Abstract
Before environmental DNA (eDNA) can establish itself as a robust tool for biodiversity monitoring, comparison with existing approaches is necessary, yet is lacking for terrestrial mammals. Moreover, much is unknown regarding the nature, spread and persistence of DNA shed by animals into terrestrial environments, or the optimal experimental design for understanding these potential biases. To address some of these challenges, we compared the detection of terrestrial mammals using eDNA analysis of soil samples against confirmed species observations from a long-term (approx. 9-year) camera-trapping study. At the same time, we considered multiple experimental parameters, including two sampling designs, two DNA extraction kits and two metabarcodes of different sizes. All mammals regularly recorded with cameras were detected in eDNA. In addition, eDNA reported many unrecorded small mammals whose presence in the study area is otherwise documented. A long metabarcode (≈220 bp) offering a high taxonomic resolution, achieved a similar efficiency as a shorter one (≈70 bp) and a phosphate buffer-based extraction gave similar results as a total DNA extraction method, for a fraction of the price. Our results support that eDNA-based monitoring should become a valuable part of ecosystem surveys, yet mitochondrial reference databases need to be enriched first.
Keywords: environmental DNA, camera trapping, mammals, Illumina MiSeq, biodiversity monitoring
1. Introduction
Biodiversity loss due to human activities has been documented by numerous studies and calls for improved evaluations of species diversity, distribution and abundance [1,2]. Ideally, frequent surveys would be used to obtain an unbiased evaluation of faunal diversity or to document changes over time. But faunal diversity surveys remain time-consuming, expensive, invasive and usually limited in terms of taxa covered [3,4]. Moreover, distinguishing cryptic species (i.e. species that look identical but that are genetically distinct) remains an arduous task because surveys are based either on photographs or measurements of phenotypic traits, requiring substantial expertise and comprehensive sampling [5]. For these reasons, biodiversity surveys remain difficult to perform despite being crucial for the successful implementation of large-scale conservation efforts.
For terrestrial mammals in particular, camera trapping is an increasingly used approach but needs long temporal coverage, remains expensive, demands substantial maintenance and generally does not detect small animals as large animals are usually targeted [6,7]. In addition, post-processing remains laborious as manual tagging of images is still required. For small mammals, live trapping is more common but the type of trap, bait and sampling design can strongly affect the detection probability of particular species [8,9]. Moreover, trapping is highly invasive, labour-intensive and generally requires onerous permitting.
Terrestrial environmental DNA (eDNA) is poised to become an effective alternative to existing monitoring approaches [4]. For animals, the premise of eDNA is that pieces of skin, hair, faeces or saliva are shed in the environment and that, by collecting environmental samples such as water or soil, we should be able to identify to which species the extracted DNA belongs. In addition, vertebrates can be detected indirectly through DNA that was ingested by other species, such as sanguivorous species or carrion scavengers [10,11]. Yet comprehensive comparisons of species diversity identified through eDNA or ingested DNA (iDNA) alongside traditional surveys are required prior to using eDNA for biodiversity monitoring across ecosystems [4]. These comparisons are, however, rare, and usually conducted in aquatic ecosystems [12,13]. For terrestrial mammals, existing eDNA studies starting from soil or water samples are primarily proof-of-concept studies, either done in enclosed environments (e.g. fenced reserve, zoo) [14–16], which may not be directly transferrable to natural environments, or on a restricted panel of species [17,18]. Among the few eDNA studies that have compared their findings to other monitoring techniques, we note a study on salt licks [16] and several iDNA studies, which have reported a higher sensitivity and a lower sampling effort compared to camera traps, yet still do not report the entire diversity that is present [10,16].
Many questions remain unanswered about the potential of eDNA in natural environments. For example, we do not know how frequently an animal must pass by a given area to be detectable in an eDNA sample, or how recent that passage must be. The size and behaviour of an animal probably affects the amount of DNA it leaves in the environment [19], meaning that some animals may only rarely be sampled while others may be over-represented. On the methodological side, unanswered questions include the volume and number of environmental samples that should be collected, which environmental source is the most versatile, and most importantly, whether all target species are detectable. Previous studies have provided partial answers to these questions. For example, Andersen et al. [14] found that the detection rate was higher when combining subsamples in a large grid rather than from one unique point, but the amount of soil they collected per site was low (6.5 g). Their study did, however, show that topsoil is a relevant source of mammal DNA, with the advantage that it is unlikely to move over long distances, in contrast to aquatic sources. The inconvenience, however, is that extracellular DNA, the most abundant component of soil DNA, is adsorbed by soil particles, but can be extracted with a saturated phosphate buffer [20]. On DNA degradation, Ushio et al. [15] successfully recovered 200 bp long fragments in pond water samples, and recovery of much larger fragments (i.e. 15 kb) has been reported, also from water samples [21]. In general, eDNA is detected for a longer period of time and tends to be of smaller size in soil than in water samples [22].
In this study, we assessed the reliability of eDNA-based species detection for mammalian diversity surveys. Using long-term camera-trapping data [23], we compared species identified from soil surface eDNA collected on trail segments located in front of six camera traps to species recorded by these cameras. We aimed to answer the following questions:
-
1.
Can DNA from terrestrial mammals be recovered from soil surface samples and does it reliably reflect mammal diversity of that study area?
-
2.
How does experimental design affect our ability to perform biodiversity monitoring using eDNA?
-
3.
Are eDNA and camera-trapping results comparable?
We collected large amounts of soil, extracted DNA and amplified target loci with mammal-specific primers. We then compared eDNA-detected species with the species diversity recorded by camera traps and other previous studies in the study area. As no experimental protocols have been yet defined, we compared different experimental options. In particular, we collected soil samples with two different sampling strategies: extracted DNA with two different extraction kits and PCR amplification performed with two metabarcodes of different sizes. We further evaluated quantitative relationships between eDNA and camera traps and looked at the temporal and spatial accuracy by comparing eDNA results with species distribution and abundance over 3 years.
2. Methods
(a). Study area and camera trapping
Our study was conducted at the Jasper Ridge Biological Preserve (JRBP), California, USA, where we took advantage of a long-term camera-trapping effort initiated in 2009. As of October 2017, the date of soil sampling, 18 wireless camera traps were installed, mostly along trails, to monitor wildlife (see Leempoel et al. [23]). Out of these, we selected six cameras in contrasting habitats (i.e. oak woodland, riparian, grassland) that recorded wildlife continuously for 1132 days before soil sampling (electronic supplementary material, figure S1).
Images from these cameras were manually examined by volunteers who identified animals and entered requisite metadata. We used these images for comparison with eDNA results. We first counted the number of cameras at which species were detected (i.e. occupancy) and calculated their relative abundance index (RAI) across the six cameras for periods of times ranging from 30 to 1132 days by steps of 60 days. We also gathered species presence/absence on a site-by-site basis for the same periods of time. Finally, we listed all the mammals recorded by any of the cameras since 2009 for further comparison with eDNA data.
(b). Soil sampling and DNA extraction
Two soil samples were collected at each collection site (i.e. camera). For the first sample, we collected 20 cross-sections of the trail soil surface (depth of 2 cm), every 2 m for 10 m in each direction from the camera, each filling a 50 ml Falcon tube, for a total of 1 l mixed in a 1.6 l sterile sampling bag (Fisher Scientific). For the second, we collected 80 subsamples of soil surface in 12.5 ml tubes every 2 m for 80 m in each direction from the camera, for a total of 1 l. These subsamples consisted of random points either on the centre or a side of the trail. Shovels were cleaned with bleach after each sample to prevent cross-contamination. Soil samples were frozen and stored before extraction.
For DNA extraction, we used a dedicated pre-PCR laboratory room designed for low quality DNA samples that is separated from downstream PCR products. To avoid contamination, personnel were both physically and temporally separated from amplifications. We used two soil DNA extraction kits on the 12 soil samples collected. The first extraction protocol is based on the PowerMax (hereafter referred as PM) Soil DNA isolation kit (Qiagen GmbH), as in Andersen et al. [14]. Mixed soils (10 g) were processed following the manufacturer's instructions. The second extraction protocol, developed by Taberlet et al. [20], aimed to extract extracellular DNA by adding a saturated phosphate buffer to the soil sample (hereafter referred as PB), followed by a filtration and elution using the NucleoSpin Soil kit (Macherey-Nagel, Düren, Germany), skipping the lysis step. We followed the proposed protocol [20], adding 1.97 g of NaH2PO4 and 14.7 g of Na2HPO4 to 1 l of sterile water (Corning Cell Culture Grade Water, 25-055-CM) before mixing with the remaining of the soil sample (≈1 l) in two sterile 1 l bottles, and regularly shaking for 30 min. We then sampled a 1.5 ml aliquot from each bottle. Two negative extractions were performed per extraction kit. The eluted DNA was quantified on a Nanodrop 2000 (Thermo Fisher Scientific Inc.).
(c). DNA amplification and sequencing
We amplified mammal DNA with two partial mitochondrial rRNA genes of different size: a short ≈70 bp metabarcode from the 16S, and a longer ≈210 bp metabarcode from the 12S. The 16S was developed by Rasmussen et al. [24] for human coprolite analysis and generally reaches its highest taxonomic resolution at the genus or family rank. This metabarcode was used by Andersen et al. [14] to detect large mammals in both surface and core soil samples. The 12S has a higher resolution consistent with its longer size, reaching species rank for most sequences. It corresponds to the MiMammal-U developed by Ushio et al. [15], who tested it on extracted DNA from 25 species representing major groups of mammals before testing it on pond water samples from zoo cages. For both metabarcodes, we used a 2-step PCR similar to Ushio et al. [15]. The primers are combined with six random bases and an adaptor in the first PCR. Then the P5/P7 Miseq adaptors and dual-index barcodes (10 different forward and 13 reverse) were added to amplified sequences in a second PCR. Two negative PCRs were added for each primer pair during the first PCR. For the first PCR, we used 10 µl of Amplitaq Gold 360 Master Mix, 1 µl of each primer (5 µM), 8 µl mix template with H2O (PM: 8 µl Template, PB: 4 µl Template + 4 µl H2O). Cycles: Holding 10 min at 95°C, 45 cycles, denature 30 s at 96°C, annealing for 30 s at 60°C for 12S and 54°C for 16S, extension at 72°C for 60 s for the 12S and 30 s for the 16S, hold 10 s at 72°C, and a final hold at 4°C. 3 µl of each PCR product were visualized on a 2% agarose gel and the remaining product was purified using the QIAquick PCR Purification Kit (Qiagen GmbH). For the second PCR, we used 10 µl Amplitaq Gold 360 Master Mix, 1 µl of each index primer, 3 µl of template and 5 µl H2O. Cycles: holding 10 min at 95°C, 12 cycles, denature 30 s at 96°C, annealing 30 s at 65°C, extension 60 s at 72°C, hold 10 s at 72°C, hold at 4°C. The indexed second PCR products were quantified and assessed for quality control using a Fragment Analyzer (AATI), normalized to equimolar concentrations and pooled together before purification using QIAquick PCR Purification Kit. However, sequencing was performed in two separate runs, to separate the 12S and 16S PCR products, on a MiSeq platform with other unrelated projects using the MiSeq Reagent Kit v3 (2 × 150-cycle) (Illumina, San Diego, CA, USA) with 30% PhiX and ran at the Stanford University PAN Facility. To summarize, we had 12 PCR products for each of the six collection sites: two soil samples per site, three DNA extractions per soil sample (one PM and two PB), two PCRs per DNA extract (one with the 12S, one with the 16S).
(d). Sequence filtering and taxonomic assignment
We chose a series of filtering steps to be as conservative as possible while also attempting to retain as much of the ‘true’ eDNA diversity detected in our soil samples. DNA sequences were automatically sorted (MiSeq post-processing) by amplicon pool using exact matches to the dual index barcodes. Then, sequences were filtered using the OBITools software [25]. Forward and reverse reads were aligned using illuminapairedend, and only sequences with a joined-alignment score above 40 were kept. Quality scores of paired sequences were checked using FastQC, prior to adapter trimming (a maximum mismatch of 10% with the primers was tolerated) in Cutadapt [26]. At the same time, low quality sequences (quality score < 30) were removed. Afterwards, sequences shorter or longer than expected from the databases (see next paragraph) were removed using Obigrep (min. 24 bp and max. 52 bp for 16S, 150 and 192 for 12S). All samples were then pooled in a single fasta file and dereplicated using Obiuniq. Next, sequences occurring less than 10 times were removed before applying Obiclean to identify PCR and sequencing errors. To do so, Obiclean classifies sequences either as head, internal or singleton. Head sequences, the most common ones, correspond to true sequences or chimera product and can have multiple variants. Similarly, singletons are either true sequences or chimeras but are not related to any other sequences. Finally, internal sequences correspond to amplification/sequencing errors. See Boyer et al. [25] for a detailed explanation. Obiclean was applied sample by sample, with a maximum of one difference between two variant sequences and a threshold ratio between counts of one, meaning that all less abundant sequences are considered as variants. Only sequences with head or singleton status in at least one sample were kept. Further, sequences whose status in the global dataset was more commonly ‘internal’ than ‘head’ or ‘singleton’ were discarded [27].
Afterwards, remaining sequences were matched against reference databases built using EcoPCR [28]. To do so, we downloaded the EMBL database of standard sequences (http://ftp.ebi.ac.uk/pub/databases/embl/release/std/, release 135) of mammals (mam), vertebrates (vrt), mouse (mus) and human (hum), before converting it to the EcoPCR database format with Obiconvert. We then used EcoPCR to find sequences amplified by the primer pairs, using a maximum of three mismatches, and a minimum and maximum length identical to those mentioned above. Each resulting database was then dereplicated with Obiuniq. Expected mammal species were searched for in the database and their presence at species, genus or family rank was recorded to inform interpretation of results (see electronic supplementary material). Thereafter, sequences were matched to the databases using Ecotag, and only sequences with an identity above 95% and 90% for the 12S and 16S respectively were kept. In addition, sequences that did not attain the rank of class or lower were deleted, and sequences assigned at species rank but whose identity was lower than 99% were ranked at genus level. Finally, sequences matching to the same reference sequence were grouped and their read count updated. After these steps, we proceeded on a case-by-case basis for regrouping or removal of sequences. For example, sequences assigned to the same species were grouped, or species not found in the Americas were removed. See electronic supplementary material for detailed decisions on each of these sequences.
To discard potentially contaminant sequences, molecular operational taxonomic units (MOTUs) whose relative read abundance (RRA) was higher on average in the negative controls and/or negative PCRs than in true samples were removed. Finally, any MOTUs with abundances representing less than 0.05% of the total MOTU abundance across samples were removed to correct for potential cross-contamination. The detailed number of sequences, reads and taxa discarded at each step can be found in the electronic supplementary material tables.
(e). Data analysis
We first compared the list of MOTUs reported by both metabarcodes, by extraction method and by sampling design at family level, as well as the number of sites at which they were detected. We then calculated accumulation curves of the number of MOTUs as a function of the number of soil samples, again comparing metabarcodes, extraction methods and sampling design, with the specaccum function from the package vegan [29] in R (R Core Team, 2018), using 1000 permutations in the random method.
We then compared the presence/absence of species from the eDNA survey with the camera trapping records. We performed linear regressions with the function ‘lm’ in R between species occupancy and the RAI on the camera trap side, and the number of positive PCR, positive soil samples and positive sites on the eDNA side. Only species recorded by camera traps were kept for the comparison (electronic supplementary material, table S1). Note, however, that although we are confident with species-level identity in the camera images, not all these species can be identified down to the species level with either metabarcode, and this is denoted by our use of open nomenclature.
Finally, we calculated the similarity between eDNA and camera traps between matrices of species presence/absence per site (matrices of species X sites) from both camera traps and 12S eDNA using Mantel tests (Pearson method, 999 permutations) on dissimilarity indices calculated with vegdist, using the Jaccard method. With the same method, we calculated the similarity between the 12S and 16S by grouping MOTUs at the family level.
3. Results
We obtained 9 795 610 and 6 008 091 paired reads for the 16S and 12S, respectively. After initial filtering, we identified 25 MOTUs with 16S and 44 MOTUs with 12S but many were removed or regrouped during our data processing pipeline. For example, all birds identified were removed as they were not part of the target taxon. One MOTU was discarded based on a geographical criterion (Capreolus detected in 16S, a strictly Eurasian genus of deer, probably PCR error originating from Cervidae). We further detail the filtering procedure in the electronic supplementary material. The proportion of human sequences was high, representing 49.14% in 16S and 35.30% in 12S. Humans are the most common species in camera trap records but there is also a high probability of human contamination during laboratory work. Because of their high RRA in negative extractions, all human sequences were considered as contaminants. Similarly, the grey fox (Urocyon cf. cinereoargenteus) was discarded from the 12S results as its RRA in negative controls was above the defined threshold. After these final filtering steps, we detected 19 and 17 MOTUs with the short (16S) and long (12S) metabarcode respectively (table 1).
Table 1.
eDNA detections at each six sampling sites as reported by the number of positive PCRs per family identified with the short and long metabarcodes (16S/12S), sorted in alphabetical order. Six PCRs were performed per site and per metabarcode, or 36 per metabarcode in total. MOTUs grouped by family are given in columns 16S and 12S at their highest taxonomic rank as obtained in OBITools. Note that the highest rank obtained can be family. Column CT reports the number of independent photographic events recorded during the year preceding sampling for all species of a given family and summed over the six sites. Urocyon sp. was removed from 12S as it was considered as a contaminant. There is no category for horses in the camera trap dataset, as they are merged with humans. Species only recorded by camera traps can be found in electronic supplementary material, table S11. Detailed results of the 16S and 12S can be found in electronic supplementary material, tables S1–S6.
| family name | 16S | 12S | A | B | C | D | E | F | total | CT |
|---|---|---|---|---|---|---|---|---|---|---|
| Bovidae | Bovinaea | 0/0 | 0/0 | 0/0 | 5/0 | 0/0 | 0/0 | 5/0 | ||
| Canidae | Urocyonb, Canidaeb | Canisb | 2/0 | 1/0 | 0/1 | 1/2 | 0/1 | 4/0 | 8/4 | 364 |
| Cervidae | Cervidaeb | Odocoileus hemionus | 4/1 | 0/0 | 2/2 | 0/2 | 0/3 | 4/2 | 10/10 | 562 |
| Cricetidae | Arvicolinaec, Neotominaed, Neotoma fuscipesa, Peromyscusb | Arvicolinaec, Peromyscusc, Neotomad | 3/3 | 4/2 | 3/3 | 6/3 | 1/2 | 4/3 | 21/16 | 3 |
| Equidae | Equusa | Equusd | 0/1 | 0/1 | 1/0 | 0/1 | 1/0 | 0/2 | 2/5 | |
| Felidae | Felidaeb | Puma concolor, Lynx rufus | 1/2 | 0/1 | 2/2 | 5/2 | 1/3 | 1/5 | 10/15 | 292 |
| Leporidae | Leporidaec | Sylvilagus bachmani, Lepusd | 4/3 | 1/1 | 0/0 | 0/1 | 0/1 | 1/3 | 6/9 | 1136 |
| Muridae | Murinaeb | Rattusb | 0/0 | 5/0 | 0/2 | 0/0 | 0/1 | 0/0 | 5/3 | |
| Sciuridae | Tamiasd, Sciuridaec, Marmotinid | Sciurusc | 0/0 | 0/0 | 0/1 | 6/0 | 2/1 | 6/2 | 14/4 | 623 |
| Suidae | Suidaeb | 0/0 | 3/0 | 0/0 | 0/0 | 0/0 | 0/0 | 3/0 | ||
| Talpidae | Talpidaec | Scapanus latimanus, Neurotrichusd | 2/0 | 2/1 | 0/1 | 0/2 | 0/0 | 0/1 | 4/5 | |
| Vespertilionidae | Myotisb,e, Myotisb,e | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 3/0 | 3/0 | ||
| Didelphidae | Didelphisd | 0/1 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/1 | 81 | |
| Geomyidae | Thomomysd | 0/2 | 0/6 | 0/0 | 0/0 | 0/0 | 0/0 | 0/8 | ||
| Mephitidae | Mephitis mephitis | 0/1 | 0/0 | 0/1 | 0/1 | 0/1 | 0/0 | 0/4 | 280 |
aSequence matched with multiple species in the reference database but only one is known in the region at that taxonomic level.
bSequence matched with multiple species in the reference database and more than one is known in the region at that taxonomic level.
cNo matching sequence in the reference database and more than one species known in the region at that taxonomic level.
dNo matching sequence in the reference database but only one species known in the region at that taxonomic level.
eUnique MOTUs matching at the same taxonomic level.
Both metabarcodes largely concurred at the family level: out of the 15 families detected, nine were found with both metabarcodes and three were unique to each of them. The three families only detected using 16S were Bovidae, Suidae and Vespertilionidae (Myotis spp.), but all three were detected at a single site. For the 12S, these were Mephitidae (cf. Mephitis mephitis), Didelphidae (cf. Didelphis virginianus) and Geomyidae (cf. Thomomys bottae). The maximum taxonomic resolution reached was higher for the 12S, with identification at genus rank or lower 16/17 times, compared to only 7/19 with the 16S. For example, Puma concolor and Lynx rufus identified at species level in 12S both correspond to Felidae in 16S. There is a notable exception for the Sciuridae, for which we detected only 1 MOTU in 12S (Sciurus spp.) but 3 in 16S (Tamias sp., Sciuridae, Marmotini).
MOTUs accumulation curves at the family level converged towards a maximum when accumulating the 36 PCRs per metabarcode (figure 1). MOTUs were also detected more frequently with the 16S, as the percentage of MOTUs detected more than once is twice that of 12S (15% against 8% respectively). These percentages are low in both cases, suggesting that the number of PCR per site was insufficient, and limiting the potential for comparisons between extraction kits and sub-sampling strategies. Therefore, for the purpose of these comparisons, we decided to group 12S and 16S MOTUs taxonomically at the family level. For example, detections of Lynx rufus and Puma concolor (12S) were summed with Felidae detections (16S).
Figure 1.
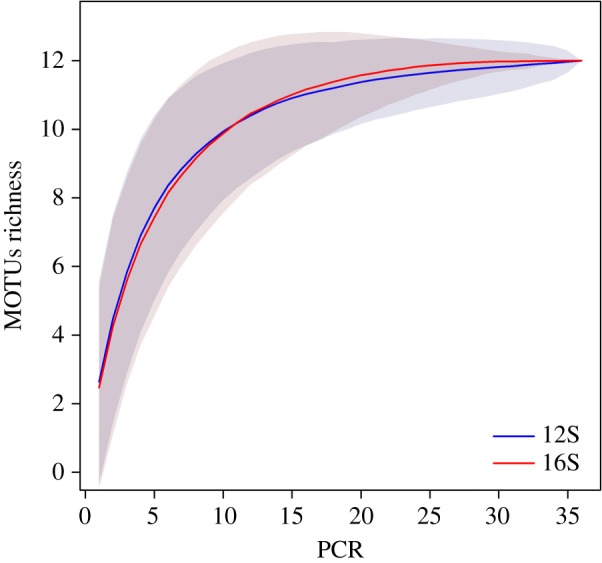
MOTUs accumulation curves of both metabarcodes (16S and 12S) by PCR product. MOTUs were merged at the family level. Grey areas correspond to the standard deviation, estimated from 1000 random permutations. (Online version in colour.)
Extraction protocols displayed similar accumulation curves (electronic supplementary material, figure S2A) and reported the same families, except for Didelphidae, which was detected in only one PCR with the second PB extraction. Similarly, MOTUs detected by just one sub-sampling strategy were among the rarest detected (Didelphidae and Vespertilionidae with SS80). Species accumulation curves comparing sampling strategies show no substantial differences between samples made of 20 subsamples (SS20) or 80 subsamples (SS80) (electronic supplementary material, figure S2B), except for the two families mentioned above and a higher standard deviation of the latter.
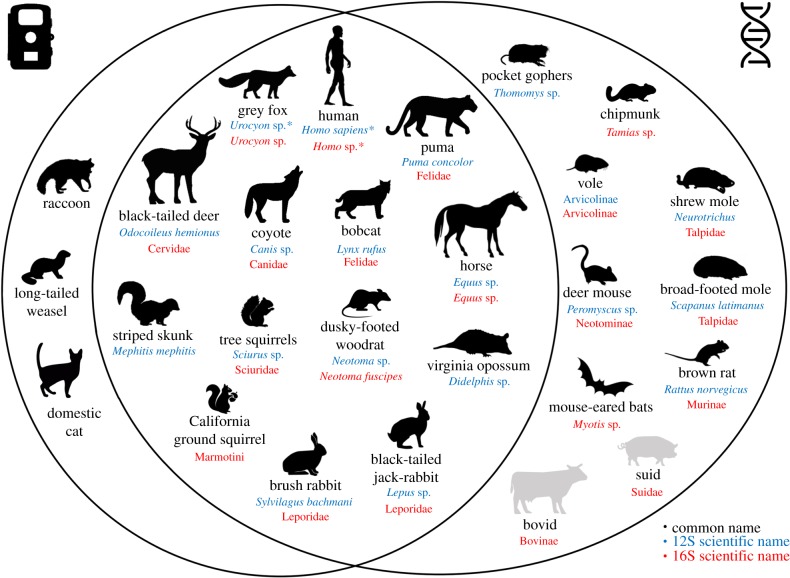
All species frequently recorded by camera traps were also detected with eDNA (figure 2). Conversely, all mid-to-large-sized mammals detected with 12S were recorded by camera traps. Most importantly, a large panel of small mammals rarely, if ever, recorded by camera traps were detected with eDNA at multiple sites. All of them can be reasonably expected to be present in the preserve or the region [30]. With the 16S, however, 2 MOTUs not known to be in the preserve were detected (i.e. Suidae and Bovinae) in two soil samples but only from one site each.
Figure 2.
Venn diagram between species recorded by camera traps and species detected with eDNA from both metabarcodes. Species detected with the 16S are marked in red and those detected with the 12S are in blue. Scientific names are given at the maximum rank reached with each metabarcode. Species known to be present in the study area are in black. Species absent from the study area but detected with eDNA are in grey. Species considered as contaminant are indicated with asterisks. Credits for illustration are in electronic supplementary material. (Online version in colour.)
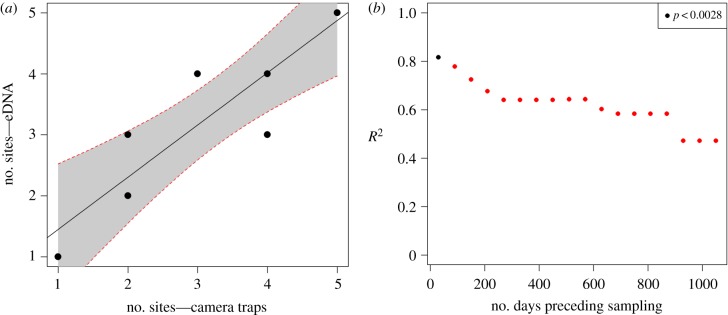
We found a strong relationship between species occupancy from eDNA and camera traps. The number of camera traps at which species are recorded correlated with the number of sites at which they were detected with the 12S (figure 3). However, this relationship was only significant when using at least 30 days of camera-trapping data, after which the regression coefficient kept decreasing with longer recording periods. Other attempts to identify quantitative relationship between the number of camera trap sites and eDNA metrics had lower regression coefficients and were not significant (electronic supplementary material, figures S2–S6).
Figure 3.
(a) Quantitative relationship between the number of sites at which species (n = 9) are recorded by camera traps and detected with eDNA (12S), using a month of camera-trapping records preceding sampling. (b) Change in the regression coefficient for the same relationship with increasing camera trap records. Camera trap records from one month to 3 years by steps of 60 days. Significant regressions (p < 0.05 with Bonferroni correction) are marked with black points. (Online version in colour.)
We also found substantial inconsistencies between the sites at which species are detected with eDNA and those at which they are recorded with camera traps. Indeed, the Mantel test between species presence/absence per site from camera trapping records against 12S detections was not significant (best correlation obtained with 180 days of camera trapping data, Mantel statistic: 0.485, p: 0.097). Similarly, the Mantel test between family presence/absence per site from the 16S against 12S was not significant either (Mantel statistic: 0.487, p: 0.056).
4. Discussion
We detected a large and similar ensemble of species with both mitochondrial metabarcodes, matching closely with the expected species composition in the studied area. Moreover, the detection of many small mammals is a considerable advantage of eDNA, as these species are generally more difficult to document than larger mammals in camera-trapping efforts [31]. Our results show that eDNA-based surveys offer a meaningful, and non-invasive, complement to the multiplicity of approaches that would have been needed to capture the same diversity.
We found several advantages to the longer metabarcode 12S (≈210 bp) compared to the shorter 16S (≈70 bp). First, the amount of long mitochondrial fragments we recovered from soil surface samples was sufficient to detect most animals at the species-level, and allowed a more precise taxonomic rank than did the same samples for 16S. For example, multiple Arvicolinae are known in the region and the 16S sequence we collected does not go further than sub-family level, with BLASTN [32] not providing additional information. With the 12S Arvicolinae sequence, however, the top ten matches in BLASTN are all from the genus Microtus, suggesting the most likely candidate is Microtus cf. californicus, given it is the only known species of Microtus in western California. Second, the 12S has the potential to discover previously undescribed haplotypes or subspecies. The Neotoma sequence we detected matched at 98% identity with existing reference sequences of Neotoma fuscipes, and the only known occurrence of Neotoma in the region is the subspecies Neotoma fuscipes annectens [33], suggesting that we may have revealed hidden diversity. Third, the high taxonomic resolution of this metabarcode can help distinguish closely related species. For example, the detection of Rattus norvegicus not only confirmed previous observations but also helped to differentiate it from another member of its genus, Rattus rattus, which is locally abundant in areas outside Jasper Ridge Biological Preserve but rare within it. Fourth, there were no unexpected taxa with the 12S, unlike with the 16S. Bovinae and Suidae, detected only with the 16S, are not present in Jasper Ridge as live animals, but might have come from sources such as nearby cattle ranches, contamination from food items, soles of human footwear, residuals of a geographically large-ranging predator diet, or laboratory supplies or reagents (e.g. BSA, which is used in the laboratory). We found that the 12S detected multiple species of birds, due to the lack of specificity of the MiMammal-U primers, suggesting that soil contains a large spectrum of above ground species that deserve further evaluation (see electronic supplementary material, table S4 for the list of birds detected). While it could be a disadvantage if one aims to maximize the number of reads for the target taxon, the number of bird-identified reads recovered was small (3%). The only major drawback to using a metabarcode of this size (≈210 bp) is that we found it less frequently than the short ≈70 bp fragments, which would be an issue if the amount or quality of the starting material is limited.
The large standard deviations in our accumulation curves and the single detections of many MOTUs suggest that PCR replicates should have been performed to reduce stochasticity, in addition to our replications at the sampling and extraction steps. We assumed that 12 PCRs per site, when merging both metabarcodes at the family level, would be sufficient to detect rare taxa [34], but the lack of convergence suggests that MOTUs could have been missed. Nevertheless, we show that collecting a large amount of surface soil is essential because of potentially high heterogeneity of soil samples and deposition rates [20,35]. Here, 12 l of surface soil was just enough to obtain a complete picture of the mammalian diversity. Finally, we sampled soil in all types of habitats in the preserve and noticed a higher number of detections at sites with shade (presumably less UV light to degrade DNA) and limited wind (sites D and F in the riparian and oak-woodland habitats; table 1), although we did not sample at enough sites per habitat to firmly support this observation.
After combining all samples and metabarcodes, we found that the two extraction kits we studied had only a marginal influence on the results, although differences on a site-by-site basis are notable. Both performed equally well despite relying on very different protocols and amount of dirt processed. These results show that even for species with an expected low deposition rate, compared to plants or insects for example, the Phosphate Buffer (PB) extraction protocol is suitable, costs a fraction of the price of the PowerMax (PM) and requires less equipment [20,35]. In terms of sampling strategy, our results do not suggest a substantial improvement of detection with more sub-samples, as opposed to Andersen et al. [14].
Spatio-temporal relationships between species detection with eDNA and camera traps showed mixed results. On one hand, the long-term camera-trapping study allowed us to test the accuracy of eDNA over time and sighting frequency. We found that eDNA best reflected species presence from camera images between 30 to 150 days before soil sampling (figure 3b). In addition, coyotes were detected in only 3 out of 12 soil samples, the least of all carnivores. Coyote, who were by far the dominant predator before the mountain lions increased substantially in 2013 [23], but now are seen an order of magnitude less frequently (984 records in 2012, 98 in 2017). This suggests that the DNA of coyotes does not stay detectable for long (4+ years) in the environment and that recent and infrequent presence does affect detection probability in eDNA. Similarly, the presence of raccoons decreased substantially over the past several years (150 records in 2012 to 24 in 2017) and we were not able to detect them at any site using our eDNA methods. We also did not detect the American badger (Taxidea taxus), the domestic cat (Felis catus) nor the long-tailed weasel (Mustela frenata). The former was not recorded since 2013, while the latter two were only recorded two and three times respectively in the 2 years preceding sampling (see electronic supplementary material, table S11). In comparison, raccoons were recorded 21 times by cameras over that same period. Therefore, it seems that both time since camera trapping and decreasing local abundance contributes to the non-detection of species using eDNA.
On the other hand, we did not find a strong spatial accuracy. We found a strong relationship between the number of sites where a species was seen by cameras over short durations and detected by eDNA, suggesting that eDNA could be used for species occupancy modelling, as is the case in camera-trapping studies [6], and as recently demonstrated with leech iDNA [10]. However, eDNA results from a single site did not directly match camera trap images there. Moreover, the comparison between the 16S and 12S on a site-by-site analysis show a lack of similarity too. For example, Felidae is not detected at site B, which is consistent with camera trap observations, but Puma concolor is detected with the 12S at that site. Another striking inconsistency is the opposite pattern of detection for Equus (table 1). These observations are in contrast to the correlation observed in figure 3, and we do not have a clear explanation for this. Pieces of skin, fur or dried scats could be transported by wind, or involuntarily by other species or through their diet. Animals may be present close by, but not in the camera trap field of view. And behavioural and ecological characteristics will also have a strong impact on eDNA detectability. In our data, some species appeared to be over-represented, such as the felines (puma, bobcat), which could be due to their habit of marking their territory via urine and faeces along trails. Both felines are 3 and 10 times less frequent, respectively, on camera trap records than black-tailed deer, but were detected more frequently with eDNA. Future work should perhaps consider collecting larger volumes of soil per site but also process subsamples independently to find better correlations between species abundance and eDNA.
While our results are promising for eDNA as a survey tool for terrestrial mammals, similar studies need to be improved and replicated in many habitats and environments before being considered for ecosystem surveys more globally. The major and critical hindrances we faced were the incompleteness of reference databases and improper amplification due to primer mismatch. At least two inconsistent results between the two metabarcodes can be attributed to these factors. For example, Didelphis and Thomomys were detected with the 12S but are not in the 16S reference database. For Didelphis, this is due to too high a mismatch with the 16S forward primer, while for Thomomys, it is due to the absence of a reference sequence for any sister species in its family (Geomyidae). If this data shortfall is an issue in a region where biodiversity is well studied, it is easy to imagine that it can only be worse in poorly covered ecosystems and/or more biodiverse regions. To investigate the utility of our approach more globally and using the same methods as described in electronic supplementary material, we found that 59% of all known mammals [36] are missing from the 12S database and 33% of these missing species have no sister species at the genus level (electronic supplementary material, table S2), which greatly hampers our ability to conduct eDNA studies worldwide. These numbers get even worse for specific orders, with 48% of missing rodents having no sister species at the genus level or 62% for missing carnivores. Axtner et al. [37] reported similar concerns for tetrapods in a tropical ecosystem. Using metabarcodes located in four different genes, they revealed vastly different coverage per Class, with no marker exceeding 85% of coverage for the targeted species, leading them to suggest using multiple genes in eDNA studies to reduce that coverage bias. Yet, even when combining their four metabarcodes for all known tetrapods, the coverage at the species level hardly reaches 50%. This preliminary analysis of detectability and database evaluation is often overlooked in eDNA papers, despite being a critical step to understand if the non-detection of species is due to their true absence in the studied area or to the shortcomings of databases and primers. In such context, taxonomic assignment by phylogenetic placement, such as with Protax [38], is recommended to obtain more accurate probabilities of assignment.
Another consequence of incomplete databases is that sequences with an identity below the defined filtering threshold will be discarded. We therefore looked at these discarded sequences to find potentially missing species. We found a sequence corresponding to the sub-family Soricinae in the 16S sequences, discarded because its best identity was lower that the decided threshold in our data processing pipeline (i.e. 90%). This sequence probably corresponds to Sorex ornatus, the only known Sorex in the preserve. Similarly, several sequences attributed to squirrels in the 12S were discarded, which could help explain why we did not successfully report their known diversity in the study area. For example, we found a sequence attributed to Callospermophilus lateralis, but instead it probably belongs to the California ground squirrel (Otospermophilus beecheyi), which is abundant in the preserve, but also is missing from the 12S database at the species and genus level. It is worth mentioning that 2 MOTUs of bats were detected out of the 14 known in the region, showing the limit of trail soil sampling for this order of mammals.
Part of the success of this study can be attributed to the habitat and topography of Jasper Ridge. Indeed, the vegetation is a dense chaparral in many areas, discouraging the movement of large mammals. Steep, uneven terrain, deep drainages, and creeks that flood in the rainy season also influence the routes animals take across the landscape. Therefore, trails represent the easiest way to move around the reserve. Thus, large mammals repeatedly pass by the same locations, increasing the concentration of DNA dropped on trails. Most of our cameras are set on trails or at trail intersections for this reason. This opens the question of whether we would have been able to detect these species with eDNA had we sampled randomly in an open grassland with no clear trail structure. Still, small mammals, who do not rely on built trails, were easily detected with eDNA.
Our study demonstrates once more that eDNA is a remarkably promising approach for ecosystem assessment, and opens new possibilities for managers and researchers to reveal the distribution and interaction of species in a single survey. From soil surface samples, we detected most species present in the study area, including those which are generally too small to trigger camera traps. As such, eDNA alone was enough to obtain a reasonable picture of species diversity without requiring previous knowledge of the study area. In terms of sampling design, neither the number of subsamples and extraction kits affected substantially the results of our survey. On the other hand, we suggest that long fragments (200 bp) are ideal for present-day biodiversity studies, as our comparison with camera traps shows that they do not last more than a couple of months and provide a finer taxonomic resolution. Nevertheless, many unknowns remain with regards to the detectability of extremely rare species, as well as the strategy to adopt for technical replication. While promising, eDNA remains currently time-consuming and cannot yet be scaled up to a landscape level.
Supplementary Material
Supplementary Material
Acknowledgements
We thank many Jasper Ridge volunteers for their contribution to the tagging of camera trap images, Hadly lab members for their suggestions, Fred Boyer for helping us troubleshoot with OBITools, Nicole Nova for providing several illustrations of mammals. Finally, we thank the three anonymous reviewers for their useful comments and for the general improvement of the manuscript.
Data accessibility
12S and 16S eDNA curated sequences are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.2547d7wmf [39]. R scripts were published on GitHub: https://github.com/kleempoel/Soil_eDNA_Leempoeletal2019.git.
Authors' contribution
K.L. designed and performed the research, analysed the data and wrote the paper. T.H. installed and maintained the camera traps, curated the image database and contributed to the paper. E.A.H. designed the research and contributed to the paper.
Competing interests
We declare we have no competing interests.
Funding
Swiss National Science Foundation Early Postdoc Mobility grant P2ELP3_175075, Stanford University Mellon grant (K.L.), HHMI Professorship (E.A.H.).
References
- 1.Barnosky AD, et al. 2012. Approaching a state shift in Earth's biosphere. Nature 486, 52–58. ( 10.1038/nature11018) [DOI] [PubMed] [Google Scholar]
- 2.Mendenhall CD, Karp DS, Meyer CFJ, Hadly EA, Daily GC. 2014. Predicting biodiversity change and averting collapse in agricultural landscapes. Nature 509, 213–217. ( 10.1038/nature13139) [DOI] [PubMed] [Google Scholar]
- 3.Stem C, Margoluis R, Salafsky N, Brown M. 2005. Monitoring and evaluation in conservation: a review of trends and approaches. Conserv. Biol. 19, 295–309. ( 10.1111/j.1523-1739.2005.00594.x) [DOI] [Google Scholar]
- 4.Deiner K, et al. 2017. Environmental DNA metabarcoding: transforming how we survey animal and plant communities. Mol. Ecol. 26, 5872–5895. ( 10.1111/mec.14350) [DOI] [PubMed] [Google Scholar]
- 5.Bickford D, Lohman DJ, Sodhi NS, Ng PKL, Meier R, Winker K, Ingram KK, Das I. 2007. Cryptic species as a window on diversity and conservation. Trends Ecol. Evol. 22, 148–155. ( 10.1016/j.tree.2006.11.004) [DOI] [PubMed] [Google Scholar]
- 6.O'Connell AF, Nichols JD, Karanth KU. 2010. Camera traps in animal ecology: methods and analyses. Tokyo, Japan: Springer Science & Business Media. [Google Scholar]
- 7.Burton AC, Neilson E, Moreira D, Ladle A, Steenweg R, Fisher JT, Bayne E, Boutin S. 2015. Wildlife camera trapping: a review and recommendations for linking surveys to ecological processes. J. Appl. Ecol. 52, 675–685. ( 10.1111/1365-2664.12432) [DOI] [Google Scholar]
- 8.Harkins KM, Keinath D, Ben-David M. 2019. It's a trap: optimizing detection of rare small mammals. PLoS ONE 14, e0213201 ( 10.1371/journal.pone.0213201) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bovendorp RS, McCleery RA, Galetti M. 2017. Optimising sampling methods for small mammal communities in Neotropical rainforests. Mamm. Rev. 47, 148–158. ( 10.1111/mam.12088) [DOI] [Google Scholar]
- 10.Abrams JF, et al. 2019. Shifting up a gear with iDNA: from mammal detection events to standardised surveys. J. Appl. Ecol. 56, 1637–1648. ( 10.1111/1365-2664.13411) [DOI] [Google Scholar]
- 11.Calvignac-Spencer S, Merkel K, Kutzner N, Kühl H, Boesch C, Kappeler PM, Metzger S, Schubert G, Leendertz FH. 2013. Carrion fly-derived DNA as a tool for comprehensive and cost-effective assessment of mammalian biodiversity. Mol. Ecol. 22, 915–924. ( 10.1111/mec.12183) [DOI] [PubMed] [Google Scholar]
- 12.Port JA, O'Donnell JL, Romero-Maraccini OC, Leary PR, Litvin SY, Nickols KJ, Yamahara KM, Kelly RP. 2016. Assessing vertebrate biodiversity in a kelp forest ecosystem using environmental DNA. Mol. Ecol. 25, 527–541. ( 10.1111/mec.13481) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomsen PF, Kielgast J, Iversen LL, Moller PR, Rasmussen M, Willerslev E. 2012. Detection of a diverse marine fish fauna using environmental DNA from seawater samples. PLoS ONE 7, e41732 ( 10.1371/journal.pone.0041732) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andersen K, Bird KL, Rasmussen M, Haile J, Breuning-Madsen H, Kjær KH, Orlando L, Gilbert MTP, Willerslev E. 2012. Meta-barcoding of ‘dirt’ DNA from soil reflects vertebrate biodiversity. Mol. Ecol. 21, 1966–1979. ( 10.1111/j.1365-294X.2011.05261.x) [DOI] [PubMed] [Google Scholar]
- 15.Ushio M, et al. 2017. Environmental DNA enables detection of terrestrial mammals from forest pond water. Mol. Ecol. Resour. 17, e63–e75. ( 10.1111/1755-0998.12690) [DOI] [PubMed] [Google Scholar]
- 16.Ishige T, Miya M, Ushio M, Sado T, Ushioda M, Maebashi K, Yonechi R, Lagan P, Matsubayashi H. 2017. Tropical-forest mammals as detected by environmental DNA at natural saltlicks in Borneo. Biol. Conserv. 210, 281–285. ( 10.1016/j.biocon.2017.04.023) [DOI] [Google Scholar]
- 17.Dalén L, Götherström A, Meijer T, Shapiro B. 2007. Recovery of DNA from footprints in the snow. Can. Field-Naturalist 121, 321–324. ( 10.22621/cfn.v121i3.482) [DOI] [Google Scholar]
- 18.Nichols RV, Cromsigt JPGM, Spong G. 2015. Using eDNA to experimentally test ungulate browsing preferences. Springerplus 4, 489 ( 10.1186/s40064-015-1285-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adams IC, Hoekstra AL, Muell RM, Janzen JF. 2019. A brief review of non-avian reptile environmental DNA (eDNA), with a case study of painted turtle (Chrysemys picta) eDNA under field conditions. Diversity 11, 50 ( 10.3390/d11040050) [DOI] [Google Scholar]
- 20.Taberlet P, et al. 2012. Soil sampling and isolation of extracellular DNA from large amount of starting material suitable for metabarcoding studies. Mol. Ecol. 21, 1816–1820. ( 10.1111/j.1365-294X.2011.05317.x) [DOI] [PubMed] [Google Scholar]
- 21.Deiner K, Renshaw MA, Li Y, Olds BP, Lodge DM, Pfrender ME. 2017. Long-range PCR allows sequencing of mitochondrial genomes from environmental DNA. Methods Ecol. Evol. 8, 1888–1898. ( 10.1111/2041-210X.12836) [DOI] [Google Scholar]
- 22.Barnes MA, Turner CR. 2016. The ecology of environmental DNA and implications for conservation genetics. Conserv. Genet. 17, 1–17. ( 10.1007/s10592-015-0775-4) [DOI] [Google Scholar]
- 23.Leempoel K, Meyer JM, Hebert T, Nova N, Hadly EA.2019. Return of an apex predator to a suburban preserve triggers a rapid trophic cascade. bioRxiv , 564294. ( ) [DOI]
- 24.Rasmussen M, Cummings LS, Gilbert MTP, Bryant V, Smith C, Jenkins DL, Willerslev E. 2009. Response to comment by Goldberg et al. on ‘DNA from pre-Clovis human coprolites in Oregon, North America’. Science 325, 148 ( 10.1126/science.1167672) [DOI] [Google Scholar]
- 25.Boyer F, Mercier C, Bonin A, Le Bras Y, Taberlet P, Coissac E. 2016. obitools: a unix-inspired software package for DNA metabarcoding. Mol. Ecol. Resour. 16, 176–182. ( 10.1111/1755-0998.12428) [DOI] [PubMed] [Google Scholar]
- 26.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. ( 10.14806/ej.17.1.200) [DOI] [Google Scholar]
- 27.Giguet-Covex C, et al. 2014. Long livestock farming history and human landscape shaping revealed by lake sediment DNA. Nat. Commun. 5, 3211 ( 10.1038/ncomms4211) [DOI] [PubMed] [Google Scholar]
- 28.Ficetola GF, Coissac E, Zundel S, Riaz T, Shehzad W, Bessière J, Taberlet P, Pompanon F. 2010. An In silico approach for the evaluation of DNA barcodes. BMC Genomics 11, 434 ( 10.1186/1471-2164-11-434) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dixon P. 2003. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930. ( 10.1111/j.1654-1103.2003.tb02228.x) [DOI] [Google Scholar]
- 30.Jameson EW, Peeters HJ. 2004. Mammals of California. Berkeley, CA: University of California Press. [Google Scholar]
- 31.Weiskopf SR, McCarthy KP, Tessler M, Rahman HA, McCarthy JL, Hersch R, Faisal MM, Siddall ME. 2018. Using terrestrial haematophagous leeches to enhance tropical biodiversity monitoring programmes in Bangladesh. J. Appl. Ecol. 55, 2071–2081. ( 10.1111/1365-2664.13111) [DOI] [Google Scholar]
- 32.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10, 421 ( 10.1186/1471-2105-10-421) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matocq MD, Atocq MADM. 2002. Morphological and molecular analysis of a contact zone in the Neotoma fuscipes species complex. J. Mammal. 83, 866–883. () [DOI] [Google Scholar]
- 34.Ficetola GF, et al. 2015. Replication levels, false presences and the estimation of the presence/absence from eDNA metabarcoding data. Mol. Ecol. Resour. 15, 543–556. ( 10.1111/1755-0998.12338) [DOI] [PubMed] [Google Scholar]
- 35.Zinger L, et al. 2016. Extracellular DNA extraction is a fast, cheap and reliable alternative for multi-taxa surveys based on soil DNA. Soil Biol. Biochem. 96, 16–19. ( 10.1016/j.soilbio.2016.01.008) [DOI] [Google Scholar]
- 36.IUCN. 2019. The IUCN Red List of Threatened Species. See http://www.iucnredlist.org (accessed 5 May 2019).
- 37.Axtner J, Crampton-Platt A, Hörig LA, Mohamed A, Xu CCY, Yu DW, Wilting A. 2019. An efficient and robust laboratory workflow and tetrapod database for larger scale environmental DNA studies. Gigascience 8, giz029 ( 10.1093/gigascience/giz029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Somervuo P, Koskela S, Pennanen J, Henrik Nilsson R, Ovaskainen O. 2016. Unbiased probabilistic taxonomic classification for DNA barcoding. Bioinformatics 32, 2920–2927. ( 10.1093/bioinformatics/btw346) [DOI] [PubMed] [Google Scholar]
- 39.Leempoel K, Hebert T, Hadly EA. 2020. Data from: A comparison of eDNA to camera trapping for assessment of terrestrial mammal diversity. Dryad Digital Repository ( 10.5061/dryad.2547d7wmf) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Leempoel K, Hebert T, Hadly EA. 2020. Data from: A comparison of eDNA to camera trapping for assessment of terrestrial mammal diversity. Dryad Digital Repository ( 10.5061/dryad.2547d7wmf) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
12S and 16S eDNA curated sequences are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.2547d7wmf [39]. R scripts were published on GitHub: https://github.com/kleempoel/Soil_eDNA_Leempoeletal2019.git.