Abstract
The Mos1-mediated Single-Copy Insertion (MosSCI) method is widely used to establish stable Caenorhabditis elegans transgenic strains. Cloning MosSCI targeting plasmids can be cumbersome because it requires assembling multiple genetic elements including a promoter, a 3′UTR and gene fragments. Recently, Schwartz and Jorgensen developed the SapTrap method for the one-step assembly of plasmids containing components of the CRISPR/Cas9 system for C. elegans. Here, we report on the adaptation of the SapTrap method for the efficient and modular assembly of a promoter, 3′UTR and either 2 or 3 gene fragments in a MosSCI targeting vector in a single reaction. We generated a toolkit that includes several fluorescent tags, components of the ePDZ/LOV optogenetic system and regulatory elements that control gene expression in the C. elegans germline. As a proof of principle, we generated a collection of strains that fluorescently label the endoplasmic reticulum and mitochondria in the hermaphrodite germline and that enable the light-stimulated recruitment of mitochondria to centrosomes in the one-cell worm embryo. The method described here offers a flexible and efficient method for assembly of custom MosSCI targeting vectors.
Keywords: MosSCI, C. elegans, mitochondria, endoplasmic reticulum, SapTrap
The rich toolbox of techniques available to manipulate gene expression in C. elegans is a major attraction of this model organism. Several approaches have been developed to introduce transgenes and to induce efficient CRISPR/Cas9 mediated gene editing (Nance and Frøkjær-Jensen 2019). The Mos1-mediated Single-Copy Insertion (MosSCI) method has been widely adopted to introduce transgenes in C. elegans because single-copy transgenes are integrated at defined chromosomal positions, thereby mitigating potential concerns of transgene integration at random positions (Frøkjær-Jensen et al. 2008; Frøkjær-Jensen et al. 2012; Frøkjær-Jensen et al. 2014). MosSCI transgene integration results from homologous recombination between a MosSCI targeting vector containing the transgene construct and one of the safe-harbor integration sites that have been engineered at defined positions in the genome.
Transgenes typically include multiple genetic elements including a promoter, one or more gene fragments and a 3′UTR. A number of strategies can be used to assemble these elements together including traditional restriction enzyme cloning, Gateway cloning (Hartley et al. 2000), in vivo recombineering (Philip et al. 2019) or Gibson cloning (Gibson et al. 2009). Each of these strategies has both advantages and disadvantages. For example, Gateway cloning allows the efficient modular “mix and match” cloning of large collections of promoter, ORF and 3′UTR cassettes (Brasch et al. 2004; Dupuy et al. 2004; Mangone et al. 2010; Zeiser et al. 2011). However, Gateway cloning can be expensive due to the required use of proprietary enzyme mixes and leaves ∼25 base pair att recombination site “scars” at each cassette junction. In contrast, Gibson cloning allows the efficient, “scar-free” assembly of multiple gene fragments but does not allow the “mix and match” cloning of existing cassettes, making this approach laborious if many constructs are needed.
Schwartz and Jorgensen recently developed the SapTrap method for efficient, modular and single step assembly of CRISPR/Cas9 vectors for C. elegans (Schwartz and Jorgensen 2016). The SapTrap method is based on the Golden Gate cloning technique (Engler et al. 2008) and takes advantage of the SapI type II restriction enzyme, which cuts DNA at defined positions adjacent to its recognition sequence to generate three-base 5′ overhangs. By designing SapI restriction fragments with complementary overhangs, multiple fragments can be assembled together in a defined order in a single digestion and ligation reaction. In this study, we report on the adaptation of the SapTrap system for the efficient, inexpensive, modular, and “scar-free” assembly of transgenes in a MosSCI targeting vector. We have developed a toolkit for expression of transgenes in the C. elegans germline, including a collection of cassettes containing tags for fluorescence imaging and for the ePDZ/LOV optogenetic system (Strickland et al. 2012; Fielmich et al. 2018). As a proof of principle, we have used this system to generate a collection of mitochondrial and endoplasmic reticulum reporter strains and a strain in which light induces the transport of mitochondria to centrosomes in the one-cell worm embryo.
Materials and Methods
C. elegans
C. elegans hermaphrodite strains were maintained at either 20° or 25° on Nematode Growth Medium (NGM) plates containing 3 g/L NaCl, 2.5 g/L peptone and 17 g/L agar supplemented with 1 mM CaCl2, 1 mM MgSO4, 1 mM KPO4 and 5 mg/L cholesterol with E. coli OP50 as a source of food. All strains used in this study are listed in Table 1.
Table 1. Strains used in this study.
| Strain | Genotype | Construction | Reference: |
|---|---|---|---|
| EG8078 | oxTi185 I; unc-119(ed3) III | Frøkjær-Jensen et al. (2014) | |
| EG8079 | oxTi179 II; unc-119(ed3) III | Frøkjær-Jensen et al. (2014) | |
| EGD329 | egxSi126 [mex-5p::hsp-3(aa 1-19)::halotag::hdel::pie-1 3′UTR + unc119(+)] I; unc-119(ed3) III | Injected pJF13 into EG8078 | This study |
| EGD412 | egxSi136 [mex-5p::tomm-20::halotag::pie-1 3′UTR + unc119(+)] II; unc-119(ed3) III | Injected pJF17 into EG8079 | This study |
| EGD496 | egxSi117 [pmex-5p::npp-20::gfp;;pie-1 3′UTR + unc119(+)] I; unc-119(ed3) III | Injected pXF253 into EG8078 | This study |
| EGD497 | egxSi118 [mex-5p::npp-20::halotag::pie-1 3′UTR + unc119(+)] II; unc-119 (ed3) III | Injected pXF255 into EG8079 | This study |
| EGD549 | egxSi144 [mex-5p::cox-4::halotag::pie-1 3′UTR + unc119(+)] II; unc-119 (ed3) III | Injected pXF266 into EG8079 | This study |
| EGD565 | egxSi145 [mex-5p::hsp-3(aa 1-19)::halotag::hdel::pie-1 3′UTR + unc119(+)] II; unc-119 (ed3) III | Injected pJF13 into EG8079 | This study |
| EGD623 | egxSi152 [mex-5p::tomm-20::gfp::pie-1 3′UTR + unc119(+)] II; unc-119(ed3) III | Injected pSM16 into EG8079 | This study |
| EGD629 | egxSi155 [mex-5p::tomm-20::mkate2::pie-1 3′UTR + unc119(+)] II; unc-119(ed3) III | Injected pSM20 into EG8079 | This study |
| EGD631 | egxSi157 [mex-5p::tomm-20::dendra2::pie-1 3′UTR + unc119(+)] II; unc-119(ed3) III | Injected pSM17 into EG8079 | This study |
| EGD633 | egxSi159 [mex-5p::tomm-20::mscarlet::pie-1 3′UTR + unc119(+)] II; unc-119(ed3) III | Injected pSM22 into EG8079 | This study |
| EGD615 | cox-4(zu476[cox-4::eGFP::3XFLAG]) I; egxSi136 [mex-5p::tomm-20::halotag::pie-1 3′UTR + unc119(+)] II; unc-119(ed3?) III | Crossed EGD412 and JJ2586 | This study |
| JJ2586 | cox-4(zu476[cox-4::eGFP::3XFLAG]) I | Raiders et al. 2018 | |
| TBD307 | dhc-1(he255[epdz::mcherry::dhc-1]) I; utdSi51(mex-5p::tomm-20(aa 1-55)::halotag::lov::tbb-2 3′UTR + unc119(+)) II | Injected pSDH68 into EG8079. Crossed to SV2095. | This study |
| SV2095 | dhc-1(he255[epdz::mcherry::dhc-1]) I; ruls57[pie-1p::gfp::tbb-2 + unc119(+)] V | Fielmich et al. 2018 |
Cloning
To generate the expression vector pXF87, the two SapI restriction sites in pCFJ350 (Frøkjær-Jensen et al. 2012) were mutated using Q5 Site-Directed Mutagenesis (New England Biolabs (NEB)) with the oligo pairs XF30F/XF30R and XF31F/XF31R. In addition, the annealed oligos Eg717 and Eg718 were cloned between the XhoI and SpeI sites of pCFJ350.
HaloTag and ceGFP containing PATC-rich endogenous introns were generated in several steps. First, genes were designed in silico to minimize germline silencing and increase expression by codon adaptation (Redemann et al. 2011), removal of homology to piRNAs (Batista et al. 2008), and inclusion of a short endogenous intron from rpl-18 and four synthetic introns (Okkema et al. 1993) using the freely available gene editor ApE (M. Wayne Davis, https://jorgensen.biology.utah.edu/wayned/ape/). Second, the synthetic genes were synthesized as gBlocks (IDT), cloned into a plasmid, and sequence verified. Third, PATC-rich introns from a gene that is resistant to germline silencing, smu-1 (Spike et al. 2001), were introduced into the synthetic genes by Golden Gate cloning as described previously (Frøkjær-Jensen et al. 2016). Finally, correct splicing and expression was verified by expression of the synthetic genes with and without PATC-rich introns using an eft-3 promoter and tbb-2 3′UTR.
Donor cassette plasmids numbered pXF, pJF and pSM were generated by cloning PCR products into the pCR BluntII vector backbone using the Zero Blunt Topo system (Thermo Fisher Scientific). pSDH donor cassette plasmids were cloned by ligating PCR products into pSDH76, a derivative of pCR BluntII containing two XcmI sites that generate T-overhangs following digestion with XcmI. pXF87 and all donor plasmids were sequence verified.
To assemble HSP-3 (aa 1-19) into the second cassette of the expression vector pJF13, 10 μM of oligos XF17F and XF17R were gradually cooled from 95° to 25° in a BioRad T1000 thermocycler. Annealed oligos were phosphorylated by T4 polynucleotide kinase (NEB) for two hours at 37°, followed by an enzyme inactivation step at 65° for 20 min. The donor plasmids and primers are listed in Tables 2 and 3, respectively.
Table 2. Donor cassette plasmids used in this study.
| Name | Description |
|---|---|
| Cassette 1 for 4-cassette or 5 cassette system (5′-TGG ...... 3′-TAC) | |
| pXF121 | mex-5 promoter |
| pSDH60 | spe-11 promoter |
| Cassette 2 for 4-cassette or 5-cassette system (5′-ATG-3′ ...... 3′-CCA-5′) | |
| Tags | |
| pXF89 | halotag (no STOP codon, PATC introns) |
| pJF5 | gfp (no STOP codon, PATC introns) |
| pXF222 | mkate2 (no STOP codon) |
| pSDH61 | epdz (no STOP codon) |
| pSM10 | mscarlet (no STOP codon) |
| pSM12 | dendra2 (no STOP codon) |
| Genes | |
| pJF7 | tomm-20 (no STOP codon) |
| pSDH50 | tomm-20 (aa 1-55) (no STOP codon) |
| pXF262 | cox-4 (no STOP codon) |
| pXF250 | npp-20 (no STOP codon) |
| Cassette 3 for 4-cassette system (5′-GGT-3′ ...... 3′-ATT-5′) | |
| Tags | |
| pXF88 | halotag (includes STOP codon, PATC introns) |
| pJF6 | gfp (includes STOP codon, PATC introns) |
| pXF130 | mkate2 (includes STOP codon) |
| pSM08 | mscarlet (includes STOP codon) |
| pSM03 | dendra2 (includes STOP codon) |
| ORFs | |
| pXF90 | halotag::hdel (includes STOP codon, PATC introns) |
| Cassette 3A for 5-cassette system (5′-GGT-3′ ...... 3′-TGC-5′) | |
| Tags | |
| pSDH51 | halotag (no STOP codon, PATC introns) |
| pSM04 | mkate2 (no STOP codon) |
| pSDH57 | mscarlet (no STOP codon) |
| Cassette 3B for 5-cassette system (5′-ACG-3′ ...... 3′-ATT-5′) | |
| Tags | |
| pXF276 | lov domain (includes STOP codon) |
| pSDH52 | epdz (includes STOP codon) |
| pSM05 | mkate2 (includes STOP codon) |
| Cassette 4 for 4-cassette or 5-cassette system (5′-TAA-3′ ...... 3′-CAT-5′) | |
| pXF85 | pie-1 3′UTR |
| pSDH54 | tbb-2 3′UTR |
| pSDH66 | unc-54 3′UTR |
Table 3. Primers used in this study.
| Name | Description | Sequence (SAP1 site and Overhang) | Corresponding plasmid |
|---|---|---|---|
| XF32F | mex-5 promoter (F) | GCAGCTCTTCGTGGATATCAGTTTTTAAAAAATTA | pXF121 |
| XF32R | mex-5 promoter (R) | GCAGCTCTTCGCATTCTCTGTCTGAAACA | |
| JF5F | tomm-20 (F) | GCAGCTCTTCGATGTCGGACACAATTCTTGG | pJF7 |
| JF5R | tomm-20 (R) | GCAGCTCTTCGACCCTCCAAGTCGTCGGTGTC | |
| JF1F | gfp (F) | GCAGCTCTTCGATGTCCAAGGTAACACTTAGTTT | pJF5 |
| JF1R | gfp (R) | GCAGCTCTTCGACCGCCGCTTCCCTTGTAGAGCTCGTCCAT | |
| JF2F | gfp (F) | GCAGCTCTTCGGGTGGAAGCGGCTCCAAGAACACTTAGTTT | pJF6 |
| JF2R | gfp (R) | GCAGCTCTTCGTTACTTGTAGAGCTCGTCCAT | |
| XF17F | hsp-3 (1-19aa) (F) | ATGAAGACCTTATTCTTGTTGGGCTTGATCGCCCTATCCGCCGTCAGTGTCTACTGC | |
| XF17R | hsp-3 (1-19aa) (R) | ACCGCAGTAGACACTGACGGCGGATAGGGCGATCAAGCCCAACAAGAATAAGGTCTT | |
| spe-11(SAP C1) F | spe-11 promoter (F) | GCAGCTCTTCGTGGGTCGACAGAACATTTTTCCGT | pSDH60 |
| spe-11(SAP C1) R | spe-11 promoter (R) | GCAGCTCTTCGCATTTTATCTAGTCGGTTTGCGA | |
| XF24F | halotag (F) | GCAGCTCTTCGATGGCCGAGGTAACACTTAGTTTTTGT | pXF89 |
| XF24R | halotag (R) | GCAGCTCTTCGACCGCCGCTTCCTCCGGAGATCTCGAGGGT | |
| XF63F | mkate2 (F) | GCAGCTCTTCGATGGTCTCCGAGCTCATTAAAGAAAACA | pXF222 |
| XF63R | mkate2 (R) | GCAGCTCTTCGACCACCTCCACCTCCACGGTGTCCGAGCTTGG | |
| ePDZ (SAP C2) F | epdz (F) | GCAGCTCTTCGATGCCAGAGCTCGGATTCTCGAT | pSDH61 |
| ePDZ (SAP C2) R | epdz (R) | GCAGCTCTTCGACCAGCTCCCGTCGCGACGGGTGGATCAC | |
| XF79F | cox-4 (F) | GCAGCTCTTCGATGATGCTGCCACGTTTG | pXF262 |
| XF79R | cox-4 (R) | GCAGCTCTTCGACCCTTCCACTTCTTGTTCTCGTAATC | |
| XF76F | npp-20 (F) | GCAGCTCTTCGATGACCACGGTCCGCCAG | pXF250 |
| XF76R | npp-20 (R) | GCAGCTCTTCGACCTCTCTGAGCTCCCGGAGCT | |
| XF23F | halotag (F) | GCAGCTCTTCGGGTGGAAGCGGCGCCGAGGTAACACTTAGTTTTTGT | pXF88 |
| XF23R | halotag (R) | GCAGCTCTTCGTTATCCGGAGATCTCGAGGGT | |
| XF53F | mkate2 (F) | GCAGCTCTTCGGGTGGAGGTGGAGGTGTCTCCGAGCTCATTAAAGAAAAC | pXF130 |
| XF53R | mkate2 (R) | GCAGCTCTTCGTTAACGGTGTCCGAGCTTGGA | |
| XF22F | halotag::hdel (F) | GCAGCTCTTCGGGTGGAAGCGGCGCCGAGGTAACACTTAGTTTTTGT | pXF90 |
| XF22R | halotag::hdel (R) | GCAGCTCTTCGTTAGAGTTCGTCATGTCCGGAGATCTCGAGGGT | |
| SIM8F | mscarlet (F) | GCAGCTCTTCGATGGTCTCCAAGGGCGAGGCA | pSM10 |
| SIM8R | mscarlet (R) | GCAGCTCTTCGACCACCTCCACCTCCCTTGTACAGCTCGTCCATTCCT | |
| SIM10F | dendra2 (F) | GCAGCTCTTCGATGAACCTTATTAAGGAAGATATG | pSM12 |
| SIM10R | dendra2 (R) | GCAGCTCTTCGACCGCCGCTTCCCCATACTTGACTTGGTAG | |
| SIM1F | dendra2 (F) | GCAGCTCTTCGGGTGGAAGCGGCAACCTTATTAAGGAAGATATG | pSM03 |
| SIM1R | dendra2 (R) | GCAGCTCTTCGTTACCATACTTGACTTGGTAG | |
| SIM2F | mkate2 (F) | GCAGCTCTTCGGGTGGAGGTGGAGGTGTCTCCGAGCTCATTAAAGAAAACA | pSM04 |
| SIM2R | mkate2 (R) | GCAGCTCTTCGCGTACCTCCACCTCCACGGTGTCCGAGCTTGGA | |
| SIM3F | mkate2 (F) | GCAGCTCTTCGACGGGAGGTGGAGGTGTCTCCGAGCTCATTAAAGAAAACA | pSM05 |
| SIM3R | mkate2 (R) | GCAGCTCTTCGTTAACGGTGTCCGAGCTTGGA | |
| SIM6F | mscarlet (F) | GCAGCTCTTCGGGTGGAGGTGGAGGTGTCTCCAAGGGCGAGGCA | pSM08 |
| SIM6R | mscarlet (R) | GCAGCTCTTCGTTACTTGTACAGCTCGTCCATTCCT | |
| mScarlet (SAPC3)F | mscarlet (F) | GCAGCTCTTCGGGTGTCTCCAAGGGCGAGGCAGTCAT | pSDH57 |
| mScarlet (SAPC3)R | mscarlet (R) | GCAGCTCTTCGCGTGGCCGCGGCTTTTGCAGCGG | |
| XF84F | lov (F) | GCAGCTCTTCGACGCCTCGTCTTGCTGCT | pXF276 |
| XF84R | lov (R) | GCAGCTCTTCGTTAGACCCAAGTGTCGACGGC | |
| XF12F | pie-1 3′UTR (F) | GCAGCTCTTCGTAATTTTGCCGTATTTTCCAT | pXF85 |
| XF12R | pie-1 3′UTR (R) | GCAGCTCTTCGTACATCATCGTTCACTTTTCAC | |
| tbb2 3′UTR (SAPC5)F | tbb-2 3′UTR (F) | GCAGCTCTTCGTAAATGCAAGATCCTTTCAAGCATTC | pSDH54 |
| tbb2 3′UTR (SAPC5)R | tbb-2 3′UTR (R) | GCAGCTCTTCGTACGACTTTTTTCTTGGCGGCAC | |
| Halo (SAP C3)F | halotag (F) | GCAGCTCTTCGGGTGGAAGC | pSDH51 |
| Halo (SAP C3)R | halotag (R) | GCAGCTCTTCGCGTTCCGGAGATCTCGAGGGTGG | |
| ePDZ (SAP C4)F | epdz (F) | GCAGCTCTTCGACGGGAGGTTCCGGAGGATCTGGC | pSDH52 |
| ePDZ (SAP C4)R | epdz (R) | GCAGCTCTTCGTTACGTCGCGACGGGTGGAT | |
| unc-54 (SAPC5)F | unc-54 3′UTR (F) | GCAGCTGTTCGTAAGAGCTCCGCATCGGCCGCTG | pSDH66 |
| unc-54 (SAPC5)R | unc-54 3′UTR (R) | GCAGCTCTTCGTACAAACAGTTATGTTTGGTATATTGGGA | |
| Eg717 | Replace pCFJ350 MCS (F) | TCGAGTGGCGAAGAGCCCATGGATCCCATATGGAATTCTGCAGGCCTGCTCTTCGGTAA | pXF87 |
| Eg718 | Replace pCFJ350 MCS (R) | CTAGTTACCGAAGAGCAGGCCTGCAGAATTCCATATGGGATCCATGGGCTCTTCGCCAC | |
| XF30F | Mutate SapI site in pCFJ350 | GATTATGGGCACTTCTTTTATCC | pXF87 |
| XF30R | Mutate SapI site in pCFJ350 | CGACAAGCAACTTTTCTATAC | |
| XF31F | Mutate SapI site in pCFJ350 | AATGGCGAAGtGCAAAGCAGAG | pXF87 |
| XF31R | Mutate SapI site in pCFJ350 | GTTTCCTGAAAATAATGTAACTTGAATTG |
Note: For the expression plasmid pJF13 annealed oligos were used to generate HSP-3(aa 1-19) in cassette 2.
Additional oligo sequences used to generate pSDH50:
TOMM-20 short forward. GCAGCTCTTCGATGTCGGACACAATTCTTGGTTTCAAcaaatcaaacgtcgttttggctgctggaattgctggagccgctttcctcggctactgcatttacttcgatcataagagaatcaacgctccagactacaaggacaagattaggcaaagtcagtgttttaacaacatatttccttcggatttttatctaaaaacaacttattttctttcagagagaCGTGCCCAGGCTGGAGCAggagctggtgcaggcgctggagccggagccGGTCGAAGAGCtgc.
TOMM-20 short reverse GCAGCTCTTCGACCggctccggctccagcgcctgcaccagctccTGCTCCAGCCTGGGCACGtctctctgaaagaaaataagttgtttttagataaaaatccgaaggaaatatgttgttaaaacactgactttgcctaatcttgtccttgtagtctggagcgttgattctcttatgatcgaagtaaatgcagtagccgaggaaagcggctccagcaattccagcagccaaaacgacgtttgatttgTTGAAACCAAGAATTGTGTCCGACATCGAAGAGCtgc.
Assembly reaction
Assembly reactions (total final volume of 50 μL) included 1 nM of pXF87 and of each donor cassette plasmid, 400 units of T4 DNA ligase (NEB), 10 units of SapI enzyme (NEB), 1x NEB CutSmart buffer and 1 mM ATP. For assemblies including annealed oligos, phosphorylated annealed oligos were used at a final concentration of 3 nM in the assembly reaction. Reactions were incubated for 22-24 hr at 25°, and transformed into Stellar Competent cells (Clontech). Four to six plasmid clones were first screened by restriction digest with XhoI and SpeI. Plasmids with the correct restriction digest pattern were sequenced across each cassette boundary. MosSCI targeting vector assembly reactions are listed in Table 4. Note that because the background of unassembled vectors in our assembly reactions was typically low, our protocol omits the counterselection restriction enzyme step described in the original SapTrap protocol (Schwartz and Jorgensen 2016).
Table 4. MosSCI targeting vectors used in this study.
| Name | Comments | Assembly | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| pXF87 | MosSCI backbone | Derived from pCFJ350 | |||||||||
| Donor vectors used for assembly | Assembly efficiency | ||||||||||
| Cassettes | Digestion | Sequencing | |||||||||
| 1 | 2 | 3 | 4 | ||||||||
| pJF13 | ER lumen, Halotag | pXF121 | XF17F/Ra | pXF90 | pXF85 | 4/5 | 2/2 | ||||
| pJF17 | Mitochondrial OM, Halotag | pXF121 | pJF7 | pXF88 | pXF85 | 4/5 | 1/2 | ||||
| pXF253 | ERES + nuclear pores (NPP-20), GFP | pXF121 | pXF 250 | pJF6 | pXF85 | 4/6 | 2/2 | ||||
| pXF255 | ERES + nuclear pores (NPP-20), Halotag | pXF121 | pXF 250 | pXF88 | pXF85 | 5/6 | 2/2 | ||||
| pXF266 | Mitochondrial matrix, Halotag | pXF121 | pXF 262 | pXF88 | pXF85 | 1/4 | 1/1 | ||||
| pSM20 | Mitochondrial OM, mKate2 | pXF121 | pJF7 | pXF130 | pXF85 | 4/5 | 2/2 | ||||
| pSM22 | Mitochondrial OM, mScarlet | pXF121 | pJF7 | pSM08 | pXF85 | 4/5 | 2/2 | ||||
| pSM17 | Mitochondrial OM, Dendra2 | pXF121 | pJF7 | pSM03 | pXF85 | 4/5 | 2/2 | ||||
| pSM16 | Mitochondrial OM, GFP | pXF121 | pJF7 | pJF6 | pXF85 | 2/5 | 2/2 | ||||
| 1 | 2 | 3A | 3B | 4 | |||||||
| pSDH68 | Mitochondrial OM, Halotag, LOV | pXF121 | pSDH50 | pSDH51 | PCR fragment | pSDH54 | 11/15 | 2/2 | |||
Annealed oligos.
Transgenesis
Double-stranded breaks at Mos1 landing sites were generated using CRISPR/Cas9. With the exception of strains EGD623, EGD629, EGD631 and EGD633, injection mixes contained 50 ng/μL of each of the following vectors: an assembled MosSCI targeting vector, pXW7.01 and pXW7.02 sgRNA/Cas9 vectors (gifts from Katya Voronina, University of Montana), which direct Cas9 to generate double-stranded breaks at the ttTi5605 universal MosSCI insertion site. For strains EGD623, EGD629, EGD631 and EGD633, injection mixes contained 0.25 μg/μL Cas9 protein, 0.1 μg/μL tracrRNA, 0.028 μg/μL crRNA BH0278 (GCGUCUUCGUACCUUUUUGGGUUUUAGAGCUAUGCUGUUUUG), 0.028 μg/μL crRNA BH0279 (GUCCCAUCGAAGCGAAUAGGGUUUUAGAGCUAUGCUGUUUUG) (Dharmacon) and 0.1 μg/μL assembled MosSCI targeting plasmid. The universal MosSCI strains EG8078 or EG8079 (Frøkjær-Jensen et al. 2014) were injected, singled and incubated for 10 days at 20°. ∼10 worms from plates containing non-Unc animals were transferred to new plates. Plates that stably gave rise to non-unc progeny were visually screened for fluorescent transgene expression.
HaloTag staining
20 to 30 L4 worms were stained in 25 μL S medium containing concentrated OP50 bacteria and 2.5 μM of either JF549 HaloTag ligand or JF646 HaloTag ligand (Grimm et al. 2015) in a darkened 96-well plate shaking at 150 rpm for 19 hr at 23°. Water was placed in the neighboring wells to help prevent evaporation. Animals were recovered on NGM plates for up to two hours before imaging.
MitoTracker deep red staining
L4 worms were fed overnight on an NGM plate that had been seeded with 100 μL concentrated OP50 bacteria mixed with 1 μL of 1 mM MitoTracker Deep Red FM dye (Cell Signaling Technology, Cat #8778S).
Imaging
With the exceptions of the TOMM-20::Dendra2 strain and optogenetic strains (Figure 4), all images were collected on a spinning-disk microscope built on a Nikon Eclipse Ti base and equipped with an Andor CSU-W1 two camera spinning disk module, Zyla sCMOS cameras, an Andor ILE laser module and a Nikon 100X Plan Apo 1.45 NA oil immersion objective (Micro Video Instruments, Avon, MA).
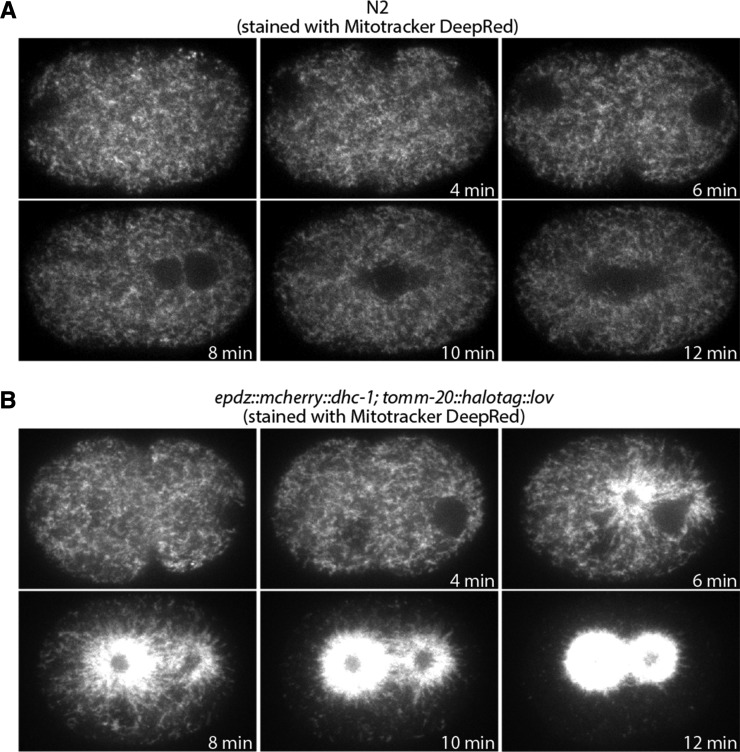
Figure 4.
Optogenetic control of mitochondrial distribution in the 1-cell embryo. A. Control embryo stained with Mitotracker DeepRed and imaged with 488 nm and 640 nm illumination (640 nm channel shown). B. 1-cell epdz::mcherry::dhc-1; tomm-20::halotag::lov embryo stained with Mitotracker DeepRed and imaged with 488 nm and 640 nm illumination (640 nm channel shown). The 488 nm illumination was used to stimulate the interaction between the ePDZ and LOV domains.
TOMM-20::Dendra2 was imaged on a Marianas spinning disk microscope (Intelligent Imaging Innovations) built around a Zeiss Axio Observer Z.1 equipped with a Photometrics Evolve EMCCD camera, 50 mW 488 and 561 nm solid state lasers, a CSU-X1 spinning disk (Yokogawa, Tokyo Japan) and a Zeiss 100X Plan-Apochromat objective. Photoconversion was performed by 5 sec illumination with a 405 epifluorescent light source.
To stimulate the relocalization of mitochondria (Figure 4), embryos were illuminated with a 50 mW 640 nm solid-state laser used to excite MitoTracker DeepRed (20% laser power, 100 msec exposure, camera gain of 1) and a 50 mW 488 nm solid-state laser used to stimulate the interaction between ePDZ and LOV domains (80% laser power and 100 msec exposure). A Plan-Apochromat 100x/1.4 NA oil immersion DIC objective (Zeiss) was used and Z-stacks (one micrometer step size, 11 steps) were collected at 60-second intervals. The images displayed in Figure 4 are maximum intensity projections of three Z planes from the cell midplane.
Data availability
With the exception of EGD633, the C. elegans strains generated in this study have been deposited at the Caenorhabditis Genetics Center (CGC; https://cgc.umn.edu). The plasmids listed in Figures 1 and 3 have been deposited at Addgene (http://www.addgene.org). Other donor plasmids, assembled expression plasmids and EGD633 are available upon request. Supplemental materials describing the sequence of tag donor cassettes are available through the GSA figshare portal: https://doi.org/10.25387/g3.9978611.
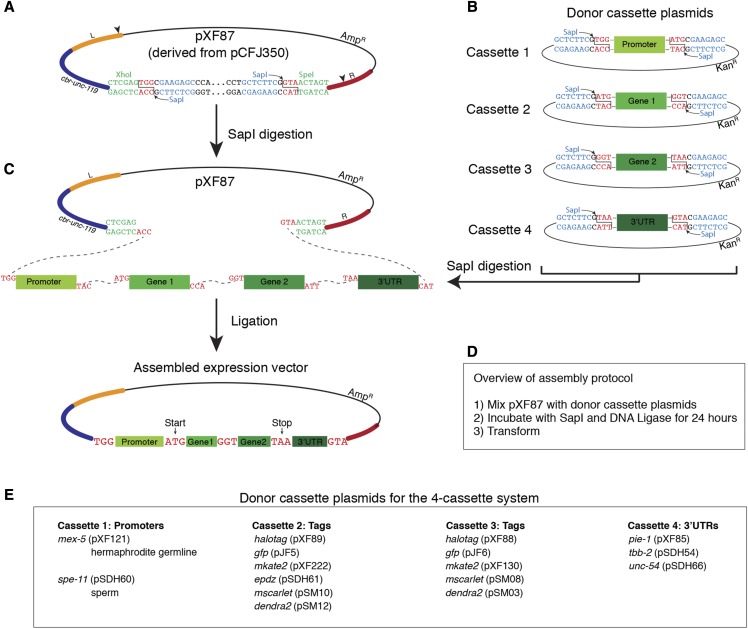
Figure 1.
SapTrap assembly of MosSCI targeting vectors using the four-cassette system. A. The MosSCI targeting vector pXF87 was derived from pCFJ350 by mutating two SapI restriction sites (indicated by arrowheads in the “Left” (L) and “Right” (R) homology arms) and introducing two SapI sites (blue text) between the XhoI and SpeI sites (green text). SapI cleavage sites are in red text. The SapI recognition sites are oriented such that upon digestion they are removed from the vector backbone. The cbr-unc-119 gene is used as a positive selection marker to facilitate the identification of transgenic animals. B. Design of the donor cassette vectors used for the 4-cassette cloning strategy. C. The curved dotted lines indicate the overhangs that anneal during the ligation reaction. D. Overview of the assembly protocol. For a detailed protocol, see the Materials and Methods section. E. Summary of available promoter, gene tag and 3′UTR donor cassette plasmids.
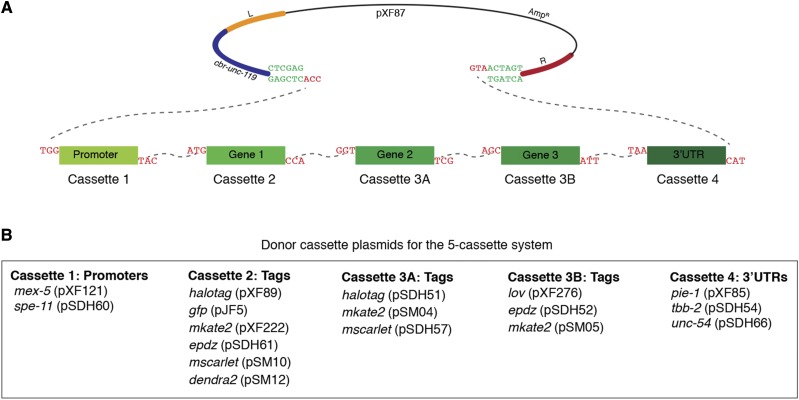
Figure 3.
SapTrap assembly of MosSCI targeting vectors using the five-cassette system. A. Schematic of pXF87 and the donor cassettes following SapI digestion. The dotted lines indicate the overhangs that anneal during ligation. B. Summary of available promoter, gene tag and 3′UTR donor cassette plasmids for the five-cassette system.
Results
Adaptation of the SapTrap system for cloning MosSCI targeting vectors
To adapt the SapTrap approach (Schwartz and Jorgensen 2016) for the assembly of MosSCI targeting vectors, we started by making two changes to the universal MosSCI targeting vector pCFJ350 (Frøkjær-Jensen et al. 2012), which targets transgenes for insertion at the commonly used ttTi5605 site (Frøkjær-Jensen et al. 2008). First, we introduced single base pair changes to disrupt the two SapI restriction sites located in the “Left” and “Right” homology arms of pCFJ350. Second, we inserted two SapI sites into the multiple cloning site that were oriented such that they are removed from the vector backbone by digestion with SapI. The resulting MosSCI targeting vector was named pXF87 (Figure 1A). Although pXF87 is compatible with the standard Mos1-mediated transgenesis protocol, the transgenic strains described in this study were isolated using CRISPR/Cas9 to generate double-stranded breaks in MosSCI integration sites (described in the Methods section).
We next cloned a series of plasmids that contain donor cassettes flanked by SapI restriction sites (Figure 1B). Following digestion with SapI, the cassettes are liberated from the vector backbone and are flanked by 5′ overhangs that direct their order of assembly in pXF87 (Figure 1C). A four-insert cassette system was designed with a promoter in cassette 1, gene fragments in cassettes 2 and 3 (typically a gene and a tag) and a 3′UTR in cassette 4 (Figure 1B). To minimize the inclusion of extraneous sequences, the junction between the first and second cassettes is the translation start (ATG), between second and third cassettes is glycine (GGT) and between the third and fourth cassettes is the ochre translation stop codon (TAA) (Figure 1C). Donor cassettes encoding tags (such as fluorescent proteins) include short flexible linkers at the protein fusion site (the carboxy terminus of cassette 2 and the amino terminus of cassette 3) (Supplemental Figure S1- S7). The currently available promoter, tag and 3′UTR donor cassette plasmids are listed in Figure 1E and Table 2.
The C. elegans germline is a notoriously difficult tissue in which to achieve stable transgene expression due to silencing of multi-copy extra-chromosomal arrays (Kelly et al. 1997), single-copy insertions generated by MosSCI (e.g., (Shirayama et al. 2012; Frøkjær-Jensen et al. 2016)) or endogenous genes tagged using CRISPR/Cas9 gene editing (e.g., (Fielmich et al. 2018)). Each of our tag donor cassettes encoding gene tags incorporates at least one modification that buffers against silencing, including the inclusion of PATC introns in HaloTag and ceGFP (Frøkjær-Jensen et al. 2016), the elimination of piRNA binding sites in mScarlet, mKate2 and Dendra2 (Seth et al. 2018; Zhang et al. 2018) and the use of sequence motifs found in native germline genes in ePDZ and the LOV domain (Fielmich et al. 2018).
Similar to the SapTrap method developed by Schwartz and Jorgensen (Schwartz and Jorgensen 2016), MosSCI targeting vectors were assembled in a single tube by incubating pXF87, four donor cassette plasmids, SapI enzyme, ATP and T4 DNA ligase at 25° for 22 - 24 hr (Figure 1D and Materials and Methods). This reaction was then transformed into E. coli and plasmid clones were screened by restriction enzyme digestion followed by sequencing. We assembled nine vectors using the 4-cassette system and 32 of 46 (69.6%) of the plasmids screened had the correct restriction digest pattern (Table 4). Of the vectors with the correct restriction digest pattern, 22 of 23 were correct based on Sanger sequencing analysis of the cassette junctions. Therefore, the SapTrap method provides an efficient method for the assembly of MosSCI targeting vectors.
A collection of fluorescent ER and mitochondria strains
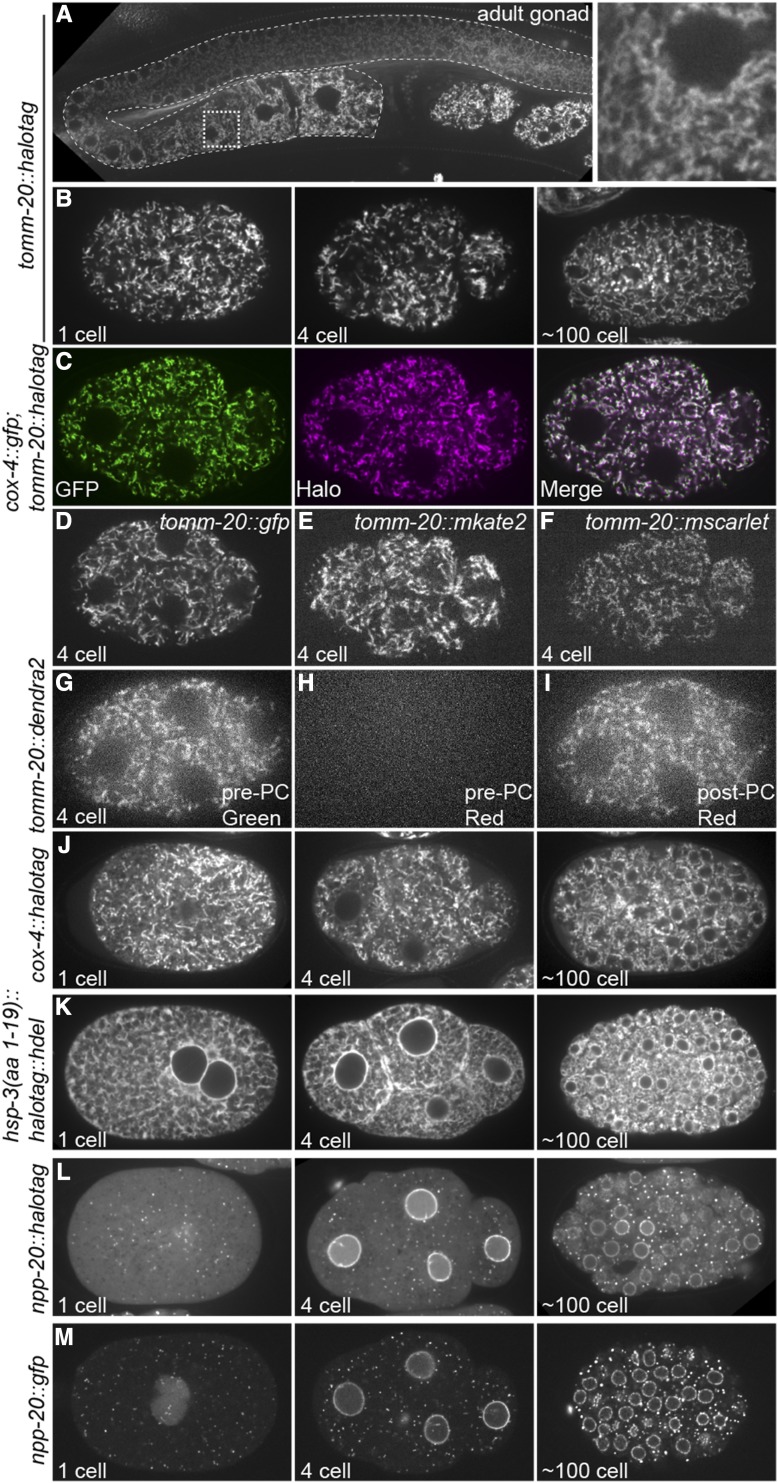
We used SapTrap-assembled MosSCI targeting vectors to generate a collection of transgenic strains for analysis of endoplasmic reticulum and mitochondrial dynamics. We first targeted GFP, mKate2, mScarlet, Dendra2 and HaloTag to the cytoplasmic face of the mitochondrial outer membrane by fusing them to the carboxy terminus of TOMM-20. The expression of these transgenes was controlled by the mex-5 promoter and by the pie-1 3′UTR, which results in germline expression that increases around the bend of the adult hermaphrodite gonad (Merritt et al. 2008) (Figure 2A). Strains expressing TOMM-20 fused to HaloTag were labeled with the fluorescent JF646 HaloTag ligand (Grimm et al. 2015) by feeding hermaphrodites bacteria mixed with the ligand. Each TOMM-20 fusion protein exhibited the expected tubular localization pattern in the early embryo (Figure 2B-I). We confirmed that TOMM-20::HaloTag colocalized to the same organelle as the mitochondrial matrix protein COX-4::GFP (Raiders et al. 2018) (Figure 2C). We additionally generated strains in which the HaloTag was targeted to the mitochondrial matrix (COX-4::HaloTag) (Figure 2J) and the lumen of the endoplasmic reticulum (HSP-3(aa 1-19)::HaloTag::HDEL) (Figure 2K) (Lee et al. 2016). We fused both GFP and HaloTag to NPP-20, the worm homolog of SEC13, which is both a component of the COPII coat that concentrates to ER exit sites (ERES) (D’Arcangelo et al. 2013) and a component of nuclear pore complexes (Siniossoglou et al. 1996) (Figure 2L, M).
Figure 2.
Images of transgenic strains. A. Images of TOMM-20::HaloTag labeled with JF646 HaloTag ligand in the adult gonad (outlined with curved dotted line), including an inset of the region in the stippled box. B. Images of embryos expressing TOMM-20::HaloTag labeled with JF646 HaloTag ligand at the 1-cell, 4 cell and ∼100 cell stages. C. Images of a 4 cell embryo expressing TOMM-20::HaloTag labeled with JF646 HaloTag ligand (magenta) and COX-4::GFP (green) (Raiders et al., 2018). D – F. Images of embryos expressing the indicated transgenes at the 4-cell stage. G – I. Images of a 4 cell embryo expressing TOMM-20::Dendra2 before and after photoconversion (PC). Dendra2 switches from green to red fluorescence upon photoconversion. J – M. Images of embryos expressing the indicated transgenes at the 1-cell, 4 cell and ∼100 cell stages.
Five-cassette system
One of the advantages of the SapTrap approach is that it can be easily expanded to include additional insert fragments to create more complex transgenes. To establish a five-cassette system, we used the cassettes 1, 2 and 4 from the four-cassette system and replaced cassette 3 with cassettes 3A and 3B (Figure 3A and 3B). We used this approach to generate an optogenetic system to control the localization of mitochondria in the early embryo based on the light induced interaction between the ePDZ and LOV domains (Strickland et al. 2012; Fielmich et al. 2018). We assembled a MosSCI targeting vector that directed expression of TOMM-20::HaloTag::LOV, which targets the LOV domain to the mitochondrial outer membrane. 11 of 15 assembled plasmids had the corrected restriction digest pattern and 2 of 2 of these plasmids were correct by Sanger sequence analysis of the cassette junctions. A TOMM-20::HaloTag::LOV strain was crossed with a strain in which the dynein heavy chain DHC-1 was fused to ePDZ (Fielmich et al. 2018). Whereas mitochondria in wild-type embryos are dispersed through the cytoplasm (Figure 4A), upon the recruitment of ePDZ::mCherry::DHC-1 to mitochondria by stimulation with 488 nm light, mitochondria were transported onto centrosomes, leaving the peripheral cytoplasm largely devoid of mitochondria (Figure 4B).
Discussion
The SapTrap system described here provides an efficient and simple method for the assembly of MosSCI targeting vectors. This approach is similar to the Gateway assembly system (ThermoFisher Scientific) in that once donor cassette plasmids are cloned, they can be assembled in any modular combination. The Gateway system has been widely used to generate MosSCI transgenes and is attractive because there are large collections of promoter, ORF, and 3′UTR donor plasmids available (Brasch et al. 2004; Dupuy et al. 2004; Mangone et al. 2010; Zeiser et al. 2011). However, the Gateway system has disadvantages, including i) ∼25 bp att recombination sites present between each cassette after assembly, ii) the cost of proprietary enzyme mixes, and iii) the difficulty in assembling more than four cassettes together. In contrast, the SapTrap system i) uses three-base pair junctions, two of which are designed to encode the translation start and STOP codons, ii) is relatively inexpensive, and iii) can efficiently assemble at least 5 cassettes. In principle, the number of cassettes could be increased if desired. The most significant consideration in generating new donor cassette plasmids for SapTrap assembly is that internal SapI sites cannot be present within the donor cassette sequence. Gibson cloning also allows the “scar-free” cloning of transgene vectors, but the specific cloning strategies must be designed for each unique vector. While we have focused on generating transgenes expressed in the hermaphrodite germline, the MosSCI targeting vector pXF87, the gene tag donor cassettes and cloning approach described here should be readily adaptable to expressing transgenes in other tissues.
The advantages of tagging and fluorescently labeling proteins with the HaloTag include increased brightness and photostability (especially compared to red fluorescent proteins) and excellent optical pairing with green fluorescent proteins for 2-color imaging. Additionally, HaloTag labeling offers the flexibility to label a single strain with either JF549 HaloTag ligand or JF646 HaloTag ligand (Grimm et al. 2015). The disadvantages of HaloTag labeling include the need to introduce the fluorescent ligand (for example, using small scale liquid culture) and the cost of the ligand. Additionally, care should be taken to optimize labeling procedures for each protein to maximize labeling efficiency and minimize background from free ligand. In practice, we find that HaloTag labeling is particularly useful when photobleaching of conventional fluorescent proteins is limiting and/or when imaging in far red is advantageous.
Acknowledgments
We thank Bing He and members of the Griffin lab for comments on the manuscript. We thank Katya Voronina (U. of Montana) for the plasmids pXW7.01 and pXW7.02. We thank Ann Lavanway and Zdenek Svindrych of the Dartmouth Life Sciences Imaging Facility for assistance with imaging. The Molecular Biosciences core facility is supported by the Norris Cotton Cancer Center and by NCI grant 5P30CA023108-40. This work was supported by grants from the NIH (R01GM110194 to EEG), baseline funding from KAUST (to CFJ) and the NWO (016.Veni.181.051 to SDH). Erik Jorgensen (U. of Utah, HHMI) contributed resources to generate some of the transgenes from NIH grant R01GM095817.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25387/g3.9978611.
Communicating editor: K. Gunsalus
Literature Cited
- Batista P. J., Ruby J. G., Claycomb J. M., Chiang R., Fahlgren N. et al. , 2008. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol. Cell 31: 67–78. 10.1016/j.molcel.2008.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasch M. A., Hartley J. L., and Vidal M., 2004. ORFeome cloning and systems biology: standardized mass production of the parts from the parts-list. Genome Res. 14: 2001–2009. 10.1101/gr.2769804 [DOI] [PubMed] [Google Scholar]
- D’Arcangelo J. G., Stahmer K. R., and Miller E. A., 2013. Vesicle-mediated export from the ER: COPII coat function and regulation. Biochim. Biophys. Acta 1833: 2464–2472. 10.1016/j.bbamcr.2013.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy D., Li Q. R., Deplancke B., Boxem M., Hao T. et al. , 2004. A first version of the Caenorhabditis elegans Promoterome. Genome Res. 14: 2169–2175. 10.1101/gr.2497604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C., Kandzia R., and Marillonnet S., 2008. A one pot, one step, precision cloning method with high throughput capability. PLoS One 3: e3647 10.1371/journal.pone.0003647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielmich L. E., Schmidt R., Dickinson D. J., Goldstein B., Akhmanova A. et al. , 2018. Optogenetic dissection of mitotic spindle positioning in vivo. eLife 7: e38198 10.7554/eLife.38198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Davis M. W., Ailion M., and Jorgensen E. M., 2012. Improved Mos1-mediated transgenesis in C. elegans. Nat. Methods 9: 117–118. 10.1038/nmeth.1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Davis M. W., Hopkins C. E., Newman B. J., Thummel J. M. et al. , 2008. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40: 1375–1383. 10.1038/ng.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Davis M. W., Sarov M., Taylor J., Flibotte S. et al. , 2014. Random and targeted transgene insertion in Caenorhabditis elegans using a modified Mos1 transposon. Nat. Methods 11: 529–534. 10.1038/nmeth.2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Jain N., Hansen L., Davis M. W., Li Y. et al. , 2016. An Abundant Class of Non-coding DNA Can Prevent Stochastic Gene Silencing in the C. elegans Germline. Cell 166: 343–357. 10.1016/j.cell.2016.05.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. G., Young L., Chuang R. Y., Venter J. C., Hutchison C. A. 3rd et al. , 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6: 343–345. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- Grimm J. B., English B. P., Chen J., Slaughter J. P., Zhang Z. et al. , 2015. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods 12: 244–250. 10.1038/nmeth.3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley J. L., Temple G. F., and Brasch M. A., 2000. DNA cloning using in vitro site-specific recombination. Genome Res. 10: 1788–1795. 10.1101/gr.143000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly W. G., Xu S., Montgomery M. K., and Fire A., 1997. Distinct requirements for somatic and germline expression of a generally expressed Caenorhabditis elegans gene. Genetics 146: 227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Z. Y., Prouteau M., Gotta M., and Barral Y., 2016. Compartmentalization of the endoplasmic reticulum in the early C. elegans embryo. J. Cell Biol. 214: 665–676. 10.1083/jcb.201601047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangone M., Manoharan A. P., Thierry-Mieg D., Thierry-Mieg J., Han T. et al. , 2010. The landscape of C. elegans 3′UTRs. Science 329: 432–435. 10.1126/science.1191244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt C., Rasoloson D., Ko D., and Seydoux G., 2008. 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr. Biol. 18: 1476–1482. 10.1016/j.cub.2008.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nance J., and Frøkjær-Jensen C., 2019. The Caenorhabditis elegans Transgenic Toolbox. Genetics 212: 959–990. 10.1534/genetics.119.301506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkema P. G., Harrison S. W., Plunger V., Aryana A., and Fire A., 1993. Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics 135: 385–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip N. S., Escobedo F., Bahr L. L., Berry B. J., and Wojtovich A. P., 2019. Mos1 Element-Mediated CRISPR Integration of Transgenes in Caenorhabditis elegans. G3 (Bethesda) 9: 2629–2635. 10.1534/g3.119.400399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiders S. A., Eastwood M. D., Bacher M., and Priess J. R., 2018. Binucleate germ cells in Caenorhabditis elegans are removed by physiological apoptosis. PLoS Genet. 14: e1007417 10.1371/journal.pgen.1007417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redemann S., Schloissnig S., Ernst S., Pozniakowsky A., Ayloo S. et al. , 2011. Codon adaptation-based control of protein expression in C. elegans. Nat. Methods 8: 250–252. 10.1038/nmeth.1565 [DOI] [PubMed] [Google Scholar]
- Schwartz M. L., and Jorgensen E. M., 2016. SapTrap, a Toolkit for High-Throughput CRISPR/Cas9 Gene Modification in Caenorhabditis elegans. Genetics 202: 1277–1288. 10.1534/genetics.115.184275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth M., Shirayama M., Tang W., Shen E. Z., Tu S. et al. , 2018. The Coding Regions of Germline mRNAs Confer Sensitivity to Argonaute Regulation in C. elegans. Cell Reports 22: 2254–2264. 10.1016/j.celrep.2018.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M., Seth M., Lee H. C., Gu W., Ishidate T. et al. , 2012. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell 150: 65–77. 10.1016/j.cell.2012.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S., Wimmer C., Rieger M., Doye V., Tekotte H. et al. , 1996. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell 84: 265–275. 10.1016/S0092-8674(00)80981-2 [DOI] [PubMed] [Google Scholar]
- Spike C. A., Shaw J. E., and Herman R. K., 2001. Analysis of smu-1, a gene that regulates the alternative splicing of unc-52 pre-mRNA in Caenorhabditis elegans. Mol. Cell. Biol. 21: 4985–4995. 10.1128/MCB.21.15.4985-4995.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland D., Lin Y., Wagner E., Hope C. M., Zayner J. et al. , 2012. TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nat. Methods 9: 379–384. 10.1038/nmeth.1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiser E., Frøkjær-Jensen C., Jorgensen E., and Ahringer J., 2011. MosSCI and gateway compatible plasmid toolkit for constitutive and inducible expression of transgenes in the C. elegans germline. PLoS One 6: e20082 10.1371/journal.pone.0020082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Tu S., Stubna M., Wu W. S., Huang W. C. et al. , 2018. The piRNA targeting rules and the resistance to piRNA silencing in endogenous genes. Science 359: 587–592. 10.1126/science.aao2840 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
With the exception of EGD633, the C. elegans strains generated in this study have been deposited at the Caenorhabditis Genetics Center (CGC; https://cgc.umn.edu). The plasmids listed in Figures 1 and 3 have been deposited at Addgene (http://www.addgene.org). Other donor plasmids, assembled expression plasmids and EGD633 are available upon request. Supplemental materials describing the sequence of tag donor cassettes are available through the GSA figshare portal: https://doi.org/10.25387/g3.9978611.
Figure 1.
SapTrap assembly of MosSCI targeting vectors using the four-cassette system. A. The MosSCI targeting vector pXF87 was derived from pCFJ350 by mutating two SapI restriction sites (indicated by arrowheads in the “Left” (L) and “Right” (R) homology arms) and introducing two SapI sites (blue text) between the XhoI and SpeI sites (green text). SapI cleavage sites are in red text. The SapI recognition sites are oriented such that upon digestion they are removed from the vector backbone. The cbr-unc-119 gene is used as a positive selection marker to facilitate the identification of transgenic animals. B. Design of the donor cassette vectors used for the 4-cassette cloning strategy. C. The curved dotted lines indicate the overhangs that anneal during the ligation reaction. D. Overview of the assembly protocol. For a detailed protocol, see the Materials and Methods section. E. Summary of available promoter, gene tag and 3′UTR donor cassette plasmids.
Figure 3.
SapTrap assembly of MosSCI targeting vectors using the five-cassette system. A. Schematic of pXF87 and the donor cassettes following SapI digestion. The dotted lines indicate the overhangs that anneal during ligation. B. Summary of available promoter, gene tag and 3′UTR donor cassette plasmids for the five-cassette system.