Abstract
Marine coastal habitats are complex cyclic environments as a result of sun and moon interactions. In contrast with the well-known circadian orchestration of the terrestrial animal rhythmicity (approx. 24 h), the mechanism responsible for the circatidal rhythm (approx. 12.4 h) remains largely elusive in marine organisms. We revealed in subtidal field conditions that the oyster Crassostrea gigas exhibits tidal rhythmicity of circadian clock genes and clock-associated genes. A free-running (FR) experiment showed an endogenous circatidal rhythm. In parallel, we showed in the field that oysters' valve behaviour exhibited a strong tidal rhythm combined with a daily rhythm. In the FR experiment, all behavioural rhythms were circatidal, and half of them were also circadian. Our results fuel the debate on endogenous circatidal mechanisms. In contrast with the current hypothesis on the existence of an independent tidal clock, we suggest that a single ‘circadian/circatidal’ clock in bivalves is sufficient to entrain behavioural patterns at tidal and daily frequencies.
Keywords: circatidal clock, circadian clock, tidal rhythm, biological timing, oyster, bimodal behaviour
1. Introduction
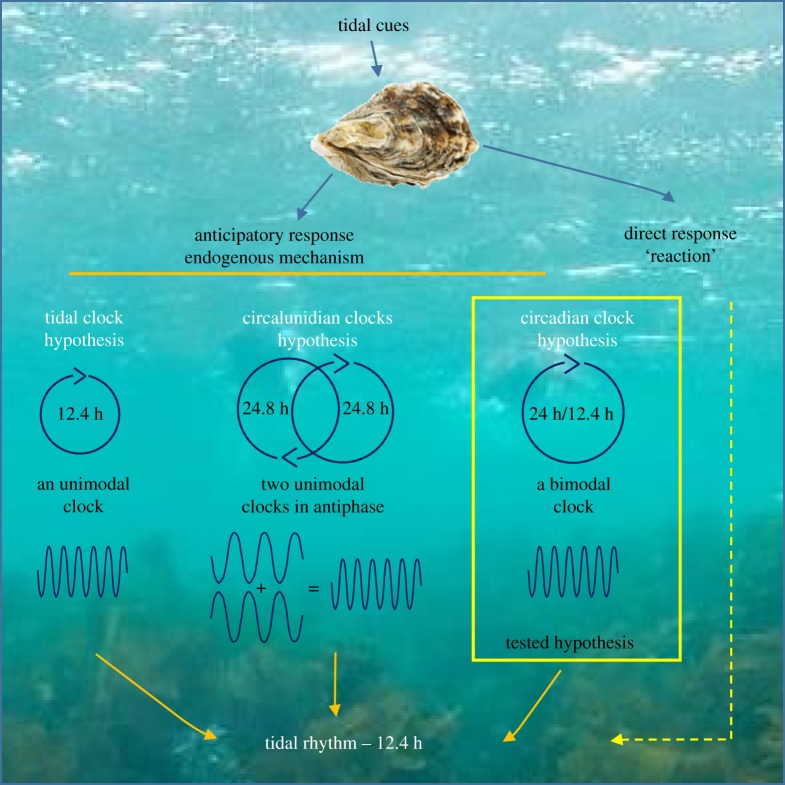
In a littoral zone, composed of intertidal and superior subtidal zones, multiple environmental cycles drive the behaviour of the inhabiting species. In addition to the well-known circadian rhythm synchronized to the solar day at the origin of the light and temperature cycles, organisms have to deal and be in harmony with tidal cycles (12.4 h). Tidal cycles occurring once or twice per lunar day (24.8 h) are the result of the combination of the Moon's orbit around the Earth, for approximately 29.5 days (synodic lunar month cycle), and the 24 h Earth rotation (solar day cycle). To anticipate and take advantage of environmental oscillations, marine organisms have developed endogenous mechanisms. Internal clock(s), nested in cells, enable organisms to account for time and are specifically synchronized by environmental cues associated with tides and solar day cycles [1–5]. Since the famous Cold Spring Harbor Symposium in 1960 [6], a debate persists on the existence and nature of one or several clocks to explain both circadian and circatidal rhythmicity [7]. Based on crustacean behavioural studies, three major hypotheses have been advanced (figure 1) to explain tidal rhythm. Naylor [8] hypothesized the existence of two separate and unimodal circatidal and circadian clocks. A second hypothesis, by Palmer [9,10], argued the existence of two unimodal circalunidian clocks (24.8 h lunar day cycle) coupled in antiphase. Finally, a third hypothesis advanced by Enright [11] suggested a single bimodal oscillator that governs both circadian and circatidal patterns. Unfortunately, most of these hypotheses are based on behavioural actograms of free-running (FR) experiments often characterized by high individual variability patterns as well as lability of rhythmicity, typical of the species living in the littoral zone [10]. Moreover, according to the time window and duration of the chronobiological analyses in FR experiments after entrained/field conditions, results can be differently interpreted to support the different hypotheses [4,7]. Since the 1980s for terrestrial organisms [12] and the 2000s for marine organisms [13], sequencing of clock genes has provided remarkable progress to understanding circadian rhythms. Knockdown approaches to study circadian clock genes in the crustacean isopod Eurydice pulchra [14,15] and the mangrove cricket Apteronemobius asahinai [16,17] have provided strong results indicating that the circatidal rhythm is independent of the circadian rhythm. However, although the results of these studies are robust, these findings do not definitively rule out the possibility of another driving mechanism. Moreover, no specific circatidal clock gene has been cloned in any species to date.
Figure 1.
Three main molecular clock hypothesis proposed to explain the tidal rhythm of organisms living in littoral areas. In the yellow rectangle is the tested hypothesis using the mollusc bivalve, the oyster Crassostrea gigas.
Bivalve molluscs such as the oyster Crassostrea gigas occupy a key position in marine benthic food webs in terms of interactions with pelagic phytoplankton food resources [18]. Crassostrea gigas exhibits the largest geographical distribution among shellfishes, probably related to the extensive exploitation of oysters for human consumption. Locally, C. gigas could be considered as an invasive species and represents a threat to ecosystems [19]. This mollusc inhabits intertidal and superior subtidal zones, making this species a key model for investigating mechanisms underlying the multiple rhythms of littoral organisms. Recently, the complete genome of this species was published [20]. Moreover, circadian clock genes and the cyclic transcriptome have been studied in C. gigas [21,22], making it an ideal candidate to investigate molecular clockwork and test hypotheses to explain circatidal rhythmicity.
2. Material and methods
(a). Experimental model and field details
All investigations were performed on Pacific oysters, C. gigas, of comparable age (approx. 1.5 years old, 65–75 mm shell length). All oysters were from natural recruitment in Arcachon Bay. Before each experiment, oysters were translocated and kept either at the study site (Eyrac Pier, Arcachon Bay, France, latitude 44.66°, longitude −1.16°) or at the Marine station of Arcachon (Arcachon Bay, France) with a continuous flow of natural sea water from the bay and a natural photoperiod. Detailed conditions are available in the electronic supplementary material, methods S1, table S1 and figure S1.
(b). Valve activity behaviour
For the valve activity behaviour in the field experiment, 15 oysters were placed in a bag fixed on a permanently immersed oyster table on 5 March 2018. The valve activity behaviour of these oysters was recorded for 18 days (5–23 March 2018; 00 corresponds to 17 h, local time, UTC + 1) using a high-frequency non-invasive (HFNI) valvometer field technology [23]. For the FR behaviour laboratory experiment, 14 oysters were pre-equipped with lightweight electrodes and placed in the study site (subtidal conditions, Eyrac Pier, Arcachon Bay, France) for 3.5 weeks. Then, the oysters were placed in the laboratory under constant darkness (dark–dark, DD) and homogenized filtrated seawater, to record valve behaviour with the HFNI valvometer laboratory technology over 7 days. Details of HFNI valvometry technology and behavioural studies conditions are available in the electronic supplementary material, methods S1. Hourly individual valve opening amplitude (VOA) data from field and laboratory experiments are accessible in the electronic supplementary material, Dataset S1.
(c). Sampling for gene expression
For the gene expression field experiment, 112 oysters were placed in the study site in 16 oysters bags. Gill tissues were sampled from seven oysters every 3.1 h. The experimental sampling details are in the electronic supplementary material, table S1. The oysters were quickly dissected (2–3 min) in a shelter on the Eyrac Pier under dim red light during the night. Gill tissues were immediately and individually transferred into 700 µl of RNA lysis buffer (Kit SV Total RNA Isolation System, Promega) with 0.5 g of ceramic beads and homogenized before storage at −80°C. The experimental protocols for the FR experiment (DD) and the experiment under light/dark (LD) entrainment are described in [21,22], respectively, from where raw data were extracted and used for the rhythmicity analysis incorporating nonparametric methods (RAIN). Details of sampling for gene expressions and experimental conditions are available in the electronic supplementary material, methods S1.
(d). Gene expression
For the field study, total RNA from the gills was extracted from samples using SV Total RNA Isolation System kits (Promega). Total RNA quantity and quality were assessed by spectrophotometry and reverse transcribed using GoScript™ Reverse Transcription System kits (Promega). Real-time quantitative polymerase chain reaction (qPCR) reactions were performed on individual samples using the GoTaqR qPCR Master Mix kit (Promega). For experiments in controlled environments (DD and LD), methods for total RNA extraction from the gills and gene expression quantification are explained in [21,22], respectively. Primer sets of circadian clock genes (Cgclock, Cgbmal1, Cgper, Cgtim1, Cgcry2, Cgcry1, Cgrev-erb, Cgror), clock-associated genes (Cghiomt, CgRhodopsin 1, 2 and 3) and housekeeping genes applied in this study are listed in the electronic supplementary material, table S3. The comparative Ct method 2−ΔΔCt [24] was used to determine the relative transcript levels of candidate genes, where ΔCt = Ct (gene) – Ct (housekeeping gene). The mRNA sequences of the investigated genes can be found via the GenBank accession numbers given in the electronic supplementary material, table S3. Gene expression data from field studies, LD laboratory experiments and DD laboratory experiments are accessible in the electronic supplementary material, Dataset S2. The detailed gene expressions procedure is available in the electronic supplementary material, methods S1.
(e). Statistical and algorithmic analysis
Field and laboratory valve behaviour data were analysed with LabView 8.0 software (National Instruments). For valve behaviour rhythmicity, chronobiological analyses were performed using TSA Serial Cosinor 6.3 software (Expert Soft Tech). Several steps were required to validate a significant rhythm [25,26]. Datasets of gene expressions were investigated for tidal and circadian periodicities in R [27] (32-bit, v. 3.2.2) using the RAIN package [28]. The RAIN algorithm is a robust non-parametric method for the detection of rhythms of specified periods in biological data that can detect arbitrary wave forms. Circatidal/tidal periodicities were defined by a significant period range of 12.4 ±3.1 h, and circadian/daily periodicities were defined by a significant period range of 24 ± 4 h.
To account for multiple testing of behavioural or genes expression, the Benjamini–Hochberg adjusted p< 0.05 were considered significant [28]. Details of chronobiological analysis using Cosinor or RAIN are available in the electronic supplementary material, methods S1.
3. Results
(a). Bimodal valve activity behaviour in the field
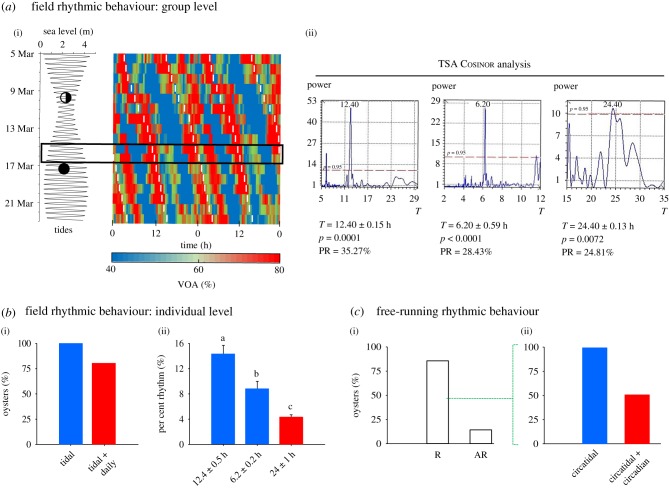
To determine oyster behaviour in the natural environment, VOA was monitored by an HFNI valvometer biosensor at Eyrac, Arcachon Bay, France (44°66′ N, 1°9′ W). This biosensor was deployed in March 2018 to record the behaviour of 15 native oysters of Arcachon Bay that are exposed to tidal and daily cycles (electronic supplementary material, table S1 and figure S1). In our subtidal conditions, the oysters were always submersed, avoiding drastically changing temperatures, oxygen levels and food supply at low tides (approx. 2 m under sea level depending on tide intensity). Valvometer data showed clear rhythmic VOA activities. Actograms (figure 2a) indicated a maximal VOA at high tide and a minimal VOA at low tide. A Cosinor analysis of the mean VOA data (figure 2b) significantly validated a 12.4 h tidal rhythm correlated to the sea-level cycle, as well as a 6.2 h rhythm correlated to the maximum water current at each midebb and midflow of a tidal cycle. Moreover, oysters also expressed a significant but less pronounced 24.4 h daily rhythm. The individual VOA data (figure 2b; electronic supplementary material, figure S2 and table S2) showed that 100% of the oysters had a tidal rhythm of which 80% were also bimodal (tidal and daily patterns). Mainly, the tidal component (12.4 and 6.2 h cycles) expressed by the per cent rhythm (PR) are more pronounced that the daily one (24 h).
Figure 2.
Valve activity to determine tidal and daily influences on oyster behaviour. (a(i)). Coloured panel shows a double-plotted actogram of the field mean VOA on 15 oysters at 4 m depths (±tides) over 18 days in Arcachon Bay, France. On the left, tide amplitudes are represented by sea level during this period. Black and half-black circles represent the new moon and the third-quarter moon, respectively. White dashed lines on the actogram represent the high tides. The rectangle shows the period of gene expression sampling times. Experimental conditions, sunset, sunrise, moon illumination and position, sea level, light intensity and water temperature are detailed in the electronic supplementary material table S1 and figure S1. (a(ii)) Chronobiological analysis of the mean valve behaviour during the field experiment. Time-series analysis Cosinor analysis was used to detect tidal periodicities (12.4 and 6.2 h periods) and daily periodicity (24 h). Significant rhythms were determined by the periods found with Lomb and Scargle periodograms (above the dotted line, corresponding to p > 0.95, peaks of significant periods T, the power corresponds to the intensity of the period) and by the adjusted p-value of the cosinor model. PR corresponds to the per cent rhythm (%) of the model. The second periodicities correspond to the second significant periodicities after residue injections. (b) Field chronobiological analysis of individual oyster VOAs. (b(i)) Percentage of rhythmic oysters (n = 15) expressing solely a tidal pattern or expressing at the time a tidal and a daily pattern. (b(ii)) The mean PR (i.e. the percentage of rhythmicity explained by the Cosinor model) according to the tidal periodicities (12.4 ± 0.5 and 6.2 ± 0.2 h) and the daily one (24.0 ± 1.0 h). Different letters represent significant differences at adjusted p < 0.05 (one-way ANOVA test and Bonferroni post hoc test). Detailed analysis are presented in the electronic supplementary material, figure S2 and table S2. (c) Rhythmic analysis on individual VOA over 7 days under laboratory FR conditions (constant darkness, DD). (c(i)) TSA Cosinor analysis was used to detect a significant rhythm (n = 14 oysters; R, rhythmic individuals (p < 0.05); AR, arrhythmic individuals). (c(ii)) Percentage of rhythmic individuals with solely circatidal periodicity (approx. 12.4 h) or expressing at time a circatidal and a circadian periodicity (approx. 24 h). Individual profiles and periodograms are detailed in the electronic supplementary material, figure S3.
(b). Bimodal and circatidal behaviour in free-running conditions
To address the issue of the genetic origin of bimodal behaviour, C. gigas individuals were collected at the same location and at the same time of year (March 2014). Oysters were placed immediately under constant darkness (DD conditions). The VOA behaviour of 14 oysters was recorded during the 7 following days, and 85.7% of the animals had significant rhythmic behaviour (figure 2c; electronic supplementary material, figure S3). At an individual level, 100% exhibited a clear circatidal rhythm with a period of 13.31 ± 0.41 h (mean ± s.e.m.), whereas 50% of the individuals also exhibited a circadian pattern with a period of 24.70 ± 1.32 h (mean ± s.e.m.).
(c). Tidal expression of circadian clock genes in the field
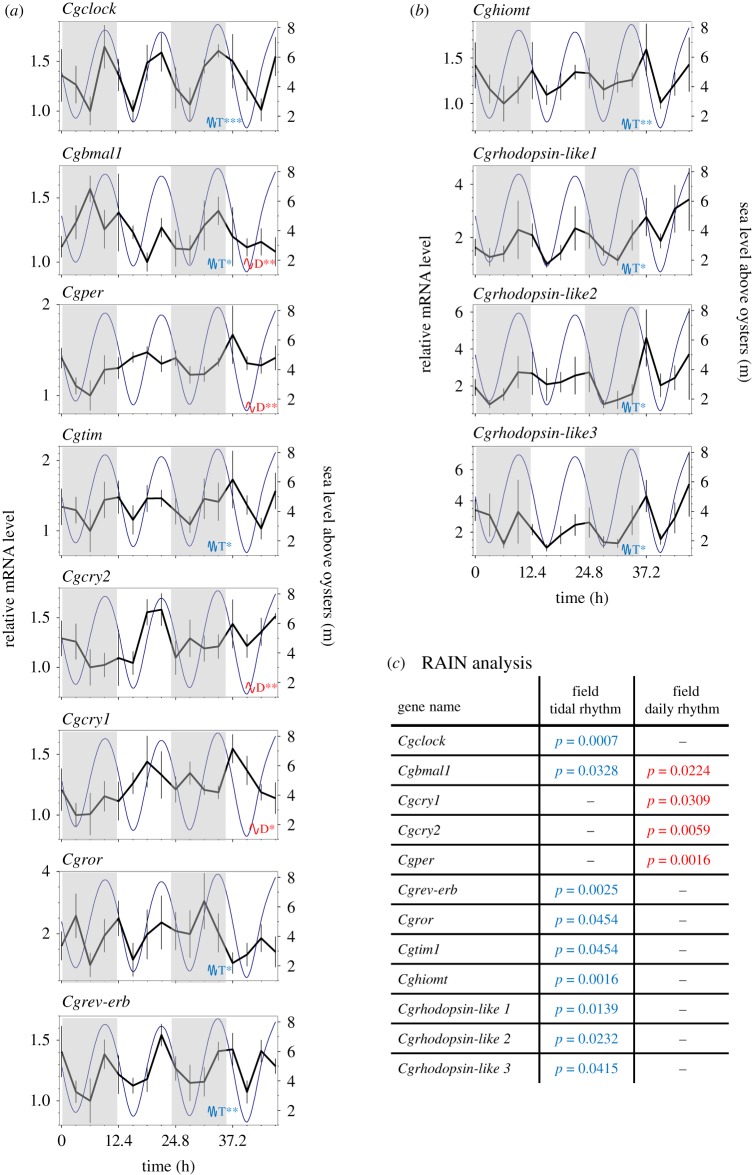
To investigate the expression of the circadian clock genes in the field, oysters were collected at the same location as for the behavioural study (figure 3). Sampling was performed from 14 to 16 March 2018, close to the new moon to avoid any potential moonlight effect [29,30]. The experimental sampling details are in the electronic supplementary material, table S1. Transcriptional variations in the eight circadian core clock genes (figure 3a) as well as putative clock-associated genes (cags, figure 3b) were investigated (electronic supplementary material, table S3) [21]. Among these candidates, Cghiomt is involved in the synthesis of melatonin [31], and three rhodopsin-likes genes (Cgrhodopsin-like 1, 2, 3) are known to be associated with light perception [32]. The statistical analysis (figure 3c) using the RAIN algorithm [28] indicated a significant tidal rhythm for five circadian clock genes: Cgclock, Cgbmal1, Cgtim1, Cgror and Cgrev-erb, which peaked during high tide slack. The cyclic expression of Cgbmal1 was bimodal, with a significant daily oscillation. Finally, Cgcry1, Cgper and Cgcry2 exhibited only a significant daily rhythm. The peak expression for Cgbmal1 was nocturnal, whereas the peak expressions of Cgper, Cgcry1 and Cgcry2 were diurnal. In addition to the core circadian genes, all cags (Cghiomt and Cgrhodopsin-like 1, 2, 3) showed a significant tidal expression, with a peak of expression between high tides and ebb tides (figure 3b).
Figure 3.
Cyclic expression patterns of C. gigas clock genes in the field. Temporal profiles of core clock genes (a) and clock-associated genes (b) mRNA levels were measured in C. gigas gills sampled in the field. Grey areas indicate night phases. Dark blue lines indicate sea level above oysters in metres (tides) on the right axis. Values are expressed in relative mRNA levels, means ± s.e.m., n = 5–7 individual replicates per time point (16 sampling times every 3.1 h for 46.5 h). For clock genes full names, see the electronic supplementary material, table S3. Data were analysed for rhythmic expression using the R package RAIN. The blue T indicates significant tidal rhythm (approx. 12.4 h), and the red D indicates significant daily rhythm (approx. 24 h). Asterisks indicate the power of significance: *p < 0.05; **p < 0.01; ***p < 0.001. (c) Inserted table of exact adjusted p-values using RAIN analysis. For details of the experimental sampling and environmental conditions, see the electronic supplementary material table S1 and figure S1.
(d). Circatidal endogenous oscillations and daily expression of circadian clock genes
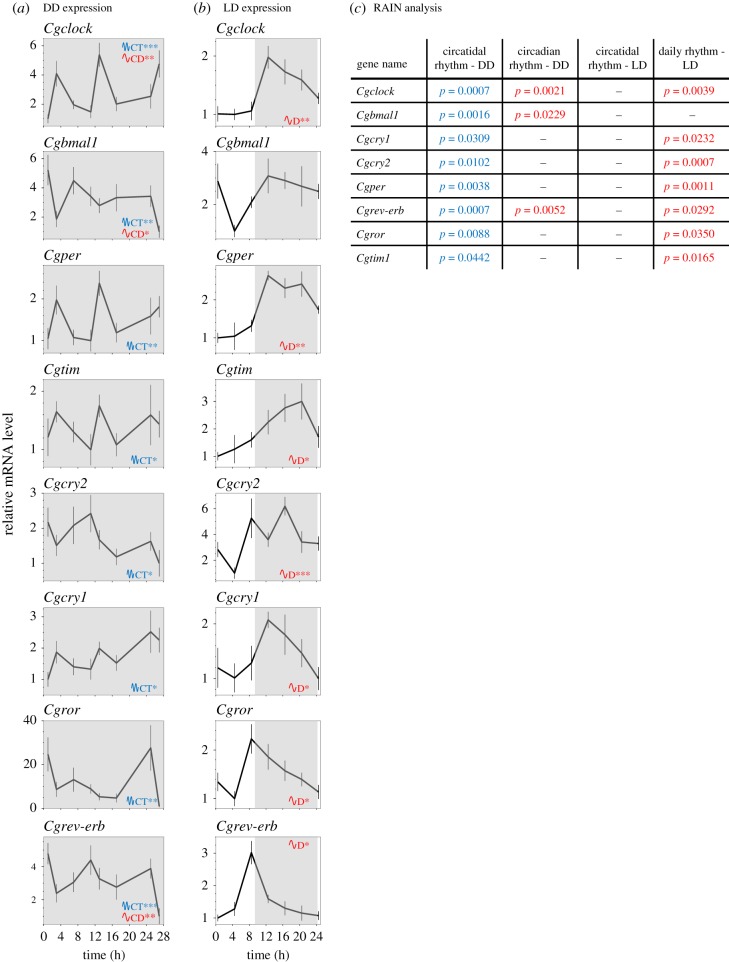
Raw data for clock gene expression from two previous experiments [21,22] were analysed using the RAIN algorithm [28]. We first reanalysed cyclic gene expression in FR conditions in DD from previous experiments [21] (figure 4a), and second, we reanalysed clock gene expression in LD (9 : 15) entrainment, mimicking the photoperiod in winter, in laboratory experiment [22] (figure 4b). The statistical analysis (figure 4c) using the RAIN algorithm showed that in DD, all core clock genes exhibited significant circatidal profiles. However, three transcripts, Cgclock, Cgbmal1 and Cgrev-erb, showed bimodal expression with a significant circadian profile as well.
Figure 4.
Cyclic expression of C. gigas core clock genes in controlled conditions. Temporal profiles of clock gene mRNA levels were measured in C. gigas gills sampled in the laboratory (a) under constant darkness (DD) and (b) LD alternating regimen 9 : 15 h. Grey areas indicate darkness. Values are expressed as relative mRNA levels, means ± s.e.m., n = 9 oysters per time point in the DD and n = 5–6 oysters per time point in LD. For clock genes full names, see the electronic supplementary material, table S3. The blue CT indicates significant circatidal rhythm (approx. 12.4 h), and the red CD/D indicates significant circadian/daily rhythm (approx. 24 h). Asterisks indicate the power of significance: *p < 0.05; **p < 0.01; ***p < 0.001. (c) Inserted table of exact adjusted p-values using RAIN analysis for DD and LD experimental data.
To verify that the circadian genes could oscillate at circadian frequency, their cyclic profiles were analysed under LD conditions (figure 4c). We showed that all the clock genes expression exhibited a strong daily pattern, except for Cgbmal1. No circatidal expression was significant for any transcripts.
4. Discussion
In coastal and subtidal field conditions, the oyster C. gigas expresses bimodal valve behaviour with a strong tidal pattern modulated by a daily rhythm. This bimodal phenotype has often been described in intertidal organisms, such as insects, crustaceans [4] and molluscs, such as the gastropod Cellana rota [33], but less for organisms settled in the subtidal zone of the littoral, excepted in the horseshoe crab Limulus polyphemus [34]. These results that oyster behaviour exhibits both tidal and daily rhythms were validated by previous 1 year studies at the same field location [25,35]. By contrast, in the absence of strong tidal forces in the Mediterranean Sea, the mussel Pinna nobilis expresses cyclic behaviour related only to light entrainment [36]. Results from FR experiments show that either circatidal or bimodal (circatidal and circadian) behavioural rhythms are observed in DD conditions. This result for C. gigas is confirmed by previous results by Perrigault & Tran [21]. Bimodal swimming activity under DD conditions has also been reported in the isopod E. pulchra and is characterized by robust circatidal behaviour, modulated by stronger activity during the subjective night [14]. The persistence of circatidal and circadian phenotypes in C. gigas valve activity behaviour under constant conditions clearly shows an overt underlying clock mechanism(s) for both rhythms. However, these phenotypes do not predetermine the nature of the mechanism(s) that may involve one or two independent clockworks. Thus, we tested the hypothesis that a unique clockwork is able to generate both circatidal and circadian rhythms. To test this hypothesis in ecological field conditions [37,38], we studied the ability of circadian core clock genes to oscillate at tidal frequency. The expression of core circadian clock genes and putative clock-associated genes (cags) was analysed in the same field location as the behavioural study. It turns out that in natural conditions, five of the eight circadian clock genes express significant tidal rhythmicity, as well as the four analysed putative cags. Interestingly, one of the clock genes (Cgbmal1) showed both tidal and daily expression patterns. Finally, the three other clock genes exhibited only daily oscillations. The proteins CgCLOCK and CgBMAL1 form a heterodimer that acts as the principal transcriptional activator in the clock mechanism [39]. If Cgclock runs at tidal frequency and Cgbmal1 is bimodal, then this scenario suggests that the protein complex CgCLOCK–CgBMAL1 could have bimodal action on generated rhythms allowing either a circatidal or circadian rhythm or both for the gene expression under their control [40]. According to the model proposed for the oyster C. gigas clockwork [21], which is closely related to the monarch butterfly Danaus plexippus clockwork [41], the protein CgCRY2, associated with CgPER and CgTIM1, could act as a transcriptional repressor of clock functioning. Cgcry2 and Cgper have daily expression, whereas Cgtim1 is tidally expressed. We propose that the protein heterotrimer CgCRY2–CgPER–CgTIM1 could also have a bimodal circadian/circatidal pattern to its transcriptional repressor action on Cgclock and Cgbmal1. The protein REV-ERB is known to repress the action of the positive transcriptional factor ROR on bmal1 expression [42,43]. Our findings suggest that tidal oscillations in Cgrev-erb and Cgror could contribute to the tidal pattern of Cgbmal1 expression. Finally, the weak diurnal Cgcry1 oscillations are consistent with its putative involvement in light synchronization of the clock [44] and consequently with the weak daily rhythm of VOA shown (figure 2). To validate the transcriptional regulatory actions of each clock gene, functional approaches are necessary. Unfortunately, the use of the Drosophila S2 cell transcriptional assay [14] is adapted for light entrainment experiments but not for investigations of the effects of tidal zeitgeber. The knockout gene technology, using CRISPR–Cas9 [45] gene-editing, is not currently operational for bivalves. Finally, gene interference that allows the knockdown of clock gene expression [46] does not give a clear and definitive response, as gene transcripts that might be sufficient to maintain the transcriptional feedback loops of the clock machinery always remain.
Cghiomt is a homologue of the mammal hiomt, which is involved in the final step of melatonin synthesis [31]. Melatonin, the hormone of the day/night cycle, is under control of the circadian clock and plays a key role in the circadian rhythms of many physiological and hormonal processes [47]. Melatonin could also have a feedback effect on the clock as an inhibitor of the proteasome associated with the degradation of clock transcriptional factors [48]. Tidal expression of Cghiomt may shape the temporal action of melatonin in a tidal manner. Similarly, a tidal rhythmicity of the Cgrhodopsin-likes 1–3, which are homologues of the rhodopsin involved in light signal transduction [32], is observed. One can be argued that this tidal rhythmicity is explained by a tidal periodicity of light availability to the gills inside the palleal cavity, as a result of the valve opening tidal rhythm.
In the FR experiment, we showed a clear endogenous circatidal rhythm for all clock genes studied, of which three genes also had circadian rhythms. Our results clearly indicate that in the absence of zeitgeber, clock genes could run at tidal frequency and bimodal patterns were exhibited for some of the genes. In comparison, only tim1 (EpTim) in the head of the crustacean E. pulchra expressed a circadian rhythm under DD exposure, whereas other clock genes did not express cyclic patterns [14]. However, the profile of EpBmal1 strongly suggests an approximately 12 h circatidal expression that should be confirmed by statistical analysis. To control the ability of these circadian clock genes to run on a daily rhythm, we placed the oysters in LD entrainment conditions, which showed that C. gigas clock genes can express daily oscillations. These results suggest that oysters should have only a daily pattern in field conditions without tidal effects, such as in the Mediterranean Sea. Our data indicate that the circadian clock can run at a tidal frequency as well as at a daily frequency according to the field locations and could be the origin of bimodal behaviour in oysters.
Controversies and hypotheses still exist in terms of explaining the tidal rhythms of organisms living in the coastal environments of oceans [1–10]. Three major hypotheses are opposed based either on the existence of two unimodal clocks or either on a single bimodal clock (figure 1). In this study, we demonstrated that some clock genes are tidally expressed when oysters are subjected to both tidal and daily environmental cues occurring in the field. This finding suggests that a single clockwork could be entrained by two different zeitgebers, i.e. LD synchronization for the daily rhythm and mechanical vibration and/or water pressure for tidal rhythm [49].
Recent findings on crustaceans and insects [14,16,17] note the existence of two independent circadian and circatidal clockworks that could share some components, such as casein kinase ɛ. This assumption is based on the fact that the interference of circadian clock gene expression (dsRNAi) lowers or removes the circadian rhythmicity but not circatidal rhythmicity in FR experiments. Despite their merits, the limits of these studies are that RNAi applications lead to a decreased-but-not-abolished expression of the targeted genes [14,16,17]. These knockdown studies do not preclude the possibility that interference in clock gene expression was not sufficient to affect tidal oscillation. Depending on the species and the ecological niches they occupy, circadian rhythm is probably more labile than circatidal rhythm. Thus, the partial knockdown of the clock gene would at least partially disrupt the circadian pattern but would have no effect or less of an effect on the tidal pattern, which would be more robust in the littoral species. Finally, although the strong arguments of this study are in favour of a bimodal clock, the possibility to have a ‘master tidal clock’ that drives a ‘slave circadian clock’ to run at tidal frequency might be raised. To validate or refute the unimodal tidal clock hypothesis (dissociated from the circadian one), the use of a complete knockout technique by mutagenesis, whose availability is still limited in marine organisms, might be a promising approach.
5. Conclusion
In natural conditions, we tested the hypothesis that the tidal rhythm of oysters might be driven by the circadian oscillator. Our results showed that the circadian clock genes could run at tidal frequency. This finding suggests that a single clock strategy could be selected to manage and interconnect the multi-temporal organization of biological processes in marine organisms. Integrating the tidal cues with the daily cues might occur in a single clock that would give bimodal or unimodal oscillation outputs according to the balance between tidal and daily cues in each specific location inhabited by oysters. However, no one can assume that all marine species share the same circatidal timing mechanism. Evolutionary selection across phyla could have resulted in the existence of separate clocks that share some components or single clocks that interact with different intermediate cogs and gears dedicated to the specific rhythms. Since the first ancestral clock emerged in aquatic environments where organisms first appeared [50], clockworks have faced both tidal and daily cues. In oysters, efficient evolutionary processes have been selected to keep the same components to deal with both rhythms.
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
The authors thank M. Sow, G. Durier, C. Portier and J.-C. Massabuau for their technical help and discussion.
Ethics
All procedures using bivalves were approved and carried out in accordance with international ethical standards and French guidelines of the universities.
Data accessibility
The mRNA sequences of the investigated genes can be found via the GenBank accession numbers given in the electronic supplementary material, table S3. Hourly individual VOA data from field studies and laboratory experiments are accessible in the electronic supplementary material, Dataset 1. Gene expression data from field studies, LD laboratory experiments and DD laboratory experiments are accessible in the electronic supplementary material, Dataset 2.
Authors' contributions
D.T. was the principal investigator and performed the study design, fieldwork, laboratory experiments, behavioural analysis, rhythmic analysis, interpretation and manuscript preparation and review, funding. L.P. performed the study design, fieldwork, laboratory experiments, behavioural analysis, gene expression analysis, rhythmic analysis, interpretation and manuscript preparation and review. M.P. performed the study design, fieldwork, laboratory experiments, gene expression analysis, interpretation and manuscript review. P.C. performed the study design, fieldwork and manuscript review.
Competing interests
We declare we have no competing interests.
Funding
The funds were provided by the French National Research Agency (WAQMOS project 15-CE04-0002).
References
- 1.Tessmar-Raible K, Raible F, Arboleda E. 2011. Another place, another timer: marine species and the rhythms of life. Bioessays 33, 165–172. ( 10.1002/bies.201000096) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De la Iglesia HO, Johnson CH. 2013. Biological clocks: riding the tides. Curr. Biol. 23, R921–R923. ( 10.1016/j.cub.2013.09.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bulla M, Oudman T, Bijleveld AI, Piersma T, Kyriacou CP. 2017. Marine biorhythms: bridging chronobiology and ecology. Phil. Trans. R. Soc. B 372, 20160253 ( 10.1098/rstb.2016.0253) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neumann D. 2014. Timing in tidal, semilunar, and lunar rhythms. In Annual, lunar, and tidal clocks: patterns and mechanisms of nature's enigmatic rhythms (eds Numata H, Helm B), pp. 3–24. Tokyo, Japan: Springer; ( 10.1007/978-4-431-55261-1_1) [DOI] [Google Scholar]
- 5.Wilcockson D, Zhang L. 2008. Circatidal clocks. Curr. Biol. 18, R753–R755. ( 10.1016/j.cub.2008.06.041) [DOI] [PubMed] [Google Scholar]
- 6.Fingerman M. 1960. Tidal rhythmicity in marine organisms. Cold Spring Harb. Symp. Quant. Biol. 25, 481–489. ( 10.1101/SQB.1960.025.01.050) [DOI] [PubMed] [Google Scholar]
- 7.Webb HM. 1976. Interactions of daily and tidal rhythms. In Biological rhythms in the marine environment (ed. DeCoursey DJ), pp. 129–135. Columbia, SC: University of South Carolina. Press. [Google Scholar]
- 8.Naylor E. 2010. Chronobiology of marine organisms. Cambridge, UK: Cambridge University Press; ( 10.1017/CBO9780511803567) [DOI] [Google Scholar]
- 9.Palmer JD. 2000. The clocks controlling the tide-associated rhythms of intertidal animals. Bioessays 22, 32–37. () [DOI] [PubMed] [Google Scholar]
- 10.Palmer JD. 1995. Review of the dual-clock control of tidal rhythms and the hypothesis that the same clock governs both circatidal and circadian rhythms. Chronobiol. Int. 12, 299–310. ( 10.3109/07420529509057279) [DOI] [Google Scholar]
- 11.Enright JT. 1976. Plasticity in an isopod's clockworks: shaking shapes form and affects phase and frequency. J. Comp. Physiol. 107, 13–37. ( 10.1007/BF00663916) [DOI] [Google Scholar]
- 12.Reddy P, Zehring WA, Wheeler DA, Pirrotta V, Hadfield C, Hall JC, Rosbash M. 1984. Molecular analysis of the period locus in Drosophila melanogaster and identification of a transcript involved in biological rhythms. Cell 38, 701–710. ( 10.1016/0092-8674(84)90265-4) [DOI] [PubMed] [Google Scholar]
- 13.Constance CM, Green CB, Tei H, Block GD. 2002. Bulla gouldiana period exhibits unique regulation at the mrnA and protein levels. J. Biol. Rhythms 17, 413–427. ( 10.1177/074873002237136) [DOI] [PubMed] [Google Scholar]
- 14.Zhang L, Hastings MH, Green EW, Tauber E, Sladek M, Webster SG, Kyriacou CP, Wilcockson DC. 2013. Dissociation of circadian and circatidal timekeeping in the marine crustacean Eurydice pulchra. Curr. Biol. 23, 1863–1873. ( 10.1016/j.cub.2013.08.038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Neill JS, Lee KD, Zhang L, Feeney K, Webster SG, Blades MJ, Kyriacou CP, Hastings MH, Wilcockson DC. 2015. Metabolic molecular markers of the tidal clock in the marine crustacean Eurydice pulchra. Curr. Biol. 25, R326–R327. ( 10.1016/j.cub.2015.02.052) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takekata H, Numata H, Shiga S, Goto SG. 2014. Silencing the circadian clock gene Clock using RNAi reveals dissociation of the circatidal clock from the circadian clock in the mangrove cricket. J. Insect Physiol. 68, 16–22. ( 10.1016/j.jinsphys.2014.06.012) [DOI] [PubMed] [Google Scholar]
- 17.Takekata H, Matsuura Y, Goto SG, Satoh A, Numata H. 2012. RNAi of the circadian clock gene period disrupts the circadian rhythm but not the circatidal rhythm in the mangrove cricket. Biol. Lett. 8, 488–491. ( 10.1098/rsbl.2012.0079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bayne BL. 2017. Biology of oysters. New York, NY: Academic Press. [Google Scholar]
- 19.FAO Fisheries & Aquaculture. 2019. Species fact sheets, Crassostrea gigas (Thunberg, 1793). See http://www.fao.org/fishery/species/3514/en.
- 20.Zhang G, Fang X, Wang J. 2012. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490, 49–54. ( 10.1038/nature11413) [DOI] [PubMed] [Google Scholar]
- 21.Perrigault M, Tran D. 2017. Identification of the molecular clockwork of the oyster Crassostrea gigas. PLoS ONE 12, e0169790 ( 10.1371/journal.pone.0169790) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Payton L, et al. 2017. Remodeling of the cycling transcriptome of the oyster Crassostrea gigas by the harmful algae Alexandrium minutum. Sci. Rep. 7, 3480 ( 10.1038/s41598-017-03797-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andrade H, Massabuau J-C, Cochrane S, Ciret P, Tran D, Sow M, Camus L. 2016. High frequency non-invasive (HFNI) bio-sensors as a potential tool for marine monitoring and assessments. Front. Mar. Sci. 3, 187 ( 10.3389/fmars.2016.00187) [DOI] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. ( 10.1006/meth.2001.1262) [DOI] [PubMed] [Google Scholar]
- 25.Tran D, Nadau A, Durrieu G, Ciret P, Parisot J-P, Massabuau J-C. 2011. Field chronobiology of a molluscan bivalve: how the Moon and Sun cycles interact to drive oyster activity rhythms. Chronobiol. Int. 28, 307–317. ( 10.3109/07420528.2011.565897) [DOI] [PubMed] [Google Scholar]
- 26.Gouthiere L, Mauvieux B, Davenne D, Waterhouse J. 2005. Complementary methodology in the analysis of rhythmic data, using examples from a complex situation, the rhythmicity of temperature in night shift workers. Biol. Rhythm Res. 36, 177–193. ( 10.1080/09291010400026298) [DOI] [Google Scholar]
- 27.R Core Team. 2013. R: the R project for statistical computing. See https://www.r-project.org/ (accessed 5 March 2019).
- 28.Thaben PF, Westermark PO. 2014. Detecting rhythms in time series with RAIN. J. Biol. Rhythms 29, 391–400. ( 10.1177/0748730414553029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zantke J, Ishikawa-Fujiwara T, Arboleda E, Lohs C, Schipany K, Hallay N, Straw AD, Todo T, Tessmar-Raible K. 2013. Circadian and circalunar clock interactions in a marine annelid. Cell Rep. 5, 99–113. ( 10.1016/j.celrep.2013.08.031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Last KS, Hobbs L, Berge J, Brierley AS, Cottier F. 2016. Moonlight drives ocean-scale mass vertical migration of zooplankton during the Arctic winter. Curr. Biol. 26, 244–251. ( 10.1016/j.cub.2015.11.038) [DOI] [PubMed] [Google Scholar]
- 31.Pévet P, Balemans MGM, Legerstee WC, Vivien-Rœls B. 1980. Circadian rhythmicity of the activity of hydroxyindole-o-methyl transferase (HIOMT) in the formation of melatonin and 5-methoxytryptophol in the pineal, retina, and harderian gland of the golden hamster. J. Neural Transm. 49, 229–245. ( 10.1007/BF01252128) [DOI] [PubMed] [Google Scholar]
- 32.Wu C, et al. 2018. A rhodopsin-like gene may be associated with the light-sensitivity of adult Pacific oyster Crassostrea gigas. Front. Physiol. 9, 221 ( 10.3389/fphys.2018.00221) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schnytzer Y, Simon-Blecher N, Li J, Ben-Asher HW, Salmon-Divon M, Achituv Y, Hughes ME, Levy O. 2018. Tidal and diel orchestration of behaviour and gene expression in an intertidal mollusc. Sci. Rep. 8, 4917 ( 10.1038/s41598-018-23167-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anderson RL, Watson WH, Chabot CC. 2017. Local tidal regime dictates plasticity of expression of locomotor activity rhythms of American horseshoe crabs, Limulus polyphemus. Mar. Biol. 164, 63 ( 10.1007/s00227-017-3098-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Payton L, Sow M, Massabuau J-C, Ciret P, Tran D. 2017. How annual course of photoperiod shapes seasonal behavior of diploid and triploid oysters, Crassostrea gigas. PLoS ONE 12, e0185918 ( 10.1371/journal.pone.0185918) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.García-March JR, Sanchís Solsona MÁ, García-Carrascosa AM. 2008. Shell gaping behaviour of Pinna nobilis L., 1758: circadian and circalunar rhythms revealed by in situ monitoring. Mar. Biol. 153, 689–698. ( 10.1007/s00227-007-0842-6) [DOI] [Google Scholar]
- 37.Helm B, Visser ME, Schwartz W, Kronfeld-Schor N, Gerkema M, Piersma T, Bloch G. 2017. Two sides of a coin: ecological and chronobiological perspectives of timing in the wild. Phil. Trans. R. Soc. B 372, 20160246 ( 10.1098/rstb.2016.0246) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van der Veen DR, Riede SJ, Heideman PD, Hau M, van der Vinne V, Hut RA.. 2017. Flexible clock systems: adjusting the temporal programme. Phil. Trans. R. Soc. B 372, 20160254 ( 10.1098/rstb.2016.0254) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dunlap JC. 1999. Molecular bases for circadian clocks. Cell 96, 271–290. ( 10.1016/S0092-8674(00)80566-8) [DOI] [PubMed] [Google Scholar]
- 40.Yoshitane H, et al. 2014. CLOCK-controlled polyphonic regulation of circadian rhythms through canonical and noncanonical E-boxes. Mol. Cell. Biol. 34, 1776–1787. ( 10.1128/MCB.01465-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu H, Yuan Q, Froy O, Casselman A, Reppert SM. 2005. The two CRYs of the butterfly. Curr. Biol. 15, R953–R954. ( 10.1016/j.cub.2005.11.030) [DOI] [PubMed] [Google Scholar]
- 42.Guillaumond F, Dardente H, Giguère V, Cermakian N. 2005. Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J. Biol. Rhythms 20, 391–403. ( 10.1177/0748730405277232) [DOI] [PubMed] [Google Scholar]
- 43.Vogeler S, Galloway TS, Lyons BP, Bean TP. 2014. The nuclear receptor gene family in the Pacific oyster, Crassostrea gigas, contains a novel subfamily group. BMC Genomics 15, 369 ( 10.1186/1471-2164-15-369) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oliveri P, et al. 2014. The cryptochrome/photolyase family in aquatic organisms. Mar. Genomics 14, 23–37. ( 10.1016/j.margen.2014.02.001) [DOI] [PubMed] [Google Scholar]
- 45.Cong L, et al. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. ( 10.1126/science.1231143) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Payton L, Perrigault M, Bourdineaud J-P, Marcel A, Massabuau J-C, Tran D. 2017. Trojan horse strategy for non-invasive interference of clock gene in the oyster Crassostrea gigas. Mar. Biotechnol. 19, 361–371. ( 10.1007/s10126-017-9761-9) [DOI] [PubMed] [Google Scholar]
- 47.Hardeland R. 2008. Melatonin, hormone of darkness and more: occurrence, control mechanisms, actions and bioactive metabolites. Cell. Mol. Life Sci. 65, 2001–2018. ( 10.1007/s00018-008-8001-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vriend J, Reiter RJ. 2014. Melatonin as a proteasome inhibitor. Is there any clinical evidence? Life Sci. 115, 8–14. ( 10.1016/j.lfs.2014.08.024) [DOI] [PubMed] [Google Scholar]
- 49.Chandrashekara MK, Sharma VK. 2008. Tidal rhythms. In Ultradian rhythms from molecules to mind: a new vision of life (eds Lloyd D, Rossi E), pp. 201–226. Berlin, Germany: Springer Science & Business Media; ( 10.1007/978-1-4020-8352-5_9) [DOI] [Google Scholar]
- 50.Tauber E, Last KS, Olive PJW, Kyriacou CP. 2004. Clock gene evolution and functional divergence. J. Biol. Rhythms 19, 445–458. ( 10.1177/0748730404268775) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mRNA sequences of the investigated genes can be found via the GenBank accession numbers given in the electronic supplementary material, table S3. Hourly individual VOA data from field studies and laboratory experiments are accessible in the electronic supplementary material, Dataset 1. Gene expression data from field studies, LD laboratory experiments and DD laboratory experiments are accessible in the electronic supplementary material, Dataset 2.