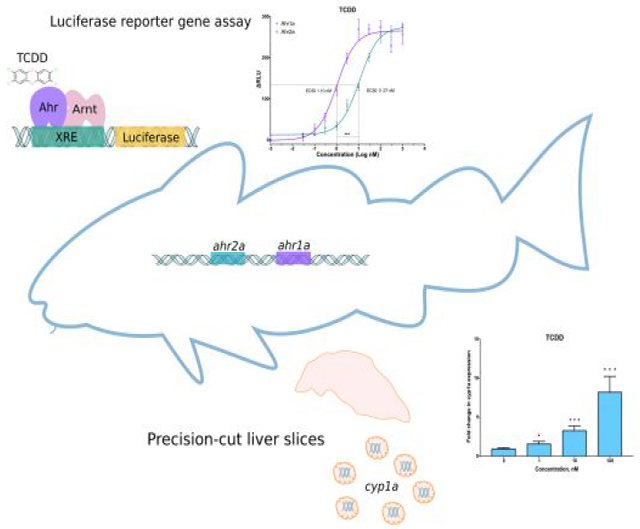
Abstract
The aryl hydrocarbon receptor (Ahr) is a ligand-activated transcription factor that mediates the toxicity of halogenated and polycyclic aromatic hydrocarbons in vertebrates. Atlantic cod (Gadus morhua) has recently emerged as a model organism in environmental toxicology studies, and increased knowledge of Ahr-mediated responses to xenobiotics is imperative. Genome mining and phylogenetic analyses revealed two Ahr-encoding genes in the Atlantic cod genome, gmahr1a and gmahr2a. In vitro binding assays showed that both gmAhr proteins bind to TCDD, but stronger binding to gmAhr1a was observed. Transactivation studies with a reporter gene assay revealed that gmAhr1a is one order of magnitude more sensitive to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) than gmAhr2a, but the maximal response of the receptors were similar. Other well-known Ahr agonists, such as β-naphthoflavone (BNF), 3,3′,4,4′,5-pentachlorobiphenyl (PCB126) and 6-formylindolo[3,2-b]carbazole (FICZ), also activated the gmAhr proteins, but gmAhr1a was in general the more sensitive receptor and produced the highest efficacies. Induction of cyp1a in exposed precision-cut cod liver slices confirmed the activation of the Ahr signaling pathway ex vivo. In conclusion, the differences in transcriptional activation by gmAhrs with various agonists, the distinct binding properties with TCDD and BNF, and the distinct tissue-specific expression profiles, indicate different functional specializations of the Atlantic cod Ahrs.
Graphical Abstract
Introduction
The ligand-activated transcription factor aryl hydrocarbon receptor (AHR) is a member of the basic helix-loop-helix PER-ARNT-SIM (bHLH-PAS) superfamily, and has been widely studied because of its important role in mediating cellular responses to environmental pollutants. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) has been established as the most potent exogenous AHR ligand, but also planar polycyclic aromatic hydrocarbons (PAHs), dioxin-like polychlorinated biphenyls (dl-PCBs) such as 3,3′,4,4′,5-pentachlorobiphenyl (PCB126), the synthetic flavonoid β-naphthoflavone (BNF), and certain endogenous compounds such as the tryptophan derivative 6-formylindolo[3,2-b]carbazole (FICZ) have been shown to bind to and activate AHR1–5. Recently, many investigations have focused on elucidating the physiological roles of AHR. It is now known that besides acting as a xenosensor and modulating the transcription of genes that encode proteins involved in the biotransformation of xenobiotic compounds, AHR participates in different signalling pathways and functions in physiological systems such as the cardiovascular-, reproductive- and immune systems6–8. In fact, it has been suggested that the original function of AHR started as a developmental regulatory gene in invertebrates, and that the ability to mediate xenobiotic responses is an evolved adaptative response mechanism present in vertebrates9,10.
The unliganded AHR is located in the cytoplasm in a protein complex with two HSP90 proteins, a co-chaperone protein (p23), and AHR-interacting protein (AIP)11–14. Upon ligand activation, AIP dissociates from the protein complex and it is suggested that only the AHR-HSP90 complex translocates into the nucleus15–19. AHR disassociates from HSP90 and heterodimerizes with the aryl hydrocarbon receptor nuclear translocator (ARNT). This heterodimer binds to xenobiotic response elements (XRE) upstream of AHR target genes, modulating the transcription of a battery of genes encoding enzymes involved in the biotransformation of xenobiotics, including cytochrome P450 1A (CYP1A)14,20–23.
A tandem gene duplication prior to the divergence of cartilaginous and bony fish lineages led to the appearance of two Ahr clades; Ahr1 and Ahr210,24. Further, due to the teleost-specific whole genome duplication event, fish may posses both an ahr1a-ahr2a and an ahr1b-ahr2b tandem pair, although the number of paralogous genes that have been retained throughout evolution varies among different fish species25. In contrast to teleosts, humans and rodents have retained only one AHR-encoding gene that was thought to be the ortholog of the teleost ahr1, but it is now thought to represent a different evolutionary lineage10. Ahr has previously been described in several fishes, including zebrafish (Danio rerio)24,26,27, mummichog (Fundulus heteroclitus)28, red seabream (Pagrus major)29, Atlantic salmon (Salmo salar)30–32, Japanese medaka (Oryzias latipes)33, Japanese pufferfish (Takifugu rubripes)34 and Atlantic tomcod (Microgadus tomcod)35. Expression patterns of ahr genes vary among fishes, and ahr2 is often more abundant and has wider tissue distribution, whereas expression of ahr1 is mainly found in brain and heart28–30,36. Loss-of-function studies using morpholino-modified antisense oligonucleotides or genome editing have shown that Ahr2 has a primary role in mediating toxic responses to TCDD and dl-PCBs in some fishes37–45. The exact function of Ahr1 has not yet been elucidated. A possible physiological role in early development of zebrafish has been hypothesized24, but loss-of-function studies have not yet revealed such a role44–46.
Atlantic cod (Gadus morhua) is an ecologically and commercially important species that is widely distributed in the North Atlantic Ocean. Atlantic cod has also commonly been used as an indicator species in marine pollution monitoring programs, such as the Protection of the Marine Environment of the North-East Atlantic (OSPAR) convention, water column monitoring of offshore petroleum activities in Norway, and recently in a waste dumping site outside the city of Bergen (Norway)47–50. The Ahr target gene cyp1a has been extensively studied in cod and used as a biomarker of exposure to environmental pollutants, including PAHs, dioxins and dl-PCBs51–57. These studies describing cyp1a gene expression, as well as Cyp1a protein synthesis, immunohistochemistry, and enzymatic activity, point to a functional Ahr pathway in cod. However, the molecular basis by which this species senses and responds to contaminants is not completely understood. Our group has recently described the lack of the xenobiotic sensor, pregnane X receptor (Pxr), belonging to the nuclear receptor superfamily of transcription factors, in Atlantic cod and other members of the Gadiformes order58. A hypothesis that Ahr has evolved a broader compensatory functional role as a xenosensor in Atlantic cod was therefore raised (ibid), emphasizing the need for a better understanding of the diversity and functional properties of the Ahr signalling pathway in this species. In the present study, we describe for the first time the primary structure, synteny, phylogeny, ligand binding affinities and agonist activation, as well as tissue specific expression profiles of the Atlantic cod Ahr1a and Ahr2a (denoted gmAhr1a and gmAhr2a). Our results show distinct differences in ligand binding affinities, agonist activation, and tissue-specific expression profiles of the gmAhr proteins, which indicate functional specialization.
Material and methods
Fish. Atlantic cod used in these studies were farmed fish from Austevoll Research Station (Institute of Marine Research, Bergen, Norway) and from Havbruksstasjonen in Tromsø (Nofima, Norway). All fish were kept at the Industrial and Aquatic Laboratory (ILAB, Bergen, Norway) in 500 L tanks in natural seawater at 9 °C, with a 12:12 h light/dark cycle regime and fed ad libitum with a commercial diet (Harmony Nature 500, EWOS, Bergen, Norway). Juvenile fish of both sexes (approx. 1.5–2 years old) were used for preparing ex vivo liver slices, whereas only female juvenile fish (approx. 1.5–2 years old) were used in the tissue-specific expression study. The fish were maintained and treated in accordance with the guidelines of the Norwegian Board of Biological Experiments with Living Animals.
RNA isolation and cloning of gmahr1a, gmahr2a, gmarnt1. Total RNA was extracted from Atlantic cod (Gadus morhua) heart and liver tissue following the protocol from the manufacturer (TriReagent; Sigma-Aldrich, Oslo, Norway). Complementary DNA (cDNA) was synthesized using Invitrogen Random hexamer, oligo(dT)12–18, and Superscript III/IV Reverse Transcriptases (Fisher Scientific, Oslo, Norway). gmarnt1 and gmahr1a were amplified as single fragments from cDNA prepared from cod liver and heart, respectively. gmahr2a was amplified as two overlapping fragments from liver cDNA. Detailed information on cloning and primers are presented in Supplementary material and Table S1. Atlantic cod Ahr1a, Ahr2a, and Arnt1 cDNA sequences were deposited in GenBank with the following accession numbers: Ahr1a; MN329012, Ahr2a; MN329013, and Arnt1; MN329014.
Synteny mapping, sequence alignments, and phylogenetic analyses. Synteny analyses of the genomic regions containing ahr genes of different fishes were based on genome data present in Ensembl. Multiple sequence alignments of N-terminal Ahr regions were performed in Clustal-Omega (EMBL-EBI) and edited in Jalview. The phylogenetic tree was inferred using AHR N-terminal amino acid sequences of fishes, mammals, reptiles and birds obtained from Genbank. Amino acid sequences were aligned with MUSCLE, and Bayesian inference analysis was conducted in MrBayes v3.2.7a (see Supplementary material for details).
In vitro protein expression and velocity sedimentation assays. [35S]methionine-labeled gmAhr proteins and Fundulus heteroclitus Ahr2a were synthesized in vitro using the TnT-Quick Coupled Reticulocyte Lysate System (Promega, Madison, WI) according to the manufactureŕs instructions. The [35S]methionine-labeled TnT reactions were assessed with SDS-polyacrylamide gel electrophoresis and the proteins were visualized by radiography. [3H]TCDD (2 nM) and [3H]BNF (10 nM) binding affinity to gmAhr1a, gmAhr2a and fhAhr2a was measured by velocity sedimentation with sucrose gradients in a vertical tube rotor as described in Karchner et al.28 (details in Supplementary material).
Transfection, exposure and luciferase reporter gene assay. COS-7 simian kidney cells were cotransfected with pcDNA3.1/Zeo(+) based gmAhr1a, gmAhr2a and gmArnt1 plasmids, luciferase reporter plasmid (pGudLuc6.1) and Renilla luciferase or β-galactosidase normalization plasmids (pRT-TK and pCMV- βGAL, respectively). For details on cell culturing, transfections and ligand exposure, see Table S2 and Supplementary material. Cytotoxicity was evaluated with two fluorescent dyes, resazurin and 5-carboxyfluorescein diacetate acetoxymethyl ester (5-CFDA-AM), as described by Pérez-Albaladejo et al.59 (Fig. S1, Fig. S2, protocol details in Supplementary material).
Tissue-specific expression of ahr1a, ahr2a, arnt1 and arnt2. Tissue samples from ovaries, muscle, head kidney, skin, mid intestine, spleen, heart, stomach, liver, brain, gill and eye (n=3) were collected and snap frozen in liquid nitrogen. RNA was extracted from tissue samples using the TRI Reagent® protocol, and 500 ng of RNA was reverse transcribed to cDNA using the iScript™ cDNA Synthesis Kit (Bio-Rad, California, USA). Quantitative real-time polymerase chain reaction analyses were performed using SYBR Green Master I (Roche Applied Sciences, Basel, Switzerland) and a CFX96 Touch Real-Time PCR Detection System (Bio-Rad) (for primers and protocol details, see Table S3 and Supplementary material). Expression of gmahr1a, gmahr2a, gmarnt1 and gmarnt2 was normalized across tissues by using beta-actin (actb) as the reference gene (GenBank: EX739174)60 and the method described by Livak et al.61.
Ex vivo exposure assays with precision-cut liver slices (PCLS) and analyses of cyp1a expression. PCLS were prepared as described previously with some modifications (details in Supplementary material)62. Liver slices were exposed to TCDD (n=5), FICZ (n=6), B[a]P (n=6) and PCB126 (n=7) (DMSO 0.01%). The viability and cytotoxicity of the liver slices were assessed with the 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) assay63 and the Cytotoxicity Detection Kit (LDH) (Sigma-Aldrich, Missouri, United States) (Fig. S3 and Fig. S4, protocol details in Supplementary material). Total RNA was isolated from frozen slices (two slices pooled per sample) using the RNeasy® Plus Universal Mini Kit (QIAGEN, Hilden, Germany). cDNA synthesis and qPCR analyses were performed as described in Supplementary material. Reference gene primers were reported previously60.
Results
Sequencing, phylogenetic analyses, and synteny of cod Ahrs.
Homology searches in the Atlantic cod genome (Ensembl, gadMor1) identified two putative Ahr-encoding genes organized in a tandem pair (ENSGMOG00000004709 and ENSGMOG00000004692). However, when compared to ahrs from other teleost species, neither ENSGMOG00000004709 nor ENSGMOG00000004692 appeared to encode complete Ahr protein sequences. In addition, sequence gaps introduced from inadequate genome sequencing and/or genome assembly were present in both gene models. To obtain the full protein encoding sequences, transcripts encoded by ENSGMOG00000004709 and ENSGMOG00000004692 were cloned from cDNA prepared from Atlantic cod heart and liver tissue, respectively. Sequencing of the cloned DNA revealed that ENSGMOG00000004709 and ENSGMOG00000004692 constitute two open reading frames consisting of 2874 bp and 3384 bp, encoding proteins with calculated molecular weights of 104.3 and 122.7 kDa. These cDNA sequences have been deposited in the National Center for Biotechnology information (NCBI) with accession numbers MN329012 and MN329013.
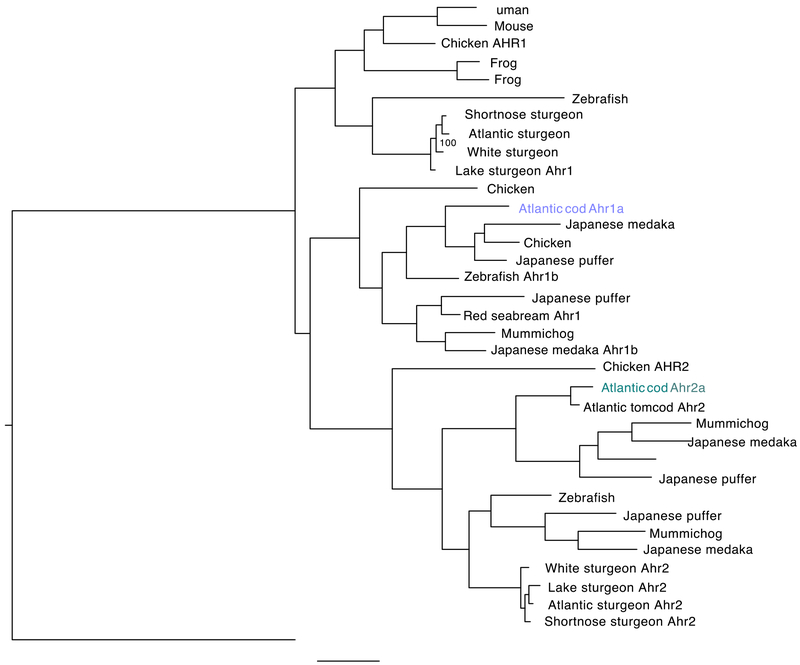
A phylogenetic tree was made based on the deduced amino acid sequences and representative Ahr sequences obtained from a diverse set of vertebrates, including several teleost species (Fig. 1). Importantly, the phylogeny shows a distinct clustering of MN329012 and MN329013 in clade 1a and clade 2a of the Ahr protein family, respectively. Based on the phylogenetic clustering, and supported by the tandem organization in the Atlantic cod genome, we have classified and named ENSGMOG00000004709 and ENSGMOG00000004692 as gmahr1a and gmahr2a, respectively. Analyses of the genomic region surrounding gmahr1a and gmahr2a revealed that the genes ndufa (NADH dehydrogenase ubiquinone 1 alpha subcomplex subunit 10; ENSGMOG00000004781) and cntnap5 (contactin-associated protein like 5; ENSGMOG00000004709) are localized adjacent and upstream of gmahr2a, oriented in the opposite and same direction, respectively (Fig. S5a). However, no genes were found immediately downstream of gmahr1a. Although the individual directions of the genes may vary, this syntenic relationship is well conserved among many teleost species, including mummichog (Fundulus heteroclitus), medaka (Oryzias latipes), green spotted puffer (Tetraodon nigroviridis), and Japanese puffer (Takifugu rubripes) (Fig. S5a). Genome mining in a recent and more comprehensive cod genome sequence assembly (gadMor2) revealed that both gmahr1a and gmahr2a consist of 11 exons and confirmed their co-localisation in a tandem pair in linkage group 20 in the Atlantic cod genome (Fig. S5b)64,65.
Figure 1. Phylogenetic analyses of Atlantic cod Ahr proteins and other vertebrate Ahr homologs.
The phylogenetic tree was made with MrBayes v3.2.7a using a BLOSUM substitution model. A selected set of AHR N-terminal amino acid sequences from fish, birds, reptiles and mammalian species were used. Alignment positions with gaps were not included. Bayesian inference analysis was conducted and Markov chain Monte Carlo (MCMC) analysis was run for 300,000 generations for each 1000 samples with a 25% burn-in. Four chains were used with a heating parameter of 0.1.
gmAhr1a and gmAhr2a have sequence identity and sequence similarity of 39.24% and 46.34%, respectively (Fig. S6). The sequence identity in the N-terminal part is high, while the C-terminal part suggested to be responsible for transcriptional transactivation is poorly conserved among the gmAhrs. Furthermore, the N-terminal parts of gmAhr1a and gmAhr2a were compared with Ahr1 and Ahr2 sequences obtained from a selected set of other teleosts (Fig. S7). The multiple sequence alignments revealed a high degree of conservation among these species, including the bHLH and PAS domains, which participate in ligand binding, HSP90 and AHR/ARNT dimerization, as well as DNA binding. The N-terminal part of Ahr1a from Japanese pufferfish (Takifugu rubripes) has the greatest sequence identity to gmAhr1a (84.35%) (Fig. S7a), while Ahr2 from another gadiform species, the Atlantic tomcod (Microgadus tomcod), shares extensive sequence identity (95.33%) with gmAhr2a in the N-terminal region (Fig. S7b). Interestingly, the identity between tomcod Ahr2 and gmAhr2 extends further C-terminally and includes an apparently repetitive sequence region present from amino acid 735 to 804 in gmAhr2, which may be characteristic for members of the gadiform order (Fig. S8). Furthermore, the nuclear localization signal (NLS), as well as the nuclear export signal 1 (NES1) and NES2 appear to be well-conserved in both gmAhr proteins. The LxxLL motif present before NES1 and the cysteine residue present in NES2, which are known to be critical for nuclear localization, are also conserved (Fig. S7)66–68. All of the amino acid residues part of the “TCDD-binding-fingerprint” characteristic of mammalian AHR, in addition to amino acids previously shown to be involved in binding of TCDD in other teleost species, including A388, H296, and Q388, are conserved in gmAhr1a and gmAhr2a (Fig. S7, Fig. S9, Fig. S10)69,70. Furthermore, the amino acid residues (P34, S35, R37, H38 and R39) found in the basic region 2 essential for binding to xenobiotic response elements (XRE) are conserved in both proteins (Fig. S7)22,71–73. The C-terminal part of mammalian AHR contains the transactivation domain that has an acidic (D/E) domain, as well as a high content of glutamine (Q), and proline, serine, and threonine-rich (P/S/T-rich) subdomains74. To reveal such putative subdomains in the gmAhr proteins, the percentages of these characteristic amino acids were plotted for every 20 bases as previously done for Ahr1 and Ahr2 from red seabream (Pagrus major) (Fig. S11)29. A putative Q-rich domain is present in both gmAhr1a and gmAhr2a, but the frequencies of glutamine were not as high as observed in mammalian AHRs. The other transactivation domains, such as acidic and P/S/T-rich domains, were found to be present in the C-terminal half of both gmAhr proteins.
Tissue-specific expression profiles of Atlantic cod ahrs and arnts.
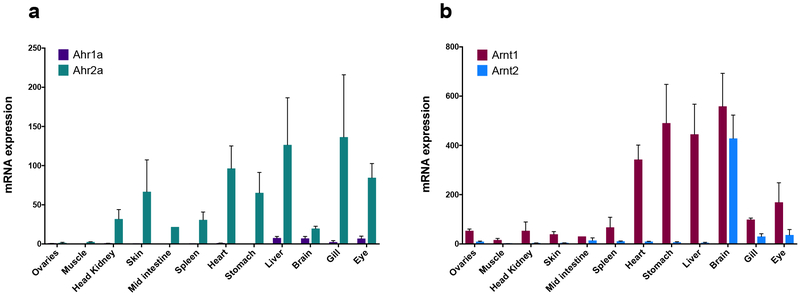
The tissue-specific expression of the two gmahr genes was assessed in juvenile Atlantic cod with quantitative real-time PCR (qPCR). The expression levels in the examined tissues were quite different between the paralogous genes, suggesting that their expression profiles appear to be gene-specific (Fig. 2a). The mRNA expression of gmahr2 was detectable in all tissues, albeit in lower levels in ovaries and muscle. Heart, liver, gill, and eye had the highest levels of ahr2a. In contrast, expression of ahr1a was not measurable in most tissues, and only notable in liver, brain, gill, and eye. The cod genome harbours two Arnt-encoding genes, representing members of both the arnt1 and arnt2 subfamily. The tissue-specific expression was also assessed for the arnt genes, demonstrating that arnt1 was ubiquitously expressed in most tissues, with the highest levels found in brain, liver stomach, and heart (Fig. 2b). Similarly, arnt2 was also expressed in high levels in brain, but to a lesser extent in other tissues with only notable mRNA levels in gill and eye (Fig. 2b).
Figure 2. Tissue-specific expression of Atlantic cod ahrs and arnts.
Expression levels of ahrs (a) and arnts (b) were assessed in gonads, muscle, head kidney, skin, mid intestine, spleen, heart, stomach, liver, brain, gill and eye obtained from female juvenile Atlantic cod (n=3). Expression levels were assessed with qPCR and normalized against the reference gene actb. Original data were multiplied by 10000; results are expressed as mean±SEM.
In vitro synthesis of gmAhr and specific binding of [3H]TCDD and [3H]BNF.
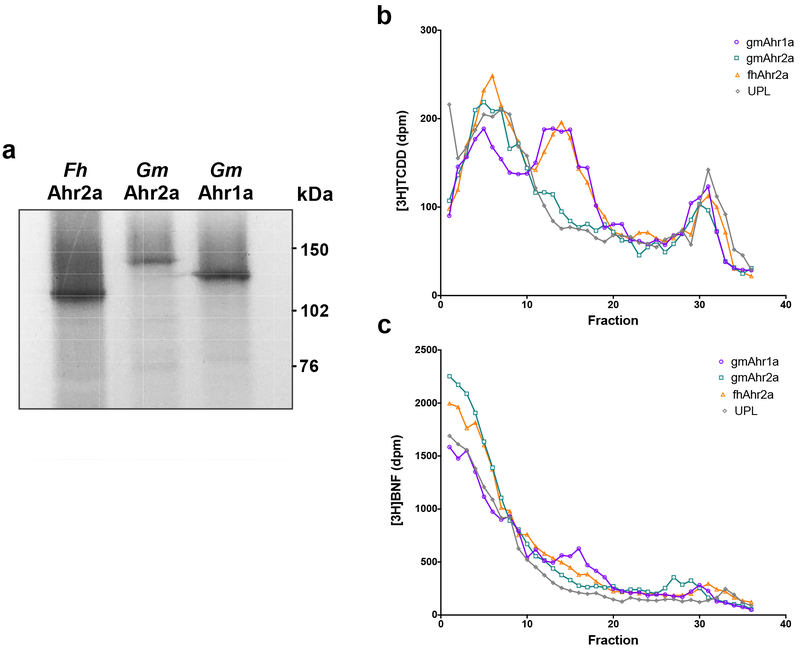
Protein syntheses of gmAhr1a and gmAhr2a, as well as F. heteroclitus Ahr2a, were carried out in an in vitro transcription and translation system. [35S]methionine-labelled Ahr proteins were successfully produced and migrated corresponding to their predicted molecular weight in SDS-PAGE (Fig. 3a). Unlabelled Ahr proteins were then synthesized and their ability to bind [3H]TCDD (2 nM) and [3H]BNF (10 nM) were determined by velocity sedimentation analyses, in which Ahr proteins are separated by sedimentation properties (size and shape) and specific binding to radioligand is measured (Fig. 3b and 3c). Specific binding of [3H]TCDD was observed for all Ahrs assessed, with a distinct peak, between fractions 10 and 20, corresponding to the sedimentation behaviour typical of Ahr proteins28. gmAhr1a and fhAhr2a exhibited a very similar binding profile to [3H]TCDD. However, gmAhr2a demonstrated significantly lower binding to [3H]TCDD than both gmAhr1a and fhAhr2a. Although the reduced signal may partly reflect a weaker expression of gmAhr2a, the significant lower specific binding suggests a lower affinity of gmAhr2a for [3H]TCDD. Notably, [3H]BNF specific binding was also different between the Ahrs; gmAhr1a demonstrated the highest amount of specific binding, while only weak or no specific binding to [3H]BNF was observed for both gmAhr2a and fhAhr2a (Fig. 3c). Although these results are suggestive of differences in binding affinity and specificity among cod Ahr proteins, the binding assay using a single ligand concentration is only qualitative. We therefore performed more quantitative assays for ligand-dependent transactivation in cell culture.
Figure 3. In vitro protein expression and velocity sedimentation assays of Atlantic cod Ahrs.
(a) Autoradiogram of in vitro translated mummichog (Fh) Ahr2a, Atlantic cod (Gm) gmAhr1a and gmAhr2a constructs labelled with [35S]methionine. (b, c) velocity sedimentation assays on sucrose gradients using [3H]TCDD (b) or [3H]BNF (c). Ahrs proteins were expressed in vitro and incubated over night with [3H]TCDD (2 nM) or [3H]BNF (10 nM). Gradients were fractionated and counted in a scintillation counter. Specific binding is the difference between total binding (expressed protein) and nonspecific binding (UPL). Mummichog Ahr2a was used as a positive control and the unprogrammed lysate containing an empty pcDNA3.1 vector (UPL) as negative control. [14C]catalase was added as an internal sedimentation marker.
Ahrs transcriptional activity in COS7-cells.
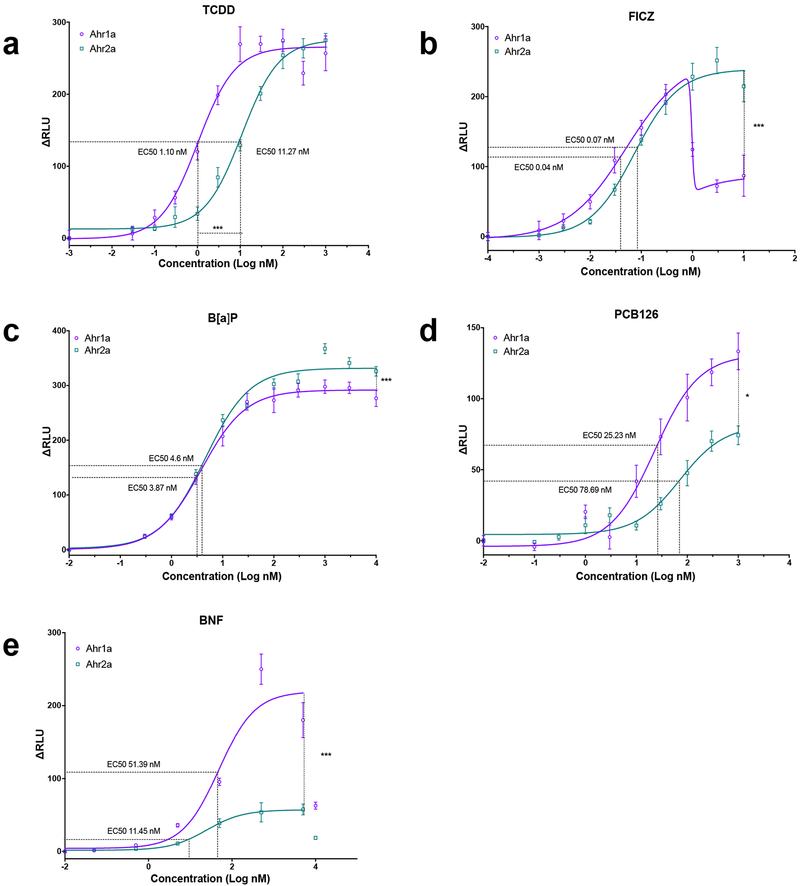
The abilities of gmAhr1a and gmAhr2a to activate transcription of an XRE-controlled luciferase reporter gene were assessed in COS-7 cells in which the gmAhrs were transiently expressed along with the Atlantic cod Arnt1 protein. Five well-known mammalian and piscine Ahr agonists, including TCDD, BNF, B[a]P, PCB126, and FICZ, were tested at increasing concentrations. Both gmAhr1a and gmAhr2a were activated by each of these compounds, but distinct differences in Emax, and EC50 were observed between the receptors (Fig. 4, Table S4). FICZ was found to be the most potent compound, with EC50 in the picomolar range for both gmAhr proteins (Fig. 4b). However, at higher FICZ concentrations, activation of gmAhr1a was significantly lowered, suggesting a bi-phasic concentration-response to this endogenous compound. Interestingly, the activation by TCDD also differed between the two receptors. Although the Emax was very similar, the EC50 values differed by one order of magnitude, where gmAhr1a displayed the lowest EC50 of approximately 1 nM (Fig. 4a). Differences in the activation patterns were revealed also for B[a]P, PCB126 and BNF (Fig. 4c,d,e). For these three compounds, Emax was significantly higher for gmAhr1a. Similarly, as for FICZ, gmAhr1a showed tendencies to a biphasic concentration-response at higher BNF concentrations.
Figure 4. Luciferase reporter gene assays of Atlantic cod Ahr1a and Ahr2a exposed to FICZ, TCDD, B[a]P, PCB126 and BNF.
COS-7 cells were transfected with either the Atlantic cod Ahr1a or Ahr2a, Atlantic cod Arnt1, pGudLuc6.1 Luciferase and pRL-TK Renilla (control) constructs. Cells were exposed to TCDD (0.03–1000 nM) (a) FICZ (0.001–10 nM) (b) B[a]P (0.3–10000 nM) (c) PCB126 (0.1–1000 nM) (d) and (e) BNF (0.05–10000 nM). Relative luciferase units (ΔRLU) was calculated by normalizing the firefly luciferase activity to the transfection control Renilla luciferase activity and to the DMSO average of each assay. The data are presented as mean ± SEM at the different concentrations. EC50 values are indicated as dotted lines in the graphs. Non-linear regression analyses were performed in Prism v7. Statistical differences between EC50 values and maximal activation were obtained using the dose-response analyses drc package in RStudio v1.2.1335. Level of significance is indicated with * (p< 0.05) or *** (p<0.001). With the exception of a slight reduction in cell viability with the highest concentration of TCDD, no significant alterations in cell viabilities were observed (Fig. S2).
Activation of the Ahr signaling pathway in precision-cut liver slices.
The compounds tested for transcriptional activation of gmAhrs in vitro were also tested ex vivo in liver slices using a similar concentration range as used in the in vitro transactivation assay (BNF presented in62). Activation of the Ahr signalling pathway was assessed by measuring altered mRNA expression of cyp1a with qPCR. The two most potent compounds in the in vitro transactivation assay, TCDD and FICZ, also induced cyp1a expression at the lowest concentrations used (1 nM) ex vivo, while B[a]P induced expression of cyp1a at 10 nM (Fig. S12a and S12b). When tested in concentrations up to 10 μM, B[a]P produced very strong transcriptional responses with approx. 400-fold increase in cyp1a expression (Fig. S12c). On the other hand, PCB126 was the least potent compound; inducing cyp1a expression only at 200 and 2000 nM (Fig. S12d). Alterations in the expression of gmahr1a and gmahr2a was also assessed in liver slices exposed to increasing concentrations of TCDD and B[a]P, but no significant differences in transcript levels were observed (Fig. S13).
Discussion
Genome mining identified two divergent gmAhr-encoding genes organized in the same orientation in a tandem pair in linkage group 20 in the Atlantic cod genome. Phylogenetic analyses revealed that these genes belong to the Ahr1a and Ahr2a clades. No other AHR genes were found in the cod genome, indicating that Atlantic cod has not retained the Ahr1b and Ahr2b paralogs that are found in some other fish. Tandem positioning of ahr genes exists in several different fish species, but the retention of the individual Ahr paralogs varies greatly, such as in zebrafish (ahr1b-ahr2(b) tandem, chromosome 22; ahr1a, chromosome 16, GRCz11, Ensembl), Japanese pufferfish (ahr1a-ahr2a tandem, chromosome 1, ahr1b-ahr2b tandem, chromosome 8; ahr2c, chromosome 6, FUGU5, Ensembl), Japanese medaka (ahr1a-ahr2a tandem, chromosome 21; ahr1b-ahr2b tandem, chromosome 2, ASM223467v1, Ensembl) and mummichog (ahr1a-ahr2a tandem, chromosome 1; ahr1b-ahr2b tandem, chromosome 18), Fundulus_heteroclitus-3.0.2,75). The syntenic relationship in the genomic region surrounding the gmAhr genes was also well conserved among Atlantic cod and several other fish species possessing an ahr-tandem pair arrangement. Characteristic Ahr features were present in both gmAhr protein sequences, including the bHLH and PAS domains, as well as functionally important amino acid residues involved in cellular translocation, ligand binding and XRE binding, supporting that the basic functions of Ahr are conserved in both proteins. As observed in Ahr in other teleosts, the sequence similarity varied greatly in the C-terminal transactivation domain, although the different subdomains, including Q-rich, acidic and P/S/T-rich, were identified in both gmAhr1a and gmAhr2a.
All of the common AHR ligands tested in the reporter gene assay bound to and activated both gmAhr proteins. The endogenous compound FICZ was the most potent ligand, although a decrease in gmAhr1a activity at the highest concentrations was observed. High sensitivity to this tryptophan derivative is not surprising since FICZ has been shown to be among the most potent ligands for AHR in many animals, including rodents, human, birds, amphibians and fish40,76–80. Differences in transactivation between gmAhr1a and gmAhr2a were observed for TCDD, B[a]P, PCB126 and BNF. While the two Atlantic cod paralogs had comparable sensitivity to FICZ and B[a]P, gmAhr1a demonstrated the highest sensitivity to TCDD and PCB126. The EC50 values of TCDD differed by one order of magnitude and were calculated to be ~1 nM and ~10 nM for gmAhr1a and gmAhr2a, respectively. The TCDD EC50 value of gmAhr1a is comparable to those observed for Ahr1 in white sturgeon, lake sturgeon, and red seabream, as well as for zebrafish Ahr2 and all of the Atlantic salmon Ahr2 proteins24,30,70,81. A greater sensitivity of Ahr1 to TCDD in fish possessing both Ahr1 and Ahr2 has previously been reported in lake sturgeon, Atlantic sturgeon and red seabream. Significant differences in Emax values between gmAhr1a and gmAhr2a were observed for B[a]P, PCB126, and BNF, where gmAhr1a produced higher maximum responses for PCB126 and BNF. Higher transactivation activity of Ahr1a compared to Ahr2a after PCB126 exposure is in line with previous findings in Atlantic sturgeon and shortnose sturgeon82.
Differences in gmAhr1a and gmAhr2a binding to [3H]TCDD and [3H]BNF were found in in vitro expressed proteins. In accordance with the luciferase reporter gene assay with TCDD and BNF, gmAhr1a was the receptor with the highest binding to both compounds. Differences in binding to [3H]TCDD have been reported previously with Ahr in other teleosts, and appear to vary among different species and Ahr paralogs. Zebrafish Ahr2 and Ahr1b bound with similar magnitude to [3H]TCDD, while the zebrafish Ahr1a did not bind to this compound24,26. Both mummichog Ahr1a and Ahr2a bound [3H]TCDD in a similar manner, but there were some differences in sensitivity to Ahr activation among the four Atlantic salmon Ahr2 proteins24,28,30. Andreasen et al. also showed that zebrafish Ahr2 was capable of binding to [3H]BNF, in contrast to zebrafish Ahr1a that was not26.
Homology modelling and in silico ligand docking of the zebrafish Ahr1a, Ahr1b and Ahr2 suggested that H296 and A386 are important amino acids for binding of TCDD69,83. These predictions were confirmed with in vitro mutagenesis, where replacement of Y296H and T386A restored the ability of zebrafish Ahr1a to bind to both TCDD and DNA69. In silico protein modelling and ligand docking analyses with Ahr1 and Ahr2 from lake sturgeon and white sturgeon suggested a higher sensitivity of white sturgeon Ahr2 to TCDD and dioxin-like compounds due to the presence of the amino acid A388 in its ligand-binding domain70. The presence of A388 would result in a larger binding cavity, which was suggested to provide a more optimal orientation for such compounds. Notably, the residues important for binding and coordination of TCDD in zebrafish and white sturgeon Ahr2, in addition to the amino acids constituting the mammalian “TCDD binding-fingerprint”84, are positionally conserved in gmAhr1a and gmAhr2a. Thus, the discrepancies in ligand binding, sensitivities, and efficacies of gmAhr1a and gmAhr2a observed in this study must be attributed to other structural features present in these protein sequences.
While in vitro ligand activation assays demonstrated that Atlantic cod Ahrs could be ligand activated, activation of the Ahr signalling pathway by FICZ, TCDD, B[a]P, and PCB126 was also confirmed ex vivo in Atlantic cod liver slices. These data are in agreement with previous in vivo and in vitro studies that have reported induced CYP1A protein activity and expression in Atlantic cod exposed to BNF, TCDD, B[a]P and PCB10553,55–57,85,86. In line with the low EC50 values determined for FICZ and TCDD in the Ahr transactivation assay, induction of cyp1a was observed in slices exposed to these two ligands in the low nanomolar range. Interestingly, a greater fold change induction of cyp1a was observed in liver slices exposed to B[a]P and PCB126 as compared to TCDD. The reduced viability of liver slices exposed to 100 nM TCDD may explain the lower induction of cyp1a (Fig. S3). However, the liver of Atlantic cod differs from other fishes due to its high content of lipids87. Lipid droplets present in the hepatocytes might sequester lipophilic compounds and make them less available for the cytosolic receptors55,56. Hence, different distribution and accumulation of these compounds in liver slices may contribute to the differences in cyp1a induction produced by TCDD, B[a]P, and PCB126.
In general, gmahr2a was the most abundant and widely expressed gene in the different tissues sampled, while gmahr1a expression was only detectable in liver, brain, gill and eye. In other fishes, ahr2 is also the most abundant and widely distributed gene, whereas expression of ahr1 is mainly found in brain and heart28–30,36. The opposite case was seen for the arnt transcripts, where arnt1 was the most abundant and detectable gene in many tissues. The almost absence of arnt2 in the liver indicates that gmArnt1 is most likely the heterodimer partner of gmAhr2. The presence of ~10 times more gmahr2a transcripts than gmarh1a in liver tissue suggests an important role of gmahr2a in the recognition of xenobiotics and controlling transcription of biotransformation enzymes.
Toxic responses to TCDD, PCB126 and B[a]P in zebrafish and mummichog have been demonstrated to be mediated by Ahr237–45,88. Other studies also suggested a role of Ahr2 in mediating TCDD toxicity in red seabream, medaka, and Atlantic salmon30,33,89. In spite of gmahr2a being the highest expressed gene in the liver, gmAhr1a was the receptor that demonstrated the strongest binding and highest sensitivity for most of the compounds tested in vitro. This suggests that gmAhr1a may be involved in mediating toxicity responses in tissues where this receptor is expressed. Studies with red seabream and white sturgeon suggested also a role for both Ahr proteins in mediating dioxin toxicity81,90. Moreover, in birds and white sturgeon, a greater sensitivity to dioxin-like compounds has been linked to Ahr1 activity91–93. However, the involvement of Ahr1 in mediating toxicities is still not clear. A recent study compared Ahr1 and Ahr2 EC50 values obtained from transactivation studies and early life stage mortality data from fishes and birds exposed to dioxin-like compounds. Importantly, only a significant linear relationship between Ahr2 activation and early life stage mortality was revealed94.
It is suggested that the ability of AHR to regulate transcription of xenobiotic-metabolizing enzymes in vertebrates is an adaptive function evolved in the vertebrate lineage10. In invertebrates, AHR is involved in development of sensory structures and neural systems (ibid). The physiological roles of AHR in vertebrates is less well understood, but it is now known that AHR participates in different signalling pathways and physiological systems such as the cardiovascular-, reproductive- and immune system6–8. In addition, the multiple Ahr paralogs found in some non-mammalian vertebrates, like fish, may have acquired Ahr protein specialization through subfunction partitioning. Tissue-specific expression patterns, and ligand and target gene specificity are among some partitioning mechanisms. Zebrafish is an example of evolution leading to development of different physiological roles of the Ahrs. As discussed earlier, zfAhr2 has been shown to be involved in mediating toxicity of dioxin-like compounds and PAHs, as well as a suggested a role of in the development of the nervous system43,44,95. On the other hand, Karchner et al. found high expression levels of zfahr1b in zebrafish embryos compared to the other two zfahrs, suggesting a role of zfAhr1b in embryonic development24.
Although zfAhr1a was originally thought to be non-functional (unable to bind TCDD or activate transcription in vitro), it was later shown to be involved in responses to a variety of compounds, including leflunomide, pyrene, and xanthone42,96,97. In a study in zebrafish embryos, the endogenous compound FICZ was shown to bind both zfAhr1b and zfAhr2, but cyp1a induction was largely mediated by zfAhr240. Atlantic cod gmAhr1a and gmAhr2a receptors are also very sensitive to FICZ, which could indicate that both receptors are involved in controlling physiological responses. Importantly, the different tissue expression profiles, ligand binding affinities, and transactivation activities also support the idea that subfunction partitioning of Ahr has occurred in Atlantic cod. Higher levels of gmahr2a expression in the liver indicate that this protein is most likely involved in mediating xenobiotic responses in Atlantic cod, as it is in other fishes. Moreover, the high sensitivity of gmAhr1a to the different ligands tested does not exclude the possibility of gmAhr1a activity being modulated by certain pollutants. Atlantic cod’s chemical defensome is slightly different compared to several other fishes because of the lack of Pxr. Hence, a role of both paralogs in mediating responses to pollutants may be a compensatory functional role to modulate xenobiotic responses, as previously suggested58. Further studies, such as additional Ahr transactivation studies using PXR ligand compounds, expression and localization patterns in Atlantic cod embryos, as well as mutagenesis and gene knock-out studies, may help to elucidate the specific roles of the gmAhrs.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Bergen Marine Research Cluster for the Ocean Outlook fellowship to LA-A, and the Biology Department of Woods Hole Oceanographic Institution for hosting LA-A as part of this study and providing research facilities to conduct velocity sedimentation- and luciferase reporter gene assays. We thank the national cod breeding program in Norway, Nofima, for providing farmed Atlantic cod for this project. We also wish to thank Pål Olsvik for helping with the analyses of the qPCR data, Knut Helge Jensen for helping with statistical analysis, Pernille Iden for technical assistance and Aintzane Santaquiteria-Gil for making the phylogenetic tree.
Funding
This study was funded by Research Council of Norway through the iCod 2.0: Integrative environmental genomics of Atlantic cod project (project no. 244564) and the dCod 1.0: decoding systems toxicology of Atlantic cod project (Center for Digital Life Norway project no. 248840). This research was also supported by an Ocean Outlook fellowship to LA-A and by the U.S. National Institute of Environmental Health Sciences (Superfund Research Program at Boston University, grant P42ES007381) which provided funding to carry out part of this study at WHOI.
Footnotes
Supporting information
Method details, cloning primers, COS-7 transfection details, cytotoxicity/viability measurements, multiple sequence alignments, amino acid composition analyses, and quantitative real-time PCR analyses.
The authors declare no competing financial interest.
REFERENCES
- (1).Denison MS; Nagy SR Activation of the Aryl Hydrocarbon Receptor by Structurally Diverse Exogenous and Endogenous Chemicals. Annu. Rev. Pharmacol. Toxicol 2003, 43 (1), 309–334. 10.1146/annurev.pharmtox.43.100901.135828. [DOI] [PubMed] [Google Scholar]
- (2).Fernandez-Salguero PM; Hllbert DM; Rudikoff S; Ward JM; Gonzalez FJ Aryl-Hydrocarbon Receptor-Deficient Mice Are Resistant to 2,3,7,8-Tetrachlorodibenzop-Dioxin-Induced Toxicity. Toxicol. Appl. Pharmacol 1996, 140 (1), 173–179. 10.1006/taap.1996.0210. [DOI] [PubMed] [Google Scholar]
- (3).Mimura J; Fujii-Kuriyama Y Functional Role of AhR in the Expression of Toxic Effects by TCDD. Biochim. Biophys. Acta - Gen. Subj 2003, 1619 (3), 263–268. 10.1016/S0304-4165(02)00485-3. [DOI] [PubMed] [Google Scholar]
- (4).Mimura J; Yamashita K; Nakamura K; Morita M; Takagi TN; Nakao K; Ema M; Sogawa K; Yasuda M; Katsuki M; Fujii-Kuriyama Y Loss of Teratogenic Response to 2,3,7,8-Tetrachlorodibenzo-p-Dioxin (TCDD) in Mice Lacking the Ah (Dioxin) Receptor. Genes to cells 1997, 2 (10), 645–654. 10.1046/j.1365-2443.1997.1490345.x. [DOI] [PubMed] [Google Scholar]
- (5).Soshilov AA; Denison MS Ligand Promiscuity of Aryl Hydrocarbon Receptor Agonists and Antagonists Revealed by Site-Directed Mutagenesis. Mol. Cell. Biol 2014, 34 (9), 1707–1719. 10.1128/mcb.01183-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Nebert DW Aryl Hydrocarbon Receptor (AHR): “Pioneer Member” of the Basic-Helix/Loop/Helix per-Arnt-Sim (BHLH/PAS) Family of “Sensors” of Foreign and Endogenous Signals. Prog. Lipid Res 2017, 67 (June), 38–57. 10.1016/j.plipres.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Mulero-Navarro S; Fernandez-Salguero PM New Trends in Aryl Hydrocarbon Receptor Biology. Front. Cell Dev. Biol 2016, 4 (May), 1–14. 10.3389/fcell.2016.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Esser C; Rannug A The Aryl Hydrocarbon Receptor in Barrier Organ Physiology, Immunology, and Toxicology. Pharmacol. Rev 2015, 67 (2), 259–279. 10.1124/pr.114.009001. [DOI] [PubMed] [Google Scholar]
- (9).Hahn ME Aryl Hydrocarbon Receptors : Diversity and Evolution 1. Chem. Biol. Interact 2002, 141, 131–160. [DOI] [PubMed] [Google Scholar]
- (10).Hahn ME; Karchner SI; Merson RR Diversity as Opportunity: Insights from 600 Million Years of AHR Evolution. Curr. Opin. Toxicol 2017, 2, 58–71. 10.1016/j.cotox.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Perdew GH Association of the Ah Receptor with the 90-KDa Heat Shock Protein *. J. Biol. Chem 1988, 263 (27), 13802–13805. [PubMed] [Google Scholar]
- (12).Carver LA; Bradfield CA Ligand-Dependent Interaction of the Aryl Hydrocarbon Receptor with a Novel Immunophilin Homolog in Vivo*. J. Biol. Chem 1997, 272 (17), 11452–11456. [DOI] [PubMed] [Google Scholar]
- (13).Kazlauskas A; Poellinger L; Pongratz I Evidence That the Co-Chaperone P23 Regulates Ligand Responsiveness of the Dioxin (Aryl Hydrocarbon) Receptor. J. Biol. Chem 1999, 274 (19), 13519–13524. 10.1074/jbc.274.19.13519. [DOI] [PubMed] [Google Scholar]
- (14).Denison MS; Soshilov AA; He G; Degroot DE; Zhao B Exactly the Same but Different : Promiscuity and Diversity in the Molecular Mechanisms of Action of the Aryl Hydrocarbon ( Dioxin ) Receptor. Toxicol. Sci 2011, 124 (1), 1–22. 10.1093/toxsci/kfr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Pongratz I; Mason GF; Poellinger L Dual Roles of the 90-KDa Heat Shock Protein Hsp90 in Modulating Functional Activities of the Dioxin Receptor RECEPTORS WHICH REQUIRE Hsp90 BOTH FOR LIGAND BINDING ACTIVITY AND REPRESSION OF. J. Biol. Chem 1992, 267 (19), 13728–13734. [PubMed] [Google Scholar]
- (16).Meyer BK; Perdew GH Characterization of the AhR-Hsp90-XAP2 Core Complex and the Role of the Immunophilin-Related Protein XAP2 in AhR Stabilization. Biochemistry 1999, 38 (28), 8907–8917. 10.1021/bi982223w. [DOI] [PubMed] [Google Scholar]
- (17).Kudo I; Hosaka M; Haga A; Tsuji N; Nagata Y; Okada H; Fukuda K; Kakizaki Y; Okamoto T; Grave E; Itoh H The Regulation Mechanisms of AhR by Molecular Chaperone Complex. J. Biochem 2018, 163 (3), 223–232. 10.1093/jb/mvx074. [DOI] [PubMed] [Google Scholar]
- (18).Antonsson C; Whitelaw ML; McGuire J; Gustafsson JA; Poellinger L Distinct Roles of the Molecular Chaperone Hsp90 in Modulating Dioxin Receptor Function via the Basic Helix-Loop-Helix and PAS Domains. Mol. Cell. Biol 2015, 15 (2), 756–765. 10.1128/mcb.15.2.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Rothhammer V; Quintana FJ The Aryl Hydrocarbon Receptor: An Environmental Sensor Integrating Immune Responses in Health and Disease. Nat. Rev. Immunol 2019, 19 (3), 184–197. 10.1038/s41577-019-0125-8. [DOI] [PubMed] [Google Scholar]
- (20).Pollenz RS; Sattler CA; Poland A The Aryl Hydrocarbon Receptor and Aryl Hydrocarbon Receptor Nuclear Translocator Protein Show Distinct Subcellular Localizations in Hepa 1 Ci C7 Cells by Immunfluorescence Microscopy. Mol. Pharmacol 1993, 45, 428–438. [PubMed] [Google Scholar]
- (21).Soshilov A; Denison MS Role of the Per/Arnt/Sim Domains in Ligand-Dependent Transformation of the Aryl Hydrocarbon Receptor. J. Biol. Chem 2008, 283 (47), 32995–33005. 10.1074/jbc.M802414200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Swanson HI DNA Binding and Protein Interactions of the AHR/ARNT Heterodimer That Facilitate Gene Activation. Chem. Biol. Interact 2002, 141 (1–2), 63–76. 10.1016/S0009-2797(02)00066-2. [DOI] [PubMed] [Google Scholar]
- (23).Whitlock JPJ Induction of Cytochrome P4501a1. Annu. Rev. Pharmacol. Toxicol 1999, 39 (1), 103–125. 10.1146/annurev.pharmtox.39.1.103. [DOI] [PubMed] [Google Scholar]
- (24).Karchner SI; Franks DG; Hahn ME AHR1B, a New Functional Aryl Hydrocarbon Receptor in Zebrafish: Tandem Arrangement of Ahr1b and Ahr2 Genes. Biochem. J 2005, 392 (1), 153–161. 10.1042/BJ20050713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Glasauer SMK; Neuhauss SCF Whole-Genome Duplication in Teleost Fishes and Its Evolutionary Consequences. Mol. Genet. Genomics 2014, 289 (6), 1045–1060. 10.1007/s00438-014-0889-2. [DOI] [PubMed] [Google Scholar]
- (26).Andreasen EA; Hahn ME; Heideman W; Peterson RE; Tanguay RL The Zebrafish (Danio Rerio) Aryl Hydrocarbon Receptor Type 1 Is a Novel Vertebrate Receptor. Mol. Pharmacol 2002, 62 (2), 234–249. 10.1124/mol.62.2.234. [DOI] [PubMed] [Google Scholar]
- (27).Tanguay RL; Abnet CC; Heideman W; Peterson RE Cloning and Characterization of the Zebrafish (Danio Rerio) Aryl Hydrocarbon Receptor. Biochim. Biophys. Acta 1999, 1444 (1), 35–48. 10.1016/S0167-4781(98)00252-8. [DOI] [PubMed] [Google Scholar]
- (28).Karchner SI; Powell WH; Hahn ME Identification and Functional Characterization of Two Highly Divergent Aryl Hydrocarbon Receptors ( AHR1 and AHR2 ) in the Teleost Fundulus Heteroclitus. J. Biol. Chem 1999, 274 (47), 33814–33824. [DOI] [PubMed] [Google Scholar]
- (29).Yamauchi M; Kim EY; Iwata H; Tanabe S Molecular Characterization of the Aryl Hydrocarbon Receptors (AHR1 and AHR2) from Red Seabream (Pagrus Major). Comp. Biochem. Physiol. - C Toxicol. Pharmacol 2005, 141 (2), 177–187. 10.1016/j.cca.2005.06.003. [DOI] [PubMed] [Google Scholar]
- (30).Hansson MC; Hahn ME Functional Properties of the Four Atlantic Salmon (Salmo Salar) Aryl Hydrocarbon Receptor Type 2 (AHR2) Isoforms. Aquat. Toxicol 2008, 86 (2), 121–130. 10.1016/j.aquatox.2007.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Hansson MC; Wittzell H; Persson K; Schantz T Von. Unprecedented Genomic Diversity of AhR1 and AhR2 Genes in Atlantic Salmon (Salmo Salar L.). Aquat. Toxicol 2004, 68, 219–232. 10.1016/j.aquatox.2004.02.006. [DOI] [PubMed] [Google Scholar]
- (32).Hansson MC; Wittzell H; Persson K; Von Schantz T Characterization of Two Distinct Aryl Hydrocarbon Receptor (AhR2) Genes in Atlantic Salmon (Salmo Salar) and Evidence for Multiple AhR2 Gene Lineages in Salmonid Fish. Gene 2003, 303, 197–206. 10.1016/S0378-1119(02)01178-2. [DOI] [PubMed] [Google Scholar]
- (33).Hanno K; Oda S; Mitani H Effects of Dioxin Isomers on Induction of AhRs and CYP1A1 in Early Developmental Stage Embryos of Medaka (Oryzias Latipes). Chemosphere 2010, 78, 830–839. 10.1016/j.chemosphere.2009.11.043. [DOI] [PubMed] [Google Scholar]
- (34).Karchner SI; Hahn ME Pufferfish (Fugu Rubripes) Aryl Hydrocarbon Receptors: Unusually High Diversity in a Compact Genome. Mar. Environ. Res 2004, 58, 139–140. 10.1016/j.marenvres.2004.03.105. [DOI] [Google Scholar]
- (35).Roy NK; Wirgin I Characterization of the Aromatic Hydrocarbon Receptor Gene and Its Expression in Atlantic Tomcod. Arch. Biochem. Biophys 1997, 344 (2), 373–386. 10.1006/abbi.1997.0238. [DOI] [PubMed] [Google Scholar]
- (36).Abnet CC; Tanguay RL; Hahn ME; Heideman W; Peterson RE Two Forms of Aryl Hydrocarbon Receptor Type 2 in Rainbow Trout ( Oncorhynchus Mykiss). J. Biol. Chem 1999, 274 (21), 15159–15166. 10.1074/jbc.274.21.15159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Prasch AL; Teraoka H; Carney SA; Dong W; Hiraga T; Stegeman JJ; Heideman W; Peterson RE Aryl Hydrocarbon Receptor 2 Mediates 2,3,7,8-Tetrachlorodibenzo-p-Dioxin Developmental Toxicity in Zebrafish. Toxicol. Sci 2003, 76 (1), 138–150. 10.1093/toxsci/kfg202. [DOI] [PubMed] [Google Scholar]
- (38).Jönsson ME; Jenny MJ; Woodin BR; Hahn ME; Stegeman JJ Role of AHR2 in the Expression of Novel Cytochrome P450 1 Family Genes, Cell Cycle Genes, and Morphological Defects in Developing Zebra Fish Exposed to 3,31’,4,4’,5-Pentachlorobiphenyl or 2,3,7,8-Tetrachlorodibenzo-p-Dioxin. Toxicol. Sci 2007, 100 (1), 180–193. 10.1093/toxsci/kfm207. [DOI] [PubMed] [Google Scholar]
- (39).Yin HC; Tseng HP; Chung HY; Ko CY; Tzou WS; Buhler DR; Hu CH Influence of TCDD on Zebrafish CYP1B1 Transcription during Development. Toxicol. Sci 2008, 103 (1), 158–168. 10.1093/toxsci/kfn035. [DOI] [PubMed] [Google Scholar]
- (40).Jönsson ME; Franks DG; Woodin BR; Jenny MJ; Garrick RA; Behrendt L; Hahn ME; Stegeman JJ The Tryptophan Photoproduct 6-Formylindolo[3,2-b]Carbazole (FICZ) Binds Multiple AHRs and Induces Multiple CYP1 Genes via AHR2 in Zebrafish. Chem. Biol. Interact 2009, 181 (3), 447–454. 10.1016/j.cbi.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Clark BW; Matson CW; Jung D; Di Giulio RT AHR2 Mediates Cardiac Teratogenesis of Polycyclic Aromatic Hydrocarbons and PCB-126 in Atlantic Killifish (Fundulus Heteroclitus). Aquat. Toxicol 2010, 99 (2), 232–240. 10.1016/j.aquatox.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Goodale BC; la Du JK; Bisson WH; Janszen DB; Waters KM; Tanguay RL AHR2 Mutant Reveals Functional Diversity of Aryl Hydrocarbon Receptors in Zebrafish. PLoS One 2012, 7 (1), 1–13. 10.1371/journal.pone.0029346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Garcia GR; Bugel SM; Truong L; Spagnoli S; Tanguay RL AHR2 Required for Normal Behavioral Responses and Proper Development of the Skeletal and Reproductive Systems in Zebrafish. PLoS One 2018, 13 (3), 1–21. 10.1371/journal.pone.0193484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Souder JP; Gorelick DA Ahr2, But Not Ahr1a or Ahr1b, Is Required for Craniofacial and Fin Development and TCDD- Dependent Cardiotoxicity in Zebrafish. Toxicol. Sci 2019, 170 (1), 25–44. 10.1093/toxsci/kfz075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Sugden WW; Leonardo-Mendonça RC; Acuña-Castroviejo D; Siekmann AF Genetic Dissection of Endothelial Transcriptional Activity of Zebrafish Aryl Hydrocarbon Receptors (AHRs). PLoS One 2017, 12 (8), 1–22. 10.1371/journal.pone.0183433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Karchner S; Jenny M; Aluru N; Franks D; Laub L; Linney E; Williams L; Teraoka H; Hahn M Evidence for Developmental versus Toxicological Roles for Zebrafish AHR1b. Toxicol. Sci. (The Toxicologist Suppl 2017, 156 (S39), Abstract #1165. 10.1016/j.jhsa.2018.06.116. [DOI] [Google Scholar]
- (47).Hylland K; Tollefsen KE; Ruus A; Jonsson G; Sundt RC; Sanni S; Røe Utvik TI; Johnsen S; Nilssen I; Pinturier L; Balk L; Baršiene J; Marigòmez I; Feist W; Børseth JF. Water Column Monitoring near Oil Installations in the North Sea 2001–2004. Mar. Pollut. Bull 2008, 56 (3), 414–429. 10.1016/j.marpolbul.2007.11.004. [DOI] [PubMed] [Google Scholar]
- (48).OSPAR Assessment of Impacts of Offshore Oil and Gas Activities in the North-East Atlantic Offshore Industry Series; London, 2009. [Google Scholar]
- (49).Sundt RC; Ruus A; Jonsson H; Skarphédinsdóttir H; Meier S; Grung M; Beyer J; Pampanin DM Biomarker Responses in Atlantic Cod (Gadus Morhua) Exposed to Produced Water from a North Sea Oil Field: Laboratory and Field Assessments. Mar. Pollut. Bull 2012, 64 (1), 144–152. 10.1016/j.marpolbul.2011.10.005. [DOI] [PubMed] [Google Scholar]
- (50).Dale K; Müller MB; Tairova Z; Khan EA; Hatlen K; Grung M; Yadetie F; Lille-Langøy R; Blaser N; Skaug HJ; Lyche JL; Arukwe A; Hylland K; Karlsen OA; Goksøyr A Contaminant Accumulation and Biological Responses in Atlantic Cod (Gadus Morhua) Caged at a Capped Waste Disposal Site in Kollevåg, Western Norway. Mar. Environ. Res 2019, 145, 39–51. 10.1016/j.marenvres.2019.02.003. [DOI] [PubMed] [Google Scholar]
- (51).Lie KK; Meier S; Olsvik PA Effects of Environmental Relevant Doses of Pollutants from Offshore Oil Production on Atlantic Cod (Gadus Morhua). Comp. Biochem. Physiol. - C Toxicol. Pharmacol 2009, 150 (2), 141–149. 10.1016/j.cbpc.2009.04.004. [DOI] [PubMed] [Google Scholar]
- (52).Sturve J; Hasselberg L; Fälth H; Celander M; Förlin L Effects of North Sea Oil and Alkylphenols on Biomarker Responses in Juvenile Atlantic Cod (Gadus Morhua). Aquat. Toxicol 2006, 78, 73–78. 10.1016/j.aquatox.2006.02.019. [DOI] [PubMed] [Google Scholar]
- (53).Goksøyr A; Beyer J; Husøy A-M; Larsen HE; Westrheim K; Wilhelmsen S; Klungsøyr J Accumulation and Effects of Aromatic and Chlorinated Hydrocarbons in Juvenile Atlantic Cod (Gadus Morhua) Caged in a Polluted Fjord (Sørfjorden, Norway). Aquatic Toxicology. 1994, pp 21–35. 10.1016/0166-445X(94)90045-0. [DOI] [Google Scholar]
- (54).Husøy A-M; Myers MS; Goksøyr A Cellular Localization of Cytochrome P450 (CYPl A) Induction and Histology in Atlantic Cod (Gadus Morhua L.) and European Flounder (Platichthys Flesus) after Environmental Exposure to Contaminants by Caging in Sarrfiorden, Norway. Aquat. Toxicol 1996, 36, 53–74. [Google Scholar]
- (55).Hektoen H; Bernhoft A; Ingebrigtsen K; Utne Skaare J; Goksøyr A Response of Hepatic Xenobiotic Metabolizing Enzymes in Rainbow Trout (Oncorhynchus Mykiss) and Cod (Gadus Morhua) to 2,3,7,8-Tetrachlorodibenzo-p-Dioxin (2,3,7,8-TCDD). Aquat. Toxicol 1994, 28 (1–2), 97–106. 10.1016/0166-445X(94)90023-X. [DOI] [Google Scholar]
- (56).Bernhoft A; Hektoen H; Utne Skaare J; Ingebrigtsen K Tissue Distribution and Effects on Hepatic Xenobiotic Metabolising Enzymes of 2,3,3′,4,4′-Pentachlorobiphenyl (PCB-105) in COD (Gadus Morhua) and Rainbow Trout (Oncorhynchus Mykiss). Environ. Pollut 1994, 85 (3), 351–359. 10.1016/0269-7491(94)90058-2. [DOI] [PubMed] [Google Scholar]
- (57).Husøy A-M; Myers MS; Willis ML; Collier TK; Celander M; Goksoyr A Immunohistochemical Localization of CYP1A-Like and CYP3A-like Isozymes in Hepatic and Extrahepatic Tissues of Atlantic Cod (Gadus Morhua L), a Marine Fish. Toxicol. Appl. Pharmacol 1994, 129, 294–308. 10.1006/taap.1994.1254. [DOI] [PubMed] [Google Scholar]
- (58).Eide M; Rydbeck H; Tørresen OK; Lille-Langøy R; Puntervoll P; Goldstone JV; Jakobsen KS; Stegeman J; Goksøyr A; Karlsen OA Independent Losses of a Xenobiotic Receptor across Teleost Evolution. Sci. Rep 2018, 8 (1), 1–13. 10.1038/s41598-018-28498-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Pérez-Albaladejo E; Rizzi J; Fernandes D; Lille-Langøy R; Karlsen OA; Goksøyr A; Oros A; Spagnoli F; Porte C Assessment of the Environmental Quality of Coastal Sediments by Using a Combination of in Vitro Bioassays. Mar. Pollut. Bull 2016, 108, 54–61. 10.1016/j.marpolbul.2016.04.063. [DOI] [PubMed] [Google Scholar]
- (60).Olsvik PA; Søfteland L; Lie KK Selection of Reference Genes for QRT-PCR Examination of Wild Populations of Atlantic Cod Gadus Morhua. BMC Res. Notes 2008, 1, 1–9. 10.1186/1756-0500-1-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Livak KJ; Schmittgen TD Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2–ΔΔCT Method. Methods 2001, 25 (4), 402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- (62).Eide M; Karlsen OA; Kryvi H; Olsvik PA; Goksøyr A Precision-Cut Liver Slices of Atlantic Cod ( Gadus Morhua ): An in Vitro System for Studying the Effects of Environmental Contaminants. Aquat. Toxicol 2014, 153, 110–115. 10.1016/j.aquatox.2013.10.027. [DOI] [PubMed] [Google Scholar]
- (63).Moronvalle-Halley V; Sacré-Salem B; Sallez V; Labbe G; Gautier JC Evaluation of Cultured, Precision-Cut Rat Liver Slices as a Model to Study Drug-Induced Liver Apoptosis. Toxicology 2005, 207 (2), 203–214. 10.1016/j.tox.2004.09.014. [DOI] [PubMed] [Google Scholar]
- (64).Hubert S; Higgins B; Borza T; Bowman S Development of a SNP Resource and a Genetic Linkage Map for Atlantic Cod (Gadus Morhua). BMC Genomics 2010, 11 (1). 10.1186/1471-2164-11-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Tørresen OK; Star B; Jentoft S; Reinar WB; Grove H; Miller JR; Walenz BP; Knight J; Ekholm JM; Peluso P; et al. An Improved Genome Assembly Uncovers Prolific Tandem Repeats in Atlantic Cod. BMC Genomics 2017, 1–23. 10.1186/s12864-016-3448-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Ikuta T; Watanabe J; Kawajiri K Characterization of the LxxLL Motif in the Aryl Hydrocarbon Receptor: Effects on Subcellular Localization and Transcriptional Activity. J. Biochem 2002, 131 (1), 79–85. [DOI] [PubMed] [Google Scholar]
- (67).Ikuta T; Eguchi H; Tachibana T; Yoneda Y; Kawajiri K Nuclear Localization and Export Signals of the Human Aryl Hydrocarbon Receptor. J. Biol. Chem 1998, 273 (5), 2895–2904. 10.1074/jbc.273.5.2895. [DOI] [PubMed] [Google Scholar]
- (68).Berg P; Pongratz I Differential Usage of Nuclear Export Sequences Regulates Intracellular Localization of the Dioxin (Aryl Hydrocarbon) Receptor. J. Biol. Chem 2001, 276 (46), 43231–43238. 10.1074/jbc.M105261200. [DOI] [PubMed] [Google Scholar]
- (69).Fraccalvieri D; Soshilov AA; Karchner SI; Franks DG; Pandini A; Bonati L; Hahn ME; Denison MS Comparative Analysis of Homology Models of the Ah Receptor Ligand Binding Domain: Verification of Structure-Function Predictions by Site-Directed Mutagenesis of a Nonfunctional Receptor. Biochemistry 2013, 52 (4), 714–725. 10.1021/bi301457f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Doering JA; Farmahin R; Wiseman S; Beitel SC; Kennedy SW; Giesy JP; Hecker M Differences in Activation of Aryl Hydrocarbon Receptors of White Sturgeon Relative to Lake Sturgeon Are Predicted by Identities of Key Amino Acids in the Ligand Binding Domain. Environ. Sci. Technol 2015, 49 (7), 4681–4689. 10.1021/acs.est.5b00085. [DOI] [PubMed] [Google Scholar]
- (71).Swanson HI; Yang JH Mapping the Protein/DNA Contact Sites of the Ah Receptor and Ah Receptor Nuclear Translocator. J. Biol. Chem 1996, 271 (49), 31657–31665. 10.1074/jbc.271.49.31657. [DOI] [PubMed] [Google Scholar]
- (72).Bacsi SG; Hankinson O Functional Characterization of DNA-Binding Domains of the Subunits of the Heterodimeric Aryl Hydrocarbon Receptor Complex Imputing Novel and Canonical Basic Helix-Loop-Helix Protein-DNA Interactions. J. Biol. Chem 1996, 271 (15), 8843–8850. 10.1074/jbc.271.15.8843. [DOI] [PubMed] [Google Scholar]
- (73).Dong L; Ma Q; Whitlock JPJ DNA Binding by the Heterodimeric Ah Receptor. J. Biol. Chem 1996, 271 (14), 7942–7948. 10.1074/jbc.271.14.7942. [DOI] [PubMed] [Google Scholar]
- (74).Rowlands JC; McEwan IJ; Gustafsson J-Å Trans-Activation by the Human Aryl Hydrocarbon Receptor and Aryl Hydrocarbon Receptor Nuclear Translocator Proteins: Direct Interactions with Basal Transcription Factors. Mol. Pharmacol 1996, 50 (3), 538–548. [PubMed] [Google Scholar]
- (75).Miller JT; Reid N; Nacci D; Whitehead A Developing a High-Quality Linkage Map for the Atlantic Killifish Fundulus Heteroclitus. G3 Genes, Genomes, Genet 2019, 1–12. 10.1534/g3.119.400262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Eide M; Rusten M; Male R; Jensen KHM; Goksøyr A A Characterization of the ZFL Cell Line and Primary Hepatocytes as in Vitro Liver Cell Models for the Zebrafish (Danio Rerio). Aquat. Toxicol 2014, 147, 7–17. 10.1016/j.aquatox.2013.11.023. [DOI] [PubMed] [Google Scholar]
- (77).Rannug A; Rannug U; Rosenkranz HS; Winqvist L; Westerholm R; Agurell E; Grafstrom A-K Certain Photooxidized Derivatives of Tryptophan Bind with Very High Affinity to the Ah Receptor and Are Likely to Be Endogenous Signal Substances. J. Biol. Chem 1987, 262 (32), 15–427. [PubMed] [Google Scholar]
- (78).Schmidt JV; Su GH; Reddy JK; Simon MC; Bradfield CA; Schwarz T; Hübenthal U; Cline JE; Hajimiragha H; Schroeder P; Klotz LO; Rannug A; Fürst P; Hanenberg H; Abel J; Krutmann J Characterization of a Murine Ahr Null Allele: Involvement of the Ah Receptor in Hepatic Growth and Development. Proc. Natl. Acad. Sci 1996, 93 (13), 6731–6736. 10.1073/pnas.93.13.6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Laub LB; Jones BD; Powell WH Responsiveness of a Xenopus Laevis Cell Line to the Aryl Hydrocarbon Receptor Ligands 6-Formylindolo[3,2-b]Carbazole (FICZ) and 2,3,7,8-Tetrachlorodibenzo-p-Dioxin (TCDD). Chem. Biol. Interact 2010, 183 (1), 202–211. 10.1016/j.cbi.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Farmahin R; Crump D; Kennedy SW Sensitivity of Avian Species to the Aryl Hydrocarbon Receptor Ligand 6-Formylindolo [3,2-b] Carbazole (FICZ). Chem. Biol. Interact 2014, 221, 62–69. 10.1016/j.cbi.2014.07.009. [DOI] [PubMed] [Google Scholar]
- (81).Bak SM; Iida M; Hirano M; Iwata H; Kim EY Potencies of Red Seabream AHR1 and AHR2-Mediated Transactivation by Dioxins: Implication of Both AHRs in Dioxin Toxicity. Environ. Sci. Technol 2013, 47 (6), 2877–2885. 10.1021/es304423w. [DOI] [PubMed] [Google Scholar]
- (82).Roy NK; DellaTorre M; Candelmo A; Chambers RC; Habeck E; Wirgin I Characterization of AHR1 and Its Functional Activity in Atlantic Sturgeon and Shortnose Sturgeon. Aquat. Toxicol 2018, 205 (September), 25–35. 10.1016/j.aquatox.2018.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Bisson WH; Koch DC; O’Donnell EF; Khalil SM; Kerkvliet NI; Tanguay RL; Abagyan R; Kolluri SK Modeling of the Aryl Hydrocarbon Receptor (AhR) Ligand Binding Domain and Its Utility in Virtual Ligand Screening to Predict New AhR Ligands. J. Med. Chem 2009, 52 (18), 5635–5641. 10.1021/jm900199u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Pandini A; Soshilov AA; Song Y; Zhao J; Bonati L; Denison MS Detection of the TCDD Binding-Fingerprint within the Ah Receptor Ligand Binding Domain by Structurally Driven Mutagenesis and Functional Analysis. Biochemistry 2009, 48 (25), 5972–5983. 10.1021/bi900259z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Yadetie F; Zhang X; Hanna EM; Aranguren-Abadía L; Eide M; Blaser N; Brun M; Jonassen I; Goksøyr A; Karlsen OA RNA-Seq Analysis of Transcriptome Responses in Atlantic Cod (Gadus Morhua) Precision-Cut Liver Slices Exposed to Benzo[a]Pyrene and 17A-Ethynylestradiol. Aquat. Toxicol 2018, 201, 174–186. 10.1016/j.aquatox.2018.06.003. [DOI] [PubMed] [Google Scholar]
- (86).Søfteland L; Holen E; Olsvik PA Toxicological Application of Primary Hepatocyte Cell Cultures of Atlantic Cod (Gadus Morhua) — Effects of BNF, PCDD and Cd. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol 2010, 151 (4), 401–411. 10.1016/J.CBPC.2010.01.003. [DOI] [PubMed] [Google Scholar]
- (87).Fujita H; Tatsumi H; Ban T; Tamura S Fine-Structural Characteristics of the Liver of the Cod (Gadus Morhua Macrocephalus), with Special Regard to the Concept of a Hepatoskeletal System Formed by Ito Cells. Cell Tissue Res 1986, 244 (1), 63–67. 10.1007/BF00218382. [DOI] [Google Scholar]
- (88).Incardona JP; Linbo TL; Scholz NL Cardiac Toxicity of 5-Ring Polycyclic Aromatic Hydrocarbons Is Differentially Dependent on the Aryl Hydrocarbon Receptor 2 Isoform during Zebrafish Development. Toxicol. Appl. Pharmacol 2011, 257, 242–249. 10.1016/j.taap.2011.09.010. [DOI] [PubMed] [Google Scholar]
- (89).Yamauchi M; Kim EY; Iwata H; Shima Y; Tanabe S Toxic Effects of 2,3,7,8-Tetrachlorodibenzo-p-Dioxin (TCDD) in Developing Red Seabream (Pagrus Major) Embryo: An Association of Morphological Deformities with AHR1, AHR2 and CYP1A Expressions. Aquat. Toxicol 2006, 80, 166–179. 10.1016/j.aquatox.2006.08.006. [DOI] [PubMed] [Google Scholar]
- (90).Doering JA; Farmahin R; Wiseman S; Kennedy SW; Giesy JP; Hecker M Functionality of Aryl Hydrocarbon Receptors (AhR1 and AhR2) of White Sturgeon (Acipenser Transmontanus) and Implications for the Risk Assessment of Dioxin-like Compounds. Environ. Sci. Technol 2014, 48 (14), 8219–8226. 10.1021/es502054h. [DOI] [PubMed] [Google Scholar]
- (91).Doering JA; Wiseman S; Beitel SC; Giesy JP; Hecker M Identification and Expression of Aryl Hydrocarbon Receptors (AhR1 and AhR2) Provide Insight in an Evolutionary Context Regarding Sensitivity of White Sturgeon (Acipenser Transmontanus) to Dioxin-like Compounds. Aquat. Toxicol 2014, 150, 27–35. 10.1016/j.aquatox.2014.02.009. [DOI] [PubMed] [Google Scholar]
- (92).Karchner SI; Franks DG; Kennedy SW; Hahn ME The Molecular Basis for Differential Dioxin Sensitivity in Birds: Role of the Aryl Hydrocarbon Receptor. Proc. Natl. Acad. Sci. U. S. A 2006, 103 (16), 6252–6257. 10.1073/pnas.0509950103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (93).Head JA; Hahn ME; Kennedy SW Key Amino Acids in the Aryl Hydrocarbon Receptor Predict Dioxin Sensitivity in Avian Species. Environ. Sci. Technol 2008, 42 (19), 7535–7541. 10.1021/es801082a. [DOI] [PubMed] [Google Scholar]
- (94).Doering JA; Wiseman S; Giesy JP; Hecker M A Cross-Species Quantitative Adverse Outcome Pathway for Activation of the Aryl Hydrocarbon Receptor Leading to Early Life Stage Mortality in Birds and Fishes. Environ. Sci. Technol 2018, 52 (13), 7524–7533. 10.1021/acs.est.8b01438. [DOI] [PubMed] [Google Scholar]
- (95).Andreasen EA; Spitsbergen JM; Tanguay RL; Stegeman JJ; Heideman W; Peterson RE Tissue-Specific Expression of AHR2, ARNT2, and CYP1A in Zebrafish Embryos and Larvae : Effects of Developmental Stage and 2, 3, 7, 8-Tetrachlorodibenzop -Dioxin Exposure. Toxicol. Sci 2002, 68, 403–419. [DOI] [PubMed] [Google Scholar]
- (96).Incardona JP; Day HL; Collier TK; Scholz NL Developmental Toxicity of 4-Ring Polycyclic Aromatic Hydrocarbons in Zebrafish Is Differentially Dependent on AH Receptor Isoforms and Hepatic Cytochrome P4501A Metabolism. Toxicol. Appl. Pharmacol 2006, 217, 308–321. 10.1016/j.taap.2006.09.018. [DOI] [PubMed] [Google Scholar]
- (97).Knecht AL; Goodale BC; Truong L; Simonich MT; Swanson AJ; Matzke MM; Anderson KA; Waters KM; Tanguay RL Comparative Developmental Toxicity of Environmentally Relevant Oxygenated PAHs. Toxicol. Appl. Pharmacol 2013, 271, 267–275. 10.1016/j.taap.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.