Abstract
Comprehensive genomic profiling using next-generation sequencing (NGS) enables the identification of multiple genomic biomarkers established in advanced gastrointestinal (GI) cancers. However, tissue-based NGS has limitations, such as long turnaround time and failure to detect tumour heterogeneity. Recently, the analysis of circulating tumour DNA (ctDNA) using polymerase chain reaction-based or NGS-based methods has demonstrated the capability to detect genomic alterations with high accuracy compared with tumour tissue analysis with short turnaround time and identify heterogeneous resistance mechanisms. Furthermore, ctDNA analysis can be repeatedly performed on disease progression to clarify resistant clones. Clinical trials that test the outcome of a selected targeted therapy based on a ctDNA result are ongoing to prospectively evaluate the clinical utility of ctDNA analysis. Furthermore, the improvement of ctDNA analysis beyond current technical limits of mutation-based ctDNA detection methods has expanded the potential for detecting the presence of tumours in patients with no clinically evident disease, such as minimal residual disease and early cancer. Although a careful understanding of the advantages and limitations are required and further prospective studies are needed, the ctDNA analysis has the potential to overcome several challenges in the treatment of various types of cancers at all stages, including GI cancers.
Keywords: circulating tumor DNA, gastrointestinal cancer
Introduction
Preclinical studies and clinical trials involved in the treatment of solid tumours, including gastrointestinal (GI) cancers, have established several predictive genomic biomarkers, such as RAS and BRAF mutations for metastatic colorectal cancer (mCRC), HER2 (ERBB2) amplification for metastatic gastro-oesophageal cancer, germline BRCA mutations for metastatic pancreatic cancer and microsatellite instability (MSI) and NTRK fusions for advanced solid tumours.1–7 Furthermore, advances in next-generation sequencing (NGS) technologies have enabled large-scale genomic profiling in GI cancers and have elucidated potentially targetable alterations. Tumour genomic profiling using NGS testing is now widely used in clinical practices for patients with advanced GI cancers to identify genomic alterations that can be therapeutically targeted.
Tissue-based NGS, however, has inherent limitations. For example, the long turnaround time between the receipt of tissue samples and reporting results may delay the start of treatment. Furthermore, recent studies have revealed that multiple genomic changes can arise because of clonal evolution under treatment pressure in cancer, resulting in acquired secondary resistance.8 Therefore, the longitudinal surveillance of clonal evolution is required to identify secondary resistance or adequately select subsequent treatments. However, repeat tissue biopsies are sometimes challenging to perform because of the inherent risk of complications. Moreover, biopsies or tissue sections represent only a single snapshot of the tumour in time and space; therefore, they often fail to detect intratumoural genetic heterogeneity, which is a great challenge for the optimal treatment selection for GI cancers.
Recent technical advances have enabled the analysis of tumour materials obtained from the blood or other body fluids. These liquid biopsies can be used to assess intratumoural genetic heterogeneity and overcome the limitations of tissue analyses. Specifically, the analysis of circulating tumour DNA (ctDNA), which is tumour-derived fragmented DNA released into the bloodstream, has been suggested to have clinical utility in detecting genomic alterations in various types of cancers. Furthermore, evolving technologies of ctDNA analysis using NGS-based methods have expanded the potential of ctDNA analysis for genomic profiling as an alternative for tissue-based NGS and for detecting the presence of a tumour in patients with no clinically evident disease. In this review, we focus on the utility of ctDNA analyses for the identification of predictive genomic biomarkers for GI cancers and the potential for detecting minimal residual disease (MRD) and early cancers with no clinically evident disease.
Comparison between tumour and ctDNA analysis for validated biomarkers in advanced GI cancers
Assays available for ctDNA analysis can be categorised into two general classes: those targeted for a single or small number of variants and those aimed at a broader coverage.9 Targeted assays generally use one of several polymerase chain reaction (PCR)-based strategies, such as BEAMing (beads, emulsion, amplification and magnetics) or droplet digital PCR (ddPCR) method, for the detection of specific known variants, often at very low levels within the circulating cell-free DNA (cfDNA) composed of germline DNA from normal cells.10 11 On the contrary, broad-coverage assays use NGS-based approaches and have the capability of detecting a larger number of variants in multiple genes, often examining parts of >50 genes to be applied to multiple different tumour types. Modified NGS-based approaches incorporating deep sequencing coverage, molecular barcoding methods and error-suppression algorithms have improved the limits of detection.12–15 Currently available PCR-based or NGS-based clinical ctDNA assays are listed in table 1.
Table 1.
Currently available PCR or NGS-based clinical ctDNA assays
| Method | Assay | Cancer | Gene | Company |
| PCR-based | Cobas EGFR Mutations Test v2 | NSCLC | EGFR del19, EGFR L858R and EGFR T790M | Roche Molecular Diagnostics |
| Therascreen EGFR RGQ Plasma PCR kit | NSCLC | EGFR del19 and EGFR L858R | Qiagen Inc | |
| AmoyDx Super-ARMS EGFR mutation test | NSCLC | EGFR del19, EGFR L858R and EGFR T790M | AmoyDx | |
| OncoBEAM RAS CRC Kit | CRC | KRAS and NRAS mutations | Sysmex Inostics | |
| Idylla ctKRAS Mutation Test and Idylla ctNRAS-BRAF Mutation Test | CRC | KRAS, NRAS and BRAF mutations | Biocartis, Inc. | |
| NGS-based | Guardant360 | Solid tumours | 74 genes and MSI | Guardant Health |
| FoundationOne Liquid | Solid tumours | 70 genes and MSI | Foundation Medicine Inc. | |
| PlasmaSELECT | Solid tumours | 64 genes and MSI | Personal Genome Diagnostics | |
| Oncomine Lung cfDNA Assay | NSCLC | 11 genes | ThermoFisher Scientific | |
| Reveal ctDNA 28 Kit | Solid tumours | 28 genes | ArcherDX | |
| OptiSeq NGS Pan-Cancer Panel | Solid tumours | 65 genes | DiaCarta |
CRC, colorectal cancer; ctDNA, circulating tumour DNA; MSI, microsatellite instability; NGS, next-generation sequencing; NSCLC, non-small cell lung cancer; PCR, polymerase chain reaction.
Mutations in RAS are negative predictive biomarkers that have been validated in prospective–retrospective or retrospective analyses in randomised studies with anti-epidermal growth factor receptor (EGFR) antibodies for mCRC. Several studies on PCR-based ctDNA assays have detected RAS mutations with high accuracy compared with validated tissue RAS testing.16–20 In addition to retrospective studies that compared with the progression-free survival (PFS) of patients with mCRC treated with an anti-EGFR therapy using tissue RAS versus ctDNA, RAS testing results indicated a similar PFS following first-line19 and second/third-line treatments.21 The OncoBEAMTM RAS CRC Kit, which detects RAS mutations in ctDNA derived from mCRC by a BEAMing method, is the only ctDNA assay approved in Europe and Japan for ctDNA genomic biomarker testing for GI cancers.
Recently, NGS-based ctDNA assays have demonstrated a high correlation with ddPCR assays for the measurement of mutant allele fractions (MAF) of genes, including RAS mutations in mCRC.22 23 Furthermore, multiple NGS-based ctDNA assays have been shown to detect genomic alterations in advanced GI cancers, such as RAS and BRAF mutations in CRC and HER2 amplification in gastro-oesophageal cancer, with high accuracy compared with tissue NGS analysis (table 2).23–28 One of these investigations showed that the turnaround time was 18 days and 7 days for tumour tissue and ctDNA analysis in this study, respectively.25
Table 2.
Concordance of genomic alteration statuses between tumour tissue and NGS-based ctDNA analysis
| Author | Platform | N | Cancer | Alterations | Concordance | Sensitivity | Specificity |
| Bettegowda et al, 201424 |
Safe-SeqS | 206 | CRC | KRAS codon 12 to 13 | 95% | 87.2% | 99.2% |
| Bachet et al, 201825 | BPER | 330 | CRC | Extended RAS | 85.2% | 76.0% | 98.2% |
| Demuth et al, 201823 | Not specified | 28 | CRC | KRAS codon 12 to 13 | 79% | NA | NA |
| Wang et al, 201826 | Not specified | 56 | Gastro-oesophageal | HER2 amplification | 91.1% | 92% | 90.3% |
| Zill et al, 201527 | Guardant360 | 26 | Pancreatobiliary | KRAS, TP53, APC, FBXW7 and SMAD4 mutations | 97.7% | 92.3% | 100% |
| Schrock et al, 201828 | Not specified | 25 | GI | Not specified | 95% for mutations, 50% for amplifications | NA | NA |
CRC, colorectal cancer; ctDNA, circulating tumour DNA; GI, gastrointestinal; NA, not available.
MSI is a predictive biomarker of the response to immune checkpoint inhibitors in unresectable or metastatic solid tumours, including GI cancers.6 29 MSI is the archetypical manifestation of a defective DNA mismatch repair, which leads to dramatically increased mutation loads throughout the genome, including the gain and/or loss of nucleotides within repeating motifs, known as microsatellite tracts. Currently, MSI testing is most commonly performed via PCR and/or immunohistochemistry (IHC) analysis of tumour tissue samples, and NGS can also accurately characterise MSI status in tumours by assessing the microsatellite length, allowing for a comprehensive profiling of targetable genomic biomarkers and MSI status via a single NGS testing.30–33 Recent publications have demonstrated that ctDNA-based NGS testing can assess the MSI status in various types of cancers, including GI cancers, with a high concordance with tissue-based testing.34 35
Germline BRCA mutations are associated with the efficacy of the poly (adenosine diphosphate-ribose) polymerase (PARP) inhibitors in patients with ovarian or breast cancer.36 37 A randomised phase III trial, POLO, demonstrated that patients who had a germline BRCA mutation and metastatic pancreatic cancer that had not progressed during first-line platinum-based chemotherapy had significantly longer PFS with maintenance olaparib than with placebo.5 Although germline mutations are generally tested through the analysis of genomic DNA extracted from whole blood, broad NGS-based approaches also incidentally identify germline variants. Tissue-based NGS testing cannot definitively distinguish germline from somatic mutations without a comparison with healthy tissues. However, ctDNA-based NGS testing may identify germline mutations at approximately 50% of the MAF, which are distinguishable from somatic mutations that typically occur at lower MAFs in ctDNA. Data from more than 10 000 patients with advanced solid tumours who underwent Guardant360, an NGS-based ctDNA assay receiving Food and Drug Administration (FDA) breakthrough device designation, showed suspected hereditary cancer mutations in 1.4% of the total population.38 In this study, putative germline BRCA1/2 mutations were found in 8 out of 332 (2.4%) patients with pancreatic cancer.
Utility of ctDNA analysis for assessing intratumoural genomic heterogeneity
Intratumoural genomic heterogeneity has been shown in various types of cancers and can contribute to treatment failure and drug resistance.8 As ctDNA is shed from tumour cells throughout the body, ctDNA analysis can potentially identify multiple concurrent heterogeneous resistance mechanisms in individual patients that single-lesion tumour biopsies may miss. Pectasides et al revealed that 42% of mutations and 67% of gene amplifications were discordant between primary tumours and synchronous metastasis in metastatic gastro-oesophageal cancer.39 Of note, NGS analyses of matched metastatic lesions and ctDNA showed 87.5% concordance for genomic alterations in samples for which discordance between the primary tumour and the metastases were observed.
Most recently, Parikh et al directly compared genomic alterations between ctDNA and the tumour biopsy in a prospective cohort of patients with advanced GI cancers.40 NGS analyses of matched ctDNA and biopsy specimens of the brain, liver and subcutaneous metastases from patients with BRAF V600E mCRC showed that only subclonal alterations were detected in each metastasis, such as KRAS and NRAS mutations, and EGFR amplification was represented in ctDNA. Similarly, multiple FGFR2 mutations detected in 17 autopsy specimens of a patient with FGFR2 fusion-positive metastatic gastric cancer who was treated with an FGFR inhibitor were also detected by ctDNA analysis.
These findings suggest that ctDNA analysis may be more useful in identifying heterogeneous clinically relevant subclones than a single-lesion tumour biopsy, although treatment strategies have not been established for subclones with alterations identified by ctDNA analysis but not by tissue analysis.
Evaluation of ctDNA-based patient selection in prospective clinical trials
Recently, the association between the efficacy of targeted agents and genomic alterations in ctDNA has been evaluated in prospective clinical trials, in which ctDNA was retrospectively analysed using blood samples collected at baseline. Sym004 is a mixture of two monoclonal antibodies, futuximab and modotuximab, that bind to nonoverlapping epitopes on the EGFR extracellular domain (ECD) III. In a randomised phase II trial comparing Sym004 with the investigator’s choice of treatments for patients with KRAS exon2 wild-type mCRC, the significant difference in overall survival (OS) was not shown, but in a subgroup by ctDNA (RAS/BRAF/EGFR ECD-mutation negative), the OS was longer in Sym004-treated patients compared with those in the investigator’s choice of treatment.41
As the plasma copy number of gene amplifications in ctDNA is often misleadingly low in samples with a low tumour fraction, the plasma copy number needs to be adjusted by the ctDNA amount. NGS-based ctDNA analyses allow adjustments to the plasma copy number using the maximum MAF as a surrogate for the tumour content. The HERACLES study of trastuzumab plus lapatinib for HER2-positive mCRC showed that an adjusted plasma copy number of HER2 amplification in pre-treatment ctDNA was correlated with the best objective response and PFS.42 A Korean umbrella trial, VIKTORY, also showed that an increased adjusted MET plasma copy number was significantly associated with prolonged PFS on savolitinib to a significantly greater degree than the tissue NGS MET copy number.43 These findings have indicated that the adjustment of plasma copy number assessed by NGS-based ctDNA assays can be more predictive for the efficacy of targeted agents than the absolute plasma copy number.
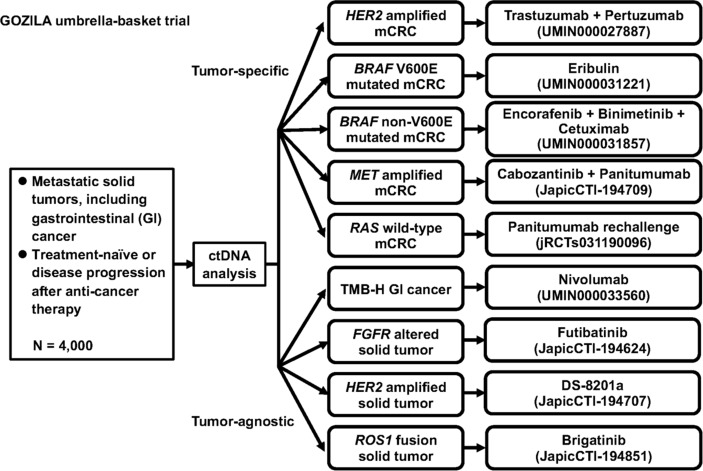
Although these studies suggest that genomic profiling by ctDNA analysis can identify patients who may benefit from an optimal targeted therapy, clinical trials that prospectively test the outcome of selected targeted therapy based on a ctDNA result are very limited. A molecular profiling programme, TARGET, matched patients with advanced cancers to early phase clinical trials based on an analysis using a 641 cancer-associated gene panel in a single ctDNA assay.44 Actionable mutations were identified in 41 of 100 patients, and 11 of these patients received a matched therapy with some tumour responses. We are now conducting an umbrella/basket project, the GOZILA study (UMIN000029315), for patients with advanced solid malignancies, including GI cancer. In the GOZILA study, genomic alterations in the ctDNA have been analysed using Guardant360 for 4000 patients with advanced solid malignancies, and clinical trials have concurrently been conducted for rare fractions of patients with advanced solid malignancies (figure 1). Patients with metastatic GI cancers having specific genomic alterations are enrolled in organ-specific trials. Tumour-agnostic basket trials are also conducted for rare gene alterations identified in various types of cancers, such as FGFR alterations. Most recently, we reported an interim result of a clinical trial for the HER2-amplified mCRC, TRIUMPH, conducted as an arm in the GOZILA study.45 In the TRIUMPH study, patients with treatment-refractory RAS wild-type mCRC with HER2 amplification confirmed by tissue or ctDNA analysis were treated with the combination of trastuzumab and pertuzumab. The primary endpoint of the confirmed objective response rate was 35% in the tissue-positive group and 33% in the ctDNA-positive group, respectively. This response rate suggested that ctDNA analysis can equally identify patients with HER2-amplified mCRC who benefit from the dual HER2-targeted therapy as the tissue analysis. For advanced gastro-oesophageal cancer, a randomised phase III trial to evaluate the efficacy of the addition of bemarituzumab, a monoclonal antibody against FGFR2b, to modified FOLFOX6 for disease with FGFR2 amplification by ctDNA analysis or FGFR2b overexpression by IHC analysis is ongoing.46 Other ongoing ctDNA-based clinical trials for advanced GI cancers are listed in table 3.
Figure 1.
Schema of the GOZILA study, an umbrella/basket project for patients with advanced solid malignancies, including GI cancers. GI, gastrointestinal; mCRC, metastatic colorectal cancer.
Table 3.
Ongoing clinical trials incorporating ctDNA analysis for patient selection in advanced GI cancer
| Trial identifier (Title acronym) |
Phase | Estimated no. of patients | Criteria for patient selection | Intervention | Primary endpoint | Study location |
|
NCT03343301 (FIGHT) |
I/III | 10+548 | Metastatic gastro-oesophageal cancer with FGFR2 amplification by ctDNA analysis or FGFR2b overexpression by IHC analysis | mFOLFOX6 +bemarituzumab vs mFOLFOX6 | Safety and OS | Global |
|
NCT02980510 (PANIRINOX) |
II | 209 | RAS/BRAF wild-type mCRC | mFOLFOX6 +panitumumab vs FOLFIRINOX +panitumumab | CR rate in FOLFIRINOX+ Panitumumab |
France |
|
NCT03227926 (CHRONOS) |
II | 129 | mCRC with subsequent decay of RAS mutant clones | Rechallenge with panitumumab | ORR | Italy |
| NCT03087071 | II | 84 | mCRC according to RAS/BRAF/EGFR mutation status | Panitumumab vs panitumumab +trametinib | ORR | USA |
| UMIN000027887 (TRIUMPH) |
II | 25 | ERBB2-amplified mCRC | Trastuzumab +pertuzumab | ORR | Japan |
CR, complete response; CRC, colorectal cancer; ctDNA, circulating tumour DNA; IHC, immunohistochemistry; ORR, objective response rate; OS, overall survival.
Longitudinal ctDNA surveillance to identify acquired resistance mechanisms
Genomic alterations of each tumour change over time as a result of the Darwinian clonal evolution imposed on cancer cells by selective pressures, including targeted therapy. Longitudinal ctDNA surveillance potentially interrogates the clonal evolution with minimal invasiveness. RAS mutant clones have been identified as drivers of acquired resistance to anti-EGFR therapy in clinical and preclinical studies.47–51 Acquired KRAS mutations have been suggested to emerge from the selection of pre-existing KRAS-mutant subclones and also as a result of ongoing mutagenesis in cancer during anti-EGFR therapy.47 Previous studies have identified the emergence of RAS mutations by the analysis of plasma collected after anti-EGFR therapy.52–55 Furthermore, a longitudinal surveillance of ctDNA using an NGS-based ctDNA assay during anti-EGFR therapy indicated the emergence of acquired RAS mutations and alterations in other genes, including MET, ERBB2, FLT3, EGFR and MEK.56 Interestingly, the temporal withdrawal of an anti-EGFR antibody resulted in a decline in KRAS-mutant alleles, suggesting that the sensitivity to anti-EGFR antibody was restored.52 57 Indeed, the CRICKET study of a rechallenge with anti-EGFR therapy for the RAS wild-type mCRC indicated that it can achieve tumour responses only for patients with the absence of KRAS-mutant clones in ctDNA before the rechallenge.58 59 These findings warrant prospective trials evaluating the strategy of monitoring genomic alterations, including RAS mutations, by ctDNA analyses and temporarily stopping or rechallenging anti-EGFR therapy.
Trastuzumab, in combination with chemotherapy, significantly improved OS in patients with HER2-positive gastro-oesophageal cancer in a randomised phase III trial, the ToGA study.4 However, the unsatisfactory gain of the median PFS suggests acquired resistance to HER2-targeted therapies. Wang et al conducted a longitudinal surveillance of serial plasma samples using an NGS-based ctDNA assay from 24 patients with HER2-positive metastatic gastro-oesophageal cancer who were treated with trastuzumab plus chemotherapy.60 In this study, NF1 mutation was newly identified in ctDNA at disease progression and was confirmed to be related to the resistance to trastuzumab in vitro and in vivo.
The potential of ctDNA to identify various resistance mechanisms has now been used in the development of new drugs. In early clinical trials, the emergence of FGFR2 mutations in patients with FGFR2 fusion-positive cholangiocarcinoma treated with FGFR inhibitors,61 62 BRCA reverse mutations in patients with BRCA-mutated pancreatic cancer treated with PARP inhibitors,63 and NTRK mutations in patients with TRK fusion-positive solid tumours, including GI cancers, treated with TRK inhibitors7 64 were identified as secondary resistance mechanisms by longitudinal ctDNA surveillance.
Limitations of ctDNA analysis
Although the findings reviewed above suggest that ctDNA analysis can be a potential surrogate for standard-of-care tumour tissue analysis, some limitations for ctDNA analysis have been observed. First, preanalytical variables for ctDNA, such as sample collection, handling, transport, processing and storage conditions, may affect ctDNA analysis. However, limited data are available regarding the effect of preanalytical variables on ctDNA results. Patient-related factors, including diurnal or other biological influences, smoking, pregnancy, exercise and numerous non-malignant disorders, such as inflammatory conditions, anaemia, heart disease, metabolic syndrome and autoimmune disorders, may also contribute to the release of cfDNA. Future studies using blood samples with well-documented preanalytical variables are required to address which variables affect the quality of the samples and results of the ctDNA analysis.65
Second, the release of ctDNA may also be affected by tumour-associated factors. For example, the ctDNA concentration varies with the tumour type. Most patients with advanced solid tumours harbour detectable levels of ctDNA, whereas those with primary brain tumours have only low levels of ctDNA probably owing to the presence of the blood–brain barrier.24 Among GI malignancies, mCRC tends to have higher ctDNA levels. In patients with mCRC, undetectable ctDNA is associated with several factors, such as those related to low tumour burden, including primary tumour resection and metachronous metastases, the absence of liver metastases, presence of lung metastases or peritoneal carcinomatosis, low leucocyte counts, low lactate dehydrogenase, alkaline phosphatase, CA19-9 and CEA levels and high albumin levels.19 20 25
Third, healthy haematopoietic cells accumulate somatic mutations during ageing, which can drive clonal expansions of haematopoietic cells in the absence of dysplasia. These mutations are referred to as clonal haematopoiesis of indeterminate potential (CHIP).66 Although no definitive method can discriminate these somatic mutations in healthy cells released into cfDNA and ctDNA mutations, CHIP does not affect the care of advanced cancers because the allele frequencies are generally low compared with those of ctDNA in advanced cancers. However, CHIP may cause false-positive ctDNA results in cases where high sensitivity is required, such as MRD or early cancer detection.
New application of ctDNA analysis: MRD and early cancer detection
Analysis of ctDNA evaluates biomarkers as an alternative for tissue analysis and potentially detects the presence of tumours in patients with no clinically evident disease. This potential capability may be useful for the detection of MRD after surgery or screening in the early detection of new cancers, which can be achieved with high sensitivity assays. In a landmark study, Diehl et al identified somatic mutations of resected tumours in 18 patients with CRC by conventional Sanger sequencing and then analysed ctDNA using a BEAMing method to measure allele fractions as low as 0.01% of a given gene that was identified in each tumour.67 Most patients in whom ctDNA was detectable at the first follow-up visit (13–56 days after the surgery) had a recurrence, while no patients with undetectable ctDNA had a recurrence (p=0.006). In a prospective study involving 178 patients with stage II CRC who did not receive adjuvant chemotherapy, the postoperative ctDNA levels assessed by measuring allele fractions of mutations identified in the primary tumour using an NGS-based method (Safe-SeqS) were significantly associated with an 18 times higher risk of recurrence in patients with positive ctDNA than in those with negative ctDNA.68 Most recently, Reinert et al and Wang et al corroborated the poor prognostic implications of ctDNA positivity in patients with resected stages I–III CRC, respectively.69 70 In Reinerts’ study, Signatera assay, a custom-designed ctDNA assay tracking 16 tumour-specific variants, which received FDA breakthrough device designation, was used. All studies mentioned above used a tumour-directed identification of somatic mutations to personalise ctDNA probes and determine the ctDNA status. This approach substantially reduces confounders, such as CHIP, although additional complexity exists by requiring the sequencing of the resected tumour before generating probes specific for the individual patient. Assays with a fixed-gene panel that analyse only plasma and do not require tissue analysis have also been studied as another approach to assess the MRD. The DYNAMIC study (ACTRN1261500381583) is an ongoing Australian trial of stage II colon cancer that randomly assigned patients to usual care versus ctDNA-informed adjuvant therapy. The NRG Oncology/National Clinical Trials Network phase II/III trial NRG GI-005 (COBRA) will randomise US and Canadian patients with resected low-risk stage II colon cancer to standard surveillance or adjuvant therapy in accordance with ctDNA results.
Implementation of ctDNA analysis for cancer screening requires a degree of analytical sensitivity beyond current technical limits of mutation-based ctDNA detection methods. A blood test, called CancerSEEK, which simultaneously evaluates levels of cancer proteins and the presence of cancer gene mutations from circulating DNA in the blood, was applied to 1005 patients with non-metastatic colorectal, gastric, oesophageal, pancreatic, liver, lung, breast and ovarian cancers.71 The test was positive in a median of 70% of the 1005 patients with sensitivities ranging from 33% for breast cancer to 98% for ovarian cancer and a specificity of greater than 99%. An analysis of tumour-specific methylation in cfDNA has been conducted to identify ctDNA in previous studies. A recent study using immunoprecipitation-based profiling of methylation patterns in cfDNA examined 388 blood samples from patients with early and late-stage cancer of different tumour types and demonstrated receiver operating characteristic area under the curve values of 0.97 in detecting lung cancer, 0.92 for pancreatic cancer and 0.96 for healthy controls.72 Fragmentation patterns of cfDNA are also known to be different between healthy individuals and patients with cancer. Cristiano et al revealed various fragment sizes at different genomic regions in patients with cancer using a low-coverage whole-genome sequencing method. Incorporating this method with mutation-based ctDNA analysis demonstrated a sensitivity of 91% and a specificity of 98% for cancer detection.73
Conclusion
ctDNA analysis has emerged as a potential tool for evaluating the complicated biological processes involved in GI cancers, represented by intratumoural genomic heterogeneity and clonal evolution, given the limited availability of precision therapy for patients with advanced GI cancers. The practical advantages, such as short turnaround time, enable medical oncologists to guide patients to an optimal therapy promptly and survey the potential therapeutic resistance using multiple sampling to implement changes during therapy. In addition, improvement in the limit of detection by evolving technologies has demonstrated the potential of ctDNA analysis to stratify adjuvant chemotherapy through the detection of postoperative MRD and identification of early cancers in patients with no clinically evident disease. Although a careful understanding of the advantages and limitations are required and further prospective study is needed, ctDNA analysis has the potential to overcome challenges for all stages in various types of cancers, including GI cancers.
Patients with advanced solid malignancies, including GI cancers, and those who were not previously treated or do not have disease progression after anticancer therapy were screened by an NGS-based ctDNA assay.
Footnotes
Contributors: YN wrote the manuscript with support from KS. All authors discussed the results and contributed to the final manuscript.
Funding: The authors have not declared a specific grant for this research from any funding agency in the public, commercial or not-for-profit sectors.
Competing interests: YN reports research funding from Taiho Pharmaceutical. KS reports paid consulting or advisory roles for Astellas, Lilly, Bristol-Myers Squibb, Takeda, Pfizer, Ono and MSD; honoraria from Novartis, AbbVie and Yakult and research funding from Astellas, Lilly, Ono, Sumoitomo Dainippon, Daiichi Sankyo, Taiho, Chugai, MSD and Medi Science.
Patient consent for publication: Not required.
Provenance and peer review: Commissioned; externally peer reviewed.
References
- 1. Douillard J-Y, Oliner KS, Siena S, et al. . Panitumumab–FOLFOX4 Treatment and RAS Mutations in Colorectal Cancer. N Engl J Med Overseas Ed 2013;369:1023–34. 10.1056/NEJMoa1305275 [DOI] [PubMed] [Google Scholar]
- 2. Van Cutsem E, Lenz H-J, Köhne C-H, et al. . Fluorouracil, Leucovorin, and Irinotecan Plus Cetuximab Treatment and RAS Mutations in Colorectal Cancer. JCO 2015;33:692–700. 10.1200/JCO.2014.59.4812 [DOI] [PubMed] [Google Scholar]
- 3. Kopetz S, Grothey A, Van Cutsem E, et al. . LBA-006BEACON CRC: a randomized, 3-Arm, phase 3 study of encorafenib and cetuximab with or without binimetinib vs. choice of either irinotecan or FOLFIRI plus cetuximab in BRAF V600E–mutant metastatic colorectal cancer. Ann Oncol 2019;30 10.1093/annonc/mdz183.004 [DOI] [Google Scholar]
- 4. Bang Y-J, Van Cutsem E, Feyereislova A, et al. . Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet 2010;376:687–97. 10.1016/S0140-6736(10)61121-X [DOI] [PubMed] [Google Scholar]
- 5. Golan T, Hammel P, Reni M, et al. . Maintenance Olaparib for Germline BRCA -Mutated Metastatic Pancreatic Cancer. N Engl J Med 2019;381:317–27. 10.1056/NEJMoa1903387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. DT L, Uram JN, Wang H, et al. . Pd-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Drilon A, Laetsch TW, Kummar S, et al. . Efficacy of Larotrectinib in TRK Fusion–Positive Cancers in Adults and Children. N Engl J Med 2018;378:731–9. 10.1056/NEJMoa1714448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell 2017;168:613–28. 10.1016/j.cell.2017.01.018 [DOI] [PubMed] [Google Scholar]
- 9. Oxnard GR, Paweletz CP, Sholl LM. Genomic analysis of plasma cell-free DNA in patients with cancer. JAMA Oncol 2017;3:740–1. 10.1001/jamaoncol.2016.2835 [DOI] [PubMed] [Google Scholar]
- 10. Diehl F, Li M, He Y, et al. . Beaming: single-molecule PCR on microparticles in water-in-oil emulsions. Nat Methods 2006;3:551–9. 10.1038/nmeth898 [DOI] [PubMed] [Google Scholar]
- 11. Hindson BJ, Ness KD, Masquelier DA, et al. . High-Throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem 2011;83:8604–10. 10.1021/ac202028g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kinde I, Wu J, Papadopoulos N, et al. . Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A 2011;108:9530–5. 10.1073/pnas.1105422108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Newman AM, Bratman SV, To J, et al. . An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548–54. 10.1038/nm.3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Newman AM, Lovejoy AF, Klass DM, et al. . Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol 2016;34:547–55. 10.1038/nbt.3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lanman RB, Mortimer SA, Zill OA, et al. . Analytical and clinical validation of a digital sequencing panel for quantitative, highly accurate evaluation of cell-free circulating tumor DNA. PLoS One 2015;10:e0140712 10.1371/journal.pone.0140712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Taly V, Pekin D, Benhaim L, et al. . Multiplex picodroplet digital PCR to detect KRAS mutations in circulating DNA from the plasma of colorectal cancer patients. Clin Chem 2013;59:1722–31. 10.1373/clinchem.2013.206359 [DOI] [PubMed] [Google Scholar]
- 17. Thierry AR, Mouliere F, El Messaoudi S, et al. . Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med 2014;20:430–5. 10.1038/nm.3511 [DOI] [PubMed] [Google Scholar]
- 18. Schmiegel W, Scott RJ, Dooley S, et al. . Blood-based detection of RAS mutations to guide anti-EGFR therapy in colorectal cancer patients: concordance of results from circulating tumor DNA and tissue-based RAS testing. Mol Oncol 2017;11:208–19. 10.1002/1878-0261.12023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vidal J, Muinelo L, Dalmases A, et al. . Plasma ctDNA Ras mutation analysis for the diagnosis and treatment monitoring of metastatic colorectal cancer patients. Ann Oncol 2017;28:1325–32. 10.1093/annonc/mdx125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bando H, Kagawa Y, Kato T, et al. . A multicentre, prospective study of plasma circulating tumour DNA test for detecting Ras mutation in patients with metastatic colorectal cancer. Br J Cancer 2019;120:982–6. 10.1038/s41416-019-0457-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grasselli J, Elez E, Caratù G, et al. . Concordance of blood- and tumor-based detection of Ras mutations to guide anti-EGFR therapy in metastatic colorectal cancer. Ann Oncol 2017;28:1294–301. 10.1093/annonc/mdx112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Odegaard JI, Vincent JJ, Mortimer S, et al. . Validation of a plasma-based comprehensive cancer genotyping assay utilizing orthogonal tissue- and plasma-based methodologies. Clin Cancer Res 2018;24:3539–49. 10.1158/1078-0432.CCR-17-3831 [DOI] [PubMed] [Google Scholar]
- 23. Demuth C, Spindler K-LG, Johansen JS, et al. . Measuring KRAS mutations in circulating tumor DNA by droplet digital PCR and next-generation sequencing. Transl Oncol 2018;11:1220–4. 10.1016/j.tranon.2018.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bettegowda C, Sausen M, Leary RJ, et al. . Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24 10.1126/scitranslmed.3007094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bachet JB, Bouché O, Taieb J, et al. . Ras mutation analysis in circulating tumor DNA from patients with metastatic colorectal cancer: the AGEO RASANC prospective multicenter study. Ann Oncol 2018;29:1211–9. 10.1093/annonc/mdy061 [DOI] [PubMed] [Google Scholar]
- 26. Wang H, Li B, Liu Z, et al. . Her2 copy number of circulating tumour DNA functions as a biomarker to predict and monitor trastuzumab efficacy in advanced gastric cancer. Eur J Cancer 2018;88:92–100. 10.1016/j.ejca.2017.10.032 [DOI] [PubMed] [Google Scholar]
- 27. Zill OA, Greene C, Sebisanovic D, et al. . Cell-Free DNA next-generation sequencing in pancreatobiliary carcinomas. Cancer Discov 2015;5:1040–8. 10.1158/2159-8290.CD-15-0274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schrock AB, Pavlick D, Klempner SJ, et al. . Hybrid Capture–Based genomic profiling of circulating tumor DNA from patients with advanced cancers of the gastrointestinal tract or anus. Clin Cancer Res 2018;24:1881–90. 10.1158/1078-0432.CCR-17-3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Le DT, Durham JN, Smith KN, et al. . Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409–13. 10.1126/science.aan6733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Salipante SJ, Scroggins SM, Hampel HL, et al. . Microsatellite instability detection by next generation sequencing. Clin Chem 2014;60:1192–9. 10.1373/clinchem.2014.223677 [DOI] [PubMed] [Google Scholar]
- 31. Hause RJ, Pritchard CC, Shendure J, et al. . Classification and characterization of microsatellite instability across 18 cancer types. Nat Med 2016;22:1342–50. 10.1038/nm.4191 [DOI] [PubMed] [Google Scholar]
- 32. Bonneville R, Krook MA, Kautto EA, et al. . Landscape of microsatellite instability across 39 cancer types. JCO Precision Oncology 2017:1–15. 10.1200/PO.17.00073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Middha S, Zhang L, Nafa K, et al. . Reliable pan-cancer microsatellite instability assessment by using targeted next-generation sequencing data. JCO Precision Oncology 2017:1–17. 10.1200/PO.17.00084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mayrhofer M, De Laere B, Whitington T, et al. . Cell-Free DNA profiling of metastatic prostate cancer reveals microsatellite instability, structural rearrangements and clonal hematopoiesis. Genome Med 2018;10:85 10.1186/s13073-018-0595-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Willis J, Lefterova MI, Artyomenko A, et al. . Validation of microsatellite instability detection using a comprehensive plasma-based genotyping panel. Clin Cancer Res 2019. [DOI] [PubMed] [Google Scholar]
- 36. Robson M, Im S-A, Senkus E, et al. . Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. N Engl J Med 2017;377:523–33. 10.1056/NEJMoa1706450 [DOI] [PubMed] [Google Scholar]
- 37. Moore K, Colombo N, Scambia G, et al. . Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med 2018;379:2495–505. 10.1056/NEJMoa1810858 [DOI] [PubMed] [Google Scholar]
- 38. Slavin TP, Banks KC, Chudova D, et al. . Identification of incidental germline mutations in patients with advanced solid tumors who underwent cell-free circulating tumor DNA sequencing. J Clin Oncol 2018:3459–65. 10.1200/JCO.18.00328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pectasides E, Stachler MD, Derks S, et al. . Genomic heterogeneity as a barrier to precision medicine in gastroesophageal adenocarcinoma. Cancer Discov 2018;8:37–48. 10.1158/2159-8290.CD-17-0395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Parikh AR, Leshchiner I, Elagina L, et al. . Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat Med 2019;25:1415–21. 10.1038/s41591-019-0561-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Montagut C, Argilés G, Ciardiello F, et al. . Efficacy of Sym004 in patients with metastatic colorectal cancer with acquired resistance to anti-EGFR therapy and molecularly selected by circulating tumor DNA analyses. JAMA Oncol 2018;4:e175245 10.1001/jamaoncol.2017.5245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Siravegna G, Sartore-Bianchi A, Nagy RJ, et al. . Plasma HER2 (ERBB2) Copy Number Predicts Response to HER2-targeted Therapy in Metastatic Colorectal Cancer. Clin Cancer Res 2019;25:3046–53. 10.1158/1078-0432.CCR-18-3389 [DOI] [PubMed] [Google Scholar]
- 43. Lee J, Kim ST, Kim K, et al. . Tumor genomic profiling guides metastatic gastric cancer patients to targeted treatment: the VIKTORY umbrella trial. Cancer Discov 2019:CD-19-0442 10.1158/2159-8290.CD-19-0442 [DOI] [PubMed] [Google Scholar]
- 44. Rothwell DG, Ayub M, Cook N, et al. . Utility of ctDNA to support patient selection for early phase clinical trials: the target study. Nat Med 2019;25:738–43. 10.1038/s41591-019-0380-z [DOI] [PubMed] [Google Scholar]
- 45. Nakamura Y, Okamoto W, Kato T, et al. . Triumph: primary efficacy of a phase II trial of trastuzumab (T) and pertuzumab (P) in patients (PTS) with metastatic colorectal cancer (mCRC) with HER2 (ErbB2) amplification (AMP) in tumour tissue or circulating tumour DNA (ctDNA): a GOZILA sub-study. Annals of Oncology 2019;30:v199–200. 10.1093/annonc/mdz246.004 [DOI] [Google Scholar]
- 46. Catenacci DVT, Tesfaye A, Tejani M, et al. . Bemarituzumab with modified FOLFOX6 for advanced FGFR2-positive gastroesophageal cancer: fight phase III study design. Future Oncol 2019;15:2073–82. 10.2217/fon-2019-0141 [DOI] [PubMed] [Google Scholar]
- 47. Misale S, Yaeger R, Hobor S, et al. . Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 2012;486:532–6. 10.1038/nature11156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Diaz Jr LA, Williams RT, Wu J, et al. . The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 2012;486:537–40. 10.1038/nature11219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Andreou A, Kopetz S, Maru DM, et al. . Adjuvant chemotherapy with FOLFOX for primary colorectal cancer is associated with increased somatic gene mutations and inferior survival in patients undergoing hepatectomy for metachronous liver metastases. Ann Surg 2012;256:642–50. 10.1097/SLA.0b013e31826b4dcc [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Otsuka K, Satoyoshi R, Nanjo H, et al. . Acquired/intratumoral mutation of KRAS during metastatic progression of colorectal carcinogenesis. Oncol Lett 2012;3:649–53. 10.3892/ol.2011.543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Misale S, Arena S, Lamba S, et al. . Blockade of EGFR and MEK intercepts heterogeneous mechanisms of acquired resistance to anti-EGFR therapies in colorectal cancer. Sci Transl Med 2014;6:224ra26 10.1126/scitranslmed.3007947 [DOI] [PubMed] [Google Scholar]
- 52. Morelli MP, Overman MJ, Dasari A, et al. . Characterizing the patterns of clonal selection in circulating tumor DNA from patients with colorectal cancer refractory to anti-EGFR treatment. Ann Oncol 2015;26:731–6. 10.1093/annonc/mdv005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Laurent-Puig P, Pekin D, Normand C, et al. . Clinical relevance of KRAS-mutated subclones detected with picodroplet digital PCR in advanced colorectal cancer treated with anti-EGFR therapy. Clin Cancer Res 2015;21:1087–97. 10.1158/1078-0432.CCR-14-0983 [DOI] [PubMed] [Google Scholar]
- 54. Van Emburgh BO, Arena S, Siravegna G, et al. . Acquired Ras or EGFR mutations and duration of response to EGFR blockade in colorectal cancer. Nat Commun 2016;7:13665 10.1038/ncomms13665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shitara K, Yamanaka T, Denda T, et al. . REVERCE: a randomized phase II study of regorafenib followed by cetuximab versus the reverse sequence for previously treated metastatic colorectal cancer patients. Ann Oncol 2019;30:259–65. 10.1093/annonc/mdy526 [DOI] [PubMed] [Google Scholar]
- 56. Siravegna G, Mussolin B, Buscarino M, et al. . Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med 2015;21:795–801. 10.1038/nm.3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Parseghian CM, Loree JM, Morris VK, et al. . Anti-EGFR-resistant clones decay exponentially after progression: implications for anti-EGFR re-challenge. Ann Oncol 2019;30:243–9. 10.1093/annonc/mdy509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cremolini C, Antoniotti C, Lonardi S, et al. . Activity and Safety of Cetuximab Plus Modified FOLFOXIRI Followed by Maintenance With Cetuximab or Bevacizumab for RAS and BRAF Wild-type Metastatic Colorectal Cancer. JAMA Oncol 2018;4:529–36. 10.1001/jamaoncol.2017.5314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rossini D, Cremolini C, Conca E, et al. . Liquid biopsy to predict benefit from rechallenge with cetuximab (cet) + irinotecan (IRI) in RAS/BRAF wild-type metastatic colorectal cancer patients (PTS) with acquired resistance to first-line cet+iri: final results and translational analyses of the cricket study by GONO. JCO 2018;36:12007–07. 10.1200/JCO.2018.36.15_suppl.12007 [DOI] [Google Scholar]
- 60. Wang D-S, Liu Z-X, Lu Y-X, et al. . Liquid biopsies to track trastuzumab resistance in metastatic HER2-positive gastric cancer. Gut 2019;68:1152–61. 10.1136/gutjnl-2018-316522 [DOI] [PubMed] [Google Scholar]
- 61. Goyal L, Saha SK, Liu LY, et al. . Polyclonal Secondary FGFR2 Mutations Drive Acquired Resistance to FGFR Inhibition in Patients with FGFR2 Fusion–Positive Cholangiocarcinoma. Cancer Discov 2017;7:252–63. 10.1158/2159-8290.CD-16-1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Goyal L, Shi L, Liu LY, et al. . TAS-120 overcomes resistance to ATP-competitive FGFR inhibitors in patients with FGFR2 Fusion–Positive intrahepatic cholangiocarcinoma. Cancer Discov 2019;9:1064–79. 10.1158/2159-8290.CD-19-0182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shroff RT, Hendifar A, McWilliams RR, et al. . Rucaparib Monotherapy in Patients With Pancreatic Cancer and a Known Deleterious BRCA Mutation. JCO Precision Oncology 2018:1–15. 10.1200/PO.17.00316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Russo M, Misale S, Wei G, et al. . Acquired resistance to the Trk inhibitor entrectinib in colorectal cancer. Cancer Discov 2016;6:36–44. 10.1158/2159-8290.CD-15-0940 [DOI] [PubMed] [Google Scholar]
- 65. Merker JD, Oxnard GR, Compton C, et al. . Circulating tumor DNA analysis in patients with cancer: American Society of clinical oncology and College of American pathologists joint review. JCO 2018;36:1631–41. 10.1200/JCO.2017.76.8671 [DOI] [PubMed] [Google Scholar]
- 66. Steensma DP, Bejar R, Jaiswal S, et al. . Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood 2015;126:9–16. 10.1182/blood-2015-03-631747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Diehl F, Schmidt K, Choti MA, et al. . Circulating mutant DNA to assess tumor dynamics. Nat Med 2008;14:985–90. 10.1038/nm.1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tie J, Wang Y, Tomasetti C, et al. . Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med 2016;8:346ra92 10.1126/scitranslmed.aaf6219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Reinert T, Henriksen TV, Christensen E, et al. . Analysis of plasma cell-free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol 2019;5:1124 10.1001/jamaoncol.2019.0528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wang Y, Li L, Cohen JD, et al. . Prognostic potential of circulating tumor DNA measurement in postoperative surveillance of nonmetastatic colorectal cancer. JAMA Oncol 2019;5:1118 10.1001/jamaoncol.2019.0512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cohen JD, Li L, Wang Y, et al. . Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018;359:926–30. 10.1126/science.aar3247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Shen SY, Singhania R, Fehringer G, et al. . Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 2018;563:579–83. 10.1038/s41586-018-0703-0 [DOI] [PubMed] [Google Scholar]
- 73. Cristiano S, Leal A, Phallen J, et al. . Genome-Wide cell-free DNA fragmentation in patients with cancer. Nature 2019;570:385–9. 10.1038/s41586-019-1272-6 [DOI] [PMC free article] [PubMed] [Google Scholar]