Abstract
Virus filtration with nanometer size exclusion membranes (“nanofiltration”) is effective for removing infectious agents from biopharmaceuticals. While the virus removal capability of virus removal filters is typically evaluated based on calculation of logarithmic reduction value (LRV) of virus infectivity, knowledge of the exact mechanism(s) of virus retention remains limited. Here, human parvovirus B19 (B19V), a small virus (18–26 nm), was spiked into therapeutic plasma protein solutions and filtered through Planova™ 15N and 20N filters in scaled‐down manufacturing processes. Observation of the gross structure of the Planova hollow fiber membranes by transmission electron microscopy (TEM) revealed Planova filter microporous membranes to have a rough inner, a dense middle and a rough outer layer. Of these three layers, the dense middle layer was clearly identified as the most functionally critical for effective capture of B19V. Planova filtration of protein solution containing B19V resulted in a distribution peak in the dense middle layer with an LRV >4, demonstrating effectiveness of the filtration step. This is the first report to simultaneously analyze the gross structure of a virus removal filter and visualize virus entrapment during a filtration process conducted under actual manufacturing conditions. The methodologies developed in this study demonstrate that the virus removal capability of the filtration process can be linked to the gross physical filter structure, contributing to better understanding of virus trapping mechanisms and helping the development of more reliable and robust virus filtration processes in the manufacture of biologicals.
Keywords: downstream processing, material science
1. INTRODUCTION
Human plasma products carry the risk of contamination by infectious agents, such as viruses and prions, even when manufactured from plasma obtained from apparently healthy donors.1 In order to reduce this risk, a set of safety measures are implemented and rigorously monitored, including careful screening of donors, testing of individual plasma donations for various viral markers, testing of manufacturing plasma pools by genomic assays, and, most importantly, use of various dedicated virus inactivation or removal steps during the manufacturing process.2, 3, 4 The virus safety of products is evaluated by virus validation studies performed according to international guidelines using a scaled‐down version of the manufacturing processes.5, 6 These studies assess the extent of inactivation or removal of spiked model viruses, based on logarithmic reduction value (LRV), typically using infectivity assays in cellular models.7 However, although these studies evaluate virus removal, the data obtained do not unveil the specific mechanisms leading to virus removal. Therefore, in our current study of scaled‐down industrial processes, we conducted both a determination of virus LRV data and an actual visualization of virus entrapment in the filter membrane of a particular type of virus removal filter. Size‐based exclusion is recognized as the primary mechanism of virus removal taking place in the filter membrane.8 However, various types of filter with different structures are commercially available. The Planova™ series were chosen for the present study as they are among the most established and widely used filters for virus removal by so‐called “nanofiltration”. Planova filters have a precisely defined, nanometer‐scale nominal pore size and there are many reports describing their structure.9, 10, 11, 12, 13, 14, 15, 16, 17, 18
Planova virus removal filters have hollow fiber membranes made of cuprammonium‐regenerated cellulose that are effective for removing virus particles from biopharmaceuticals,8, 9, 10, 11 achieving high virus LRV in protein solutions under appropriate operating conditions.12, 13 In filtration processes with Planova filters, a protein solution potentially contaminated with viruses enters the filter and passes through the hollow fiber membranes under pressure, wherein any virus larger than the nominal pore size of the membrane is captured. Indeed, the hollow fiber membrane of Planova filters has a three‐dimensional (3D) microporous structure characterized as a network of interconnected voids and capillaries that act as a multilayered membrane in which virus particles are effectively captured while protein molecules pass through.12 Planova 15N and 20N filters have a characteristic nominal pore size that is calculated by Hagen–Poiseuille's formula using flow rate, wall thickness, fluid viscosity, pressure, and porosity14, 15 as the complex 3D structure makes direct measurements of pore size extremely challenging. Using a prototype Planova filter (the “BMM” filter; Asahi Kasei Corp, Tokyo, Japan), Tsurumi et al15 were the first to achieve high Japanese encephalitis virus LRV under conditions that also enabled excellent permeability and recovery of various proteins. This simultaneously effective virus capture and good protein filterability was enabled by the unique 3D capillary and void pore structure of the BMM hollow fiber membranes. Scanning electron microscopy (SEM) revealed a membrane structure with uniform capillary size, decreasing gradient of void size, and connections between the voids from the inner to outer surfaces.15 Indeed, SEM of cross‐sectional images revealed various layers within the filter membrane15 while transmission electron microscopy (TEM) analysis performed by Yamaguchi et al16 after filtration of gold particles of different sizes showed that larger particles are captured in a layer closer to the inner surface.
Subsequent experimental work on virus entrapment in virus removal filters was conducted using virus or gold particles and various visualization methods. It should be noted that the physicochemical conditions of filtrations using different buffers or solutions, with or without proteins, are expected to impact the virus entrapment in the hollow fiber membrane while gold particle behavior may differ from that of viruses, which limits the comparability of experimental results. Hongo‐Hirasaki et al17, 18 observed that gold colloidal particles of about 18–21 nm (average diameter, 19.5 nm) were trapped and accumulated as a layer around the entire cross‐sectional circumference of Planova 20N filter hollow fiber membranes. In filtrations conducted in the absence of proteins and particularly in the presence of sodium dodecyl sulfate (SDS) surfactant, Kosiol et al19 identified size exclusion as the main mechanism for nanoparticle capture based on TEM and spectrophotometric quantification of gold particles. Using the latest SEM technology to visualize captured gold nanoparticles, Nazem‐Bokaee et al20 developed a methodology for characterizing the internal pore size and structure of different virus filtration membranes and demonstrated different capture positions depending on the membrane material and structure for Planova 20N, Planova BioEX, Viresolve® Pro, and Ultipor® DV20. Yamaguchi et al16 used TEM to show the capture of human parvovirus B19 (B19V) in Planova filter membranes of different pore sizes and corroborated virus localization by visualizing the capture of similarly sized gold nanoparticles. Bakhshayeshi et al21 established a method using confocal scanning laser microscopy for visualizing fluorescent dye‐conjugated bacteriophages and demonstrated capture in virus filters (Ultipor DV20 and Viresolve Pro) with sheet membranes. Yamamoto et al22 showed that migration of viruses in the Planova filter membrane is affected by Brownian motion of the particles and is dependent on the hydrodynamic forces occurring during filtration. Consequently, the pattern of entrapped viruses in the filter membrane is the result of the interaction of hydrodynamic forces and filter structure. However, none of these studies investigated the relationship between membrane structure, particularly gross structure, and the virus retention mechanism. Further, the experiments were not conducted under conditions considered to be representative of those encountered in manufacturing processes of protein products using virus removal filters, or were not relevant to virus safety. B19V has a single‐stranded DNA genome within a small nonenveloped capsid of ca. 18–26 nm diameter. Among the smallest of known human plasma‐borne viruses,23 B19V can be transmitted to humans through blood and plasma products23, 24, 25 and is a primary target for manufacturing‐scale removal by nanometer‐size virus removal filtration utilizing a size‐exclusion mechanism.26
In the present study, Planova filter microporous membranes were shown to have a rough inner, a dense middle and a rough outer layer. The dense middle layer was identified by TEM as the primary site for B19V capture. Laboratory‐scale filtrations with various proteins and B19V were conducted on Planova 15N and 20N filters, and those filters were additionally visualized by fluorescence microscopy to detect B19V capture. This study is the first to simultaneously report gross structural analysis of a virus removal filter and visualization of virus entrapment following a filtration process performed under conditions used in the biopharmaceutical industry.
2. MATERIALS AND METHODS
2.1. B19V preparation for TEM
B19V DNA‐positive plasma collected from a compensated anti‐B19V immunoglobulin G‐negative donor in the United States was purchased from Alpha Therapeutic Corp. (Los Angeles, CA). Virus was purified from this B19V‐positive plasma by cesium chloride density gradient ultracentrifugation at 150,000g for 26 hr at 4°C according to the methods of Kajigaya et al.27 B19V viral particle‐enriched fractions (density, 1.48 g/cm3) were identified by real‐time polymerase chain reaction (PCR) using a quantitative PCR assay kit (QuantiTect Probe PCR Kit; Qiagen, Inc., Germantown, MD) with the primer/probe set B1 (nt 3,187–3,206; 5′‐CAAAAGCATGTGGAGTGTGAGG‐3′), B2 (nt 3,558–3,539; 5′‐GTGCTGTCAGTAACCTGTAC‐3′) and probe FB19V‐1 (nt 3,310–3,339; 5′‐TAGCTGCCACAATGCCAGTGGAAAGGAGGC‐3′). This primer/probe set detects superchain DNA of intact B19V particle. B19V copy numbers were used to calculate B19V LRV for the virus clearance studies. B19V viral particle‐enriched fractions were then dialyzed overnight at 4°C against phosphate‐buffered saline (Dulbecco's Phosphate Buffered Saline [DPBS]; Invitrogen Corp., Carlsbad, CA).
2.2. B19V preparation for optical microscopy
B19V‐positive plasma, obtained as described above, was ultracentrifuged at 150,000g for 3 hr at 4°C. The pellet was resuspended in one of the following protein process solutions obtained in manufacturing runs immediately before the filtration step: antithrombin (Neuart®; Benesis Corp., Osaka, Japan), albumin (Albumin; Benesis Corp.), haptoglobin (Haptoglobin Injection‐Yoshitomi; Benesis Corp.), or heat/PEG‐treated intravenous immunoglobulin (Venoglobulin‐IH; Benesis Corp.). The solutions were sonicated to disperse the virus particles and then prefiltered through a 0.22‐μm sterilized filter (Millex‐GP, 0.22 μm; Millipore Corp., Billerica, MA).
2.3. Membrane structure analysis
2.3.1. Filters and virus filtration
To evaluate the gross structure of the hollow fibers, 5 to 10 mL of B19V preparation for TEM (viral load: 11 log10 IU) in DPBS prepared by cesium chloride ultracentrifugation was filtered on specifically manufactured experimental Planova 15N and 20N filter modules with an effective surface area of 10−1 cm2 and mean pore sizes of 15 ± 2 and 19 ± 2 nm, respectively (Asahi Kasei Medical Co. Ltd., Tokyo, Japan). The loading dose of B19V in these specific TEM experiments was thus close to 12 log10 IU/cm2 and was selected to ensure the presence of enough visible virus particles in the observation field. These experimental Planova modules were designed to decrease the effective surface area while using the same hollow fibers as those used in commercial Planova filters. All filtrations were carried out at 25°C and 80 kPa.
2.3.2. Virus fixation and inactivation in the membrane
After virus filtration, the hollow fibers were cut out from each filter module and fixed in 4% (w/v) paraformaldehyde in 100 mM phosphate buffer (pH 7.4) for 1 hr followed by washing with 100 mM phosphate buffer (pH 7.4) for 10 min.
2.3.3. Analysis of membrane structure by TEM
Each hollow fiber was then prepared for TEM visualization according to the method of Yamaguchi et al16 Ultrathin cross‐sections (50 nm) were stained with 2% uranyl acetate and observed using a JEM‐1200EX TEM (JEOL Ltd., Tokyo, Japan) at 80 kV. The images were captured using a Morada charge‐coupled device (CCD) camera with iTEM software (Soft Imaging Systems; Olympus Corp., Tokyo, Japan) and digitized.
Void size at the inner and outer membrane surfaces and on sections spanning from the inner to outer membrane surfaces (cut perpendicular to the fiber axis) was calculated, for both Planova 15N and Planova 20N filters, as follows. First, the membrane ultrathin cross‐sections were scanned at a magnification high enough to visualize the B19V particles from the inner to the outer membrane surfaces at positions corresponding to the cardinal directions. These serial images were then combined into a panoramic image. Four panoramic images from one section were prepared for further analysis. To differentiate void and capillary structures from the cellulose substrate, the combined images were binarized by ImageJ (version 1.44; https://imagej.nih.gov/ij/plugins/download.html). Each binarized image was analyzed for void size and number at locations within intervals spanning the distance between the inner and outer membrane surfaces. Each of the 20 intervals covered 5% of the distance from inner to outer membrane surface. Although the voids did not appear as perfect circles, void diameter was calculated based on their area on the ultrathin cross‐section images with the assumption that the void cross‐sectional profile was a perfect circle. Across all intervals, the smallest calculated void diameter was set at 1, calculated void diameter at each interval was represented as void diameter ratio, and void number and void diameter ratio were plotted together for all intervals of the hollow fiber membrane.
2.3.4. B19V localization and quantification in the membrane
The localization of the B19V particles within the membrane was determined as follows. The B19V particles identified on the combined panoramic images were marked with black dots. The images were then binarized to clearly visualize the marked B19V particles. The density profile of the B19V particles in the binarized image was then analyzed and plotted on a graph from inner to outer surface. The void size and B19V particle distributions were evaluated in four regions selected at each of the cardinal directions of each membrane ultrathin cross‐section. All imaging analyses were performed using Adobe Photoshop (version 7.0.1; Adobe Systems, San Jose, CA) and ImageJ (version 1.44).
2.4. Process evaluation
2.4.1. Virus clearance study
To evaluate virus removal in processes using Planova filters, filtration with process solution was performed on commercially available Planova 15N and 20N filters (effective surface area, 10 cm2; Asahi Kasei Medical Co., Ltd.) with mean pore sizes of 15 ± 2 and 19 ± 2 nm, respectively. For process evaluation, B19V preparations for optical microscopy were spiked into antithrombin or albumin solutions and filtered on Planova 15N filters or spiked into haptoglobin or (anti‐B19V immunoglobulin‐free) immunoglobulin solutions and filtered on Planova 20N filters. As the pooled plasma contains anti‐B19V immunoglobulin G, the immunoglobulin solution was subjected to anti‐B19V immunoglobulin G removal to prevent formation of virus‐antibody aggregates.7 Solution pH was adjusted to 5.5 following the addition of B19V preparation for optical microscopy at a titer of 9 log10 IU/ml. After incubation for 1 hr, the immunoglobulin solution was passed through a 0.22‐μm filter then subjected to pH adjustment to 4.2 to produce anti‐B19V immunoglobulin G‐free immunoglobulin solution. The scaled‐down filtration conditions using Planova filters mimicked actual manufacturing conditions for each product according to published guidelines.5, 6 Two independent filtration experiments were conducted for each protein/virus preparation using Planova 15N and 20N filters as shown in Table 1.
Table 1.
Removal of B19 virus particles from protein solutions by filtration through Planova filters
| Virus titer (log10 IU) | |||||
|---|---|---|---|---|---|
| Filter type | Protein | Distribution peak positiona | Load | Filtrate | B19 LRV (log10) |
| Planova 15N (10 cm2) | Antithrombin | 39% | 11.0/10.8 | <4.3/<4.3 | ≥6.7/≥6.5 |
| Albumin | 50% | 10.7/10.7 | 5.4/5.4 | 5.3/5.3 | |
| Planova 20N (10 cm2) | Haptoglobin | 39% | 10.9/11.0 | 6.5/7.1 | 4.4/3.9 |
| Immunoglobulin | 39% | 10.6/10.8 | 6.4/6.0 | 4.2/4.8 | |
LRV, logarithmic reduction value.
Expressed as a percentage (%) of the distance between the inner and outer membrane surfaces.
2.4.2. Visualization of B19V localization by fluorescence microscopy
Hollow fibers cut out from Planova filters were fixed as described above, dipped in Optimal Cutting Temperature (OCT) compound (SAKURA Seiki Corp., Tokyo, Japan) for 30 min and then flash‐frozen at −80°C. The frozen‐embedded hollow fibers were cut in 6‐μm thick sections perpendicular to the long axis of the hollow fiber using a Cryostat (CM3050 S, Leica Microsystems GmbH, Wetzlar, Germany) and attached to glass slides. The following procedures were performed at room temperature. Immunostaining was performed according to the method of Seto et al28 using polyclonal rabbit anti‐parvovirus B19 (DakoCytomation, Carpinteria, CA) as the primary antibody and fluorescein isothiocyanate (FITC)‐conjugated polyclonal mouse anti‐rabbit immunoglobulin (DakoCytomation) as the secondary antibody. Prior to primary antibody treatment, Planova sections were immersed in 2% bovine serum albumin in DPBS on glass slides as a blocking step. After 1 hr, the blocking solution was replaced by primary antibody diluted in blocking solution (1/1,000 primary antibody). The sections were incubated for at least 1 hr then washed 10 times with DPBS. Secondary antibody solution (1/1,000 secondary antibody) was added and the sections were incubated for 1 hr, washed 10 times with DPBS, and finally covered with VectaShield Mounting Medium (H‐1000; Vector Laboratories, INC., Burlingame, CA) and a cover glass. Each specimen was observed under a fluorescence microscope (TE2000; Nikon Corp., Tokyo, Japan). Images were captured digitally using a CCD camera (AQUACOSMOS/Basic System; Hamamatsu Photonics K.K., Shizuoka, Japan).
2.4.3. Fluorescence microscopy image processing
The digital images of hollow fiber cross‐sections were processed and analyzed using ORCA (Hamamatsu Photonics), Adobe Photoshop (version 7.0.1) and ImageJ (version 1.44). The distribution of the fluorescence intensity around the entire circumference of the membrane was calculated for each pixel in terms of the distance ratio between the inner and outer surfaces of the hollow fibers.
3. RESULTS
3.1. Planova 15N and Planova 20N membrane structure
TEM images of ultrathin (50 nm) cross sections of Planova 15N and 20N membranes (Figure 1a,b) were analyzed for void diameter ratio and number from binarized images (Figure 1c) The purpose of void diameter ratio analysis was to show the transition in diameter across the Planova filter membrane in order to understand the gross structure and to relate the structure to virus distribution during virus filtration. As shown in Figure 1b, the void structures were so complex that actual measurements of void diameter could not be used for comparison. Although the voids did not appear as perfect circles, void diameter was calculated based on their area on the ultrathin cross‐section images with the assumption that the void cross‐sectional profile was a perfect circle. Calculated in this way, the diameter does not reflect the entire filter nominal pore size (15 ± 2 and 19 ± 2 nm for Planova 15N and 20N, respectively) as part of the entire filter structure, which functions to actually filter unwanted particles. Void diameter was expressed and plotted as void diameter ratio with the smallest void diameter set at 1 to clearly demonstrate void diameter transition from the inner to the outer membrane surface.
Figure 1.
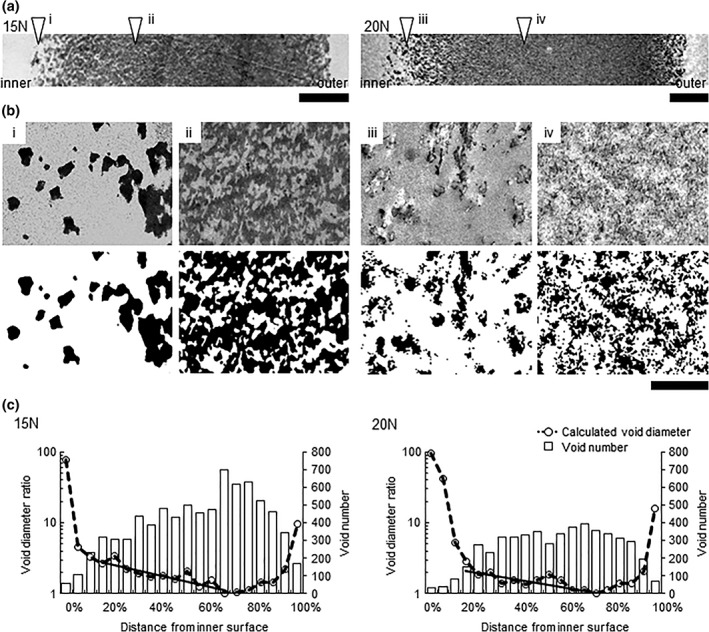
Analysis of the gross structure of Planova 15N and 20N membranes. (a) Transmission electron microscopy (TEM) images of membrane ultrathin cross‐sections. Bar: 5 μm. (b) (i)–(iv) Binarization of TEM images. White and black areas indicate voids and regenerated cellulose, respectively. The left and right side of the images are the inner and outer side of the membranes, respectively. Bar: 500 nm. (c) Void diameter ratio (dashed lines) and void number (open bars) with increasing distance from the inner membrane surface. The smallest calculated void diameters (210 and 383 nm for Planova 15N and 20N, respectively) across all intervals (n = 20) were set to 1. The solid line represents the exponential regression curve for the dense middle layer (corresponding to the regions at 20–70% and 20–75% from the inner surface in Planova 15N and 20N filter membranes, respectively)
Void diameter ratio in Planova 15N and 20N membranes showed that the membrane comprised a rough inner, a dense middle and a rough outer layer. It decreased sharply from the inner surface up to 20% of the distance between the inner and outer membrane surfaces, this area of the membrane corresponding to the rough inner layer. Void diameter ratio then gradually decreased up to 70% and 75% of the distance from the inner surface for Planova 15N and 20N membranes, respectively, corresponding to the dense middle layer. In both membranes, calculated void diameter then increased with closer proximity to the outer membrane surface; this area corresponds to the rough outer layer. From the inner surface through to the dense middle layer, the decrease in void diameter ratio tended to be inversely correlated with the increase in the number of voids (Figure 1c).
Practically, the mean pore size of Planova filters reflects the diameters of the capillaries as observed previously in fractured SEM “thick” cross‐sectional images of a “void replica” of Planova hollow fiber membranes.15 Capillaries were not visualized in the ultrathin cross‐section TEM images of hollow fiber membranes in the present study. However, narrow paths of several tens of nm were observed in the complex voids on TEM images (Figure 1b). Virus distribution was analyzed by fluorescent microscopy, as discussed below, for several modules used to filter B19V. Similar virus distributions were observed after filtration using different modules of the same type of Planova filters under the same conditions (data not shown). These findings suggest that the gross structure of the Planova filters is quite similar across different modules.
3.2. Functional substructure of Planova 15N and 20N membranes for capture of B19V
The present study used quantitative PCR to calculate the LRV of B19V in order to completely eliminate the influence of any immunoglobulin‐mediated neutralization of B19V infectivity on LRV calculation. Virus was purified from B19V‐positive plasma by cesium chloride density gradient ultracentrifugation followed by sonication, allowing pure, infectious, monodispersed B19V to be obtained. This was important due to the potential difficulty of differentiating the tiny B19V particles from any impurities in the B19V spike on TEM.
To evaluate the functional substructure of the Planova filters for B19V particle capture, 5–10 mL of B19V preparation for TEM (viral load: 11 log10 IU) in DPBS was filtered on Planova 15N and 20N filters with an effective surface area of 10−1 cm2. In conventional virus clearance studies, the virus is usually applied to the Planova module at about 10 log10 IU/cm2. Here, an extreme virus load of 12 log10 IU/cm2 was applied to ensure the presence of enough virus particles in the observation field. Broadly speaking, the mechanism for virus capture in Planova 15N and 20N membranes appeared to be related to void size as most of the B19V capture occurred in the region with the smallest void size. In TEM images of Planova 15N and 20N membrane ultrathin cross‐sections following filtration of B19V without protein (Figure 2a), the major distribution peaks were in the region of the filter membrane with the smallest mean void size (Figure 2b). Differences in the distribution of B19V within the Planova 15N and 20N hollow fiber membranes reflected differences in membrane structure and effective B19V capture regions.
Figure 2.
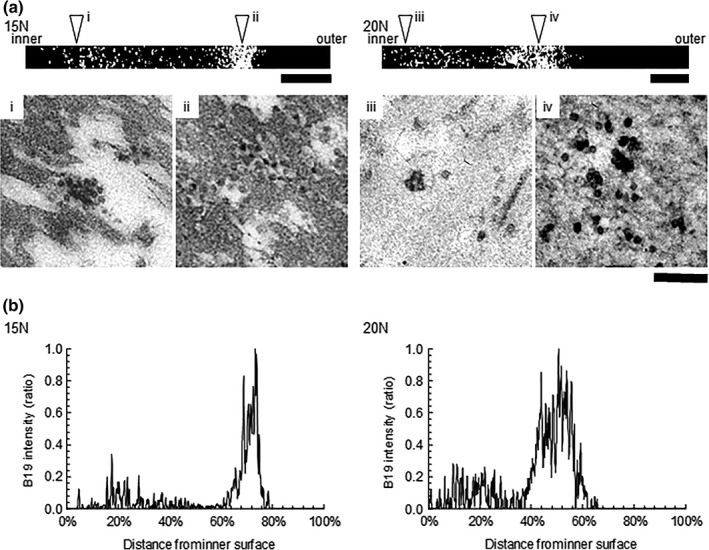
Distribution of B19V virus particles as assessed by transmission electron microscopy. (a) B19V localization in binarized transmission electron microscopy (TEM) images. Upper panels (i)–(iv): Four merged binarized TEM images showing B19V localization between the inner and outer membrane surfaces. B19V particles are visible as white dots. Lower panels (i)–(iv): Magnified images corresponding to the regions highlighted by the arrowheads in the upper panels. Bar: 5 μm and 200 nm in images in the upper and lower panels, respectively. The Planova 15N and 20N membrane thicknesses are about 30 and 40 μm, respectively. (B) Localization of B19V particles in the filter membranes as visualized by TEM. The x‐ and y‐axes represent the distance from the inner surface (0%) to the outer surface (100%) with B19V signal intensity observed on binarized TEM images, (A) top panels
TEM images taken in the cardinal directions of the ultrathin cross‐section of each hollow fiber membrane, binarized to visualize only B19V and then merged to show the distribution from the inner to outer surfaces, showed that capture of B19V particles of approximately 20 nm in diameter was limited to the region of 0–80% from the inner surface for Planova 15N and 0–70% from the inner surface for Planova 20N filters. No virus particles were identified within the regions corresponding to 80–100% and 70–100% of the distance from the inner surface of Planova 15N and 20N membranes, respectively (Figure 2b).
B19V localization within the capture region differed between the two filters. Minor and major peaks were observed at around 20% and 75%, respectively, from the inner membrane surface of Planova 15N and at around 20% and 55%, respectively, for Planova 20N membrane (Figure 2b). Based on TEM visualization, aggregated B19V particles appeared to be captured at the boundary region between the rough inner and dense middle layers (Figure 2a, i and iii), for the minor peak, while monodispersed B19V particles appeared to be captured in the dense middle layer (Figure 2a, ii and iv), for the major peak. The differences in localization between the two filters reflected the differences in their structures.
3.3. B19V capture during process filtration
Virus was purified from B19V‐positive plasma by ultracentrifugation and sonicated to obtain infectious, monodispersed B19V. B19V preparation for optical microscopy was spiked in process solutions of antithrombin, albumin, haptoglobin and (anti‐B19V immunoglobulin G‐free) immunoglobulin at 10.6 to 11.0 log10 IU and then filtered through 10 cm2 Planova 15N and 20N modules (Table 1). Prior to fluorescence microscopy observation of the Planova filters used for process evaluation, the validity of the planned immunofluorescence staining method was verified. Specifically, immunofluorescence staining was applied to Planova 15N and 20N hollow fiber membranes in the absence of B19V and these specimens were examined under fluorescence microscopy to provide negative controls. As shown in Figure 3a,b, no fluorescence signals were observed, demonstrating the validity of the planned immunofluorescence staining method. Subsequent fluorescence microscopy assessment of Planova filters used for process evaluation revealed that the functional regions for effective B19V capture following filtrations of antithrombin and albumin solutions on Planova 15N and haptoglobin and immunoglobulin solutions on Planova 20N were centered at 39%, 50%, 39% and 39%, respectively, of the distance from the inner surface, corresponding to the dense middle layer of the membrane (Figure 3; Table 1). The peak signals of the fluorescence from B19V capsid proteins in Planova filters loaded with antithrombin, haptoglobin and immunoglobulin, but not albumin, overlapped with the B19V capture peak occurring in regions with the smallest voids, at 70% and 75% from the inner membrane surfaces of Planova 15N and 20N filters, respectively. For albumin, however, the major peak was shifted for 11% outer, and a minor shoulder was also observed around 80% from the inner membrane surface (Figure 3d). The capture location for Planova 20N was the same for both haptoglobin and immunoglobulin.
Figure 3.
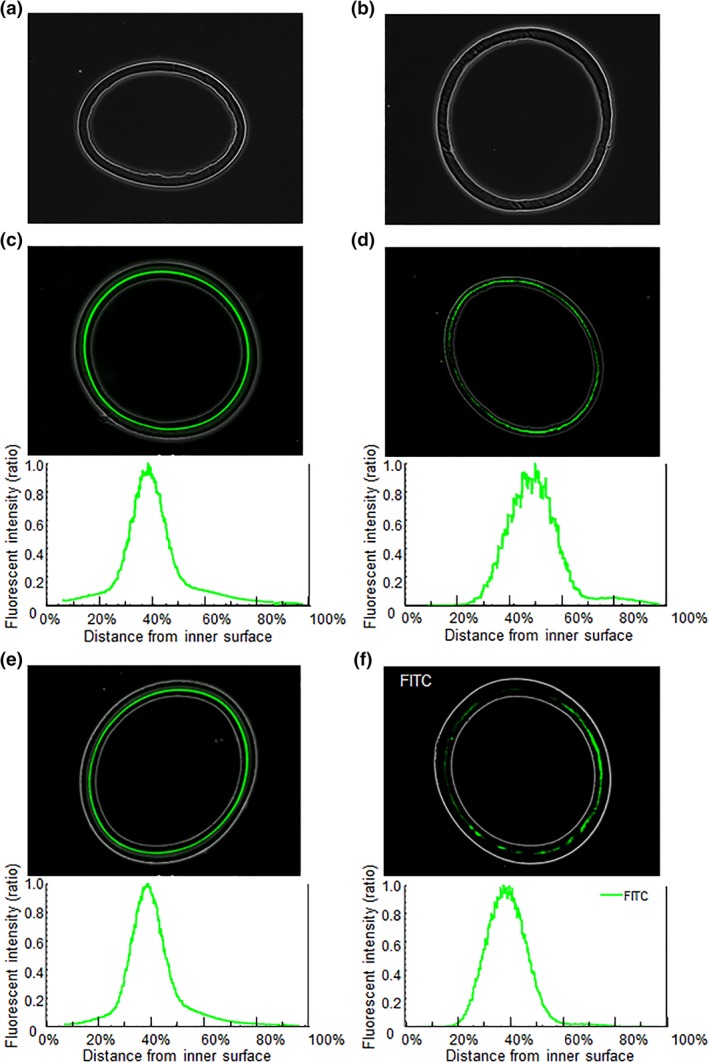
Distribution of B19V particles within Planova filters used for process evaluation as assessed by fluorescence microscopy. Negative controls for (a) Planova 15N and (b) Planova 20N filters showing no fluorescence signal by immunostaining in the absence of B19V. Fluorescence microscopy images and representation of fluorescence intensity ratios showing the distribution of B19V within the filter membrane on filtration of solutions containing (c) antithrombin and (d) albumin (Planova 15N) and (e) haptoglobin and (f) immunoglobulin (Planova 20N). Immunostaining and fluorescence microscopy were performed under the same conditions for all filter membrane specimens from (a) to (f)
The B19V clearance study results showed higher B19V LRV for all Planova 15N runs than for Planova 20N runs (Table 1). B19V LRV was slightly lower for the filtration of albumin than for that of antithrombin, possibly matching the observation of the B19V signal reaching the rough outer layer at the 80% position in Figure 3d B19V LRV on Planova 20N were both similarly high for haptoglobin and immunoglobulin filtration, showing a robustness similar to that of Planova 15N filters. Further experiments are needed to clarify the relationship between the shift of capture peak and B19V LRV for albumin.
4. DISCUSSION
4.1. Structural analysis of filter membrane
In a previous study, prototype Planova (Asahi Kasei Corp) filter membranes were filled with resin then processed to remove the membrane, producing a “void replica”. SEM imaging of this replica demonstrated a pore structure comprising 3D networks of voids interconnected with capillaries.15 Evaluation of the structure of several prototype hollow membranes revealed an asymmetric structure of mean void diameter distribution with a slight decrease in the radial direction from the inner to outer surfaces.15 Gold colloid (20–60 nm diameter) localization in the filter membrane demonstrated that larger gold particles were captured closer to the inner surface than smaller particles, thereby showing that both mean size and number of voids differed in different layers of the membranes.15 In the present study, TEM images of ultrathin cross‐sections (thickness, approximately 50 nm) of the hollow fibers were used to quantify the size and distribution of voids in the hollow fiber but did not show capillaries due the thickness of the specimen being insufficient to include capillaries of various orientations.
The Planova 15N and 20N membranes are about 30 and 40 μm thick, respectively. Visualization in the present study revealed that both membranes are comprised of a rough inner, a dense middle and a rough outer layer. However, there were several differences between the membranes: (i) void diameter decreased more rapidly from the inner toward the outer surface in the dense middle layer of Planova 15N compared to Planova 20N filter membranes (Figure 1c); (ii) magnified images of specific regions of the membranes indicated a much simpler pore structure, especially with respect to the voids, in Planova 15N than in Planova 20N filter membranes (Figure 1b); and (iii) calculated void diameters in the region with the smallest voids were larger in Planova 20N than in Planova 15N filter membranes.
4.2. Relationship between membrane structure and B19V localization
In the present study, B19V localization was also considered in the context of the functional structure of Planova membranes. Filtration preparations comprised mainly nonaggregated (monodispersed) and a few aggregated virus particles. Those monodispersed and aggregated B19V particles were captured in different regions, resulting in distinct inner and outer distribution peaks, respectively (Figure 2b). Conversely, only one B19V distribution peak was observed in the dense middle layer in the Planova filters used to filter protein solutions (Figure 3). This suggests there were fewer aggregated B19V particles in the B19V‐spiked protein solutions used for optical microscopy than in that used for TEM. Alternatively, the B19V in those protein solutions may have been less prone to aggregation. This is consistent with the report by Hongo‐Hirasaki et al,18 demonstrating that differences in virus stock preparations affect the filterability of process solutions, although high porcine parvovirus LRV could be obtained for all solutions. Figure 4 shows a schematic model of the pore composition within the membrane and the virus capture mechanism based on experimental data obtained here.
Figure 4.
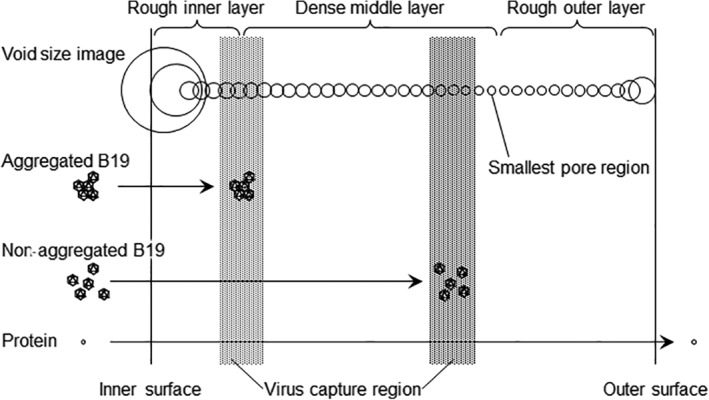
Schematic image showing membrane structure and relationship between virus particle and protein sizes. The void diameter transition from the inner to outer membrane surfaces of the membranes, the regions that capture the B19V particles and the passage of proteins through the membrane are indicated. This schematic image does not show void and capillary structures; however, the positions and diameters of the circles reflect the void diameter ratio (derived from data shown in Figure 1c)
In previous TEM observation of the capture of B19V in protein‐free suspensions by Planova 15N and 20N membranes by Yamaguchi et al,16 B19V peaks were localized at around 30% from the inner surfaces of both Planova 15N and 20N membranes. This corresponds to the innermost edge of the dense middle layer as defined in the present study. Also, consistent with our observations, TEM analyses showed that the distribution of B19V particles in Planova 20N, compared to Planova 15N, membranes was broader and closer to the inner surface of the membrane. The narrower B19V localization band observed with Planova 15N filters may be due to the sharp decline in void size from the inner to the outer membrane surfaces (Figure 1c). However, in the present study, there was a major peak of monodispersed B19V at 75% and 55% from the inner surface of Planova 15N and 20N membranes, respectively. These findings suggest that the solution in the previous study16 likely contained relatively few monodispersed B19V particles. Thus, the present observation of the broad, major B19V distribution peak closer to the inner membrane surface in Planova 20N filters indicates that a more complex microporous filter membrane provides a broader capture region.
The present analysis of filter membrane structure and observation of captured virus suggest that the rate of void size transition and complexity of the internal void surfaces may determine the localization and rate of virus migration through the membrane. These important points require elucidation as part of a more comprehensive research article.
4.3. Protein product process evaluation and virus distribution
Previous experiments have demonstrated virus capture based on distribution analyses of virus filtered through virus removal filter membranes in protein‐free solutions.16, 19 The present fluorescence microscopy observation of the B19V‐spiked antithrombin, albumin, haptoglobin, and immunoglobulin process solutions identified B19V capture peaks at 50% or less of the distance from the inner membrane surfaces (Figure 3c–f). Conversely, the capture peaks observed after filtration of B19V‐spiked DPBS were localized at 75% and 55% from the inner surfaces of Planova 15N and 20N membranes, respectively (Figure 2b). Thus, B19V capture localization is affected by (a) the presence of proteins and (b) the type of protein. These are important factors affecting virus capture regions in Planova membranes. Nonspecific absorption of proteins in solution onto the membrane substrate may reduce the actual inner volume of pores.16, 18 Proteins may also interact with B19V particles with different degree of affinity, potentially affecting virus migration. These interactions are affected by the isoelectric point of proteins and buffer pH as well as by the ionic strength of the buffer.29, 30
Process evaluation of the virus filtration step is traditionally evaluated in terms of virus LRV5, 6; however, this does not provide insight into the types of membrane structure that are effective for virus capture or the robustness of the filtration process. Experimental studies have demonstrated that small viruses can pass through some virus filters, resulting in a potential risk of infectivity.23, 29 However, virus breakthrough is unlikely when using validated conditions that are known to have a virus distribution peak located close to the inner surface of the membrane.
Omar et al31 and Kreil et al32 demonstrated that the presence of anti‐B19V immunoglobulin G in the immunoglobulin solution enhances the B19V clearance of Planova filters even up to Planova 35N (pore size, 35 ± 2 nm). Anti‐B19V immunoglobulin G was removed from the immunoglobulin solution used in the present study in order to exclude any effect of the immuno‐complex and avoid LRV overestimation. Prior to performing the experiment shown in Figure 3d and Table 1, 9.0 log10 IU of B19V preparation for optical microscopy was spiked into immunoglobulin solution (70 mg/mL immunoglobulin in 5% sorbitol, pH 4.2) and directly applied to 10 cm2 Planova 15N, 20N, and 35N modules. B19V distribution peaks were observed at around 9%, 7%, and 16% of the distance between the inner and outer membrane surfaces with LRV of 6.0, 5.5, and 4.4, respectively. These results suggest that B19V and anti‐B19V immunoglobulin G had marked affinity, creating immuno‐complexes that were trapped mainly in the rough inner layer of the Planova filters. On the contrary, when B19V was spiked in immunoglobulin solution depleted of anti‐B19V immunoglobulin G, the virus signal peak was not found in the rough inner layer but in the dense middle layer (Figure 3f and Table 1). These data indicate that optical microscopic observation provides a reliable methodology for the assessment of spike virus or spike sample preparation.
The B19V loads were 10.6 to 11.0 log10 IU in the four protein‐product filtration experiments (Table 1). The B19V capture regions within Planova 15N filters loaded with antithrombin or albumin were located at 39% and 50% from the inner surface, with B19V LRV of ≥6.7/≥6.5 and 5.3/5.3, respectively. This implies that B19V localization in the membrane may correlate with B19V LRV. Each process solution could interact with virus particles and/or with the membrane substrate. Roush et al33 reported that the impurity in the virus spike source affects virus removal capacity. Additionally, using three different monoclonal antibodies in comprehensive design space experiments, Strauss et al,34 demonstrated that solution conditions, as well as filtration pressure, could impact minute virus of mice LRV. Virus spike volume, filter area, protein concentration, and transmembrane pressure should all be covered in further experimental studies.
The present experiments and data were designed to demonstrate effective methodologies for observing filter structure and virus distribution under conditions typically used for conventional clearance studies and mimicking the particular manufacturing processes. Further applied and practical studies are required to understand how filtration conditions affect virus distribution and how this distribution correlates with LRV. Fluorescent microscopy may also be useful to visualize protein distribution within the filter membrane and clarify whether a protein layer forms on the filter, cause a decrease in void size. Data on virus and/or protein distribution within the membrane could be used to support protein permeability and LRV data from the viewpoints of membrane structure and virus removal mechanisms. Such findings could lead to further improvements in membrane manufacturing and use.
5. CONCLUSION
Here, visualization of virus in the hollow fiber membrane and determination of B19V LRV for the same process provided information from different perspectives. Applying this strategy to targets other than B19V, such as viruses with different surface properties and prion proteins, and conducting studies on other protein‐containing products or different types of virus filters may provide insights that lead to further improvements in membrane manufacturing as well as developing robust filtration processes.
ACKNOWLEDGMENTS
We are grateful to Dr. Kazuo Ikuta (Research Institute for Microbial Diseases, Osaka University) and Dr. Lin Linyen (Research Center for Ultrahigh Electron Microscopy, Osaka University) for help with the EM observations. We are also grateful to Tetsuo Sato (Asahi Kasei Medical Co., Ltd.), Dr. Mikihiro Yunoki (Japan Blood Products Organization), and other colleagues who provided valuable comments and encouragement. We are deeply appreciative of Linda Gudex (Asahi Kasei Bioprocess America Inc.) and Dr. Masaharu Marcus Inouye (Asahi Kasei Medical Co., Ltd.) who edited and reviewed this manuscript, respectively, and to Dr. Thierry Burnouf at Taipei Medical University, College of Biomedical Engineering, for critical reviews of the manuscript prior to submission. This study was part of a collaborative research project between Osaka University and Japan Blood Products Organization and between Asahi Kasei Medical Co., Ltd. and the Japan Blood Products Organization. Part of this work was also supported by the “Nanotechnology Network Project” of the Ministry of Education, Culture, Sports, Science, and Technology, Japan, based at the Research Center for Ultrahigh Electron Microscopy, Osaka University (Handai Multi‐Functional Nano‐Foundry).
Adan‐Kubo J, Tsujikawa M, Takahashi K, Hongo‐Hirasaki T, Sakai K. Microscopic visualization of virus removal by dedicated filters used in biopharmaceutical processing: Impact of membrane structure and localization of captured virus particles. Biotechnol Progress. 2019;35:e2875 10.1002/btpr.2875
Funding information Ministry of Education, Culture, Sports, Science, and Technology, Japan
REFERENCES
- 1. de Mendoza C, Altisent C, Aznar JA, Batlle J, Soriano V. Emerging viral infections—a potential threat for blood supply in the 21st century. AIDS Rev. 2012;14(4):279‐289. [PubMed] [Google Scholar]
- 2. Yunoki M, Urayama T, Tsujikawa M, et al. Inactivation of parvovirus B19 by liquid heating incorporated in the manufacturing process of human intravenous immunoglobulin preparations. Br J Haematol. 2005;128(3):401‐404. [DOI] [PubMed] [Google Scholar]
- 3. Hattori S, Yunoki M, Tsujikawa M, Urayama T, Tachibana Y, Yamamoto I. Variability of parvovirus B19 to inactivation by liquid heating in plasma products. Vox Sang. 2007;92(2):121‐124. [DOI] [PubMed] [Google Scholar]
- 4. Farcet MR, Lackner C, Antoine G, et al. Hepatitis E virus and the safety of plasma products: investigations into the reduction capacity of manufacturing processes. Transfusion. 2016;56(2):383‐391. 10.1111/trf.13343. [DOI] [PubMed] [Google Scholar]
- 5. EMA/CHMP/BWP/706271/2010. EMA/CHMP . Guideline: Guideline on plasma‐derived medicinal products. July 21, 2011.
- 6. Pharmaceutical and Food Safety Bureau , Ministry of Health, Labour and Welfare, Tokyo, Japan. Guideline on the look‐back review of blood products. Ref. No. 0310011 [in Japanese]. Published March 10, 2005.
- 7. Soltis RD, Hasz D. Studies on the nature of intermolecular bonding in antigen‐antibody complexes. Immunology. 1982;46:175‐181. [PMC free article] [PubMed] [Google Scholar]
- 8. Gefroh E, Dehghani H, McClure M, Connell‐Crowley L, Vedantham G. Use of MMV as a single worst‐case model virus in viral filter validation studies. PDA J Pharm Sci Technol. 2014;68:297‐311. [DOI] [PubMed] [Google Scholar]
- 9. Burnouf T, Radosevich M. Nanofiltration of plasma‐derived biopharmaceutical products. Haemophilia. 2003;9(1):24‐37. [DOI] [PubMed] [Google Scholar]
- 10. Burnouf T, Radosevich M, Goubran HA, Willkommen H. Place of nanofiltration for assuring viral safety of biologicals. Curr Nanosci. 2005;1:189‐201. [Google Scholar]
- 11. Caballero S, Diez JM, Belda FJ, et al. Robustness of nanofiltration for increasing the viral safety margin of biological products. Biologicals. 2014;42:79‐85. [DOI] [PubMed] [Google Scholar]
- 12. Hamamoto Y, Harada S, Kobayashi S, et al. A novel method for removal of human immunodeficiency virus: filtration with porous polymeric membranes. Vox Sang. 1989;56(4):230‐236. [DOI] [PubMed] [Google Scholar]
- 13. Radosevich M, Appourchaux P, Huart JJ, Burnouf T. Nanofiltration, a new specific virus elimination method applied to high‐purity factor IX and factor XI concentrates. Vox Sang. 1994;67(2):132‐138. [DOI] [PubMed] [Google Scholar]
- 14. Ishikawa G, Hirasaki T, Manabe S, Yamamoto N. Novel determination method of size of virus in solution using cuprammonium regenerated cellulose membrane (BMM). Membranes. 1991;16(6):376‐386. [Google Scholar]
- 15. Tsurumi T, Sato T, Osawa N, Hitaka H. Structure and filtration performances of improved cuprammonium regenerated cellulose hollow fiber (improved BMM hollow fiber) for virus removal. Polym J. 1990;22(12):1085‐1100. [Google Scholar]
- 16. Yamaguchi K, Miyagawa E, Takahashi H, Miyazaki T, Ikeda H. Electron microscopic estimation of removal of parvovirus B19 (HPVB19) by nanofiltration with a novel filter membrane. J Memb Sci. 2007;298(1–2):99‐109. [Google Scholar]
- 17. Hongo‐Hirasaki T, Yamaguchi K, Yanagida K, Okuyama K. Removal of small viruses (parvovirus) from IgG solution by virus removal filter Planova® 20N. J Memb Sci. 2006;278(1–2):8. [Google Scholar]
- 18. Hongo‐Hirasaki T, Yamaguchi K, Yanagida K, Hayashida H, Ide S. Effects of varying virus‐spiking conditions on a virus‐removal filter Planova 20N in a virus validation study of antibody solutions. Biotechnol Prog. 2011;27(1):162‐169. [DOI] [PubMed] [Google Scholar]
- 19. Kosiol P, Hansmann B, Ulbricht M, Thom V. Determination of pore size distributions of virus filtration membranes using gold nanoparticles and their correlation with virus retention. J Memb Sci. 2017;533:289‐301. [Google Scholar]
- 20. Nazem‐Bokaee H, Fallahianbijan F, Chen D, et al. Probing pore structure of virus filters using scanning electron microscopy with gold nanoparticles. J Memb Sci. 2018;551:144‐152. [Google Scholar]
- 21. Bakhshayeshi M, Jackson N, Kuriyel R, Mehta A, Reis R, Zydney AL. Use of confocal scanning laser microscopy to study virus retention during virus filtration. J Memb Sci. 2011;379(1–2):260‐267. [Google Scholar]
- 22. Yamamoto A, Hongo‐Hirasaki T, Uchi Y, Hayashida H, Nagoya F. Effect of hydrodynamic forces on virus removal capability of Planova™ filters. AIChE J. 2014;60(6):2286‐2297. [Google Scholar]
- 23. Blümel J, Burger R, Drosten C, et al. Parvovirus B19—Revised. Transfus Med Hemother. 2010;37(6):339‐350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Koenderman AHL, ter Hart HGJ, Prins‐de Nijs IMM, et al. Virus safety of plasma products using 20 nm instead of 15 nm filtration as virus removing step. Biologicals. 2012;40:473‐481. [DOI] [PubMed] [Google Scholar]
- 25. Soucie JM, De Staercke C, Monahan PE, et al. Evidence for the transmission of parvovirus B19 in patients with bleeding disorders treated with plasma‐derived factor concentrates in the era of nucleic acid test screening. Transfusion. 2013;53(6):1217‐1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Korneyeva M, Rosenthal S. Virus removal by nanofiltration. Methods Mol Biol. 2005;308:221‐231. [DOI] [PubMed] [Google Scholar]
- 27. Kajigaya S, Shimada T, Fujita S, Young NS. A genetically engineered cell line that produces empty capsids of B19 (human) parvovirus. Proc Natl Acad Sci U S A. 1989;86(19):7601‐7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Seto S, Miyata M. Attachment organelle formation represented by localization of cytadherence proteins and formation of the electron‐dense core in wild‐type and mutant strains of Mycoplasma pneumoniae . J Bacteriol. 2003;185(3):1082‐1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Willkommen H, Blümel J, Brorson K, et al. Meeting report‐workshop on spike characterizations and virus removal by filtration: trends and new developments. PDA J Pharm Sci Technol. 2014;68(3):215‐220. [DOI] [PubMed] [Google Scholar]
- 30. Hongo‐Hirasaki T, Komuro M, Ide S. Effect of antibody solution conditions on filter performance for virus removal filter Planova™ 20N. Biotechnol Prog. 2010;26(4):1080‐1086. [DOI] [PubMed] [Google Scholar]
- 31. Omar A, Kempf C. Removal of neutralized model parvoviruses and enteroviruses in human IgG solutions by nanofiltration. Transfusion. 2002;42:1005‐1010. [DOI] [PubMed] [Google Scholar]
- 32. Kreil TR, Wieser A, Berting A, et al. Removal of small nonenveloped viruses by antibody‐enhanced nanofiltration during the manufacture of plasma derivatives. Transfusion. 2006;46:1143‐1151. [DOI] [PubMed] [Google Scholar]
- 33. Roush DJ, Myrold A, Burnham MS, Hughes JV. Limits in virus filtration capability? Impact of virus quality and spike level on virus removal with xenotropic murine leukemia virus. Biotechnol Prog. 2015;31(1):135‐144. [DOI] [PubMed] [Google Scholar]
- 34. Strauss D, Goldstein J, Hongo‐Hirasaki T, et al. Characterizing the impact of pressure on virus filtration processes and establishing design spaces to ensure effective parvovirus removal. Biotechnol Prog. 2017;33(5):1294‐1302. [DOI] [PMC free article] [PubMed] [Google Scholar]


