Abstract
The evolutionary diversification of animals is one of Earth’s greatest marvels, yet its earliest steps are shrouded in mystery. Animals, the monophyletic clade known as Metazoa, evolved wildly divergent multicellular life strategies featuring ciliated sensory epithelia. In many lineages epithelial sensoria became coupled to increasingly complex nervous systems. Currently, different phylogenetic analyses of single-copy genes support mutually-exclusive possibilities that either Porifera or Ctenophora is sister to all other animals. Resolving this dilemma would advance the ecological and evolutionary understanding of the first animals and the evolution of nervous systems. Here we describe a comparative phylogenetic approach based on gene duplications. We computationally identify and analyze gene families with early metazoan duplications using an approach that mitigates apparent gene loss resulting from the miscalling of paralogs. In the transmembrane channel-like (TMC) family of mechano-transducing channels, we find ancient duplications that define separate clades for Eumetazoa (Placozoa + Cnidaria + Bilateria) vs. Ctenophora, and one duplication that is shared only by Eumetazoa and Porifera. In the Max-like protein X (MLX and MLXIP) family of bHLH-ZIP regulators of metabolism, we find that all major lineages from Eumetazoa and Porifera (sponges) share a duplicated gene pair that is sister to the single-copy gene maintained in Ctenophora. These results suggest a new avenue for deducing deep phylogeny by choosing rather than avoiding ancient gene paralogies.
Keywords: animal evolution, gene duplication, Max-like X (MLX), MLX interacting protein (MLXIP), transmembrane channel-like proteins (TMCs)
The branching order of the major metazoan lineages has received much attention due to its importance in understanding how animals and their sensory and nervous systems evolved (Jékely et al. 2015). To date, most phylogenetic analyses have used single-copy orthologs, with different genes and approaches finding support for either Ctenophora (Ryan et al. 2013; Moroz et al. 2014; Borowiec et al. 2015; Whelan et al. 2015; Shen et al. 2017) or Porifera (Pisani et al. 2015; Feuda et al. 2017; Simion et al. 2017) as the sister taxon of all other animals. In contrast to sequence-based phylogenies, comparative analysis of single-cell transcriptomes from different lineages and cell types is consistent with an independent origin of neuron-like cells in ctenophores (Sebé-Pedrós et al. 2018). Another recent study that does not rely directly on sequence analysis provides evidence that the sponge choanocyte does not correspond to the sponge cell type most similar transcriptomically to the choanoflagellate cell type (Sogabe et al. 2019). Thus to date, few studies (Sebé-Pedrós et al. 2018; Sogabe et al. 2019) have addressed the question of early animal branching without using single-copy genes for phylogenetic analysis.
Single-copy genes are preferred for phylogenetic inference of lineage branching order for several reasons (Fitch and Margoliash 1967; Woese and Fox 1977). For example, single-copy genes more closely approximate clock-like divergence (Zuckerkandl and Pauling 1965; Fitch and Margoliash 1967; Woese and Fox 1977; Pett et al. 2019) compared to duplicated genes, which frequently experience neofunctionalization and/or uneven subfunctionalization and evolutionary rate asymmetries (Walsh 1995; Lynch et al. 2001; Holland et al. 2017). An organismal tree of animals can be constructed from a single-copy gene, or from a set of concatenated single-copy genes, provided orthologous outgroup genes are included in the analysis as an aid for rooting.
Likewise, single-copy genes (strict orthologs across all lineages) offer a practical advantage in eliminating the ambiguity associated with gene duplications (paralogs). Some paralogs represent recent lineage-specific duplications while others stem from deeper duplications contributing to a larger gene super-family. Nonetheless, a gene tree of a pair of genes produced by a duplication in the stem-metazoan lineage can depict lineage-branching order doubly so, once in each paralog’s subtree. Furthermore, a gene tree of paralogs offers a unique advantage unavailable in single-copy gene trees: a tree of paralogs captures the duplication itself and unites lineages sharing the duplication relative to outgroup lineages with a single gene.
A majority of gene orthologs are part of larger super-families (Ohno 1970; Taylor and Raes 2004) as has been well documented for the Hox gene family (Holland et al. 2017). Thus, many “single-copy” genes are only ostensibly so because they can be evaluated separately from their ancient paralogs; and because in principle single-copy genes diverged lineally from a single ancestral gene present in the latest common ancestor (LCA) of a taxonomic clade. However, the choice of homologs is a poorly examined aspect of modern phylogenetic analysis even though various ortholog-calling errors associated with ancient paralogy have been noted (Noutahi et al. 2016).
Here we identify candidate gene paralogies established prior to the evolution of Eumetazoa (Bilateria + Cnidaria + Placozoa) for the purpose of determining early animal branching order. The majority of these paralogies predate the metazoan LCA and/or experienced apparent gene losses in either candidate first animal sister lineage (Porifera or Ctenophora) and are not informative (see Table S1). We also find a smaller number of paralogies that possibly support a proposed clade of “Benthozoa” (Porifera + Eumetazoa) while we have found none that support the traditional grouping of Ctenophora with Eumetazoa. The benthozoic hypothesis is premised on the latest common ancestor (LCA) of Choanozoa (Choanoflagellatea + Metazoa) being holopelagic, and the LCA of Benthozoa having evolved a biphasic pelagic larva and a benthic adult form. Other alternative life cycle scenarios have been proposed that correspond to different branching patterns (Nielsen 2008, 2013; Jékely et al. 2015), some of which are based on fossil interpretation (Zhao et al. 2019).
Among the most intriguing of our findings are the recently identified family of multimeric transmembrane mechanosensitive channel proteins, the transmembrane channel-like (TMC) proteins (Keresztes et al. 2003; Ballesteros et al. 2018), in which family we find definitive independent duplications in Eumetazoa (Tmc487 + Tmc56, where “Tmc487” and “Tmc56” represent the ancestral sister genes eventually giving rise to the five genes TMC4–TMC8 in jawed vertebrates for example), Ctenophora (Tmc-α + Tmc-β + Tmc-γ + Tmc-δ), and possibly Benthozoa (Tmc48756 + a neofunctionalized Tmc123 clade). In summary, our identification and analysis of genes duplicating and diversifying in the stem-benthozoic lineage will help to outline the extent of a shared biology for Benthozoa and the nature of independent evolutionary neuralization in Eumetazoa and Ctenophora.
Materials and Methods
Comparative genomic orthology counting and screening
To identify sets of candidate gene paralogies for further phylogenetic analysis, we used the BioMart query tool (Durinck et al. 2005; Haider et al. 2009; Smedley et al. 2009) and the EnsemblCompara orthology calls (Vilella et al. 2009) for MetazoaEnsembl Genomes Release 41. We also used these same tools to identify 2146 unique protein-coding genes in the placozoan Trichoplax adhaerens, which share the following properties: (1) these genes all can be grouped into a smaller number of paralogy groups; (2) these genes all have homologs in the cnidarian Nematostella vectensis; (3) the cnidarian homologs can also all be grouped into a smaller number of paralogy groups; (4) these genes all have homologs in the molluscan genome of Lottia gigantea, representing Lophotrochozoa, as well as in Drosophila melanogaster, representing Ecdysozoa; and (5) these genes all have homologs in the sponge Amphimedon queenslandica (a sponge in class Demospongiae). To ensure that our results would not be skewed by errors in gene annotation and curation, we focused on genes for which we could identify homologs in other cnidarians (the anthozoan Stylophora pistillata and the hydrozoan Hydra vulgaris), another sponge (the homoscleromorphan sponge Oscarella carmela), and throughout Bilateria. We then constructed phylogenetic trees for several different gene families from this list. An additional but similar search strategy is described in the text.
Sequence alignment
We identified and curated sequences only from representative taxa with whole-genome sequence assemblies. We obtained initial (pre-full-length curation) sequences from NCBI’s non-redundant protein database using the BLAST query tool with taxonomic specification (Altschul et al. 1990), and/or from the Ensembl Release 97 and Metazoa Ensembl Release 45 databases using the ComparaEnsembl orthology calls (Vilella et al. 2009). For additional ctenophore sequences from the Pleurobrachia and Beroë genomes, we queried the Neurobase transcriptome databases using BLAST (Moroz et al. 2014). For additional sequences and transcripts from cnidarian and sponge genomes we also queried the Compagen databases (Hemmrich and Bosch 2008). In addition to BLASTP queries of protein or translated transcriptome databases, we also used the TBLASTN tool to search translated genomic sequences using a protein sequence. We did this for two reasons. First, we used TBLASTN to verify that certain gene absences were not simply due to a failure to annotate a gene. Second, we used TBLASTN on several occasions when curating missing exons. We identified several genes from genome assemblies that were initially predicted by computational annotation and for which we then hand-curated for this study, typically to identify missing terminal exons. These are indicated in the tree figures and in the FASTA headers (Supplementary files) by the accession numbers with “CUR” appended. Alignment of protein-coding sequence was conducted using MUSCLE alignment option in MEGA7 (Kumar et al. 2016). The multiple sequence alignment (MSA) was conducted using default parameters that were adjusted as follows: The gap existence parameter was changed to -1.6 and the gap extension parameter was changed to -0.01. Excessively long protein sequences were trimmed at the N- and C- termini so that they began and ended on either side of the ten transmembrane domains. Lengthy, fast-evolving, loop segments and/or repetitive amino acid sequences occurring in between transmembrane domains were trimmed. Supplementary Files for curated data sets are provided as explained under “Data availability”.
Phylogenetic analyses
Candidate gene family screening:
To screen through a large number of candidate gene trees, we first constructed draft trees using Maximum Parsimony and Neighbor-Joining using sequences from the principal animal lineages and from choanoflagellates when available using MEGA7 (Kumar et al. 2016). For potentially informative trees, we then selected sequences from additional lineages, performed more extensive curation, and constructed additional more detailed tree versions. These early versions were typically of sufficient quality to identify whether or not genes were present in the correct lineages to be informative to this study’s focus on animal lineage branching.
Bayesian inference of phylogeny:
To conduct metropolis-coupled Markov chain Monte Carlo (MC) Bayesian phylogenetic analysis we used the MrBayes (version 3.2) software package (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003; Ronquist et al. 2012). All runs used two heated chains (“nchains = 2”, “temperature = 0.08”) that were run for at least 1.2 M generations with a burn-in fraction of 20–25%. Initial runs for all gene families sampled all substitution models, but we always found 100% percent posterior probability assigned to the WAG substitution model (Whelan and Goldman 2001). Subsequently all finishing runs used the WAG model with invariant-gamma rates modeling. Double precision computing was enabled using BEAGLE toolkit (Ayres et al. 2012; Ronquist et al. 2012).
All trees were computed multiple times during the process of sequence curation and annotation. The final cornichon/cornichon-related gene family tree finished with 0.009 average standard deviation of split frequencies from two heated chains. The final Tmc gene family tree finished with 0.005 average standard deviation of split frequencies from the two heated chains. The final MLX/MLXIP gene family tree finished with 0.007 average standard deviation of split frequencies from the two heated chains. The final version of the MAX super-family tree finished at 0.015 average standard deviation of split frequencies after 3.2 M generations. Tree graphics were rendered with FigTree version 1.4.4 and annotated using Adobe Photoshop software.
Data availability
All data files, including sequence files (*.fas), multiple sequence alignment files (*.masx and *.nexus), and curation documents, are provided with the Supporting Information as a zipped archive. Four files are provided for each of the three gene families shown for a total of 12 files. Supplemental material available at figshare: https://doi.org/10.25387/g3.11444400.
Results
Gene duplications from the early metazoan radiation
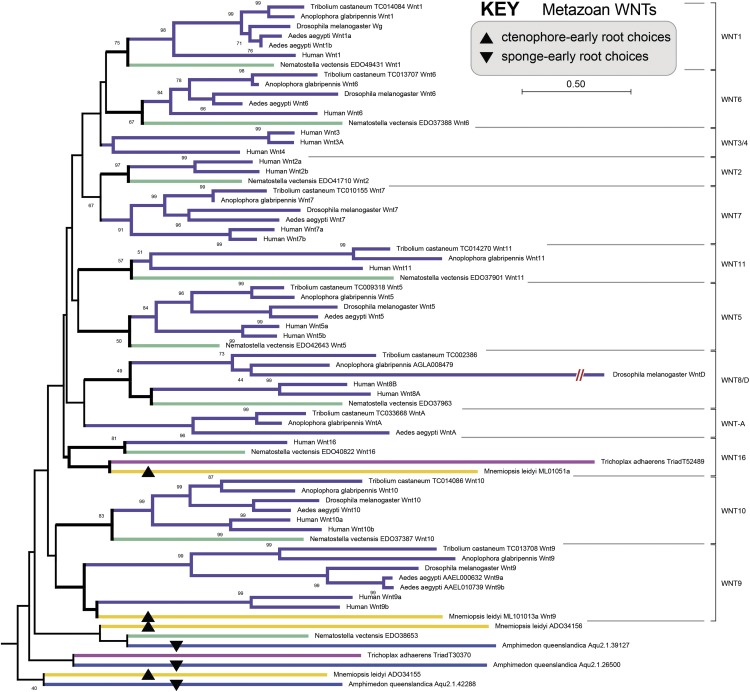
Given the preponderance and constancy of gene duplication (and gene loss) throughout evolution, one should in principle be able to find a gene duplication shared by all major animal lineages except the one true sister animal lineage. Therefore, ever since the elucidation of the first non-bilaterian animal genomes, mainly those from Cnidaria (Putnam et al. 2007), Placozoa (Srivastava et al. 2008), Porifera (Srivastava et al. 2010), and Ctenophora (Ryan et al. 2013; Moroz et al. 2014), we have sought to identify gene duplications that definitively order early metazoan phylogeny. For example, to identify a diagnostic gene family with a signature duplication occurring just after the early metazoan radiation, we have searched for Amphimedon (sponge) or Mnemiopsis (ctenophore) genes with predicted 1-to-Many relationships with the other metazoan genomes. Remarkably, this strategy has not been productive, and at best has led to the identification of gene families with many possible root choices consistent with either a sponge-early or ctenophore-early model. A good example of this root intractability with an increasing number of deep gene duplications is the phylogeny of the metazoan WNT gene family (Figure 1).
Figure 1.
Early metazoan branching and the root of the WNT gene family. Example gene tree for the metazoan WNT ligand in which sponge and ctenophore genes exist in 1-to-Many relationship with other metazoan genes. A major problem in extracting a phylogenetic signal from a large gene family defined by many paralogs is the increasing ambiguity in identifying the true root of the tree, particularly when lacking a non-metazoan outgroup gene. For the WNT family tree shown here, there are four ctenophore-early (up arrowheads) and three sponge-early (down arrowheads) choices for rooting. In addition, there are other root choices for either model in combination with inferred gene losses. For example, here this tree is rooted between a weakly supported clade (40%) containing only sponge and ctenophore genes and the clade containing all the remaining genes. If this was the correct root, then there would have been a loss of a metazoan WNT paralog in the stem-eumetazoan lineage regardless of whether the true tree is a ctenophore-early or sponge-early tree. This tree was constructed using a distance-based method (Neighbor-Joining) with 100 boot strap replicate samples. Numbers indicate boot strap supports for values 40%. The same color scheme is used throughout this study as follows: violet = Bilateria, cyan = Cnidaria, magenta = Placozoa, blue = Porifera (sponges), and mustard/yellow/orange = Ctenophora (different hues for different species).
As a possible explanation for our failure to find unambiguous, early metazoan, gene paralogies, we hypothesized the existence of a methodological bias associated with the various ortholog-calling pipelines on which we were relying. This hypothesis of an inherent bias in resolving ancient gene paralogy motivated an alternative comparative genomic screen that we present here. We begin by illustrating the problems that can arise by not accounting for ancient paralogy.
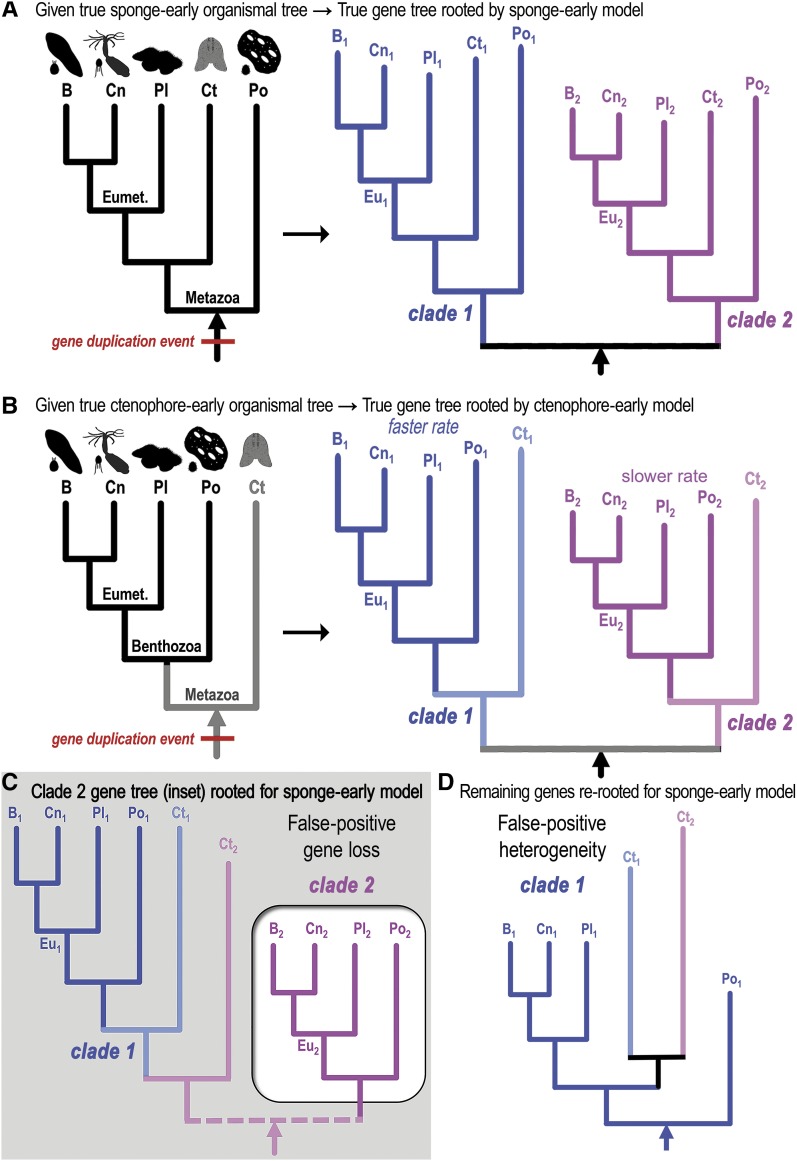
We consider the consequences of a rooting procedure defined by a sponge-early model given an early-stem metazoan gene duplication event in either a true sponge-early world (Figure 2A) or a true ctenophore-early world (Figure 2B). These arguments are meant to illustrate that human and machine-predictions of orthology will encounter unique problems specific to the true animal sister lineage of all other animals particularly when phylogenies are constructed as “single copy” genes. (Many single-copy genes are actually members of much larger super-families and so are single-copy only in the sense that an analysis restricts itself to a sub-clade of genes).
Figure 2.
Dangers and promise of ancient paralogy for deducing the deep phylogeny of animals. (A) Given a true sponge-early phylogeny (left organismal tree with animal icons), a true gene tree for a gene duplicated in the stem-metazoan lineage (red bar) and maintained in each lineage without any gene loss is composed of two separate gene clades (blue clade one and magenta clade two), depicted here as evolving at different clade-specific rates. The principal metazoan lineages correspond to Bilateria (B), Cnidaria (Cn), Placozoa (Pl), Ctenophora (Ct), and Porifera (Po, sponges). Eumetazoa is defined here as Bilateria + Cnidaria + Placozoa. The true gene tree is rooted easily between the two clades and recapitulates the sponge-early model in each clade (Po and Po are the deepest branching lineages). (B–D) Depicted below the horizontal line are the consequences of forcing gene trees from a ctenophores-first world into a sponge-early model. (B) Shown is the competing ctenophore-early model in which Ctenophora is sister to all other animals (here referred to as Benthozoa as explained in the text). For the same stem-metazoan gene duplication depicted in A, we now have a corresponding true gene tree in which the ctenophore genes (Ct and Ct) are the deepest branching lineages given the true ctenophore-early organismal tree. Asymmetric rates (faster or slower) are depicted in the gene phylogram, typical after divergent functionalization of paralogs. (C) The true gene tree in B is redrawn here with a different rooting procedure that isolates the more slowly-evolving paralog clade two (magenta clade in inset box) such that the sponge (Po) lineage appears as the outgroup lineage within that subclade. This sponge-early re-rooting procedure maintains the topology of the true gene tree. The dotted line anticipates the analyses of the improperly partitioned subclades as separate “single-copy” gene clades. The partial clade two tree (inset) is missing the Ct ctenophore gene and is associated with a false-positive signature of gene loss. (D) The sponge-early model is applied to the remaining genes as shown. This tree includes the ctenophore Ct gene that was previously flipped into the faster evolving clade one, which also has a higher propensity for long branch attraction than clade two. In contrast to the false-positive gene loss associated with the slowly-evolving clade two, the faster evolving clade one manifests false-positive heterogeneity of evolutionary rates and amino acid content due to the mixture of ctenophore paralogs.
In a sponge-early world, the sponge-early rooting procedure is simple and defines two gene clades (blue and magenta clades in Figure 2A). The same is true if we use a ctenophore-early rooting procedure given a ctenophore-early world (Figure 2B). We now describe the consequences of a forced sponge-early rooting given a ctenophore-early world, which has been the subject of much debate (Ryan et al. 2013; Moroz et al. 2014; Borowiec et al. 2015; Whelan et al. 2015; Shen et al. 2017; Jékely et al. 2015). For ease of reference, and for reasons explained in the Discussion, we will refer to the hypothetical sister clade composed of Eumetazoa + Porifera as “Benthozoa” (Figure 2B).
By choosing the most closely-related homologs to the more conserved member of a pair of duplicated genes (the more slowly-evolving magenta subclade two of Figure 2), or by internally re-rooting the true gene tree so that the subclade in question fits the standard model in which Porifera is sister to all other animals (Figure 2C), we end up with an isolated subclade with a false-positive gene loss (inset box in Figure 2C). Furthermore, the ctenophoran true outgroup sequence (“Ct”) is easily collected into the more divergent sub-clade (blue subclade one in Figure 2C). When the fast-evolving gene sub-clade is rooted separately with sponge as outgroup (“Po”), the inherent topology unites both ctenophore paralogs in an apparent ctenophore-specific duplication (bottom phylogram in Figure 2D). This second fast-evolving subclade would feature false-positive rate heterogeneity and compositional heterogeneity. False-positive compositional heterogeneity is consistent with recent attributions of (true) evolutionary sequence bias in ctenophores (Feuda et al. 2017).
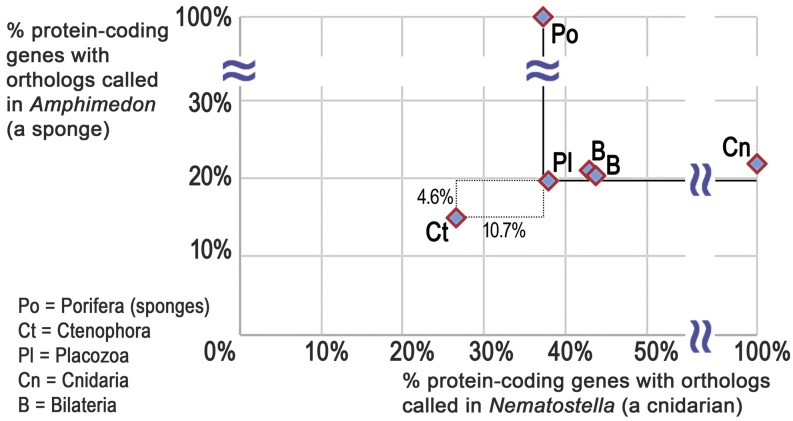
As predicted by the ctenophore-early hypothesis (Figure 2B) and deep paralogy-induced miscalling of orthologs (Figure 2C–D), we find that relatively fewer metazoan orthologs are called in the ctenophore Mnemiopsis leidyi relative to sponge (Amphimedon queenslandica), placozoan (Trichoplax adhaerens), cnidarian (Nematostella vectensis), and bilaterian (the lophotrochozoans Lottia gigantea and Capitella teleta) genomes when orthologies are predicted according to a sponge-early model (Figure 3). This finding supports the ctenophore-early hypothesis and is consistent with a similar approach in estimating deep animal phylogeny using gene content (Ryan et al. 2013).
Figure 3.
Deficit of metazoan homologs in ctenophores. Consistent with the predicted false-positive gene losses depicted in Fig. 2C, comparative genomic orthology data sets based on a sponge-early model result in far fewer orthologs called in a ctenophore relative to other metazoans. Shown are the percentages of each organism’s protein-coding gene set that have orthologs called in a cnidarian (Nematostella, x-axis) or a sponge (Amphimedon, y-axis) as inferred from the Metazoa ComparaEnsembl orthology calls (see Materials and Methods). Dotted box shows the distance from ctenophore value to the lowest value of the other animals along each axis (solid lines).
A model-agnostic screen for early metazoan duplications
Based on the above observations and rationale, we devised a comparative genomic screen in which orthology calls to the ctenophore Mnemiopsis and the sponge Amphimedon are not considered during the initial candidate paralogy group screen. In other words, we relinquished our previous reliance on 1-to-Many predictions for sponge or ctenophore genes relative to other metazoans. We began with the 16,590 protein-coding genes from the beetle Tribolium castaneum (Tcas5.2 genes), which we chose as a model bilaterian that has not experienced extensive gene loss as in nematodes (Erives 2015), nor extensive gene duplications as in vertebrates. The Tribolium gene set becomes 8,733 genes if we only consider those that exist in paralogous relationship(s) with other Tribolium genes. Of these only 2,617 have orthologs called in the mosquito Aedes aegypti, the lophotrochozoan Capitella teleta, and the placozoan Trichoplax adhaerens. Then we discard those paralogous Tribolium genes which are related by a common ancestor in more recent taxonomic group such as Cucujiformia (an infraorder of beetles), Holometabola, Hexapoda, Mandibulata, Arthropoda, Pancrustacea, Protostomia, Bilateria, and Eumetazoa. We then grouped 2,038 candidate paralogous beetle genes into families of various sizes.
We considered the optimal size for a gene family to be used in resolving early animal branching order. First and foremost, having more duplications is likely to increase the probability that some of the duplications occurred during the early metazoan radiation that produced the extant animal phyla. However, choosing extremely large gene families containing many paralogs introduces other problems in a classic trade-off scenario. For example, large gene families often present intractable root choice problems, particularly when genes are absent in the outgroup lineage of choanoflagellates, as is the case for animal-only genes (e.g., Figure 1). Extremely large gene families seem to also have a greater rate of gene loss. This ‘easily-duplicated, easily-lost’ pattern further exacerbates the choice of root. We thus concluded that it is more effective to screen for gene paralogies defined by the smallest possible number of early metazoan gene duplications (two or three).
To avoid the added complexity from greatly expanded gene families, we set aside 56% of the beetle genes from the 153 largest paralogy groups, containing 4 to 41 genes each (totalling 1,146/2038 genes). We were then left with 892 genes grouped in < 382 small paralogy groups containing two to three genes each (Supporting Table S1). We sampled 16% (60 gene families) from these small candidate paralogy groups to find gene families that had at least one ortholog called in Amphimedon queenslandica and at least one in Mnemiopsis leidyi. We note that we cannot rule out the possibility that metazoan-relevant paralogies were excluded because true orthologs failed to be computationally called in sponges and ctenophores. Many of the candidate paralogy groups have paralogs called in sponges and ctenophores, indicating that the duplications occurred prior to the latest common ancestor (LCA) of Metazoa.
We constructed draft phylogenies using different approaches, including an explicit evolutionary model (Maximum Parsimony) and a distance-based method (Neighbor-Joining). If either of these initial trees indicated a possible early metazoan duplication, we then constructed trees using a more sophisticated and informative approach with metropolis-coupled-MCMC Bayesian phylogenetic inference in conjunction with more extensive gene curation of unannotated exons (see Materials and Methods). As detailed below, this new approach begins to identify choanozoan genes with early metazoan duplications. We present our first results for several gene families as demonstration of the value of our approach and in anticipation of finding more examples as additional genome assemblies from diverse metazoans become available.
Duplication of cornichon-like transmembrane chaperones
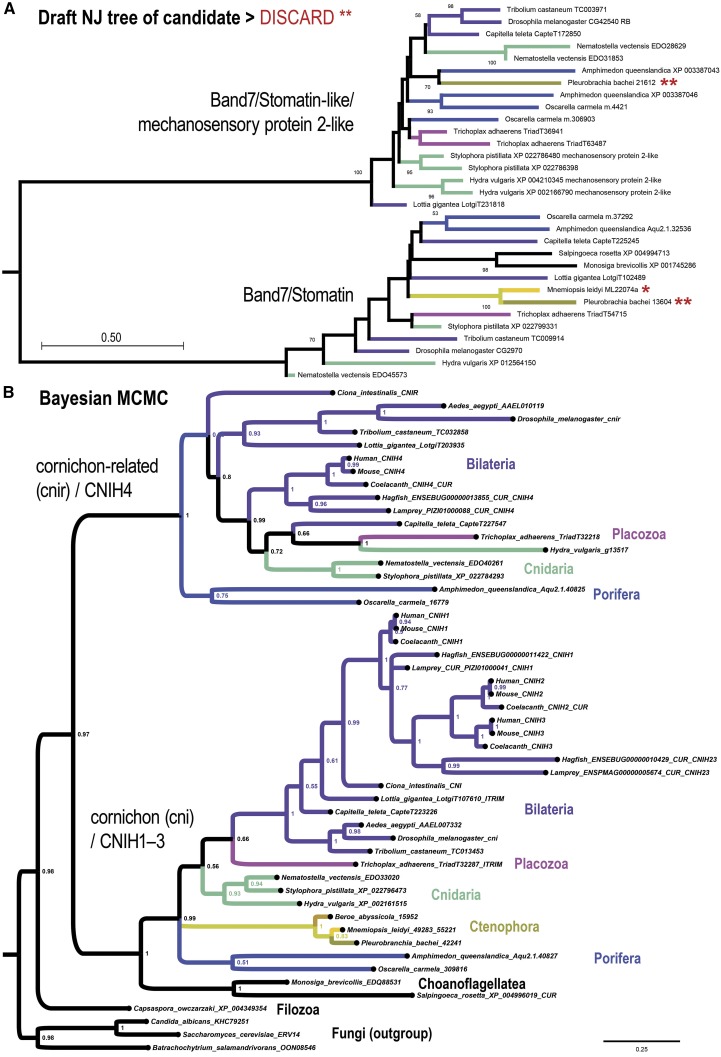
As expected, many of the candidate gene families, which were identified without regard to presence or absence in Amphimedon or Mnemiopsis, were either: (i) missing in both ctenophores and sponges, or (ii) present in both ctenophores and sponges. The latter sometimes occurred when the ctenophore Mnemiopsis did not actually have the full set of duplications but a related ctenophore did. For example, in Figure 4A we show one of our initial draft trees for the Band 7/Stomatin and Stomatin-like paralogs. These trees are consistent with a signature of gene absence for Stomatin-like in the ctenophore Mnemiopsis (Order Lobata). However, subsequent follow-up shows that a ctenophore from a different order (Order Cydippida) has both paralogs (double asterisks). The clear presence of each paralog in at least one ctenophore means that this candidate could be set aside as being an uninformative duplication that occurred in an earlier choanozoan ancestor. The Stomatin/Stomatin-like example (Figure 4A) also shows that our approach is likely to improve as additional genome assemblies become available for sponges and ctenophores. Nonetheless, other trees show from our screen, which was based on an agnostic preference for gene homologs in sponges and ctenophores, were in range to start being informative.
Figure 4.
Example choanozoan duplications: Stomatin/Stomatin-like, and cornichon/cornichon-related. (A) Shown is the draft NJ tree for the Band 7/Stomatin and Stomatin-like paralogs, which were identified in our screen due to the signature of gene loss for Stomatin-like in the ctenophore Mnemiopsis (Order Lobata), which was indicated to have only a single Stomatin gene (single asterisk). Subsequent follow-up showed that a ctenophore from a different order (Order Cydippida) has both paralogs (double astrerisks). The clear presence of each paralog in at least one ctenophore means that this candidate can be set aside as being an uninformative duplication. Boot strap support is shown for values 40% from 500 replicates. (B) Shown is another example gene tree from our screen to identify candidate duplications based on a signature of gene loss (either true or false-positive gene loss). This gene family encodes a eukaryotic chaperone of transmembrane receptors. This tree is equivocal for two scenarios: (1) loss of one of a pair of paralogs in choanoflagellates and ctenophores (see cnir part of tree) as shown, or (2) a duplication occurring later than is shown in the stem-benthozoan lineage with sufficient divergence of the cnir progenitor such that this clade is misplaced in the tree. Tree was generated from a multiple sequence alignment composed of 167 alignment columns and is rooted with Holomycota (Fungi) as outgroup.
We find that the cornichon (cni/CNIH1–CNIH3) and cornichon-related (cnir/CNIH4) paralogy groups, which encode eukaryotic chaperones of transmembrane receptors (Herring et al. 2013), was either duplicated in the stem-choanozoan lineage with subsequent gene loss only in choanoflagellates and ctenophores, or else duplicated in the stem-benthozoan lineage with subsequent neofunctionalization. We find that both cni and cnir clades exist only in Porifera, Placozoa, Cnidaria, and Bilateria but not Ctenophora and Choanoflagellatea (Figure 4B). This tree is based on a protein that is only 150 amino acids long and suggests that the ancestral cni/cnir gene was duplicated in the stem-choanozoan lineage with two subsequent independent losses in the stem-ctenophore lineage and the stem-choanoflagellate lineage (Figure 4B).
The alternative interpretation of the cni/cnir pair is that the progenitor gene was duplicated in the stem-benthozoic lineage with the cnir clade undergoing neofunctionalization to an extent promoting artifactual basal branching. In this interpretation there is no need to invoke independent gene losses for Ctenophora and Choanoflagellatea. Furthermore, key differences in client proteins for the Cornichon chaperones (CNIH1/2/3) vs. the Cornichon-related chaperone (CNIH4) support the neo-functionalization interpretation for the evolution of the CNIH4 paralog. Vertebrate CNIH1 and Drosophila Cornichon were found to mediate chaperone-like export of the transforming growth factor alpha (TGF-α)/Gurken precursor from the endoplasmic reticulum (ER) to the plasma membrane (Castro et al. 2007; Bökel et al. 2006), while the other vertebrate homologs of Drosophila Cornichon, CNIH2 and CNIH3, were found to mediate similar chaperone roles for AMPA receptor subunits (Schwenk et al. 2009). Both the TGF-α precursor and the individual AMPA receptor subunits have single transmembrane passes. In contrast, the vertebrate ortholog of Cornichon-related, CNIH4, has been found to mediate chaperone-like ER exit roles for certain G-protein coupled receptors (GPCRs), which possess seven transmembrane passes (Sauvageau et al. 2014). Thus, the Cornichon homologs, which are also present in choanoflagellates, may have evolved to help ER export of clients with only single transmembrane passes, while the Cornichon-related homolog CNIH4 may represent a neofunctionalization specialized for plasma membrane-bound proteins with multiple transmembrane passes.
In either case the progenitor cni/cnir gene was present as a single copy gene in the LCA of Holozoa as shown by the basal branching of the single gene from the filozoan Capsaspora owczarzaki, which branches before the duplication (Figure 4B).
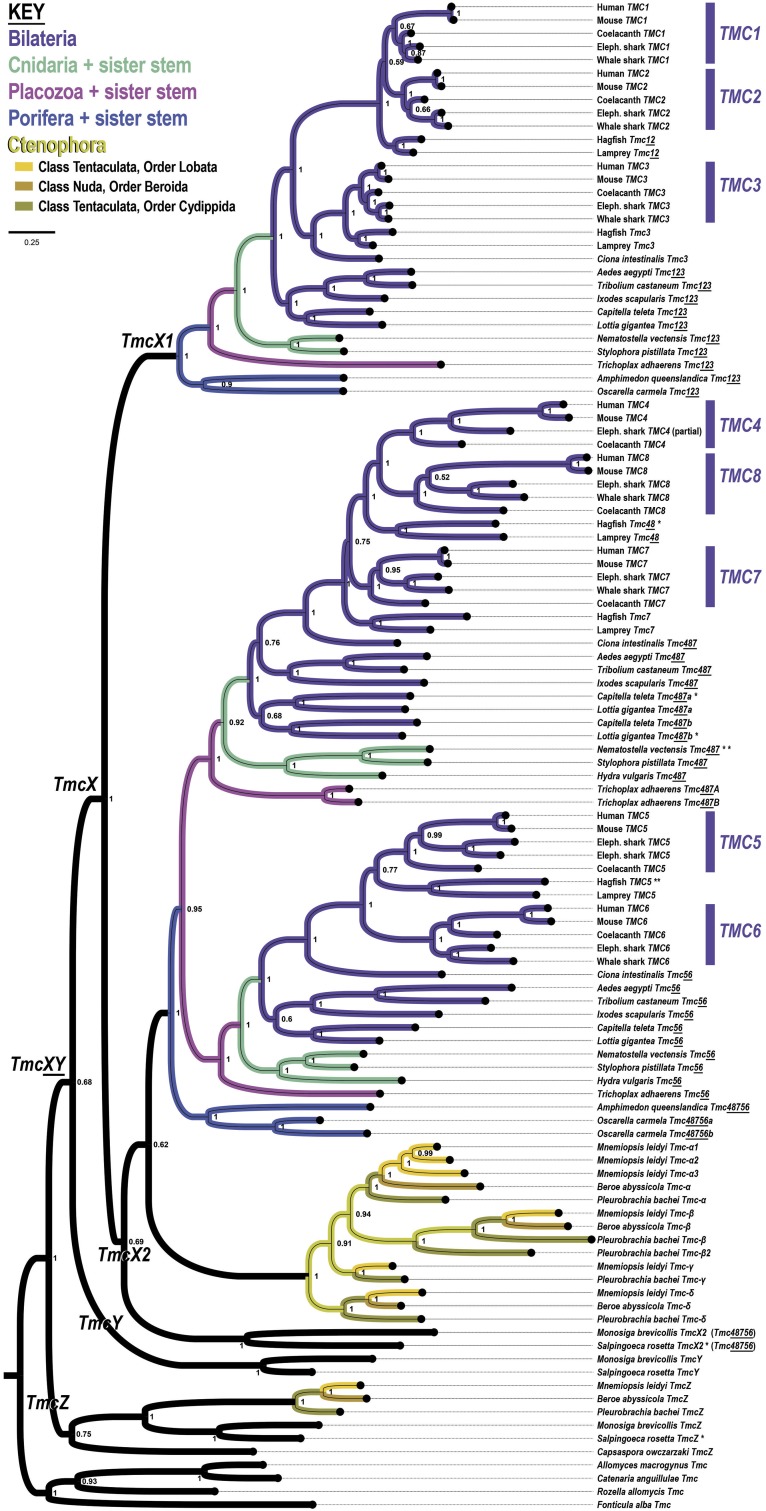
TMC sub-clades define Eumetazoa, Porifera, and Ctenophora
We find that a phylogenetic analysis of gene duplications of the transmembrane channels (TMCs), a large family of ion leak channels with roles in mechanotransduction (Delmas and Coste 2013; Pan et al. 2013, 2018; Qiu and Muller 2018; Ballesteros et al. 2018; Jia et al. 2019), excludes ctenophores from a eumetazoan super-clade composed of Bilateria, Cnidaria, and Placozoa (Figure 5). We describe this gene family’s origin and diversification beginning with the deepest duplications in this family and then proceeding to those most relevant to early metazoan evolution.
Figure 5.
A phylogenetic tree of the mechanotransducing transmembrane channel-like proteins (TMCs) resolves early metazoan diversification as a branching of only three lineages: Eumetazoa (magenta = Placozoa, teal = Cnidaria, and violet = Bilateria), Porifera (blue), and Ctenophora (mustard shades). Each of the eight TMC ortho-groups specific to gnathostomes are indicated on the right. This tree is based on a trimmed data set composed of 777 alignment columns and is rooted between unikonts (labeled “Opisthokonta” due to the apparent loss in Amoebozoa) and bikonts. Eumetazoan lineages share a duplication corresponding to the Tmc487 and Tmc56 TMC clades, while ctenophores share independent duplications of an ancestral Tmc48756 gene. Ctenophores share with choanoflagellates the more ancient Tmc-X paralog.
Early eukaryotic origin as a single-copy Tmc gene:
In our TMC phylogeny, we identify genes from both Unikonta and Bikonta, which are the main branches of the eukaryotic tree (Figure 5). An affinity of the TMC proteins with OSCA/TMEM proteins has been proposed (Murthy et al. 2018) despite low sequence similarity (Ballesteros et al. 2018), but we find that the percent identity between predicted TMC proteins is higher than between TMC proteins and TMEM. For example, the gap-normalized percent amino acid identity in a pairwise alignment of human Tmc1 with the single Naegleria gruberi Tmc sequence is 17.3%, vs. 12.7% (5% less) with human Ano3/TMEM16C (Supporting File S2). Moreover the identity between Tmc1 and TMEM16C goes away in a multiple sequence alignment suggesting it is inflated by the extra degree of freedoms allowed in a gapped pair-wise alignment.
Single-copy Tmc in Fungi and gene loss in clades with cell walls:
In the context of this ancient eukaryotic provenance of the TMC family, we find an intriguing and suggestive pattern of TMC gene loss that is likely relevant to animal evolution. We find that the Tmc gene exists as a single copy in the genome of the non-amoebozoan slime mold Fonticula alba and a few early-branching fungal lineages, altogether representing the clade of Holomycota. One of these fungal lineages, Rozella allomycis, belongs to the Cryptomycota, a phylum notable for its absence of a chitinous cell wall typically present in most other fungi (Jones et al. 2011). The absence of a cell wall allows the Cryptomycota to maintain a phagotrophic lifestyle (Jones et al. 2011). The only other fungal lineages with a TMC gene are from Blastocladiomycota, suggesting that TMC genes were lost in all of Ascomycota, Basidiomycota, and Zygomycota, for which many genomes have been sequenced.
Single-copy Tmc in Viridiplantae and gene loss With increasing complexity of cell walls:
Physiological incompatibility of Tmc function with certain cell walls is further supported by a similar distribution of Tmc genes in Viridiplantae (Figure 5 green clade). We find that a single Tmc gene can be found in a chlorophyte (Coccomyxa subellipsoidea), a charophyte (Klebsormidium nitens), and in the multicellular plants of a liverwort (Marchantia polymorpha), a moss (Physcomitrella patens), and a lycophyte (Selaginella moellendorfii). The moss Physcomitrella and the lycophyte Selaginella represent a nonvascular land plant and a vascular land plant most closely-related to the clade composed of ferns, gymnosperms, and angiosperms, which correspond to the crown group having evolved lignin-based secondary cell walls (Sarkar et al. 2009). In addition, both the Physcomitrella and Selaginella genomes lack many of the cellulose synthase (CesA) genes responsible for the synthesis of plant cell wall components (Sørensen et al. 2010). Thus these peculiar TMC gene distributions across plants and fungi suggest that rigid cell walls potentially interfere with physical environmental coupling to TMCs.
Choanozoan duplications of Tmc:
While we find Tmc genes as a single copy in some lineages of the fungal and plant kingdoms, in Naegleria gruberi, and in the filozoan Capsaspora owczarzaki, we find that these genes underwent duplication only during the holozoan radiation (or if earlier only with corresponding losses in fungi). In short, definitive Tmc gene duplications can be found only within Choanozoa (choanoflagellates + Metazoa) (Figure 5). The choanoflagellates Monosiga brevicollis and Salpingoeca rosetta have at least three Tmc genes each, which we have labeled “Tmc-X”, “Tmc-Y”, and “Tmc-Z” (Figure 5). Remarkably, of all the animals, only ctenophores appear to have genes from the Tmc-X clade, which is a basally-branching clade that is sister to all of the remaining Tmc genes from all of Opisthokonta. Only choanoflagellates and Holomycota have genes in the Tmc-Y clade. Last, except Capsaspora, whose single Tmc gene is of uncertain affinity to the well supported Tmc-X/Y/Z clades, all of the remaining Tmc genes are choanozoan genes from the Tmc-Z clade.
The Tmc duplications within Choanozoa are informative for early choanozoan and metazoan branching. Tmc-Z apparently underwent a key duplication (“Tmc123” + “Tmc48756”) in the stem-metazoan lineage with a corresponding loss of one of the paralogs (“Tmc123”) in ctenophores. Alternatively, this duplication is a stem-benthozoan duplication that occurred after ctenophores split off from the rest of Metazoa. These two Tmc-Z subclades are so named here for their relationship to the vertebrate TMC1/2/3 (“Tmc-Z1”) and TMC4/8/7/5/6 (“Tmc-Z2”) paralogy groups. We find that this Tmc-Z2 gene (Tmc48756) underwent a Eumetazoa-defining duplication that unites Placozoa, Cnidaria, and Bilateria (Figure 5). Thus, in this stem-eumetazoan lineage we see that the Tmc-Z2 bifurcates into the sister-clades of Tmc487 and Tmc56. This duplication makes it unlikely that Porifera or Ctenophora are more closely related to any single lineage within Eumetazoa. This contrasts with the stem-ctenophore lineage, in which the single Tmc-Z2 gene independently duplicated several-fold to produce four ctenophore-specific duplications (Tmc-α, Tmc-β, Tmc-γ, and Tmc-δ) (Figure 5, lineages in shades of mustard). This ctenophore specific repertoire is present in two different classes and three different orders of ctenophores. In Class Tentaculata, we have four Tmc48756 genes from each of Mnemiopsis leidyi (Order Lobata) and Pleurobrachia bachei (Order Cydippida). In class Nuda, so named because of a complete (derived) loss of tentacles, we have only three Tmc48756 genes from Beroë abyssicola (Order Beroida) because a representative gene from the Tmc-γ clade could not be identified.
Many metazoan sub-clades (e.g., Ctenophora, Lophotrochozoa, and Vertebrata) can be defined by clade-specific duplications. For example, lophotrochozoans share a duplication of Tmc487 into Tmc487a + Tmc487b, while gnathostomes share a duplication of vertebrate Tmc12 into TMC1 + TMC2 and vertebrate Tmc48 into TMC4 + TMC8. If we include the cyclostomes (hagfish + lamprey), then all vertebrates share the duplication of Tmc123 into Tmc12 + TMC3, and Tmc487 into Tmc48 + TMC7. Thus, the “canonical” vertebrate repertoire of eight genes represented by TMC1–TMC8 is more correctly characterized as a gnathostome-specific TMC repertoire because cyclostomes are united in sharing only some of the duplications seen in humans (see Figure 5).
Considering the Tmc gene duplications that are ancestral to Choanozoa and those specific to metazoan phyla, this phylogeny, based on the large TMC protein spanning ten transmembrane domains, unites Placozoa, Cnidaria, and Bilateria into a single Eumetazoa in the following ways. First, of greatest significance, is the shared derived eumetazoan duplication of Tmc48756 into Tmc487 and Tmc56. Second, is the close placement of Porifera as the sister group of Eumetazoa in both the Tmc123 and Tmc48756 subclades. Third is the unique situation that ctenophores possess TMC genes from an ancestral Tmc-X clade that were apparently lost in a stem-benthozoan lineage while also possessing an expansive repertoire via ctenophore-specific duplications (Tmc-α, Tmc-β, Tmc-γ, and Tmc-δ). These latter duplications are most parsimonious if they occurred in a metazoan lineage that itself branched off prior to the eumetazoan duplication within the Tmc-Z2 clade. In the Discussion, we speculate on the significance of the evolution of mechanotransduction in connection with the evolution of diverse ’body plans’ during the metazoan radiation.
A bHLH-ZIP duplication unites Benthozoa
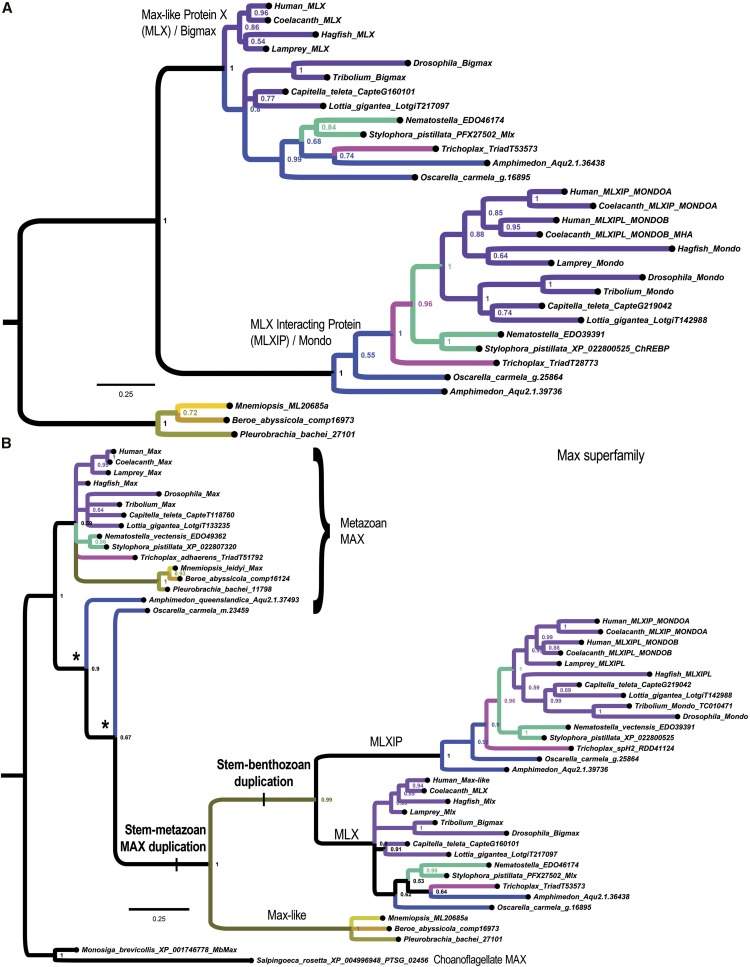
We find that a new gene family from the bHLH-ZIP superfamily originated in the stem-metazoan lineage most likely from a duplication of the more distantly-related Max gene (Figure 6). This gene occurs as one gene copy in all ctenophores but as a pair of duplicated genes in Porifera and Eumetazoa (Figure 6A). The pair of paralogous genes corresponds to Max-like protein X (MLX)/bigmax and MLX Interacting Protein (MLXIP)/Mondo. Mondo paralogs are also known as the carbohydrate response element binding protein (ChREBP) (Iizuka et al. 2004; Havula and Hietakangas 2018; Yamashita et al. 2001). The progenitor gene likely encoded a bHLH-ZIP homodimeric transcription factor (TF) in the stem-metazoan lineage, continuing as such into modern ctenophores, but evolving into the MLX:MLXIP heterodimer found in all other animals (Bigmax:Mondo in Drosophila, MLX:MondoA or MLX:MondoB in gnathostomes). To identify the origin of the MLX and MLXIP bHLH-ZIP paralogs, we then conducted phylogenetic analyses with the most closely-related bHLH-ZIP sequences, which correspond to the MAX proteins. We find that the MLX family likely originated from an earlier stem-metazoan duplication of MAX that is absent in choanoflagellates and other holozoans (Figure 6B). The Myc:Max gene regulatory network is known to have evolved in an early choanozoan stem-lineage (Brown et al. 2008; Young et al. 2011). When we root with choanoflagellate MAX as the out-group, we see that the MLX and MLXIP paralogs emerge from an earlier and presumably neofunctionalized duplication within the MAX family (see “stem-metazoan MAX duplication” in Figure 6B). However, this duplication appears to have occurred after the lineage leading to modern ctenophores branched away from all other animals as ctenophores appear to have only the progenitor Max-like protein (see “stem-benthozoan duplication” in Figure 6B).
Figure 6.
MLX/bigmax and MLXIP/Mondo encode a bHLH-ZIP heterodimer originating in a stem-benthozoan gene duplication. (A) Phylogenetic analysis of a MLX/MLXIP bHLH-ZIP gene family originating in the stem-metazoan lineage (possibly from a duplication of the distantly-related Max gene) followed by a second duplication after the divergence of the ctenophore lineage. Color coding of lineages follows Figures 4 and 5 except when topology precludes coloring sister stem lineages. This tree is rooted with ctenophores as the outgroup lineage. This tree is based on a data set composed of 263 alignment columns. (B) Shown is a phylogenetic tree constructed by Bayesian MC-MCMC and including sequences from the most closely-related and presumed progenitor MAX family (207 alignment columns). This tree places the Max-like sequences within the Max superfamily and furthermore puts the MLX and MLXIP duplications as occurring in a stem-benthozoan lineage that is sister to the single Max-like gene of ctenophores. While the Max-like/MLX/MLXIP sub-family likely originated as a stem-metazoan duplication, its sequences have diverged sufficiently to pull the sponge MAX sequences in a long-branch attraction (LBA) artifact (asterisks). This LBA is not the result of root choice because rooting between the choanozoan MAX sub-clade and the remaining sequences would result in sponges diverging prior to choanoflagellates, which would also correspond to LBA.
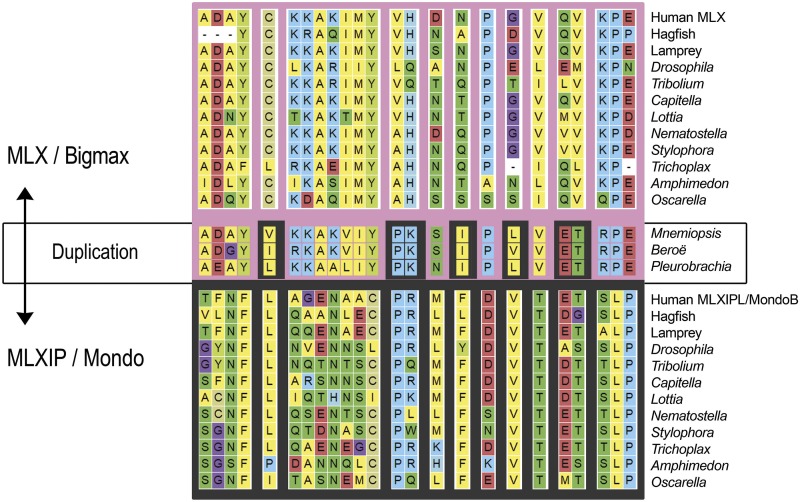
We find that the bHLH domain from ctenophores shares residues with both subfamilies (pink vs. black background in Figure 7). In the Discussion we speculate on a possible role for this gene duplication in a life cycle synapomorphy for the proposed clade of Benthozoa.
Figure 7.
Shown are a subset of alignment columns in which the ctenophore residues more closely resemble either the MLX sequences (top pink) or the MLXIP/Mondo sequences (bottom dark gray). The MLXIPL/MondoB representative sequence is shown for humans.
Discussion
By identifying and phylogenetically analyzing small paralogy groups that predate a eumetazoan super-clade composed of Bilateria, Cnidaria, and Placozoa, we find we can identify gene duplications that are consistent with Ctenophora being the sister metazoan lineage to all other animals (Figures 4, 5, 6). We discuss the significance of the different gene families that we identified and the possible relevance to metazoan biology.
Mechanosensation for animal body plans
We find that the transmembrane channel-like (TMC) gene family continued to duplicate and diversify for several phyla throughout metazoan evolution. For example, we see that the cyclostomes (hagfish and lampreys) branched early in the vertebrate tree prior to a number of duplications that occurred only in the stem-gnathostome lineage (Tmc12 → TMC1 + TMC2; Tmc48 → TMC4 + TMC8; Mondo → MondoA/MLXIP + MondoB/MLXIPL; and CNIH23 → CNIH2 + CNIH3). Nonetheless, cyclostomes and gnathostomes share the vertebrate-specific duplications corresponding to Tmc12, and Tmc48 + TMC7. Thus, like the vertebrate clade and its subclades, many other metazoan phyla can be defined solely on the basis of Tmc gene duplications (Figure 5). We therefore propose that the evolutionary diversification of the Tmc channels throughout Metazoa, including the independent diversification within Ctenophora (Figure 5), must have been under selection of two principal forces. The first is the changing physical constraints and sensorial opportunities associated with the evolutionary diversification of animal body plans themselves. The second factor is the evolutionary diversification of specialized cell types within their epithelial sensoria.
Mechanosensory, chemosensory and photosensory responses are universal among single and multicellular organisms and can be related to the evolution of specific proteins enabling already single cells to respond to mechanical, chemical and photic stimuli. For example, opsin proteins evolved in single-celled ancestors of metazoans (Arendt 2017; Feuda et al. 2012) and many single cell organisms can sense light, gravity and several chemical stimuli with dedicated sensors (Swafford and Oakley 2018). While the history of chemical and photic senses and the formation of specialized cell types and integration into appropriate organs in metazoans has been driven by the molecular insights into the molecular transducers (Arendt et al. 2016), mechanosensory transduction has seen less progress due to uncertainty of consistent association of a specific mechanotransducer channel across phyla (Beisel et al. 2010). On the one hand, mechanical sensation is clearly present in all single cell organisms to function as safety valves to release intracellular pressure sensed as tension in the lipid bilayer and was proposed as a possibly unifying principle of mechanosensation (Kung 2005). Follow up work showed a multitude of channels associated with mechanosensation (Beisel et al. 2010; Delmas and Coste 2013) arguing against a single unifying evolution of mechanotransduction. Indeed, several families of mechanosensory channels have been identified whereby pores open as a function of lipid stretch or tethers attached to extra-or intracellular structures (Cox et al. 2018a,b).
Simply speaking, no single molecule has been identified to be associated with all mechanotransduction across phyla such as the ecdysozoan TRP channels also found in bony fish but absent in mammals (Beisel et al. 2010; Qiu and Muller 2018; Cox et al. 2018a). Likewise, the ubiquitous Piezo mechanotransduction channels in Merkel cells (Delmas and Coste 2013; Ranade et al. 2015; Cox et al. 2016) were hypothesized to be the mechanotransduction channel of hair cells (Arendt et al. 2016) but have meanwhile been found not to be directly associated with the mechanotransduction process (Wu et al. 2017). Molecular analysis of mammalian mutations meanwhile has focused on the family of TMC (transmembrane channel-like proteins) as possibly involved in hair cell mechanotransduction (Delmas and Coste 2013; Pan et al. 2013) and replacement of mutated TMC can restore hearing (Yoshimura et al. 2018; Shibata et al. 2016; Askew et al. 2015).
More recently, molecular analysis has established that the TMC forms a mechanosensory pore as a homodimer with each subunit having ten predicted transmembrane domains (Pan et al. 2018). However, the detailed transmembrane protein dimer is unclear as other proteins are needed to transport the TMC channels to the tip (Pacentine and Nicolson 2019) and the entire complex of the vertebrate mechanosensory channel and its attachment to intra- and extracellular tethers remains unresolved (Qiu and Muller 2018). To what extent TMC family channel evolution aligns with mechanosensory cell and organ evolution remains to be seen but is already indicating unique features of ctenophores at every level (Fritzsch et al. 2015, 2006).
Recent work also establishes that ecdysozoan Tmc123 paralogs function in body kinesthesia (proprioception), sensory control of locomotion or egg-laying behavior via membrane depolarization, and nociception (Guo et al. 2016; Yue et al. 2018; Wang et al. 2016). In summary, our study further lays the groundwork for understanding the molecular history of a sensory channel family in the context of the evolution of developmental gene regulatory circuits in different animal lineages (Beisel et al. 2010; Fritzsch et al. 2000; Fritzsch and Elliott 2017; Corbo et al. 1997; Cox et al. 2018a).
Regulation of metabolism in larval and adult morphologies
We speculate on the possible connection of the MLX/MLXIP gene duplication (Figure 6) to an evolutionary transition from a holopelagic to a biphasic pelago-benthic life cycle in the stem-benthozoan lineage. This finding adds to a growing picture of metabolic and chaperone gene loss and gene innovation in early animal evolution (Erives and Fassler 2015; Richter et al. 2018). Basic-helix-loop-helix (bHLH) TFs function as obligate dimers for DNA-binding (Murre et al. 1989; Lassar et al. 1991; Murre et al. 1991) and their combinatorial dimerizations are a dominant aspect of their regulatory interactions (Murre et al. 1989; Neuhold and Wold 1993) and of their evolution (Brown et al. 2008). Thus, it seems unlikely that a second MLX-related gene encoding a heterodimeric partner to the single gene found in ctenophores is artifactually missing in multiple sequenced genomes and transcriptomes (Ryan et al. 2013; Moroz et al. 2014). It is possible then that the single MLX-like gene corresponds to an evolutionary intermediate state in which a single gene encodes a homodimeric bHLH TF. But this idea needs to be tested.
The MLX:MLXIP/MLXIPL heterodimeric TF acts as a transcriptional regulator of metabolic pathways (e.g., lipogenesis genes) in response to variation in intracellular sugar concentrations (Yamashita et al. 2001; Iizuka et al. 2004; Sans et al. 2006; Havula et al. 2013; Havula and Hietakangas 2018). In this regard it is interesting to speculate that the duplication evolved in connection with the evolution of a biphasic pelago-benthic life cycle featuring a pelagic feeding larva and a benthic feeding adult (whether it was a motile planula or sessile adult). Pelagic larval and benthic adult feeding forms would have distinct nutritional intakes associated with dissimilar feeding strategies and dissimilar nominal parameters. This bimodal variation would have evolved on top of variation associated with just a single feeding strategy. Thus, a stem-benthozoic ancestor may have demanded additional complexity in metabolic regulation that was subsequently afforded by duplicated paralogs to expand regulation of life cycle cell forms into differentiation of different cell types (Fritzsch et al. 2015).
Methodological pitfalls in using paralogy to infer phylogeny
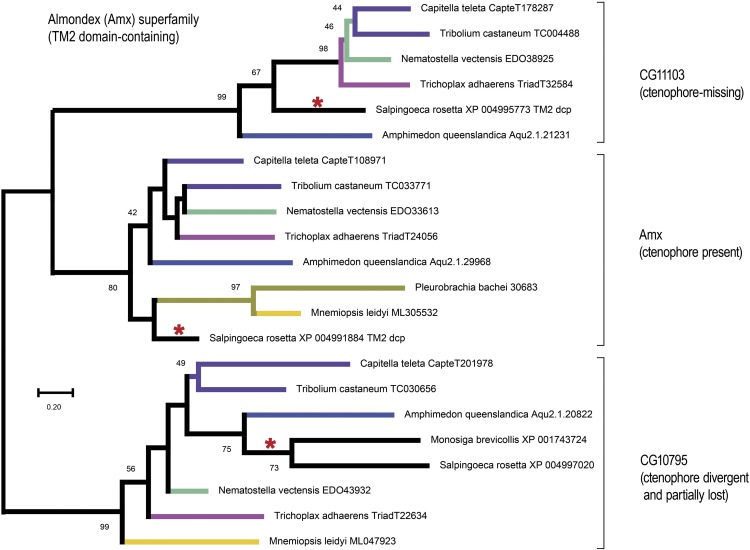
While we may have identified some of the best candidates to date depicting early metazoan branching via gene duplications, we want to point out one possible pitfall in this approach. One of our candidate paralogies encodes the Almondex (Amx) superfamily, which persisted in our screen through the stages where we required a missing orthology call for either the sponge Amphimedon or the ctenophore Mnemiopsis (see gene family 23 in size group three of Table S1). Almondex is a neurogenic TM2-domain containing protein characterized as a genetic modifier of Notch signaling in Drosophila (Shannon 1972, 1973; Michellod et al. 2003; Michellod and Randsholt 2008). Thus, as a neurogenic locus this candidate gene family could have made biological sense if it linked Ctenophora as a sister-group to Eumetazoa in the proposed Neuralia clade (Nielsen 2008). However, when we analyzed the Amx super-family in more depth, we found that one of the three paralogs was missing in the ctenophore Mnemiopsis (see the CG11103 clade in Figure 8), while all three paralogs were present in the sponge Amphimedon (Figure 8).
Figure 8.
The neurogenic Almondex superfamily. Shown is a gene phylogeny composed of three paralogy groups (labeled by their Drosophila gene names), which originated in a choanozoan ancestor given the presence in both choanoflagellates (red asterisks) and animals. Given this ancient provenance, this tree may be rooted with any one of the three sub-clades as out-group. As explained in the text, ctenophores appear to be missing some of these paralogs (e.g., CG11103), and were it not for the presence of each paralog in at least one choanoflagellate, alternate tree rooting strategies suggestive of gene duplications could have been possible. This tree was constructed using Neighbor-Joining (distance-based) with 1000 boot-strap replicates sampled from an alignment with 207 alignment columns. Boot-strap supports are shown only for values 40
To rule out Mnemiopsis-specific gene losses, we also searched for orthologs of each paralogy group in other ctenophores, and were indeed able to find Amx in the ctenophore Pleurobrachia (middle clade in Figure 8). Superficially this tree began to resemble the MLX-MLXIP tree (Figure 6B) except in its important relation to choanoflagellates. Unlike the single choanoflagellate MAX gene (Figure 6B), the Amx superfamily tree shows that the choanoflagellate Salpingoeca rosetta has each of the three paralogs (red asterisks in Figure 8). This result implies that ctenophores have lost the CG11103 ortholog, and have either lost or are missing the CG10795 ortholog (Pleurobrachia) or else possess a divergent version of this gene (Mnemiopsis).
In the case of the Amx superfamily, the evolutionary maintenance of Amx paralogs in at least one choanoflagellate is critical to ruling out duplicated genes, which would be possible with alternate root-choices. With the absence of paralogs for the choanoflagellate Salpingoeca in the Amx and CG11103 sub-clades, which already appear to be lost in the choanoflagellate Monosiga, we may justifiably have rooted the tree in Figure 8 so that the choanoflagellate CG10795 clade is the outgroup. A tree rooted in this way (in the hypothetical absence of the two lone Salpingoeca sequences) allows the possibility that CG11103 could be a Benthozoa-specific duplication. Thus, in using gene duplications to infer phylogenetic branching it is important to be mindful of unchecked misinterpretations facilitated by true gene loss. If this type of error is ubiquitous, that we should be able to find examples showing duplications shared by ctenophores and bilaterians even if they are false-positive duplications caused by unconstrained root choices. Further examples will be necessary to see if this is the case. But in summary, our comparative genomic screen designed to identify candidate families duplicated prior to the evolution of Eumetazoa has only identified examples consistent with a clade of Benthozoa that unites Porifera and Eumetazoa as the sister-clade to Ctenophora.
Acknowledgments
Funding support from the NIH (R01 AG060504 to BF) and an anonymous Iowa Foundation grant (to AE).
Footnotes
Supplemental material available at figshare: https://doi.org/10.25387/g3.11444400.
Communicating editor: R. Kulathinal
Literature Cited
- Altschul S. F., Gish W., Miller W., Myers E. W., and Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Arendt D., 2017. The enigmatic xenopsins. eLife 6: 1–3. 10.7554/eLife.31781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt D., Musser J. M., Baker C. V., Bergman A., Cepko C. et al. , 2016. The origin and evolution of cell types. Nat. Rev. Genet. 17: 744–757. 10.1038/nrg.2016.127 [DOI] [PubMed] [Google Scholar]
- Askew C., Rochat C., Pan B., Asai Y., Ahmed H. et al. , 2015. Tmc gene therapy restores auditory function in deaf mice. Sci. Transl. Med. 7: 295ra108 10.1126/scitranslmed.aab1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayres D. L., Darling A., Zwickl D. J., Beerli P., Holder M. T. et al. , 2012. BEAGLE: an application programming interface and high-performance computing library for statistical phylogenetics. Syst. Biol. 61: 170–173. 10.1093/sysbio/syr100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros A., Fenollar-Ferrer C., and Swartz K. J., 2018. Structural relationship between the putative hair cell mechanotransduction channel TMC1 and TMEM16 proteins. eLife 7: 1–26. 10.7554/eLife.38433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel K., He D., Hallworth R., and Fritzsch B., 2010. Genetics of mechanoreceptor evolution and development, Elsevier Inc., Amsterdam. [Google Scholar]
- Bökel C., Dass S., Wilsch-Bräuninger M., and Roth S., 2006. Drosophila Cornichon acts as cargo receptor for ER export of the TGFalpha-like growth factor Gurken. Development 133: 459–470. 10.1242/dev.02219 [DOI] [PubMed] [Google Scholar]
- Borowiec M. L., Lee E. K., Chiu J. C., and Plachetzki D. C., 2015. Extracting phylogenetic signal and accounting for bias in whole-genome data sets supports the Ctenophora as sister to remaining Metazoa. BMC Genomics 16: 987 10.1186/s12864-015-2146-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S. J., Cole M. D., and Erives A. J., 2008. Evolution of the holozoan ribosome biogenesis regulon. BMC Genomics 9: 442 10.1186/1471-2164-9-442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro C. P., Piscopo D., Nakagawa T., and Derynck R., 2007. Cornichon regulates transport and secretion of TGFalpha-related proteins in metazoan cells. J. Cell Sci. 120: 2454–2466. 10.1242/jcs.004200 [DOI] [PubMed] [Google Scholar]
- Corbo J. C., Erives A., Di Gregorio A., Chang A., and Levine M., 1997. Dorsoventral patterning of the vertebrate neural tube is conserved in a protochordate. Development 124: 2335–2344. [DOI] [PubMed] [Google Scholar]
- Cox C. D., Bae C., Ziegler L., Hartley S., Nikolova-Krstevski V. et al. , 2016. Removal of the mechanoprotective influence of the cytoskeleton reveals PIEZO1 is gated by bilayer tension. Nat. Commun. 7: 10366 10.1038/ncomms10366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox C. D., Bavi N., and Martinac B., 2018a Bacterial mechanosensors. Annu. Rev. Physiol. 80: 71–93. 10.1146/annurev-physiol-021317-121351 [DOI] [PubMed] [Google Scholar]
- Cox C. D., Bavi N., and Martinac B., 2018b Cytoskeleton-associated proteins modulate the tension sensitivity of Piezo1. Biophys. J. 114: 111a 10.1016/j.bpj.2017.11.641 [DOI] [Google Scholar]
- Delmas P., and Coste B., 2013. Mechano-gated ion channels in sensory systems. Cell 155: 278–284. 10.1016/j.cell.2013.09.026 [DOI] [PubMed] [Google Scholar]
- Durinck S., Moreau Y., Kasprzyk A., Davis S., De Moor B. et al. , 2005. Biomart and bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 21: 3439–3440. 10.1093/bioinformatics/bti525 [DOI] [PubMed] [Google Scholar]
- Erives A. J., 2015. Genes conserved in bilaterians but jointly lost with Myc during nematode evolution are enriched in cell proliferation and cell migration functions. Dev. Genes Evol. 225: 259–273. 10.1007/s00427-015-0508-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erives A. J., and Fassler J. S., 2015. Metabolic and chaperone gene loss marks the origin of animals: evidence for Hsp104 and Hsp78 chaperones sharing mitochondrial enzymes as clients. PLoS One 10: e0117192 10.1371/journal.pone.0117192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuda, R., M. Dohrmann, W. Pett, H. Philippe, O. Rota-Stabelli, et al., 2017 Improved modeling of compositional heterogeneity supports sponges as sister to all other animals. Curr Biol 27: 3864–3870 e4. 10.1016/j.cub.2017.11.008 [DOI] [PubMed]
- Feuda R., Hamilton S. C., McInerney J. O., and Pisani D., 2012. Metazoan opsin evolution reveals a simple route to animal vision. Proc. Natl. Acad. Sci. USA 109: 18868–18872. 10.1073/pnas.1204609109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch W. M., and Margoliash E., 1967. Construction of phylogenetic trees. Science 155: 279–284. 10.1126/science.155.3760.279 [DOI] [PubMed] [Google Scholar]
- Fritzsch B., Beisel K. W., and Bermingham N. A., 2000. Developmental evolutionary biology of the vertebrate ear: conserving mechanoelectric transduction and developmental pathways in diverging morphologies. Neuroreport 11: R35–R44. 10.1097/00001756-200011270-00013 [DOI] [PubMed] [Google Scholar]
- Fritzsch B., and Elliott K. L., 2017. Gene, cell, and organ multiplication drives inner ear evolution. Dev. Biol. 431: 3–15. 10.1016/j.ydbio.2017.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B., Jahan I., Pan N., and Elliott K. L., 2015. Evolving gene regulatory networks into cellular networks guiding adaptive behavior: an outline how single cells could have evolved into a centralized neurosensory system. Cell Tissue Res. 359: 295–313. 10.1007/s00441-014-2043-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B., Pauley S., and Beisel K. W., 2006. Cells, molecules and morphogenesis: the making of the vertebrate ear. Brain Res. 1091: 151–171. 10.1016/j.brainres.2006.02.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Wang Y., Zhang W., Meltzer S., Zanini D. et al. , 2016. Transmembrane channel-like (Tmc) gene regulates Drosophila larval locomotion. Proc. Natl. Acad. Sci. USA 113: 7243–7248. 10.1073/pnas.1606537113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider S., Ballester B., Smedley D., Zhang J., Rice P. et al. , 2009. BioMart Central Portal–unified access to biological data. Nucleic Acids Res. 37: W23–W27. 10.1093/nar/gkp265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havula E., and Hietakangas V., 2018. Sugar sensing by ChREBP/Mondo-Mlx-new insight into downstream regulatory networks and integration of nutrient-derived signals. Curr. Opin. Cell Biol. 51: 89–96. 10.1016/j.ceb.2017.12.007 [DOI] [PubMed] [Google Scholar]
- Havula E., Teesalu M., Hyotylainen T., Seppala H., Hasygar K. et al. , 2013. Mondo/ChREBP-Mlx-regulated transcriptional network is essential for dietary sugar tolerance in Drosophila. PLoS Genet. 9: e1003438 10.1371/journal.pgen.1003438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmrich G., and Bosch T. C., 2008. Compagen, a comparative genomics platform for early branching metazoan animals, reveals early origins of genes regulating stem-cell differentiation. BioEssays 30: 1010–1018. 10.1002/bies.20813 [DOI] [PubMed] [Google Scholar]
- Herring B. E., Shi Y., Suh Y. H., Zheng C. Y., Blankenship S. M. et al. , 2013. Cornichon proteins determine the subunit composition of synaptic AMPA receptors. Neuron 77: 1083–1096. 10.1016/j.neuron.2013.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland P. W., Marlétaz F., Maeso I., Dunwell T. L., and Paps J., 2017. New genes from old: asymmetric divergence of gene duplicates and the evolution of development. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372: 20150480 10.1098/rstb.2015.0480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck J. P., and Ronquist F., 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Iizuka K., Bruick R. K., Liang G., Horton J. D., and Uyeda K., 2004. Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proc. Natl. Acad. Sci. USA 101: 7281–7286. 10.1073/pnas.0401516101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jékely G., Paps J., and Nielsen C., 2015. The phylogenetic position of ctenophores and the origin(s) of nervous systems. Evodevo 6: 1 10.1186/2041-9139-6-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y., Zhao Y., Kusakizako T., Wang Y., Pan C. et al. , 2019. TMC1 and TMC2 proteins are pore-forming subunits of mechanosensitive ion channels. Neuron. 10.1016/j.neuron.2019.10.017 [DOI] [PubMed] [Google Scholar]
- Jones M. D., Forn I., Gadelha C., Egan M. J., Bass D. et al. , 2011. Discovery of novel intermediate forms redefines the fungal tree of life. Nature 474: 200–203. 10.1038/nature09984 [DOI] [PubMed] [Google Scholar]
- Keresztes G., Mutai H., and Heller S., 2003. TMC and EVER genes belong to a larger novel family, the TMC gene family encoding transmembrane proteins. BMC Genomics 4: 24 10.1186/1471-2164-4-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., and Tamura K., 2016. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33: 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung C., 2005. A possible unifying principle for mechanosensation. Nature 436: 647–654. 10.1038/nature03896 [DOI] [PubMed] [Google Scholar]
- Lassar A. B., Davis R. L., Wright W. E., Kadesch T., Murre C. et al. , 1991. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell 66: 305–315. 10.1016/0092-8674(91)90620-E [DOI] [PubMed] [Google Scholar]
- Lynch M., O’Hely M., Walsh B., and Force A., 2001. The probability of preservation of a newly arisen gene duplicate. Genetics 159: 1789–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michellod M. A., Forquignon F., Santamaria P., and Randsholt N. B., 2003. Differential requirements for the neurogenic gene almondex during Drosophila melanogaster development. Genesis 37: 113–122. 10.1002/gene.10233 [DOI] [PubMed] [Google Scholar]
- Michellod M. A., and Randsholt N. B., 2008. Implication of the Drosophila beta-amyloid peptide binding-like protein AMX in Notch signaling during early neurogenesis. Brain Res. Bull. 75: 305–309. 10.1016/j.brainresbull.2007.10.060 [DOI] [PubMed] [Google Scholar]
- Moroz L. L., Kocot K. M., Citarella M. R., Dosung S., Norekian T. P. et al. , 2014. The ctenophore genome and the evolutionary origins of neural systems. Nature 510: 109–114. 10.1038/nature13400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., and Baltimore D., 1989. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 56: 777–783. 10.1016/0092-8674(89)90682-X [DOI] [PubMed] [Google Scholar]
- Murre C., Voronova A., and Baltimore D., 1991. B-cell- and myocyte-specific E2-box-binding factors contain E12/E47-like subunits. Mol. Cell. Biol. 11: 1156–1160. 10.1128/MCB.11.2.1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy S. E., Dubin A. E., Whitwam T., Jojoa-Cruz S., Cahalan S. M. et al. , 2018. OSCA/TMEM63 are an evolutionarily conserved family of mechanically activated ion channels. eLife 7: 1–17. 10.7554/eLife.41844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhold L. A., and Wold B., 1993. HLH forced dimers: tethering MyoD to E47 generates a dominant positive myogenic factor insulated from negative regulation by Id. Cell 74: 1033–1042. 10.1016/0092-8674(93)90725-6 [DOI] [PubMed] [Google Scholar]
- Nielsen C., 2008. Six major steps in animal evolution: Are we derived sponge larvae? Evol. Dev. 10: 241–257. 10.1111/j.1525-142X.2008.00231.x [DOI] [PubMed] [Google Scholar]
- Nielsen C., 2013. Life cycle evolution: Was the eumetazoan ancestor a holopelagic, planktotrophic gastraea? BMC Evol. Biol. 13: 171 10.1186/1471-2148-13-171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noutahi E., Semeria M., Lafond M., Seguin J., Boussau B. et al. , 2016. Efficient gene tree correction guided by genome evolution. PLoS One 11: e0159559 10.1371/journal.pone.0159559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S., 1970. Evolution by gene duplication. Allen Unwin; Springer-Verlag, London, New York: 10.1007/978-3-642-86659-3 [DOI] [Google Scholar]
- Pacentine I. V., and Nicolson T., 2019. Subunits of the mechano-electrical transduction channel, Tmc1/2b, require Tmie to localize in zebrafish sensory hair cells. PLoS Genet. 15: e1007635 10.1371/journal.pgen.1007635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, B., N. Akyuz, X. P. Liu, Y. Asai, C. Nist-Lund, et al., 2018 TMC1 forms the pore of mechanosensory transduction channels in vertebrate inner ear hair cells. Neuron 99: 736–753 e6. 10.1016/j.neuron.2018.07.033 [DOI] [PMC free article] [PubMed]
- Pan B., Geleoc G. S., Asai Y., Horwitz G. C., Kurima K. et al. , 2013. TMC1 and TMC2 are components of the mechanotransduction channel in hair cells of the mammalian inner ear. Neuron 79: 504–515. 10.1016/j.neuron.2013.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pett W., Adamski M., Adamska M., Francis W. R., Eitel M. et al. , 2019. The role of homology and orthology in the phylogenomic analysis of metazoan gene content. Mol. Biol. Evol. 36: 643–649. 10.1093/molbev/msz013 [DOI] [PubMed] [Google Scholar]
- Pisani D., Pett W., Dohrmann M., Feuda R., Rota-Stabelli O. et al. , 2015. Genomic data do not support comb jellies as the sister group to all other animals. Proc. Natl. Acad. Sci. USA 112: 15402–15407. 10.1073/pnas.1518127112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam N. H., Srivastava M., Hellsten U., Dirks B., Chapman J. et al. , 2007. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317: 86–94. 10.1126/science.1139158 [DOI] [PubMed] [Google Scholar]
- Qiu X., and Muller U., 2018. Mechanically gated ion channels in mammalian hair cells. Front. Cell. Neurosci. 12: 100 10.3389/fncel.2018.00100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranade S. S., Syeda R., and Patapoutian A., 2015. Mechanically activated ion channels. Neuron 87: 1162–1179. Erratum: 433. 10.1016/j.neuron.2015.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter D. J., Fozouni P., Eisen M. B., and King N., 2018. Gene family innovation, conservation and loss on the animal stem lineage. eLife 7: 1–43. 10.7554/eLife.34226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., and Huelsenbeck J. P., 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A. et al. , 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61: 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan J. F., Pang K., Schnitzler C. E., Nguyen A. D., Moreland R. T. et al. , 2013. The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science 342: 1242592 10.1126/science.1242592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sans C. L., Satterwhite D. J., Stoltzman C. A., Breen K. T., and Ayer D. E., 2006. MondoA-Mlx heterodimers are candidate sensors of cellular energy status: Mitochondrial localization and direct regulation of glycolysis. Mol. Cell. Biol. 26: 4863–4871. 10.1128/MCB.00657-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar P., Bosneaga E., and Auer M., 2009. Plant cell walls throughout evolution: Towards a molecular understanding of their design principles. J. Exp. Bot. 60: 3615–3635. 10.1093/jxb/erp245 [DOI] [PubMed] [Google Scholar]
- Sauvageau E., Rochdi M. D., Oueslati M., Hamdan F. F., Percherancier Y. et al. , 2014. CNIH4 interacts with newly synthesized GPCR and controls their export from the endoplasmic reticulum. Traffic 15: 383–400. 10.1111/tra.12148 [DOI] [PubMed] [Google Scholar]
- Schwenk J., Harmel N., Zolles G., Bildl W., Kulik A. et al. , 2009. Functional proteomics identify cornichon proteins as auxiliary subunits of AMPA receptors. Science 323: 1313–1319. 10.1126/science.1167852 [DOI] [PubMed] [Google Scholar]
- Sebé-Pedrós A., Chomsky E., Pang K., Lara-Astiaso D., Gaiti F. et al. , 2018. Early metazoan cell type diversity and the evolution of multicellular gene regulation. Nat. Ecol. Evol. 2: 1176–1188. 10.1038/s41559-018-0575-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon M. P., 1972. Characterization of the female-sterile mutant almondex of Drosophila melanogaster. Genetica 43: 244–256. 10.1007/BF00123632 [DOI] [PubMed] [Google Scholar]
- Shannon M. P., 1973. The development of eggs produced by the female-sterile mutant almondex of Drosophila melanogaster. J. Exp. Zool. 183: 383–400. 10.1002/jez.1401830312 [DOI] [PubMed] [Google Scholar]
- Shen X. X., Hittinger C. T., and Rokas A., 2017. Contentious relationships in phylogenomic studies can be driven by a handful of genes. Nat. Ecol. Evol. 1: 126 10.1038/s41559-017-0126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata S. B., Ranum P. T., Moteki H., Pan B., Goodwin A. T. et al. , 2016. RNA interference prevents autosomal-dominant hearing loss. Am. J. Hum. Genet. 98: 1101–1113. 10.1016/j.ajhg.2016.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simion P., Philippe H., Baurain D., Jager M., Richter D. J. et al. , 2017. A large and consistent phylogenomic dataset supports sponges as the sister group to all other animals. Curr. Biol. 27: 958–967. 10.1016/j.cub.2017.02.031 [DOI] [PubMed] [Google Scholar]
- Smedley D., Haider S., Ballester B., Holland R., London D. et al. , 2009. BioMart–biological queries made easy. BMC Genomics 10: 22 10.1186/1471-2164-10-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogabe S., Hatleberg W. L., Kocot K. M., Say T. E., Stoupin D. et al. , 2019. Pluripotency and the origin of animal multicellularity. Nature 570: 519–522. 10.1038/s41586-019-1290-4 [DOI] [PubMed] [Google Scholar]
- Sørensen I., Domozych D., and Willats W. G. T., 2010. How have plant cell walls evolved? Plant Physiol. 153: 366–372. 10.1104/pp.110.154427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M., Begovic E., Chapman J., Putnam N. H., Hellsten U. et al. , 2008. The Trichoplax genome and the nature of placozoans. Nature 454: 955–960. 10.1038/nature07191 [DOI] [PubMed] [Google Scholar]
- Srivastava M., Simakov O., Chapman J., Fahey B., Gauthier M. E. et al. , 2010. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466: 720–726. 10.1038/nature09201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swafford A. J., and Oakley T. H., 2018. Multimodal sensorimotor system in unicellular zoospores of a fungus. J. Exp. Biol. 221: jeb163196 10.1242/jeb.163196 [DOI] [PubMed] [Google Scholar]
- Taylor J. S., and Raes J., 2004. Duplication and divergence: the evolution of new genes and old ideas. Annu. Rev. Genet. 38: 615–643. 10.1146/annurev.genet.38.072902.092831 [DOI] [PubMed] [Google Scholar]
- Vilella A. J., Severin J., Ureta-Vidal A., Heng L., Durbin R. et al. , 2009. EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 19: 327–335. 10.1101/gr.073585.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh J. B., 1995. How often do duplicated genes evolve new functions? Genetics 139: 421–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Li G., Liu J., Liu J., and Xu X. Z., 2016. TMC-1 mediates alkaline sensation in C. elegans through nociceptive neurons. Neuron 91: 146–154. 10.1016/j.neuron.2016.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan N. V., Kocot K. M., Moroz L. L., and Halanych K. M., 2015. Error, signal, and the placement of Ctenophora sister to all other animals. Proc. Natl. Acad. Sci. USA 112: 5773–5778. 10.1073/pnas.1503453112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan S., and Goldman N., 2001. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18: 691–699. 10.1093/oxfordjournals.molbev.a003851 [DOI] [PubMed] [Google Scholar]
- Woese C. R., and Fox G. E., 1977. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl. Acad. Sci. USA 74: 5088–5090. 10.1073/pnas.74.11.5088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Grillet N., Zhao B., Cunningham C., Harkins-Perry S. et al. , 2017. Mechanosensory hair cells express two molecularly distinct mechanotransduction channels. Nat. Neurosci. 20: 24–33. 10.1038/nn.4449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita H., Takenoshita M., Sakurai M., Bruick R. K., Henzel W. J. et al. , 2001. A glucose-responsive transcription factor that regulates carbohydrate metabolism in the liver. Proc. Natl. Acad. Sci. USA 98: 9116–9121. 10.1073/pnas.161284298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura H., Shibata S. B., Ranum P. T., and Smith R. J., 2018. Enhanced viral-mediated cochlear gene delivery in adult mice by combining canal fenestration with round window membrane inoculation. Sci. Rep. 8: 2980 10.1038/s41598-018-21233-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young S. L., Diolaiti D., Conacci-Sorrell M., Ruiz-Trillo I., Eisenman R. N. et al. , 2011. Premetazoan ancestry of the Myc-Max network. Mol. Biol. Evol. 28: 2961–2971. 10.1093/molbev/msr132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue X., Zhao J., Li X., Fan Y., Duan D., et al. , 2018. TMC proteins modulate egg laying and membrane excitability through a background leak conductance in C. elegans Neuron 97: 571–585 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Vinther J., Parry L. A., Wei F., Green E. et al. , 2019. Cambrian sessile, suspension feeding stem-group ctenophores and evolution of the comb jelly body plan. Curr. Biol. 29: 1112–1125.e2. 10.1016/j.cub.2019.02.036 [DOI] [PubMed] [Google Scholar]
- Zuckerkandl E., and Pauling L., 1965. Molecules as documents of evolutionary history. J. Theor. Biol. 8: 357–366. 10.1016/0022-5193(65)90083-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data files, including sequence files (*.fas), multiple sequence alignment files (*.masx and *.nexus), and curation documents, are provided with the Supporting Information as a zipped archive. Four files are provided for each of the three gene families shown for a total of 12 files. Supplemental material available at figshare: https://doi.org/10.25387/g3.11444400.