Abstract
Migratory animals respond to environmental heterogeneity by predictably moving long distances in their lifetime. Migration has evolved repeatedly in animals, and many adaptations are found across the tree of life that increase migration efficiency. Life-history theory predicts that migratory species should evolve a larger body size than non-migratory species, and some empirical studies have shown this pattern. A recent study analysed the evolution of body size between diadromous and non-diadromous shads, herrings, anchovies and allies, finding that species evolved larger body sizes when adapting to a diadromous lifestyle. It remains unknown whether different fish clades adapt to migration similarly. We used an adaptive landscape framework to explore body size evolution for over 4500 migratory and non-migratory species of ray-finned fishes. By fitting models of macroevolution, we show that migratory species are evolving towards a body size that is larger than non-migratory species. Furthermore, we find that migratory lineages evolve towards their optimal body size more rapidly than non-migratory lineages, indicating body size is a key adaption for migratory fishes. Our results show, for the first time, that the largest vertebrate radiation on the planet exhibited strong evolutionary determinism when adapting to a migratory lifestyle.
Keywords: adaptive landscape, macroevolution, adaptation
1. Introduction
Migration is a widespread phenomenon among animals in which individuals predictably move long distances at various stages in their life cycle [1]. During migration, species often move hundreds to thousands of kilometres to exploit heterogeneous resources for fitness gains [2–4]. Traversing such great distances imposes extreme physiological demands [5–7], and presumably to offset these demands, the evolution of migration is often linked to phenotypes that increase energy efficiency [6,8]. The repeated evolution of migration throughout the radiation of animals predicts common patterns in trait evolution associated with long-distance migration [2,9].
Long-distance migration selects for efficient locomotion and high energy storage [10,11], and many species across the tree of life have adaptations that minimize migration costs [7,12,13]. For instance, many migratory insects evolved large body sizes and wings that allow for more efficient long-distance movement [7,14], and many avian migrants evolved longer and more pointed wings than non-migrants [15,16]. Some adaptions, such as increased body sizes, have apparently evolved across multiple clades of animals including birds, fishes, mammals and insects [17,18], indicating shared selective pressures and a common phenotypic response when adapting to a migratory behaviour [6,7]. However, few studies have explicitly investigated phenotypic diversification across broad lineages of migratory and non-migratory species in a comparative phylogenetic framework.
Ray-finned fishes (Actinopterygii) are an ideal group to test whether disparate lineages share common selective pressures and phenotypic patterns when adapting to a migratory behaviour. Comprising over 29 000 species [19], fishes are the most diverse vertebrate radiation, representing approximately half of all vertebrates. Fishes have transitioned between migratory and non-migratory behaviour repeatedly and have evolved different modes of migration that include both freshwater and marine migrants, and diadromous migrants that move between freshwater and marine environments at different life-history stages [20,21]. Fishes also exhibit remarkable variation in body size across phylogenetic scales [22–24], with individuals ranging from species of sturgeon that reach multiple metres in length, to a coral reef dwarf goby, which reach only a few centimetres [19]. Most studies on ecomorphological adaptation to migration have focused on intraspecific variation in diadromous fishes, and found that migratory populations have significantly larger body sizes than non-migratory populations [25–27]. Selection is hypothesized to favour larger body size to increase migration distance by improving energetic efficiency [7] and allowing individuals to bypass natural barriers to movement [28]. For example, diadromous rainbow trout are significantly larger than landlocked populations, presumably because larger body size allows adults to migrate hundreds of kilometres to natal freshwater spawning grounds on a limited energy budget [26,29,30]. Larger body sizes in migratory fishes are not restricted to intraspecific populations. A study investigating body size across lineages of diadromous and non-diadromous fishes found that directional selection was driving diadromous Clupeiformes (shads, herrings, anchovies and allies) towards a larger body size than non-diadromous lineages [31]. The results of these studies suggest that a larger body size may represent an adaptive peak [32,33] in migratory fishes, and suggests there may be a high degree of evolutionary determinism [34–36] across large phylogenetic scales in migratory fishes.
For this study, we examined the tempo and mode of body size evolution across more than 4500 migratory and non-migratory ray-finned fishes to determine whether natural selection drives larger body sizes in migratory fishes. To reconstruct the macroevolutionary landscape, we synthesized data from the primary literature on the migratory status and maximum body size for individual species across ray-finned fishes and tested body size evolution using phylogenetic comparative methods. Specifically, we assessed (1) how many times migration has evolved across ray-finned fishes, (2) whether migratory fishes had a significantly larger body size than non-migratory lineages, (3) the hypothesis that migratory lineages share a deterministic pattern of evolving towards a larger body size optimum than the non-migratory lineages optimum, and (4) whether there was an increase in the rate of body size evolution in migratory lineages, as predicted when traits are under strong natural selection. Our macroevolutionary study offers critical insights into how migration can shape phenotypic evolution across the largest radiation of vertebrates.
2. Material and methods
(a). Ecological classification and body size
Any classification of migratory species requires a clear definition of migration. We follow Bowlin et al. [1] and define migratory species as any species that cyclically and predictably move long distances from one area to another using active transport. We obtained an initial migration classification of ray-finned fish species in the Rabosky et al. [37] phylogeny from FishBase [38] using the R package rfishbase [39]. We then performed a primary literature review of fish migration to compare the classifications from the FishBase database with our definition of migration. We further examined species descriptions, field guides and checklists to correctly classify migratory species and remove the inclusion of false positives in our dataset. We removed species from the database that were classified as migratory in FishBase for which we could not confirm with another source through our search of the primary literature, personal observations or communication with taxonomic experts.
The classification of non-migratory species proved more difficult because species were rarely labelled as non-migratory in the primary literature unless the non-migratory species was being compared to a closely related migratory species. To classify non-migratory species, we made non-migratory the default classification for species for which we could not find primary sources stating the species were migratory and in which FishBase classified the species as either ‘non-migratory’ or ‘unknown’. Once we finalized the migration database, we assembled maximum body size data for each species in the database from FishBase [38] using the R package rfishbase [39]. We then log10-transformed the body size data to correct for normality as many of the statistical tests we use assume an underlying normal distribution. We had a total of 4648 number of species, with 590 species classified as migratory and 4058 classified as non-migratory. We trimmed the Rabosky et al. [37] phylogeny to the 4648 number of species we had ecological and body size data for using the drop.tip function in the R package ape [40]. The trimmed phylogeny was used in all subsequent analyses. Not all tips in the Rabosky et al. [37] phylogeny are placed based on underlying genetic data; many species in the phylogeny are placed using taxonomic back-filling of taxa without molecular data. However, the majority of the taxa (n = 4635) used in our tree were placed using genetic data, with only 13 species being imputed based on taxonomy.
(b). Ancestral state reconstructions
We inferred the number of times that migration evolved using stochastic character mapping with the function make.simmap in the R package phytools [41]. The evolutionary history of migration was reconstructed 100 times on all 100 distribution of trees from Rabosky et al. [37] to account for phylogenetic uncertainty in the position of the imputed taxa. We visualized the uncertainty in number of times migration evolved across all 100 trees using a histogram. To assess the best model for the transition matrix, we fitted a model with an equal rate of transition between migratory and non-migratory lineages and a model with all rates different using the function ace in the R package ape [40]. We compared the two models using a likelihood ratio test and found that an all rates equal model was the best model. We used the ‘equal rates’ (ER) model and estimated the prior distribution of the states at the root of the tree and used the MCMC option to set the parameters of the Q matrix. The distribution of maximum body size and migration status was visualized across the MCC from [37] using the function plotTree.wBars in the R package phytools [41].
(c). Phylogenetic ANOVA and evolutionary covariation
We used phylogenetic ANOVA to test for differences in body size between migratory and non-migratory species. We performed a phylogenetic ANOVA in the R package geiger v2.0 using the function phy.anova [42]. For the phylogenetic ANOVA, we performed 10 000 simulations under a Brownian motion model of evolution. We visualized differences in the body size between migratory and non-migratory species using a boxplot in R.
We tested for evolutionary covariation between a species's body size and migratory status using the threshold model [43,44] as implemented in the function threshBayes of the R package phytools [41]. We ran threshBayes for 1.0 × 106 generations, sampling every 100 generations. Under the threshold model, a discrete character evolves as a function of underlying continuously varying attribute called ‘liability’. Once ‘liability’ crosses a value, the state of the discrete character changes in value [43,44]. The threshold model can be used to test for evolutionary covariation between continuous and discrete traits [44].
(d). Macroevolutionary model fitting
We tested seven evolutionary models in the R package OUwie [45] to determine whether migratory and non-migratory fishes evolved towards different adaptive peaks in body size. The evolutionary models were run on all 100 SIMMAP reconstructions from our ancestral character reconstruction analysis across all 100 trees (see above) to account for phylogenetic uncertainty. The first two evolutionary models we tested were models of Brownian motion, which assumes no trait differences between migratory and non-migratory lineages, with trait variation accruing randomly as a proportion of time. The simplest Brownian motion model was a single-rate Brownian motion model (BM1), while the more complex Brownian motion model (BMS) allows different rates of stochastic variance for migratory and non-migratory lineages. The next model, a single Ornstein–Uhlenbeck model (OU1), assumes that migratory and non-migratory lineages are evolving towards a shared trait optimum. The next sets of models were multi-peak OU models, with increasing parameter complexity. The simplest multiple peak OU model was OUM, which assumes different trait optima (θ) for migratory and non-migratory lineages, but each lineage has the same strength of selection towards the optimal trait value (α) and the same rate of stochastic variance (σ2). OUMA allows α to vary between lineages, OUMV allows lineages to differ σ2 values and OUMVA allows both α and σ2 to vary between lineages.
Model fit was evaluated using the Akaike information criterion for small sample size (AICc). We calculated the relative strength of support for each model in each iteration using Akaike weights and averaged across all iterations for each model. The model with the best AIC weight was selected as the best model. Eigen decomposition of the Hessian matrix provides an indication of whether the model search returned the maximum-likelihood estimate [45]. If the eigenvalues are positive, then the results are considered reliable. To ensure that all maximum-likelihood results were reliable, we removed any model run that returned a negative eigenvalue prior to evaluating the model fit.
OUwie uses complex OU models that can be incorrectly favoured over models of Brownian evolution [46,47]. To determine whether we had significant power to accurately detect the complex models, we performed 100 OUwie simulations for max body size using the function OUwie.sim. The simulated datasets were performed with the parameter estimates for each of the six models in our empirical dataset. Each set of simulations corresponding to a given model was then run through all seven models in OUwie to determine if each simulated model could be accurately recovered.
Models of continuous trait evolution have been shown to produce misleading results when the ecological states of taxa are misclassified [48,49]. Large-scale comparative studies may contain a few species with erroneous ecological states, especially among poorly studied taxonomic groups. To determine whether the results of the OUwie analyses were influenced by possible ecological misclassifications, we used the R package l1ou [50] to capture evolutionary shifts in body size optima without a pre-defined classification of behaviour. l1ou detects shifts in trait evolution under a model-selection and evaluation approach implemented using the least absolute shrinkage and selection operator (LASSO) method [51,52]. The relative position of the adaptive optima and shift magnitudes was evaluated using AICc.
3. Results
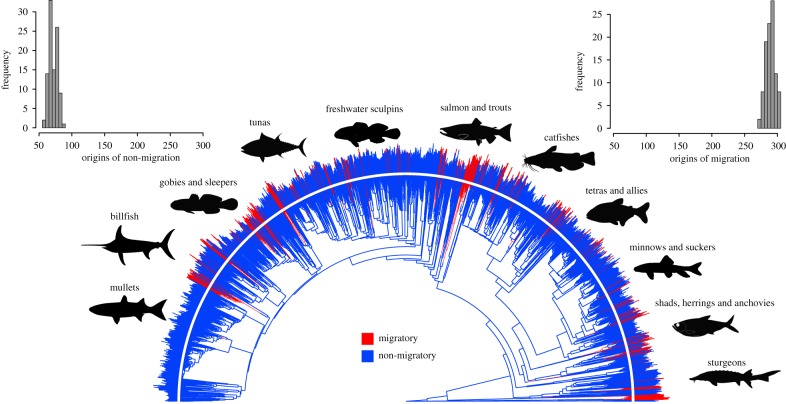
Our SIMMAP reconstructions showed that the ancestor of ray-finned fishes was non-migratory in more than half of the reconstructions, but that migration evolved early at the base of ray-finned fishes (electronic supplementary material, figure S1). Transitions between a migratory and non-migratory life-history strategy occurred at least 434 times across ray-finned fishes with at least 503 different transition events possible (figure 1). Migration independently evolved 289 times on average across ray-finned fishes including multiple origins at the base of the tree and many more independent evolutions occurring throughout crown groups (figure 1). Transitions back to a non-migratory behaviour were much less common, only occurring 70 times on average in ray-finned fishes (figure 1).
Figure 1.
SIMMAP reconstructions of migratory and non-migratory behaviour in ray-finned fishes. Select migratory lineages are represented by an image placed near the represented tip in the phylogeny. Lines projecting from the phylogeny represent maximum log body size for each species and line lengths are scaled to the total tree height. Inset histograms represent the number of origins of migratory and non-migratory behaviour inferred from our SIMMAP reconstructions across the entire distribution of trees from the Rabosky et al. [37] phylogeny. These distributions illustrate the variability in the number of transitions owing to the uncertainty in the character mapping, as well as the tree topology and branch lengths. (Online version in colour.)
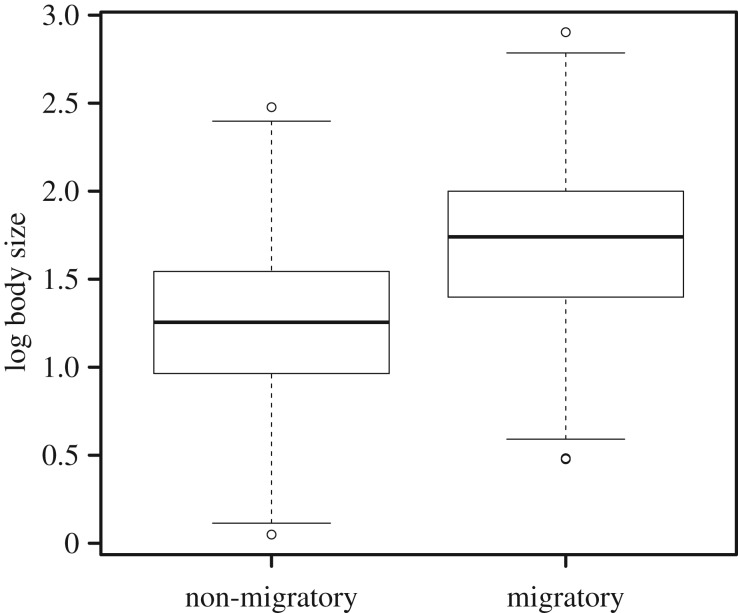
Migratory species generally have larger body sizes than non-migratory species (figure 2). The largest body sizes achieved in ray-finned fishes are found in migratory species (figure 2). About 85% of non-migratory species are less than 1.5 log-transformed size while about 60% of migratory species are larger than 1.5 log-transformed size. Migratory species can vary anywhere from 0.5 to 2.8 log-transformed maximum body size. Even though migratory lineages vary significantly in maximum body size, most migratory lineages have a larger body size than their nearest non-migratory sister taxa (figure 1). The results of the Phylogenetic ANOVA revealed that migratory lineages have statistically larger body sizes than non-migratory lineages regardless of phylogenetic placement (F = 664.1, p = 1.0 × 10−4). Under the threshold model, we found strong evolutionary covariation between body size and migratory behaviour (r = 0.93).
Figure 2.
Boxplot of log body size between migratory and non-migratory species. Points represent outliers.
The OUwie analyses of body size for migratory versus non-migratory lineages find the best-fit model of evolution is OUMV, a model supporting different optimal trait value (θ) and different rates of stochastic variance (σ2) for migratory and non-migratory lineages (table 1; electronic supplementary material, figure S2). Parameter estimates show migratory lineages have larger θ and σ2 than non-migratory lineages, which indicates a higher rate of variance around the adaptive optima than non-migratory lineages (table 1; electronic supplementary material, figure S3). The next best model selected in OUwie was OUMA, a model supporting different optimal trait values (θ and different strength of selection towards the optimal trait value (α) (table 1). The OUMV model found that migratory lineages were evolving towards a larger body size with a stronger pull of adaptation than non-migratory lineages. No other model of evolution was closely supported, with the next nearest model over 28 AIC larger than either the OUMV or OUMA model (table 1; electronic supplementary material, figure S2).
Table 1.
Comparison of average model fits and parameter values for body size between migratory (m) and non-migratory (nm) species.
| model | rank | AICc | ΔAICc | AICw | θ (m) | θ (nm) | α (m) | α (nm) | σ2 (m) | σ2 (nm) |
|---|---|---|---|---|---|---|---|---|---|---|
| OUMV | 1 | 4961 | 0 | 0.98 | 1.77 | 1.23 | 0.96 | 0.96 | 0.39 | 0.33 |
| OUMA | 2 | 4970 | 9 | 0.02 | 1.78 | 1.22 | 0.98 | 0.94 | 0.33 | 0.33 |
| OUM | 3 | 4989 | 28 | 0 | 1.79 | 1.25 | 0.96 | 0.96 | 0.34 | 0.34 |
| OU1 | 4 | 5538 | 577 | 0 | 1.16 | 1.16 | 0.96 | 0.96 | 0.38 | 0.38 |
| BMS | 5 | 10 329 | 5368 | 0 | — | — | — | — | 0.032 | 0.012 |
| BM1 | 6 | 10 500 | 5539 | 0 | — | — | — | — | 0.17 | 0.17 |
| OUMVAa | — | — | — | — | — | — | — | — | — | — |
aModel could not converge.
The results of our simulations show that our dataset has enough statistical power to differentiate between the different models of evolution (electronic supplementary material, figure S4). The BM1 and BMS models were recovered as the best model during their simulation sets, indicating that model overfitting was not causing the complex OU models to be favoured in our empirical OUwie analyses. The more complex multi-peak OU models had consistently lower AIC scores than either of the Brownian motion models (BM1 and BMS) or the single peak OU model (OU1) across their sets of simulations. There was a slight overlap among the OUMA and OUMV models during their respective simulation sets, but each model was consistently recovered as the best model in their respective set of simulations. There was a degree of variance in our simulated estimates of θ, α and σ2 (electronic supplementary material, figure S5), but the simulated parameters were closely aligned, with the empirical parameter values indicating enough statistical power to recover relatively accurate parameter estimates (electronic supplementary material, figure S5).
The l1ou analysis recovered 100 adaptive shifts in body size across the phylogeny (electronic supplementary material, figure S6). Most of the adaptive shifts occurred on branches subtending families and larger clades across ray-finned fishes. Radiations dominated by migratory species, represented by arrows in electronic supplementary material, figure S6, tended to exhibit an adaptive shift in body size. However, lineages that evolved migration in only a few species, dispersed among a larger clade of non-migratory species, tended to not exhibit adaptive shifts in body size. The lack of adaptive shifts in these migratory lineages probably results from the lack of power to distinguish between the subtle shifts in body size occurring in a few migratory taxa and the more pronounced body size shift of the larger clade.
4. Discussion
Migration has evolved hundreds of times across ray-finned fishes, including numerous independent radiations in both freshwater and marine lineages. Our analyses showed that migratory species vary in size but were consistently larger than their non-migratory sister group. Migratory species are larger than non-migratory relatives in nearly all clades and across all modes of migration, indicating shared selective pressure and strong evolutionary determinism in the pattern of body size evolution regardless of habitat, migratory strategy or evolutionary history. Our OUwie parameter estimates clarify the macroevolutionary processes underlying these patterns and suggest that migratory lineages experienced strong selection that resulted in more rapid evolution towards a larger theta value. Taken together our results demonstrate that migratory fishes are predictably larger than their non-migratory relatives.
Migration is a widespread behaviour among animals and is thought to be rapidly lost and regained among many species and populations [9]. Our ancestral state reconstructions show that migration is a common and highly evolvable behaviour in fishes. Migration has evolved almost 290 times, including in almost every major clade of modern ray-finned fishes. Although the evolution of migratory behaviour was relatively common in fishes, migratory lineages rarely give rise to non-migratory descendants; migratory lineages only transitioned to a non-migratory behaviour 25% of the time. Many migratory fishes have life-history specializations that increase migration ability but also reduce fitness through decreased fecundity [12,53,54] and increased mortality [55,56]. The reduced rate of transition back to a non-migratory behaviour is probably because specialization for migration creates an evolutionary dead end and limits further evolution [33,57–59].
Although migration has evolved across a remarkable number of clades throughout the past 200 million years, our results show that adaptation to a migratory behaviour exhibits strong evolutionary determinism; migratory species consistently evolve a larger body size than non-migratory species despite some selective forces acting against large body sizes [60,61]. Roff [7] suggested that a larger body size in migratory fishes results in higher swimming efficiency and reduced energetic requirements, which allows for longer migrations and a better ability to bypass barriers to movement [7,28]. This argument is supported by physiological data, for example, a study on the body mass and bioenergetics of 15 migratory species and populations found increasing body mass resulted in an exponential decrease in energetic cost per unit distance [62]. Moreover, intraspecific studies have demonstrated that phenotypic traits can rapidly evolve in response to the evolution of migration, or the loss of migration [63,64], rather than predisposing larger species to evolve migratory a behaviour. These studies offer a compelling theoretical framework and functional explanation for why larger body size is adaptive in migratory fishes and suggest a link between intraspecific processes [28–30,64] and widespread macroevolutionary patterns. We find that directional selection is driving migratory and non-migratory lineages towards different body size optima, with migratory fishes converging on an optimum that is three times larger than the optimum for non-migratory fishes. Migratory fishes are also rapidly ascending the adaptive peak, with body sizes in migratory fishes evolving faster than non-migratory lineages. These results suggest that the evolution of a larger body size is a key adaptation for migratory behaviour in fishes and provide a compelling link between theoretical hypotheses, intraspecific processes and broad macroevolutionary patterns across ray-finned fishes.
Natural selection favouring larger body sizes in migratory lineages is likely not unique to fishes and our results may be applicable across animal lineages. An energetic and biomechanical model of 200 migratory birds, mammals, fishes and invertebrates found that body size influenced maximum migration distance because larger body size increased energetics and locomotion efficiency [65]. Their model shows that the same selective pressure driving body size patterns in migratory fishes is likely favouring large body sizes across all migratory animals [65]. Many empirical studies, including examples from insects [7,14], birds [15,66], mammals [67] and amphibians [68], have found larger body sizes in migratory lineages. Our study of ray-finned fishes, which represent nearly half of all vertebrates, suggests a highly deterministic evolution of body size, resulting in a common topography in the adaptive landscape for migratory animals.
Supplementary Material
Acknowledgements
We thank Kathryn Docherty for access to a computing cluster to run many of these analyses. This research was carried out using computational resources and services provided by the Michigan State University iCER facility and the Cornell University BioHPC facility. We thank Thaddaeus Buser for access to computing resources for preliminary analyses that helped shape much of the early analytical framework for the study. M.D.B. thanks the Gilbert Ichthyological Society for insightful discussions on the difficulty of classifying migratory species across broad fish clades. We thank Matt Girard with help classifying the migratory status of polynemids. We thank Aaron Chapman, Nicholas McBryan, and Adam Channell who helped assemble the migration database. Four anonymous reviewers and Ricardo Betancur provided helpful reviews that improved our study.
Data accessibility
The tree files and body size data used in this study are available from the Dryad Digital Repository: https://dx.doi.org/10.5061/dryad.3tx95x6bm [69].
Authors' contributions
Both authors conceived and designed the study, collected the data and wrote the manuscript. M.D.B. analysed the data.
Competing interests
We declare we have no competing interests.
Funding
Funding was provided by NSF DEB (grant no. 1754627). M.D.B. was partially funded by an Edward W. Rose Postdoctoral Fellowship from the Cornell Lab of Ornithology, Cornell University.
References
- 1.Bowlin MS, et al. 2010. Grand challenges in migration biology. Integr. Comp. Biol. 50, 261–279. ( 10.1093/icb/icq013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alerstam T, Hedenström A, Åkesson S. 2003. Long-distance migration: evolution and determinants. Oikos 103, 247–260. ( 10.1034/j.1600-0706.2003.12559.x) [DOI] [Google Scholar]
- 3.Christie MR, McNickle GG, French RA, Blouin MS. 2018. Life history variation is maintained by fitness trade-offs and negative frequency-dependent selection. PNAS 115, 4441–4446. ( 10.1073/pnas.1801779115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaw AK. 2016. Drivers of animal migration and implications in changing environments. Evol. Ecol. 30, 991–1007. ( 10.1007/s10682-016-9860-5) [DOI] [Google Scholar]
- 5.Piersma T. 2007. Using the power of comparison to explain habitat use and migration strategies of shorebirds worldwide. J. Ornithol. 148, 45 ( 10.1007/s10336-007-0240-3) [DOI] [Google Scholar]
- 6.Piersma T, Perez-tris J, Mouritsen H, Bauchinger U, Bairlein F. 2005. Is there a ‘migratory syndrome’ common to all migrant birds? Ann. NY Acad. Sci. 1046, 282–293. ( 10.1196/annals.1343.026) [DOI] [PubMed] [Google Scholar]
- 7.Roff DA. 1991. Life history consequences of bioenergetic and biomechanical constraints on migration. Am. Zool. 31, 205–216. ( 10.1093/icb/31.1.205) [DOI] [Google Scholar]
- 8.Salewski V, Bruderer B. 2007. The evolution of bird migration: a synthesis. Naturwissenschaften 94, 268–279. ( 10.1007/s00114-006-0186-y) [DOI] [PubMed] [Google Scholar]
- 9.Alerstam T, Bäckman J. 2018. Ecology of animal migration. Curr. Biol. 28, R968–R972. ( 10.1016/j.cub.2018.04.043) [DOI] [PubMed] [Google Scholar]
- 10.Tsukamoto K. 1994. Origin of diadromous fishes and mechanism of migration. In Freshwater fishes migrating between rivers and the sea (eds Goto A, Tsukamoto K, Maekawa K), pp. 2–17. Tokyo, Japan: Tokai University Press. [Google Scholar]
- 11.Tsukamoto K, Miller MJ, Kotake A, Aoyama J, Uchida K. 2009. The origin of fish migration: the random escapement hypothesis. In Challenges for diadromous fishes in a dynamic global environment (eds Haro AJ, et al.), pp. 45–62. Bethesda, MD: American Fisheries Society. [Google Scholar]
- 12.Crossin GT, Hinch S, Farrell A, Higgs D, Lotto A, Oakes J, Healey M. 2004. Energetics and morphology of sockeye salmon: effects of upriver migratory distance and elevation. J. Fish Biol. 65, 788–810. ( 10.1111/j.0022-1112.2004.00486.x) [DOI] [Google Scholar]
- 13.Roff DA. 1988. The evolution of migration and some life history parameters in marine fishes. Environ. Biol. Fishes 22, 133–146. ( 10.1007/BF00001543) [DOI] [Google Scholar]
- 14.Roff DA, Fairbairn DJ. 1991. Wing dimorphisms and the evolution of migratory polymorphisms among the Insecta. Am. Zool. 31, 243–251. ( 10.1093/icb/31.1.243) [DOI] [Google Scholar]
- 15.Hamilton T. 1961. The adaptive significances of intraspecific trends of variation in wing length and body size among bird species. Evolution 15, 180–195. ( 10.1111/j.1558-5646.1961.tb03142.x) [DOI] [Google Scholar]
- 16.Mönkkönen M. 1995. Do migrant birds have more pointed wings? A comparative study. Evol. Ecol. 9, 520–528. ( 10.1007/BF01237833) [DOI] [Google Scholar]
- 17.Dingle H. 2006. Animal migration: is there a common migratory syndrome? J. Ornithol. 147, 212–220. ( 10.1007/s10336-005-0052-2) [DOI] [Google Scholar]
- 18.Ramenofsky M, Wingfield JC. 2007. Regulation of migration. Bioscience 57, 135–143. ( 10.1641/B570208) [DOI] [Google Scholar]
- 19.Nelson JS, Grande TC, Wilson MVH. 2016. Fishes of the world. Hoboken, NJ: John Wiley. [Google Scholar]
- 20.Bemis WE, Kynard B. 1997. Sturgeon rivers: an introduction to acipenseriform biogeography and life history. Environ. Biol. Fishes 48, 167–183. ( 10.1023/A:1007312524792) [DOI] [Google Scholar]
- 21.McDowall RM. 1997. The evolution of diadromy in fishes (revisited) and its place in phylogenetic analysis. Rev. Fish Biol. Fish 7, 443–462. ( 10.1023/A:1018404331601) [DOI] [Google Scholar]
- 22.Albert JS, Johnson DM. 2012. Diversity and evolution of body size in fishes. Evol. Biol. 39, 324–340. ( 10.1007/s11692-011-9149-0) [DOI] [Google Scholar]
- 23.Romanuk TN, Hayward A, Hutchings JA. 2011. Trophic level scales positively with body size in fishes. Glob. Ecol. Biogeogr. 20, 231–240. ( 10.1111/j.1466-8238.2010.00579.x) [DOI] [Google Scholar]
- 24.Griffiths D. 2012. Body size distributions in North American freshwater fish: large-scale factors. Glob. Ecol. Biogeogr. 21, 383–392. ( 10.1111/j.1466-8238.2011.00680.x) [DOI] [Google Scholar]
- 25.VanGerwen-Toyne M, Tallman RF, Gillis D. 2012. Comparison of life history traits between anadromous and lacustrine stocks of broad whitefish (Coregonus nasus): an intra-specific test of Roff's hypothesis. Adv. Limnol. 63, 159–173. ( 10.1127/advlim/63/2012/159) [DOI] [Google Scholar]
- 26.Kendall NW, McMillan JR, Sloat MR, Buehrens TW, Quinn TP, Pess GR, Kuzishchin KV, McClure MM, Zabel RW. 2014. Anadromy and residency in steelhead and rainbow trout (Oncorhynchus mykiss): a review of the processes and patterns. Can. J. Fish. Aquat. Sci. 72, 319–342. ( 10.1139/cjfas-2014-0192) [DOI] [Google Scholar]
- 27.Snyder RJ. 1991. Migration and life histories of the threespine stickleback: evidence for adaptive variation in growth rate between populations. Environ. Biol. Fishes 31, 381–388. ( 10.1007/BF00002363) [DOI] [Google Scholar]
- 28.Hendry AP, Bohlin T, Jonnson B, Berg OK. 2004. To sea or not to sea? Anadromy versus non-anadromy in salmonids. In Evolution illuminated: salmon and their relatives (eds Hendry AP, Stearns SB), pp. 92–125. Oxford, UK: Oxford University Press. [Google Scholar]
- 29.Jonsson B, Jonsson N. 2006. Life-history effects of migratory costs in anadromous brown trout. J. Fish Biol. 69, 860–869. ( 10.1111/j.1095-8649.2006.01160.x) [DOI] [Google Scholar]
- 30.Ohms HA, Sloat MR, Reeves GH, Jordan CE, Dunham JB. 2014. Influence of sex, migration distance, and latitude on life history expression in steelhead and rainbow trout (Oncorhynchus mykiss). Can. J. Fish. Aquat. Sci. 71, 70–80. ( 10.1139/cjfas-2013-0274) [DOI] [Google Scholar]
- 31.Bloom DD, Burns MD, Schriever TA. 2018. Evolution of body size and trophic position in migratory fishes: a phylogenetic comparative analysis of Clupeiformes (anchovies, herring, shad and allies). Biol. J. Linn. Soc. 125, 302–314. ( 10.1093/biolinnean/bly106) [DOI] [Google Scholar]
- 32.Simpson GG. 1944. Tempo and mode in evolution. New York, NY: Columbia University Press. [Google Scholar]
- 33.Simpson GG. 1953. The major features of evolution. New York, NY: Columbia University Press. [Google Scholar]
- 34.Losos JB, Jackman TR, Larson A, de Queiroz K, Rodríguez-Schettino L. 1998. Contingency and determinism in replicated adaptive radiations of island lizards. Science 279, 2115–2118. ( 10.1126/science.279.5359.2115) [DOI] [PubMed] [Google Scholar]
- 35.Conway MS. 2003. Life's solution: inevitable humans in a lonely universe. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 36.Blount ZD, Lenski RE, Losos JB. 2018. Contingency and determinism in evolution: replaying life's tape. Science 362, eaam5979 ( 10.1126/science.aam5979) [DOI] [PubMed] [Google Scholar]
- 37.Rabosky DL, et al. 2018. An inverse latitudinal gradient in speciation rate for marine fishes. Nature 559, 392–395. ( 10.1038/s41586-018-0273-1) [DOI] [PubMed] [Google Scholar]
- 38.Froese R, Pauly D. 2000. FishBase2000: concepts designs and data sources. Laguna, Philippines: ICLARM. [Google Scholar]
- 39.Boettiger C, Lang DT, Wainwright P. 2012. rfishbase: exploring, manipulating and visualizing FishBase data from R. J. Fish Biol. 81, 2030–2039. ( 10.1111/j.1095-8649.2012.03464.x) [DOI] [PubMed] [Google Scholar]
- 40.Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290. ( 10.1093/bioinformatics/btg412) [DOI] [PubMed] [Google Scholar]
- 41.Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223. ( 10.1111/j.2041-210x.2011.00169.x) [DOI] [Google Scholar]
- 42.Pennell MW, Eastman JM, Slater GJ, Brown JW, Uyeda JC, FitzJohn RG, Alfaro ME, Harmon LJ. 2014. geiger v2.0: an expanded suite of methods for fitting macroevolutionary models to phylogenetic trees. Bioinformatics 30, 2216–2218. ( 10.1093/bioinformatics/btu181) [DOI] [PubMed] [Google Scholar]
- 43.Felsenstein J. 2011. A comparative method for both discrete and continuous characters using the threshold model. Am. Nat. 179, 145–156. ( 10.1086/663681) [DOI] [PubMed] [Google Scholar]
- 44.Revell LJ. 2014. Ancestral character estimation under the threshold model from quantitative genetics. Evolution 68, 743–759. ( 10.1111/evo.12300) [DOI] [PubMed] [Google Scholar]
- 45.Beaulieu JM, Jhwueng DC, Boettiger C, O'Meara BC. 2012. Modeling stabilizing selection: expanding the Ornstein–Uhlenbeck model of adaptive evolution. Evolution 66, 2369–2383. ( 10.1111/j.1558-5646.2012.01619.x) [DOI] [PubMed] [Google Scholar]
- 46.Ho L.ST, Ané C. 2014. Intrinsic inference difficulties for trait evolution with Ornstein-Uhlenbeck models. Methods Ecol. Evol. 5, 1133–1146. ( 10.1111/2041-210X.12285) [DOI] [Google Scholar]
- 47.Cooper N, Thomas GH, FitzJohn RG. 2016. Shedding light on the ‘dark side'of phylogenetic comparative methods. Methods Ecol. Evol. 7, 693–699. ( 10.1111/2041-210X.12533) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Silvestro D, Kostikova A, Litsios G, Pearman PB, Salamin N. 2015. Measurement errors should always be incorporated in phylogenetic comparative analysis. Methods Ecol. Evol. 6, 340–346. ( 10.1111/2041-210X.12337) [DOI] [Google Scholar]
- 49.Pennell MW, FitzJohn RG, Cornwell WK, Harmon LJ. 2015. Model adequacy and the macroevolution of angiosperm functional traits. Am. Nat. 186, E33–E50. ( 10.1086/682022) [DOI] [PubMed] [Google Scholar]
- 50.Khabbazian M, Kriebel R, Rohe K, Ané C. 2016. Fast and accurate detection of evolutionary shifts in Ornstein–Uhlenbeck models. Methods Ecol. Evol. 7, 811–824. ( 10.1111/2041-210X.12534) [DOI] [Google Scholar]
- 51.Tibshirani R. 1996. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. B 58, 267–288. ( 10.1111/j.2517-6161.1996.tb02080.x) [DOI] [Google Scholar]
- 52.Tibshirani R. 2011. Regression shrinkage and selection via the lasso: a retrospective. J. R. Stat. Soc. B 73, 273–282. ( 10.1111/j.1467-9868.2011.00771.x) [DOI] [Google Scholar]
- 53.Beacham T, Murray C. 1993. Fecundity and egg size variation in North American Pacific salmon (Oncorhynchus). J. Fish Biol. 42, 485–508. ( 10.1111/j.1095-8649.1993.tb00354.x) [DOI] [Google Scholar]
- 54.Kinnison MT, Unwin MJ, Hendry AP, Quinn TP. 2001. Migratory costs and the evolution of egg size and number in introduced and indigenous salmon populations. Evolution 55, 1656–1667. ( 10.1111/j.0014-3820.2001.tb00685.x) [DOI] [PubMed] [Google Scholar]
- 55.Gross MR. 1987. Evolution of diadromy in fishes. American Fisheries Society Symposium 1, 14–25. [Google Scholar]
- 56.Northcote TG. 1997. Potamodromy in Salmonidae—living and moving in the fast lane. N. Am. J. Fish Manage. 17, 1029–1045. () [DOI] [Google Scholar]
- 57.Mayr E. 1963. Animal species and evolution. Cambridge, MA: Belknap Press. [Google Scholar]
- 58.Bloom DD, Lovejoy NR. 2014. The evolutionary origins of diadromy inferred from a time-calibrated phylogeny for Clupeiformes (herring and allies). Proc. R. Soc. B 281, 20132081 ( 10.1098/rspb.2013.2081) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Feutry P, Castelin M, Ovenden JR, Dettaï A, Robinet T, Cruaud C, Keith P. 2012. Evolution of diadromy in fish: insights from a tropical genus (Kuhlia species). Am. Nat. 181, 52–63. ( 10.1086/668593) [DOI] [PubMed] [Google Scholar]
- 60.Carlson SM, Rich J, Harry B, Quinn TP. 2009. Does variation in selection imposed by bears drive divergence among populations in the size and shape of sockeye salmon? Evolution 63, 1244–1261. ( 10.1111/j.1558-5646.2009.00643.x) [DOI] [PubMed] [Google Scholar]
- 61.Carlson S, Quinn T, Hendry A. 2011. Eco-evolutionary dynamics in Pacific salmon. Heredity 106, 438–447. ( 10.1038/hdy.2010.163) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bernatchez L, Dodson JJ. 1987. Relationship between bioenergetics and behavior in anadromous fish migrations. Can. J. Fish. Aquat. Sci. 44, 399–407. ( 10.1139/f87-049) [DOI] [Google Scholar]
- 63.Post DM, Palkovacs EP, Schielke EG, Dodson SI. 2008. Intraspecific variation in a predator affects community structure and cascading trophic interactions. Ecology 89, 2019–2032. ( 10.1890/07-1216.1) [DOI] [PubMed] [Google Scholar]
- 64.Palkovacs EP, Dion KB, Post DM, Caccone A. 2008. Independent evolutionary origins of landlocked alewife populations and rapid parallel evolution of phenotypic traits. Mol. Ecol. 17, 582–597. ( 10.1111/j.1365-294X.2007.03593.x) [DOI] [PubMed] [Google Scholar]
- 65.Hein AM, Hou C, Gillooly JF. 2012. Energetic and biomechanical constraints on animal migration distance. Ecol. Lett. 15, 104–110. ( 10.1111/j.1461-0248.2011.01714.x) [DOI] [PubMed] [Google Scholar]
- 66.Phillips AG, Töpfer T, Böhning-Gaese K, Fritz SA. 2018. Evidence for distinct evolutionary optima in the morphology of migratory and resident birds. J. Avian Biol. 49, e01807 ( 10.1111/jav.01807) [DOI] [Google Scholar]
- 67.Goldbogen J, Madsen P. 2018. The evolution of foraging capacity and gigantism in cetaceans. J. Exp. Biol. 221, jeb166033 ( 10.1242/jeb.166033) [DOI] [PubMed] [Google Scholar]
- 68.Tucker JK. 2000. Body size and migration of hatchling turtles: inter-and intraspecific comparisons. J. Herpetol. 34, 541–546. ( 10.2307/1565269) [DOI] [Google Scholar]
- 69.Burns MD, Bloom DD. 2019. Data from: Migratory lineages rapidly evolve larger body sizes than non-migratory relatives in ray-finned fishes. Dryad Digital Repository ( 10.5061/dryad.3tx95x6bm) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Burns MD, Bloom DD. 2019. Data from: Migratory lineages rapidly evolve larger body sizes than non-migratory relatives in ray-finned fishes. Dryad Digital Repository ( 10.5061/dryad.3tx95x6bm) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
The tree files and body size data used in this study are available from the Dryad Digital Repository: https://dx.doi.org/10.5061/dryad.3tx95x6bm [69].