Abstract
Transmissible spongiform encephalopathy (TSE) surveillance in goats relies on tests initially approved for cattle, subsequently assessed for sheep, and approval extrapolated for use in “small ruminants.” The current EU-approved immunodetection tests employ antibodies against various epitopes of the prion protein PrPSc, which is encoded by the host PRNP gene. The caprine PRNP gene is polymorphic, mostly at codons different from the ovine PRNP. The EU goat population is much more heterogeneous than the sheep population, with more PRNP-related polymorphisms, and with marked breed-related differences. The ability of the current tests to detect disease-specific PrPSc generated against these different genetic backgrounds is currently assumed, rather than proven. We examined whether common polymorphisms within the goat PRNP gene might have any adverse effect on the relative performance of EU-approved rapid tests. The sample panel comprised goats from the UK, Cyprus, France, and Italy, with either experimental or naturally acquired scrapie at both the preclinical and/or unknown and clinical stages of disease. Test sensitivity was significantly lower and more variable when compared using samples from animals that were preclinical or of unknown status. However, all of the rapid tests included in our study were able to correctly identify all samples from animals in the clinical stages of disease, apart from samples from animals polymorphic for serine or aspartic acid at codon 146, in which the performance of the Bio-Rad tests was profoundly affected. Our data show that some polymorphisms may adversely affect one test and not another, as well as underline the dangers of extrapolating from other species.
Keywords: ELISA, goats, prion protein, scrapie, transmissible spongiform encephalopathy
Introduction
Classical scrapie, the archetype of the transmissible spongiform encephalopathies (TSE) or prion diseases, was first recognized in sheep almost 300 y ago and became endemic in the national flocks of several countries.10,16 Classical scrapie was first reported in goats in France in the 1940s.6 Active surveillance for scrapie in small ruminants has been compulsory in the EU since 2002, with the requirement that EU member states test a random sample of healthy slaughtered animals and fallen stock (TSE Regulation (EC) 999/2001),12 complementing passive surveillance, that requires the detection of bovine spongiform encephalopathy (BSE) in cattle and scrapie in sheep and goats since 1998 (Decision 98/272/EC),11 focusing on animals showing clinical signs compatible with TSE. This TSE Regulation, which evolved through the need to control BSE in cattle, sets out the framework for all the measures related to the prevention and control of scrapie in the EU, including the age of animals to be tested and the tests that are listed as approved for this purpose.
When mandatory active surveillance of small ruminant populations was first introduced, EU-approved rapid tests that performed satisfactorily on bovine tissues (Decision 2000/374/EC)13 were provisionally approved for small ruminants and used for the active and passive surveillance of TSE in sheep and goats during 2002–2004. In 2007, an EU evaluation of the diagnostic and analytical sensitivity of these rapid postmortem tests was undertaken using samples from cases of naturally occurring classical and atypical scrapie.14 At this point, some tests were not able to meet all of these requirements for atypical scrapie and were subsequently delisted. All of these evaluations were based on sheep samples, and approval extrapolated to use in “small ruminants.”
The currently approved rapid and confirmatory tests are all based on immunodetection methods, using antibodies that recognize PrPSc, the prion protein, an abnormal isoform of the host protein PrP that is encoded by the PRNP gene. In sheep, polymorphisms in the PRNP gene at codons 136, 141, 154, and 171 are demonstrated to be associated with disease susceptibility.4,7,20 The goat PRNP gene is also polymorphic, but the polymorphisms are largely located at different codons from those in sheep, and many of the polymorphisms have been observed in only 1 or 2 countries, and associated with specific breeds. At least 5 of these changes have been suggested to be associated with TSE susceptibility.17,32 At present, the EU goat population is much more heterogeneous than the sheep population, with more PRNP-related polymorphisms, and with marked breed-related differences.
The ability of the current tests to detect disease-specific PrPSc generated against these different genetic backgrounds is assumed rather than tested. Many TSE-affected goats have been identified through active surveillance using these tests, and these cases have been readily confirmed by alternative methods. There is no evidence, therefore, to suggest that test specificity is an issue. There has been no systematic assessment, however, of the sensitivity of these tests in goats, to our knowledge.
Data indicate that the performance of both rapid and confirmatory tests for the detection of disease in goats experimentally infected with either scrapie or BSE can be variable as a consequence of host allelic variations.23,24,28 Similar data have been reported previously for sheep.31
The experimental challenge study28 of scrapie in goats polymorphic at codon 146 showed that the presence of aspartic acid (D) at codon 146 resulted in false-negative results using an EU-approved rapid test. We explored the relative performance of the commonly used commercial tests in scrapie-positive goats with a range of PRNP polymorphisms, to establish whether any other polymorphisms might have an adverse effect on test performance.
Materials and methods
Composition of sample panel
Brain samples were obtained from 36 goats that had been experimentally inoculated intracerebrally with brain tissue from naturally occurring classical scrapie cases originating in Cyprus, as described previously.17,28 These goats represented all of the polymorphisms at codon 146 (NN, ND, DD, NS, SS), while being homozygous for the wild-type arginine (RR) at codon 154.
In addition, EU member states were invited to provide examples of positive caprine scrapie cases representative of the genetic composition recognized within their national herd. These (referred to as the mixed EU panel) comprised both field cases and experimental animals from France, Italy, Cyprus, and the UK (Supplementary Table 1). Disease had been confirmed by the contributing laboratory based on the presence of PrPSc in the brainstem and/or the lymphoreticular tissues using statutory confirmatory tests (immunohistochemistry [IHC] or western blot [WB]). Five goats experimentally challenged with BSE22 completed the set.
The full PRNP sequence was not available for any animal, so the “wild-type” genotype has been assumed for codons that were not specifically reported by the donating laboratory. The panel comprised samples from 162 animals, representing (in various proportions) the polymorphisms G127S, I142M, N146DS, R154H, R211Q, Q222K, and P240S.
Limitations as a result of the amount of tissue available from single animals mean that no dilution series were used as part of our analysis and therefore no analytical sensitivity estimates were possible. Consequently, our results are not directly comparable to previous formal European Food Safety Authority (EFSA) evaluations of test performance on bovine and ovine samples, but rather give a qualitative assessment of the possible field (i.e., diagnostic) sensitivity of the tests when they are used for disease detection in an active surveillance context. In addition, some animals were at clinical end-stage and others were apparently healthy, thereby representing the range of disease states that might be encountered in the field during active surveillance activities (Supplementary Table 1).
Testing of samples
The mixed EU panel samples were finely chopped and divided into 4 aliquots; the Cypriot experimental panel samples were chopped and divided into 3 aliquots. Each aliquot was tested using 1 of the following 3 rapid test kits: IDEXX HerdChek Small Ruminant conjugate SRB-CC (IDEXX SR; IDEXX Laboratories, Westbrook, ME), IDEXX HerdChek Bovine conjugate CC (IDEXX BOV), and Bio-Rad Sheep/Goat (Bio-Rad SG; Bio-Rad Laboratories, Hercules, CA). In addition, the Bio-Rad TeSeE was applied to all samples except the Cypriot experimental panel, for which data were already available.28 All assays were carried out according to the manufacturers’ instructions. All tests were performed at the Animal and Plant Health Agency (APHA; Weybridge, UK), thereby minimizing operator variability.
Calculation of test sensitivity
The samples in our study represent goats with polymorphisms at codons 127, 142, 146, 154, 211, 222, and 240. Of these polymorphisms, only I142M and R211Q were represented by numbers of cases that were sufficient to use for sensitivity calculations. Estimation of sensitivity of the rapid tests was conducted considering the result of the confirmatory tests (either IHC or WB) as the true disease status of the animal. The sensitivity of the 4 rapid tests considered, namely IDEXX SR, IDEXX BOV, Bio-Rad SG, and Bio-Rad TeSeE, were calculated according to clinical status and genotypes at codon 142 and 211 for the mixed EU panel, and at codon 146 for the Cypriot experimental panel. The 95% confidence intervals (CIs) of the proportion were calculated by applying the Wilson score interval. Comparisons of the sensitivity were conducted by applying the 2-sample test for equality of proportions with continuity correction, and calculation of the CIs using the R package “binom” (Dorai-Raj S. Binom: binomial confidence intervals for several parameterizations. R package v.3.6.0. 2014. Available at: https://rdrr.io/cran/binom/).
Results
Cypriot experimental panel
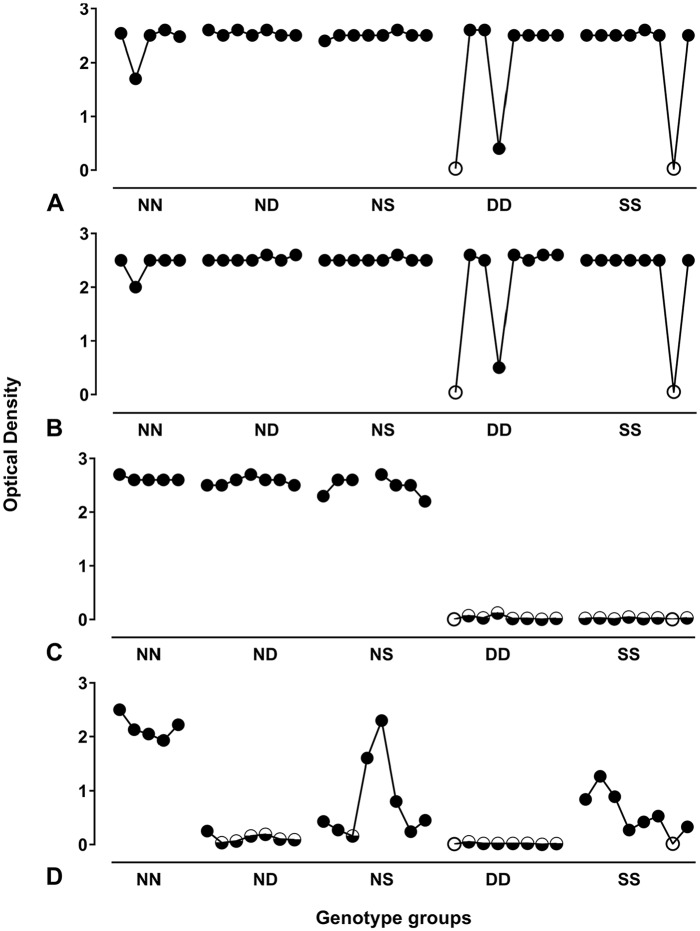
The 36 positive cases of the Cypriot experimental panel were subject to the IDEXX HerdChek test (both conjugates) and to the Bio-Rad SG. The IDEXX HerdChek test (both conjugates) performed well regardless of the host genotype (Figs. 1A, 1B), and were able to correctly detect all cases except 2 animals that were preclinical intercurrent deaths (one DD and one SS). The Bio-Rad SG (Fig. 1C) detected all cases of ND, NN, and NS genotypes but not those of DD (0 of 8) and SS (0 of 8) genotypes. Using the Bio-Rad TeSeE (Fig. 1D),28 only the NN animals resulted in a consistently high optical density (OD); the other genotypes gave more variable OD results. The presence of aspartic acid (D), and to a lesser extent serine (S), at codon 146, appeared to be a limiting factor for the performance of this test.
Figure 1.
Comparative test performance by genotype for the Cypriot experimental sample panel. The IDEXX SR (A) and the IDEXX BOV (B) tests both performed well regardless of the host genotype, and only failed to detect disease-specific levels of PrP in the 2 animals that were preclinical intercurrent deaths. Bio-Rad SG (C) and Bio-Rad TeSeE (D) data25 are shown for comparison. Filled circles indicate positive test results from transmissible spongiform encephalopathy (TSE)-positive animals; open circles indicate TSE-negative test results derived from TSE-negative animals; half-filled circles indicate TSE-negative test results associated with TSE-positive animals.
European surveillance panel
The sensitivity of each test by clinical status was compared with the observed sensitivity of these tests as reported by other studies (Table 1). This initial comparison did not take genotype into account.
Table 1.
Test sensitivity (when applied to brain tissue) relative to confirmatory methods. Published data from other studies are summarized in the lower half of the table for ease of comparison.
| Reference | n | Test sensitivity | |||
|---|---|---|---|---|---|
| IDEXX BOV | IDEXX SR | Bio-Rad SG | Bio-Rad TeSeE | ||
| Current study | |||||
| Total sample set* | 128 | 108/128† (84.4%) (77.1–89.6%‡) |
106/128 (82.8%) (75.3–88.4%) |
94/128 (73.4 %) (65.1–80.3%) |
55/73 (75.3%) (64.3–83.8%) |
| Clinically affected | 41 | 41/41 (100%) (91.4–100%) |
41/41 (100%) (91.4–100%) |
41/41 (100%) (91.4–100%) |
40/41 (97.4%) (87.4–99.5%) |
| Preclinical animals or of unknown clinical status | 87 | 65/87 (74.7%) (64.6–82.6%) |
65/87 (74.7%) (64.6–82.6%) |
53/87 (60.9%) (50.4–70.5%) |
15/32 (46.8%) (30.8–63.5%) |
| UK herd cull16 | 34 | 56%* | |||
| 68 | 27.9%§ | ||||
| French surveillance | |||||
| Lab 18 | 180 | 47.3%* § | |||
| Lab 28 | 58.4%* § | ||||
| European scrapie cases21 | 61 | 96.7%¦ | 83.6%¦ | 85.2%¦ | |
All positive samples from clinically normal as well as suspect and unknown clinical status animals.
No. of test-positive animals/no. of TSE-positive animals.
Confidence intervals of the proportions (sensitivity).
§ Includes animals positive only in lymphoreticular tissues.
¦ Sensitivity is reported as 100% for animals showing clinical signs.
We included 128 positive samples in this analysis: 49 from Cyprus (38%), 42 from the UK (33%), 31 from France (24%), and 6 (representing 4 animals) from Italy (5%). Of the 128 positive samples, 25 were clinically negative (from the UK), 41 were clinically positive (31 from France and 10 from the UK), 7 inconclusive (from the UK), and 55 were of unknown clinical status (49 from Cyprus and 6 from Italy; Supplementary Table 1).
A total of 106 samples were positive by IDEXX SR (82.8%; 95% CI: 75.3–88.4%), 108 by IDEXX BOV (84.4%; 95% CI: 77.1–89.6%), and 94 by Bio-Rad SG (73.4%; 95% CI: 65.1–80.3%). For the Bio-Rad TeSeE, there were results from 73 samples, of which 55 were positive (75.3%. 95% CI: 64.3–83.8%).
When looking at data from all animals, the sensitivity of the 4 tests is not significantly different pairwise, except when comparing IDEXX BOV and Bio-Rad SG (chi-squared test [χ2] = 3.97, df = 1, p = 0.046), albeit marginally. The sensitivity of the 4 tests when applied to samples from clinical positive animals (41) was very high, with all samples correctly classified by both IDEXX tests and the Bio-Rad SG, and only 1 positive sample missed by the Bio-Rad TeSeE (40 of 41). However, test performance varied between rapid tests in samples from clinically negative animals, or those of unknown status, in which the sensitivity of all the tests was consistently lower than in samples from clinical cases. The sensitivity of the Bio-Rad TeSeE in this subset of samples (n = 87) was significantly lower than that of the IDEXX BOV and IDEXX SR (χ2 = 7, df = 1, p = 0.008; 2-sample test for equality of proportions with continuity correction). Although the sensitivity of the Bio-Rad SG in this subset of samples (87) was lower than that of the IDEXX BOV and IDEXX SR, it was not statistically significantly lower, albeit marginally (p = 0.007). There was no significant difference in the sensitivity between the 2 Bio-Rad tests (p = 0.2).
The sensitivity of the 4 tests across genotypes at codons 142 and 211 (Table 2) was not significantly different, despite the absolute differences. For example, between IDEXX conjugates and Bio-Rad in the I142M genotype, there is no significant difference in the proportion of positive samples detected by the 2 IDEXX conjugates (92% and 100%, respectively) and the Bio-Rad conjugates (68% and 72%, respectively).
Table 2.
Estimation of test sensitivity (brain).
| Genotype | IDEXX BOV | IDEXX SR | Bio-Rad SG | Bio-Rad TeSeE |
|---|---|---|---|---|
| Codon 142 (all goats: 128) | ||||
| II142 | 78/95 (82.1%) (73.2–88.5%) |
76/95 (80%) (70.8–86.8%) |
71/95 (74.7%) (65.1–82.4%) |
33/40 (82.5%) (68–91.2%) |
| IM142 | 23/25 (92%) (75–97.7%) |
25/25 (100%) (86.6–100%) |
18/25 (72%) (52.4–85.7%) |
17/25 (68%) (48.4–82.8%) |
| MM142 | 7/8 (87.5%) (52.9–97.7%) |
7/8 (87.5%) (52.9–97.7%) |
5/8 (62.5%) (30.5–86.3%) |
5/86 (2.5%) (30.5–86.3%) |
| Codon 211 (all goats: 128) | ||||
| QQ211 | 8/8 (100%) (67.5–100%) |
8/8 (100%) (67.5–100%) |
8/8 (100%) (67.5–100%) |
8/8 (100%) (67.5–100%) |
| RQ211 | 4/4(100%) (51–100%) |
4/4 (100%) (51–100%) |
4/4 (100%) (51–100%) |
4/4 (100%) (51–100%) |
| RR211 | 59/74 (79.7%) (69.2–87.3%) |
57/74 (77%) (66.2–85.1%) |
55/74 (74.3%) (63.3–82.9%) |
19/19 (100%) (83.1–100%) |
| Codon 146 (Cyprus goats: 36) | ||||
| DD146 | 7/8 (87.5%) (52.9–97.7%) |
7/8 (87.5%) (52.9–97.7%) |
0/8 (0%) (0–32.4%) |
Data for this test published elsewhere25 |
| ND146 | 7/7 (100%) (64.5–100%) |
7/7 (100%) (64.5–100%) |
7/7 (100%) (64.5–100%) |
|
| NN146 | 5/5 (100%) (56.5–100%) |
5/5 (100%) (56.5–100%) |
5/5 (100%) (56.5–100%) |
|
| NS146 | 8/8 (100%) (67.5–100%) |
8/8 (100%) (67.5–100%) |
7/7 (100%) (64.5–100%) |
|
| SS146 | 7/8 (87.5%) (52.9–97.7%) |
7/8 (87.5%) (52.9–97.7%) |
0/8 (0%) (0–32.4%) |
Data presented as no. of test-positive samples/total positive samples (clinical and preclinical) by genotypes, with 95% confidence intervals in parentheses below.
Discussion
The 4 rapid tests included in our study were able to detect scrapie in goats in the clinical stages of the disease. However, test performance was more variable for animals that were preclinical or of unknown status.
The early detection of disease in animals with preclinical infection (i.e., field diagnostic sensitivity) relies on the correct sampling of brain target areas in which the initial accumulation of PrPSc is known to occur, regardless of the test that is applied. The analytical sensitivity aspect of previous formal test evaluations15 recognized this problem by the inclusion of sample dilution series as a proxy for early disease and/or less accurate sampling. Because of the very long incubation period in TSE, and the reliance on PrPSc accumulation as a marker of infection, it is accepted that all active surveillance programs in healthy animals, regardless of species, will miss a proportion of animals incubating disease.
When samples from the preclinical cases in our study are considered (i.e., a sample that might be considered representative of field active surveillance), there is a spread of apparent test sensitivity, with the IDEXX HerdChek test formats displaying greater sensitivity than the Bio-Rad SG, and with the Bio-Rad TeSeE giving a lower sensitivity against a gold standard of either IHC or WB. This general ranking is supported by the data from other studies in both sheep and goats.5,21,24,25
Our results confirm that the stage of the incubation period at which an infected animal is tested determines the amount of PrPSc available for detection and consequently the likelihood that any rapid test will correctly identify an infected animal.1–3,8,19,27, 29,30,33 A considerable proportion of preclinical animals are likely to be missed in large-scale surveillance. We compared sub-aliquots of the same finely chopped tissue samples, allowing the consideration of test performance specifically, without the confounding effect of sample location.
Each test uses a capture antibody or ligand, and then a detection antibody raised against a different epitope of the PrP molecule. Historically, many monoclonal antibodies (mAbs) have been raised against PrP from several animal species. Given the difficulty of raising an antibody against a host protein, synthetic peptides have often been used as the antigen. This means that some antibodies recognize linear epitopes, whereas others, raised against whole protein, may recognize conformational epitopes based on the 3D structure of the abnormal isoform of the protein. Test manufacturers do not publicly declare which antibodies are used in their tests. It is possible that if a polymorphism is present within the epitope recognized by a test antibody, the binding of that antibody and the subsequent sensitivity of the test may be compromised. This effect has already been documented in the context of mAb F99/97.6.1, an antibody that has a binding epitope that aligns with PRNP codons 220–225,9,23 and is widely used for TSE confirmation in a range of species. Chronic wasting disease–infected mule deer homozygous for phenylalanine at codon 225 (F225F), showed relatively poor staining when immunolabeled with F99,34 as did goats polymorphic for lysine at codon 222 (K222).23,32 Polymorphism at codon 143 has also been reported to result in a reduced signal when the mAb F89/160.1.5 (which is raised against bovine PRNP epitope 146–15926) was used.
Seven of the 29 reported amino acid changes in the goat PRNP gene appear to have worldwide distribution by haplotype ranking, from most common: proline at codon 240 (P240), arginine at codon 143 (R143), serine at codon 127 (S127), histidine at codon 154 (H154), lysine at codon 222 (K222), glutamine at codon 211 (Q211), methionine at codon 142 (M142), and serine or aspartic acid at codon 146 (S/D 146). All of these polymorphisms were represented in the sample panel, but only 2 (M142 and Q211) were available in sufficient numbers to allow statistical analysis. Although polymorphisms at codon 211 had no effect on test outcome, relative test performance was different in animals homozygous or heterozygous for M codon 142 compared to the wild type (II), but this effect was not absolute, or statistically significant. The presence of M at codon 142 does appear to compromise to some extent the sensitivity of the Bio-Rad tests.
The only absolute effect on test performance was seen in the samples from the goats of the experimentally challenged Cypriot panel that were polymorphic for serine or aspartic acid at codon 146, in which the performance of the Bio-Rad tests was profoundly affected. Aspartic acid, in particular, prevented the binding of at least one test antibody and resulted in false-negative results for samples from animals of this genotype. When these samples were further analyzed by western blot (data not shown; Chaplin, Papasavva-Stylianou, Simmons, Georgiadou, pers. comm., 2012) the blots showed a loss of binding, with substantial reduction of immunoreactivity with the N-terminal mAb P4 89–104 amino acid sequence of ovine PrP (which is a BSE-like outcome) in animals that are heterozygous at codon 146 or are homozygous for SS. In principle, this could confound discriminatory testing, but to date no naturally occurring cases of scrapie have been identified in animals of this genotype.18
Our data show that a polymorphism may adversely affect one test and not another, depending on how the polymorphism might affect the linear or conformational epitopes recognized by the mAbs or ligands used in the test, and underline the dangers of extrapolating from other studies. TSE tests should always be assessed specifically for the populations in which they are to be applied, regarding species and the full range of host genetics within a species (where known), to minimize the possibility of false-negative test results.
Supplemental Material
Supplemental material, Supplemental_material for Transmissible spongiform encephalopathy in goats: is PrP rapid test sensitivity affected by genotype? by Marion M. Simmons, Leigh Thorne, Angel Ortiz-Pelaez, John Spiropoulos, Soteria Georgiadou, Penelope Papasavva-Stylianou, Olivier Andreoletti, Stephen A.C. Hawkins, Daniela Meloni and Claire Cassar in Journal of Veterinary Diagnostic Investigation
Acknowledgments
A. Ortiz-Pelaez is employed by the European Food Safety Authority (EFSA). The present article is published under the sole responsibility of the authors and may not be considered as an EFSA scientific output. The positions and opinions presented in this article are those of the authors alone and do not necessarily represent the views or scientific works of EFSA.
Footnotes
Declaration of conflicting interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: This work was funded by the EC and Defra through the activities of the EURL for TSE at APHA. The UMR INRA ENVT 1225 was funded for this work by the EU FEDER/INTERREG (EFA148/16 REDPRION)
ORCID iD: Claire Cassar  https://orcid.org/0000-0001-6823-184X
https://orcid.org/0000-0001-6823-184X
Supplementary material: Supplementary material for this article is available online.
References
- 1. Arnold ME, et al. Estimating the temporal relationship between PrPSc detection and incubation period in experimental bovine spongiform encephalopathy of cattle. J Gen Virol 2007;88:3198–3208. [DOI] [PubMed] [Google Scholar]
- 2. Arnold ME, et al. Pathogenesis of experimental bovine spongiform encephalopathy (BSE): estimation of tissue infectivity according to incubation period. Vet Res 2009;40:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Balkema-Buschmann A, et al. Pathogenesis of classical and atypical BSE in cattle. Prev Vet Med 2011;102:112–117. [DOI] [PubMed] [Google Scholar]
- 4. Benestad SL, et al. Atypical/Nor98 scrapie: properties of the agent, genetics, and epidemiology. Vet Res 2008;39:19. [DOI] [PubMed] [Google Scholar]
- 5. Bozzetta E, et al. Comparative performance of three TSE rapid tests for surveillance in healthy sheep affected by scrapie. J Virol Methods 2011;173:161–168. [DOI] [PubMed] [Google Scholar]
- 6. Chelle PLP. Un cas de tremblante chez la chèvre [A case of scrapie in the goat]. Bull Acad Vet Fr 1942;15:294–295. French. [Google Scholar]
- 7. Clouscard C, et al. Different allelic effects of the codons 136 and 171 of the prion protein gene in sheep with natural scrapie. J Gen Virol 1995;76:2097–2101. [DOI] [PubMed] [Google Scholar]
- 8. Corbière F, et al. The limits of test-based scrapie eradication programs in goats. PLoS One 2013;8 e54911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dassanayake RP. Accumulation profiles of PrP(Sc) in hemal nodes of naturally and experimentally scrapie-infected sheep. BMC Vet Res 2013;9:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Detwiler LA. Scrapie. Rev Sci Tech 1992;11:491–537. [DOI] [PubMed] [Google Scholar]
- 11. European Commission. 98/272/EC: Commission Decision of 23 April 1998 on epidemio-surveillance for transmissible spongiform encephalopathies and amending Decision 94/474/EC. Off J Eur Commun 1998;L122:59–63. [Google Scholar]
- 12. European Commission. Regulation (EC) No 999/2001 of the European Parliament and of the Council of 22 May 2001 laying down rules for the prevention, control and eradication of certain transmissible spongiform encephalopathies. Off J Eur Commun 2001;L147:1–40. [Google Scholar]
- 13. European Commission. 2000/374/EC: Commission Decision of 5 June 2000 amending Decision 98/272/EC on epidemio-surveillance for transmissible spongiform encephalopathies. Off J Eur Commun 2000;L135:27–35. [Google Scholar]
- 14. European Food Safety Authority (EFSA) BIOHAZ Panel (EFSA Panel on Biological Hazards). Protocol for the evaluation of rapid post mortem tests to detect TSE in small ruminants. EFSA J 2007;509:1–31. Question no. EFSA-Q-2007-055. [Google Scholar]
- 15. European Food Safety Authority (EFSA) BIOHAZ Panel (EFSA Panel on Biological Hazards). Scientific opinion on analytical sensitivity of approved TSE rapid tests. EFSA J 2009;7:1436. Question no. EFSA-Q-2009-00687. [Google Scholar]
- 16. European Food Safety Authority (EFSA) BIOHAZ Panel (EFSA Panel on Biological Hazards). Scientific opinion on the scrapie situation in the EU after 10 years of monitoring and control in sheep and goats. EFSA J 2014;12:3781. Question no. EFSA-Q-2012-00646. [Google Scholar]
- 17. European Food Safety Authority (EFSA) BIOHAZ Panel (EFSA Panel on Biological Hazards). Scientific opinion on genetic resistance to transmissible spongiform encephalopathies (TSE) in goats. EFSA J 2017;15:4962. Question no. EFSA-Q-2016-00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Georgiadou S, et al. Goats with aspartic acid or serine at codon 146 of the PRNP gene remain scrapie-negative after lifetime exposure in affected herds in Cyprus. Epidemiol Infect 2017;145:326–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. González L, et al. High prevalence of scrapie in a dairy goat herd: tissue distribution of disease-associated PrP and effect of PRNP genotype and age. Vet Res 2009;40:65. [DOI] [PubMed] [Google Scholar]
- 20. Hunter N, et al. Natural scrapie in a closed flock of Cheviot sheep occurs only in specific PrP genotypes. Arch Virol 1996;141:809–824. [DOI] [PubMed] [Google Scholar]
- 21. Kittelberger R, et al. Evaluation of two commercial, rapid, ELISA kits testing for scrapie in retro-pharyngeal lymph nodes in sheep. N Z Vet J 2014;62:343–350. [DOI] [PubMed] [Google Scholar]
- 22. Konold T, et al. Pruritus is a common feature in sheep infected with the BSE agent. BMC Vet Res 2008;4:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Madsen-Bouterse SA, et al. PRNP variants in goats reduce sensitivity of detection of PrP(Sc) by immunoassay. J Vet Diagn Invest 2015;27:332–343. [DOI] [PubMed] [Google Scholar]
- 24. Meloni D, et al. EU-approved rapid tests might underestimate bovine spongiform encephalopathy infection in goats. J Vet Diagn Invest 2017;29:232–236. [DOI] [PubMed] [Google Scholar]
- 25. Nappi R, et al. Interlaboratory trial on TSE rapid tests for the control of the Italian scrapie surveillance network. Vet Microbiol 2009;39:126–131. [DOI] [PubMed] [Google Scholar]
- 26. O’Rourke KI, et al. Monoclonal antibody F89/160.1.5 defines a conserved epitope on the ruminant prion protein. J Clin Microbiol 1998;36:1750–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ortiz-Pelaez A, et al. Allelic variants at codon 146 in the PRNP gene show significant differences in the risk for natural scrapie in Cypriot goats. Epidemiol Infect 2015;143:1304–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Papasavva-Stylianou P, et al. The effect of polymorphisms at codon 146 of the goat PRNP gene on susceptibility to challenge with classical scrapie by different routes. J Virol 2017;91:e01142–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reckzeh C, et al. Rapid testing leads to the underestimation of the scrapie prevalence in an affected sheep and goat flock. Vet Microbiol 2007;123:320–327. [DOI] [PubMed] [Google Scholar]
- 30. Tongue SC, et al. Prevalence of scrapie infection in cull animals from 14 scrapie-affected flocks in Great Britain. Vet Rec 2005;157:480–482. [DOI] [PubMed] [Google Scholar]
- 31. Tongue SC, et al. The importance of the PrP genotype in active surveillance for ovine scrapie. Epidemiol Infect 2008;136:703–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vaccari G, et al. State-of-the-art review of goat TSE in the European Union, with special emphasis on PRNP genetics and epidemiology. Vet Res 2009;40:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wells GA, et al. Preliminary observations on the pathogenesis of experimental bovine spongiform encephalopathy (BSE): an update. Vet Rec 1998;142:103–106. [DOI] [PubMed] [Google Scholar]
- 34. Wolfe LL, et al. “Atypical” chronic wasting disease in PRNP genotype 225FF mule deer. J Wildl Dis 2014;50:660–665. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supplemental_material for Transmissible spongiform encephalopathy in goats: is PrP rapid test sensitivity affected by genotype? by Marion M. Simmons, Leigh Thorne, Angel Ortiz-Pelaez, John Spiropoulos, Soteria Georgiadou, Penelope Papasavva-Stylianou, Olivier Andreoletti, Stephen A.C. Hawkins, Daniela Meloni and Claire Cassar in Journal of Veterinary Diagnostic Investigation