Abstract
Reduced numbers of carpal and tarsal bones (wrist and ankle joints) are extensively observed in the clade of Cetacea and Ruminantia (Cetruminantia). Homebox D11 (Hoxd11) is one of the important genes required for limb development in mammals. Mutations in Hoxd11 can lead to defects in particular bones of limbs, including carpus and tarsus. To test whether evolutionary changes in Hoxd11 underlie the loss of these bones in Cetruminantia, we sequenced and analyzed Hoxd11 coding sequences and compared them with other 5′ HoxA and HoxD genes in a taxonomic coverage of Cetacea, Ruminantia and other mammalian relatives. Statistical tests on the Hoxd11 sequences found an accelerated evolution in the common ancestor of cetaceans and ruminants, which coincided with the reduction of carpal and tarsal bones in this clade. Five amino acid substitutions (G222S, G227A, G229S, A240T and G261V) and one amino acid deletion (G254Del) occurred in this lineage. In contrast, other 5′ HoxA and HoxD genes do not show this same evolutionary pattern, but instead display a highly conserved pattern of evolution in this lineage. Accelerated evolution of Hoxd11, but not other 5′ HoxA and HoxD genes, is probably related to the reduction of the carpal and tarsal bones in Cetruminantia. Moreover, we found two amino acid substitutions (G110S and D223N) in Hoxd11 that are unique to the lineage of Cetacea, which coincided with hindlimb loss in the common ancestor of cetaceans. Our results give molecular evidence of Hoxd11 adaptive evolution in cetaceans and ruminants, which could be correlated with limb morphological adaptation.
Keywords: Limb, carpal, tarsal, 5′Hox genes, Hoxd11, mutation
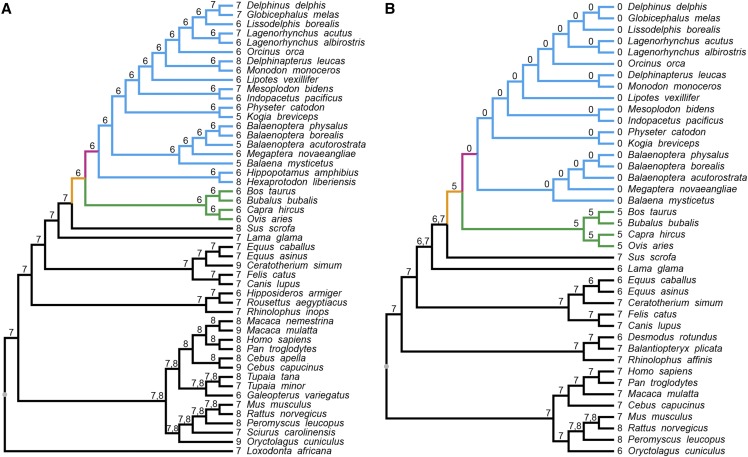
The numbers of carpal and tarsal bones in limbs have evolved differently among species (Hinchliffe and Johnson 1980) (Figure 1). The carpus of mammalian species normally have eight bones, consisting of a proximal row that includes the scaphoid, lunate, triquetral, and pisiform bones and a distal row that contains the trapezium, trapezoid, capitate, and hamate (Yalden 1971; Sadan and Misk 2014; Netter and Colacino 1989). The centrale is located between the two rows; However, this bone is fused with the trapezoid or with the scaphoid and lunate in some mammalian species (Kivell and Begun 2007; Stafford and Thorington 1998). The tarsus of mammals usually has seven bones, with a proximal row of the calcaneus and the talus and a distal row of a cuboid and three cuneiform bones, with a navicular bone between the two rows (Lewis 1980).
Figure 1.
Evolution of carpal and tarsal bone numbers in mammals. (A) Carpus. (B) Tarsus. The numbers of carpal and tarsal bones in extant species are shown to the left side of the names of each species. Numbers for the ancestors are shown on the branches of the species tree. The lineage leading to Cetruminantia is shown in orange and the lineage leading to Cetacea is in pink. Clades for cetaceans and ruminants are indicated in blue and green, respectively.
The Cetruminantia belong to the order Cetartiodactyla and consists of diverse species, including cetaceans, hippopotamuses and ruminants. In species of this clade, the carpus normally has six bones due to the loss of the trapezium, although not in specific toothed whales, and the fusion of the trapezoid and magnum, with the exception of certain whale and hippopotamus (Cooper et al. 2007; Sadan and Misk 2014; Schellhorn and Pfretzschner 2014; Fisher et al. 2007; Turner 1909; Raven 1942; Turner 1885; Benham 1902). However, in the pig, which is a close relative of the Cetruminantia, the carpus has eight bones (Karan 2012). In the orders Perissodactyla and Carnivora, which are close to Cetartiodactyla, the carpus normally consists of seven bones with the rhinoceros even having nine carpal bones (Sadan and Misk 2014; Tung et al. 2010). For hindlimbs, ruminants normally have five tarsal bones, with the fusion of cuneiform bones and also cuboid and navicular (Rajtová 1974; Ehlert et al. 2011). Cetaceans even lost their hindlimbs to adapt to the aquatic lifestyle (Bejder and Hall 2002). In contrast to ruminants, the pig still has seven tarsal bones (Akers and Denbow 2008). Species in orders Perissodactyla and Carnivora have either six or seven tarsal bones (Alam El Din et al. 1986; Galateanu et al. 2014; Akers and Denbow 2008) (Figure 1).
Homeobox (Hox) genes encode transcription factors that control developmental patterning in animals (Pearson et al. 2005; He et al. 2018). In mammals, 39 Hox genes have been identified, which belong to the HoxA, HoxB, HoxC and HoxD clusters (Mallo 2018). Among these genes, some within the HoxA and HoxD clusters are especially critical for early limb patterning (Young et al. 2019; Moreau et al. 2019). Deletion of these two clusters causes severe limb truncation during development (Kmita et al. 2005). The 5′Hox genes (groups 9-13) from these two clusters regulate limb patterning. Paralogous genes of each cluster are expressed in a sequential pattern during development (Zákány and Duboule 1999; Kmita et al. 2002). The 5′Hox genes then participate in regulating the development of the most proximal stylopod, the middle zeugopod and the most distal autopod of limbs (Zákány and Duboule 2007).
Among the Hox genes, Hoxd11 and Hoxa11 are particularly important for distal limb development (Zákány and Duboule 1999). Expression of Hoxa11 is mainly restricted to the zeugopod, while Hoxd11 is expressed not only in the zeugopod, but also in the autopod (Koyama et al. 2010). Hoxd11 is highly expressed at carpal, tarsal and digital regions of mammals during limb development (Wang et al. 2014; Booker et al. 2016). Mutations in Hoxd11 can lead to abnormal ulna, radius, carpals, metacarpals and phalanges in forelimbs (Boulet and Capecchi 2004; Fleischman et al. 2002; Boulet and Capecchi 2002). More interestingly, although no obvious morphological changes happen in hindlimbs, Hoxd11 does affect part of the tarsal bones (Davis and Capecchi 1994). Similarly, mutants of Hoxa11 cause malformations of both forelimbs, as well as the ulna, radius and carpal bones, and tibia and fibula of the hindlimbs (Small and Potter 1993). Double mutations of Hoxa11 and Hoxd11 together caused even more severe morphological malformations, including the disappearance of some tarsal bones (Davis et al. 1995).
Thus, the Hoxd11 gene is of particular interest in studies of the evolution of the mammalian carpal and tarsal bones. The evolution of the Hoxd12 and Hoxd13 genes, which are more related to the development of digits (Davis and Capecchi 1996), has been studied in better detail (Wang et al. 2009). Molecular evolutionary studies on mammalian Hoxd12 and Hoxd13 genes has been reported that the genes were associated with changes in digit and phalange numbers in cetaceans (Wang et al. 2009). Recent evolutionary studies provided further evidences that different Hox genes are important for the diversified morphologies during mammalian evolution (Liang et al. 2013; Nery et al. 2016; Li et al. 2018). Considering that Hoxd11 is more specifically involved in carpal and tarsal bone development (Davis and Capecchi 1994; Koyama et al. 2010), we conducted a molecular evolutionary study of this gene to determine whether evolution of Hoxd11 might explain the bone number variations in Cetruminantia.
Materials and Methods
Ethics approval
The collection procedure of mammalian samples in this study followed ethical principles and approved by the Animal Ethics Committee of Shenyang Agricultural University.
Mammalian species coverage for Hoxd11
Tissue samples from 12 mammals, including 10 cetartiodactyls, were obtained. The samples cover eight cetacean species (Sowerby’s beaked whale, Mesoplodon bidens; Blainville’s beaked whale, Mesoplodon densirostris; Northern bottlenose whale, Hyperoodon ampullatus; Atlantic white-sided dolphin, Lagenorhynchus acutus; pygmy sperm whale, Kogia breviceps; humpback whale, Megaptera novaeangliae; fin whale, Balaenoptera physalus; common minke whale, Balaenoptera acutorostrata), two artiodactyls (goat, Capra hircus; pig, Sus scrofa), one perissodactyl (donkey, Equus asinus), and one bat (great leaf-nosed bat, Hipposideros armiger).
To trace the evolutionary history of Hoxd11 in mammals, whole coding sequences for this gene were obtained from other 41 species representing orders Cetartiodactyla, Perissodactyla, Carnivora, Chiroptera, Primates, Rodentia, Lagomorpha and Afrosoricida. Whole coding regions of Hoxd11 for these taxa were identified in GenBank (www.ncbi.nlm.nih.gov) using BLAST. The hippopotamus, the species in Camelidae and some other species were not included, because their Hoxd11 sequences cannot be found in the genome data using the BLAST method or they were partial coding sequences.
Sequences from all 53 species were used to perform the evolutionary analyses, including comparisons of the highly variable region of Hoxd11 and to reconstruct the ancestral states for amino acid sites. Detailed species information and GenBank accession numbers are listed in Supplemental Table S1.
Hoxd11 gene amplification, cloning and sequencing
Genomic DNA was isolated from the 12 tissue samples using DNeasy Blood & Tissue Kits (Qiagen) and then used as template to amplify the Hoxd11 coding regions. PCR were conducted for exon 1 and for most part of exon 2 separately. For exon 1, we designed one set of primers for 11 species (F1: 5′ ATG AAC GAC TTT GAC GAG TGC 3′ and R1: 5′ CCC TTC GAA CGC TTA TAA AGT A 3′) and another set of primers specifically for the great leaf-nosed bat (F2: 5′ GTG TGG GGA AT[G/C] GGA ACC TC 3′ and R2: 5′ CCA CCC ACC CGT AAA A[A/G]C AG 3′). For exon 2, we used one set of primers (F3: 5′ AAA GCG CTG TCC CTA CAC C 3′ and R3: 5′ CAG TGA CCC ATG CCT TGA TA 3′) for all 12 mammals. PCR products were purified using QIAquick Gel Extraction Kits (Qiagen), ligated into pGEM-T easy vectors (Promega) and then cloned using DH5α competent cells (Tiangen). Positive clones were sequenced on an ABI 3730 sequencer (Applied Biosystems).
Species tree construction
We constructed a species tree for the analyses of the Hox genes, evolutionary history of bone numbers, and the focal amino acid sites according to previous studies. In detail, relationships of the orders was determined according to Springer et al. (Springer et al. 2003). For the relationships of the various species in each order, several references were used as none included all of the species used in this study. The Cetartiodactyla tree was constructed according to the following five references (McGowen et al. 2009; Hernández Fernández and Vrba 2005; Chen et al. 2019; Chen et al. 2018; O’Leary et al. 2013), the Perissodactyla tree was constructed according to Steiner and Ryder (Steiner and Ryder 2011), the Carnivora tree was constructed according to Nyakatura and Bininda-Emonds (Nyakatura and Bininda-Emonds 2012), the Chiroptera tree was constructed according to the following three references (Kawai et al. 2003; Teeling et al. 2005; Shi and Rabosky 2015), the Primates tree was constructed according to Perelman et al. (Perelman et al. 2011), the Rodentia tree was constructed according to the following three references (Romanenko et al. 2012; Fabre et al. 2017; Fabre et al. 2012), and the Afrotheria tree was constructed according to Kuntner et al. (Kuntner et al. 2011).
Evolutionary analysis of the number of carpal and tarsal bones
Based on the published data for extant mammalian species (see details and references in the first two paragraphs of Introduction section), the evolutionary histories and the numbers of carpal and tarsal bones in the ancestral lineages were estimated using the parsimony method using Mesquite 2 (Maddison and Maddison 2001).
Molecular evolution analyses on Hoxd11
Our new Hoxd11 sequences and those from databases were aligned together using ClustalW in MEGA 5 software (Thompson et al. 1994; Tamura et al. 2011). To examine the possibility that Hoxd11 experienced adaptive evolution in Cetruminantia, selective pressures acting on the coding sequences were estimated using the Codeml program in PAML 4 (Yang 2007). ω values (the nonsynonymous substitution rate [dN]/synonymous substitution rate [dS]) significantly greater than one represents positive selection; those equal to one are neutral evolution; and those less than one indicated purifying selection.
To test whether Hoxd11 underwent natural selection in Cetruminantia, we carried out several “branch models” in Codeml. The one-ratio model assumes all branches have the same ω value, but the free-ratio model assumes variable ω values in different lineages (Yang 1998). The two-ratio model allows ω to have different (but same within each group) values in the foreground and background branches (Yang 1998). Foreground lineage tested was the ancestor leading to Cetruminantia (termed branch “C” in this study). Codeml was also used to infer ancestral sequences and amino acid substitutions.
To visualize variation in the ω values along the Hoxd11 coding region, we undertook a sliding window analysis using SWAAP 1 (Pride 2000). Parameters for window size was set as 75 bp with a step size of 12 bp, and ω values were calculated by the Nei and Gojobori’s method (Nei and Gojobori 1986). Based on the 53 species dataset of Hoxd11, evolutionary histories of the focal amino acid sites in the ancestral lineage of Cetruminantia were estimated using the parsimony method in Mesquite 2 (Maddison and Maddison 2001).
Molecular evolution of other 5′ HoxA and HoxD genes
The two-ratio and one-ratio models were also applied to other 5′ HoxA and HoxD genes to determine whether these genes underwent the same evolutionary pattern as Hoxd11 in the ancestor of Cetruminantia. The coding sequences for the genes were retrieved from GenBank using BLAST with accession numbers shown in Supplemental Table S2. The datasets were aligned by MEGA 5 (Tamura et al. 2011; Thompson et al. 1994) and analyzed by PAML 4 as mentioned above (Yang 2007, 1998).
Data availability
The newly sequenced Hoxd11 used in this study are available in GenBank with accession numbers KU842353-KU842364. We have uploaded supplementary material to figshare. File S1 contains species trees for the evolutionary analyses of other 5′ HoxA and HoxD genes (Figure S1) and GenBank accession numbers of Hoxd11 sequences from 53 mammalian species analyzed in this study (Table S1). File S2 contains GenBank accession numbers of other 5′ HoxA and HoxD genes analyzed in this study (Table S2). Supplemental material available at figshare: https://doi.org/10.25387/g3.11278271.
Results
Variation in carpus and tarsus bone number in mammals
Although the numbers of carpal and tarsal bones vary in extant mammalian species, they only changed once (carpus) or twice to four times (tarsus) from remote ancestors. The numbers of bones in both the carpus and tarsus has been reduced by at least one bone in the common ancestor of cetaceans and ruminants (Figure 1). In the common ancestor of all modern cetaceans, the numbers of tarsal bones was reduced from five to zero (Figure 1B).
Mammalian Hoxd11 gene sequences
We successfully cloned and sequenced ∼93% of the protein coding region of Hoxd11 from ten Cetartiodactyla and two other mammalian species. Taken together with whole coding sequences downloaded from public databases, a total of 53 mammalian Hoxd11 sequences, including sequences from 18 species of Cetruminantia, were analyzed in this study. The full set of sequences from 53 species was used for the PAML analyses, reconstruction of ancestral states for amino acid sites in Cetruminantia, and an alignment of the highly variable region of Hoxd11. Detailed taxonomic and gene information are listed in Supplemental Table S1.
Molecular evolution of mammalian Hoxd11
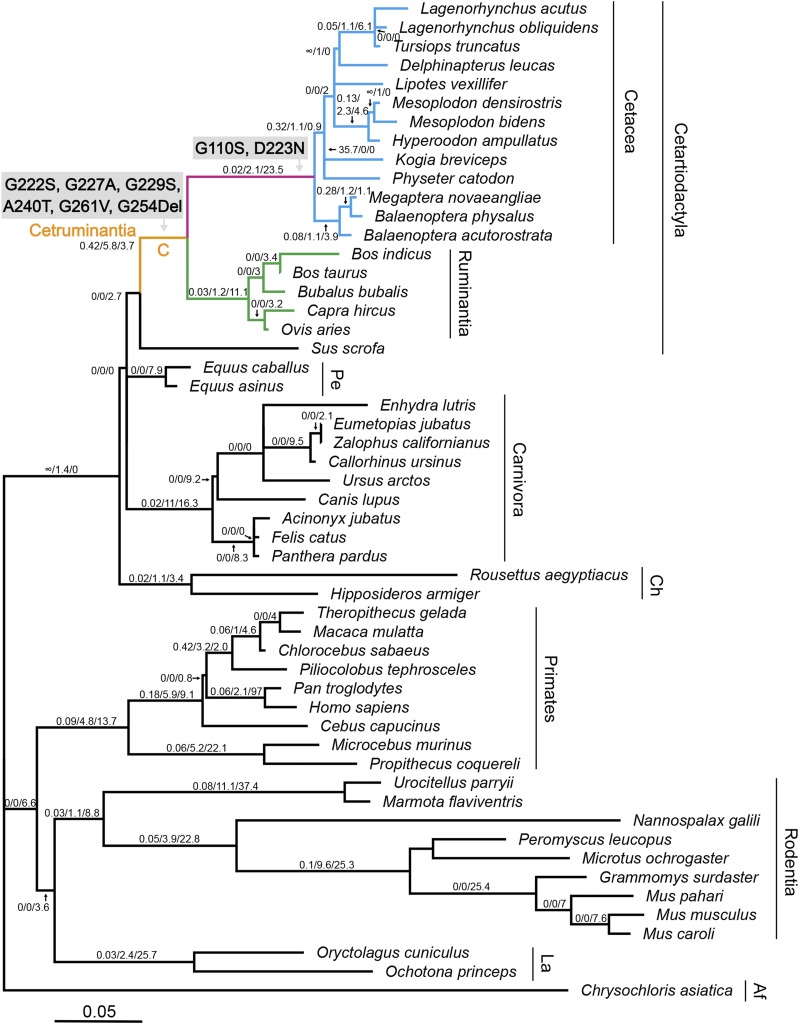
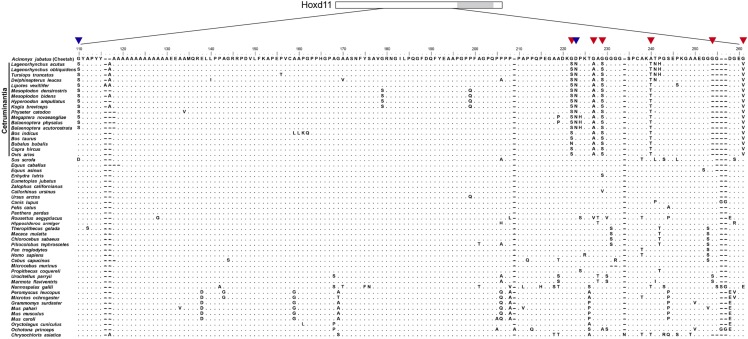
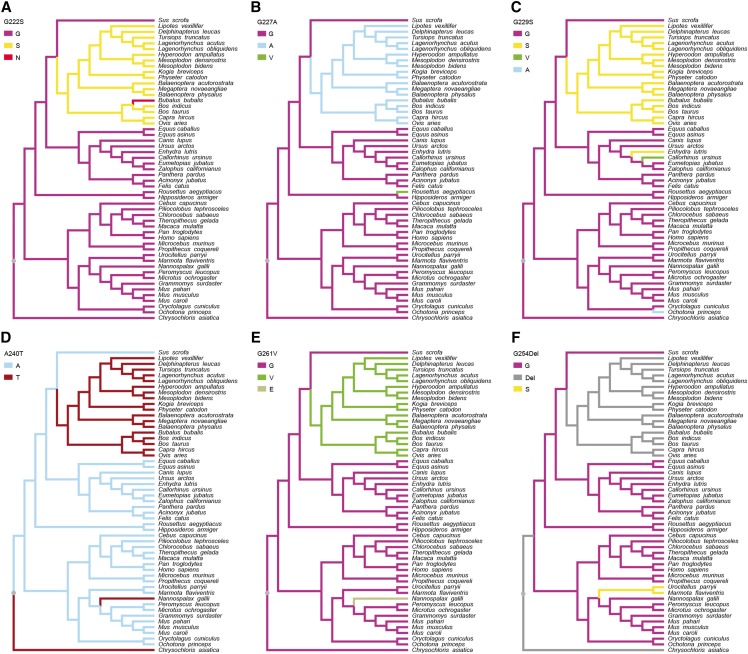
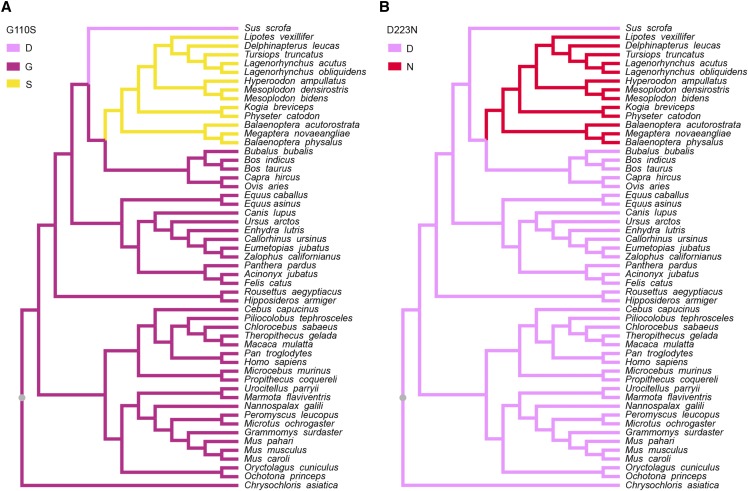
As a first step in examining the evolution of Hoxd11 we assessed the levels of selection that had acted upon this sequence using PAML (Yang 2007). For the branch models, the free-ratio model was significantly better than the one-ratio model (P = 0.0008), indicating that selective pressures differed among the mammalian lineages. We identified five amino acid substitutions (G222S, G227A, G229S, A240T and G261V) and one amino acid deletion (G254Del), located adjacent to the conserved homeodomain, which occurred on the branch leading to Cetruminantia, and two amino acid substitutions (G110S and D223N) that occurred on the branch leading to Cetacea (Figures 2 and 3). To further investigate the evolution of these eight amino acid substitutions or deletions, we aligned the Hoxd11 protein sequences from the 53 species (Figure 3) and separately traced the evolutionary history of each substitution site (Figures 4 and 5). Of the eight amino acid substitutions, three (G222S, G227A and G261V) are exclusive to Cetruminantia, and two (G110S and D223N) are exclusive to Cetacea, while the remaining three occur not only in cetaceans and ruminants, but also in several other mammalian species.
Figure 2.
Molecular evolution of Hoxd11 in mammalian ancestors. Branch lengths are proportional to nucleotide substitutions per codon. The ω values, numbers of nonsynonymous and synonymous substitutions inferred from the free-ratio model are shown on the species tree (the three numbers on the branches). The five amino acid substitutions on the lineage leading to Cetruminantia (branch “C”), shown in orange, and the two amino acid substitutions on the lineage leading to Cetacea, shown in pink, were mapped onto the branches. The clades for cetaceans and ruminants are indicated in blue and green respectively. Abbreviations for taxonomy are “Pe” (Perissodactyla), “Ca” (Carnivora), “Ch” (Chiroptera), “La” (Lagomorpha) and “Af” (Afrosoricida).
Figure 3.
Alignment of mammalian Hoxd11 protein sequences. Partial amino acid sequences from 53 mammalian species are aligned. The five amino acid substitutions and one amino acid deletion that occurred in the ancestor of Cetruminantia are highlighted by the red arrows. The two amino acid substitutions that occurred in the ancestor of Cetacea are highlighted by the blue arrows. The gray box in the schematic of Hoxd11 is homeodomain.
Figure 4.
Evolution of the six amino acids in Hoxd11 proteins with mutations that occurred on the Cetruminantia branch. Evolutionary changes in the 53 mammals at the sites of the amino acid substitutions or deletion G222S, G227A, G229S, A240T, G261V and G254Del are shown in A, B, C, D, E and F, respectively.
Figure 5.
Evolution of the two amino acids that are substituted on the branch leading to Cetacea in the Hoxd11 protein. Evolutionary changes in the 53 mammals at the sites of the amino acid substitutions G110S and D223N are shown in A and B, respectively.
Detailed ω values, nonsynonymous and synonymous substitution numbers inferred from the free-ratio model for each branch are shown in Figure 2. The branch leading to Cetruminantia shows the highest ω value (ω = 0.42) among the ancestral lineages that have more than 0 synonymous substitutions. Specifically, on this branch the number of nonsynonymous substitutions (dN = 5.8) is higher than on most ancestral lineages, and the number of synonymous substitutions (dS = 3.7) is much lower than on other ancestral lineages (dS = 9.1 to 37.4) that have higher numbers of nonsynonymous substitutions.
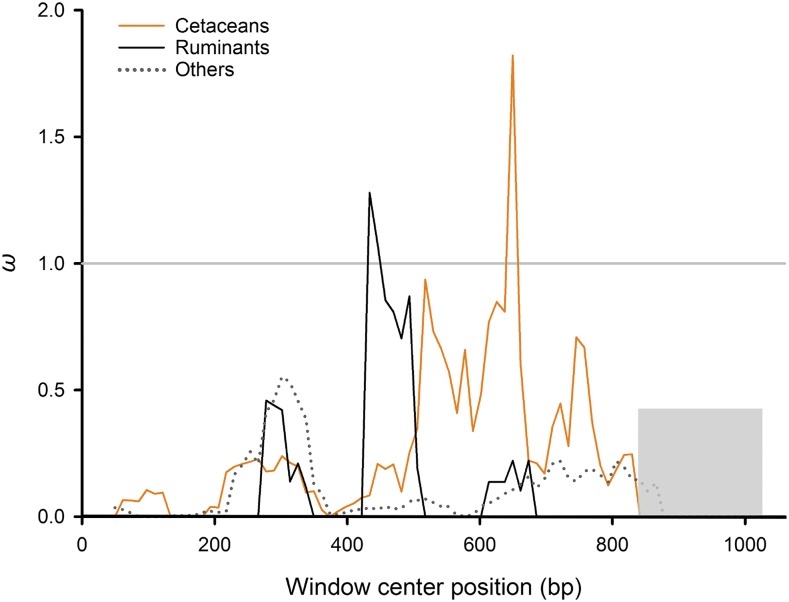
When the two-ratio test was applied, it revealed that the lineage leading to Cetruminantia has a greater than five times larger ω value (0.3839) than those for other branches (0.0712; P = 0.0215; Table 1). A sliding window analyses across the protein-coding region of Hoxd11 shows that the ω value changes along the gene. This result showed that there is a highly variable region (ω > 1) in Hoxd11 sequences of cetaceans and ruminants, but not in other mammals, which is adjacent to the conserved homeodomain (Figure 6).
Table 1. Branch model parameters for 5′ HoxA and HoxD genes.
| Gene | Model | ω0 | ωC | 2Δℓ | P value |
|---|---|---|---|---|---|
| Hoxa9 | One-ratio (ω0) | 0.0483 | = ω0 | ||
| Two-ratio (ω0, ωC) | 0.0483 | 0.0993 | 0.3076 | 0.5791 | |
| Hoxa10 | One-ratio (ω0) | 0.0863 | = ω0 | ||
| Two-ratio (ω0, ωC) | 0.0863 | 0.2286 | 0.4007 | 0.5267 | |
| Hoxa11 | One-ratio (ω0) | 0.0675 | = ω0 | ||
| Two-ratio (ω0, ωC) | 0.0675 | 0.4016 | 0.0002 | 0.9885 | |
| Hoxa13 | One-ratio (ω0) | 0.0136 | = ω0 | ||
| Two-ratio (ω0, ωC) | 0.0136 | 0.0001 | 0.3530 | 0.5524 | |
| Hoxd9 | One-ratio (ω0) | 0.1073 | = ω0 | ||
| Two-ratio (ω0, ωC) | 0.1073 | 0.1558 | 0.0596 | 0.8071 | |
| Hoxd10 | One-ratio (ω0) | 0.1230 | = ω0 | ||
| Two-ratio (ω0, ωC) | 0.1230 | 0.0001 | 0.6032 | 0.4374 | |
| Hoxd11 | One-ratio (ω0) | 0.0712 | = ω0 | ||
| Two-ratio (ω0, ωC) | 0.0712 | 0.3839 | 5.28 | 0.0215* | |
| Hoxd12 | One-ratio (ω0) | 0.1050 | = ω0 | ||
| Two-ratio (ω0, ωC) | 0.1050 | 0.0001 | 1.0701 | 0.3009 | |
| Hoxd13 | One-ratio (ω0) | 0.0417 | = ω0 | ||
| Two-ratio (ω0, ωC) | 0.0417 | 0.0001 | 0.7609 | 0.3830 |
ωC is for the common ancestor of Cetruminantia in Figure 2 (the foreground branch). ω0 is for all other branches (the background branches). *, P value of Hoxd11 is significant (<0.05). Significantly different ω values for Hoxd11 are shown in bold.
Figure 6.
Sliding window analysis of Hoxd11. ω values were calculated in a sliding window across the Hoxd11 sequence. The orange line represents ω values within cetaceans, the black line is ruminants, and the dotted gray line is other mammals. The homeobox is shown in the gray rectangle.
Molecular evolution of other 5′ HoxA and HoxD genes
For other 5′ HoxA and HoxD genes, complete coding sequences from representative species were analyzed. Two-ratio tests specifically on the ancestral branch leading to Cetruminantia indicate none of the genes shown an ω value significantly different from background branches (Table 1 and Supplemental Figure S1).
Discussion
In mammals, reduced numbers of carpal and tarsal bones are extensively observed in species of Cetruminantia (Cooper et al. 2007; Benham 1902; Schellhorn and Pfretzschner 2014; Sadan and Misk 2014; Rajtová 1974; Ehlert et al. 2011). To uncover the genetic basis and evolution of the bone reductions, we reconstructed a phylogenetic tree of the phenotypic changes (Figure 1) and performed molecular evolutionary analyses focusing on the Hoxd11 gene (Figure 2 and Table 1). The two-ratio model analysis shows that Hoxd11 had a significantly higher ω value in the common ancestral lineage of Cetruminantia than those in other lineages. Five nonsynonymous substitutions (G222S, G227A, G229S, A240T and G261V; four of them are related to the loss of glycine) and one amino acid deletion (G254Del; loss of glycine) occurred on the ancestral branch for Cetruminantia. Two mutations (G110S and D223N) occurred on the ancestral branch for Cetacea that are unique to this clade. These eight mutations are located adjacent to the conserved homeodomain in Hoxd11, and three of them transform hydrophilic amino acids to hydrophobic residues (G227A and G261V) or vice versa (A240T). In addition, the sliding window analysis show dramatic differences between Cetruminantia and other mammals. As Hoxd11 is a transcription factor that is typically composed of a DNA-binding domain and an activator domain, the area containing the mutations might correspond to the latter domain and thus influence the efficiency of DNA binding and thus the expression of its own gene and of downstream genes (Berg et al. 2002). These results indicate that the mutations might be due to positive selection and result in the bone losses or due to relaxed selective constraint. Given high expression of Hoxd11 in mammalian limbs and functional importance of 5′HoxD genes on limb development (Kmita et al. 2005; Gehrke et al. 2015; Fabre et al. 2018; Bickelmann et al. 2017; Booker et al. 2016), it is unlikely that Hoxd11 underwent relaxed purifying selection during the origin of Cetruminantia, however, it possible that this occurred in the evolution of the ancestral cetaceans, as this is when hindlimb loss occurred. Hoxd11 is required for positioning of the pelvic girdle in mice (Davis and Capecchi 1994). Reductions in the hindlimb and pelvic girdle development in Cetaceans may also reflect changes in the Hoxd11 amino acid sequence and expression patterns. The consequences of changes in gene expression cannot be directly investigated in developing embryos of Cetruminantia, however, deregulated premature expression of Hoxd11-13 in mice can induce massive defects in the proximal limbs of mice, including the carpus (Zákány et al. 2007), indicating that relaxed selective constraint might be associated with the loss of the hind limbs in Cetacea.
In a previous study we reported that the Hoxd12 and Hoxd13 genes underlie the origin and diversification of the cetacean flipper (Wang et al. 2009). However, little consideration was paid to Hoxd11 evolution and its role in the patterning of carpal and tarsal bones. In this study, we provided evidence on the adaptive evolution of Hoxd11 and suggested a role for this gene in the reduction in the number of carpal and tarsal bones in Cetruminantia.
Mice with mutations in Hoxd11 have been reported to possess fusions between specific carpal bones (Davis and Capecchi 1994; Favier et al. 1995). However, since the development of the limb involves the complex of Hox genes and their interactions, Hoxd11 also likely works together with other genes to implement the reduction in the number of carpal and tarsal bones (Zákány and Duboule 2007). For example, the mice with double mutations in Hoxa11 and Hoxd11 can also have carpal bone fusions (Davis et al. 1995). Mutant mice involving both the Hoxd11 and Hoxd13 genes display a fusion of distal carpal bones (Davis and Capecchi 1996). In this study, the analyses on other 5′ HoxA and HoxD genes shown the genes are highly conservative in mammals, and none of them underwent accelerated evolution in the common ancestor of Cetruminantia. The results suggest that only Hoxd11, rather than other 5′ HoxA and HoxD genes, may underlay critical morphological modification during the evolution of Cetruminantia limb patterning.
In conclusion, the Hoxd11 gene sequences exhibit a significant accelerated evolution during the origin of Cetruminantia. Moreover, other 5′ HoxA and HoxD genes show highly conserved evolution in this lineage, which supports a potentially important role of Hoxd11 in Cetruminantia evolution. Accelerated evolution of Hoxd11 in the ancestor of Cetruminantia was probably associated with the reduction of the carpal and tarsal bones in this lineage, whereas the mutations that are unique to the ancestor of Cetacea are probably related to Cetacean hindlimb loss. Further experimental analyses would be necessary to determine the exact function of the eight mutations observed in Cetruminantia.
Acknowledgments
We thank Yang Liu, Lu Fang, Fanxing Meng, Haijian Sun and Chunzheng He for data analysis and experimental assistance. This work was supported by grants from the National Natural Science Foundation of China (Nos. 31672274 and 31570382).
Footnotes
Supplemental material available at fisshare: https://doi.org/10.25387/g3.11278271.
Communicating editor: A. Sethuraman
Literature Cited
- Akers R. M., and Denbow D. M., 2008. Anatomy and Physiology of Domestic Animals, Blackwell Publishing, Ames. [Google Scholar]
- Alam El Din M. A., Aly M. A., Youssef H. A., and Abdel Moneim M. E., 1986. Surgical anatomy of the tarsal joint in donkey. Assiut Veterinary Medical Journal 17: 25–30. [Google Scholar]
- Bejder L., and Hall B. K., 2002. Limbs in whales and limblessness in other vertebrates: mechanisms of evolutionary and developmental transformation and loss. Evol. Dev. 4: 445–458. 10.1046/j.1525-142X.2002.02033.x [DOI] [PubMed] [Google Scholar]
- Benham W. B., 1902. Notes on the osteology of the short-nosed sperm whale. Proc. Zool. Soc. Lond. 1: 54–62. [Google Scholar]
- Berg J. M., Tymoczko J. L., and Stryer L., 2002. The control of gene expression, pp. 1280–1319 in Biochemistry, edited by Moran S., Hadler G. L., and Zimmerman P.. W.H. Freeman and Company, New York. [Google Scholar]
- Bickelmann C., van der Vos W., de Bakker M. A., Jimenez R., Maas S. et al. , 2017. Hox gene expression in the specialized limbs of the Iberian mole (Talpa occidentalis). Evol. Dev. 19: 3–8. 10.1111/ede.12216 [DOI] [PubMed] [Google Scholar]
- Booker B. M., Friedrich T., Mason M. K., VanderMeer J. E., Zhao J. et al. , 2016. Bat Accelerated Regions Identify a Bat Forelimb Specific Enhancer in the HoxD Locus. PLoS Genet. 12: e1005738 10.1371/journal.pgen.1005738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulet A. M., and Capecchi M. R., 2002. Duplication of the Hoxd11 gene causes alterations in the axial and appendicular skeleton of the mouse. Dev. Biol. 249: 96–107. 10.1006/dbio.2002.0755 [DOI] [PubMed] [Google Scholar]
- Boulet A. M., and Capecchi M. R., 2004. Multiple roles of Hoxa11 and Hoxd11 in the formation of the mammalian forelimb zeugopod. Development 131: 299–309. 10.1242/dev.00936 [DOI] [PubMed] [Google Scholar]
- Chen L., Qin Q., Jiang Y., Wang K., Lin Z. S. et al. , 2019. Large-scale ruminant genome sequencing provides insights into their evolution and distinct traits. Science 364: eaav6202 10.1126/science.aav6202 [DOI] [PubMed] [Google Scholar]
- Chen N. B., Cai Y. D., Chen Q. M., Li R., Wang K. et al. , 2018. Whole-genome resequencing reveals world-wide ancestry and adaptive introgression events of domesticated cattle in East Asia. Nat. Commun. 9: 2337 10.1038/s41467-018-04737-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper L. N., Berta A., Dawson S. D., and Reidenberg J. S., 2007. Evolution of hyperphalangy and digit reduction in the cetacean manus. Anat. Rec. (Hoboken) 290: 654–672. 10.1002/ar.20532 [DOI] [PubMed] [Google Scholar]
- Davis A. P., and Capecchi M. R., 1994. Axial homeosis and appendicular skeleton defects in mice with a targeted disruption of hoxd-11. Development 120: 2187–2198. [DOI] [PubMed] [Google Scholar]
- Davis A. P., and Capecchi M. R., 1996. A mutational analysis of the 5′ HoxD genes: dissection of genetic interactions during limb development in the mouse. Development 122: 1175–1185. [DOI] [PubMed] [Google Scholar]
- Davis A. P., Witte D. P., Hsieh-Li H. M., Potter S. S., and Capecchi M. R., 1995. Absence of radius and ulna in mice lacking hoxa-11 and hoxd-11. Nature 375: 791–795. 10.1038/375791a0 [DOI] [PubMed] [Google Scholar]
- Ehlert A., Ferguson J., and Gerlach K., 2011. Magnetic resonance imaging and cross-sectional anatomy of the normal bovine tarsus. Anat. Histol. Embryol. 40: 234–240. 10.1111/j.1439-0264.2011.01062.x [DOI] [PubMed] [Google Scholar]
- Fabre P. H., Hautier L., Dimitrov D., and Douzery E. J. P., 2012. A glimpse on the pattern of rodent diversification: a phylogenetic approach. BMC Evol. Biol. 12: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre P. H., Upham N. S., Emmons L. H., Justy F., Leite Y. L. R. et al. , 2017. Mitogenomic Phylogeny, Diversification, and Biogeography of South American Spiny Rats. Mol. Biol. Evol. 34: 613–633. [DOI] [PubMed] [Google Scholar]
- Fabre P. J., Leleu M., Mascrez B., Lo Giudice Q., Cobb J. et al. , 2018. Heterogeneous combinatorial expression of Hoxd genes in single cells during limb development. BMC Biol. 16: 101 10.1186/s12915-018-0570-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favier B., Le Meur M., Chambon P., and Dolle P., 1995. Axial skeleton homeosis and forelimb malformations in Hoxd-11 mutant mice. Proc. Natl. Acad. Sci. USA 92: 310–314. 10.1073/pnas.92.1.310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. E., Scott K. M., and Naples V. L., 2007. Forelimb myology of the pygmy hippopotamus (Choeropsis liberiensis). Anat. Rec. (Hoboken) 290: 673–693. 10.1002/ar.20531 [DOI] [PubMed] [Google Scholar]
- Fleischman R. A., Letestu R., Mi X., Stevens D., Winters J. et al. , 2002. Absence of mutations in the HoxA10, HoxA11 and HoxD11 nucleotide coding sequences in thrombocytopenia with absent radius syndrome. Br. J. Haematol. 116: 367–375. 10.1046/j.1365-2141.2002.03263.x [DOI] [PubMed] [Google Scholar]
- Galateanu G., Hermes R., Saragusty J., Goritz F., Potier R. et al. , 2014. Rhinoceros Feet Step Out of a Rule-of-Thumb: A Wildlife Imaging Pioneering Approach of Synchronized Computed Tomography-Digital Radiography. PLoS One 9: e100415 10.1371/journal.pone.0100415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrke A. R., Schneider I., de la Calle-Mustienes E., Tena J. J., Gomez-Marin C. et al. , 2015. Deep conservation of wrist and digit enhancers in fish. Proc. Natl. Acad. Sci. USA 112: 803–808. 10.1073/pnas.1420208112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S., Del Viso F., Chen C. Y., Ikmi A., Kroesen A. E. et al. , 2018. An axial Hox code controls tissue segmentation and body patterning in Nematostella vectensis. Science 361: 1377–1380. 10.1126/science.aar8384 [DOI] [PubMed] [Google Scholar]
- Hernández Fernández M., and Vrba E. S., 2005. A complete estimate of the phylogenetic relationships in Ruminantia: a dated species-level supertree of the extant ruminants. Biol. Rev. Camb. Philos. Soc. 80: 269–302. 10.1017/S1464793104006670 [DOI] [PubMed] [Google Scholar]
- Hinchliffe J. R., and Johnson D. R., 1980. The Development of the Vertebrate Limb, Oxford University Press, New York. [Google Scholar]
- Karan M., 2012. Macro-anatomical study of ossa membri thoracici in the feral pigs (Sus scrofa). Firat Uni. Vet. J. Health Sci. 26: 17–20. [Google Scholar]
- Kawai K., Nikaido M., Harada M., Matsumura S., Lin L. K. et al. , 2003. The status of the Japanese and East Asian bats of the genus Myotis (Vespertilionidae) based on mitochondrial sequences. Mol. Phylogenet. Evol. 28: 297–307. 10.1016/S1055-7903(03)00121-0 [DOI] [PubMed] [Google Scholar]
- Kivell T. L., and Begun D. R., 2007. Frequency and timing of scaphoid-centrale fusion in hominoids. J. Hum. Evol. 52: 321–340. 10.1016/j.jhevol.2006.10.002 [DOI] [PubMed] [Google Scholar]
- Kmita M., Fraudeau N., Herault Y., and Duboule D., 2002. Serial deletions and duplications suggest a mechanism for the collinearity of Hoxd genes in limbs. Nature 420: 145–150. 10.1038/nature01189 [DOI] [PubMed] [Google Scholar]
- Kmita M., Tarchini B., Zákány J., Logan M., Tabin C. J. et al. , 2005. Early developmental arrest of mammalian limbs lacking HoxA/HoxD gene function. Nature 435: 1113–1116. 10.1038/nature03648 [DOI] [PubMed] [Google Scholar]
- Koyama E., Yasuda T., Wellik D. M., and Pacifici M., 2010. Hox11 paralogous genes are required for formation of wrist and ankle joints and articular surface organization. Ann. N. Y. Acad. Sci. 1192: 307–316. 10.1111/j.1749-6632.2009.05234.x [DOI] [PubMed] [Google Scholar]
- Kuntner M., May-Collado L. J., and Agnarsson I., 2011. Phylogeny and conservation priorities of afrotherian mammals (Afrotheria, Mammalia). Zool. Scr. 40: 1–15. 10.1111/j.1463-6409.2010.00452.x [DOI] [Google Scholar]
- Lewis O. J., 1980. The joints of the evolving foot. Part II. The intrinsic joints. J. Anat. 130: 833–857. [PMC free article] [PubMed] [Google Scholar]
- Li K., Sun X., Chen M., Sun Y., Tian R. et al. , 2018. Evolutionary changes of Hox genes and relevant regulatory factors provide novel insights into mammalian morphological modifications. Integr. Zool. 13: 21–35. 10.1111/1749-4877.12271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L., Shen Y. Y., Pan X. W., Zhou T. C., Yang C. et al. , 2013. Adaptive evolution of the Hox gene family for development in bats and dolphins. PLoS One 8: e65944 10.1371/journal.pone.0065944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison W. P., and Maddison D. R., 2001. Mesquite: a modular system for evolutionary analysis. http://mesquiteproject.org.
- Mallo M., 2018. Reassessing the Role of Hox Genes during Vertebrate Development and Evolution. Trends Genet. 34: 209–217. 10.1016/j.tig.2017.11.007 [DOI] [PubMed] [Google Scholar]
- McGowen M. R., Spaulding M., and Gatesy J., 2009. Divergence date estimation and a comprehensive molecular tree of extant cetaceans. Mol. Phylogenet. Evol. 53: 891–906. 10.1016/j.ympev.2009.08.018 [DOI] [PubMed] [Google Scholar]
- Moreau C., Caldarelli P., Rocancourt D., Roussel J., Denans N. et al. , 2019. Timed Collinear Activation of Hox Genes during Gastrulation Controls the Avian Forelimb Position. Curr Biol 29: 35–50 e34. 10.1016/j.cub.2018.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., and Gojobori T., 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3: 418–426. [DOI] [PubMed] [Google Scholar]
- Nery M. F., Borges B., Dragalzew A. C., and Kohlsdorf T., 2016. Selection on different genes with equivalent functions: the convergence story told by Hox genes along the evolution of aquatic mammalian lineages. BMC Evol. Biol. 16: 113 10.1186/s12862-016-0682-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netter F. H., 2018. Atlas of human anatomy, 7th edition Elsevier, Philadelphia. [Google Scholar]
- Nyakatura K., and Bininda-Emonds O. R. P., 2012. Updating the evolutionary history of Carnivora (Mammalia): a new species-level supertree complete with divergence time estimates. BMC Biol. 10: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary M. A., Bloch J. I., Flynn J. J., Gaudin T. J., Giallombardo A. et al. , 2013. The placental mammal ancestor and the post-K-Pg radiation of placentals. Science 339: 662–667. 10.1126/science.1229237 [DOI] [PubMed] [Google Scholar]
- Pearson J. C., Lemons D., and McGinnis W., 2005. Modulating Hox gene functions during animal body patterning. Nat. Rev. Genet. 6: 893–904. 10.1038/nrg1726 [DOI] [PubMed] [Google Scholar]
- Perelman P., Johnson W. E., Roos C., Seuanez H. N., Horvath J. E. et al. , 2011. A molecular phylogeny of living primates. PLoS Genet. 7: e1001342 10.1371/journal.pgen.1001342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pride, D.T., 2000 Pride, D. T. A tool for analyzing substitutions and similarity in multiple alignments in Distributed by the author.
- Rajtová V., 1974. Postnatal development of the bones of the limbs in sheep and goat. Anat. Histol. Embryol. 3: 29–39. [Google Scholar]
- Raven H. C., 1942. On the structure of Mesoplodon densirostris, a rare braked whale. Bull. Am. Mus. Nat. Hist. 80: 23–50. [Google Scholar]
- Romanenko S. A., Perelman P. L., Trifonov V. A., and Graphodatsky A. S., 2012. Chromosomal evolution in Rodentia. Heredity 108: 4–16. 10.1038/hdy.2011.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadan M. A., and Misk N. A., 2014. Radiographic interpretation of bones of carpal joints in horses, donkeys, buffaloes, cattle and camels. Indian J. Vet. Surg. 35: 131–133. [Google Scholar]
- Schellhorn R., and Pfretzschner H. U., 2014. Biometric study of ruminant carpal bones and implications for phylogenetic relationships. Zoomorphology 133: 139–149. 10.1007/s00435-013-0209-0 [DOI] [Google Scholar]
- Shi J. J., and Rabosky D. L., 2015. Speciation dynamics during the global radiation of extant bats. Evolution 69: 1528–1545. 10.1111/evo.12681 [DOI] [PubMed] [Google Scholar]
- Small K. M., and Potter S. S., 1993. Homeotic transformations and limb defects in Hox A11 mutant mice. Genes Dev. 7: 2318–2328. 10.1101/gad.7.12a.2318 [DOI] [PubMed] [Google Scholar]
- Springer M. S., Murphy W. J., Eizirik E., and O’Brien S. J., 2003. Placental mammal diversification and the Cretaceous-Tertiary boundary. Proc. Natl. Acad. Sci. USA 100: 1056–1061. 10.1073/pnas.0334222100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford B. J., and Thorington R. W., 1998. Carpal development and morphology in archontan mammals. J. Morphol. 235: 135–155. [DOI] [PubMed] [Google Scholar]
- Steiner C. C., and Ryder O. A., 2011. Molecular phylogeny and evolution of the Perissodactyla. Zool. J. Linn. Soc. 163: 1289–1303. 10.1111/j.1096-3642.2011.00752.x [DOI] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M. et al. , 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeling E. C., Springer M. S., Madsen O., Bates P., O’brien S. J. et al. , 2005. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science 307: 580–584. 10.1126/science.1105113 [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G., and Gibson T. J., 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung W. L., Zhao C., Yoshii Y., Su F. C., An K. N. et al. , 2010. Comparative study of carpal tunnel compliance in the human, dog, rabbit, and rat. J. Orthop. Res. 28: 652–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner W., 1885. The anatomy of a second specimen of Sowerby’s whale (Mesoplodon bidens) from Shetland. J Anat Physiol 20: 144–188. [PMC free article] [PubMed] [Google Scholar]
- Turner W., 1909. The skeleton of a Sowerby’s whale (Mesoplodon bidens) stranded at St. Andrews, and the morphology of the manus in Mesoplodon, Hyperoodon and the Delphinidae. Proc. R. Soc. Edinb. 29: 687–720. 10.1017/S0370164600009184 [DOI] [Google Scholar]
- Wang Z., Dai M., Wang Y., Cooper K. L., Zhu T. et al. , 2014. Unique expression patterns of multiple key genes associated with the evolution of mammalian flight. Proc. Biol. Sci. 281: 20133133 10.1098/rspb.2013.3133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Yuan L., Rossiter S. J., Zuo X., Ru B. et al. , 2009. Adaptive evolution of 5′HoxD genes in the origin and diversification of the cetacean flipper. Mol. Biol. Evol. 26: 613–622. 10.1093/molbev/msn282 [DOI] [PubMed] [Google Scholar]
- Yalden D. W., 1971. The functional morphology of the carpus in ungulate mammals. Acta Anat. (Basel) 78: 461–487. 10.1159/000143609 [DOI] [PubMed] [Google Scholar]
- Yang Z., 1998. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol. Biol. Evol. 15: 568–573. 10.1093/oxfordjournals.molbev.a025957 [DOI] [PubMed] [Google Scholar]
- Yang Z., 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24: 1586–1591. 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- Young J. J., Grayson P., and Tabin C. J., 2019. Developmental Biology: Hox Timing Determines Limb Placement. Curr. Biol. 29: R52–R54. 10.1016/j.cub.2018.11.068 [DOI] [PubMed] [Google Scholar]
- Zákány J., and Duboule D., 1999. Hox genes in digit development and evolution. Cell Tissue Res. 296: 19–25. 10.1007/s004410051262 [DOI] [PubMed] [Google Scholar]
- Zákány J., and Duboule D., 2007. The role of Hox genes during vertebrate limb development. Curr. Opin. Genet. Dev. 17: 359–366. 10.1016/j.gde.2007.05.011 [DOI] [PubMed] [Google Scholar]
- Zákány J., Zacchetti G., and Duboule D., 2007. Interactions between HOXD and Gli3 genes control the limb apical ectodermal ridge via Fgf10. Dev. Biol. 306: 883–893. 10.1016/j.ydbio.2007.03.517 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The newly sequenced Hoxd11 used in this study are available in GenBank with accession numbers KU842353-KU842364. We have uploaded supplementary material to figshare. File S1 contains species trees for the evolutionary analyses of other 5′ HoxA and HoxD genes (Figure S1) and GenBank accession numbers of Hoxd11 sequences from 53 mammalian species analyzed in this study (Table S1). File S2 contains GenBank accession numbers of other 5′ HoxA and HoxD genes analyzed in this study (Table S2). Supplemental material available at figshare: https://doi.org/10.25387/g3.11278271.