Abstract
Plasmodium vivax is responsible for most of the malaria infections outside Africa and is currently the predominant malaria parasite in countries under elimination programs. P. vivax preferentially enters young red cells called reticulocytes. Advances in understanding the molecular and cellular mechanisms of entry are hampered by the inability to grow large numbers of P. vivax parasites in a long‐term in vitro culture. Recent progress in understanding the biology of the P. vivax Reticulocyte Binding Protein (PvRBPs) family of invasion ligands has led to the identification of a new invasion pathway into reticulocytes, an understanding of their structural architecture and PvRBPs as targets of the protective immune response to P. vivax infection. This review summarises current knowledge on the role of reticulocytes in P. vivax infection, the function of the PvRBP family of proteins in generating an immune response in human populations, and the characterization of anti‐PvRBP antibodies in blocking parasite invasion.
Keywords: antibodies, host–pathogen interactions, Plasmodium falciparum, Plasmodium vivax, structure biology
1. Plasmodium vivax INVASION OF RETICULOCYTES
Malaria parasites are exquisitely adapted for invasion into red blood cells. The merozoite, an ovoid‐shaped cell with an apical prominence, is the form of the malaria parasite that invades blood cells. Merozoites express parasite adhesins at the apical tip, where secretory organelles such as rhoptires and micronemes are present, which bind to specific red blood cell receptors to initiate a series of molecular events that commit the parasite to invasion and successful entry (Cowman, Tonkin, Tham, & Duraisingh, 2017; Tham, Beeson, & Rayner, 2017). After entry, the merozoite grows and replicates within the blood cell to produce 16–32 new merozoites that rupture out of the infected cell to invade other healthy red blood cells. This blood stage cycle of infection results in the clinical symptoms observed in malaria infection.
Understanding how malaria parasites recognize and enter blood cells provide opportunities to block invasion and stop the cycle of blood stage infection. There are six Plasmodium species that commonly infect humans: Plasmodium falciparum, Plasmodium knowlesi, Plasmodium vivax, Plasmodium ovale curtisi, Plasmodium ovale wallikeri, and Plasmodium malariae. P. falciparum and P. vivax are responsible for the majority of malaria infections in humans. P. falciparum, P. knowlesi, and P. malariae invade mature red blood cells called normocytes, although P. falciparum and P. knowlesi may also preferentially enter reticulocytes (Gruner et al., 2004; Lim et al., 2013; Moon et al., 2016). In contrast, P. vivax and P. ovale are more restricted in their host cell preference than P. falciparum and will generally invade reticulocytes. Since the establishment of a continuous in vitro culture for P. falciparum in the late 1970s, the field of malaria parasite invasion has been dominated by studies of P. falciparum invading normocytes. Collectively, these studies have provided insights into the step‐wise nature of parasite entry, have identified parasite and host factors involved in invasion, and led to the development of inhibitors and antibodies that can block parasite invasion and provide protection from clinical disease (Paul et al., 2015; Cowman et al., 2017; Draper et al., 2018). In contrast, P. vivax invasion into reticulocytes is poorly understood due to the lack of a long‐term in vitro culture system for this parasite species (Kanjee, Rangel, Clark, & Duraisingh, 2018; Tham et al., 2017).
The preference of P. vivax for reticulocytes has implications in infection dynamics, parasite reservoirs, and potential parasite killing mechanisms. There are two distinct classes of reticulocytes that are present within the bone marrow compartment and in peripheral circulation (Griffiths et al., 2012). In the bone marrow compartment, R1 reticulocytes that have expelled the nucleus, but retain residual reticulum and are motile and multi‐lobular. R2 reticulocytes are released from the bone marrow to the peripheral circulation and are non‐motile and mechanically stable. As these reticulocytes mature in the bone marrow and in peripheral circulation, they remove all their organelles and lose 20% of their plasma membrane surface area (Moras, Lefevre, & Ostuni, 2017). Reticulocytes express several surface proteins that are lost as they mature into normocytes. In particular, CD71 (Transferrin Receptor 1, TfR1), CD49d, CD151, CD81, and CD82 are present only on young reticulocytes compared with mature red blood cells (Thomson‐Luque et al., 2018). Using short‐term ex vivo cultures, P. vivax has been observed to have higher invasion rates into reticulocytes with high levels of TfR1 compared with reticulocytes with lower levels of TfR1 (Malleret et al., 2014). In the same study, P. vivax invasion into TfR1 high‐reticulocytes caused a more rapid loss of TfR1 and expulsion of the residual reticulum compared with uninfected reticulocytes. However, a study using Indian P. vivax strains showed large differences in reticulocyte preferences (Lim et al., 2016). Although there was a low prevalence of circulating schizonts (the mature replicative form of the parasite), there was an association between increased reticulocyte preference and the number of schizonts, suggesting a potential link between invasion of younger reticulocytes and effective parasite development. This study also showed the detection of early‐stage P. vivax infection in reticulocytes with visible reticulum staining, suggesting that modifications to reticulocytes as observed ex vivo may not happen as rapidly in vivo (Lim et al., 2016). In a separate study, it was also shown that P. vivax had normal growth and development in TfR1‐high reticulocytes in G6PD‐Mahidol mutants suggesting an advantage to invasion of reticulocytes in these settings (Bancone et al., 2017).
2. THE Plasmodium vivax RETICULOCYTE BINDING PROTEIN FAMILY
P. vivax invasion into reticulocytes is mediated by the P. vivax Reticulocyte Binding Protein (PvRBP) family. Genome sequencing of several P. vivax isolates identified 11 PvRBP family members that comprises of five full‐length genes (pvrbp1a, pvrbp1b, pvrbp2a, pvrbp2b, and pvrbp2c), three partial genes (pvrbp1p1, pvrbp2p1, and pvrbp2p2), and three pseudogenes (pvrbp2d, pvrbp2e, and pvrbp3) based on sequence homology to existing P. vivax RBP and P. yoelli Py235 members (Carlton et al., 2008; Gruner et al., 2004; Hester et al., 2013; Rayner et al., 2005; Rayner, Galinski, Ingravallo, & Barnwell, 2000). Full‐length genes encode large molecular weight proteins of over 280 kDa. Almost all PvRBPs have a signal peptide at the N‐terminus and a putative transmembrane domain at the C‐terminus. Transcriptome analyses show that several of the PvRBPs are expressed in field isolates (Bozdech et al., 2008).
The first functional study on PvRBPs showed that native PvRBP1a and PvRBP2c are expressed at the apical tip of P. vivax merozoites and form a high molecular weight complex that binds reticulocytes (Galinski, Medina, Ingravallo, & Barnwell, 1992). Recent studies using recombinant PvRBP proteins have further described their binding characteristics and defined regions of the proteins involved in binding red blood cells (summarised in Table 1). In particular, PvRBP1a has been particularly well characterized and is shown to bind preferentially to reticulocytes (Franca et al., 2016; Gupta et al., 2017; Han et al., 2016; Ntumngia et al., 2018). Although several studies have purified different PvRBP1a fragments, the collective results show that two of the best binding fragments range from residues 30–778 and 157–650 (Gupta et al., 2017; Ntumngia et al., 2018). In contrast, it has also been shown that PvRBP1a (residues 160–1170) binds normocytes (Franca et al., 2016). These conflicting results may indicate that in comparison with shorter recombinant fragments, PvRBP1a (residues 160–1170) may not be folded correctly and therefore not reflect its true binding properties. Recombinant PvRBP1b (residues 339–587), PvRBP2c (residues 464–876), and PvRBP2b (residues 168–1124) also showed preferential binding to reticulocytes (Franca et al., 2016; Gruszczyk et al., 2018; Gupta et al., 2017; Han et al., 2016), whereas recombinant PvRBP2a (residues 160–1135) has been shown to bind to both normocytes and reticulocytes (Franca et al., 2016).
Table 1.
Binding Characteristics of PvRBPs
| PvRBP | aa | Construct | Binding profile | Enzyme treatment | Reference |
|---|---|---|---|---|---|
| PvRBP1a | 160–1170 | Normocytes | Franca et al. (2016) | ||
| Native protein | Reticulocytes | Galinski et al. (1992) | |||
| Native protein | Reticulocytes | Nr, Ts, Cs | Gupta et al. (2017) | ||
| 30–778 | rRBP1.1 | Reticulocytes (34%) | Nr, Ts, Cs | Gupta et al. (2017) | |
| 30–351 | rRBP1.2 | No binding | Gupta et al. (2017) | ||
| 352–778 | rRBP1.3 | Reticulocytes (~10%) | Gupta et al. (2017) | ||
| 352–599 | rRBP1.4 | Reticulocytes (~10%) | Gupta et al. (2017) | ||
| 1956–2315 | rRBP1.5 | No binding | Gupta et al. (2017) | ||
| 351–599 | Reticulocytes | Han et al. (2016) | |||
| 352–599 | rRBP1.4 | Reticulocytes | Gupta et al. (2018) | ||
| 157–481 | rRBP1:F7 | No binding | Ntumngia et al. (2018) | ||
| 157–650 | rRBP1:F8 | Reticulocytes (~50%), Normocytes | Nr, Ts, Cs | Ntumngia et al. (2018) | |
| 461–976 | rRBP1:F4 | Reticulocytes (~10–20%) | Ntumngia et al. (2018) | ||
| 632–976 | rRBP1:F6 | Reticulocytes (~10–20%) | Ntumngia et al. (2018) | ||
| 632–1078 | rRBP1:F5 | No binding | Ntumngia et al. (2018) | ||
| 950–1569 | rRBP1:F1 | Reticulocytes (~10–20%) | Ntumngia et al. (2018) | ||
| 1542–2192 | rRBP1:F2 | Reticulocytes (~10–20%) | Ntumngia et al. (2018) | ||
| 2162–2662 | rRBP1:F3 | Reticulocytes (~10–20%) | Ntumngia et al. (2018) | ||
| PvRBP1b | 140–1275 | Normocytes | Franca et al. (2016) | ||
| 339–587 | Reticulocytes | Han et al. (2016) | |||
| PvRBP2a | 160–1135 | Normocytes and reticulocytes | Franca et al. (2016) | ||
| PvRBP2b | 161–1454 | Reticulocytes | Franca et al. (2016) | ||
| PvRBP2c | 501–1300 | No binding | Franca et al. (2016) | ||
| Native protein | Reticulocytes | Galinski et al. (1992) | |||
| Native protein | Reticulocytes | Nr, Tr, Cr | Gupta et al. (2017) | ||
| 168–524 | rRBP2.1 | Reticulocytes (10%) | Gupta et al. (2017) | ||
| 464–876 | rRBP2.2 | Reticulocytes (34%) | Nr, Tr, Cr | Gupta et al. (2017) | |
| 2398–2736 | rRBP2.3 | No binding | Gupta et al. (2017) | ||
| PvRBP2‐P2 | 161–641 | Normocytes and reticulocytes | Franca et al. (2016) |
The use of recombinant PvRBP proteins have facilitated the determination of their functional properties; however, it would be important to examine if the respective native parasite proteins also display similar binding characteristics. Gupta and colleagues have also determined that the native PvRBP1a and PvRBP2c demonstrate the same binding profiles as has been previously shown by Galinski et al. (1992, Gupta et al., 2017). Experiments with native parasite proteins are extremely challenging for P. vivax research due to the absence of a long‐term in vitro culture system and limited access to clinical P. vivax samples. Nevertheless, it will be crucial to validate the binding profiles obtained with recombinant proteins with what is observed by using parasite lysates in which the PvRBP proteins are folded and functional for invasion.
3. NEW INVASION PATHWAY INTO RETICULOCYTES
Since the 1970s, it has been known that P. vivax invasion into red blood cells is dependent on Duffy Antigen Receptor for Chemokine (DARC; Miller, Mason, Clyde, & McGinniss, 1976). DARC is recognised by P. vivax Duffy Binding Protein (PvDBP) to mediate an essential step in P. vivax invasion, and many studies have contributed to the development of PvDBP as the lead P. vivax vaccine candidate, including the characterisation of anti‐PvDBP inhibitory antibodies that block invasion. However, DARC is present on both normocytes and reticulocytes and therefore cannot govern specific recognition of reticulocytes by P. vivax. Although it has been proposed that there is exposure of PvDBP binding site on DARC in young reticulocytes that allows it to selectively bind reticulocytes, this mechanism remains to be tested further by other laboratories (Ovchynnikova et al., 2017). Recent studies show that Transferrin Receptor 1 (TfR1, CD71) binds to PvRBP2b to mediate a critical pathway into reticulocytes (Gruszczyk et al., 2018; Gruszczyk, Kanjee, et al., 2018). TfR1 binds to its human ligand, iron‐loaded transferrin (Tf), and this complex regulates one of the main mechanisms for transporting iron into cells (Kawabata, 2019). TfR1 is highly expressed on reticulocytes and is selectively lost as they mature into normocytes. TfR1 mutant cells that are deficient in TfR1 expression have a strong defect in P. vivax invasion, showing that TfR1 mediates a critical invasion pathway into reticulocytes (Gruszczyk, Kanjee, et al., 2018). Mouse monoclonal antibodies against PvRBP2b also inhibit PvRBP2b binding to reticulocytes and block complex formation with TfR1. These anti‐PvRBP2b monoclonal antibodies also resulted in a reduction in parasite invasion using Thai and Brazilian clinical isolates (Gruszczyk, Kanjee, et al., 2018).
A 3.7 Å cryo‐electron microscopy (cryo‐EM) structure of the ternary complex of PvRBP2b, TfR1, and Tf has shed light on the critical residues involved in mediating complex formation (Gruszczyk, Huang, et al., 2018). The ternary complex is composed of homodimeric TfR1 (residues 120–760) bound to two molecules of iron‐loaded Tf (residues 1–679), with two molecules of PvRBP2b (residues 168–633) bound on either side. The most extensive interaction site is between PvRBP2b and TfR1, which has a surface buried area of ~1271 Å2, whereas the interaction site with Tf has a surface buried area of ~386 Å2. The region of PvRBP2b from 169 to 470 is highly polymorphic and under balancing selection (Gruszczyk, Huang, et al., 2018). Although this region binds to TfR1 and Tf, none of the prevalent field polymorphisms map to the amino acid residues on PvRBP2b that form critical contacts with either TfR1 or Tf as observed by the cryo‐EM structure (Figure 1). Extensive mutagenesis experiments identified three critical pairs of interacting residues for PvRBP2b‐TfR1 complex formation: those formed by PvRBP2b(Y542) and TfR1(Y211), a salt bridge formed between PvRBP2b(K600) and TfR1(E294), and a second salt bridge between TfR1(E149) and PvRBP2b(R359) (Figure 1).
Figure 1.
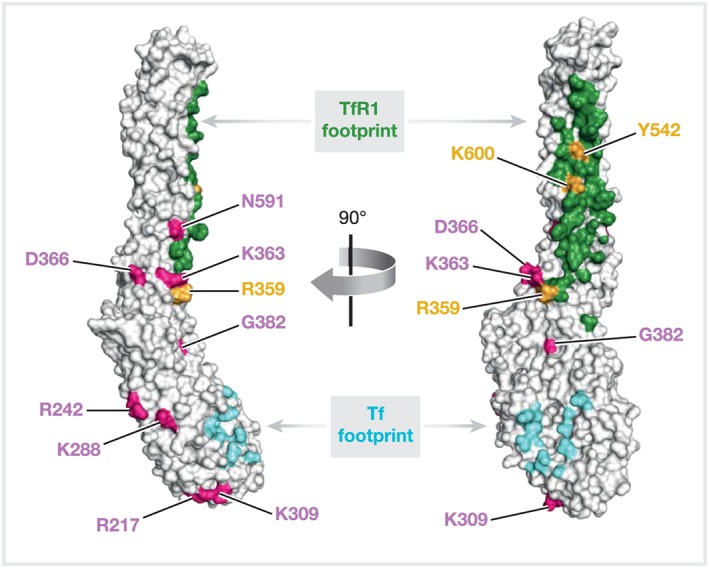
Surface representation of the cryo‐EM structure of PvRBP2b (168 to 633) shown in two orthogonal views. Regions interacting with TfR1 and Tf are shown in green and cyan, respectively. Field polymorphisms are labelled in pink and residues critical for TfR1 binding are labelled in orange
At present, apart from TfR1, there has been no additional reticulocyte receptors that have been implicated as cognate receptors for PvRBPs. PvRBP1a binding is sialic acid independent but trypsin and chymotrypsin sensitive (Ntumngia et al., 2018), whereas PvRBP2c binding is insensitive to all three enzyme treatments (Gupta et al., 2017). These results strongly suggest that other reticulocyte receptors will be important for P. vivax invasion
4. TfR1 AS A CELLULAR RECEPTOR FOR NEW WORLD HAEMORRHAGIC FEVER ARENAVIRUSES
TfR1 is also a cellular receptor for human New World haemorrhagic fever arenaviruses, including Machupo (MACV), Junin, Guanarito, and Sabiá viruses (Abraham, Corbett, Farzan, Choe, & Harrison, 2010; Radoshitzky et al., 2007). Residues 208 to 212 of the TfR1 apical domain provide a critical recognition site for these viruses (Abraham et al., 2010). Of particular interest, Y211 that is localized in the apical domain of TfR1 is a critical residue for entry of these New World haemorrhagic arenaviruses and P. vivax (Abraham et al., 2010; Gruszczyk et al., 2018). Furthermore, the binding of recombinant PvRBP2b can compete with the binding of recombinant MACV Glycoprotein 1 to TfR1, showing that these two pathogens have co‐opted a similar site on the apical domain of TfR1 for entry into their target cells (Gruszczyk, Kanjee, et al., 2018).
5. STRUCTURAL SCAFFOLDS OF PvRBPs
The first description of a family of Reticulocyte or Normocyte binding proteins (RBP or NBP) was described in P. yoelli as the Py235 proteins (reviewed in Gruner et al., 2004). The description of the Py235 and the PvRBP family led to the identification of the homologous P. falciparum Reticulocyte‐binding Homolog (PfRh) family by gene structure and sequence similarity (Rayner et al., 2000; Triglia et al., 2001). The PfRh family consists of several members PfRh1, PfRh2a, PfRh2b, PfRh4, PfRh5, and the pseudogene PfRh3, and many of these members have been shown to be important in red blood cell invasion (Chen et al., 2011; Crosnier et al., 2011; Lopaticki et al., 2011; Tham et al., 2010; Weiss et al., 2015). Two red blood cell receptors have been identified to bind to PfRh proteins; PfRh5 binds to Basigin (otherwise known as CD147, EMMPRIN) and PfRh4 binds to Complement Receptor 1 (CR1) (Crosnier et al., 2011; Tham et al., 2010).
PfRh5 cannot be deleted in any strain (Baum et al., 2009; Hayton et al., 2008), suggesting that it would be an ideal component targeting the blood stages in an effective P. falciparum vaccine. PfRh5 is smaller than the other PfRh members and does not contain a transmembrane region (Baum et al., 2009; Hayton et al., 2008). During parasite invasion, PfRh5 forms a complex with P. falciparum PfRh5‐interacting protein (PfRipr; Chen et al., 2011) and the cysteine‐rich protective antigen (PfCyRPA; Dreyer et al., 2012; Reddy et al., 2015). The PfRipr/CyRPA/PfRh5‐Basigin complex is required for triggering the release of Ca2+ and establishing the tight junction (Volz et al., 2016). These observations show that the PfRh5/PfRipr/CyRPA complex is essential in the sequential molecular events leading to P. falciparum invasion of human red blood cells.
In P. vivax and P. knowlesi, the exact homologue to PfRh5 remains elusive, but the homologs for PvRipr and PvCyRPA have been identified (Hoo et al., 2016). In a recent study, P. knowlesi PkRipr and PkCyRPA were shown to be essential for parasite viability (Knuepfer et al., 2019). Furthermore, PkRipr did not form a complex with PkCyRPA, but instead formed a trimeric complex with thrombospondin‐related anonymous protein (PkPTRAMP) and cysteine‐rich small secreted protein (PkCSS). Conditional knockout of any component of the trimeric complex resulted in merozoites that could attach to human erythrocytes, but were unable to invade. These results suggest that Ripr and CyRPA have different roles in P. knowlesi than in P. falciparum and may also have different roles in P. vivax invasion.
The crystal structure of the N‐terminal domain of PvRBP2b closely resembles the structures of the homologous domains of PvRBP2a and PfRh5 (Chen et al., 2014; Wright et al., 2014a; Gruszczyk et al., 2016; Gruszczyk, Kanjee, et al., 2018). Structural comparisons of PfRh5, PvRBP2a and PvRBP2b show that the placements of two disulfide bridges are conserved (Figure 2; Chen et al., 2014, Gruszczyk et al., 2016, Wright, Hjerrild, Bartlett, Douglas, et al., 2014a). The first bonded pair of cysteine residues between C345 and C351 in PfRh5 overlaps with C299 and C303 in PvRBP2a and with C312 and C316 in PvRBP2b and links a loop at the tip of the domain. The second bonded pair cysteine residues between C224 and C317 in PfRh5 overlap with C227 and C271 in PvRBP2a and with C240 and C284 in PvRBP2b and connects two antiparallel α‐helices (α2b, α3a) in the middle of the domain in each case. Although these domains have similar α‐helical scaffolds, they bind different receptors: PvRBP2b to TfR1 and PfRh5 to Basigin (Crosnier et al., 2011; Gruszczyk, Kanjee, et al., 2018; Wright, Hjerrild, Bartlett, Douglas, et al., 2014a). In contrast to PfRh5 that receptor recognition site is located on the tip of the domain, binding of PvRBP2b to TfR1‐Tf involves residues located at the side of the domain and residues of the adjacent α‐helical domain of this protein. The receptor for PvRBP2a is currently unknown.
Figure 2.
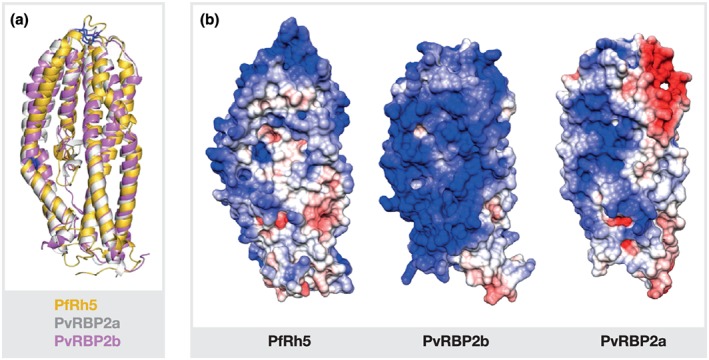
Structural comparison of PvRBP2a, PvRBP2b and PfRh5. (a) Superposition of PvRBP2a (PDB ID 4Z8N, grey), PvRBP2b (PDB ID 5W53, pink), and PfRh5 (PDB ID 4WAT, yellow) structures shown in ribbon representation. The position of two disulfide bridges (indicated by blue) is conserved in all three structures. (b) Surface charge distribution of PvRBP2a, PvRBP2b, and PfRh5. The electrostatic surface potentials were calculated using the programs PDB2PQR and APBS in Chimera with the non‐linear Poisson–Boltzmann equation and contoured at ±5 kT/e. Negatively charged surface areas are coloured in red, positively charged surface areas in blue
The surface charge distribution of PvRBP2a, PvRBP2b, and PfRh5 is different (Figure 2). In particular, PvRBP2a has a distinctive negative patch on the apex of the molecule that corresponds to the Basigin‐binding area of PfRh5, whereas PfRh5 as well as the N‐terminal domain of PvRBP2b mostly displays a positive charged surface (Gruszczyk, Kanjee, et al., 2018; Wright, Hjerrild, Bartlett, Douglas, et al., 2014a). The differences in surface charge may be important in mediating the specific interactions between these parasite adhesins and their respective red blood cell receptors.
6. ANTIBODIES TO PvRBP2b AND PfRh5
Antibodies against PvRBPs and antibodies against PfRh family members are known to inhibit parasite invasion. Several antibody binding regions critical for receptor–ligand interactions have been identified on PfRh5 and PvRBP2b by X‐ray crystallography and small‐angle X‐ray scattering (SAXS; Figure 3; Wright, Hjerrild, Bartlett, Douglas, et al., 2014a; Gruszczyk et al., 2018). Crystal structures of PfRh5 complexed with the antigen binding fragments (Fab) of mouse antibodies QA1 and 9AD4, respectively, revealed distinct conformational epitopes (Wright, Hjerrild, Bartlett, Douglas, et al., 2014a). A linear epitope of another inhibitory antibody, QA5, was identified by peptide mapping, and the binding region was confirmed by SAXS. Binding sites of QA1 and QA5 that are able to inhibit Rh5‐Basigin binding in vitro overlap with different parts of the receptor binding site on PfRh5. The 9AD4 antibody does not interfere with in vitro PfRh5‐binding to Basigin, and its epitope is next to the receptor binding site but does not overlap. The ability of this antibody to inhibit parasite growth is probably caused by a steric hindrance of the interaction of membrane‐tethered PfRh5 and Basigin.
Figure 3.
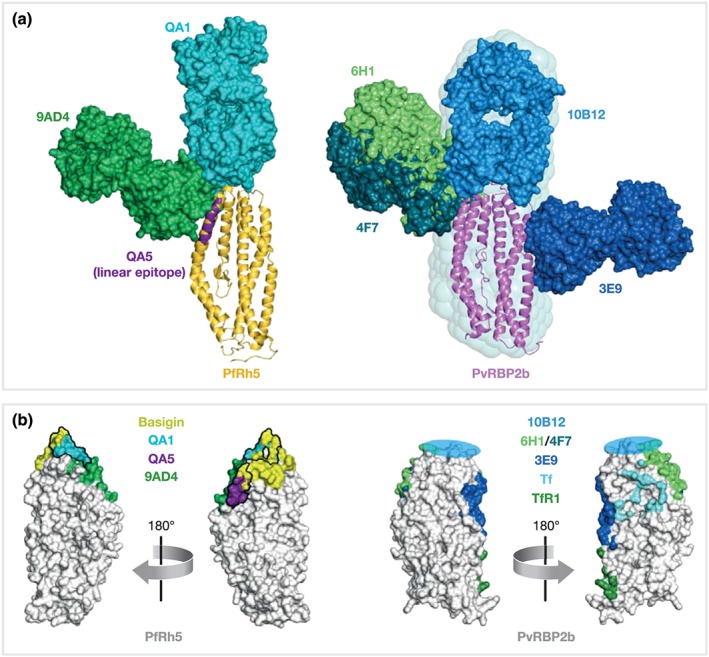
Epitopes of inhibitory mouse antibodies against PfRh5 and PvRBP2b. Structures of antigen‐Fab complexes are superimposed on the corresponding antigen. (a) Antigens PfRh5 (PDB ID 4WAT, yellow) and PvRBP2b (PDB ID 5W53, pink) are shown in ribbon representation, Fabs (each with heavy and light chains in the same colour) are in surface representation. The linear epitope of inhibitory antibody QA5 (PfRh5 residues 201–213) is indicated in purple. The SAXS‐derived model of the PvRBP2b‐10B12 complex is surrounded by its SAXS‐envelope. (b) Surface representation of PfRh5 and PvRBP2b (white). Antibody and receptor footprints are coloured with a 6 Å‐distance cut‐off. The rough binding region of 10B12 is indicated by a blue circle. Antibody epitopes of QA1 and QA5 directly overlap with the Basigin binding site of PfRh5
For PvRBP2b, four mouse monoclonal antibodies (3E9, 4F7, 6H1, and 10B12) are able to inhibit PvRBP2b binding to reticulocytes. Three of these 3E9, 6H1, and 10B12 were able to block PvRBP2b‐TfR1 complex formation and P. vivax invasion in Thai and Brazilian clinical isolates (Gruszczyk et al., 2018). Co‐crystal structures of 3E9, 4F7, and 6H1 with PvRBP2b, as well as a SAXS derived model of 10B12 binding to PvRBP2b, revealed that these antibodies recognise different structural epitopes, all located in the N‐terminal domain of PvRBP2b (Gruszczyk et al., 2018). The antibodies 4F7 and 6H1 have overlapping epitopes, both bind to the side of PvRBP2b. Their binding sites on PvRBP2b map to a similar area as the binding site of 9AD4 on PfRh5 (Figure 3). All four anti‐PvRBP2b antibodies do not completely overlap with the receptor recognition site but would either sterically clash with TfR1‐Tf binding of PvRBP2b (3E9) or inhibit receptor engagement through steric hindrance with the reticulocyte membrane (4F7, 6H1, and 10B12).
Similar to anti‐PfRh5 antibody QA1, antibody 10B12 engages its antigen at the tip of the N terminal domain, probably also facing the loop which is stabilised by a disulfide bond (Gruszczyk et al., 2018). The presence of the corresponding disulfide bridge and loop of PvRBP2a is also required for the protein function as mutation of C299 and C303 to serine residues results in a loss of the capability of PvRBP2a to bind erythrocytes while not altering the overall structural conformation of the protein (Gruszczyk et al., 2016). Collectively, these results highlight the importance of the disulfide linked loop at the apex of PvRBP and PfRh red blood cell binding domains as a target for inhibitory antibodies. Future investigations should explore conformational epitopes as immunogens that may generate cross‐inhibitory antibodies that inhibit PvRBP and PfRh function in parasite invasion.
In addition to the anti‐PvRBP2b antibodies, several studies have examined the role of antibodies to other PvRBPs in blocking parasite invasion or red blood cell binding. Natural human antibodies against PvRBP1a and PvRBP2c have been shown to inhibit reticulocyte binding (Gupta et al., 2017), but the evidence for inhibition of P. vivax invasion is lacking. Rabbit antibodies targeting PvRBP1a (residues 352–599) were recently shown to have no significant impact on P. vivax invasion (Gupta et al., 2018); however, the failure to reach statistical significance may be due to the large variation in the inhibition efficacy across the few biological replicates. The large variation of results obtained from P. vivax invasion assays from different clinical isolates remains the one of the main challenges in understanding the importance of any particular P. vivax invasion ligand.
7. NEW MODEL SYSTEMS
New model systems using other malaria species may facilitate anti‐PvRBP antibody screening and accelerate the pace of vaccine development for these proteins (Martinelli & Culleton, 2018). Recently, transgenic P. knowlesi parasites genetically modified to express PvDBP were used successfully to assess the efficacy of anti‐PvDBP human monoclonal antibodies (Mohring et al., 2019; Rawlinson et al., 2019). However, the respective RBP ligands in P. knowlesi, which are P. knowlesi Normocyte Binding Proteins (NBPXa and NBPXb), are strongly divergent from PvRBPs. PkNBPXa is required for the P. knowlesi growth and replication in human red blood cells, as deletion of this gene restricts the growth of P. knowlesi to monkey red blood cells (Moon et al., 2013; Moon et al., 2016). It should be noted that PkNBPXa and PkNBPXb are both found as pseudogenes in the reference genomes of P. vivax as PvRBP3 and PvRBP2e. However, sequencing of a P. vivax field isolate has identified an intact copy of PvRBP2e (Hester et al., 2013). The distinct RBPs or NBP that are expressed in P. vivax or P. knowlesi may actually govern why one is restricted to reticulocytes and the other not. It would be interesting if the transgenic lines expressing different PvRBPs in a PkNBPXa‐null background could “complement” the deletion and thus allow entry of P. knowlesi into human red blood cells. If any expressed transgenic PvRBP is able to rescue the PkNBPxa‐null phenotype, this would imply that the respective PvRBP is expressed and functional in P. knowlesi and provide an important model to determine the functional activity of anti‐PvRBP antibodies. P. cynomolgi is the sister taxon to P. vivax and possesses multiple PvRBP homologues with most showing a high degree of sequence identity (PcRBPs; Tachibana et al., 2012). Substantial progress has been made to establish a robust P. cynomolgi culture in monkey cells that would allow a short‐term invasion assays into human reticulocytes (Kosaisavee et al., 2017; Zeeman, der Wel, & Kocken, 2013). Although P. cynomolgi is able to enter monkey red cells of all ages, it has a strict preference for TfR1 and DARC‐positive reticulocytes in the presence of human red blood cells (Kosaisavee et al., 2017). To determine if P. cynomolgi would be a useful system to examine the function of anti‐PvRBP antibodies, it would be important to determine if the PcRBP homologs have the same molecular function as their respective PvRBPs and whether anti‐PvRBP antibodies recognise different members of PcRBPs. Both transgenic P. knowlesi and P. cynomolgi cultures are promising models to complement the characterization of inhibitory antibodies to P. vivax ligands and facilitate the understanding of the mechanisms of P. vivax invasion.
8. NATURALLY ACQUIRED ANTIBODY RESPONSE TO PvRBPs
The study of the human immune response to malaria infection is central to the discovery of new vaccine candidates and biological markers of disease. The availability of the P. vivax parasite genome since 2008 has enabled researchers to examine a much larger panel of proteins (Baum et al., 2016; Finney et al., 2014; Franca et al., 2017; Hostetler et al., 2015; Longley et al., 2017), not least the entire family of PvRBPs. Although these studies show that many other parasite antigens that are associated with protection, we will only focus on the results for PvRBPs for this review. To date, all PvRBPs have been subjected to serological examinations and are all shown to be targets of natural antibody responses (Franca et al., 2016; Longley, White, et al., 2017).
Plasma samples from different endemic regions have been used to study natural acquisition of antibodies to PvRBPs, from the relatively high transmission areas in India (Gupta et al., 2017) and Papua New Guinea (PNG; Franca et al., 2016; Ntumngia et al., 2018) to the lower transmission sites in Brazil (Ferreira et al., 2014; Longley et al., 2018; Ntumngia et al., 2018; Tran et al., 2005), Cambodia (Hostetler et al., 2015), Colombia (Rojas Caraballo, Delgado, Rodriguez, & Patarroyo, 2007), Korea (Han et al., 2015; Han et al., 2016), Solomon Islands (Longley et al., 2018), and Thailand (Hietanen et al., 2015; Longley et al., 2017; Longley et al., 2018). These samples encompassed specimens from uninfected endemic residents, infected asymptomatic P. vivax carriers, and acute P. vivax patients. The diversity of these samples provides rich data for identifying new vaccine candidates as well as serological markers of P. vivax malaria.
PvRBP1a was the first protein in the PvRBP family whose antibody response was systematically examined (Tran et al., 2005). In this initial study published in 2005, plasma specimens from the Brazilian Amazon state of Rondonia were tested against five recombinant PvRBP1a fragments, which together span nearly the entire extracellular domain (>2,600 amino acids). The study revealed key features of the natural antibody response that appear to be common among PvRBPs, including (a) positive correlation between the antibody level and parasite exposure, (b) IgG subtypes being biased towards cytophilic IgG1 and IgG3, and (c) association between the antibody level and clinical protection. To date, PvRBP1a is the only full‐length PvRBP whose entire extracellular sequence has been mapped serologically. Comparison of the IgG responses across different subdomains of PvRBP1a revealed stronger reactivity towards the N‐terminal half, a finding which was later confirmed in a separate study (Ntumngia et al., 2018). This N‐terminal bias coincides with a higher degree of genetic diversity of the N‐terminal domain (Rayner et al., 2005), suggesting a stronger immunogenic selection pressure on this region of the protein.
In the endemic populations, the magnitudes of IgG responses to different PvRBPs are generally correlated (Gupta et al., 2017; Hietanen et al., 2015) and tend to increase with age (Hietanen et al., 2015; Longley et al., 2017). This co‐acquisition of antibodies to different PvRBPs is unlikely due to cross‐reactivity given the limited sequence similarity between these proteins. Not surprisingly, human IgG obtained by affinity purification with PvRBP1a showed little cross‐reactivity against PvRBP2c, and vice‐versa (Gupta et al., 2017). Rabbit immunizations with recombinant PvRBPs also generated antibodies specific to the respective antigen (Franca et al., 2016). Therefore, the co‐development of IgG responses to different PvRBPs is likely due to repeated P. vivax infections that expose the human body to all proteins simultaneously. In addition to age, other measures of parasite exposure such as the number of previous malaria episodes (Tran et al., 2005), years of residence in endemic areas (Tran et al., 2005), and the estimated number of new infections acquired in the lifetime (Franca et al., 2016) have been shown to positively associate with anti‐PvRBP reactivities.
9. SEROLOGICAL MARKERS OF EXPOSURE
People with concurrent P. vivax infection, both symptomatic and asymptomatic, tend to have higher levels and breadth of IgG responses to PvRBPs than do uninfected individuals (Franca et al., 2017; Longley, França, et al., 2017). The heightened sero‐reactivities in infected individuals suggest that PvRBPs may be viable markers for comparing malaria burdens in endemic populations. Indeed, the IgG responses to PvRBP1a and PvRBP2c fragments have been shown to map spatial heterogeneity in western Thailand (Longley et al., 2017) and be able to differentiate different endemic communities in Brazilian Amazon (Tran et al., 2005). The magnitudes of IgG responses to several PvRBPs were also found to decline after clearance of infection, with estimated half‐lives of 2–18 months (Longley et al., 2017). Therefore, antibodies to some of these proteins, especially the short‐lived ones, may be good surrogates of recent exposure. In a recent attempt to develop a panel of serological markers of recent infections (Longley et al., 2018), a recombinant fragment of PvRBP2b was the best performing antigen among the >300 P. vivax proteins tested. The IgG response to this protein has the sensitivity and selectivity of 74% in detecting P. vivax infection within the previous 9 months. Combining PvRBP2b with other proteins further improved the diagnostic performance, making antibody‐based surveillance an attractive approach for malaria control and elimination programs (Greenhouse et al., 2019).
10. SEROLOGICAL MARKERS OF PROTECTION
Identification of parasite proteins associated with clinical protection is an approach often taken in malaria vaccine discovery (Cutts et al., 2014; Franca et al., 2017). Because naturally acquired immunity against malaria is gained through repeated infections, an association between the antibody level and protection is often confounded by exposure, a factor that has to be accounted for in sero‐epidemiological analysis. The most comprehensive study to examine association between the IgG reactivities to PvRBPs and clinical protection (Franca et al., 2016) came from a cohort of young children in East Sepik, PNG, whose exposure to P. vivax was defined by molecular force of blood stage infection (molFOB, the incidence of new blood infection). In this study, children 1–3 years old were followed for up to 16 months, with blood sampling every 8 weeks and at clinical malaria episodes. P. vivax infections were detected by PCR, and the parasites were genotyped for determination of molFOB. After adjusting for exposure, IgG for each of the six PvRBPs tested (PvRBP1a, PvRBP1b, PvRBP2a, PvRBP2b, PvRBP2c, and PvRBP2p2) was associated with protection, with a reduction of 30–50% in risk of P. vivax malaria observed in children who had medium to high antibody levels. However, in multivariate analysis to account for co‐acquisition of antibodies to different PvRBPs, only IgG to PvRBP1a and PvRBP2b remained, with IgG1 being the underlying antibody subclass associated with protection. Notably, these two proteins were amongst the strongest protective antigens in the panel of 38 P. vivax antigens tested with the same methodology (Franca et al., 2017). Nevertheless, it is important to note that these studies were only performed with recombinant proteins, which may have inherent caveats such as the use of a single variant and that the protein folding may be different from native protein. In particular, for proteins that are highly polymorphic, immune responses to a different variant may be very different. Consistently, the magnitude of IgG to PvRBP1a fragments had previously been associated with duration since the last known malaria episode, a proxy of protection, in a Brazilian cohort (Tran et al., 2005). The magnitude of IgG response to PvRBP2b had also been associated with reduced parasitaemia in Thai adult patients independently of age (Hietanen et al., 2015). Thus, among the PvRBPs, PvRBP1a and PvRBP2b appear to be the strongest vaccine candidates.
11. CONCLUSIONS
The PvRBP family of P. vivax parasite adhesins is clearly emerging to be important for reticulocyte invasion and is the target of naturally acquired immunity in many different transmission settings. Future studies on the identification of other reticulocyte receptors for PvRBP and the characterization of inhibitory antibodies will provide insightful information on the multiple invasion pathways of P. vivax and how to target them to block entry into reticulocytes.
ACKNOWLEDGEMENTS
W.H.T. is a Howard Hughes Medical Institute‐Wellcome Trust International Research Scholar supported by the Wellcome Trust (208693/Z/17/Z) and supported by the National Health and Medical Research Council (GNT1143187, GNT1160042, GNT1160042, and GNT1154937).
Chan L‐J, Dietrich MH, Nguitragool W, Tham W‐H. Plasmodium vivax Reticulocyte Binding Proteins for invasion into reticulocytes. Cellular Microbiology. 2020;22:e13110 10.1111/cmi.13110
REFERENCES
- Abraham, J. , Corbett, K. D. , Farzan, M. , Choe, H. , & Harrison, S. C. (2010). Structural basis for receptor recognition by New World hemorrhagic fever arenaviruses. Nature Structural & Molecular Biology, 17, 438–444. 10.1038/nsmb.1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancone, G. , Malleret, B. , Suwanarusk, R. , Chowwiwat, N. , Chu, C. S. , McGready, R. , … Russell, B. (2017). Asian G6PD‐Mahidol reticulocytes sustain normal Plasmodium vivax development. The Journal of Infectious Diseases, 216, 263–266. 10.1093/infdis/jix278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum, E. , Sattabongkot, J. , Sirichaisinthop, J. , Kiattibutr, K. , Jain, A. , Taghavian, O. , … Yan, G. (2016). Common asymptomatic and submicroscopic malaria infections in Western Thailand revealed in longitudinal molecular and serological studies: A challenge to malaria elimination. Malaria Journal, 15, 333 10.1186/s12936-016-1393-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum, J. , Chen, L. , Healer, J. , Lopaticki, S. , Boyle, M. , Triglia, T. , … Cowman, A. F. (2009). Reticulocyte‐binding protein homologue 5 – An essential adhesin involved in invasion of human erythrocytes by Plasmodium falciparum. International Journal for Parasitology, 39, 371–380. 10.1016/j.ijpara.2008.10.006 [DOI] [PubMed] [Google Scholar]
- Bozdech, Z. , Mok, S. , Hu, G. , Imwong, M. , Jaidee, A. , Russell, B. , … Preiser, P. R. (2008). The transcriptome of Plasmodium vivax reveals divergence and diversity of transcriptional regulation in malaria parasites. Proceedings of the National Academy of Sciences of the United States of America, 105, 16290–16295. 10.1073/pnas.0807404105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton, J. M. , Adams, J. H. , Silva, J. C. , Bidwell, S. L. , Lorenzi, H. , Caler, E. , … Fraser‐Liggett, C. M. (2008). Comparative genomics of the neglected human malaria parasite Plasmodium vivax . Nature, 455, 757–763. 10.1038/nature07327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L. , Lopaticki, S. , Riglar, D. T. , Dekiwadia, C. , Uboldi, A. D. , Tham, W.‐H. , … Cowman, A. F. (2011). An EGF‐like protein forms a complex with PfRh5 and is required for invasion of human erythrocytes by Plasmodium falciparum . PLoS Pathogens, 7, e1002199 10.1371/journal.ppat.1002199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L. , Xu, Y. , Healer, J. , Thompson, J. K. , Smith, B. J. , Lawrence, M. C. , & Cowman, A. F. (2014). Crystal structure of PfRh5, an essential P. falciparum ligand for invasion of human erythrocytes. eLife, 3 10.7554/eLife.04187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowman, A. F. , Tonkin, C. J. , Tham, W.‐H. , & Duraisingh, M. T. (2017). The molecular basis of erythrocyte invasion by malaria parasites. Cell Host & Microbe, 22, 232–245. 10.1016/j.chom.2017.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosnier, C. , Bustamante, L. Y. , Bartholdson, S. J. , Bei, A. K. , Theron, M. , Uchikawa, M. , … Wright, G. J. (2011). Basigin is a receptor essential for erythrocyte invasion by Plasmodium falciparum . Nature, 480, 534–537. 10.1038/nature10606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutts, J. C. , Powell, R. , Agius, P. A. , Beeson, J. G. , Simpson, J. A. , & Fowkes, F. J. (2014). Immunological markers of Plasmodium vivax exposure and immunity: A systematic review and meta‐analysis. BMC Medicine, 12, 150 10.1186/s12916-014-0150-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper, S.J. , Sack, B.K. , King, C.R. , Nielsen, C.M. , Rayner, J.C. , Higgins, M.K. , etal. (2018). Malaria Vaccines: Recent Advances and New Horizons. Cell HostMicrobe, 24, 43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyer, A. M. , Matile, H. , Papastogiannidis, P. , Kamber, J. , Favuzza, P. , Voss, T. S. , … Pluschke, G. (2012). Passive immunoprotection of Plasmodium falciparum‐infected mice designates the CyRPA as candidate malaria vaccine antigen. The Journal of Immunology Baltim Md 1950, 188, 6225–6237. [DOI] [PubMed] [Google Scholar]
- Ferreira, A. R. , Singh, B. , Cabrera‐Mora, M. , Magri De Souza, A. C. , Queiroz Marques, M. T. , Porto, L. C. S. , … Moreno, A. (2014). Evaluation of naturally acquired IgG antibodies to a chimeric and non‐chimeric recombinant species of Plasmodium vivax Reticulocyte Binding Protein‐1: Lack of association with HLA‐DRB1*/DQB1* in malaria exposed individuals from the Brazilian Amazon. PLoS ONE, 9, e105828 10.1371/journal.pone.0105828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finney, O. C. , Danziger, S. A. , Molina, D. M. , Vignali, M. , Takagi, A. , Ji, M. , … Wang, R. (2014). Predicting antidisease immunity using proteome arrays and sera from children naturally exposed to malaria. Molecular & Cellular Proteomics, 13, 2646–2660. 10.1074/mcp.M113.036632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franca, C. T. , He, W. Q. , Gruszczyk, J. , Lim, N. T. , Lin, E. , Kiniboro, B. , … Mueller, I. (2016). Plasmodium vivax Reticulocyte Binding Proteins Are key targets of naturally acquired immunity in young Papua New Guinean children. PLoS Neglected Tropical Diseases, 10, e0005014 10.1371/journal.pntd.0005014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franca, C. T. , White, M. T. , He, W. Q. , Hostetler, J. B. , Brewster, J. , Frato, G. , … Kiniboro, B. (2017). Identification of highly‐protective combinations of Plasmodium vivax recombinant proteins for vaccine development. eLife, 6 https://www.ncbi.nlm.nih.gov/pubmed/28949293, 10.7554/eLife.28673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinski, M. R. , Medina, C. C. , Ingravallo, P. , & Barnwell, J. W. (1992). A reticulocyte‐binding protein complex of Plasmodium vivax merozoites. Cell, 69, 1213–1226. 10.1016/0092-8674(92)90642-P [DOI] [PubMed] [Google Scholar]
- Greenhouse, B. , Daily, J. , Guinovart, C. , Goncalves, B. , Beeson, J. , Bell, D. , … The Malaria Serology Convening (2019). Priority use cases for antibody‐detecting assays of recent malaria exposure as tools to achieve and sustain malaria elimination [version 1; peer review: 1 approved]. Gates Open Res, 3 https://gatesopenresearch.org/articles/3-131/v1, 10.12688/gatesopenres.12897.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths, R. E. , Kupzig, S. , Cogan, N. , Mankelow, T. J. , Betin, V. M. S. , Trakarnsanga, K. , … Lane, J. D. (2012). The ins and outs of human reticulocyte maturation: Autophagy and the endosome/exosome pathway. Autophagy, 8, 1150–1151. 10.4161/auto.20648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruner, A. , Snounou, G. , Fuller, K. , Jarra, W. , Renia, L. , & Preiser, P. (2004). The Py235 proteins: Glimpses into the versatility of a malaria multigene family. Microbes and Infection, 6, 864–873. 10.1016/j.micinf.2004.04.004 [DOI] [PubMed] [Google Scholar]
- Gruszczyk, J. , Huang, R. K. , Chan, L.‐J. , Menant, S. , Hong, C. , Murphy, J. M. , … Tham, W. H. (2018). Cryo‐EM structure of an essential Plasmodium vivax invasion complex. Nature, 559, 135–139. 10.1038/s41586-018-0249-1 [DOI] [PubMed] [Google Scholar]
- Gruszczyk, J. , Kanjee, U. , Chan, L.‐J. , Menant, S. , Malleret, B. , Lim, N. T. Y. , … Tham, W. H. (2018). Transferrin receptor 1 is a reticulocyte‐specific receptor for Plasmodium vivax . Science, 359, 48–55. 10.1126/science.aan1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruszczyk, J. , Lim, N. T. Y. , Arnott, A. , He, W.‐Q. , Nguitragool, W. , Roobsoong, W. , … Tham, W. H. (2016). Structurally conserved erythrocyte‐binding domain in Plasmodium provides a versatile scaffold for alternate receptor engagement. Proceedings of the National Academy of Sciences of the United States of America, 113, E191–E200. 10.1073/pnas.1516512113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, E. D. , Anand, G. , Singh, H. , Chaddha, K. , Bharti, P. K. , Singh, N. , … Gaur, D. (2017). Naturally acquired human antibodies against reticulocyte‐binding domains of Plasmodium vivax proteins, PvRBP2c and PvRBP1a, exhibit binding‐inhibitory activity. The Journal of Infectious Diseases, 215, 1558–1568. 10.1093/infdis/jix170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, S. , Singh, S. , Popovici, J. , Roesch, C. , Shakri, A. R. , Guillotte‐Blisnick, M. , … Chitnis, C. E. (2018). Targeting a Reticulocyte Binding Protein and Duffy Binding Protein to inhibit reticulocyte invasion by Plasmodium vivax . Scientific Reports, 8, 10511 10.1038/s41598-018-28757-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, J.‐H. , Lee, S.‐K. , Wang, B. , Muh, F. , Nyunt, M. H. , Na, S. , … Tsuboi, T. (2016). Identification of a reticulocyte‐specific binding domain of. Scientific Reports, 6, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, J.‐H. , Li, J. , Wang, B. , Lee, S.‐K. , Nyunt, M. H. , Na, S. , … Han, E. T. (2015). Identification of immunodominant B‐cell epitope regions of Reticulocyte Binding PROTEINS in Plasmodium vivax by protein microarray based immunoscreening. The Korean Journal of Parasitology, 53, 403–411. 10.3347/kjp.2015.53.4.403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayton, K. , Gaur, D. , Liu, A. , Takahashi, J. , Henschen, B. , Singh, S. , … Wellems, T. E. (2008). Erythrocyte binding protein PfRH5 polymorphisms determine species‐specific pathways of Plasmodium falciparum invasion. Cell Host & Microbe, 4, 40–51. 10.1016/j.chom.2008.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hester, J. , Chan, E. R. , Menard, D. , Mercereau‐Puijalon, O. , Barnwell, J. , Zimmerman, P. A. , & Serre, D. (2013). De novo assembly of a field isolate genome reveals novel Plasmodium vivax erythrocyte invasion genes. PLoS Neglected Tropical Diseases, 7, e2569 10.1371/journal.pntd.0002569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietanen, J. , Chim‐Ong, A. , Chiramanewong, T. , Gruszczyk, J. , Roobsoong, W. , Tham, W.‐H. , … Nguitragool, W. (2015). Gene models, expression repertoire, and immune response of Plasmodium vivax Reticulocyte Binding Proteins. Infection and Immunity, 84, 677–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoo, R. , Zhu, L. , Amaladoss, A. , Mok, S. , Natalang, O. , Lapp, S. A. , … Preiser, P. R. (2016). Integrated analysis of the Plasmodium species transcriptome. eBioMedicine, 7, 255–266. 10.1016/j.ebiom.2016.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hostetler, J. B. , Sharma, S. , Bartholdson, S. J. , Wright, G. J. , Fairhurst, R. M. , & Rayner, J. C. (2015). A library of Plasmodium vivax recombinant merozoite proteins reveals new vaccine candidates and protein‐protein interactions. PLoS Neglected Tropical Diseases, 9, e0004264 10.1371/journal.pntd.0004264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanjee, U. , Rangel, G. W. , Clark, M. A. , & Duraisingh, M. T. (2018). Molecular and cellular interactions defining the tropism of Plasmodium vivax for reticulocytes. Current Opinion in Microbiology, 46, 109–115. 10.1016/j.mib.2018.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabata, H. . (2019). Transferrin and transferrin receptors update. Free Radic Biol Med, 133, 46–54. [DOI] [PubMed] [Google Scholar]
- Knuepfer, E. , Wright, K. E. , Kumar Prajapati, S. , Rawlinson, T. A. , Mohring, F. , Koch, M. , … Holder, A. A. (2019). Divergent roles for the RH5 complex components, CyRPA and RIPR in human‐infective malaria parasites. PLoS Pathogens, 15, e1007809 10.1371/journal.ppat.1007809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaisavee, V. , Suwanarusk, R. , Chua, A. C. Y. , Kyle, D. E. , Malleret, B. , Zhang, R. , … Russell, B. (2017). Strict tropism for CD71+/CD234+ human reticulocytes limits the zoonotic potential of Plasmodium cynomolgi . Blood, 130, 1357–1363. 10.1182/blood-2017-02-764787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, C. , Hansen, E. , DeSimone, T. M. , Moreno, Y. , Junker, K. , Bei, A. , … Duraisingh, M. T. (2013). Expansion of host cellular niche can drive adaptation of a zoonotic malaria parasite to humans. Nature Communications, 4, 1638 10.1038/ncomms2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, C. , Pereira, L. , Saliba, K. S. , Mascarenhas, A. , Maki, J. N. , Chery, L. , … Duraisingh, M. T. (2016). Reticulocyte preference and stage development of Plasmodium vivax isolates. The Journal of Infectious Diseases, 214, 1081–1084. 10.1093/infdis/jiw303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longley, R. J. , França, C. T. , White, M. T. , Kumpitak, C. , Sa‐Angchai, P. , Gruszczyk, J. , … Rayner, J. C. (2017). Asymptomatic Plasmodium vivax infections induce robust IgG responses to multiple blood‐stage proteins in a low‐transmission region of western Thailand. Malaria Journal, 16, 178 10.1186/s12936-017-1826-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longley, R.J. , White, M.T. , Takashima, E. , Brewster, J. , Morita, M. , Harbers, M. , Robinson LJ, Matsuura F, Liu SJ, Li‐Wai‐Suen CS, Tham WH (2018) Development and validation of serological markers for detecting recent exposure to Plasmodium vivax infection. bioRxiv 481168. [DOI] [PubMed]
- Longley, R. J. , White, M. T. , Takashima, E. , Morita, M. , Kanoi, B. N. , Li Wai Suen, C. S. N. , … Mueller, I. (2017). Naturally acquired antibody responses to more than 300 Plasmodium vivax proteins in three geographic regions. PLoS Neglected Tropical Diseases, 11, e0005888 10.1371/journal.pntd.0005888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopaticki, S. , Maier, A. G. , Thompson, J. , Wilson, D. W. , Tham, W.‐H. , Triglia, T. , … Cowman, A. F. (2011). Reticulocyte and erythrocyte binding‐like proteins function cooperatively in invasion of human erythrocytes by malaria parasites. Infection and Immunity, 79, 1107–1117. 10.1128/IAI.01021-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malleret, B. , Li, A. , Zhang, R. , Tan, K. S. W. , Suwanarusk, R. , Claser, C. , … Ng, M. L. (2014). Plasmodium vivax: Restricted tropism and rapid remodelling of CD71 positive reticulocytes. Blood, 125(8), 1314–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli, A. , & Culleton, R. (2018). Non‐human primate malaria parasites: Out of the forest and into the laboratory. Parasitology, 145, 41–54. 10.1017/S0031182016001335 [DOI] [PubMed] [Google Scholar]
- Miller, L. H. , Mason, S. J. , Clyde, D. F. , & McGinniss, M. H. (1976). The resistance factor to Plasmodium vivax in blacks. The Duffy‐blood‐group genotype, FyFy. The New England Journal of Medicine, 295, 302–304. 10.1056/NEJM197608052950602 [DOI] [PubMed] [Google Scholar]
- Mohring, F. , Hart, M. N. , Rawlinson, T. A. , Henrici, R. , Charleston, J. A. , Diez Benavente, E. , … Moon, R. W. (2019). Rapid and iterative genome editing in the malaria parasite Plasmodium knowlesi provides new tools for P. vivax research. eLife, 8 10.7554/eLife.45829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon, R. W. , Hall, J. , Rangkuti, F. , Ho, Y. S. , Almond, N. , Mitchell, G. H. , … Blackman, M. J. (2013). Adaptation of the genetically tractable malaria pathogen Plasmodium knowlesi to continuous culture in human erythrocytes. PNAS, 110, 531–536. 10.1073/pnas.1216457110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon, R. W. , Sharaf, H. , Hastings, C. H. , Ho, Y. S. , Nair, M. B. , Rchiad, Z. , … Holder, A. A. (2016). Normocyte‐binding protein required for human erythrocyte invasion by the zoonotic malaria parasite Plasmodium knowlesi . Proceedings of the National Academy of Sciences of the United States of America, 113, 7231–7236. 10.1073/pnas.1522469113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moras, M. , Lefevre, S. D. , & Ostuni, M. A. (2017). From erythroblasts to mature red blood cells: Organelle clearance in mammals. Frontiers in Physiology, 8, 1076 10.3389/fphys.2017.01076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntumngia, F. B. , Thomson‐Luque, R. , Galusic, S. , Frato, G. , Frischmann, S. , Peabody, D. S. , … Adams, J. H. (2018). Identification and immunological characterization of the ligand domain of Plasmodium vivax Reticulocyte Binding Protein 1a. The Journal of Infectious Diseases, 218, 1110–1118. 10.1093/infdis/jiy273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchynnikova, E. , Aglialoro, F. , Bentlage, A. E. H. , Vidarsson, G. , Salinas, N. D. , von Lindern, M. , … van den Akker, E. (2017). DARC extracellular domain remodeling in maturating reticulocytes explains Plasmodium vivax tropism. Blood, 130, 1441–1444. 10.1182/blood-2017-03-774364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul, A.S. , Egan, E.S. , Duraisingh, M.T. . (2015). Host‐parasite interactions that guide redblood cell invasion by malaria parasites. Curr Opin Hematol, 22, 220–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radoshitzky, S. R. , Abraham, J. , Spiropoulou, C. F. , Kuhn, J. H. , Nguyen, D. , Li, W. , … Choe, H. (2007). Transferrin receptor 1 is a cellular receptor for New World haemorrhagic fever arenaviruses. Nature, 446, 92–96. 10.1038/nature05539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlinson, T. A. , Barber, N. M. , Mohring, F. , Cho, J. S. , Kosaisavee, V. , Gérard, S. F. , … Quinkert, D. (2019). Structural basis for inhibition of Plasmodium vivax invasion by a broadly neutralizing vaccine‐induced human antibody. Nature Microbiology, 4, 1497–1507. http://www.nature.com/articles/s41564-019-0462-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner, J. C. , Galinski, M. R. , Ingravallo, P. , & Barnwell, J. W. (2000). Two Plasmodium falciparum genes express merozoite proteins that are related to Plasmodium vivax and Plasmodium yoelii adhesive proteins involved in host cell selection and invasion. Proceedings of the National Academy of Sciences of the United States of America, 97, 9648–9653. 10.1073/pnas.160469097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner, J. C. , Tran, T. M. , Corredor, V. , Huber, C. S. , Barnwell, J. W. , & Galinski, M. R. (2005). Dramatic difference in diversity between Plasmodium falciparum and Plasmodium vivax reticulocyte binding‐like genes. The American Journal of Tropical Medicine and Hygiene, 72, 666–674. 10.4269/ajtmh.2005.72.666 [DOI] [PubMed] [Google Scholar]
- Reddy, K. S. , Amlabu, E. , Pandey, A. K. , Mitra, P. , Chauhan, V. S. , & Gaur, D. (2015). Multiprotein complex between the GPI‐anchored CyRPA with PfRH5 and PfRipr is crucial for Plasmodium falciparum erythrocyte invasion. Proceedings of the National Academy of Sciences of the United States of America, 112, 1179–1184. 10.1073/pnas.1415466112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas Caraballo, J. , Delgado, G. , Rodriguez, R. , & Patarroyo, M. A. (2007). The antigenicity of a Plasmodium vivax reticulocyte binding protein‐1 (PvRBP1) recombinant fragment in humans and its immunogenicity and protection studies in Aotus monkeys. Vaccine, 25, 3713–3721. 10.1016/j.vaccine.2006.12.041 [DOI] [PubMed] [Google Scholar]
- Tachibana, S.‐I. , Sullivan, S. A. , Kawai, S. , Nakamura, S. , Kim, H. R. , Goto, N. , … Tanabe, K. (2012). Plasmodium cynomolgi genome sequences provide insight into Plasmodium vivax and the monkey malaria clade. Nature Genetics, 44, 1051–1055. 10.1038/ng.2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tham, W.‐H. , Beeson, J. G. , & Rayner, J. C. (2017). Plasmodium vivax vaccine research – we've only just begun. International Journal for Parasitology, 47, 111–118. 10.1016/j.ijpara.2016.09.006 [DOI] [PubMed] [Google Scholar]
- Tham, W.‐H. , Wilson, D. W. , Lopaticki, S. , Schmidt, C. Q. , Tetteh‐Quarcoo, P. B. , Barlow, P. N. , … Cowman, A. F. (2010). Complement receptor 1 is the host erythrocyte receptor for Plasmodium falciparum PfRh4 invasion ligand. Proceedings of the National Academy of Sciences of the United States of America, 107, 17327–17332. 10.1073/pnas.1008151107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson‐Luque, R. , Wang, C. , Ntumngia, F. B. , Xu, S. , Szekeres, K. , Conway, A. , … Jiang, R. H. Y. (2018). In‐depth phenotypic characterization of reticulocyte maturation using mass cytometry. Blood Cells, Molecules & Diseases, 72, 22–33. 10.1016/j.bcmd.2018.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, T. M. , Oliveira‐Ferreira, J. , Moreno, A. , Santos, F. , Yazdani, S. S. , Chitnis, C. E. , … Galinski, M. R. (2005). Comparison of IgG reactivities to Plasmodium vivax merozoite invasion antigens in a Brazilian Amazon population. The American Journal of Tropical Medicine and Hygiene, 73, 244–255. 10.4269/ajtmh.2005.73.244 [DOI] [PubMed] [Google Scholar]
- Triglia, T. , Thompson, J. , Caruana, S. R. , Delorenzi, M. , Speed, T. , & Cowman, A. F. (2001). Identification of proteins from Plasmodium falciparum that are homologous to reticulocyte binding proteins in Plasmodium vivax. Infection and Immunity, 69, 1084–1092. 10.1128/IAI.69.2.1084-1092.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz, J. C. , Yap, A. , Sisquella, X. , Thompson, J. K. , Lim, N. T. , Whitehead, L. W. , … Nebl, T. (2016). Essential role of the PfRh5/PfRipr/CyRPA complex during Plasmodium falciparum invasion of erythrocytes. Cell Host & Microbe, 20, 60–71. 10.1016/j.chom.2016.06.004 [DOI] [PubMed] [Google Scholar]
- Weiss, G. E. , Gilson, P. R. , Taechalertpaisarn, T. , Tham, W.‐H. , de Jong, N. W. M. , Harvey, K. L. , … Crabb, B. S. (2015). Revealing the sequence and resulting cellular morphology of receptor‐ligand interactions during Plasmodium falciparum invasion of erythrocytes. PLoS Pathogens, 11, e1004670 10.1371/journal.ppat.1004670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, K. E. , Hjerrild, K. A. , Bartlett, J. , Douglas, A. D. , Jin, J. , Brown, R. E. , … Higgins, M. K. (2014a). Structure of malaria invasion protein RH5 with erythrocyte Basigin and blocking antibodies. Nature, 515, 427–430. 10.1038/nature13715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeman, A.‐M. , der Wel, A. V. , & Kocken, C. H. M. (2013). Ex vivo culture of Plasmodium vivax and Plasmodium cynomolgi and in vitro culture of Plasmodium knowlesi blood stages. Methods in Molecular Biology Clifton NJ, 923, 35–49. [DOI] [PubMed] [Google Scholar]


