Abstract
Background
Rhodosporidium toruloides has emerged as a promising host for the production of bioproducts from lignocellulose, in part due to its ability to grow on lignocellulosic feedstocks, tolerate growth inhibitors, and co-utilize sugars and lignin-derived monomers. Ent-kaurene derivatives have a diverse range of potential applications from therapeutics to novel resin-based materials.
Results
The Design, Build, Test, and Learn (DBTL) approach was employed to engineer production of the non-native diterpene ent-kaurene in R. toruloides. Following expression of kaurene synthase (KS) in R. toruloides in the first DBTL cycle, a key limitation appeared to be the availability of the diterpene precursor, geranylgeranyl diphosphate (GGPP). Further DBTL cycles were carried out to select an optimal GGPP synthase and to balance its expression with KS, requiring two of the strongest promoters in R. toruloides, ANT (adenine nucleotide translocase) and TEF1 (translational elongation factor 1) to drive expression of the KS from Gibberella fujikuroi and a mutant version of an FPP synthase from Gallus gallus that produces GGPP. Scale-up of cultivation in a 2 L bioreactor using a corn stover hydrolysate resulted in an ent-kaurene titer of 1.4 g/L.
Conclusion
This study builds upon previous work demonstrating the potential of R. toruloides as a robust and versatile host for the production of both mono- and sesquiterpenes, and is the first demonstration of the production of a non-native diterpene in this organism.
Keywords: Rhodotorula, Mevalonate pathway, Diterpene, Geranylgeranyl pyrophosphate synthase, Mutant farnesyl pyrophosphate synthase, Metabolic engineering
Background
The diverse terpenoid family contains over 70,000 unique compounds that are predominantly produced from two fundamental isoprenoid building blocks, the C5 prenyl phosphates, dimethylallyl diphosphate (DMAPP) and isopentenyl diphosphate (IPP), via the mevalonate (MVA) pathway or the 1-deoxyxylulose 5-phosphate (DXP) pathway [1–3]. These precursors are combined to generate longer prenyl phosphates such as the C10 monoterpene precursor, geranyl diphosphate (GPP), the C15 sesquiterpene precursor, farnesyl diphosphate (FPP), and C20 diterpene precursor, geranylgeranyl diphosphate (GGPP) [4, 5].
Diterpenes are less volatile than monoterpenes and sesquiterpenes, and have various industrial and biological applications from resin-based adhesives to potential new drugs [6–8]. In plants, diterpenes play diverse roles such as protection against pathogens and herbivores [9] and production of growth regulators such as gibberellins [10–12]. Plants, fungi and bacteria produce gibberellins from a universal precursor, ent-kaurene, which may be synthesized from GGPP in a single step or in two steps, via the intermediate ent-copalyl diphosphate (CDP) [13]. Gibberellins are used as a sustainable means of increasing yields and stress-tolerance in agriculture and floriculture but are currently expensive, leading to calls for biotechnological approaches to reduce cost and to increase the diversity of commercially available gibberellins [13, 14]. Besides serving as the precursor to gibberellins, ent-kaurene may also be transformed into a variety of bioactive derivatives, with a range of potential applications. Anti-microbial, anti-cancer, and anti-inflammatory activities are among the properties ascribed to kaurene-derived diterpenoids, such as ent-kaur-16-en-19-oic acid (kaurenoic acid) [15]. In traditional Chinese medicine, pharmacologically active ent-kaurene diterpenoids from Isodon plants (Lamiaceae) are used to treat inflammation and cancers [16]. Finally, kaurene-derivatives may play a key role in the development of new renewable polymers [17].
Only a few studies have reported on engineering microbes to produce ent-kaurene. For example, kaurene synthase (KS) from Gibberella fujikuroi was expressed in Aspergillus nidulans and although ent-kaurene production was demonstrated, it was not quantified [18]. In another study, CDP synthase and KS from Stevia rebaudiana were expressed in Escherichia coli along with three genes from the native DXP pathway, yielding 578 mg/L ent-kaurene from a glycerol-based (20 g/L) medium in a 1 L bioreactor [19]. To develop a truly sustainable solution for microbial production of bioproducts, the next step is to transition preliminary findings from studies like these to microbial hosts capable of utilizing cheap renewable carbon sources that do not compete with our food supply, such as lignocellulose.
Lignocellulose poses a challenge in that it is difficult to deconstruct into fermentable carbon, and many deconstruction technologies can produce toxic molecules that inhibit microbial growth and productivity. Much work has been done to develop technologies for efficient lignocellulose deconstruction and generation of non-toxic hydrolysates that are easily converted into bioproducts by microbial hosts capable of consuming lignocellulosic sugars. For example, a process was recently developed that generates clean lignocellulosic hydrolysates, called DMR-EH, with low concentrations of microbial growth inhibitors such as acetate, furfural, and 5-hydroxymethylfurfural and up to 230 g/L of monomeric sugars [20, 21]. This hydrolysate has been used to produce other terpene bioproducts, such as 1,8-cineole in a robust microbial host, Rhodosporidium toruloides [22].
Rhodosporidium toruloides, has emerged as a promising host for the utilization of lignocellulosic feedstocks, in part because it can withstand osmotic stress [23] and potential growth inhibitors found in biomass hydrolysates [24]. In addition, R. toruloides adapts well to the mixed carbon sources in low-cost lignocellulosic feedstocks, utilizing mixtures of C5 and C6 sugars in combination with lignin-derived compounds such as p-coumarate [25, 26]. R. toruloides can be grown to high cell densities, surpassing 150 g/L dry cell weight in high-gravity fermentation [27]. To date, R. toruloides has been engineered to produce a number of bioproducts, including lipids, the terpene biofuel candidates 1,8-cineole and bisabolene, and the non-ribosomal peptide, indigoidine [22, 26, 28, 29]. This work explores the potential for R. toruloides to produce diterpene products from lignocellulosic biomass, targeting the production of ent-kaurene from corn stover DMR-EH hydrolysate.
Results
To produce ent-kaurene in R. toruloides, we selected kaurene synthase from Gibberella fujikuroi (GfKS) [30, 31] because it generates ent-kaurene directly from GGPP (Fig. 1) [13]. In plant and bacterial systems, ent-kaurene is synthesized from GGPP in a two-step process, via CDP, while G. fujikuroi and other fungi contain bifunctional CDP/KS enzymes that generate ent-kaurene directly from GGPP (Fig. 1) [13]. The native promoters GAPDH (glyceraldehyde 3-phosphate dehydrogenase) and ANT (adenine nucleotide translocase) were chosen for heterologous expression of GfKS based on analysis of RNAseq data from a previous study that suggests that they both are constitutive and drive a high level of gene expression [32].
Fig. 1.
Production of ent-kaurene from acetyl-CoA via the mevalonate pathway. Genes expressed in R. toruloides are listed in Table 1. Mutant FPP synthases employed in this study are designed to alter prenyl phosphate product chain length, resulting in enzymes that generate mainly GGPP from IPP and DMAPP, instead of the native FPP product
Constructs were transformed into R. toruloides by Agrobacterium tumefaciens mediated transformation (ATMT), which results in random chromosomal integration of transgenes. Typically, 40 transformants were selected for initial measurement of ent-kaurene production and then the three highest-titer strains for each construct were compared in triplicate. Following screening of strains transformed with constructs harboring PANT-GfKS (construct 1) and PGAPDH-GfKS (construct 2), we found that similar maximum ent-kaurene titers of 15-20 mg/L were reached in YPD10 medium (YPD containing 100 g/L glucose; Fig. 2; constructs are listed in Table 1). The low ent-kaurene titers observed relative to other heterologous terpenes produced in this organism (typically several hundred mg/L) suggests that the availability of the GfKS substrate, GGPP, may be limiting. Although R. toruloides produces carotenoids, they are produced at relatively low levels, indicating that native carbon flux to GGPP may be low [26].
Fig. 2.

Expression of kaurene synthase from Gibberella fujikuroi (GfKS) in R. toruloides. Ent-kaurene titer at 9 days is shown for the three highest-titer strains transformed with constructs 1 (PANT-GfKS) and 2 (PGAPDH-GfKS). (N = 3, data shown as average ± standard deviation, from a single experiment in YPD10 medium. ABFPUB identification numbers are listed in Table 1.)
Table 1.
Overview of constructs used to engineer ent-kaurene production in R. toruloides
| Plasmids | Genotypes/features | ABFPUB registry ID |
|---|---|---|
| Construct 1 | PANT-GfKS-HYGR | ABFPUB_1 |
| Construct 2 | PGAPDH-GfKS-NATR | ABFPUB_3 |
| Construct 3 | PTEF1-RtERG20(F91C)-KanMXR | ABFPUB_6 |
| Construct 4 | PTEF1-TcGGPPS(S239C,G29D)-KanMXR | ABFPUB_7 |
| Construct 5 | PTEF1-GgFPS(F112A)-KanMXR | ABFPUB_8 |
| Construct 6 | PTEF1-RtBTS1-KanMXR | ABFPUB_9 |
| Construct 7 | PANT-GfKS-PTEF1-RtERG20(F91C)-NATR | ABFPUB_10 |
| Construct 8 | PANT-GfKS-PTEF1-TcGGPPS(S239C, G295D)-NATR | ABFPUB_11 |
| Construct 9 | PANT-GfKS-PTEF1-GgFPS(F112A)-NATR | ABFPUB_12 |
| Construct 10 | PANT-GfKS-PTEF1-RtBTS1-NATR | ABFPUB_13 |
| Strains | IFO0880 (WT) | ABFPUB_14 |
| IFO0880/PGAPDH-GfKS-NATR | ABFPUB_15–17 | |
| IFO0880/PANT-GfKS-HYGR | ABFPUB_18, 30, 34 | |
| IFO0880/PGAPDH-GfKS-NATR/PTEF1-RtERG20(F91C)-KanMXR | ABFPUB_47–49 | |
| IFO0880/PGAPDH-GfKS-NATR/PTEF1-TcGGPPS(S239C,G29D)-KanMXR | ABFPUB_19, 20, 31 | |
| IFO0880/PGAPDH-GfKS-NATR/PTEF1-GgFPS(F112A)-KanMXR | ABFPUB_32, 33, 35 | |
| IFO0880/PGAPDH-GfKS-NATR/PTEF1-RtBTS1-KanMXR | ABFPUB_38–40 | |
| IFO0880/PANT-GfKS-PTEF1-RtERG20(F91C)-NATR | ABFPUB_41–43 | |
| IFO0880/PANT-GfKS-PTEF1-TcGGPPS(S239C,G295D)-NATR | ABFPUB_21, 22, 29 | |
| IFO0880/PANT-GfKS-PTEF1-GgFPS(F112A)-NATR | ABFPUB_23, 26, 36 | |
| IFO0880/PANT-GfKS-PTEF1-RtBTS1-NATR | ABFPUB_44–46 |
Strains and plasmids used in this study are available upon request through the Agile BioFoundry Strain Registry (http://public-registry.agilebiofoundry.org// [37])
GAPDH glyceraldehyde 3-phosphate dehydrogenase, ANT, adenine nucleotide translocase, TEF1 translational elongation factor, GfKS, Gibberella fujikuroi kaurene synthase (NCBI Accession Number, Q9UVY5), RtERG20(F91C) mutant farnesyl pyrophosphate synthase from R. toruloides (PRQ75922.1), TcGGPPS(S239C,G295D) mutant geranylgeranyl pyrophosphate synthase from Taxus canadensis (AAD16018), GgFPS(F112A) mutant farnesyl pyrophosphate synthase from Gallus gallus (P08836.2) and RtBTS1 geranylgeranyl pyrophosphate synthase BTS1 from R. toruloides (PRQ72579)
Similar maximum titers were reached following transformation of R. toruloides with the PANT-GfKS and PGAPDH-GfKS constructs, even though the ANT promoter is natively stronger, as indicated by ANT transcript levels and reporter studies [32]. To test whether this relative difference in promoter strength also applies to expression of the heterologous KS, GfKS copy number, transcript levels, and protein levels were measured for selected GfKS strains (Fig. 3). In strains harboring GfKS at similar copy numbers, GfKS transcript and protein levels were higher when expressed from PANT (strains ABFPUB_18 and 30) than from PGAPDH (strain ABFPUB_16). In one PGAPDH-GfKS strain, ABFPUB_15, GfKS transcript and protein levels are higher, but this is likely due to the two- to threefold higher GfKS copy number in ABFPUB_15.
Fig. 3.

Transcript level, protein level and copy number of GfKS. Average relative abundance of GfKS transcript (ΔCt), protein, and copy number on day 3. Samples are designated by their ABFPUB identification number and description. (N = 3, data shown as average ± standard deviation, from a single experiment in YPD10 medium.)
Comparison of transcript and protein levels of KS with ent-kaurene titers can also give an indication of whether gene expression or metabolite pools are limiting ent-kaurene production. The PGAPDH-GfKS strain ABFPUB_16 had the lowest transcript and protein levels of the strains examined. While other strains (e.g., the PANT-GfKS strain ABFPUB_30) had two- to fourfold higher transcript and protein levels, no substantial improvement in ent-kaurene titers was observed (Figs. 2 and 3). This indicates that a further increase in KS expression is not likely to significantly improve ent-kaurene production.
To investigate the possibility that GGPP levels may be a major factor limiting ent-kaurene titers, several GGPP synthases (GGPPSs) were selected for expression in a strain harboring GfKS. ABFPUB_16 was selected as a base strain, as it ranked most consistently as a low-variance, top-titer strain among the 80 GfKS transformants assayed in repeated screening trials. The native promoter TEF1 (translational elongation factor 1) was chosen to express each of four candidate GGPPSs: the native R. toruloides GGPPS (construct 6, PTEF1-RtBTS1); a mutant of the R. toruloides farnesyl pyrophosphate (FPP) synthase (construct 3, PTEF1-RtERG20(F91C)); a mutant GGPPS from Taxus canadensis (construct 4, PTEF1-TcGGPPS(S239C, G295D)); and a mutant FPP synthase from Gallus gallus (construct 5, PTEF1-GgFPS(F112A)). The mutations, F91C and F112A, introduced to the R. toruloides and G. gallus FPP synthases, respectively, are designed to alter prenyl phosphate product chain length, resulting in enzymes that predominantly generate GGPP instead of the native FPP product [33]. The mutations S239C and G295D were previously identified in a carotenoid-based screen for improvements to the T. canadensis GGPPS [34]. Overexpression of RtERG20(F91C) (construct 3) generally resulted in titers that were lower than or, at best, matching those of the parent strain ABFPUB_16, perhaps due to an unfavorable balance between FPP and GGPP biosynthesis, unintended enzyme inactivation due to mutation of the F91 residue, or feedback regulation of the native RtERG20 gene in R. toruloides (Fig. 4). However, overexpression of either the native R. toruloides GGPPS (construct 6) or the mutant GGPPS from T. canadensis (construct 4) resulted in more than two- and threefold increases in ent-kaurene titer, respectively. The most successful strategy was overexpression of the mutant FPP synthase from G. gallus (construct 5), which yielded a 17-fold increase in ent-kaurene titer, to 345 mg/L, in YPD10 medium. These results indicate that overexpression of GGPPS increases the available pool of GGPP for KS, resulting in an improvement in ent-kaurene titers.
Fig. 4.
Co-expression of kaurene synthase from Gibberella fujikuroi (GfKS) and various GGPP synthases in R. toruloides. Strains were constructed by transformation of strain ABFPUB_16 (PGAPDH-GfKS) with constructs 3 to 6. Ent-kaurene titer at 9 days is shown for the three highest-titer strains for each construct. (N = 3, data shown as average ± standard deviation, from a single experiment in YPD10 medium. ABFPUB identification numbers are listed in Table 1.)
Optimizing carbon flux through a biosynthetic pathway requires balancing the expression of each pathway enzyme. The overexpression of GGPPS alongside KS shifted the balance of the pathway and resulted in an increase in ent-kaurene titers. To test whether this shift in pathway balance has caused KS to become a limiting factor, designs were made to balance KS and GGPPS expression by incorporating each PTEF1-GGPPS and PANT-GfKS in a 1:1 ratio in the same construct. PANT was chosen over PGAPDH in an attempt to restore any potential deficit in KS expression in the stacked KS GGPPS strains. After transformation into WT R. toruloides by ATMT, relative performance of the four GGPPSs was observed to remain the same but higher absolute ent-kaurene titers were achieved with GfKS expression under control of PANT, suggesting that the KS may have become limiting as GGPP supply increased (Fig. 5). Co-transformation of PANT-GfKS with PTEF1-TcGGPPS(S239C, G295D) (construct 8) and PTEF1-GgFPS(F112A) (construct 9), resulted in ent-kaurene titers of 184 and 531 mg/L in YPD10 medium, respectively.
Fig. 5.
Co-transformation of kaurene synthase from Gibberella fujikuroi (GfKS) and various GGPP synthases into WT R. toruloides (constructs 7–10, Table 1). Ent-kaurene titer at 9 days is shown for the three highest-titer strains for each construct. (N = 3, data shown as average ± standard deviation, from a single experiment in YPD10 medium. ABFPUB identification numbers are listed in Table 1.)
In order to understand how co-expression of KS and GGPPS in a single construct affected expression of each of these enzymes relative to iteratively stacking individual constructs into R. toruloides, transgene copy number, transcript and protein abundances of GfKS and GgFPS(F112A) were compared in three high-titer strains (Fig. 6). Transgene copy number was three- to fourfold higher in strains ABFPUB_23 and ABFPUB_26, which were generated by transformation of a single construct, relative to strain ABFPUB_35, which was constructed by gene stacking—transformation of ABFPUB_16 with PTEF1-GgFPS(F112A). Interestingly, GfKS protein level is similar in all three strains, while and GgFPS(F112A) protein level is higher in strains ABFPUB_23 and ABFPUB_26 than in strain ABFPUB_35. Overall, the highest average ent-kaurene titer was achieved in strain ABFPUB_26, which had the highest GgFPS(F112A) protein level.
Fig. 6.
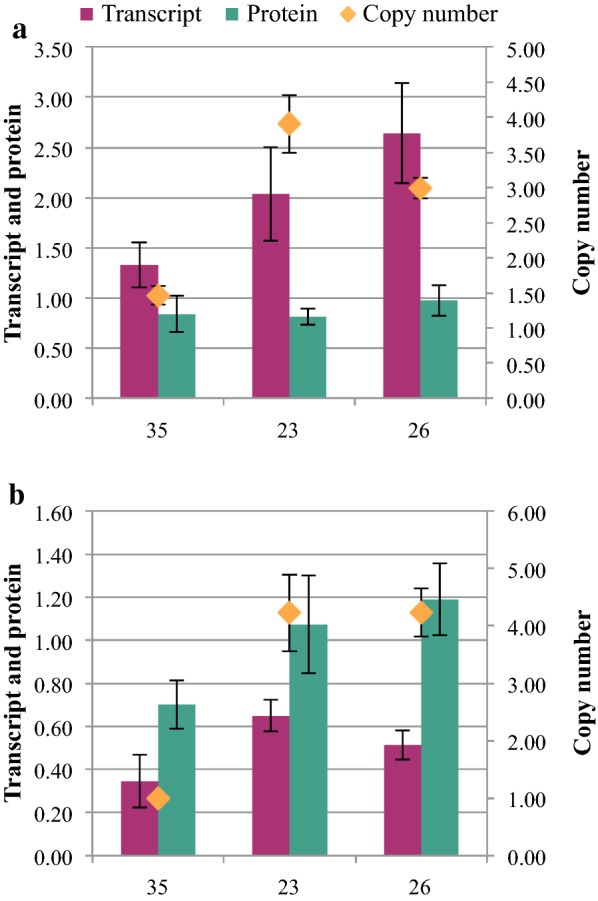
Average relative abundance of transcript (ΔCt), protein and copy number of a GfKS and b GfFPS(F112A) on day 3. Samples are designated by their ABFPUB identification number. (N = 3, data shown as average ± standard deviation, from a single experiment in YPD10 medium.)
One of the advantages of R. toruloides as a host is its ability to grow on lignocellulosic hydrolysates and to perform well when cultivated in bioreactors [26, 35]. To demonstrate production of ent-kaurene from lignocellulose, DMR-EH hydrolysate was prepared from corn stover as described previously [21]. Strain ABFPUB_26 (harboring PANT-GfKS and PTEF1-GgFPS(F112A)) was selected for scale-up to a 2 L bioreactor in a medium composed of 75% corn stover DMR-EH hydrolysate, supplemented with 10 g/L yeast extract. Under these conditions, an ent-kaurene titer of 1.44 g/L was achieved (Fig. 7). OD600 and titer increased proportionally, reaching an OD600 of 70 by 281 h; glucose was completely consumed by 161 h and xylose was fully consumed after 207 h. Strain ABFPUB_26 had produced ent-kaurene at 531 mg/L when cultivated in YPD10 medium in culture tubes, yet produced 2.7 fold more ent-kaurene when scaled up to a 2 L bioreactor and 75% DMR-EH growth medium, which contains only 13% more sugar (76 g/L glucose and 37 g/L xylose) than YPD10. This underscores the compatibility of R. toruloides with lignocellulosic feedstocks.
Fig. 7.
Sugar concentration, OD600, and ent-kaurene titer data for strain ABFPUB_26 cultivated in a 2 L bioreactor containing 75% DMR-EH hydrolysate, supplemented with 10 g/L yeast extract
Discussion
This work applies the Design, Build, Test, and Learn (DBTL) approach to engineer production of the non-native diterpene ent-kaurene in R. toruloides. The ATMT transformation method used in this study promotes random integration of constructs into the R. toruloides genome, generating strains that vary in transgene insertion site and copy number. The resulting variation in transgene expression level can be advantageous in that it adds an additional mechanism to adjust transgene expression in pathway engineering work. However, it also can confound direct comparison of different engineering strategies. Fortunately, this issue can be overcome simply by measuring transgene copy number. For example, a direct comparison between the ANT and GAPDH promoters was made in this study and we observed that PANT resulted in stronger GfKS expression than PGAPDH, in agreement with previous data comparing these promoters [32].
Interestingly, unlike previous studies that used these same strong promoters to express a sesquiterpene synthase to produce 100–500 mg/L of sesquiterpene, simply overexpressing GfKS resulted in almost tenfold lower titers of ent-kaurene [26]. Sesquiterpenes are produced from FPP (the immediate precursor of GGPP), which is a metabolite used for many cellular functions, including biosynthesis of essential sterols such as ergosterol. In contrast, GGPP is used primarily for producing low levels of non-essential carotenoids in R. toruloides. Therefore, it’s not surprising that this study found evidence that the apparent GGPP metabolite pool is lower than that of FPP. This was substantiated by the significant increase in ent-kaurene titers observed upon expression of a GGPPS. The variance in ent-kaurene titer in the GGPPS overexpression strains is relatively high, so it’s difficult to make conclusive comparisons, but the relative trend in titer suggests that higher levels of GgFPS(F112A) protein promoted higher ent-kaurene titers. These results suggest that the increased GGPPS expression level in the strains generated from a single construct in the final DBTL cycle created a better balance in the pathway, leading to higher flux toward ent-kaurene. Overall, the difference in titer in strains harboring GGPPS constructs was relatively modest, likely indicating that expression of these two terminal enzymes is reaching saturation.
While g/L titers of ent-kaurene were achieved by engineering two pathway steps, there are many improvements that can be made to further improve titer, rate and yield (TRY). Additional investigation of the impact of GGPPS expression on the balance between FPP and GGPP could prove fruitful, either from further promoter optimization for both KS and GGPPS expression or by examining other KS and GGPPS orthologs. Beyond these terminal steps in the biosynthetic pathway, optimization of the entire mevalonate pathway will likely lead to improvements in TRY. There are also several broader avenues to explore to facilitate engineering this nascent host. For example, little is known about the impact of integration locus on expression in R. toruloides and developing a better understanding of this topic is a high-priority for further investigation. Other engineering tools that would be highly beneficial for pathway optimization include methods to downregulate competing pathways, such as lipid biosynthesis. To that end, the application of RNAi has recently been demonstrated in R. toruloides, with downregulation of fatty acid synthases [36]. This tool will be valuable for future studies.
Outside of metabolic engineering approaches, process optimization of cultivation conditions will be important to improve heterologous terpene production in R. toruloides. For example, optimization of DMR-EH hydrolysate media to eliminate the use of the yeast extract in favor of a more industrially-relevant nitrogen source such as ammonium sulfate would be ideal. Also, identification of other vitamins and minerals that are limiting in DMR-EH hydrolysates will likely improve TRY. Finally, a deeper exploration of cultivation conditions such as pH, mixing, aeration, and temperature are also needed in this relatively new host organism. Even without these important optimizations, the final titer of 1.44 g/L achieved in this study is the highest reported ent-kaurene titer in any microbial cell factory.
Conclusion
This study builds upon previous work demonstrating the potential of R. toruloides as a robust and versatile host for the production of both mono- and sesquiterpenes, and is the first demonstration of the production of a non-native diterpene in this organism. It provides a roadmap for rapid high-titer diterpene production in R. toruloides; it is possible that other diterpene synthases could be combined with the GGPPS GgFPS(F112A) in a single expression construct to achieve production of g/L quantities of the diterpene. These results, in combination with previous studies on mono- and sesquiterpenes, indicate that R. toruloides is an ideal host for the production of a range of different heterologous terpene bioproducts from cheap renewable carbon sources, such as lignocellulosic biomass.
Materials and methods
Plasmid design and construction
Plasmids were based on a binary vector for Agrobacterium tumefaciens mediated transformation (ATMT) containing a bacterial expression cassette conferring resistance to kanamycin and R. toruloides expression cassettes conferring resistance to nourseothricin (NAT), G418 (KanMX) or hygromycin B (HYG) (Table 1). Coding sequences were optimized for expression in R. toruloides, synthesized, and cloned into ATMT plasmid backbones described previously [22, 28], by Genscript (Piscataway, NJ).
Transformation and screening of R. toruloides
Transformation of R. toruloides was achieved by ATMT as described previously [22]. Prior to screening, transformants were grown on yeast peptone dextrose (YPD, BD Biosciences, 242820, San Jose, CA) agar containing 300 μg/mL cefotaxime (TCI America, TCI-C2224-25G, Portland, OR) and 300 μg/mL carbenicillin (Sigma-Aldrich, C1389-5G, St. Louis, MO) to prevent the growth of A. tumefaciens. Plates were grown at 30 °C for three days and single colonies were inoculated into culture tubes containing 5 mL lysogeny broth (LB, Miller, VWR, J106-500G, Radnor, PA) and cultured overnight at the same temperature with shaking at 200 rpm (Adolf Kühner AG, SBM-X, Birsfelden, Switzerland). Optical density (OD) was determined by measuring absorbance at 600 nm (OD600) with a SpectraMax Plus 384 Microplate Reader (Molecular Devices, PLUS384, San Jose, CA). Cultures were inoculated into 10 mL YPD at a 1:100 dilution and a 20% (v/v) dodecane (Sigma-Aldrich, D221104) overlay was added to capture ent-kaurene. Strains were grown at 30 °C with shaking at 200 rpm for 9–10 days. After an initial round of screening, ent-kaurene titer of the three highest producing strains was confirmed in triplicate for each construct by growth in YPD10 (YPD containing 100 g/L glucose).
Quantification of ent-kaurene
Following growth of engineered R. toruloides cultures, the dodecane phase was sampled and diluted 1:40 into dodecane containing 40 mg/L pentadecane (Sigma-Aldrich, 76510), and analyzed by gas chromatography—mass spectrometry (GC–MS) using an Agilent 6890 Plus gas chromatograph (Agilent Technologies, G1530A, Santa Clara, CA) connected to an Agilent 5973 Network mass spectrometer (Agilent Technologies, G1099A). 1 µL of each sample was injected by a CombiPal autosampler (CTC Analytics, MXY 02-00B, Zwingen, Switzerland). Analytes were separated on a DB-5MS column (30 m long, 0.25 mm internal diameter, 0.25 μm film thickness, Agilent Technologies, 122-5532) using the following oven parameters: hold for 0.5 min at an initial temperature of 100 °C, followed by a temperature ramp of 30 °C/min to 250 °C, a ramp of 10 °C/min to 270 °C, and a ramp of 30 °C/min to 300 °C. The mass spectrometer was operated in selected ion mode, with target ions (m/z) of 70, 85, 139 and 154. A standard curve was generated by running ent-kaurene standards at concentration range of 5–80 µg/mL. Analysis was performed using Enhanced ChemStation (Agilent Technologies, MSD Chemstation E.02.00.493) with ent-kaurene peak areas normalized to peak areas for pentadecane. The ent-kaurene standard was a gift from Dr. Joe Chappell, University of Kentucky, Lexington, KY.
Cultivation at 2 L bioreactor scale
ABFPUB_26 was selected for growth in lignocellulosic hydrolysate at 2 L bioreactor scale. Lignocellulosic hydrolysate was prepared from corn stover by deacetylation, mechanical refining and enzymatic hydrolysis, as described previously, and is referred to as DMR-EH hydrolysate [21]. A BIOSTAT B® fermentation system (Sartorius AG., Goettingen, Germany) was employed in batch mode, using a jacketed 2 L borosilicate glass vessel (UniVessel®, Sartorius AG, Goettingen, Germany) equipped with two 6-blade Rushton impellers, a dissolved oxygen (DO) probe (Hamilton VisiFerm DO 225, Bonaduz, Switzerland), and a pH probe (Hamilton EasyFerm Plus VP 225, Bonaduz, Switzerland). Seed cultures were grown to exponential phase and then used to inoculate 0.75 L aqueous media (75% DMR hydrolysate, 10 g/L yeast extract, and 30 mg/L cefotaxime), to which a 20% organic solvent overlay (150 mL dodecane spiked with 200 mg/L pentadecane as an internal standard) was added to capture ent-kaurene. The culture was grown at 30 °C with aeration maintained at 0.37 LPM and agitation at 400 rpm. After initial adjustment of the growth medium pH to 5.0, the pH was not controlled during growth. Process values were monitored and recorded using the integrated Sartorius data acquisition software (BioPAT MFCS/win). Sugar consumption, OD, and ent-kaurene production were measured over a period of 12 days. Ent-kaurene was measured by sampling of the dodecane phase, dilution, and analysis by GC–MS.
Determination of glucose and xylose
Sugars were quantified on a Dionex Ultimate 3000 system UHPLC (Agilent Technologies) using an Aminex HPX-87H column (Bio-Rad, Hercules, CA) and Thermo Scientific™ RefractoMax 520 Refractive Index Detector (RID) held at 35 °C. Prior to analysis, samples were filtered through 0.45 μm filters (VWR Centrifugal Filters) by centrifugation at 3000×g for 3 min. Samples were run for 26 min using an isocratic 4 mM sulfuric acid mobile phase at 0.6 mL min−1 and 65 °C. Glucose, xylose, and arabinose standards were prepared and diluted to create a 7-point calibration curve ranging from 0.0625 to 4.0 mg mL−1. Standards were run at the beginning and end of each run, and sugar concentrations were calculated using the Chromeleon 7 software package.
Targeted proteomics
Cultures were grown in culture tubes with 5 mL YPD10 medium and a dodecane overlay. Time points were collected on day 1 and 3. Protein was extracted using a method based on a previously established protocol [38]. Each cell pellet, corresponding to 6 OD units, was diluted in 200 µL of H2O and transferred to 2 mL pre-filled Micro-Organism Lysing Mix glass bead tubes and bead beat in a Bead Ruptor Elite bead mill homogenizer (OMNI International, Kennesaw, Georgia) at speed 5.5 for 45 s. After bead beating, the lysate was immediately placed in an ice-block and then spun into a 4 mL tube at 1,000×g for 10 min at 4 °C. To separate the protein, metabolites and lipids, 1 mL cold (− 20 °C) chloroform:methanol mix (prepared 2:1 (v/v)) was pipetted into a chloroform compatible 2 mL Sorenson MulTI™ SafeSeal™ microcentrifuge tubes (Sorenson bioscience, Salt Lake City, UT) inside an ice-block. The 200 µL of sample homogenate was then added to the Sorenson tube at a ratio of 1:5 sample:chloroform mix (2:1 (v/v)) and vigorously vortexed. The sample was then placed in the ice block for 5 min and then vortexed for 10 s followed by centrifugation at 10,000×g for 10 min at 4 °C. The upper water-soluble metabolite phase and the lower lipid soluble phase was removed. The remaining protein interlayer had 1 mL of cold 100% methanol added to each, vortexed and centrifuged again at 10,000×g for 10 min at 4 °C to pellet the protein. The methanol was then decanted off and the samples were placed in a fume hood to dry for ~ 10 min. 200 µL of an 8 M urea solution was added to the protein pellet and vortexed into solution. A bicinchoninic acid (BCA) assay (Thermo Scientific, Waltham, MA) was performed to determine protein concentration. Following the assay, 10 mM dithiothreitol (DTT) was added to the samples and incubated at 60 °C for 30 min with constant shaking at 800 rpm followed by the addition of 40 mM iodoacetamide (IAA) with 30 min of room temperature incubation in the dark. Samples were then diluted eightfold for preparation for digestion with 100 mM NH4HCO3, 1 mM CaCl2 and sequencing grade trypsin (USB, Santa Clara, CA) was added to all protein samples at a 1:50 (w/w) trypsin-to-protein ratio for 3 h at 37 °C. Digested samples were desalted using a 4-probe positive pressure Gilson GX-274 ASPEC™ system (Gilson Inc., Middleton, WI) with Discovery C18 50 mg/1 mL solid phase extraction tubes (Supelco, St. Louis, MO), using the following protocol: 3 mL of methanol was added for conditioning followed by 3 mL of 0.1% trifluoroacetic acid (TFA) in H2O. The samples were then loaded onto each column followed by 4 mL of 95:5 water:acetonitrile, 0.1% TFA. Samples were eluted with 1 mL 80:20 acetonitrile:water, 0.1% TFA. The samples were concentrated down to ~ 100 µL using a Speed Vac and a final BCA was performed to determine the peptide concentration and samples were diluted to 0.20 µg/µL with nanopure water for targeted proteomics analysis.
Targeted proteomics was performed via Liquid Chromatography (LC)–Selected Reaction Monitoring (SRM) approach. Five peptides per protein were initially selected based on their SRM suitability scores predicated by CONSeQuence [39, 40] software tools. All the peptides were further blasted to ensure their uniqueness to target proteins in the organism. Crude synthetic heavy isotope-labeled (e.g., 13C/15 N on C-terminal lysine and arginine) peptides were purchased from New England Peptide (Gardner, MA). Upon receiving, the crude synthetic heavy peptides were mixed together and diluted with 0.1% formic acid in 15% acetonitrile in water to obtain a nominal concentration of 3 pmol/µL for each individual peptide. The heavy peptide mixture stock solution was aliquoted and stored at − 80 °C until further use.
To develop targeted proteomics assay, all the SRM precursor-fragment ion pairs (i.e., transitions) were first analyzed using LC-SRM by spiking heavy peptides in test samples. Three transitions per peptide and three peptides per protein were selected in a final assay based on their LC performance, MS response, transition interferences, endogenous peptide detectability. Collision energies of transitions were obtained using empirical equations provided in Skyline software [41]. The selected peptides, their transitions and collision energy in the final assay are listed in Additional file 1: Table S1.
Crude heavy peptide mixture stock solution was spiked in the 0.20 µg/µL peptide samples at a nominal concentration of 37.5 fmol/µL for each peptide. LC-SRM analysis utilized a nanoACQUITY UPLC® system (Waters Corporation, Milford, MA) coupled online to a TSQ Altis™ triple quadrupole mass spectrometer (Thermo Fisher Scientific). The UPLC® system was equipped with an ACQUITY UPLC BEH 1.7 µm C18 column (100 µm i.d. × 10 cm) and the mobile phases were (A) 0.1% formic acid in water and (B) 0.1% formic acid in acetonitrile. 2 µL of sample (i.e., 0.4 µg of peptides) were loaded onto the column and separated using a 110-min gradient profile as follows (min:flow-rate-µL/min: %B): 0:0.4:1, 6:0.6:1, 7:0.4:1, 9:0.4:6, 40:0.4:13, 70:0.4:22, 80:0.4:40, 85:0.4:95, 91:0.5:95, 92:0.5:95, 93:0.5:50, 94:0.5:95, 95:0.6:1, 98:0.4:1. The LC column is operated with a temperature of 42 °C. The TSQ Altis™ triple quadrupole mass spectrometer was operated with ion spray voltages of 2100 ± 100 V and a capillary inlet temperature of 350 °C. Tube lens voltages were obtained from automatic tuning and calibration without further optimization. Both Q1 and Q3 were set at unit resolution of 0.7 FWHM and Q2 gas pressure was optimized at 1.5 mTorr. The transitions were scanned with a 30 min retention time window and a duty cycle of 0.8 s.
All the LC-SRM data were imported into Skyline software and the peak boundaries were manually inspected to ensure correct peak assignment and peak boundaries. Peak detection and integration were determined based on two criteria: 1. The same LC retention time and 2. Approximately the same relative peak intensity ratios across multiple transitions between the light peptides and heavy peptide standards. The total peak area ratios of endogenous light peptides and their corresponding heavy isotope-labeled internal standards were then exported from Skyline software as Ratio-to-Standard. For each peptide, the total peak area ratios of individual samples were normalized to the average total peak area ratio of all the samples. For each sample, protein abundance was calculated as an average of the normalized total peak area ratios of all three peptides of a protein.
Measurement of transcript levels
RNA was extracted using the Maxwell 16 AS2000 instrument with a Maxwell RSC Plant RNA Kit (Promega, AS1500, Madison, WI). RNA was quantified with a NanoDrop™ 2000 (Thermo Scientific) and 25 ng was used once linear range had been identified. Relative abundance (ΔCt) of transcript levels for GfKS was measured using Superscript IV One-Step RT-PCR System (Thermo Fisher Scientific, 12594100, Waltham, MA) with EvaGreen (Biotium, 31000, Hayward, CA) and a CFX96 Real-Time System C1000 Touch Thermal Cycler (Bio-Rad). Relative abundance was compared with housekeeping genes histone H3 (XP_016270870.1) and actin (XP_016271443.1). Primers are listed in Addition file 2: Table S2.
Copy number quantification
1.5 mL aliquots were sampled from 3-day R. toruloides cultures, and centrifuged at 3,000×g for 5 min to pellet cells. Genomic DNA was harvested from the cell pellets using the Quick-DNA™ Fungal/Bacterial Miniprep Kit (Zymo Research) following the manufacturer’s instructions. Genomic DNA was quantified by using a NanoDrop™ 2000 (Thermo Scientific). To determine the relative copy number of the introduced transgenes, quantitative PCR was performed using the PowerUp™ SYBR™ Green Master Mix (Thermo Scientific) on a CFX384 Touch Real-Time PCR Detection System (BioRad) using the manufacturer’s instructions. Each reaction was set up in triplicate with 1 ng of genomic DNA as template. PCR products (approximately 1 kb) spanning the qPCR amplicons were amplified from genomic DNA for the native sequences and plasmid DNA for the transgenes. These PCR products were gel purified using the Qiaquick gel extraction kit (Qiagen) and used to generate standard curves for each qPCR primer set. Standard curves were used to calculate the copy number of transgenes relative to the native actin and GAPDH (EGU13160.1) genes. Primers are listed in Additional file 2: Table S2.
Supplementary information
Additional file 1: Table S1. Peptide standards used to quantify GkKs and GgFPS(F112A) protein levels in R toruloides and their fragmentation ions.
Additional file 2: Table S2. Primer sequences used for q-PCR and qRT-PCR.
Acknowledgements
We would like to thank Sam Coradetti and Joonhoon Kim for providing data analysis help, critical feedback, and resources. We would also like to thank the National Renewable Energy Lab for providing the DMR-EH hydrolysate used in this project. A portion of this research was performed using EMSL (grid.436923.9), a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory.
Authors’ contributions
XZ, JMG, JMS, MBTG, JK, and GMG conceptualized the project and designed the study. GMG, XZ, JK MBTG, JPP, ERS, YG, NMM, CDN, KEBJ, DMC, and GP conducted experiments, collected data, and performed data analysis. GMG, JK, XZ, JPP, ERS, YG, NMM, CDN, KEBJ, DMC, GP, JKM, DT, JMS and JMG composed the manuscript, gave critical feedback on the manuscript and provided resources. All authors read and approved the final manuscript.
Funding
Sandia National Laboratories is a multi-mission laboratory managed and operated by National Technology and Engineering Solutions of Sandia, LLC., a wholly owned subsidiary of Honeywell International, Inc., for the U.S. Department of Energy’s National Nuclear Security Administration under contract DE-NA0003525. This research was part of the Agile BioFoundry (https://agilebiofoundry.org) supported by the U. S. Department of Energy, Energy Efficiency and Renewable Energy, Bioenergy Technologies Office, through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U. S. Department of Energy. The views and opinions of the authors expressed herein do not necessarily state or reflect those of the United States Government or any agency thereof. Neither the United States Government nor any agency thereof, nor any of their employees, makes any warranty, expressed or implied, or assumes any legal liability or responsibility for the accuracy, completeness, or usefulness of any information, apparatus, product, or process disclosed, or represents that its use would not infringe privately owned rights. The Department of Energy will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (http://energy.gov/downloads/doe-public-access-plan).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Gina M. Geiselman and Xun Zhuang contributed equally to this work
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12934-020-1293-8.
References
- 1.Li R, Chou WKW, Himmelberger JA, Litwin KM, Harris GG, Cane DE, et al. Reprogramming the chemodiversity of terpenoid cyclization by remolding the active site contour of epi-isozizaene synthase. Biochemistry. 2014 doi: 10.1021/bi401643u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sacchettini JC, Poulter CD. Creating isoprenoid diversity. Science. 1997 doi: 10.1126/science.277.5333.1788. [DOI] [PubMed] [Google Scholar]
- 3.Christianson DW. Chemistry. Roots of biosynthetic diversity. Science. 2007 doi: 10.1126/science.1141630. [DOI] [PubMed] [Google Scholar]
- 4.Poulter CD, Rilling HC. The prenyl transfer reaction. Enzymic and mechanistic studies of the 1′-4 coupling reaction in the terpene biosynthetic pathway. Acc Chem Res. 1978 doi: 10.1021/ar50128a004. [DOI] [Google Scholar]
- 5.Poulter CD. Farnesyl diphosphate synthase. A paradigm for understanding structure and function relationships in E-polyprenyl diphosphate synthases. Phytochem Rev. 2006 doi: 10.1007/s11101-005-4887-1. [DOI] [Google Scholar]
- 6.Faustino C, Neto Í, Fonte P, Macedo A. Cytotoxicity and chemotherapeutic potential of natural rosin abietane diterpenoids and their synthetic derivatives. Curr Pharm Des. 2018 doi: 10.2174/1381612825666190112162817. [DOI] [PubMed] [Google Scholar]
- 7.Barros de Alencar Marcus Vinícius Oliveira, de Castro e Sousa João Marcelo, Rolim Hercília Maria Lins, de Medeiros Maria das Graças Freire, Cerqueira Gilberto Santos, de Castro Almeida Fernanda Regina, Citó Antônia Maria das Graças Lopes, Ferreira Paulo Michel Pinheiro, Lopes José Arimatéia Dantas, de Carvalho Melo-Cavalcante Ana Amélia, Islam Md. Torequl. Diterpenes as lead molecules against neglected tropical diseases. Phytotherapy Research. 2016;31(2):175–201. doi: 10.1002/ptr.5749. [DOI] [PubMed] [Google Scholar]
- 8.Prieto JM, Silveira D. Natural cytotoxic diterpenoids, a potential source of drug leads for melanoma therapy. Curr Pharm Des. 2018 doi: 10.2174/1381612825666190111143648. [DOI] [PubMed] [Google Scholar]
- 9.Block AK, Vaughan MM, Schmelz EA, Christensen SA. Biosynthesis and function of terpenoid defense compounds in maize (Zea mays) Planta. 2019 doi: 10.1007/s00425-018-2999-2. [DOI] [PubMed] [Google Scholar]
- 10.Xu Y, Charles MT, Luo Z, Mimee B, Tong Z, Véronneau P-Y, et al. Ultraviolet-C priming of strawberry leaves against subsequent Mycosphaerella fragariae infection involves the action of reactive oxygen species, plant hormones, and terpenes. Plant Cell Environ. 2019 doi: 10.1111/pce.13491. [DOI] [PubMed] [Google Scholar]
- 11.Ratnadass A, Wink M. The phorbol ester fraction from Jatropha curcas seed oil: potential and limits for crop protection against insect pests. Int J Mol Sci. 2012 doi: 10.3390/ijms131216157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamaguchi S. Gibberellin metabolism and its regulation. Annu Rev Plant Biol. 2008 doi: 10.1146/annurev.arplant.59.032607.092804. [DOI] [PubMed] [Google Scholar]
- 13.Salazar-Cerezo S, Martínez-Montiel N, García-Sánchez J, Pérez-Y-Terrón R, Martínez-Contreras RD. Gibberellin biosynthesis and metabolism: a convergent route for plants, fungi and bacteria. Microbiol Res. 2018 doi: 10.1016/j.micres.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 14.Camara MC, Vandenberghe LPS, Rodrigues C, de Oliveira J, Faulds C, Bertrand E, et al. Current advances in gibberellic acid (GA3) production, patented technologies and potential applications. Planta. 2018 doi: 10.1007/s00425-018-2959-x. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi JA, Gomes DC, Lyra FH, Dos Santos GF, Martins LR. The remarkable structural diversity achieved in ent-Kaurane Diterpenes by fungal biotransformations. Molecules. 2014 doi: 10.3390/molecules19021856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du G, Gong H-Y, Feng K-N, Chen Q-Q, Yang Y-L, Fu X-L, et al. Diterpene synthases facilitating production of the kaurane skeleton of eriocalyxin B in the medicinal plant Isodon eriocalyx. Phytochemistry. 2019 doi: 10.1016/j.phytochem.2018.11.015. [DOI] [PubMed] [Google Scholar]
- 17.Hillwig ML, Mann FM, Peters RJ. Diterpenoid biopolymers: new directions for renewable materials engineering. Biopolymers. 2011 doi: 10.1002/bip.21538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bromann K, Toivari M, Viljanen K, Ruohonen L, Nakari-Setälä T. Engineering Aspergillus nidulans for heterologous ent-kaurene and gamma-terpinene production. Appl Microbiol Biotechnol. 2016 doi: 10.1007/s00253-016-7517-5. [DOI] [PubMed] [Google Scholar]
- 19.Kong MK, Kang H-J, Kim JH, Oh SH, Lee PC. Metabolic engineering of the Stevia rebaudiana ent-kaurene biosynthetic pathway in recombinant Escherichia coli. J Biotechnol. 2015 doi: 10.1016/j.jbiotec.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 20.Chen X, Shekiro J, Pschorn T, Sabourin M, Tao L, Elander R, et al. A highly efficient dilute alkali deacetylation and mechanical (disc) refining process for the conversion of renewable biomass to lower cost sugars. Biotechnol Biofuels. 2014 doi: 10.1186/1754-6834-7-98. [DOI] [Google Scholar]
- 21.Chen X, Kuhn E, Jennings EW, Nelson R, Tao L, Zhang M, et al. DMR (deacetylation and mechanical refining) processing of corn stover achieves high monomeric sugar concentrations (230 g L− 1) during enzymatic hydrolysis and high ethanol concentrations (> 10% v/v) during fermentation without hydrolysate purification or concentration. Energy Environ Sci. 2016 doi: 10.1039/C5EE03718B. [DOI] [Google Scholar]
- 22.Zhuang X, Kilian O, Monroe E, Ito M, Tran-Gymfi MB, Liu F, et al. Monoterpene production by the carotenogenic yeast Rhodosporidium toruloides. Microb Cell Fact. 2019 doi: 10.1186/s12934-019-1099-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh G, Jawed A, Paul D, Bandyopadhyay KK, Kumari A, Haque S. Concomitant production of lipids and carotenoids in Rhodosporidium toruloides under osmotic stress using response surface methodology. Front Microbiol. 2016 doi: 10.3389/fmicb.2016.01686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu C, Zhao X, Zhao J, Wu S, Zhao ZK. Effects of biomass hydrolysis by-products on oleaginous yeast Rhodosporidium toruloides. Bioresour Technol. 2009 doi: 10.1016/j.biortech.2009.04.041. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez A, Ersig N, Geiselman GM, Seibel K, Simmons BA, Magnuson JK, et al. Conversion of depolymerized sugars and aromatics from engineered feedstocks by two oleaginous red yeasts. Bioresour Technol. 2019 doi: 10.1016/j.biortech.2019.121365. [DOI] [PubMed] [Google Scholar]
- 26.Yaegashi J, Kirby J, Ito M, Sun J, Dutta T, Mirsiaghi M, et al. Rhodosporidium toruloides: a new platform organism for conversion of lignocellulose into terpene biofuels and bioproducts. Biotechnol Biofuels. 2017 doi: 10.1186/s13068-017-0927-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Zhao Z, Bai F. High-density cultivation of oleaginous yeast Rhodosporidium toruloides Y4 in fed-batch culture. Enzyme Microb Technol. 2007 doi: 10.1016/j.enzmictec.2007.02.008. [DOI] [Google Scholar]
- 28.Zhang S, Skerker JM, Rutter CD, Maurer MJ, Arkin AP, Rao CV. Engineering Rhodosporidium toruloides for increased lipid production. Biotechnol Bioeng. 2016 doi: 10.1002/bit.25864. [DOI] [PubMed] [Google Scholar]
- 29.Wehrs M, Gladden JM, Liu Y, Platz L, Prahl J-P, Moon J, et al. Sustainable bioproduction of the blue pigment indigoidine: Expanding the range of heterologous products in R. toruloides to include non-ribosomal peptides. Green Chem. 2019 doi: 10.1039/C9GC00920E. [DOI] [Google Scholar]
- 30.Zhuang X. Engineering Novel Terpene Production Platforms in the Yeast Saccharomyces cerevisiae. Doctoral dissertation. University of Kentucky; 2013.
- 31.Toyomasu T, Kawaide H, Ishizaki A, Shinoda S, Otsuka M, Mitsuhashi W, et al. Cloning of a full-length cDNA encoding ent-kaurene synthase from Gibberella fujikuroi: functional analysis of a bifunctional diterpene cyclase. Biosci Biotechnol Biochem. 2000 doi: 10.1271/bbb.64.660. [DOI] [PubMed] [Google Scholar]
- 32.Nora LC, Wehrs M, Kim J, Cheng J-F, Tarver A, Simmons BA, et al. A toolset of constitutive promoters for metabolic engineering of Rhodosporidium toruloides. BioRxiv. 2019 doi: 10.1101/592774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tarshis LC, Proteau PJ, Kellogg BA, Sacchettini JC, Poulter CD. Regulation of product chain length by isoprenyl diphosphate synthases. Proc Natl Acad Sci USA. 1996 doi: 10.1073/pnas.93.26.15018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leonard E, Ajikumar PK, Thayer K, Xiao W-H, Mo JD, Tidor B, et al. Combining metabolic and protein engineering of a terpenoid biosynthetic pathway for overproduction and selectivity control. Proc Natl Acad Sci USA. 2010 doi: 10.1073/pnas.1006138107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sundstrom E, Yaegashi J, Yan J, Masson F, Papa G, Rodriguez A, et al. Demonstrating a separation-free process coupling ionic liquid pretreatment, saccharification, and fermentation with Rhodosporidium toruloides to produce advanced biofuels. Green Chem. 2018 doi: 10.1039/C8GC00518D. [DOI] [Google Scholar]
- 36.Liu X, Zhang Y, Liu H, Jiao X, Zhang Q, Zhang S, et al. RNA interference in the oleaginous yeast Rhodosporidium toruloides. FEMS Yeast Res. 2019 doi: 10.1093/femsyr/foz031. [DOI] [PubMed] [Google Scholar]
- 37.Ham TS, Dmytriv Z, Plahar H, Chen J, Hillson NJ, Keasling JD. Design, implementation and practice of JBEI-ICE: an open source biological part registry platform and tools. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakayasu ES, Nicora CD, Sims AC, Burnum-Johnson KE, Kim YM, Kyle JE, et al. MPLEx: a robust and universal protocol for single-sample integrative proteomic, metabolomic, and lipidomic analyses. Msystems. 2016 doi: 10.1128/mSystems.00043-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eyers CE, Lawless C, Wedge DC, Lau KW, Gaskell SJ, Hubbard SJ. CONSeQuence: prediction of reference peptides for absolute quantitative proteomics using consensus machine learning approaches. Mol Cell Proteomics. 2011 doi: 10.1074/mcp.M110.003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Searle BC, Egertson JD, Bollinger JG, Stergachis AB, MacCoss MJ. Using data independent acquisition (DIA) to model high-responding peptides for targeted proteomics experiments. Mol Cell Proteomics. 2015 doi: 10.1074/mcp.M115.051300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010 doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Peptide standards used to quantify GkKs and GgFPS(F112A) protein levels in R toruloides and their fragmentation ions.
Additional file 2: Table S2. Primer sequences used for q-PCR and qRT-PCR.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.






