Visual Abstract
Keywords: FSGS, INF2, podocyte, Actin, Cathepsins, cytoskeleton
Abstract
Background
Mutations in the gene encoding inverted formin-2 (INF2), a member of the formin family of actin regulatory proteins, are among the most common causes of autosomal dominant FSGS. INF2 is regulated by interaction between its N-terminal diaphanous inhibitory domain (DID) and its C-terminal diaphanous autoregulatory domain (DAD). INF2 also modulates activity of other formins, such as the mDIA subfamily, and promotes stable microtubule assembly. Why the disease-causing mutations are restricted to the N terminus and how they cause human disease has been unclear.
Methods
We examined INF2 isoforms present in podocytes and evaluated INF2 cleavage as an explanation for immunoblot findings. We evaluated the expression of INF2 N- and C-terminal fragments in human kidney disease conditions. We also investigated the localization and functions of the DID-containing N-terminal fragment in podocytes and assessed whether the FSGS-associated R218Q mutation impairs INF2 cleavage or the function of the N-fragment.
Results
The INF2-CAAX isoform is the predominant isoform in podocytes. INF2 is proteolytically cleaved, a process mediated by cathepsin proteases, liberating the N-terminal DID to function independently. Although the N-terminal region normally localizes to podocyte foot processes, it does not do so in the presence of FSGS-associated INF2 mutations. The C-terminal fragment localizes to the cell body irrespective of INF2 mutations. In podocytes, the N-fragment localizes to the plasma membrane, binds mDIA1, and promotes cell spreading in a cleavage-dependent way. The disease-associated R218Q mutation impairs these N-fragment functions but not INF2 cleavage.
Conclusions
INF2 is cleaved into an N-terminal DID–containing fragment and a C-terminal DAD–containing fragment. Cleavage allows the N-terminal fragment to function independently and helps explain the clustering of FSGS-associated mutations.
Glomerular epithelial cells, or podocytes, exhibit a polarized morphology characterized by a large cell body with extending primary processes and secondary foot processes.1,2 These foot processes interdigitate to form uniquely specialized cell-cell contacts called slit diaphragms. The unique morphology of podocyte structure is dependent on the spatial and temporal regulation of the actin cytoskeleton which, if impaired, can cause proteinuric kidney diseases.3–5
FSGS is etiologically and genetically heterogeneous.6 Highly penetrant Mendelian forms of FSGS are rare examples where we can unequivocally say we know the cause of disease.7 Although rare in absolute terms, INF2-associated FSGS is among the most common forms of inherited FSGS.8–10 Missense mutations in INF2 lead to kidney disease characterized by proteinuria, progressive kidney dysfunction, and FSGS with or without Charcot–Marie–Tooth disease (CMT).11,12 Approximately 9%–17% of patients with familial FSGS have INF2 mutations.7 INF2 is one of the 15 members of the formin family of proteins, which share a formin homology domain (FH2) involved in control of actin polymerization.13,14 However, most formins, including INF2, contain additional distinct regions of homology outside of the FH2 domain that are critical for actin assembly and interactions with regulatory binding partners. These regions include the diaphanous autoregulatory domain (DAD) near the INF2 C terminus and the diaphanous inhibitory domain (DID) near the INF2 N terminus. An intermolecular interaction between the DID and the DAD maintains INF2 in an autoinhibited state.15 All pathogenic FSGS mutations of INF2 identified to date localize to the DID.16
Cells express at least two INF2 isoforms, CAAX and non-CAAX.17,18 The CAAX isoform localizes to endoplasmic reticulum (ER)-rich regions, whereas the non-CAAX form is cytoplasmic. Over the past few years, multiple studies have increased our understanding of INF2 isoform–mediated regulation of the actin cytoskeleton.15,19–23 Many of these studies have relied on the cooperative regulation of the INF2 DID-DAD interaction to explain INF2 activity, including INF2 regulation of mitochondrial fission.23–25 INF2 also modulates activity of other formins such as the mDIA subfamily, and promotes stable microtubule assembly.19,26,27 Although disease-associated INF2 mutations appear to alter the DID-DAD interaction, disruption of this interaction does not explain why such mutations are limited to the DID. We reasoned that the DID region of INF2 might have unique functions in glomeruli independent of the DAD.
In this study, we report the existence of proteolytic cleavage of INF2 that causes the DID to localize and function independently of the DAD and FH2 regions. Furthermore, the ability of wild-type INF2, but not mutant R218Q, to counteract mDIA activity and promote cell spreading in a cleavage-dependent manner indicates that INF2 possesses unique cleavage-dependent functions mediated via the N-terminal fragment. These activities are lost in the presence of pathogenic INF2 mutations.
Methods
Cell Culture
Mouse podocytes generated from wild-type and INF2 KO C57BL/6 mice (Supplemental Figure 3) and human podocytes28 were maintained in RPMI 1640 (ThermoFisher Scientific, Waltham, MA) supplemented with 10% FBS (ThermoFisher), Insulin-Transferrin-Selenium and 1% penicillin-streptomycin (ThermoFisher). Human primary podocytes from Celprogen Inc. (Torrance, CA) were maintained per manufacturer’s instructions. Human embryonic kidney epithelial cells (HEK293T cells) were maintained in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin.
Plasmid Constructs and INF2 Expression
For overexpression experiments, human INF2-CAAX isoform sequences corresponding to amino acids 1–547 (N-fragment), 548–1249 (C-fragment), and 1–1249 (full-length) were cloned into an EGFPC1 plasmid (N-terminal GFP tag) (Takara Bio, Mountain View, CA). For cleavage site validation experiments, FLAG-tag was inserted between the CAAX motif (CVIQ) and stop codon in the full-length construct (catalog No.: 200523, QuickChange II Site-Directed Mutagenesis kit; Agilent Technologies, Lexington, MA). For removal of the INF2 cleavage site, amino acids 543–548 of INF2 were deleted in full-length CAAX constructs. Pathogenic mutations and variations in the INF2 sequence were introduced by a PCR-based mutagenesis (catalog No.: 200523, QuickChange II Site-Directed Mutagenesis kit). To achieve stable expression of fragments in human podocytes, N- and C-fragments were cloned into a lentiviral expression plasmid (catalog No.: PS100101; OriGene, Rockville, MD). For generating CRISPR KO podocytes, gRNA (sense: 5′-TGCGCGCCGTCATGAACTCG-3′; antisense: 5′-CGAGTTCATGACGGCGCGCA-3′) targeting the DID domain region of INF2 was expressed using a lentiviral plasmid (catalog No.: 5296, lentiCRISPRV2; Addgene, Boston, MA). All cloned plasmids were confirmed for the correct sequence by DNA sequencing (Genewiz, Boston, MA). For variations and mutant cleavage analysis, variations in the DID domain of INF2 were first extracted from Exome Aggregation Consortium (ExAC) and analyzed using a software tool, PROVEAN (Protein Variation Effect Analyzer), to group the variations as either structurally permissive or nonpermissive variants. Representative variants for structurally permissive and nonpermissive groups and pathogenic mutants were then cloned into EGFPC1 plasmid and overexpressed in cells.
Coimmunoprecipitation
HEK293T cells were transiently transfected with the indicated INF2 constructs using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). After incubation for 24 hours, the cells were lysed in 1% NP-40 lysis buffer (1% NP-40, 50 mM Tris-HCl, 150 mM NaCl, 5 mM EDTA, pH 7.4) supplemented with protease inhibitors. Cell lysates were then incubated with mouse anti–GFP-tag antibody (MA5–15256; ThermoFisher) for 1 hour followed by 40 μl protein A/G-magnetic beads for a further 1 hour at 4°C. The coimmunoprecipitation of DIAPH1 was then analyzed. Samples were probed for GFP or DIAPH1 using respective antibodies as follows: GFP (1:500) (sc-9996; Santa Cruz Biotechnology, Dallas, TX) or DIAPH1 antibody (1:500) (A300–078A; Bethyl Laboratories, Montgomery, TX). The membranes were then incubated with TrueBlot Anti-Rabbit IgG HRP (1:1000) (eB182; Rockland Antibodies, Limerick, PA) and developed using a chemiluminescent-based substrate (Super Signal West Dura; ThermoFisher).
Immunoblotting
Cells or glomerular preparations were lysed in a RIPA buffer (Boston BioProducts, Ashland, MA) supplemented with a cocktail of protease and phosphatase inhibitors (Roche, Pleasanton, CA) and clarified by centrifugation at approximately 13,000 × g for 15 minutes at 4°C. Equal protein loads were separated on a 4%–20% reducing gel, transferred to a polyvinylidene difluoride membrane (Bio-Rad), and probed with respective primary and secondary antibodies as follows: INF2 (N) (1:500) (A303–427A; Bethyl Laboratories); INF2 (C) (1:500) (20466–1-AP; Proteintech, Rosemont, IL); Podocin (1:1000) (P0372; Sigma, St Louis, MO); and anti-rabbit (1:4000) (Santa Cruz Biotechnology). The membranes were then developed using a chemiluminescent-based substrate (Super Signal West Dura). Total β-actin (1:4000) (sc47778; Santa Cruz) level was used as a loading control.
Chemical Screen
To assess changes in INF2 cleavage with different protease inhibitors, human podocytes were treated with small molecules from a protease inhibitor library (SelleckChem LLC, Houston, TX). Cells were treated with a concentration of 10 μM for 24 hours. Post incubation, cells were lysed in RIPA buffer and lysates examined for cleavage inhibition by INF2 immunoblot. The ratio of full-length INF2 to the cleaved C-terminal fragment was used to quantitate INF2 cleavage and inhibition of cleavage by the library compounds.
INF2 Isoform Analysis
Total RNA of human podocytes was extracted per manufacturer’s instructions (Qiagen, Germantown, MD) and 1 μg RNA was used for cDNA synthesis (Transcriptor first strand cDNA synthesis kit, Roche). INF2 was then PCR amplified from cDNA using isoform-specific primers (Supplemental Figure 1) (Accuprime DNA Polymerase system; ThermoFisher). The PCR-amplified product was gel-purified and examined for INF2 isoform–specific sequences using DNA sequencing.
Cleavage Site Mapping and In Silico INF2 Cleavage Analysis
HEK293T cells were transiently transfected with the GFP-INF2-FLAG constructs using Lipofectamine 2000. After incubation for 24 hours, the cells were lysed in RIPA buffer and the clarified supernatant was subjected to immunoprecipitation by anti-flag M2 beads (Sigma) for 2 hours, followed by washing and elution by anti-flag peptide (Sigma). The eluates were run on a 4%–20% denaturing gel, transferred to a polyvinylidene difluoride membrane, and stained using Coomassie blue. The C-fragment band was cut, digested per vendor guidelines (Alphalyse, Palo Alto, CA), and subjected to N-terminal sequencing. To assess the cleavage sites in INF2 in silico, we used the PROSPER bioinformatics tool.29
In Vitro Cleavage Assay
The in vitro cleavage assay was performed as described previously.30 Briefly, immunoprecipitated GFP-INF2 was diluted in a buffer containing 200 mM NaCl, 10 mM HEPES (pH 7.0), 2 mM EGTA, 1 mM MgCl2, and 1 mM DTT. When indicated, 100 μM E64 inhibitor (SelleckChem) was added. The reaction was initiated by addition of purified Cat L (0.1 μl), B (0.1 μl), or K (0.05 μl) enzyme (Sigma), and samples were placed at 37°C in a water bath for 15 minutes. Total assay volume was 25 μl. The reaction was terminated with the addition of 4× sample buffer (BioRad, Hercules, CA).
Cell Spreading Assays
Cells were serum starved for 2 consecutive days at 0.5% FBS in RPMI 1640 medium, then trypsinized and seeded in equal number on fibronectin-coated cover slips for 45 minutes. Cells were washed with ice-cold PBS buffer and processed for either fixation or lysis. The cell-spreading area was measured after immunostaining fixed cells with anti–F-actin.
Immunofluorescence
Cells were stained and imaged using confocal microscopy as described previously.31 Briefly, cells were fixed with 4% paraformaldehyde for 15 minutes, PBS-rinsed, and permeabilized 15 minutes with 0.5% Triton x-100. The fixed cells were then blocked with 5% BSA followed by sequential incubation with primary and secondary antibodies in blocking buffer. The following antibodies were used for immunofluorescence analysis: cortactin (ab333333, 1:50; Abcam), KDEL (ER-marker) (ab176333, 1:50; Abcam), mDIA (610848, 1:50; BD Biosciences), INF2 N terminus (SAB1401801, 1:50; Sigma-Aldrich), INF2 C terminus (20466–1 AP, 1:50; Proteintech), and synaptopodin (AP33487SU, 1:50; Origene). After primary and secondary antibody incubations, cell nuclei were counterstained with Hoechst (dsDNA) (Invitrogen), and mounted using ProLong Diamond Antifade (Invitrogen). For plasma membrane staining, cells were incubated with deep red cell mask plasma membrane stain (C10046; Thermo Fisher) and imaged by live cell imaging. All fluorescence images were collected by confocal microscopy (LSM 510; Zeiss) using ZEN lite 2.3 (black edition) and processed using ZEN lite 2.3 (blue version). For human glomerular staining of INF2, the specificities of the anti-INF2 N terminus and C terminus antibodies were confirmed using podocytes that were transduced with either empty vector or INF2 CRISPR KO constructs (Supplemental Figure 4).
Structured Illumination Microscopy and Image Analysis
Structured illumination microscopy (SIM) analysis of kidney biopsy section samples, stained for synaptopodin and INF2, was performed using a Zeiss Elyra SP.1 system, as described previously.32 Briefly, z-stacks were acquired over a volume of 75.44×75.44×4 μm3 (length×width×depth) with a slice-to-slice distance of 0.13 μm. The 34-μm-period grating was shifted five times and rotated five times on every frame. SIM processing post image acquisition was performed in 3D stacks using Zeiss ZEN software with default processing parameter settings.
The SIM-processed frames were then converted to a maximum intensity projection image using ZEN software or individually analyzed for foot process organization and INF2 localization using FIJI software. The foot processes were tracked in z-stack frames by synaptopodin staining. Foot process regions at points orthogonal to the imaging frame were defined as “regions of interest” and used for analysis. Synaptopodin staining was used to scan for effaced and noneffaced regions in disease tissues. The orthogonal sections that had a minimum x-plane length of 3 μm were used for analysis. Orthogonal sections that included at least four foot processes with similar widths as in normal glomeruli (width of foot process in normal glomeruli was noted to range<0.4 μm) were defined as noneffaced regions, with larger widths (>0.4 μm) defined as effaced regions. The profiles of synaptopodin and INF2 fluorescence intensity were then plotted and examined for overlap of peaks.
INF2 Knockout Mouse
A neomycin cassette was introduced into the mouse Inf2 gene locus by homologous recombination to generate an Inf2 knockout allele in C57Bl/6 embryonic stem cells. The cassette targets the Inf2 gene locus spanning the start site of Inf2 transcription, causing a complete loss of INF2 expression. Correctly targeted embryonic stem cells were injected into eight-cell Swiss Webster mouse embryos. Embryos were cultured overnight and transferred into pseudo pregnant female mice 2.5 days postcoitus. F0 male mice were bred with C57BL/6 mice to generate KO mice (see Supplemental Figure 5). All mice were used for experiments after breeding for at least five generations.
RNAi
Knockdown of mDIA1 in mouse podocytes was performed by transiently transfecting the siRNA (catalog No.: L-064854–01–0005; Dharmacon, CO) for mDIA1 using RNAiMAX. Reduction in mDIA protein was examined by immunoblotting at 48 and 72 hours post-transfection.
Statistical Analyses
All statistical analyses were performed using one-way ANOVA between test groups. When statistical significance was seen, Tukey’s multiple comparison test was used to find group differences. Statistical significance was set at a minimal value of P<0.05. All calculations were made using GraphPad Prism Version 7, and all values were reported as means±SD.
Human Kidney Studies
Human kidney biopsy material was obtained from Beth Israel Deaconess Medical Center or outside institutions in accordance with a protocol approved by the Institutional Review Board at Beth Israel Deaconess Medical Center.
Results
INF2 Is Cleaved into Two Fragments Separating the DID and DAD Domains
We reasoned that if the INF2 DID possesses unique functions, these might be mediated by additional splice variants of INF2 that span just the N terminus, because the annotated human genome (visualized using Ensembl or the UCSC Genome Browser) suggests the existence of such transcripts.33,34 To examine this experimentally, we looked for the splice isoforms of INF2 present in human podocytes. mRNA expression analysis showed that both an INF2 full-length isoform and an additional short isoform spanning the regions of INF2 DID are expressed in human podocytes (Supplemental Figure 1, A–C). Similar patterns of INF2 transcript expression were confirmed in human and mouse glomeruli (Supplemental Figure 1D). When we examined INF2 protein expression by immunoblot, we observed a band corresponding to full-length INF2 (approximately 170 kDa) but no polypeptide band of the size encoded by the short transcript (expected at size approximately 25–35 kDa), either in cells or human glomerular lysate samples (Supplemental Figure 1E).
However, using an antibody directed against the INF2 N terminus, we observed an additional band of approximately 60 kDa in human glomerular lysates (Figure 1A, Supplemental Figure 1E). Using an antibody directed against the INF2 C terminus, we also observed an additional band of approximately 100-kDa size (Figure 1A). Neither of these bands was present in INF2 CRISPR KO podocytes (Figure 1A). We saw similar INF2 immunoblot patterns in lysates from human glomeruli and primary human podocytes, as well as in glomeruli from wild-type mice, but not INF2 knockout mice (Figure 1, A and B). When we overexpressed the INF2-CAAX isoform as GFP-INF2 or GFP-INF2-FLAG and probed for GFP or FLAG by immunoblotting, we saw similar patterns of large and small bands recognized by anti-FLAG and anti-GFP antibodies (Figure 1C). To further confirm that these bands of INF2 in overexpression lysates are indeed the same forms of INF2 as noted in the glomerular or podocyte lysates, we probed the overexpression lysates with anti-INF2 antibodies as well (Figure 1D). We observed a matching banding pattern with the glomerular and podocyte lysates, indicating that the INF2 bands noted in glomerular and overexpression lysates reflect the same INF2 fragments. Together, these data suggested specific cleavage in INF2 into two fragments, with the C-terminal fragment migrating at approximately 100 kDa and the N-terminal fragment migrating at approximately 60 kDa (approximately 87kDa as the GFP-fusion) (Figure 1C).
Figure 1.
INF2 is cleaved into two fragments separating the DID and DAD regions. (A and B) Immunoblot analysis of INF2 in human and mouse samples. (A) Total cell lysates from human podocytes expressing empty vector (control), CRISPR INF2 KO human podocytes, primary human podocytes, and human glomerular tissue lysates were each probed with anti-INF2 antibodies directed against either the INF2 N-terminal [INF2(N)] or C-terminal [INF2(C)] regions. (B) Glomerular lysates from wild-type and Inf2 knockout mice were probed using a C-terminal–directed antibody. Arrows indicate different INF2 bands: black, full-length INF2; red, band detected only by N-terminal antibody; green, band detected only by C-terminal antibody. Actin was used as a loading control. (C and D) Expression analysis of control vector, GFP-INF2, and GFP-INF2-FLAG constructs in 293T cells. (C) Immunoblotting for GFP and FLAG tags detected similar full-length and smaller bands (red, GFP; green, FLAG). (D) Immunoblotting of overexpression lysates with INF2 antibodies confirms that the bands match with glomerular or podocyte lysate patterns (A and B), suggesting that INF2 is cleaved into two fragments (red, band detected only by N-terminal antibody; green, band detected only by C-terminal antibody; *endogenous INF2 bands). (E–G) Mapping of proteolytic cleavage site by N-terminal protein sequencing. (E) Coomassie-stained polyvinylidene difluoride membrane showing immunoprecipitated GFP-INF2-FLAG. Box indicates the immunoprecipitated C-fragment. Amino acids obtained from the first five cycles of N-terminal sequencing of the C-fragment (similar results were obtained from two independent immunoprecipitations and sequencing analyses). (F) Mapping of the INF2 cleavage site. The cleavage occurs between the amino acids highlighted in red, separating the DID and DAD regions, respectively, into N- and C-terminal fragments. (G) Validation of the cleavage site. Deletion of the cleavage site (Δ543–548) in GFP-INF2-FLAG results in complete loss of INF2 fragments. Red arrow, N-fragment; green arrow, C-fragment; *indicates loss of cleaved fragments. Immunoblots shown are representative of three independent experiments. IB, immunoblot.
To map the cleavage site, we overexpressed GFP-INF2-FLAG and immunoprecipitated the C-terminal fragment using an antibody to the FLAG tag, followed by N-terminal sequencing. The first five amino acids identified were “S-A-W-V-P” (Figure 1E). This sequence matched a unique locus on the C-terminal side of the INF2 DID (Figure 1F) and aligned with an in silico–predicted29 cathepsin K cleavage site spanning residues from 543 to 548 (DHGLG/S). Cleavage was predicted to occur between INF2 residues glycine (G) 547 and serine (S) 548. To verify this cleavage site, we overexpressed a mutated form of GFP-INF2-FLAG in which we deleted the predicted cathepsin K cleavage site (noncleavable GFP-INF2-FLAG). Cleavage fragments using this construct were no longer detectable by immunoblot (Figure 1G). This observation, along with the N-terminal sequencing results, confirms that INF2 cleavage occurs between residues glycine 547 and serine 548.
INF2 Localization in Glomeruli from Normal and Diseased Kidney Tissues Correlates with Proteolytic Cleavage
To examine whether INF2 cleavage has an in vivo human correlation, we used SIM to determine INF2 distribution within glomeruli in kidney biopsy samples from individuals without known kidney disease, as well as from individuals with INF2-mediated FSGS (Figure 2), Alport syndrome, and SLE (lupus nephritis) (Supplemental Figure 2). We stained these samples with both N-terminal–directed and C-terminal–directed INF2 antibodies to compare cleavage fragment localizations.
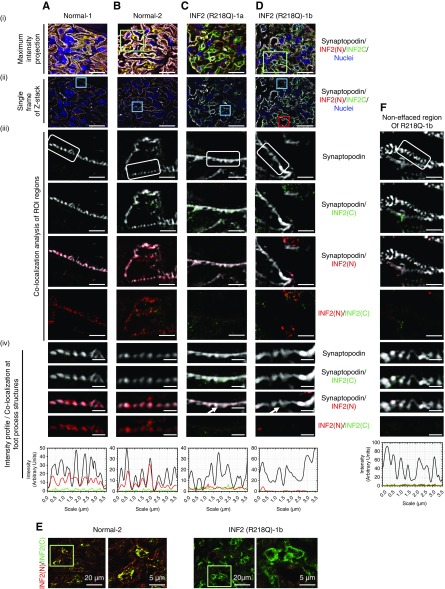
Figure 2.
SIM demonstrates that INF2 expression and localization are altered in diseased glomeruli with a R218Q mutation. Kidney sections from normal individuals (A and B) and INF2 R218Q–associated FSGS (C and D) were stained for synaptopodin (gray), INF2 N-terminal region (N, red), INF2 C-terminal region (C, green), and nuclei (blue). Two representative glomeruli from two normal individuals and two diseased glomeruli from a single patient with R218Q INF2–mediated FSGS are shown. (i and ii) SIM-processed low-magnification micrographs. (i) Maximum intensity projection of 3D z-stack optical frames. Scale bar, 20 μm. (ii) A representative single optical frame of the 3D z-stack shown (i). Blue boxes highlight region of interest (ROI) used for colocalization analysis. Scale bar, 20 μm. (iii) Colocalization analysis of INF2 with synaptopodin. Scale bar, 2.5 μm. White boxes highlight foot process structures. INF2 N-terminal and INF2 C-terminal antibody staining patterns were differentially localized. (iv) Colocalization and corresponding intensity profiles of INF2 and synaptopodin in foot process structures. Scale bar, 0.7 μm. INF2 N-terminal staining colocalized with synaptopodin in normal foot process structures (A-iv and B-iv). This colocalization is lost in effaced regions of R218Q INF2 samples (arrow highlights; C-iv and D-iv). Intensity profiles confirm colocalization of fluorescent signal intensities. (E) Overlay of INF2 N- and C-terminal staining shown in (B-i and D-i). Green boxes highlight ROI used for C-terminal staining analysis. Scale bar, 20 μm. C-terminal staining in cell body remains unaltered with the disease mutation. Additional INF2 N- and C-terminal staining of normal and disease glomeruli collected with confocal microscopy are shown in Supplemental Figure 3. (F) Colocalization analysis of INF2 with synaptopodin in noneffaced regions of R218Q disease glomeruli shown in (Ci). Red box highlights ROI used for colocalization analysis. Colocalization and corresponding intensity profiles of INF2 and synaptopodin in foot process structures. Scale bar, 0.7 μm. Colocalization of INF2 N-terminal staining with synaptopodin in foot process structures was lost in noneffaced regions of R218Q disease glomeruli as well.
Consistent with our previous observations in mice31 and as shown in Supplemental Figure 5, we noted INF2 expression exclusively in glomerular podocytes of both normal and disease samples (Figure 2, A–D). However, we observed discrete staining patterns for the different INF2 antibodies. In normal human kidney tissue, the staining pattern of C-terminal antibody showed predominant localization to the podocyte cell body, whereas the N-terminal antibody localized to both the podocyte cell body and foot process structures, colocalizing with synaptopodin (Figure 2, A and B, Supplemental Figure 3). By contrast, kidney tissue from an individual with FSGS due to an INF2 R218Q mutation was remarkable for loss of N-terminal antibody staining from foot processes (Figure 2, C and D), whereas localization of C-terminal antibody staining in the cell body remained grossly unaltered (Figure 2E, Supplemental Figure 3). We noted this pattern of N-terminal antibody staining in approximately 96% (n=24) of orthogonal sectional views of podocyte foot processes of normal glomeruli, whereas it is absent in all of the orthogonal sectional views of regions with both normal (n=6) and effaced podocyte foot processes (n=11) of glomeruli from patients with R218Q FSGS. A similar loss of staining of N-terminal antibody was observed in the effaced foot process structures of individuals with Alport syndrome (n=7) and with lupus nephritis (n=9) (Supplemental Figure 2, A and C), but it remained unaltered in regions spanning normal foot process structures (Alport syndrome, n=9; lupus nephritis glomeruli, n=6; Supplemental Figure 2, B and D). The segregation of N-terminal and C-terminal antibody staining patterns in normal kidney (Figure 2, A and B), with localization of the N-terminal INF2 fragment to foot processes in normal samples, and its loss before podocyte effacement in a sample derived from a patient with FSGS, suggests that cleavage occurs in vivo and that loss of INF2 N-fragment–associated activity may lead to altered podocyte structure and function.
INF2 Cleavage Is Mediated by Cathepsins and Is Not Altered in the Presence of FSGS-Associated INF2 Mutations
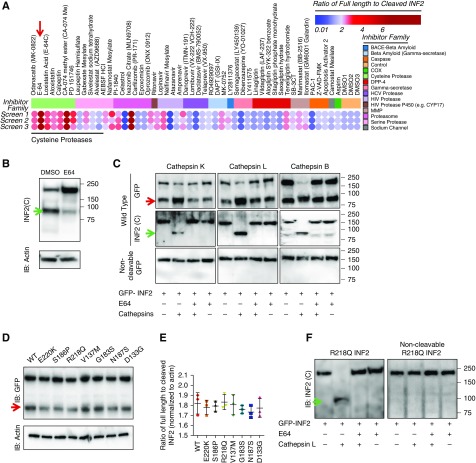
We reasoned that understanding the difference between INF2 N-terminal antibody staining in normal and diseased glomeruli might help elucidate the unique functions of the DID, as well as the mechanisms of INF2-associated FSGS. We hypothesized that the difference in N-terminal antibody staining of podocyte foot processes between normal and diseased kidney might reflect either (1) altered INF2 cleavage (e.g., mutations or other nongenetic factors affecting cleavage) or (2) events downstream of cleavage that affect N-fragment localization to foot processes. Therefore, we evaluated the regulation of INF2 cleavage. We first performed a chemical screen in podocytes using a protease inhibitor library to examine changes in the ratio of full-length INF2 to INF2 N-terminal fragment by immunoblot (Figure 3A). Most of the protease inhibitors tested had minimal effects on INF2 cleavage, whereas cathepsin inhibitors significantly increased the cleavage ratio, with greatest effect using cysteine protease inhibitor E64 (Figure 3B). To further confirm INF2 cleavage by cathepsin family members, we performed in vitro biochemical cleavage assays using immunoprecipitated wild-type full-length or noncleavable mutant full-length forms of GFP-INF2 using an anti-GFP antibody (Figure 3C). In contrast to the wild-type INF2, the noncleavable mutant was not cleaved by cathepsins. The results from the biochemical assay showed that all tested cathepsins (B, L, and K) specifically cleaved the wild-type full-length GFP-INF2, yielding approximately 100-kDa C-terminal fragments and an increase in the levels of approximately 90-kDa N-terminal GFP-fusion fragments. INF2 proteolysis after cathepsin incubation was absent in the presence of cysteine protease inhibitor E64. By contrast, none of the tested cathepsins cleaved GFP-INF2 mutated to lack the cleavage site, confirming both the specificity of the cleavage site and the role of cathepsins in mediating this cleavage.
Figure 3.
INF2 cleavage is mediated by cathepsins and is not affected by pathogenic mutations. (A) Screening of protease inhibitor library for effect on INF2 cleavage. Heat map shows the effects of different compounds on INF2 cleavage. Results from three independent sets of assays are shown. The cleavage levels with each treatment were calculated as ratios of full-length to cleaved INF2 levels from INF2 immunoblots and plotted as a heat map, with color indicating inhibitor potency. Clustering of compounds into different protease inhibitor families showed that inhibitors of the cysteine protease family shared the ability to inhibit cleavage, most potently, E-64 (red arrow). (B) Representative immunoblot of INF2 cleavage inhibition by cysteine protease inhibitor, E-64. Green arrow indicates cleaved C-fragment. (C) In vitro cleavage assay. Immunoprecipitated GFP-INF2 or noncleavable GFP-INF2 (Δ543–548) was incubated with the listed cathepsins, with or without E64. INF2 cleavage levels were then examined by immunoblot using N-terminal– (anti-GFP) or C-terminal–directed antibodies. All listed cathepsins cleaved GFP-INF2 but not the noncleavable GFP-INF2 deletion mutant. The cleavage was inhibited by E-64. Red arrow indicates cleaved N-terminal fragment. Green arrow indicates cleaved C-terminal fragment. Blots are representative of three independent experiments. (D) Immunoblot to detect cleavage of INF2 in 293T cells with different pathogenic mutations and other variants in the DID domain. (E) Quantitation of INF2 cleavage from three independent experiments is shown. No significant change in cleavage levels was noted in the presence of these INF2 mutations or variants in the DID domain (control versus mutant and control versus variant cleavages did not vary significantly (one-way ANOVA). (F) In vitro cleavage assay of INF2 with R218Q mutation. Immunoprecipitated GFP-R218Q INF2 or noncleavable GFP-R218Q INF2 was tested for cleavage using cathepsin L. Immunoblot analysis showed E-64–inhibitable cleavage of GFP-R218Q INF2, but not of noncleavable GFP-R218Q INF2. IB, immunoblot.
Next, to determine whether INF2 cleavage is altered in the presence of FSGS-associated INF2 mutations, we assayed cleavage of full-length GFP-INF2 containing either FSGS-causing mutations (R218Q, E220K, S186P) or other variants predicted in silico to be either structurally permissive (V137M, G187S) or nonpermissive (N183S, D133G), identified from the gnomAD database. Transient overexpression of these constructs showed that comparable levels of cleavage occur between control INF2 (wild-type) and all of the tested INF2 mutants and variants (Figure 3, D and E). Similar results were observed using an in vitro cleavage assay with the R218Q mutant GFP-INF2 (Figure 3F), indicating that regulation of INF2 cleavage is not altered by the presence of pathogenic mutations. These results further suggest that the altered INF2 N-terminal antibody staining in noneffaced regions of human R218Q kidney sections is not due to altered cleavage of INF2, but rather other alterations of the R218Q INF2 N-terminal fragment behavior. This is further confirmed by the presence of unaltered N-terminal antibody staining in the noneffaced regions of other disease conditions. However, the presence of altered N-terminal fragment antibody staining in effaced regions from other disease conditions suggests that altered INF2 N-terminal fragment behavior may function as a secondary mediator of the progression of other forms of glomerular disease (Supplemental Figure 2).
INF2 N-Fragment Localization to the Cell Membrane Is Altered in the Presence of Disease-Causing Mutations
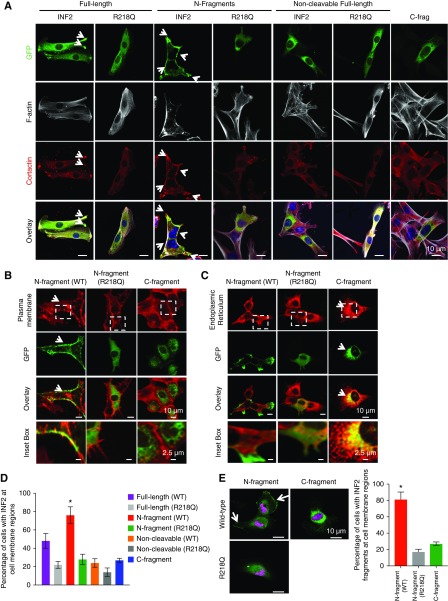
To evaluate INF2 fragment localization, we transiently overexpressed GFP-tagged INF2 N-terminal fragments (wild-type and R218Q), the INF2 C-terminal fragment, full-length INF2-CAAX (wild-type and R218Q), and noncleavable INF2-CAAX (wild-type and R218Q) in mouse INF2 KO podocytes. The wild-type N-terminal fragment showed a prominent localization to cell-boundary regions and costained with cortactin, whereas the C-terminal fragment, the wild-type full-length INF2-CAAX, and the noncleavable INF2-CAAX all showed a localization pattern largely restricted to the ER-rich cell-body regions (Figure 4A). Further costaining analysis with plasma membrane and ER markers confirmed that wild-type N-fragment and C-fragment indeed localize to plasma membrane and ER regions, respectively (Figure 4, B and C). Importantly, we noted that the pathogenic FSGS mutation, R218Q, caused a redistribution of the INF2 N-fragment from its normal plasma membrane location to a diffuse pancellular staining pattern (Figure 4, A–D). A similar difference between N-fragment and C-fragment localization was observed when we stably overexpressed the HA-tagged N- or C-fragments in human podocytes with endogenous INF2 expression, confirming that N-fragment localizes preferentially to plasma membrane regions in cells despite the existence of wild-type full-length INF2 in cells (Figure 4, E and F).
Figure 4.
INF2 fragments exhibit differential localization in podocytes. (A) Localization of INF2 fragments in mouse podocytes. GFP-tagged N-fragment, full-length, noncleavable forms of wild-type or mutant INF2 (R218Q) and C-fragment of INF2 were transfected in INF2 KO podocytes and examined for their localization by costaining with cortactin (red) and F-actin (gray). Wild-type N-fragment exhibited cell membrane colocalization with cortactin (white arrow highlights), whereas the C-fragment localizes to the cell body in an ER-like pattern. The R218Q mutation significantly altered the membrane localization of N-fragment leading to a diffuse cytoplasmic distribution, with loss of cortactin colocalization at the cell membrane. No membrane localization was observed for the noncleavable forms of INF2, with or without R218Q. Scale bar, 10 µm. (B and C) Colocalization analysis of GFP-tagged N-fragment and C-fragment with plasma membrane (cell mask staining) and ER (ER marker, KDEL). Wild-type N-fragment colocalized with plasma membrane staining (arrow highlights in [B]), whereas the C-fragment colocalized with KDEL staining (arrow highlights in [C]) The R218Q N-fragment exhibited a diffuse cytoplasmic distribution without any significant colocalization with plasma membrane and KDEL staining. Scale bar, 10 µm. White inset boxes, a magnified region of INF2 fragment staining with the respective marker proteins. Scale bar, 2.5 µm. (D) Quantitation of membrane localization of different INF2 expression forms (*P<0.01 GFP N-fragment versus all other groups; one-way ANOVA and Tukey’s multiple comparison test; n>100 for each group). (E) Localization of INF2 fragments in human podocytes. HA-tagged INF2 N-fragment or C-fragment was expressed in human podocytes and localized (green, HA tag; magenta, nucleus). N-fragments expressed in human podocytes containing endogenous INF2 exhibited similar localization to membrane regions (white arrow highlights) along with some cell body staining, whereas the C-fragment localization was in an ER-like pattern. Membrane localization of N-fragment is altered by the R218Q mutation. Scale bar, 10 µm. (F) Quantitation of cells with membrane-localized INF2 (*P<0.01, HA–N-fragment versus HA–C-fragment or HA-R218Q fragment; one-way ANOVA and Tukey’s multiple comparison test; n>100 for each group) (see Supplemental Material for version of panels using gray scale instead of red).
We also examined whether a similar effect is seen with the existence of the R218Q mutant form of full-length or N-fragment INF2 in cells. To evaluate this, we coexpressed HA-tagged wild-type INF2 N-fragment with GFP-tagged R218Q mutant full-length INF2-CAAX or N-fragment in mouse INF2 KO podocytes. We found that the R218Q mutant forms of both full-length and N-fragment can significantly impair the localization of wild-type N-fragment to plasma membrane, leading to a pancellular distribution (Figure 5, A and B). When we examined the interaction, we observed that R218Q INF2 N-fragment and full-length can both interact with wild-type N-fragment, as is the case with their wild-type equivalents (Figure 5C). Taken together, these results show that neither INF2 cleavage nor the ability of the N-terminal fragment to interact with other INF2 molecules is affected by the presence of a disease-causing mutation. However, post cleavage, presence of a pathogenic mutation dramatically alters the localization of both the wild-type and mutant N-fragments.
Figure 5.
Coexistence of different mutant forms of INF2 impairs the cell membrane localization of wild-type INF2 N-fragment. (A) Localization analysis of wild-type HA INF2 N-fragment in mouse podocytes. GFP-tagged N-fragment, full-length, and noncleavable forms of wild-type or mutant INF2 (R218Q) were cotransfected with wild-type HA INF2 N-fragment in INF2 KO podocytes and examined for their localization. Wild-type HA INF2 N-fragment (red) exhibited cell membrane localization with the copresence of GFP-tagged N-fragment, full-length, and noncleavable forms of wild-type INF2 (white arrow highlights), whereas it is significantly impaired with the copresence of GFP-tagged R218Q N-fragment, full-length, and noncleavable forms of INF2 (blue arrow highlights). Scale bar, 10 µm. White inset boxes, a magnified region of wild-type HA INF2 N-fragment staining at membrane regions. Scale bar, 2.5 µm. (B) Quantitation of membrane localization of wild-type HA INF2 N-fragment with the copresence of different INF2 expression forms (*P<0.01 GFP N-fragment versus all other groups; one-way ANOVA and Tukey’s multiple comparison test; n>100 for each group). (C) Coimmunoprecipitation of wild-type HA INF2 with different GFP-tagged INF2 expression forms. GFP-tagged INF2 N-fragment and full-length INF2 forms of wild-type or mutant INF2 (R218Q) were cotransfected with HA-tagged wild-type INF2 N-fragment in 293T cells. Cell lysates were then subjected to HA-pulldown and blotted for HA and GFP. HA immunoblot confirmed successful immunoprecipitation of transfected wild-type HA INF2 N-fragment construct. GFP immunoblot showed an interaction of HA INF2 N-fragment with both wild-type and R218Q mutant forms of N-fragment and full-length INF2. Each immunoblot is representative of three independent experiments with similar results (see Supplemental Material for version of panels using gray scale instead of red). IB, immunoblot; IP, immunoprecipitation.
INF2 N-Fragment Restores Impaired Cell Spreading by Antagonizing mDIA Signaling
We next evaluated whether the cleavage-induced differential localization of the two proteolytic fragments of INF2 regulates any specific cell functions, and whether INF2 N-terminal fragment function might be altered in the presence of INF2 mutations. Our previous studies have shown that both the mutant podocytes from R218Q knock-in mice and siRNA-silenced INF2 human podocytes exhibit impaired cell spreading.26,31 These studies suggest that INF2 has a role in cell spreading that is altered in the presence of disease-causing mutations. We hypothesized that the cleaved N-fragment may normally help mediate the cell-spreading function of INF2 and help maintain (or restore) normal podocyte structure.
To examine this, we first tested whether mouse INF2 KO podocytes and human INF2 CRISPR KO podocytes recapitulate the impaired cell spreading previously noted in other systems (Figure 6A).26,31 Consistent with previous observation, we found that both mouse INF2 KO podocytes and human INF2 CRISPR KO podocytes exhibited impaired cell spreading compared with their respective controls (Figure 6A). We next tested whether presence of the INF2 N-fragment restores normal cell spreading. As depicted in Figure 6B, we transfected mouse INF2 KO podocytes with different GFP-INF2 constructs to achieve equivalent expression levels and evaluated possible restoration of cell spreading. We found that expressing wild-type N-fragment in INF2 KO podocytes restored normal cell spreading in the INF2 KO cells. Normal cell spreading was partially restored by expression of the wild-type full-length INF2, but not by the noncleavable form of full-length INF2. In contrast, none of the R218Q mutant constructs produced any recovery in cell spreading. Similar effects on cell spreading were noted in human INF2 CRISPR KO podocytes transiently overexpressing wild-type INF2 N-fragment but not in podocytes overexpressing the R218Q mutant N-fragment (Figure 6C).
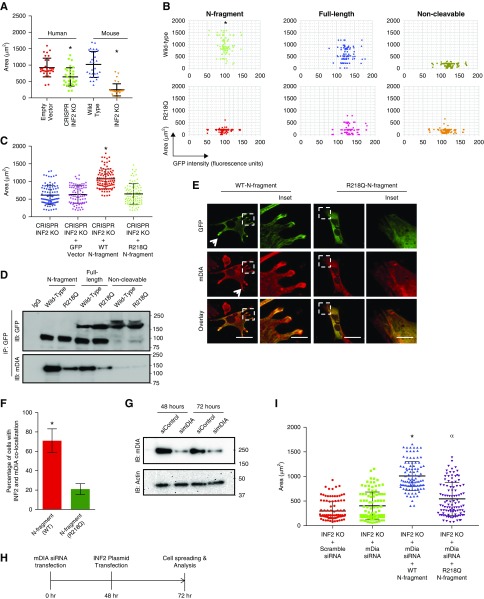
Figure 6.
INF2 N-fragment restores cell spreading in association with mDIA interaction. (A–C) Cell-spreading assays. (A) Mouse INF2 knockout podocytes and human INF2 CRISPR knockout podocytes were compared with their respective control cells for ability to spread upon attachment. The relative area covered after 45 minutes of cell spreading was calculated and analyzed for differences among groups (n>100 cells for each group). Lack of INF2 in either mouse or human podocytes was associated with impaired cell spreading (*P<0.001 EV versus CR; WT versus KO; one-way ANOVA and Tukey’s multiple comparison test; n>50 for each group). (B) Mouse INF2 knockout cells were transfected with GFP-tagged INF2 N-fragment, full-length INF2, or noncleavable forms of wild-type or mutant INF2 (R218Q) and examined for restoration of normal cell spreading. GFP-tagged INF2 N-fragment expression significantly increased relative cell-spreading area, whereas neither mutant nor noncleavable forms of INF2 restored normal cell spreading (*P<0.01, N-fragment versus other groups; one-way ANOVA and Tukey’s multiple comparison test; n>100 for each group). (C) Human INF2 CRISPR knockout podocytes were transfected with GFP-tagged wild-type or R218Q mutant N-fragment and spreading area was quantified. Wild-type N-fragment restored cell spreading, whereas the R218Q mutant form did not (*P<0.01, wild-type versus R218Q; one-way ANOVA and Tukey’s multiple comparison test; n>100 for each group). (D) Coimmunoprecipitation of INF2 with mDIA. 293T cells transfected with GFP-tagged INF2 N-fragment, full-length INF2, and noncleavable forms of wild-type or mutant INF2 (R218Q) were subjected to GFP-pulldown in cell-spreading conditions and blotted for mDIA and GFP. GFP immunoblot confirmed successful immunoprecipitation of transfected INF2 constructs. mDIA immunoblot showed an interaction of mDIA with wild-type N-fragment and full-length INF2. Mutant forms of INF2 showed significantly decreased interaction, whereas noncleavable forms of INF2 failed to interact with mDIA. Each immunoblot is representative of three independent experiments with similar results. (E and F) Colocalization of mDIA and INF2 N-fragments in podocytes. (E) Mouse INF2 KO podocytes were transfected with GFP-tagged wild-type or R218Q INF2 N-fragment and stained for mDIA. Wild-type N-fragment colocalized with mDIA in cell extension areas. Cells with the mutant R218Q N-fragment expression exhibited impaired colocalization of mDIA and R218Q N-fragment (the arrow highlights colocalization of GFP-INF2 N-fragment and mDIA). Boxed regions show a representative region for both mDIA colocalization with wild-type GFP N-fragment and its loss with R218Q GFP N-fragment. Scale bar, 10 µm. (F) Quantitation of colocalization of INF2 N-fragments with mDIA (*P<0.01 GFP-INF2 wild-type N-fragment versus GFP-INF2 R218Q N-fragment; n>100). (G) Loss of mDIA induction by RNA interference. Mouse INF2 KO podocytes were transfected with scramble or mDIA siRNA and examined for loss of mDIA. Immunoblot analysis showed loss of mDIA as examined at both 48 and 72 hours post-transfection. Each immunoblot is representative of three independent experiments with similar results. (H) Timeline of steps for examining cell spreading with INF2 N-fragment in loss-of-mDIA condition. (I) Mouse INF2 knockout cells were transfected with either mDIA siRNA alone or followed sequentially by cotransfections with GFP-tagged wild-type or R218Q INF2 N-fragment and examined for improvements in cell spreading. Loss of mDIA alone had a minimal improvement in the recovery of impaired cell spreading of INF2 KO cells, whereas with subsequent additional cotransfections with INF2 N-fragments, significant improvement was noted with wild-type N-fragment (*P<0.01, N-fragment versus other groups). The R218Q N-fragment also partially restored the impaired cell spreading in loss-of-mDIA condition (αP<0.05, R218Q fragment versus INF2 KO; one-way ANOVA and Tukey’s multiple comparison test; n>100 for each group) (see Supplemental Material for version of panels using gray scale instead of red). CR, CRISPR INF2 KO; EV, empty vector; IB, immunoblot; IP, immunoprecipitation.
Our earlier studies showed that impaired podocyte cell spreading in the presence of INF2 mutations was associated with aberrantly increased mDIA signaling,26,31 which we attributed to loss of an inhibitory interaction between INF2 DID and mDIA-DAD.19 Therefore, we examined whether the cleaved INF2 N-fragments interacted with mDIA in order to correlate our cleavage-dependent cell spreading observations with earlier findings suggesting a specific INF2-mDIA interaction.19,26 We observed that in cell-spreading conditions wild-type INF2 N-fragment interacted with mDIA, and that this interaction was significantly reduced in the presence of R218Q mutant INF2. We observed a similar trend using full-length wild-type and R218Q mutant INF2 (Figure 6D). More importantly, the noncleavable form of INF2 (both wild-type and R28Q mutant) did not interact with mDIA. When we costained overexpressed N-fragments with mDIA, we observed colocalization of mDIA with wild-type N-fragment in cell-extension areas. By contrast, with the R218Q mutant N-fragment, we no longer observed significant colocalization of R218Q N-fragment with mDIA (Figure 6, E and F). Finally, we examined whether the effect of N-fragments to promote cell spreading resulted from inhibition of mDIA activity. We coexpressed wild-type or mutant N-fragment in mouse INF2 KO podocytes with mDIA1 knockdown and found that loss of mDIA1 could enhance cell spreading with both wild-type and R218Q mutant N-fragments (Figure 6, G and H). These results indicate that INF2 cleavage mediates the antagonistic effect of INF2 DID on mDIA and that the INF2 N-fragment specifically promotes cell spreading. In the presence of an FSGS-associated INF2 point mutation, the ability of the N-terminal fragment to antagonize mDIA and promote cell spreading is impaired.
Discussion
Multiple studies have indicated that INF2, through its effects on actin and microtubules, can modulate mitochondrial fission, calcium uptake, vesicle trafficking, T cell polarization, and placental implantation, among numerous other cell processes.23,24,35–38 The mechanisms by which FSGS-associated (and CMT-associated) mutations cause disease are not well understood. In particular, explanations remain unclear for the restricted localization of these mutations to the INF2 N-terminal region, a possible manifestation of differential evolutionary pressures experienced by the N- and C-terminal portions of INF2 gene products.8,9,16
In this study, we have demonstrated that INF2 undergoes a proteolytic program that can cause the DID in the N-terminal region of INF2 to function independently of the DAD in the INF2 C-terminal region (Figure 7). Because all of the human INF2 mutations identified to date have been confined to the DID-containing N terminus, this observation suggests that N-fragment–dependent functions must be integral for glomerular structure and function, and provides a potential clue to why all mutations are limited to the DID region. In addition, we observed that INF2-induced cell spreading and the interaction of mDIA with INF2 are cleavage-dependent properties and are both significantly reduced in the presence of a disease mutation. These results are consistent with our earlier findings on INF2- and mDIA-related cell-spreading effects.26,31 In this model, INF2 acts downstream of RhoA and inhibits mDIA-related actin cytoskeletal rearrangements. By doing so, INF2 promotes formation of lamellipodial structures and facilitates the trafficking of proteins and lipid raft structures to the cell surface. Consistent with the model that increased mDIA activity leads to disease, another recent study indicated that loss of mDIA1 can preserve glomerular function in a model of diabetic kidney disease.39
Figure 7.
Summary model: Cleavage-dependent unique functions of DID domain are altered due to mutations. INF2 is cleaved by cathepsins into two fragments, separating the DID and DAD regions. The cleaved DID-containing N-fragment localizes to cell membrane regions and functions to promote cell spreading, perhaps by counteracting mDIA signaling. However, with the R218Q mutation, both cell spreading and mDIA interaction are impaired. In one possible model of INF2-mediated disease, insults that cause podocytes to lose slit diaphragm integrity may also activate cathepsins, promoting INF2 cleavage and allowing the N-terminal DID-containing region of INF2 to help restore podocyte structural integrity. With pathogenic mutations, loss of proper localization and lack of DID function may lead to persistent injury.
However, the described model, particularly for the context of mDIA, does have limitations. For instance, the studies with mDIA knock-down have been conducted solely from INF2 KO cells, where cell spreading is already compromised. Therefore, apart from being able to interact with INF2, the precise mode of mDIA influence in cell spreading remains unknown. Next, we observed a lack of response in cell spreading with mDIA KD alone in INF2 KO cells, but, with concurrent INF2 N-fragment presence, we noted the restoration of cell spreading. Although speculative, this might represent that INF2 N-fragment may have multiple functions; with one aspect it may bind and inhibit the mDIA activity, and, with the other unidentified functions, it may promote cell-spreading behavior. Future head-to-head comparative studies with normal, reduced, or increased mDIA activity in normal and INF2 KO cells will help us to refine this pathway and to delineate the precise role of mDIA in it. Nevertheless, this study suggests that these processes are all dependent on the proteolytic cleavage of INF2.
Our analysis of INF2 transcript variants indicates that podocytes may express an additional transcript that is consistent with the transcripts shown in the ENSEMB and NCBI databases and that includes just the 5′ portion of the gene, encoding only the DID. However, we note that the PCR assay used in our analysis covered only three exons, and, thus, the exact molecular identity of this short isoform has not been defined. In addition, at the protein level, we could not detect any specific immunoreactive protein product of this transcript by immunoblotting, suggesting that the PCR product may either differ from the predicted isoform or, alternatively, this short transcript may function as a regulatory RNA. Future analysis directed toward the identity of the PCR product is warranted to establish its identity from start to end and to identify possible functional roles for this transcript as well.
Prior in vitro studies of full-length INF2 have indicated that INF2 is normally kept in an autoinhibited state and is activated under spatial and temporal control.15,23 Our studies show that INF2 cleavage physically separates the N- and C-terminal portions of the polypeptide and activates at least some DID-mediated functions of INF2. Tamura et al.40 reported, using a C-terminal antibody–based immunohistochemical analysis, that INF2 in normal glomeruli is present in the podocyte cell body and major processes, with decreased or absent expression in these sites in FSGS. We have observed localization of INF2 C-terminal antibody staining in normal glomeruli. However, with the identification of INF2 cleavage, we suggest that caution is required in correlating INF2 staining intensities with disease states. The amount of INF2 cleavage and the stability of the two fragments may vary in different conditions. Nevertheless, the results presented here point to a specific role for the N-terminal region in podocyte foot processes. This activity may be lost in FSGS caused by N-terminal INF2 mutations.
All FSGS-associated mutations (with or without associated CMT) localize to the INF2 N-terminal region.41 This suggests that the N-terminal region is important for the integrity of the foot process and slit diaphragm structures, and suggests that INF2 cleavage has in vivo significance, particularly in the context of disease. We note that the loss in N-fragment immunostaining intensity in the INF2 R218Q mutant kidney is not limited to foot process structures. A grossly reduced staining intensity of this fragment is also noted in the cell body. By contrast, the staining intensity of the INF2 C-fragment remains unaltered. Although speculative, this could result from: (1) increased cleavage leading to increased fragment formation, and/or (2) presence of a mutation affecting the stability of the mutant N-fragment but not the C-fragment, thus causing a loss of total N-fragment intensity but not C-fragment intensity. Future studies designed to determine changes in INF2 cleavage levels in the course of the disease and further analysis of the stability of normal and mutant forms of INF2 (full length and fragment) should help clarify these questions.
Our experiments have shown that cathepsin proteases are primarily responsible for INF2 cleavage. A diverse set of stimuli, including transient injury events, can upregulate and activate cathepsins in the kidney.42 Multiple studies have indicated that cathepsin proteases can inactivate CD2AP and other proteins essential for podocyte structure and function.30,43 Proteolytic processing of α-actinin-4, podocin, and other proteins by cathepsins has recently been suggested to facilitate maintenance of podocyte structure and filtration barrier integrity.44 It is thus interesting to note that INF2 is also a target of cathepsin proteases. Studies of human samples indicate evidence of INF2 cleavage in normal kidney (Figure 1). Previous studies in mouse models indicate that INF2 functions as an injury response protein, facilitating podocyte recovery.31 Given that cathepsins are upregulated in transient injury conditions, it is likely that cathepsin-mediated INF2 cleavage regulates N-terminal fragment activity. Thus, taken together, it is intriguing to propose that upon glomerular injury, increased cathepsin activity causes increased proteolytic cleavage of INF2, and that increased N-fragment activity might facilitate podocyte recovery.31 This hypothesized role of INF2 in podocyte repair would be lost in the presence of FSGS-causing mutations. Further studies are needed to evaluate this model.
Our previous studies and the work presented here show that cell spreading and related cytoskeleton defects (e.g., loss of cortactin) are hallmarks of impaired INF2 function in cells.26,31 Our study provides evidence that cleavage-generated INF2 N-fragment exhibits differential localization to membrane regions and regulates cell-spreading functions, whereas C-fragment remains with an ER-like distribution throughout the cell body. Regulation of actin dynamics and cell-spreading functions by formin family members is important for proper adherens junction assembly.45 Given that slit diaphragm structures of podocytes share morphologic features similar to those of the adherens junction, the INF2 N-fragment may mediate or facilitate the actin dynamics required for proper slit diaphragm structure and function.46 Although INF2 N-fragment–mediated cell-spreading defects provide one possible mechanism for linking mutant INF2 with FSGS, changes in the full gamut of INF2 activities will need exploration.
INF2-associated kidney disease is inherited as a dominant trait. Thus, a model of disease pathogenesis should be consistent with a disease genotype consisting of one mutant and one wild-type INF2 allele. As described above, the presence of R218Q mutant INF2 alters the localization and function of the wild-type protein in the cell, consistent with a dominant-negative effect.
In summary, we have shown that INF2 undergoes proteolytic cleavage that can lead the N-terminal DID region to function independently of the C-terminal DAD region (Figure 7). In addition, we have observed cell-spreading activity and mDia interaction that are cleavage-dependent and are significantly affected by a pathogenic mutation in INF2. These data provide a plausible explanation for the altered podocyte localization of INF2 N-fragments with FSGS-associated mutations. Future studies to define the full range of INF2 N-fragment and C-fragment functions in maintenance of glomerular podocyte structure and function will increase our understanding of the physiologic roles of both INF2 cleavage and the resultant INF2 proteolytic fragments in health and disease.
Disclosures
Dr. Pollak and Dr. Schlöndorff are listed as inventors of issued patent US9499867B2 related to INF2 mutational analysis. Dr. Pollak receives research support from Vertex unrelated to this paper. All other remaining authors have nothing to disclose.
Funding
This work was supported by National Institutes of Health (NIH) grant R01DK088826. Dr. Subramanian was supported in part by NIH T32 award DK007199.
Supplementary Material
Acknowledgments
We thank the Harvard Center for Biological Imaging, Harvard University, Cambridge, MA for help with super-resolution imaging and analysis. We thank Beth Israel Deaconess Medical Center confocal core facility, Boston, MA for help with immunofluorescence imaging and analysis.
Dr. Subramanian and Dr. Pollak conceived the idea for the study, analyzed data, and wrote the manuscript. Dr. Subramanian, Dr. Chun, Ms. Perez-Gill and Mr. Yan performed experiments. Dr. Stillman analyzed the human kidney studies. Dr. Higgs, Dr. Chun, Dr. Alper, and Dr. Schlöndorff analyzed data and edited the manuscript.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
See related editorial, “New Paradigm for Cytoskeletal Organization in Podocytes: Proteolytic Fragments of INF2 Formin Function Independently of INF2 Actin Regulatory Activity,” on pages 235–236.
Supplemental Material
This article contains the following supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2019050443/-/DCSupplemental.
Supplemental Figure 1. Splice isoform analysis of INF2 in podocytes.
Supplemental Figure 2. Altered localization of INF2 N-fragment in diseased glomeruli.
Supplemental Figure 3. INF2 expression and localization analysis in normal and diseased glomeruli by confocal microscopy.
Supplemental Figure 4. Analysis of antibody specificity in human podocytes.
Supplemental Figure 5. Generation and characterization of Inf2 knockout mouse and podocytes.
Supplemental Figure 6. Alternatively colored version of Figure 4.
Supplemental Figure 7. Alternatively colored version of Figure 5.
Supplemental Figure 8. Alternatively colored version of Figure 6.
Supplemental Figure 9. Alternatively colored version of Supplemental Figure 3.
References
- 1.Scott RP, Quaggin SE: Review series: The cell biology of renal filtration. J Cell Biol 209: 199–210, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pollak MR, Quaggin SE, Hoenig MP, Dworkin LD: The glomerulus: The sphere of influence. Clin J Am Soc Nephrol 9: 1461–1469, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perico L, Conti S, Benigni A, Remuzzi G: Podocyte-actin dynamics in health and disease. Nat Rev Nephrol 12: 692–710, 2016 [DOI] [PubMed] [Google Scholar]
- 4.Reiser J, Sever S: Podocyte biology and pathogenesis of kidney disease. Annu Rev Med 64: 357–366, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Welsh GI, Saleem MA: The podocyte cytoskeleton--key to a functioning glomerulus in health and disease. Nat Rev Nephrol 8: 14–21, 2011 [DOI] [PubMed] [Google Scholar]
- 6.Rosenberg AZ, Kopp JB: Focal segmental glomerulosclerosis. Clin J Am Soc Nephrol 12: 502–517, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pollak M: Genetics of familial FSGS. Semin Nephrol 36: 467–472, 2016 [DOI] [PubMed] [Google Scholar]
- 8.Barua M, Brown EJ, Charoonratana VT, Genovese G, Sun H, Pollak MR: Mutations in the INF2 gene account for a significant proportion of familial but not sporadic focal and segmental glomerulosclerosis. Kidney Int 83: 316–322, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyer O, Benoit G, Gribouval O, Nevo F, Tête MJ, Dantal J, et al.: Mutations in INF2 are a major cause of autosomal dominant focal segmental glomerulosclerosis. J Am Soc Nephrol 22: 239–245, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gbadegesin RA, Lavin PJ, Hall G, Bartkowiak B, Homstad A, Jiang R, et al.: Inverted formin 2 mutations with variable expression in patients with sporadic and hereditary focal and segmental glomerulosclerosis. Kidney Int 81: 94–99, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown EJ, Schlöndorff JS, Becker DJ, Tsukaguchi H, Tonna SJ, Uscinski AL, et al.: Mutations in the formin gene INF2 cause focal segmental glomerulosclerosis. Nat Genet 42: 72–76, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyer O, Nevo F, Plaisier E, Funalot B, Gribouval O, Benoit G, et al.: INF2 mutations in Charcot-Marie-Tooth disease with glomerulopathy. N Engl J Med 365: 2377–2388, 2011 [DOI] [PubMed] [Google Scholar]
- 13.Goode BL, Eck MJ: Mechanism and function of formins in the control of actin assembly. Annu Rev Biochem 76: 593–627, 2007 [DOI] [PubMed] [Google Scholar]
- 14.Chesarone MA, DuPage AG, Goode BL: Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat Rev Mol Cell Biol 11: 62–74, 2010 [DOI] [PubMed] [Google Scholar]
- 15.Chhabra ES, Higgs HN: INF2 Is a WASP homology 2 motif-containing formin that severs actin filaments and accelerates both polymerization and depolymerization. J Biol Chem 281: 26754–26767, 2006 [DOI] [PubMed] [Google Scholar]
- 16.Mademan I, Deconinck T, Dinopoulos A, Voit T, Schara U, Devriendt K, et al.: De novo INF2 mutations expand the genetic spectrum of hereditary neuropathy with glomerulopathy. Neurology 81: 1953–1958, 2013 [DOI] [PubMed] [Google Scholar]
- 17.Chhabra ES, Ramabhadran V, Gerber SA, Higgs HN: INF2 is an endoplasmic reticulum-associated formin protein. J Cell Sci 122: 1430–1440, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramabhadran V, Korobova F, Rahme GJ, Higgs HN: Splice variant-specific cellular function of the formin INF2 in maintenance of Golgi architecture. Mol Biol Cell 22: 4822–4833, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun H, Schlondorff JS, Brown EJ, Higgs HN, Pollak MR: Rho activation of mDia formins is modulated by an interaction with inverted formin 2 (INF2). Proc Natl Acad Sci U S A 108: 2933–2938, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramabhadran V, Hatch AL, Higgs HN: Actin monomers activate inverted formin 2 by competing with its autoinhibitory interaction. J Biol Chem 288: 26847–26855, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gurel PS, Ge P, Grintsevich EE, Shu R, Blanchoin L, Zhou ZH, et al.: INF2-mediated severing through actin filament encirclement and disruption. Curr Biol 24: 156–164, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gurel PS, A M, Guo B, Shu R, Mierke DF, Higgs HN: Assembly and turnover of short actin filaments by the formin INF2 and profilin. J Biol Chem 290: 22494–22506, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korobova F, Ramabhadran V, Higgs HN: An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science 339: 464–467, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chakrabarti R, Ji WK, Stan RV, de Juan Sanz J, Ryan TA, Higgs HN: INF2-mediated actin polymerization at the ER stimulates mitochondrial calcium uptake, inner membrane constriction, and division. J Cell Biol 217: 251–268, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manor U, Bartholomew S, Golani G, Christenson E, Kozlov M, Higgs H, et al.: A mitochondria-anchored isoform of the actin-nucleating spire protein regulates mitochondrial division. eLife 4: 1–27, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun H, Schlondorff J, Higgs HN, Pollak MR: Inverted formin 2 regulates actin dynamics by antagonizing Rho/diaphanous-related formin signaling. J Am Soc Nephrol 24: 917–929, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bartolini F, Andres-Delgado L, Qu X, Nik S, Ramalingam N, Kremer L, et al.: An mDia1-INF2 formin activation cascade facilitated by IQGAP1 regulates stable microtubules in migrating cells. Mol Biol Cell 27: 1797–1808, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saleem MA, O’Hare MJ, Reiser J, Coward RJ, Inward CD, Farren T, et al.: A conditionally immortalized human podocyte cell line demonstrating nephrin and podocin expression. J Am Soc Nephrol 13: 630–638, 2002 [DOI] [PubMed] [Google Scholar]
- 29.Song J, Tan H, Perry AJ, Akutsu T, Webb GI, Whisstock JC, et al.: PROSPER: An integrated feature-based tool for predicting protease substrate cleavage sites. PLoS One 7: e50300, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yaddanapudi S, Altintas MM, Kistler AD, Fernandez I, Möller CC, Wei C, et al.: CD2AP in mouse and human podocytes controls a proteolytic program that regulates cytoskeletal structure and cellular survival. J Clin Invest 121: 3965–3980, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Subramanian B, Sun H, Yan P, Charoonratana VT, Higgs HN, Wang F, et al.: Mice with mutant Inf2 show impaired podocyte and slit diaphragm integrity in response to protamine-induced kidney injury. Kidney Int 90: 363–372, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siegerist F, Ribback S, Dombrowski F, Amann K, Zimmermann U, Endlich K, et al.: Structured illumination microscopy and automatized image processing as a rapid diagnostic tool for podocyte effacement. Sci Rep 7: 11473, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, et al.: Ensembl 2018. Nucleic Acids Res 46: D754–D761, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Casper J, Zweig AS, Villarreal C, Tyner C, Speir ML, Rosenbloom KR, et al.: The UCSC genome browser database: 2018 update. Nucleic Acids Res 46: D762–D769, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamm KYB, Johnson ML, Baker Phillips J, Muntifering MB, James JM, Jones HN, et al.: Inverted formin 2 regulates intracellular trafficking, placentation, and pregnancy outcome. eLife 7: 1–23, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andrés-Delgado L, Antón OM, Bartolini F, Ruiz-Sáenz A, Correas I, Gundersen GG, et al.: INF2 promotes the formation of detyrosinated microtubules necessary for centrosome reorientation in T cells. J Cell Biol 198: 1025–1037, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andrés-Delgado L, Antón OM, Madrid R, Byrne JA, Alonso MA: Formin INF2 regulates MAL-mediated transport of Lck to the plasma membrane of human T lymphocytes. Blood 116: 5919–5929, 2010 [DOI] [PubMed] [Google Scholar]
- 38.Shaye DD, Greenwald I: The disease-associated formin INF2/EXC-6 organizes lumen and cell outgrowth during tubulogenesis by regulating F-actin and microtubule cytoskeletons. Dev Cell 32: 743–755, 2015 [DOI] [PubMed] [Google Scholar]
- 39.Manigrasso MB, Friedman RA, Ramasamy R, D’Agati V, Schmidt AM: Deletion of the formin Diaph1 protects from structural and functional abnormalities in the murine diabetic kidney. Am J Physiol Renal Physiol 315: F1601–F1612, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tamura H, Nakazato H, Kuraoka S, Yoneda K, Takahashi W, Endo F: Reduced INF2 expression in nephrotic syndrome is possibly related to clinical severity of steroid resistance in children. Nephrology (Carlton) 21: 467–475, 2016 [DOI] [PubMed] [Google Scholar]
- 41.Challis RC, Ring T, Xu Y, Wong EK, Flossmann O, Roberts IS, et al.: Thrombotic microangiopathy in inverted formin 2-mediated renal disease. J Am Soc Nephrol 28: 1084–1091, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reiser J, Oh J, Shirato I, Asanuma K, Hug A, Mundel TM, et al.: Podocyte migration during nephrotic syndrome requires a coordinated interplay between cathepsin L and alpha3 integrin. J Biol Chem 279: 34827–34832, 2004 [DOI] [PubMed] [Google Scholar]
- 43.Sever S, Altintas MM, Nankoe SR, Möller CC, Ko D, Wei C, et al.: Proteolytic processing of dynamin by cytoplasmic cathepsin L is a mechanism for proteinuric kidney disease. J Clin Invest 117: 2095–2104, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rinschen MM, Hoppe AK, Grahammer F, Kann M, Völker LA, Schurek EM, et al.: N-degradomic analysis reveals a proteolytic network processing the podocyte cytoskeleton. J Am Soc Nephrol 28: 2867–2878, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kobielak A, Pasolli HA, Fuchs E: Mammalian formin-1 participates in adherens junctions and polymerization of linear actin cables. Nat Cell Biol 6: 21–30, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reiser J, Kriz W, Kretzler M, Mundel P: The glomerular slit diaphragm is a modified adherens junction. J Am Soc Nephrol 11: 1–8, 2000 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.