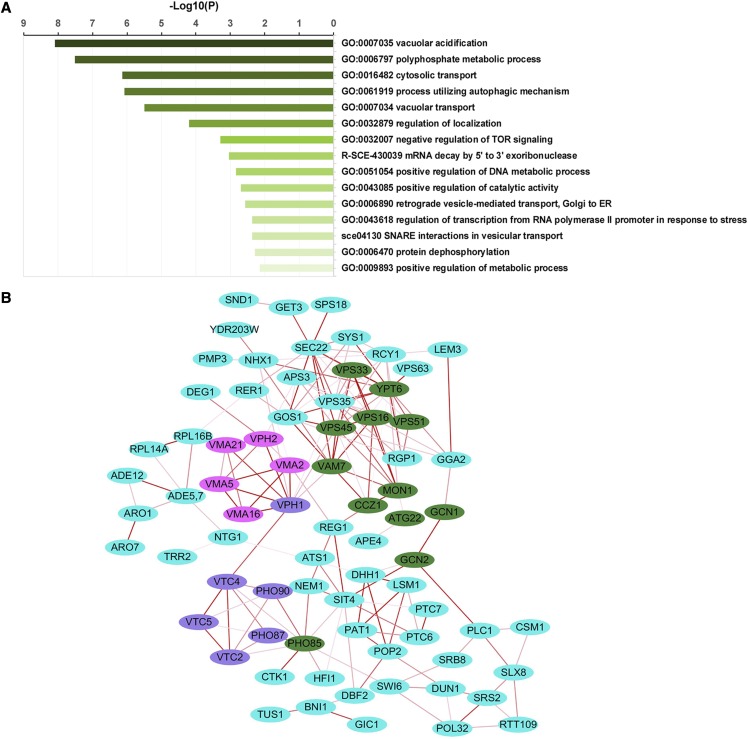
Gene Ontology (GO) enrichment analysis showed that the 108 genes were most enriched in the biological processes of vacuolar acidification, polyphosphate metabolic process, cytosolic transport, the process utilizing autophagic mechanism and vacuolar transport among the top 20 GO terms in cluster groups (Figure 1A). All the zinc-sensitive genes were then uploaded to the web-based online tool STRING (https://string-db.org/) and the information of PPI was gained and visualized by Cytoscape software (version 3.6.0) (Figure 1B). A total of 76 proteins were filtered from the 108 candidates, and a significant functional network was constructed by these 76 nodes and 168 interaction pairs.
Figure 1.
Meta-enrichment analysis summary of zinc-sensitive genes. (A) Heatmap of the top 20 enriched GO terms. For GO terms, each band represents one enriched term colored according to its -log 10 p-value. The dominant term within each group is used as a group heading. (B) The protein-protein interaction (PPI) networks of the zinc-sensitive genes. The edges represent the combined score, and the thicker the edge, the higher the similarity.
Elevated intracellular zinc levels evoked by excess zinc
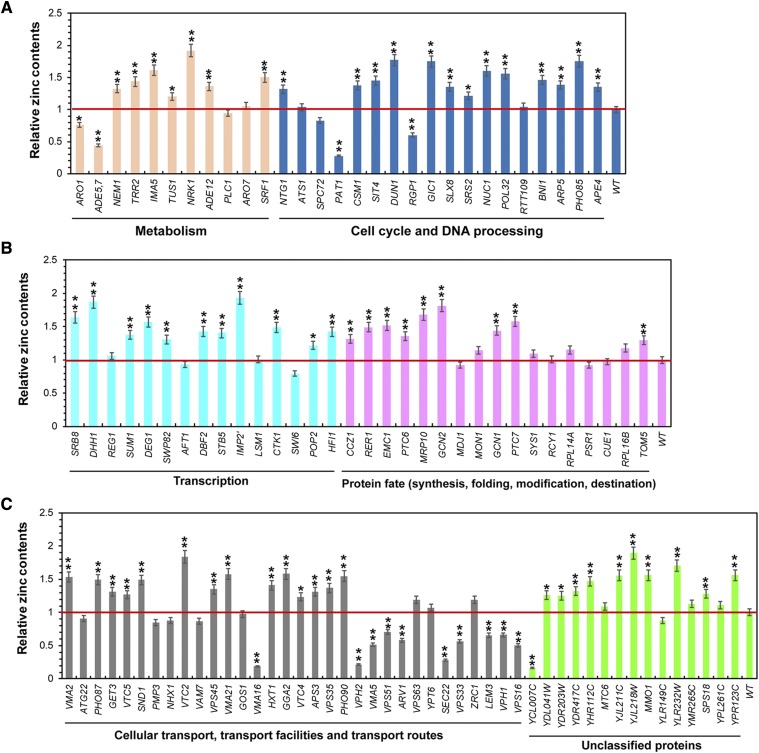
To determine the correlations between zinc sensitivity and intracellular zinc accumulations in all zinc-sensitive mutants, we analyzed the intracellular zinc concentrations in yeast cells treated with 3 mM ZnCl2. We showed that the intracellular zinc content of a number of 64 mutants (almost 59% of the total 108 genes) was significantly increased compared with wild-type cells (Figure 2). Meanwhile, 29 zinc-sensitive mutants accumulated similar intracellular zinc ions compared with the wild-type strain, and mutants for 15 genes (ARO1, ADE5,7, PAT1, RGP1, VMA16, VPH1, VMA5, VPS51, ARV1, SEC22, VPS22, LEM3, VPH1, VPS16 and YCL007C) accumulated less intracellular zinc than that of wild-type cells, respectively.
Figure 2.
Intracellular zinc levels of 108 zinc-sensitive gene mutants in response to zinc stress. Log-phase cells were grown with or without 3 mM ZnCl2 for two hours before they were collected for the measurement of intracellular zinc levels. The intracellular zinc content s of these mutants were listed according to their categories in comparison to that of wild type cell BY4743. The value is the average of three independent assays for each strain. The asterisks of “*”and “**” show statistically significant differences of P < 0.05 and P < 0.01, respectively.
Phosphate homeostasis is regulated by SPX domain proteins in eukaryotes and plays an essential role in the biosynthesis of diverse cellular components (Secco et al. 2012). Interestingly, in the present study, deletion mutants for five genes (PHO87, PHO90, VTC2, VTC4 and VTC5) encoding SPX-domain proteins, and one gene (PHO85) coding for cyclin-dependent kinase Pho85, were all sensitive to 6 mM ZnCl2 and accumulated increased intracellular zinc levels in response to excess zinc (Table 1, Figure 2). Based on these findings, we speculate that phosphate homeostasis is significantly involved in maintaining the intracellular zinc homeostasis under excess zinc conditions in budding yeast.
Yeast V-ATPase, an H+-ATPase localized in the vacuole membrane, played crucial roles in regulating the pH values, acidifying intracellular organelles and cellular homeostasis (Graham et al. 2000; Couoh-Cardel et al. 2016). In the present study, we showed that deletion of the 8 genes related to the regulation of the intracellular pH (NHX1, VMA2, VMA21, DBF2, VPH2, VMA5, VPH1 and VMA16), rendered yeast cells sensitive to 6 mM ZnCl2, although only mutants for two genes (VMA2 and VMA21) accumulated higher intracellular zinc in response to zinc stress (Figure 2C). In addition, mutants for 8 genes (VAM7, VPS45, APS3, VPS35, VPS63, VPS33 and VPS51) encoding proteins associated with the function of vacuolar protein sorting were also sensitive to zinc stress. However, only mutants for VPS45, APS3 and VPS35 accumulated higher intracellular zinc in response to zinc stress (Figure 2C).
Autophagy is a fundamental cellular process of all eukaryotic cells that aged and/or damaged cytoplasmic proteins, lipids, unwanted organelles and cytosol are translocated to the vacuole and degraded (Song and Kumar 2012; Yin et al. 2016). Interestingly, 18 mutants for ATG22, PTC6, VTC2, YPT6, VPS33, GCN2, CCZ1, VAM7, GCN1, MON1, VTC4, RPL14A, VPS51, PHO85, VPS16, SEC22, GOS1 and VPS45 enriched in the process utilizing autophagic mechanism, were sensitive to zinc (Table 1). Of these genes, mutants for 8 genes (PTC6, VTC2, GCN2, CCZ1, GCN1, VTC4, PHO85 and VPS45) accumulated higher intracellular zinc than wide-type cells, indicating their important roles in maintaining the intracellular zinc homeostasis. The only identified gene ATG gene in this study, ATG22, encodes a protein involved in transporting small molecules such as amino acids back to the cytosol for protein synthesis and other cellular functions (Tyler and Johnson 2018). Three genes of CCZ1, MON1 and YPT6 were required for the CVT pathway and the autophagy (Cabrera et al. 2014; Suda et al. 2013). VTC (Vacuolar Transporter Chaperone) complex consists of five subunits (Vtc1, Vtc2, Vtc3, Vtc4 and Vtc5), and functions in several membrane-related processes, including the microautophagic scission of vesicles into the vacuolar lumen. Three VTC genes (VTC2, VTC4 and VTC5) screened out in the present study and cyclin-dependent kinase gene PHO85 were all involved in phosphate homeostasis, while Pho85 was also a regulator of autophagy (Reggiori and Klionsky 2013). Gcn2 kinase and its positive regulator Gcn1 were both identified sensitive to high zinc. Gcn2 is involved in sensing the level of intracellular amino acids and can be regulated by transcription factor Gcn4 and the cyclin-dependent kinase Pho85 (Reggiori and Klionsky 2013).
Notably, consistent with a previous study (Pagani et al. 2007; Mesquita et al. 2016), we found that deletion of AFT1 encoding a transcription factor, ZRC1 encoding a protein transporting zinc into vacuolar, rendered yeast cells sensitive to 3 mΜ ZnCl2, although the intracellular zinc content of the two mutants reveal no significant differences in response to zinc stress (Figure 2B, C).
Oxidative stress induced by zinc stress
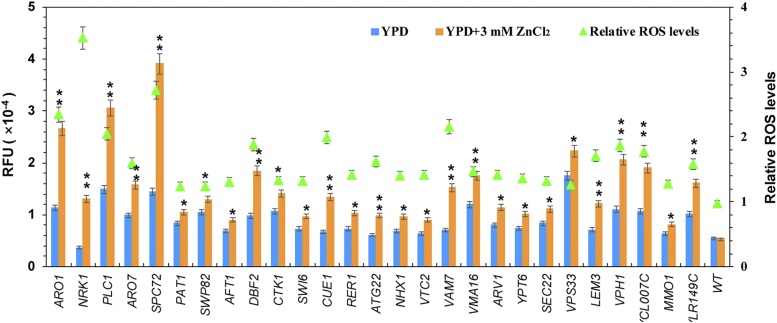
Excess zinc can generate reactive oxygen species (ROS) when it accumulates to toxic levels in cells (Pagani et al. 2007). It was reported that ROS stress was induced when the wide-type cells were treated with 10 mM. A low concentration of zinc (5 mg/L, about 77 µM) did not induce ROS production (Mesquita et al. 2016), while 5 mM zinc produced a weak ROS response in wild-type cells (Pagani et al. 2007). However, some zinc-sensitive genes might be involved in this process. To further confirm this possibility, we measured the intracellular ROS levels following 3 mM ZnCl2 or 8 mM ZnCl2 treatment, respectively. When the concentration of ZnCl2 is 3 mM, the intracellular ROS level in wild-type BY4743 cells treated with zinc showed no significant difference compared with the untreated cells. Of these 108 zinc-sensitive mutants, the intracellular ROS levels of about 27 mutants were increased by extracellular zinc stress (Figure 3), including 11 genes involved in cellular transport (ATG22, NHX1, VTC2, VAM7, VMA16, ARV1, YPT6, SEC22, VPS33, LEM3 and VPH1), five genes of transcriptional process (SWP82, AFT1, DBF2, CTK1 and SWI6), four genes of metabolism (ARO1, NRK1, PLC1 and ARO7), two gens of cell cycle and DNA processing (SPC72 and PAT1), one genes of protein fate (CUE1), and other three unidentified genes (YCL007C, MMO1 and YLR149C). It indicated that these 27 genes were all crucial for dealing with the oxidative damage induced by 3 mM ZnCl2. The other 81 mutants accumulated similar (73 mutants) or even lower (18 mutants) intracellular ROS levels when treated with zinc compared with the no treated cells (Figure S1), respectively. When these mutants were treated with 8 mM ZnCl2, it was showed that 51 mutants displayed increased ROS levels, including 23 mutants in which the intracellular Zn levels were higher than that of the wild type cells (Figure S3). These results suggested that these genes might be involved in the regulation of intracellular ROS levels induced by higher level of zinc.
Figure 3.
Increased intracellular ROS levels of zinc-sensitive gene mutants in response to 3 mM ZnCl2. The relative ROS levels of these mutants in response to zinc treatment are normalized against their related untreated cells. Log-phase cells were grown with or without 3 mM ZnCl2 for 2 hr before harvesting and measurement of intracellular ROS levels using dihydroethidium. Results are averages of three independent assays for each strain. The asterisks of “*”and “**” show statistically significant differences of P < 0.05 and P < 0.01, respectively.
The expression of genes involved in redox homeostasis is regulated by zinc stress
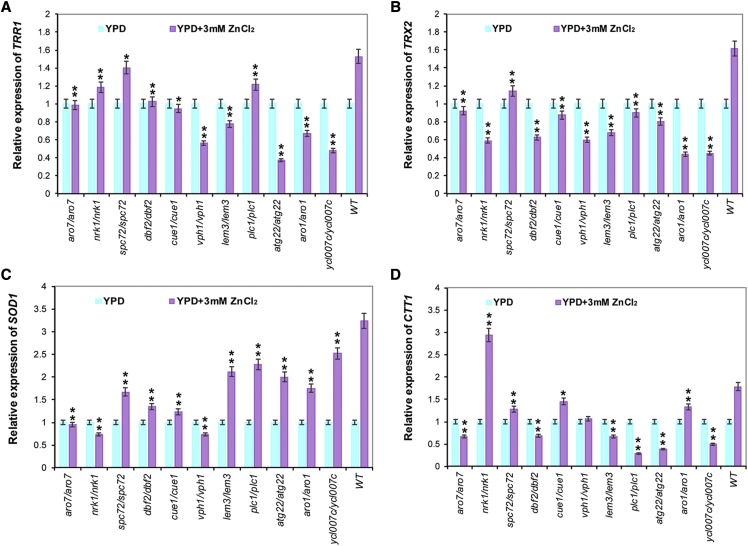
It was reported that zinc induced the expression of a number oxidative stress scavenging genes, including CTT1, GAD1, GPX1, SOD1 and SOD2, etc (Pagani et al. 2007). To investigate whether the zinc-sensitive genes were involved in regulating the expression level of genes involved in redox homeostasis, we pick up 11 mutants for ARO7, NRK1, SPC72, DBF2, CUE1, VPH1, LEM3, PLC1, ATG22, ARO1, and YCL007C accumulated the most relative ROS levels when treatment with zinc (Figure 3), to test the mechanism of oxidative damage induced by zinc. Next, we measured the expression of TRR1 (thioredoxin reductase), TRX2 (thioredoxin 2), GSH1 (glutamylcysteine synthetase), SOD1 (copper/zinc superoxide dismutase), CTT1 (cytosolic catalase T) and GPX2 (2-Cys peroxiredoxin), in these 11 mutants by qPCR assay. Four genes of TRR1, TRX2, SOD1 and CTT1 were significantly up-regulated after treatment with zinc in the wild-type cells (Figure 4), however, no significant difference was showed for the expression of GSH1 or GPX2 in the presence or absence of zinc (Figure S2). Interestingly, both of the expression levels of TRX2 and SOD1 were decreased in the 11 mutants compared with wild type cells (Figure 4B, C). Furthermore, the expressions of TRR1 and CTT1 were also reduced in these mutants except the mutants for SPC72 and NRK1, respectively (Figure 4A, D). Based on these analyses, we concluded that the decreased expression of TRR1, SOD1, CTT1 and TRX2 might be responsible for the high intracellular ROS levels evoked in these mutants.
Figure 4.
Relative expression levels of genes involved in oxidative stress response. TRR1, TRX2, SOD1, CTT1, genes in the S. cerevisiae. Gene expression is quantified using RT-qPCR and comparative critical threshold (2 -ΔΔCt) method. The PGK1 gene was used as internal control and the ratio of the fold-change without treatment was standardized to 1.0. These values represent the average of three independent experiments. The asterisks of “*”and “**” show statistically significant differences of P < 0.05 and P < 0.01, respectively.
Discussion
The main goals of this study were to identify the genes involved in zinc tolerance, and to investigate the toxicity mechanisms induced by zinc stress in yeast cells. Zinc serves as a crucial structural and catalytic cofactor for many functional proteins such as transcriptional factors containing zinc-finger (s), molecular chaperones, DNA or RNA polymerases, lipid-binding proteins, some metabolic enzymes, etc (Singh et al. 2017). However, it is toxic at high intracellular levels as zinc can generate reactive oxygen species and thus trigger several biological molecules damaged to cell growth (Howlett and Avery 1997; Chrestensen et al. 2000; Serero et al. 2008; Pagani et al. 2007). Therefore, zinc homeostasis must be tightly controlled. In the present study, we identified 108 zinc-sensitive gene deletion mutations from a genome-scale screen in budding yeast, including 60 mutants that have not been reported previously. It was reported the vacuole integrity was essential for chloride homeostasis (Jennings and Cui 2008), therefore we pick up some of the mutants to investigate whether they are sensitive to another Zn-containing compound ZnSO4, as well as NaCl. However, the results showed that these mutants shared the similar sensitivities in 6 mM of ZnCl2, ZnSO4 and 12 mM NaCl was not affected the growth of these mutants (Figure S3). The results indicated that the zinc sensitivities of these mutants were not influenced by the chloride homeostasis. Our finding might imply that excess zinc toxicity might be mainly due to the high intracellular zinc levels and/or ROS levels induced by zinc stress in yeast cells, although not all mutants revealed high intracellular zinc levels and ROS levels in response to high zinc.
In budding yeast, ten genes encode SPX domain proteins, which are named after S yg1, P ho81, X PR1 (Secco et al. 2012). Of the ten SPX domains proteins, nine have been identified to relate to Pi metabolism, including two plasma membrane Pi importers, Pho87 and Pho90 (Wykoff and O’Shea 2001; Bun-ya et al. 1996; Hürlimann et al. 2007); a vacuolar Pi exporter, Pho91 (Hürlimann et al. 2007); a cyclin-dependent kinase inhibitor Pho81, which regulate the activity of Pho80/Pho85 (Schneider et al. 1994); and a glycerophosphocholine phosphodiesterase, Gde1 (Fisher et al. 2005). Four SPX domains proteins (Vtc2, Vtc3, Vtc4 and Vtc5) involved in producing polyP, are part of the Vacuole Transporter Chaperone (VTC) complex, which play functional roles in sorting of H+-translocating ATPases, endocytosis, ER-Golgi trafficking, vacuole fusion, vacuolar polyphosphate homeostasis and the microautophagic scission of vesicles into the vacuolar lumen (Müller et al. 2002; Müller et al. 2003; Desfougères et al. 2016). The tenth SPX domain protein with unknown function is encoded by SYG1 and localizes in plasma membrane, which might have a function in exporting Pi (Vaughan et al. 2012). Interestingly, the results indicated that six genes involved in the process of phosphate mechanism were sensitive to zinc and accumulated higher intracellular zinc, indicating their crucial roles in maintaining the intracellular zinc homeostasis under excess zinc conditions in budding yeast.
Autophagy is well conserved from budding yeast to human cells and is also crucial for maintaining the cellular and organismal homeostasis, immunity and organismal development (Yin et al. 2016). There two types of autophagy in yeast, macroautophagy and microautophagy, a nonspecific or direct process, respectively (Tyler and Johnson 2018). The process of autophagy is regulated by nitrogen, glucose depletion, amino acid and phosphate starvation, mitophagy, pexophagy, transcriptional control and lipid metabolisms (Reggiori and Klionsky 2013). Zinc depletion induces non-selective autophagy in a Zap1-independent manner and this process may play a role in releasing zinc from the degraded proteins for other purposes (Kawamata et al. 2017). Interestingly, we identified 18 genes involved in the process of autophagy mechanism, were sensitive to zinc. Two groups of proteins, autophagy-related (ATG) or vacuolar protein sorting (VPS) proteins, mediate the process of autophagy. Importantly, the cytoplasm-to-vacuole targeting pathway (Cvt pathway) is used to deliver various cargos to the vacuole, and is considered a specialized form of autophagy. Furthermore, three genes related to amino acids synthesis (APE4, ARO1 and ARO7) and four genes involved in lipid homeostasis (ARV1, SRF1, NEM1 and LEM3), were all required for zinc tolerance. The SNARE (soluble N-ethylmaleimide-sensitive factor attached protein receptor) complex, a protein complex involved in membrane fusion, is also required for autophagy (Tyler and Johnson 2018). In this study, six genes (VAM7, VPS33, VPS51, SEC22, GOS1 and VPS45) encoding proteins belong to the SNARE complex were identified sensitive to zinc stress, indicating their crucial role in the process of autophagy under excess zinc conditions. Based on these analyses, we hypothesized that autophagy may play a significant role in maintaining the cellular zinc homeostasis possibly by transporting the excess zinc to vacuolar (Figure 5).
Figure 5.
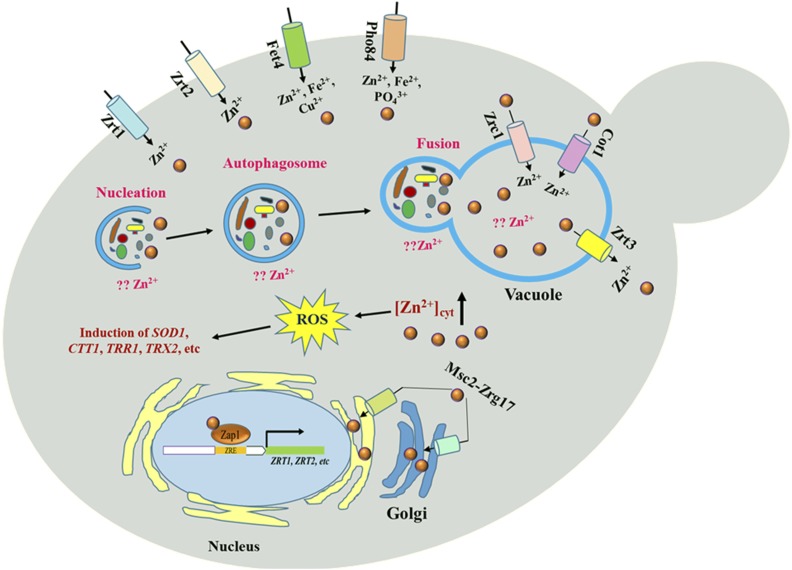
Predicted model for zinc homeostasis and its toxicity mechanisms. Yeast cells uptake the extracellular zinc through plasma membrane transporters Zrt1, Zrt2, Fet4 and Pho84. Inside the cell, two vacuolar zinc transporters Cot1, Zrt3 and Zrc1 are responsible for transporting zinc out or into the vacuolar, respectively, and the heteromeric complex formed by Msc2 and Zrg17 transports the cytoplasm zinc to the endoplasmic reticulum when it is in excess. Excess zinc can generate reactive oxygen species (ROS) induce the oxidative stress scavenging genes. The process of autophagy may play a significant role in maintaining the cellular zinc homeostasis possibly by transporting the excess zinc to vacuolar.
Under the high zinc treatment, we observed 27 mutants increased the intracellular ROS levels comparing with the wild-types, consistent with their growth defect. We pick up 11 mutants for ARO7, NRK1, SPC72, DBF2, CUE1, VPH1, LEM3, PLC1, ATG22, ARO1, and YCL007C, which accumulated the highest relative ROS levels than that of wild-type cells in response to high zinc (Figure 3), to investigate the mechanism of oxidative damage induced by zin stress. Apparently, the expression of some antioxidant defenses genes was down-regulated by zinc stress in these mutants, suggesting that these zinc-sensitive genes might involve in maintaining the redox balance in response to high zinc. However, the rest 81 mutants accumulated similar or even lower ROS levels compared wild-type cells. Since the ROS stress was induced when the zinc concentration reached 10 mM in the wide-type cells (Pagani et al. 2007), these zinc-sensitive genes might be involved in ROS process when the extracellular zinc content was higher than 3 mM. They could also play a role in detoxification of excess zinc by yeast cells, but further investigations are required.
Conclusions
In conclusion, our work has identified 108 yeast single-gene deletion mutants that are sensitive to 6 mM ZnCl2 by screening the yeast diploid nonessential gene deletion library. We have shown that 64 mutants accumulated higher intracellular zinc levels than wild-type cells in response to excess zinc, indicating their crucial role in maintaining the intracellular zinc homeostasis. Our data showed that the intracellular ROS levels in 51 mutants were significantly higher than that of the wild-type cells under high zinc stress. Our findings make it likely that excess zinc can generate oxidative damage to yeast cells through up-regulating several antioxidant defenses genes. Our current findings would provide a basis to understand molecular mechanisms of zinc toxicity in yeast cells.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (21877053 and 31601564), the Natural Science Foundation of Jiangsu Province (BK20181345), the Open Foundation of Jiangsu Key Laboratory of Industrial Biotechnology (KLIB-KF201807), the Fundamental Research Funds for the Central Universities (JUSRP51705A), and the Top-notch Academic Programs Project of Jiangsu Higher Education Institutions. The funding agencies provided financial support to generate the data, however, they played no role in the design of this study, analysis and interpretation of the data, or in writing and submitting the manuscript.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25387/g3.11365043.
Communicating editor: N. Rhind
Literature Cited
- Bird A. J., Blankman E., Stillman D. J., Eide D. J., and Winge D. R., 2004. The Zap1 transcriptional activator also acts as a repressor by binding downstream of the TATA box in ZRT2. EMBO J. 23: 1123–1132. 10.1038/sj.emboj.7600122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. J., McCall K., Kramer M., Blankman E., Winge D. R. et al. , 2003. Zinc fingers can act as Zn2+ sensors to regulate transcriptional activation domain function. EMBO J. 22: 5137–5146. 10.1093/emboj/cdg484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleackley M. R., Young B. P., Loewen C. J., and MacGillivray R. T., 2011. High density array screening to identify the genetic requirements for transition metal tolerance in Saccharomyces cerevisiae. Metallomics 3: 195–205. 10.1039/c0mt00035c [DOI] [PubMed] [Google Scholar]
- Bun-Ya M., Nishimura M., Harashima S., and Oshima Y., 1991. The PHO84 gene of Saccharomyces cerevisiae encodes an inorganic phosphate transporter. Mol. Cell. Biol. 11: 3229–3238. 10.1128/MCB.11.6.3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bun-ya M., Shikata K., Nakade S., Yompakdee C., Harashima S. et al. , 1996. Two new genes, PHO86 and PHO87, involved in inorganic phosphate uptake in Saccharomyces cerevisiae. Curr. Genet. 29: 344–351. [PubMed] [Google Scholar]
- Büttner S., Eisenberg T., Carmona-Gutierrez D., Ruli D., Knauer H. et al. , 2007. Endonuclease G regulates budding yeast life and death. Mol. Cell 25: 233–246. 10.1016/j.molcel.2006.12.021 [DOI] [PubMed] [Google Scholar]
- Cabrera M., Nordmann M., Perz A., Schmedt D., Gerondopoulos A. et al. , 2014. The Mon1-Ccz1 GEF activates the Rab7 GTPase Ypt7 via a longin-fold-Rab interface and association with PI3P-positive membranes. J. Cell Sci. 127: 1043–1051. 10.1242/jcs.140921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrestensen C. A., Starke D. W., and Mieyal J. J., 2000. Acute cadmium exposure inactivates thioltransferase (Glutaredoxin), inhibits intracellular reduction of protein-glutathionyl-mixed disulfides and initiates apoptosis. J. Biol. Chem. 275: 26556–26565. 10.1074/jbc.M004097200 [DOI] [PubMed] [Google Scholar]
- Couoh-Cardel S., Hsueh Y. C., Wilkens S., and Movileanu L., 2016. Yeast V-ATPase proteolipid ring acts as a large-conductance transmembrane protein pore. Sci. Rep. 6: 24774 10.1038/srep24774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nicola R., Hazelwood L. A., De Hulster E. A., Walsh M. C., Knijnenburg T. A. et al. , 2007. Physiological and transcriptional responses of Saccharomyces cerevisiae to zinc limitation in chemostat cultures. Appl. Environ. Microbiol. 73: 7680–7692. 10.1128/AEM.01445-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desfougères Y., Gerasimaite R. U., Jessen H. J., and Mayer A., 2016. Vtc5, a novel subunit of the vacuolar transporter chaperone complex, regulates polyphosphate synthesis and phosphate homeostasis in yeast. J. Biol. Chem. 291: 22262–22275. 10.1074/jbc.M116.746784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide D. J., 2003. Multiple regulatory mechanisms maintain zinc homeostasis in Saccharomyces cerevisiae. J. Nutr. 133: 1532S–1535S. 10.1093/jn/133.5.1532S [DOI] [PubMed] [Google Scholar]
- Eide D. J., 2009. Homeostatic and adaptive responses to zinc deficiency in Saccharomyces cerevisiae. J. Biol. Chem. 284: 18565–18569. 10.1074/jbc.R900014200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher E., Almaguer C., Holic R., Griac P., and Patton-Vogt J., 2005. Glycerophosphocholine-dependent growth requires Gde1p (YPL110c) and Git1p in Saccharomyces cerevisiae. J. Biol. Chem. 280: 36110–36117. 10.1074/jbc.M507051200 [DOI] [PubMed] [Google Scholar]
- Frey A. G., Bird A. J., Evans-Galea M. V., Blankman E., Winge D. R. et al. , 2011. Zinc-regulated DNA binding of the yeast Zap1 zinc-responsive activator. PLoS One 6: e22535 10.1371/journal.pone.0022535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitan R. S., and Eide D. J., 2000. Zinc-regulated ubiquitin conjugation signals endocytosis of the yeast ZRT1 zinc transporter. Biochem. J. 346: 329–336. 10.1042/bj3460329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitan R. S., Luo H., Rodgers J., Broderius M., and Eide D., 1998. Zinc-induced inactivation of the yeast ZRT1 zinc transporter occurs through endocytosis and vacuolar degradation. J. Biol. Chem. 273: 28617–28624. 10.1074/jbc.273.44.28617 [DOI] [PubMed] [Google Scholar]
- Graham L. A., Powell B., and Stevens T. H., 2000. Composition and assembly of the yeast vacuolar H(+)-ATPase complex. J. Exp. Biol. 203: 61–70. [DOI] [PubMed] [Google Scholar]
- Hambidge K. M., and Krebs N. F., 2007. Zinc deficiency: a special challenge. J. Nutr. 137: 1101–1105. 10.1093/jn/137.4.1101 [DOI] [PubMed] [Google Scholar]
- Hao Q., and Maret W., 2005. Imbalance between pro-oxidant and pro-antioxidant functions of zinc in disease. J. Alzheimers Dis. 8: 161–170. 10.3233/JAD-2005-8209 [DOI] [PubMed] [Google Scholar]
- Herbig A., Bird A. J., Swierczek S., McCall K., Mooney M. et al. , 2005. Zap1 activation domain 1 and its role in controlling gene expression in response to cellular zinc status. Mol. Microbiol. 57: 834–846. 10.1111/j.1365-2958.2005.04734.x [DOI] [PubMed] [Google Scholar]
- Howlett N. G., and Avery S. V., 1997. Induction of lipid peroxidation during heavy metal stress in Saccharomyces cerevisiae and influence of plasma membrane fatty acid unsaturation. Appl. Environ. Microbiol. 63: 2971–2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hürlimann H. C., Stadler-Waibel M., Werner T. P., and Freimoser F. M., 2007. Pho91 Is a vacuolar phosphate transporter that regulates phosphate and polyphosphate metabolism in Saccharomyces cerevisiae. Mol. Biol. Cell 18: 4438–4445. 10.1091/mbc.e07-05-0457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings M. L., and Cui J., 2008. Chloride homeostasis in Saccharomyces cerevisiae: high affinity influx, V-ATPase-dependent sequestration, and identification of a candidate Cl- sensor. J. Gen. Physiol. 131: 379–391. 10.1085/jgp.200709905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata T., Horie T., Matsunami M., Sasaki M., and Ohsumi Y., 2017. Zinc starvation induces autophagy in yeast. J. Biol. Chem. 292: 8520–8530. 10.1074/jbc.M116.762948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J. C., Shames D. M., and Woodhouse L. R., 2000. Zinc homeostasis in humans. J. Nutr. 130: 1360S–1366S. 10.1093/jn/130.5.1360S [DOI] [PubMed] [Google Scholar]
- Liu X., Wu J., Shi W., Shi W., Liu H. et al. , 2018. Lead induces genotoxicity via oxidative stress and promoter methylation of DNA repair genes in human Lymphoblastoid TK6 cells. Med. Sci. Monit. 24: 4295–4304. 10.12659/MSM.908425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., and Schmittgen T. D., 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Method. Methods. 25: 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- MacDiarmid C. W., Gaither L. A., and Eide D., 2000. Zinc transporters that regulate vacuolar zinc storage in Saccharomyces cerevisiae. EMBO J. 19: 2845–2855. 10.1093/emboj/19.12.2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesquita V. A., Silva C. F., and Soares E. V., 2016. Toxicity Induced by a Metal Mixture (Cd, Pb and Zn) in the Yeast Pichia kudriavzevii: the role of oxidative stress. Curr. Microbiol. 72: 545–550. 10.1007/s00284-016-0987-y [DOI] [PubMed] [Google Scholar]
- Miyabe S., Izawa S., and Inoue Y., 2000. Expression of ZRC1 coding for suppressor of zinc toxicity is induced by zinc-starvation stress in Zap1-dependent fashion in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 276: 879–884. 10.1006/bbrc.2000.3580 [DOI] [PubMed] [Google Scholar]
- Müller O., Bayer M. J., Peters C., Andersen J. S., Mann M. et al. , 2002. The Vtc proteins in vacuole fusion: coupling NSF activity to V(0) trans-complex formation. EMBO J. 21: 259–269. 10.1093/emboj/21.3.259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller O., Neumann H., Bayer M. J., and Mayer A., 2003. Role of the Vtc proteins in V-ATPase stability and membrane trafficking. J. Cell Sci. 116: 1107–1115. 10.1242/jcs.00328 [DOI] [PubMed] [Google Scholar]
- Muthukumar, K., and V. Nachiappan, 2010 Cadmium-induced oxidative stress in Saccharomyces cerevisiae. Indian j. biochem. bio. 47: 383–387. [PubMed]
- Nargund A. M., Avery S. V., and Houghton J. E., 2008. Cadmium induces a heterogeneous and caspase-dependent apoptotic response in Saccharomyces cerevisiae. Apoptosis 13: 811–821. 10.1007/s10495-008-0215-8 [DOI] [PubMed] [Google Scholar]
- North M., Steffen J., Loguinov A. V., Zimmerman G. R., Vulpe C. D. et al. , 2012. Genome-wide functional profiling identifies genes and processes important for zinc-limited growth of Saccharomyces cerevisiae. PLoS Genet. 8: e1002699 10.1371/journal.pgen.1002699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani M. A., Casamayor A., Serrano R., Atrian S., and Arino J., 2007. Disruption of iron homeostasis in Saccharomyces cerevisiae by high zinc levels: a genome-wide study. Mol. Microbiol. 65: 521–537. 10.1111/j.1365-2958.2007.05807.x [DOI] [PubMed] [Google Scholar]
- Powell S. R., 2000. The antioxidant properties of zinc. J. Nutr. 130: 1447S–1454S. 10.1093/jn/130.5.1447S [DOI] [PubMed] [Google Scholar]
- Reggiori F., and Klionsky D. J., 2013. Autophagic processes in yeast: mechanism, machinery and regulation. Genetics 194: 341–361. 10.1534/genetics.112.149013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider K. R., Smith R. L., and O’Shea E. K., 1994. Phosphate-regulated inactivation of the kinase PHO80–PHO85 by the CDK inhibitor PHO81. Science 266: 122–126. 10.1126/science.7939631 [DOI] [PubMed] [Google Scholar]
- Schothorst J., Zeebroeck G. V., and Thevelein J. M., 2017. Identification of Ftr1 and Zrt1 as iron and zinc micronutrient transceptors for activation of the PKA pathway in Saccharomyces cerevisiae. Microb. Cell 4: 74–89. 10.15698/mic2017.03.561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secco D., Wang C., Shou H., and Whelan J., 2012. Phosphate homeostasis in the yeast Saccharomyces cerevisiae, the key role of the SPX domain-containing proteins. FEBS Lett. 586: 289–295. 10.1016/j.febslet.2012.01.036 [DOI] [PubMed] [Google Scholar]
- Serero A., Lopes J., Nicolas A., and Boiteux S., 2008. Yeast genes involved in cadmium tolerance: Identification of DNA replication as a target of cadmium toxicity. DNA Repair (Amst.) 7: 1262–1275. 10.1016/j.dnarep.2008.04.005 [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T. et al. , 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13: 2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N., Yadav K. K., and Rajasekharan R., 2017. Effect of zinc deprivation on the lipid metabolism of budding yeast. Curr. Genet. 63: 977–982. 10.1007/s00294-017-0704-9 [DOI] [PubMed] [Google Scholar]
- Song Q., and Kumar A., 2012. An overview of autophagy and yeast pseudohyphal growth: integration of signaling pathways during nitrogen stress. Cells 1: 263–283. 10.3390/cells1030263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda Y., Kurokawa K., Hirata R., and Nakano A., 2013. Rab GAP cascade regulates dynamics of Ypt6 in the Golgi traffic. Proc. Natl. Acad. Sci. USA 110: 18976–18981. 10.1073/pnas.1308627110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler J. K., and Johnson J. E., 2018. The role of autophagy in the regulation of yeast life span. Ann. N. Y. Acad. Sci. 1418: 31–43. 10.1111/nyas.13549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan A. E., Mendoza R., Aranda R., Battini J. L., and Miller A. D., 2012. Xpr1 Is an Atypical G-protein-coupled receptor that mediates xenotropic and polytropic murine retrovirus neurotoxicity. J. Virol. 86: 1661–1669. 10.1128/JVI.06073-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters B. M., and Eide D. J., 2002. Combinatorial control of yeast FET4 gene expression by iron, zinc, and oxygen. J. Biol. Chem. 277: 33749–33757. 10.1074/jbc.M206214200 [DOI] [PubMed] [Google Scholar]
- Wilson S., and Bird A. J., 2016. Zinc sensing and regulation in yeast model systems. Arch. Biochem. Biophys. 611: 30–36. 10.1016/j.abb.2016.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. Y., Bird A. J., Chung L. M., Newton M. A., Winge D. R. et al. , 2008. Differential control of Zap1-regulated genes in response to zinc deficiency in Saccharomyces cerevisiae. BMC Genomics 9: 370 10.1186/1471-2164-9-370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. H., Frey A. G., and Eide D. J., 2011. Transcriptional regulation of the Zrg17 zinc transporter of the yeast secretory pathway. Biochem. J. 435: 259–266. 10.1042/BJ20102003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykoff D. D., and O’Shea E. K., 2001. Phosphate transport and sensing in Saccharomyces cerevisiae. Genetics 159: 1491–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z., Pascual C., and Klionsky D. J., 2016. Autophagy: machinery and regulation. Microb. Cell 3: 588–596. 10.15698/mic2016.12.546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Butler E., Rodgers J., Spizzo T., Duesterhoeft S. et al. , 1998. Regulation of zinc homeostasis in yeast by binding of the ZAP1 transcriptional activator to zinc-responsive promoter elements. J. Biol. Chem. 273: 28713–28720. 10.1074/jbc.273.44.28713 [DOI] [PubMed] [Google Scholar]
- Zhao H., and Eide D., 1996a. The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proc. Natl. Acad. Sci. USA 93: 2454–2458. 10.1073/pnas.93.6.2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., and Eide D., 1996b. The ZRT2 gene encodes the low affinity zinc transporter in Saccharomyces cerevisiae. J. Biol. Chem. 271: 23203–23210. 10.1074/jbc.271.38.23203 [DOI] [PubMed] [Google Scholar]
- Zhao H., and Eide D. J., 1997. Zap1p, a metalloregulatory protein involved in zinc-responsive transcriptional regulation in Saccharomyces cerevisiae. Mol. Cell. Biol. 17: 5044–5052. 10.1128/MCB.17.9.5044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Du J., Xiong B., Xu H., and Jiang L., 2013. ESCRT components regulate the expression of the ER/Golgi calcium pump gene PMR1 through the Rim101/Nrg1 pathway in budding yeast. J. Mol. Cell Biol. 5: 336–344. 10.1093/jmcb/mjt025 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at figshare: https://doi.org/10.25387/g3.11365043.